Abstract
The discovery of natural RNA enzymes (ribozymes) prompted the pursuit of artificial DNA enzymes (deoxyribozymes) by in vitro selection methods. A key motivation is the conceptual and practical advantages of DNA relative to proteins and RNA. Early studies focused on RNA-cleaving deoxyribozymes, and more recent experiments have expanded the breadth of catalytic DNA to many other reactions. Including modified nucleotides has the potential to widen the scope of DNA enzymes even further. Practical applications of deoxyribozymes include their use as sensors for metal ions and small molecules. Structural studies of deoxyribozymes are just beginning; mechanistic experiments will surely follow. From the first report 21 years ago, the field of deoxyribozymes has promise for both fundamental and applied advances in chemistry, biology, and other disciplines.
Keywords: deoxyribozyme, DNA enzyme, DNAzyme, DNA catalyst, catalytic DNA
Nucleic Acids as Catalysts
Nature has evolved a wide range of protein enzymes for biological catalysis. The notion that biomolecules other than proteins can be catalysts was largely disregarded until the early 1980s, when the enzymatic abilities of natural RNA catalysts (ribozymes; see Glossary) were discovered [1,2]. In modern biochemistry, catalysis by RNA appears restricted to RNA cleavage and ligation as well as peptide bond formation in the ribosome, although a primordial RNA World with broader RNA-based catalysis may have existed [3,4]. The properties of self-cleaving ribozymes were recently reviewed in this journal [5]. With the introduction of in vitro selection methods [6–9], synthetic ribozymes can be identified, including for particular substrates or even entire chemical reactions that are not biologically relevant. What about DNA as a catalyst?
The chemical structures of DNA and RNA are very similar, differing only by the respective absence and presence of ribose 2′-hydroxyl groups on every nucleotide monomer as well as by including T and U nucleobases. However, nature uses DNA and RNA for markedly different purposes. DNA is the long-term repository of genetic information, whereas RNA is the transient genetic messenger and is also used for a myriad of other biological functions. As the genetic material, DNA is double-stranded at nearly all times. A DNA duplex is structurally quite regular, but enzymatic activity requires irregular three-dimensional (tertiary) conformations that can bind substrates and position various functional groups for catalysis. Therefore, natural duplex DNA is unlikely to be catalytic.
Nevertheless, many experiments have shown that DNA can be a catalyst (deoxyribozyme). How can this be? The key is to consider single-stranded DNA. In its single-stranded form, DNA is similar to RNA with regard to many biochemical properties, particularly the ability to fold into secondary structures and intricate three-dimensional conformations. Indeed, synthetic RNA and DNA aptamers have been identified by in vitro selection methods for binding to a wide range of molecular targets [10], and natural RNA riboswitches that bind metabolites are important regulators of gene expression [11–13]. Upon searching through initially random sequence populations by in vitro selection methods, synthetic single-stranded deoxyribozymes can readily be found. Fundamentally, protein, RNA, and DNA enzymes are similar in that each is a well-defined sequence of monomers (amino acids or nucleotides) that adopts a tertiary structure to catalyze a chemical reaction.
