Abstract
The vast majority of microfluidic systems are molded in poly(dimethylsiloxane) (PDMS) by soft lithography due to the favorable properties of PDMS: biocompatible, elastomeric, transparent, gas-permeable, inexpensive, and copyright-free. However, PDMS molding involves tedious manual labor, which makes PDMS devices prone to assembly failures and difficult to disseminate to research and clinical settings. Furthermore, the fabrication procedures limit the 3D complexity of the devices to layered designs. Stereolithography (SL), a form of 3D-printing, has recently attracted attention as a way to customize the fabrication of biomedical devices due to its automated, assembly-free 3D fabrication, rapidly decreasing costs, and fast-improving resolution and throughput. However, existing SL resins are not biocompatible and patterning transparent resins at high resolution remains difficult. Here we report procedures for the preparation and patterning of a transparent resin based on low-MW poly(ethylene glycol) diacrylate (MW 250) (PEG-DA-250). The 3D-printed devices are highly transparent and cells can be cultured on PEG-DA-250 prints for several days. This biocompatible SL resin and printing process solves some of the main drawbacks of 3D-printed microfluidic devices: biocompatibility and transparency. In addition, it should also enable the production of non-microfluidic biomedical devices.
INTRODUCTION
Microfluidics has had a great impact in a broad range of areas, from biological analysis and basic cell biology to chemical synthesis or optics1. Microfluidic systems are usually built by replica-molding and bonding in elastomers such as poly(dimethylsiloxane) (PDMS) or in thermoplastics such as poly(methyl-methacrylate) (PMMA) or poly-styrene (PS). These polymers owe their success to four key properties: biocompatibility, transparency, low cost, and being copyright-free. PDMS, in addition, is also elastomeric2 – a property that is key for producing fluidic automation components3,4 – and gas-permeable5 – a key factor for allowing O2 and CO2 exchange in cell culture applications6,7. Although applications are not lacking1,8 and PDMS molding is able to produce micron-resolution features9, PDMS mold fabrication requires a lengthy manual procedure9. Furthermore, complex devices need to be fabricated by layering, which severely restricts the possible 3D geometries. Additionally, in order to be cost-effective, molded devices have to be produced in large numbers, require huge initial capital investments, and they cannot be customized in short time frames10. Further concerns about PDMS arise when combined with living cells, such as monomer leaching and drug absorption11, which has prompted researchers to produce devices in other materials (e.g. plastics or paper8), but designs are still layered and difficult to customize. An alternative rapid plastic fabrication method allowing for more complex geometries and multi-height structures that are not limited to layers is micromilling12; however, micromilling still requires assembly and bonding in order to produce closed channels, and the milling tool cannot cut arbitrary shapes.
Stereolithography (SL) is a form of 3D printing invented in the 1980s that allows for the assembly-free production of quasi-arbitrary 3D shapes in a single polymeric material from a photosensitive resin precursor by means of a focused laser or a digital light projector (DLP) (Figure 1A)13,14. SL has recently attracted attention as a way to custom-fabricate complex microfluidic systems for research and development15–18 due to its automated 3D fabrication, rapidly decreasing costs, and improving resolution (see Table 1 for a comparison between SL, micromilling and soft lithography). However, available SL resins do not have all the favorable physicochemical properties of the above-named polymers (e.g., biocompatibility, transparency, elasticity, and gas permeability). For cell culture studies, optical transparency and biocompatibility are two major drawbacks that SL has to overcome. Some commercial SL resins are considered clear (e.g. WaterShed, Form Labs Clear or VisijetCrystal) and only after a finishing step they became reasonably transparent. One resin that is nearly colorless and meets biocompability standards is the WaterShed XC11122 by DSM Somos18. WaterShed becomes yellow under prolonged exposure to ambient light10. Moreover, a recent systematic investigation has shown that many of the popular 3D-printing resins (including Visijet Crystal or WaterShed) release toxic leachates that inhibit growth of cells from different vertebrate and invertebrate indicator organisms19. Zebrafish embryos cultured on these resins showed developmental defects20. Finding out which components of commercial resins are responsible for the cytotoxicity and the transparency loss is very difficult because the resins have a proprietary formulation. As a result, the performance of SL-printed devices is still inferior to that of equivalent PDMS devices.
Figure 1. DLP SL setup and light absorbance.
(A) DLP SL setup. (B) Spectra of light absorbance (molar extinction coefficient) for resins composed of PEG-DA-250 + Irgacure-819 (green), plotted with the Vis-DLP spectrum w/ UV filter (red dashed) or w/o UV filter (blue dashed) and 385 nm-DLP spectrum (purple dashed).
Table 1.
Comparison between PDMS molding, micromilling and (single-photon) SL.
| PDMS molding | Micromilling | SL | |
|---|---|---|---|
| Resolution (using low-cost systems) | ~10 μm | ~50 μm | ~200 μm |
| Throughput (avg. time/device) | 4 hrs | 1 min | 30 min |
| Bonding | YES | YES | NO |
| 3D designs | HARD | MULTI-HEIGHT | EASY |
| Automated manufacturing | NO | YES | YES |
| Biocompatibility | HIGH | HIGH | LOW |
Poly(ethylene glycol) diacrylate (PEG-DA) is a highly biocompatible photocurable material, which has inspired several groups to explore the use of PEG-DA as a microfluidics building material. Previously, SL has been used in tissue engineering for the fabrication of cell-containing hydrogel structures made of high-molecular weight PEG-DA (>MW 700) 21–30; clinical applications of this technology are now emerging21. Hydrogel PEG-DA formulations are porous and therefore unsuitable for constructing impermeable microfluidic channels. Low-MW PEG-DA has been used in the past to construct microfluidic devices, but mostly by photolithography and molding. Microchannels fabricated in PEG-DA of 258 MW (PEG-DA-258) resisted swelling and were impermeable to water for up to 2 weeks, whereas PEG-DA-875 channels showed significant swelling and collapsed within 5 hrs31. PEG-DA has also been used in PDMS microfluidics to build in-channel biocompatible structures32–34 and biosensing components35. Microfluidic valves have been micromolded36 and SL-printed37 in PEG-DA-258. Various favorable properties of PEG-DA-258 have been evaluated compared to glass, PDMS, and other polymers 38. Cronin’s group was the first to SL-print PEG-DA (MW 250). However, their process, which utilized a 405 nm light source, required the addition of an opaquing agent (Sudan I) in order to increase the light absorption of the resin, resulting in orange prints37,39,40; no biocompatibility studies were shown.
Here we demonstrate the use of a monochromatic 385 nm LED-based UV light source to increase the light absorption of additive-free (colorless) PEG-DA resins. Hence we can SL-print fully transparent microfluidic devices made of PEG-DA-250; we are able to culture CHO-K1 cells and primary hippocampal neurons for 48 hrs on SL-printed PEG-DA-250 petri dishes.
RESULTS AND DISCUSSION
Choice of resin components
The goal of this study was to produce 3D-printable devices that are water-impermeable, biocompatible, transparent, and cheap (by this order of priority). The photocurable resin of choice directly affects these characteristics. Inspired by previous work on photolithographically-patterned microfluidic channels made in water-impermeable PEG-DA-25831,36, we studied SL resin formulations that consisted of a mixture of transparent PEG-DA-250 monomer with small amounts of photoinitiator. In order to achieve a high cytocompatibility in our devices, we have limited our choice of photoinitiator to commercially-available photoinitiators of the Irgacure family that have been widely used for cell encapsulation studies in combination with PEG-DA hydrogels: Irgacure-819 (BASF) and Irgacure-784 (BASF)41–45. These photoinitiators are produced in large amounts so they are inexpensive ($68/lb), making them very attractive to the cost-conscious 3D-printing community. Irgacure-784 was discarded early on because it produces a strong dark orange coloring on the prints (Figure S1).
The meaning of “resolution” in SL microfluidic fabrication
A major challenge in the development of a new resin formulation is that modern SL printers are optimized for particular resins, so optimizing resolution with a new resin is not straightforward. Most nominal resolution numbers refer to open-surface features printed with resins of proprietary composition and untested biocompatibility. Custom-built (very expensive) multi-photon optics have been used for ~1 μm resolution SL46–49. In recent years ~500 μm-wide microchannels have been reported with single-photon (very affordable) SL systems15–18. Since all the key SL patents expired in 2014, many SL machines have appeared on the market and nominal resolution is improving almost every month.
The nominal resolution numbers provided by the manufacturer can be misleading. In SL setups based on a digital light projector (DLP) (Figure 1A) such as our Ilios 3D-Printer, the ultimate XY resolution is given by the DLP’s pixel size. For this study we have compared two DLPs: a visible-light DLP (heretofore referred as “Vis-DLP”) with an XY resolution of 29.5 μm × 32.7 μm (1920 × 1080 pixels), and a 385 nm LED-based DLP (heretofore referred as “385 nm-DLP”), with a resolution of 51.2 μm × 51.2 μm (1280 × 800 pixels). However, a full characterization and optimization of the XY resolution that can be achieved with PEG-DA is beyond the scope of this manuscript, which centers on the issues of biocompatibility and transparency.
The XY resolution should not be confused with the Z resolution. The Z resolution given by the manufacturer is usually the smallest thickness of the resin layer produced by the Z stepper motor that moves the build plate. In early 2015, desktop machines became available that advertise ~20 μm XY resolution and ~6 μm Z (thickness) resolution or less50. Our SL-printer features a nominal Z resolution of 12.5 μm. Yet microchannels with a cross-section smaller than 500 μm × 500 μm are very difficult to achieve with most printers, especially in transparent resins (see below). Prior work has already addressed the diffusion of the reactants51 and the viscosity of the resin (which must be cleared from the channels after exposure)18 as factors that might be responsible for some of the loss of resolution typically observed in SL.