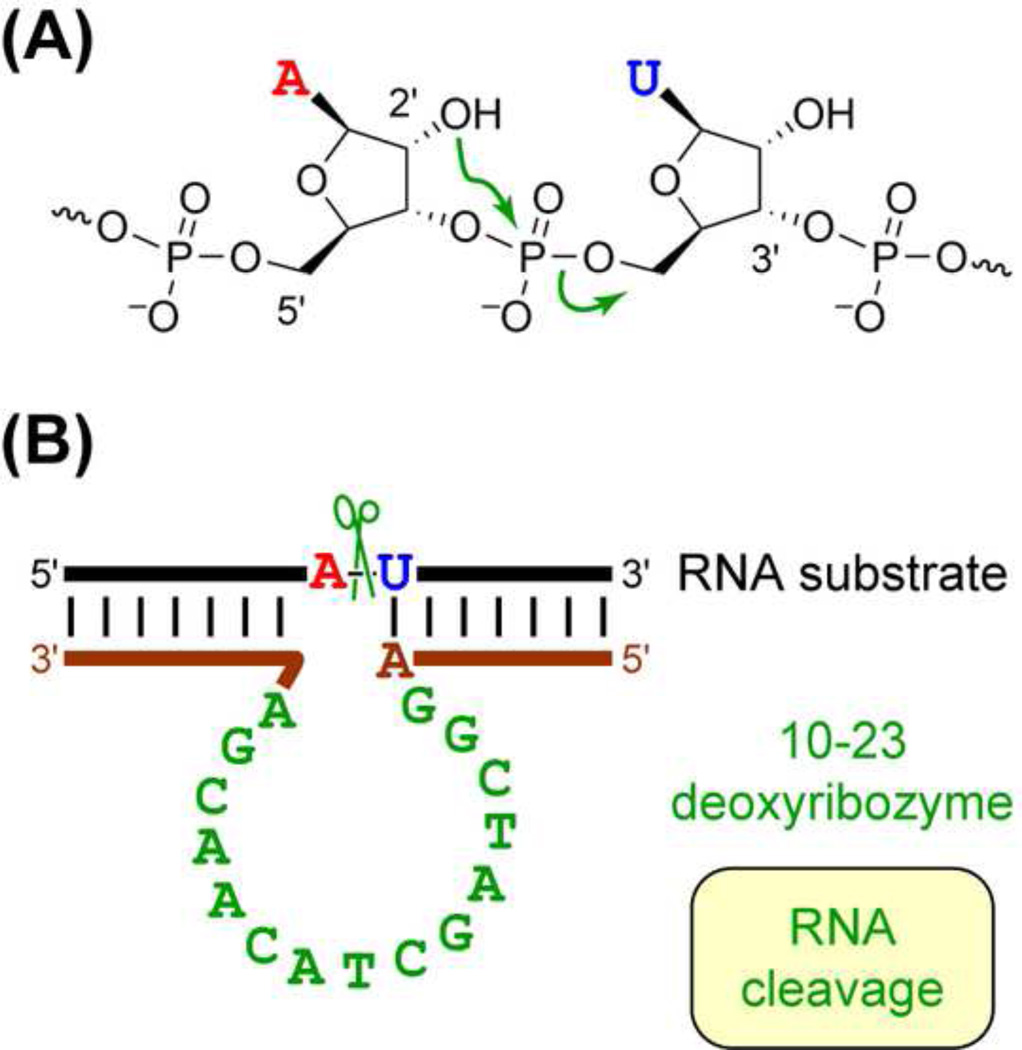
The first report of a deoxyribozyme was published in December 1994 by Breaker and Joyce [14]. Their deoxyribozyme cleaved at an RNA monomer within an oligonucleotide by modulating attack of the 2′-hydroxyl group at the adjacent phosphodiester linkage (Figure 1). The same reaction is catalyzed by protein enzymes such as ribonuclease [15]. Since that time, many labs have collectively reported hundreds and perhaps thousands of individual deoxyribozyme sequences, encompassing a broad and growing scope of catalytic activity. Several pertinent reviews have addressed the field of deoxyribozymes [16–23], including both general overviews and the relationship of DNA catalysis to other applications of DNA. This review describes the advantages of DNA as a catalyst and highlights recent advances in the scope, applications, and structural investigation of deoxyribozymes.
Figure 1. DNA-Catalyzed RNA Cleavage.
(A) RNA cleavage reaction, in which the 2′-hydroxyl group attacks the adjacent phosphodiester bond. (B) The 10–23 deoxyribozyme that cleaves RNA [37]. Note the Watson-Crick base-pairing interactions between deoxyribozyme and RNA cleavage substrate.
Advantages of Nucleic Acids Over Proteins as Catalysts
As nucleic acids, DNA and RNA enjoy many inherent advantages as catalysts relative to proteins. Entirely new synthetic nucleic acid sequences with catalytic function can be identified from random sequence populations, without any known catalytic sequence as the starting point. By contrast, performing in vitro selection from random protein sequences is generally intractable. In the context of developing entirely new synthetic catalysts, many important differences can be identified between nucleic acids and proteins.
Sequence space of random DNA/RNA populations has size 4n, where n is the random region length in nucleotides. A typical N40 random pool (40 nucleotides) comprises 440 ≈ 1024 unique sequences. For physical reasons, an in vitro selection experiment usually begins with ~1014 actual sequences, allowing only ~10−10 coverage of N40 sequence space. Nevertheless, new DNA/RNA enzymes can be identified, especially noting degeneracy (i.e., some sequences are functionally equivalent) and that not all nucleotide positions will have constrained identities within a catalytic sequence. For proteins, sequence space is 20n when all 20 standard amino acids are included. Considering that protein enzymes are usually >100 amino acids in length, this is >20100 ≈ 10130 sequences. Directed evolution of an existing protein enzyme sequence to improve rate or yield or to change substrate scope is often viable by screening variants that have mutations at a very small number of strategically chosen residues [24,25]. By contrast, identifying entirely new protein enzymes by in vitro selection from fully random amino acid sequences is generally unachievable, solely considering the numbers.
Moreover, most random protein sequences fail to adopt any particular tertiary structure due to the cooperative nature of protein folding. Protein secondary structure elements such as α-helices and β-sheets are almost always unstable by themselves [26], forming only in the context of higher-order tertiary structure. By contrast, DNA and RNA secondary structures such as stem-loops are quite stable in the absence of tertiary structure, allowing higher-order structure to form in hierarchical fashion from secondary structure elements [27].