The challenge of printing with transparent resins
Transparent resins pose an additional challenge. By definition, a transparent material does not absorb visible light. Unfortunately, most SL manufacturers – to cut costs – recently shifted the light source from UV lasers to visible light (405 nm lasers or DLP projectors). Consequently, these visible-light systems produce very poor Z resolution because the (transparent) resins do not absorb well at those wavelengths. The manufacturers of transparent resins typically advertise solid prints with surface features (no cavities), or very sparse reticles, which bear little relevance to microfluidic structures. This limitation is key for microfluidics because, if one attempts to build the roof of a microfluidic channel with a poorly absorbing resin, the light readily penetrates through the roof layer and crosslinks the whole channel.
This problem can be analyzed quantitatively. Since the monomer itself is transparent (Figure S3), the photoinitiator alone causes the vast majority of the resin’s absorbance A in a spectrophotometer, according to the Beer-Lambert law52:
| (Eqn. 1) |
where I0 and I are the intensity of incident and transmitted light, respectively, ε is the molar extinction coefficient, c is the concentration of the photoinitiator in the resin and L is the path length. Since c and L are always known during acquisition of a spectrum, measurement of A yields (a function of wavelength). The power spectrum of the Vis-DLP lamp (Figure 1B, see red dashed curve) shows a poor overlap with the spectrum of the resin’s ε, which means that the light source is rather inefficient at triggering the reaction. (For safety, DLPs contain a UV filter that filters out the small component of UV light emitted by visible bulbs; the UV filter can be removed, resulting in a slightly improved power spectrum shown in Figure 1B, blue dashed curve, but that small “tail” does not substantially increase the efficiency of the reaction.)
Transparent resins are more efficiently patterned with a 385 nm LED-based DLP (PRO4500 from Wintech Digital). At 385 nm, Irgacure-819 has 34% and 373% higher absorbance than at 405 nm and at 425 nm, respectively (see Figure 1B). From Eqn. 1, we anticipate a proportionally smaller Z penetration of light at those wavelengths and, for the same token, a Z resolution increase (not counting diffusion effects51). The power spectrum of our 385 nm-DLP is shown in Figure 1B, purple dashed curve. This wavelength overlaps with regions of higher light absorbance and is very monochromatic, so it does not overlap with regions of low light absorbance. Note that we do not claim that Irgacure-819 produces the optimum possible Z resolution; studies aimed at optimizing the Z resolution with a wiser combination of photoinitiator and light doses are ongoing.
Transparent prints
A transparent material will only produce a transparent print if its surfaces are smooth and its bulk is free of defects (Figure 2A); even if the material itself is transparent and colorless, an object printed with rough surfaces will diffract light and will appear translucent (Figure 2A). The print surface roughness is determined by the roughness of the vat surface as well as by the build plate surface. To minimize the roughness of both surfaces, we decided to use smooth glass slides for the vat and build plate surfaces. We observed that, when the resin polymerizes, it attaches to the vat glass surface instead of the build plate glass surface, making the printing process in this configuration impossible. In order to ensure that the print attaches to the top glass slide (on the build plate) and detaches from the bottom glass slide (on the vat) after each layer is printed, we derivatized the bottom vat glass slide with a passivating silane (see Methods). Additionally, derivatizing the top build plate glass slide with (adhesive) acrylate groups increased the overall efficiency of the printing process (see Methods).
Figure 2. Transparent prints with PEGDA-250 + I819 0.4%.
Disks (20 mm diameter and 1 mm thick) were printed with rough surfaces (Fig 2A) or smooth glass surfaces (Fig 2B). Other devices were printed using smooth glass surfaces, such as petri dishes for cell biocompatibility studies of 25 mm diameter and walls of 1 mm wide and 5 mm height (Fig 2C). A laminar flow device with 1 mm-wide channels (Fig 2D–F) was printed using either rough (Fig 2D) or smooth (Fig 2F) surfaces. Figure 2E is a 3D model of the laminar flow device depicted in Fig 2F.
With this surface derivatization protocol we were able to print devices that were very transparent. We compared the transparency when the devices were printed against smooth surfaces versus when the devices were printed against rough surfaces. Figure 2A–B shows a comparison of 20 mm-diam., 1 mm-thick disks; the use of rough surfaces yields a translucent disk (Figure 2A) while using smooth surfaces gives a transparent colorless disk (Figure 2B). After these initial tests, we printed more complex transparent devices, such as petri dishes (25 mm-diam. and 1 mm-wide, 5 mm-high walls, Figure 2C) for cell biocompatibility studies (see below), and a laminar flow device with 1 mm-wide channels (Figure 2E–F). We printed an additional laminar flow device with rough surfaces to better compare the transparency contribution effect of surface smoothness on a more complex print (Figure 2D). It is worth noting that by using smooth surfaces during the printing process we obtain transparent devices without any further post-processing steps. The printed disks were not only good for bright-field microscopy (Figure 3B), but also had background fluorescence comparable to polystyrene dishes (Figure S4).