De novo computational design of new protein enzyme active sites has advanced, but the designed active sites are embedded into known protein structures and optimized by directed evolution [28]; the entire enzyme is not designed from the ground up. Furthermore, the mechanism of such protein enzymes may not be as originally designed [29]. At present, and apparently for the foreseeable future, computationally designing any complete enzyme (protein or DNA/RNA) from scratch is not feasible.
Finally, all of the necessary biochemical tools to identify new deoxyribozymes and ribozymes by in vitro selection are readily obtained. This particularly includes DNA polymerase, which allows a small number of DNA sequences to be amplified into a much larger number by polymerase chain reaction (PCR); for ribozymes, reverse transcriptase and RNA polymerase are also needed. By contrast, the methodology to perform in vitro selection from random protein sequences is generally unavailable [30]. The lack of any means to copy an arbitrary amino acid sequence provides a nearly insurmountable practical roadblock to in vitro selection of protein enzymes, regardless of all other considerations.
Among the nucleic acids themselves, essentially all practical considerations favor using DNA over RNA. Such considerations include chemical and enzymatic stability, cost, and ease of synthesis. Perhaps the sole practical scenario that favors RNA over DNA is in vivo applications, where intracellular production is straightforward to achieve for RNA but typically requires a delivery mechanism for DNA. However, this advantage of RNA over DNA is negated by the need to use chemically modified variants of RNA/DNA in vivo for various reasons such as stability.
Reaction Scope of Catalysis by DNA
Since the initial report of an RNA-cleaving deoxyribozyme [14], many other catalytic activities of DNA have been reported. The scope of DNA catalysis is limited in part by the invention of suitable selection methodology for identifying catalytic sequences. In general, in vitro selection requires a way to separate active from inactive sequences without needing to screen each candidate sequence individually, as in separate wells of a plate or on separate beads. No one-at-a-time screening method is viable with 1014 candidate sequences. “In vitro compartmentalization” (IVC) [31,32] has led to several ribozyme activities [33–36] and should be applicable to deoxyribozymes as well. Nevertheless, to date all deoxyribozymes have been identified by in vitro selection using either bead-binding or polyacrylamide gel electrophoresis (PAGE) shift as the separation basis. Other articles have covered various practical considerations of in vitro selection (e.g., [16]). This section assesses use of deoxyribozymes to catalyze reactions of oligonucleotide and non-oligonucleotide substrates, and it describes inclusion of modified DNA nucleotides to expand DNA catalysis.
Deoxyribozymes for RNA Cleavage
DNA-catalyzed RNA oligonucleotide cleavage by the mechanism of Figure 1 has been found in many independent experiments and studied the most intensively of any DNA-catalyzed reaction. Soon after the initial report [14], a number of laboratories described additional RNA-cleaving deoxyribozymes. One motif, named 8–17, was identified by several different laboratories [37–40] and is likely to be the simplest possible catalytic DNA motif [41]. Indeed, the original RNA-cleaving deoxyribozyme [14] is likely an 8–17 motif. Due to its simplicity, 8–17 and a different small deoxyribozyme named 10–23 that was identified at the same time [37] have frequently been exploited for a range of practical applications, as discussed more comprehensively in a later section.
Deoxyribozymes for Other Reactions of Oligonucleotide Substrates
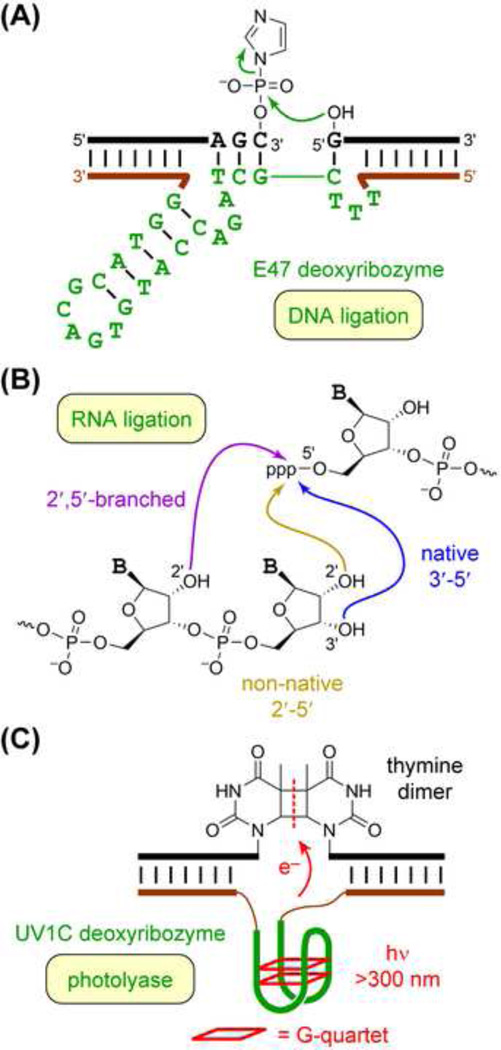
Beyond DNA-catalyzed RNA cleavage by the mechanism of Figure 1, many other reactions of oligonucleotide substrates have been catalyzed by deoxyribozymes. An early example was DNA ligation by Zn2+/Cu2+-dependent joining of 5′-hydroxyl and 3′-phosphorimidazolide groups (Figure 2A) [42]. Subsequent experiments led to a wide range of deoxyribozymes for RNA ligation using one or more divalent metal ions such as Mg2+, Mn2+, and Zn2+ along with several combinations of functional groups. These deoxyribozymes catalyzed formation of native 3′–5′ linear RNA, non-native 2′–5′ linear RNA, and 2′,5′-branched RNA (including lariat RNA) [18,43] (Figure 2B). Some of the branch-forming deoxyribozymes were later developed for utility in site-specific RNA labeling [44,45].
Figure 2. Reactions of Oligonucleotide Substrates Catalyzed by Deoxyribozymes.
(A) DNA ligation by the E47 deoxyribozyme [42]. (B) RNA ligation by reaction of 3′-hydroxyl or 2′-hydroxyl and 5′-triphosphate, with formation of various linear and branched linkages [18,43]. RNA ligase deoxyribozymes can also join the Figure 1A products of DNA-catalyzed RNA cleavage, 5′-hydroxyl and 2′,3′-cyclic phosphate. (C) Thymine dimer photoreversion by the UV1C deoxyribozyme (photolyase activity) [60].
A broad spectrum of oligonucleotide cleavage reactions has been catalyzed by DNA. These reactions include Cu2+-dependent oxidative cleavage [46–48], Cu2+/Mn2+-dependent oxidative nucleoside excision [49,50], DNA phosphodiester hydrolysis [51,52], RNA hydrolysis (involving a water molecule, a normally disfavored reaction relative to RNA cleavage by the Figure 1 mechanism) [53], and deglycosylation with subsequent backbone cleavage [54–56]. Oligonucleotide end modification has also been achieved by DNA 5′-phosphorylation (kinase activity) [57] and adenylylation (capping) [58] followed by ligation [59]. However, different deoxyribozymes and reaction conditions are required for each member of this three-reaction series.
An unusual DNA-catalyzed photochemical reaction was achieved by Sen and coworkers, who identified the UV1C deoxyribozyme that induces photoreversion of thymine dimers (Figure 2C) [60–62]. Although photochemistry has not been broadly exploited in other DNA-catalyzed reactions, this report establishes the ability of DNA to use light in catalysis. The UV-absorbing nucleobases and higher-order G-quadruplex structures, which appear to be formed by UV1C, naturally support this photolyase ability. A pterin moiety, which is a guanine nucleobase analogue, can replace any of six guanines in UV1C, allowing this deoxyribozyme to use violet light rather than UVB radiation and suggesting potential medical applications in photoreversion therapy [63]. The Sen lab also reported the Sero1C deoxyribozyme, which catalyzes thymine dimer photoreversion using serotonin as a discrete small-molecule cofactor [64]. In Sero1C, the indole ring of serotonin (rather than a G-quadruplex of the deoxyribozyme itself) serves as an antenna to enable thymine dimer photocleavage.
Deoxyribozymes for Reactions of Substrates Other Than Oligonucleotides
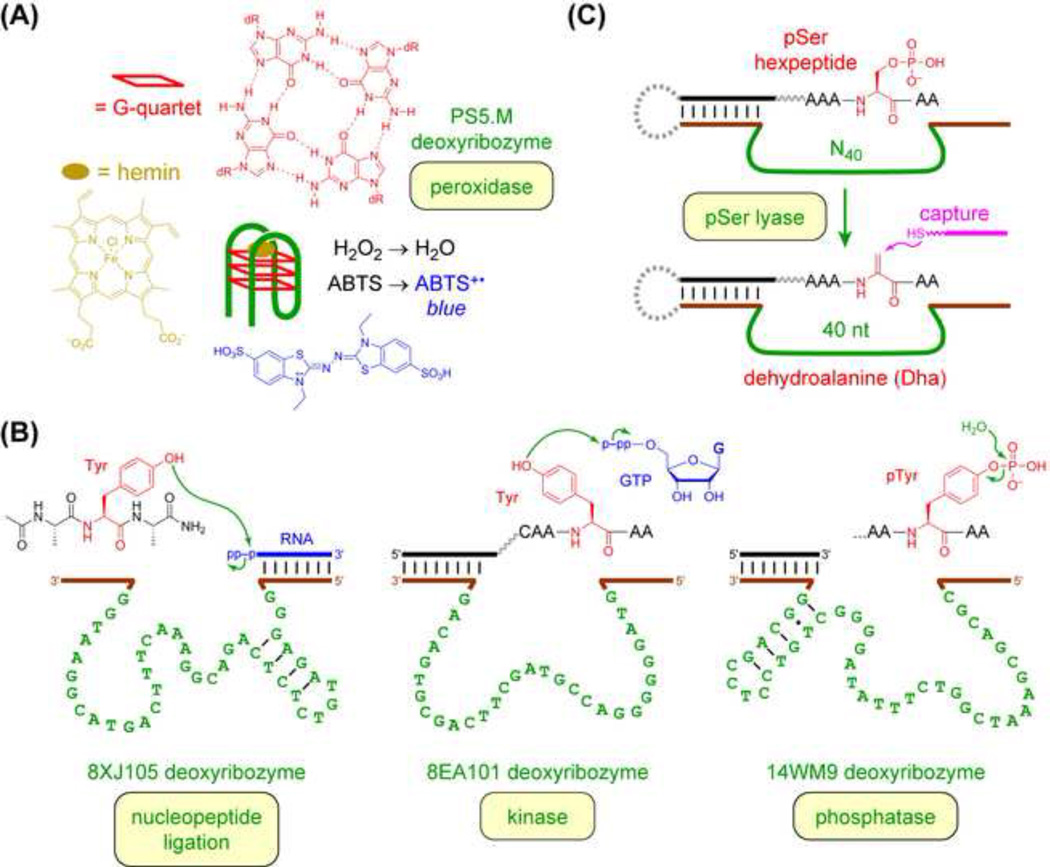
Growing efforts are revealing the capacity of DNA for catalysis involving non-oligonucleotide substrates. The first example was DNA-catalyzed porphyrin metalation, in which the deoxyribozyme was identified, unusually, by in vitro selection for binding to a transition state analogue [65]. Another example is peroxidase deoxyribozymes, which are important for practical sensor applications (see next section). Certain G-quadruplex DNA sequences can bind to the porphyrin hemin and oxidize small-molecule chromogenic substrates, leading to optical signals [66,67] (Figure 3A).
Figure 3. Reactions of Non-Oligonucleotide Substrates Catalyzed by Deoxyribozymes.
(A) Peroxidase activity by a G-quadruplex deoxyribozyme [66,67]. (B) Examples of DNA-catalyzed peptide side chain and backbone modification [22,68–74]. (C) Capture step during in vitro selection for identification of deoxyribozymes that form dehydroalanine (Dha) from phosphoserine (pSer) [74].