Figure 3. Cell cultures on printed devices.
(A) 3D model of a petri dish for cell culturing. Phase-contrast image of 2-day cultures of CHO cells seeded on (B) SL-printed PEG-DA-250 and (C) TCPS surface. Phase-contrast image of primary mouse hippocampal neurons (embryonic day 18) on (D) SL-printed PEG-DA-250 and (E) TCPS surface coated with poly-D-lysine and Matrigel. Fluorescence images of the cells stained with cell tracker green (F) or orange (G) cultured on SL-printed PEG-DA-250.
Cytocompatibility of the prints
We compared the cytocompatibility of SL-printed petri dishes with Irgacure-819 (similar to the one in Figure 2C) with that of tissue-culture polystyrene (TCPS) as a control surface. We used both Chinese Hamster Ovary (CHO) cells (a major cell line often used in biotechnology) and primary mouse (embryonic day 18) hippocampal neurons (we chose this cell type as an example of a “delicate” cell type that would be very sensitive to contaminants). Prior to seeding, we exposed the surfaces to a UV bath in water for 12 hrs so as to leach out possible un-reacted PEG-DA monomers and/or photoinitiator. We then treated the surfaces with oxygen plasma right before coating proteins or seeding cells. The surfaces that were used for primary neuron culture were coated with poly-D-lysine and Matrigel (see Methods). Both types of cells grew similarly and displayed proper morphology on both surfaces (Figure 3) for up to 2 days (CHO cells grew to confluence at that point). Since both CHO cells and neurons grow in vitro as adherent cultures, they would not have survived if the SL-printed PEG-DA-250 surfaces were protein repellant. This observation is in contrast to Woolley’s previous report that photolithographically-patterned PEG-DA-250 is protein-repellent38. We attribute this discrepancy to variations in surface chemical groups that could have resulted from differences in the polymerization process between our 3D-printed PEG-DA-250 and Woolley’s photolithographically-patterned PEG-DA-250. Importantly, the prints were optically flat and could produce nice phase-contrast and epifluorescence images almost indistinguishable from those taken on TCPS. Generally speaking Irgacure-819 is not considered as safe as LAP in terms of biocompatibility, but our results show that it is a cheap and safe alternative to using LAP photoinitiator for printing devices for cellular studies. It is important to note that the petri dishes need to be fully cured before seeding cells. Seeding cells directly in a device without post-curing resulted in acute cell death. We compared different UV post-curing times (2, 4, 8, 12 hrs) of the 3D-printed petri dishes and compared the growth and viability of CHO cells after 1 day in culture. The 12 hrs of post-curing were necessary to ensure a viability and proliferation rate of the CHO cells comparable to tissue-culture polystyrene (Figure S5).
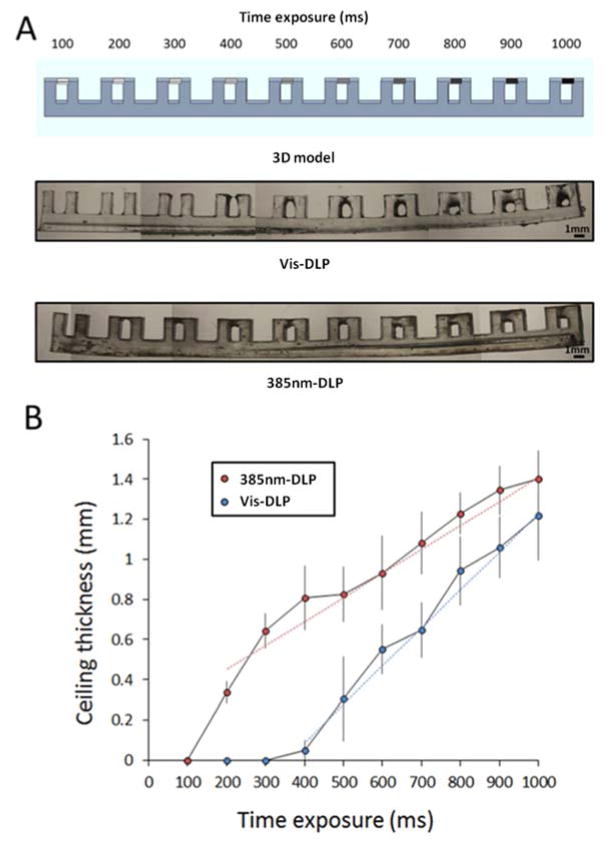
Z resolution boost
The use of the 385 nm-DLP not only enables the patterning of a transparent resin but could also help boost the Z resolution in the near future. We have compared test patterns printed with the Vis-Light DLP and with the 385 nm-DLP (Figure 4). The pattern to quantify Z resolution consisted of an array of 10 channels (2 mm height and 1 mm width). Different exposure times were set individually for each of the channel ceilings (see Methods), allowing us to quantify the effect of different time exposures on Z-resolution in one print (Figure 4A). Ceiling thickness was plotted for each exposure time with each light source. Our experimental results show that a ceiling is formed with lower exposure times at 385 nm compared to visible light (Figure 4B). Moreover, the trendline of the penetration using visible light is 47% steeper than using 385 nm light; therefore, the same increase in dose produces a larger Z penetration with visible light compared to 385 nm light. These results demonstrate that the 385 nm-DLP source produces smaller Z penetrations than the Vis-Light DLP source and could potentially help produce finer channel geometries.
Figure 4. I819 Z-resolution.
(A) 3D model of an array of 10 channels (2 mm height and 1 mm width) (upper panel). Printed design with different exposure times ceilings using an Optoma visible light projector (Vis-DLP, mid panel) or a 385 nm LED UV light projector (385 nm-DLP, lower panel). (B) Plot of the ceiling thickness for each tested time exposure for 385 nm-DLP (red dots) and Vis-DLP (blue dots). Data represent mean and standard deviation of three independent prints.
CONCLUSIONS
We have prepared and demonstrated the use of a biocompatible transparent resin for stereolithographic 3D-printing of bio-microfluidic devices. The resin is composed of low-MW PEG-DA (MW 250) and an inexpensive photoinitiator, Irgacure-819, without additional colorants. We have shown that SL prints made using the low-MW PEG-DA resin formulation supports the long-term culture of adherent mammalian cells, including sensitive cells like neurons. UV post-curing of the finished prints in a water bath is required to sufficiently remove toxic leachates. To pattern transparent resins, we implemented a UV light source that features higher light absorption (and therefore higher Z resolution) than typical (visible) light sources used in most SL printers. Since low-MW PEG-DA prints are not gas-permeable (unlike PDMS), culturing cells in enclosed 3D-printed PEG-DA microchannels would require other strategies like perfusion or hybrid device designs to facilitate better gas exchange. Most likely there is still a lot of room for improvement, as Irgacure-819 was chosen simply from a cursory literature search (not an experimental optimization). Since most SL resins have higher molar coefficient of extinction in the UV range than in the visible range, our approach should be readily applicable and beneficial to the patterning of most SL resins.
METHODS
Photoresin composition
Photoresin was based on poly(ethylene glycol) diacrylate (PEG-DA) (MW 250) (Sigma Aldrich) mixed with a photoinitiator. The photoinitiators used in this study were Irgacure-748 and Irgacure-819 (BASF Corporation). Irgacure photoinitiators were dissolved in PEG-DA at different concentrations (0.1, 0.2, 0.4, 0.8 and 1.2% wt/vol). For Irgacure-819, 0.4% was chosen as the best concentration in terms of balancing resolution and absence of color (Figure S2). All these steps were done in the dark to avoid spontaneous reaction with ambient light.
Absorbance measurements and calculations
Absorbance measurements of photoresin containing PEG-DA and Irgacure-819 were carried out by triplicate from 190 to 840 nm using a NanoDrop 2000c Spectrophotometer (Thermo Scientific). We tested three different concentrations (0.05, 0.025 and 0.0125 % wt/vol) to discard reading errors such as signal saturation by the spectrophotometer. After the measurements, the molar extinction coefficient, which is independent of concentration, was calculated using Eqn. 1.
Surface treatment
Surface roughness is a critical parameter to obtain high transparency prints. For the “rough” configuration we used a PDMS-coated vat and an aluminum build plate. For the “smooth” configuration we used a glass surface for the vat that contains the resin as well as for the build plate. We observed that prints tend to attach to the glass surface of the vat but not to the glass surface of the build plate. In order to ensure that prints attached only to the glass build plate, the vat glass surface was treated with SIGMACOTE® (Sigma-Aldrich), a hydrophobic silane. The glass surface was cleaned, dried and then covered with SIGMACOTE®. Excess of SIGMACOTE® was removed for reuse and stored at 4 °C. The treated glass was air dried in a hood. The build plate glass was treated with 3-(Trimethoxysilyl)propyl methacrylate (TMSPMA) (Sigma-Aldrich). The glass slides were cleaned, dried and placed in a closed vacuum chamber with a paper soaked in TMSPMA at 70 °C overnight.
3D-Printing setup
Our 3D printing platform consists of an ILIOS 3D printer with stepper motors with a nominal Z layer resolution of 12.5 μm, controlled by an Arduino board and a digital light projector (DLP)53. We use the ILIOS HD Kit for research that is commercial available and can be easily assembled. The printer consists of a metallic frame that once assembled has the following dimensions: ~60 cm (L) × ~50 cm (W) × ~120 cm (H). To this frame, a small VAT and build plate from the kit can be easily assembled providing an area of 100 mm × 178 mm. We mounted an Optoma HD20 HD DLP projector (“Vis-DLP”) from the kit to the frame using adaptors from the kit and adjusted its position for optimal focus and performance. This projector yields a printing area of 56.7 mm × 35.4 mm with a resolution of 29.5 μm × 32.7 μm (1920 pixels × 1080 pixels). Alternatively, we used another projector, 385 nm LED DLP projector, based on Texas Instruments’ DLP4500 chipset, the Wintech PRO4500 (“385 nm-DLP”). To mount this projector, we removed the Vis-DLP and mounted the 385nm-DLP to the same adaptors by using an inexpensive custom-made 3D-printed intermediate adaptor. The 385 nm-DLP provides a printing area of 65.6 mm × 41 mm with a resolution of 51.2 μm × 51.2 μm (1280 pixels × 800 pixels). Other electronics such as HT Stepper Motors and Arduino board were included in the kit and were easily assembled to the frame. Arduino allows the communication between the Ilios electronics (i.e. stepper motors) through USB to a computer.