Silverman and coworkers have focused on identifying deoxyribozymes for modifying peptide side chains and backbones. Some of this work was recently reviewed [22]; selected highlights are provided here (Figure 3B). Deoxyribozymes have been found for conjugation of oligonucleotides to tyrosine [68–70], tyrosine phosphorylation [71,72], phosphotyrosine (pTyr) and phosphoserine (pSer) dephosphorylation [73], formation of dehydroalanine (Dha) by elimination of phosphate from pSer [74], and lysine modification [75]. The case of tyrosine phosphorylation illustrates that DNA catalysts can distinguish among various peptide substrate sequences [72], which is important for sequence-specific modification. In most of these experiments, a crucial aspect was developing the necessary selection methodology, in particular designing a “capture step” that within each selection round is highly selective for the intended reaction product and also sufficiently fast and high-yielding. As one example, identification of Dha-forming deoxyribozymes depends critically upon a thiol-based capture by nucleophilic attack into the electrophilic Dha group, which leads to a PAGE shift solely for those initially very rare DNA sequences that catalyzed formation of Dha (Figure 3C) [74].
During many reactions, a DNA catalyst must interact with a small-molecule substrate. For example, in DNA-catalyzed adenylylation and phosphorylation of oligonucleotides [57,58], an NTP is one substrate. Few purely small-molecule “organic” reactions have been pursued with deoxyribozymes; one instance is the Diels-Alder reaction [76]. Whenever a small-molecule substrate is required, one could consider integrating aptamers with DNA catalysts. This approach was evaluated in two recent studies. Dokukin and Silverman found no practical value in providing an ATP aptamer adjacent to a random DNA pool when identifying new tyrosine kinase deoxyribozymes, although this study provided the first clear evidence of modularity in DNA catalysis [77]. Using a very different approach, Willner and coworkers improved catalysis of an existing peroxidase deoxyribozyme by appending a substrate (dopamine or arginine) aptamer [78], and they further refined their approach to improve the catalysis [79]. If broadly applicable, these results when considered together imply that finding new DNA catalysts that require small-molecule substrates may not be assisted by combining aptamers with random pools at the outset of selection, but some existing deoxyribozymes can be improved functionally by attaching aptamers.
Modified DNA Nucleotides and Other Approaches to Broaden the Scope of DNA Catalysis
Even using a suitable selection method, deoxyribozymes that have a particular catalytic activity are not always identified. In cases for which standard DNA that includes the usual A, G, T, and C nucleotides is incapable of the intended activity, incorporating suitably modified nucleotides may enable catalysis. Similarly, if standard DNA is found to be capable of only rather poor catalysis, modified nucleotides may improve important catalytic features such as rate, yield, or substrate scope (either by broadening or narrowing the scope as desired). It is important to clarify that nonstandard functional groups are not simply appended onto previously known, unmodified deoxyribozyme sequences, because there is no reason to expect that attaching such groups will convert a noncatalytic or poorly catalytic DNA sequence into a (more) functional version. Instead, modifications are included from the outset of in vitro selection, leading to modified DNA sequences that integrally make use of the nonstandard groups for catalysis.
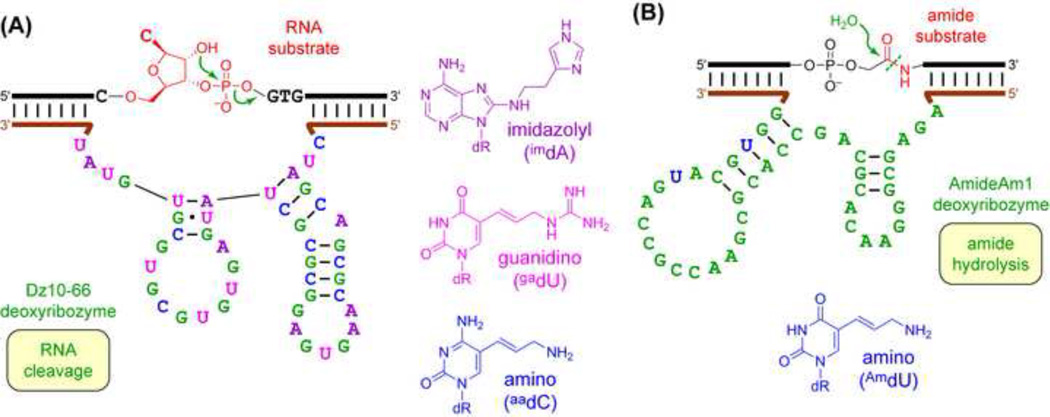
The laboratories of Perrin [80] and others [81,82] have described modified RNA-cleaving deoxyribozymes, usually with the goal of reducing or obviating the divalent metal ion (M2+) requirement. M2+-independent DNA-catalyzed RNA cleavage would be useful for in vivo mRNA cleavage (see next section). Later-generation deoxyribozymes incorporate three different kinds of modification (Figure 4A) and catalyze RNA cleavage in the complete absence of M2+ [83–85].
Figure 4. Modified DNA Nucleotides in Deoxyribozymes.
(A) M2+-independent RNA-cleaving Dz10–66 deoxyribozyme, with three kinds of modified nucleotide [84]. The substrate has a single RNA nucleotide embedded within a DNA sequence. (B) AmideAm1 deoxyribozyme for amide bond hydrolysis, with amine-modified nucleotides [86].
The Silverman laboratory recently reported the first example of modified deoxyribozymes to achieve hydrolysis of aliphatic amides (Figure 4B) [86]. Previously, unmodified DNA enzymes for hydrolysis of esters and aromatic amides were identified, but deoxyribozymes for aliphatic amide hydrolysis were not found [87]. Therefore, including modifications during the selection process enabled identification of the new DNA enzymes. One modified deoxyribozyme requires as few as one primary amino modification to achieve amide hydrolysis. Another modified deoxyribozyme was identified with primary hydroxyl groups in the form of 5-(hydroxymethyl)dU nucleotides, but unexpectedly, the same sequence in the form of entirely unmodified DNA retains substantial catalytic activity. An as-yet untested hypothesis is that an ordered water molecule can replace the 5-(hydroxymethyl) group for the unmodified DNA enzyme. These findings suggest both the utility and complexity of including modified DNA nucleotides during identification of new deoxyribozymes.
Related to DNA catalysts with modified DNA nucleotides are “XNAzymes”, which are made from xeno-nucleic acids, XNAs [88]. XNAs have unnatural backbones such as those of threose nucleic acid (TNA), 2′-fluoro-arabinonucleic acid (FANA), and several others. The finding by Holliger and coworkers of catalysis by XNAzymes is exciting conceptually. This finding may also have practical value that derives from the orthogonality of XNAs to natural biopolymers and the natural protein enzymes that interact with and degrade them.
One approach to enhance DNA catalysis is including small-molecule cofactors. One early report showed that an RNA-cleaving deoxyribozyme could be dependent on the presence of histidine [89], and a later study used periodate in DNA-catalyzed depurination [55]. Beyond that, though, this aspect of DNA catalysis remains largely unexplored. In principle, small-molecule cofactors should be able to provide functional groups that would otherwise necessitate modified nucleotides; in that regard, cofactors could allow use of standard unmodified DNA while simultaneously expanding the catalytic abilities of DNA. However, a drawback of obligatory cofactors is that the deoxyribozyme needs simultaneously to bind its substrate(s) and the cofactor as well as perform catalysis, and such multitasking may be difficult.
Are Deoxyribozymes “Good” Catalysts?