3D-Printing software
All objects were designed with Autodesk Inventor® and saved in their final form in STL format. We used Creative Workshop® software to slice the objects and convert them into an image sequence. The whole 3D printing process was controlled by custom-made control software based on Matlab (G-code) to control the Arduino Board and the DLP projectors. This software allows us to precisely control for each layer parameters such as intensity, times of exposure and thickness layer.
3D-Printing procedure
For transparent prints, the photocurable resin was poured into the vat and a glass slide was “glued” to the build plate by coating one side with uncured resin and briefly exposing with UV light using a broadband UV lamp (B-100A, UVP). (This procedure allows for easy removal of the glass slide with a scraper at the end of the printing process; mechanical methods for attaching the glass slide to the build plate should be more practical in the long term.) The build plate was then lowered until it touched the vat surface. In short the printing process was carried as follows: the DLP projects the first slice of the object for a predetermined amount of time, the build plate stage rises and then lowers, the DLP projects the second slice, the build plate stage rises again, and the process continues until the whole object is printed. Then the object is removed from the build plate, rinsed with water and cleaned with pressurized air. The print is then kept in water and exposed for an additional 2 hrs to UV light using a UV gel box (High Performance Trans-illuminator TFL-40, UVP) to ensure that all the resin is cured; when prints are used for cell culture, we take extra precautions and we extend this over-curing process overnight (the 2 hrs period was insufficient and resulted in some cell death, presumably due to leaching of cytotoxic uncured material). If the object contains a microchannel, the UV exposure is performed under perfusion with water to remove uncured material from inside the microchannel.
Cell culture and microscope analysis
Prior to plating cells, all the 3D-printed PEG-DA devices were immersed in water under UV for over 12 hrs, and then treated with oxygen plasma (75 mTorr, 10 W, 30 sec) using a Zeptoplasma cleaner (Diener).
Chinese hamster ovary cells (CHO-K1) were cultured in DMEM media (Invitrogen) supplemented with 10% fetal bovine serum (Hyclone) and grown in a 5% CO2 atmosphere at 37 °C. After 48 hrs, when the cells were confluent, they were stained with live-cell fluorescent dyes, Cell Tracker Green and Orange (Invitrogen) (5 μM in serum-free DMEM media) for 30 minutes. For fluorescent imaging of the cells, the dyes were removed and replaced with Phenol-Red free DMEM, in order to minimize auto-fluorescence. The growth and viability of the cells was measured using the Trypan Blue exclusion assay.
Before culturing the neurons, the PEG-DA surfaces were coated sequentially with poly-D-lysine (100 μg/mL) (Sigma-Aldrich) overnight and Matrigel (BD Biosciences) (diluted 1:60 with DMEM) for 1 hr at 37 °C. Primary neurons were harvested from the hippocampi of embryonic day 18 mice (Brainbits), and enzymatically dissociated using a papain dissociation kit (Worthington Biochemical), following well-established protocols. The dissociated cells were suspended in Neurobasal media (Invitrogen) supplemented with 1X B-27 (Invitrogen), 0.5 mM GlutaMax (Invitrogen) and 100 U/mL penicillin-streptomycin (Invitrogen), and plated onto the Matrigel coated PEG-DA surfaces. The neurons were cultured for over 5 days in a 37 °C, 5% CO2 incubator.
All phase-contrast and fluorescence images of the cells were taken with a Nikon TE3000 epifluorescence microscope. The background fluorescence measurements were obtained using the same exposure settings (5 sec, no binning with a 10X objective) for glass coverslips, standard polystyrene tissue culture dishes, and 3D-printed PEG-DA-250 petri dishes that have been post-cured with UV for 2, 4, 8 and 12 hrs.
Supplementary Material
Acknowledgments
The Ilios 3D-Printer is on loan from 3D-Skema, Inc. A.U. is a recipient of a “La Caixa” fellowship from Catalonia and an European Molecular Biology Organization (EMBO) short-term fellowship. C.P. is a recipient of a “Consejo Nacional de Ciencia y Tecnología” (CONACYT) Mexican fellowship. F.P. is supported by the Spanish Ministry of Economy and Competitiveness (BFU2015-64437-P and FEDER), the Catalan Government (2014 SGR 599) and an ERC Advanced Grant Number 294294 from the European Union seventh framework program (SYNCOM). F.P. is supported by Fundación Botín, by Banco Santander through its Santander Universities Global Division and recipient of an ICREA Acadèmia (Generalitat de Catalunya). For the cell culture data and cell microscopy, we acknowledge partial support from the National Institutes of Health, grant number 1R01NS064387-01A2.