Rate enhancements of deoxyribozymes can be quite large. For example, the background hydrolysis half-life for phosphoserine (at neutral pH, in the absence of metal ions, at ambient temperature) is ~1010 years, whereas DNA-catalyzed hydrolysis proceeds with half-life of ~1 hour [73]. Taking the ratio of these two half-lives leads to a calculated rate enhancement of 1014. Such calculations are complicated by the fact that divalent metal ions such as Zn2+ are required for catalysis, and separating the contributions of DNA and metal ions is tricky. The safest conclusion may be that focusing on quantitative rate enhancement values misses the point: the key observation is that DNA can perform catalysis in an interesting and potentially useful way. For ribozymes and deoxyribozymes alike, Breaker and coworkers posited that the maximum rate constants of ~1 min−1 reflect suboptimal use of catalytic strategies; using all possible modes of catalysis would allow much greater rate constants [90,91]. Therefore, the primary limitation on deoxyribozyme catalysis is not theoretical but practical. Deoxyribozymes can in theory be outstandingly good catalysts—the important question is, how can they best be identified? Using appropriately designed “capture steps”, in some cases along with including modified DNA nucleotides, illustrates how to find deoxyribozymes that catalyze difficult or disfavored reactions.
Applications of Deoxyribozymes
One major impetus for the development of new catalysts is their practical value. This section describes two specific applications for deoxyribozymes: in vivo mRNA degradation and sensors. Other applications such as site-specific RNA labeling [44,45], logic gates, computing circuits, and switches [92–94], and biofuel cell catalysis [95] are also being pursued. Many of the DNA-catalyzed reactions still under development are likely to enable an even broader range of practical applications.
Deoxyribozymes for mRNA Degradation In Vivo
Intentionally degrading specific mRNAs is an important biological approach to control gene expression, both in the laboratory and in ongoing clinical trials with RNA interference (RNAi) [96]. RNA cleavage by an introduced deoxyribozyme has long been suggested as a means to accomplish mRNA degradation [37]. However, a major concern is whether any mRNA degradation is due to DNAcatalyzed cleavage or merely to an antisense effect [97]. Two articles in the medical literature reported using the 10–23 deoxyribozyme to treat cancer and asthma, respectively [98,99], although control experiments to distinguish DNA catalysis from antisense effects were not described. Because deoxyribozymes generally exhibit little or no catalysis at the free divalent metal ion concentrations inside cells, efforts with modified deoxyribozymes are intended to improve in vivo catalysis [80–85]. In one instance with otherwise-unmodified DNA, introducing 2′-O-methyl substitutions to improve 10–23 deoxyribozyme stability was accompanied by data supporting in vivo mRNA degradation [100]. However, broadly compelling evidence for in vivo DNA-catalyzed mRNA degradation is currently unavailable.
Deoxyribozymes as Sensors
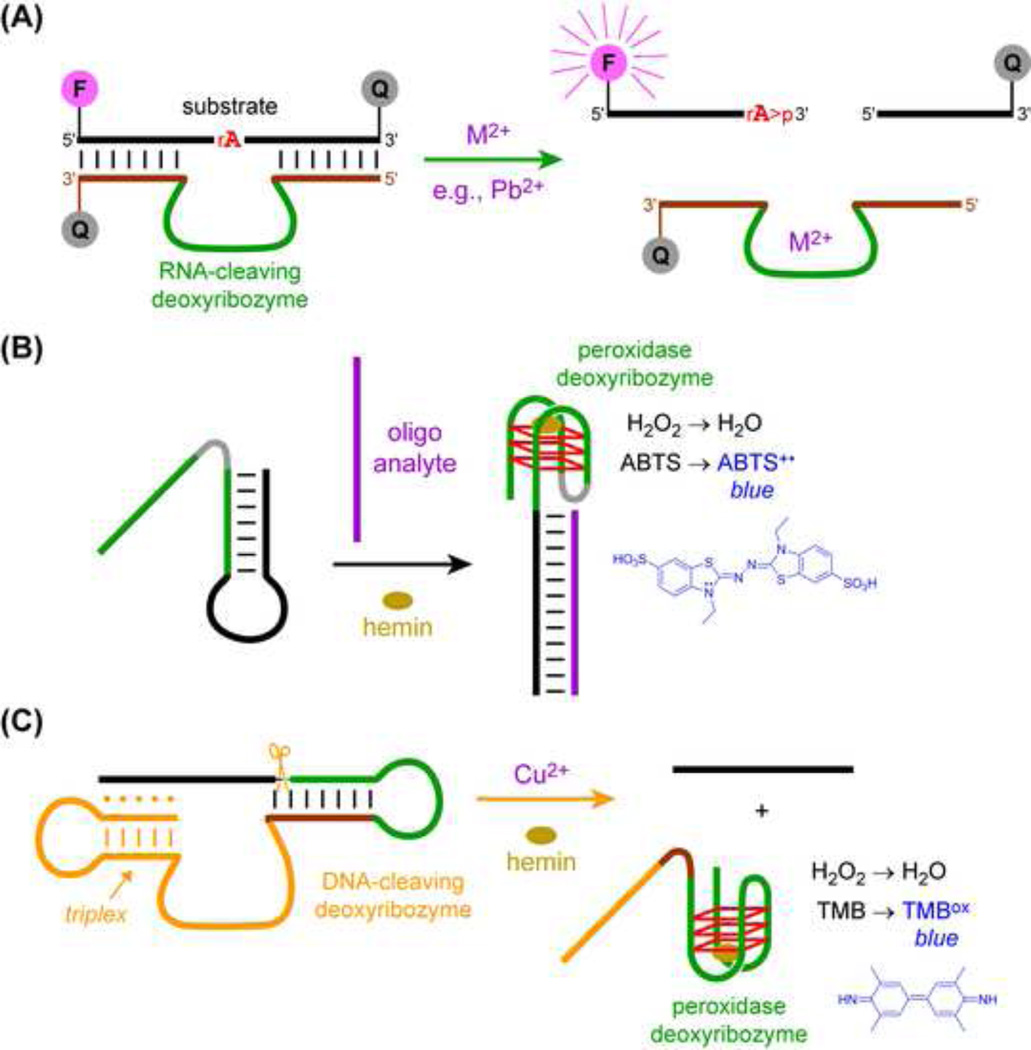