References
- 1.Whitesides GM. Nature. 2006;442:368–373. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- 2.Kim TK, Kim JK, Jeong OC. Microelectronic Engineering. 2011;88:1982–1985. [Google Scholar]
- 3.Unger MA, Chou HP, Thorsen T, Scherer A, Quake SR. Science. 2000;288:113–116. doi: 10.1126/science.288.5463.113. [DOI] [PubMed] [Google Scholar]
- 4.Thorsen T, Maerkl SJ, Quake SR. Science. 2002;298:580–584. doi: 10.1126/science.1076996. [DOI] [PubMed] [Google Scholar]
- 5.Merkel TC, Bondar VI, Nagai K, Freeman BD, Pinnau I. Journal of Polymer Science Part B: Polymer Physics. 2000;38:415–434. [Google Scholar]
- 6.Leclerc E, Sakai Y, Fujii T. Biotechnology Progress. 2004;20:750–755. doi: 10.1021/bp0300568. [DOI] [PubMed] [Google Scholar]
- 7.Kim L, Toh YC, Voldman J, Yu H. Lab on a Chip. 2007;7:681–694. doi: 10.1039/b704602b. [DOI] [PubMed] [Google Scholar]
- 8.Sackmann EK, Fulton AL, Beebe DJ. Nature. 2014;507:181. doi: 10.1038/nature13118. [DOI] [PubMed] [Google Scholar]
- 9.Folch A. Introduction to BioMEMS. CRC Press; Boca Raton, FL: 2013. [Google Scholar]
- 10.Bhattacharjee N, Urrios A, Kang S, Folch A. Lab on a Chip. 2016 doi: 10.1039/c1036lc00163g.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Regehr KJ, Domenech M, Koepsel JT, Carver KC, Ellison-Zelski SJ, Murphy WL, Schuler LA, Alarid ET, Beebe DJ. Lab on a Chip. 2009;9:2132–2139. doi: 10.1039/b903043c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guckenberger DJ, Groot TEd, Wan AMD, Beebe DJ, Young EWK. Lab on a Chip. 2015;15:2364–2378. doi: 10.1039/c5lc00234f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Waldbaur A, Rapp H, Lange K, Rapp BE. Anal Meth. 2011;3:2681–2716. [Google Scholar]
- 14.Bártolo PJ, editor. Stereolithography: Materials, Processes and Applications. Springer; 2011. [Google Scholar]
- 15.Yuen PK. Lab on a Chip. 2008;8:1374–1378. doi: 10.1039/b805086d. [DOI] [PubMed] [Google Scholar]
- 16.Yuen PK, Bliss JT, Thompson CC, Peterson RC. Lab on a Chip. 2009;9:3303–3305. doi: 10.1039/b912295h. [DOI] [PubMed] [Google Scholar]
- 17.Bhargava KC, Thompson B, Malmstadt N. Proc Nat Acad Sci (USA) 2014;111:15013–15018. doi: 10.1073/pnas.1414764111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Au AK, Lee W, Folch A. Lab Chip. 2014;14:1294. doi: 10.1039/c3lc51360b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu F, Friedrich T, Nugegoda D, Kaslin J, Wlodkowic D. Biomicrofluidics. 2015;9:061103. doi: 10.1063/1.4939031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.MacDonald NP, Zhu F, Hall CJ, Reboud J, Crosier PS, Patton EE, Wlodkowic D, Cooper JM. Lab on a Chip. 2016;16:291–297. doi: 10.1039/c5lc01374g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma B, Fermanian S, Gibson M, Unterman S, Herzka DA, Cascio B, Coburn J, Hui AY, Marcus N, Gold GE, Elisseeff JH. Science Translational Medicine. 2013;5:167ra166. doi: 10.1126/scitranslmed.3004838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan V, Jeong JH, Bajaj P, Collens M, Saif T, Kong H, Bashir R. Lab Chip. 2012;12:88–98. doi: 10.1039/c1lc20688e. [DOI] [PubMed] [Google Scholar]
- 23.Chan V, Zorlutuna P, Jeong JH, Kong H, Bashir R. Lab on a Chip. 2010;10:2062–2070. doi: 10.1039/c004285d. [DOI] [PubMed] [Google Scholar]
- 24.Han LH, Suri S, Schmidt CE, Chen S. Biomedical Microdevices. 2010;12:721–725. doi: 10.1007/s10544-010-9425-2. [DOI] [PubMed] [Google Scholar]
- 25.Han LH, Mapili G, Chen S, Roy K. Journal of Manufacturing Science and Engineering-Transactions of the Asme. 2008:130. [Google Scholar]
- 26.Hoffmann JC, West JL. Integrative Biology. 2013;5:817–827. doi: 10.1039/c3ib20280a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nguyen KT, West JL. Biomaterials. 2002;23:4307–4314. doi: 10.1016/s0142-9612(02)00175-8. [DOI] [PubMed] [Google Scholar]
- 28.Kim MH, Kumar SK, Shirahama H, Seo J, Lee JH, Cho N-J. Integrative Biology. 2016 doi: 10.1039/C5IB00269A.. [DOI] [PubMed] [Google Scholar]
- 29.Huang TQ, Qu X, Liu J, Chen SC. Biomedical Microdevices. 2014;16:127–132. doi: 10.1007/s10544-013-9812-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stampfl J, Baudis S, Heller C, Liska R, Neumeister A, Kling R, Ostendorf A, Spitzbart M. Journal of Micromechanics and Microengineering. 2008:18. [Google Scholar]
- 31.Kim P, Jeong HE, Khademhosseini A, Suh KY. Lab on a Chip. 2006;6:1432–1437. doi: 10.1039/b610503c. [DOI] [PubMed] [Google Scholar]
- 32.Dendukuri D, Gu SS, Pregibon DC, Hatton TA, Doyle PS. Lab on a Chip. 2007;7:818–828. doi: 10.1039/b703457a. [DOI] [PubMed] [Google Scholar]
- 33.Panda P, Ali S, Lo E, Chung BG, Hatton TA, Khademhosseini A, Doyle PS. Lab on a Chip. 2008;8:1056–1061. doi: 10.1039/b804234a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheung YK, Gillette BM, Zhong M, Ramcharan S, Sia SK. Lab on a Chip. 2007;7:574–579. doi: 10.1039/b700869d. [DOI] [PubMed] [Google Scholar]
- 35.Puchberger-Enengl D, Krutzler C, Keplinger F, Vellekoop MJ. Lab on a Chip. 2014;14:378–383. doi: 10.1039/c3lc50944c. [DOI] [PubMed] [Google Scholar]
- 36.Rogers CI, Oxborrow JB, Anderson RR, Tsai LF, Nordin GP, Woolley AT. Sensors and Actuators B: Chemical. 2014;191:438–444. doi: 10.1016/j.snb.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rogers CI, Qaderi K, Woolley AT, Nordin GP. Biomicrofluidics. 2015;9:016501. doi: 10.1063/1.4905840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nge PN, Rogers CI, Woolley AT. Chemical Reviews. 2013;113:2550–2583. doi: 10.1021/cr300337x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Song S, Kim MS, Lee J, Choi S. Lab on a Chip. 2015;15:1250–1254. doi: 10.1039/c4lc01417k. [DOI] [PubMed] [Google Scholar]
- 40.Gong H, Beauchamp M, Perry S, Woolley AT, Nordin GP. RSC Advances. 2015;5:106621–106632. doi: 10.1039/C5RA23855B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fairbanks BD, Schwartz MP, Bowman CN, Anseth KS. Biomaterials. 2009;30:6702–6707. doi: 10.1016/j.biomaterials.2009.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McKinnon DD, Kloxin AM, Anseth KS. Biomaterials Science. 2013;1:460–469. doi: 10.1039/c3bm00166k. [DOI] [PubMed] [Google Scholar]
- 43.Sawicki LA, Kloxin AM. Biomaterials Science. 2014;2:1612–1626. doi: 10.1039/c4bm00187g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin CC, Anseth KS. Proc Natl Acad Sci U S A. 2011;108:6380. doi: 10.1073/pnas.1014026108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Majima T, Schnabel W, Weber W. Macromol Chem Phys. 1991;192:2307–2315. [Google Scholar]
- 46.Bertsch A, Bernhard P, Vogt C, Renaud P. Rapid Prototyping Journal. 2000;6:259–266. [Google Scholar]
- 47.Nielson R, Kaehr B, Shear JB. Small. 2009;5:120–125. doi: 10.1002/smll.200801084. [DOI] [PubMed] [Google Scholar]
- 48.Kaehr B, Shear JB. Lab on a Chip. 2009;9:2632–2637. doi: 10.1039/b908119d. [DOI] [PubMed] [Google Scholar]
- 49.Lim TW, Son Y, Jeong YJ, Yang DY, Kong HJ, Lee KS, Kim DP. Lab on a Chip. 2011;11:100–103. doi: 10.1039/c005325m. [DOI] [PubMed] [Google Scholar]
- 50.Ilios. Ilios Ray 3D-Printer. http://www.ilios3d.com/en/shop/shop-3d-printing/ilios-ray/ilios-ray-printer-detail.
- 51.Fang N, Sun C, Zhang X. Appl Phys A. 2004;79:1839–1842. [Google Scholar]
- 52.Lehninger . Principles of Biochemistry. 6. Freeman, W. H. & Co; 2012. [Google Scholar]
- 53.Ilios. Ilios. http://www.ilios3d.com.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.