Two major classes of deoxyribozymes, RNA-cleaving and peroxidase, have been used to create many sensors. RNA-cleaving deoxyribozymes generally require specific metal ions, which enables the development of metal ion sensors (Figure 5A). Much of the work in this area has been performed by Lu and coworkers [101], who in 2000 reported a Zn2+-dependent RNA-cleaving deoxyribozyme [102] and more recently described fluorescence detection of UO2+ (uranyl) and Na+ in living cells [103,104]. Others reported detection of Hg2+ using deoxyribozymes with modified DNA nucleotides [105]. In general, the metal ion selectivity of RNA-cleaving deoxyribozymes can be coupled with numerous detection platforms to allow sensitive and specific responses to particular metal ions in a variety of contexts [21,106]. For example, Xiang and Lu used a personal glucose meter to create a portable Pb2+ sensor device; after Pb2+-induced RNA cleavage, a series of strand exchange events enables invertase-catalyzed conversion of sucrose to glucose, which the meter detects [107].
Figure 5. Sensor Applications of Deoxyribozymes.
(A) RNA-cleaving deoxyribozymes activated by specific metal ions (Mn+). In the illustrated example, M2+-dependent RNA cleavage separates a fluorophore (F) from its quenchers (Q), leading to a fluorescence signal [101]. (B) Oligonucleotide detection by a G-quadruplex peroxidase deoxyribozyme that functions by strand displacement [110]. In the presence of the oligonucleotide analyte and the porphyrin hemin, the deoxyribozyme uses H2O2 to oxidize the small-molecule substrate ABTS, 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid), forming a colored product. (C) Cu2+ detection by a G-quadruplex peroxidase deoxyribozyme, triggered by Cu2+-dependent DNA-catalyzed RNA cleavage [112]. Release of the DNA fragment allows Gquadruplex formation, and the peroxidase deoxyribozyme then uses H2O2 to oxidize the small-molecule substrate TMB, 3,3′,5,5′-tetramethylbenzidine, forming a colored product.
RNA-cleaving deoxyribozymes have also been applied to sense biomacromolecules. Willner and coworkers achieved sensitive detection of DNA itself, by autonomous assembly of polymeric DNA nanowires built from deoxyribozyme subunits [108]. An interesting recent application of RNA-cleaving deoxyribozymes by Li and coworkers is sensing of specific bacterial species. Using deoxyribozymes whose activity depends on the presence of the crude extracellular mixture (CEM) of a particular pathogenic strain of Clostridium difficile, fluorogenic responses to that strain were generated [109]. This approach does not require identifying the CEM component responsible for activating the deoxyribozyme, although for C. difficile the authors did determine that a truncated transcription factor in the pathogenic strain activates the deoxyribozyme.
Peroxidases are the second class of deoxyribozymes upon which many sensors are designed. Detecting a variety of molecular targets is achieved by using those targets to regulate the G-quadruplex structure of the peroxidase deoxyribozyme. For example, an oligonucleotide analyte is detected when the target strand liberates the DNA nucleotides that constitute the G-quadruplex (Figure 5B) [110], or when oligonucleotide binding triggers formation of deoxyribozyme-containing nanowires [111]. Several groups have created peroxidase deoxyribozyme sensors for metal ions and small organic compounds [101]. In one interesting example that exploits both RNA cleavage and peroxidase activity, Tan and coworkers detected Cu2+ via DNA-catalyzed RNA cleavage that enables proper folding of a peroxidase deoxyribozyme, thereby leading to a colorimetric signal (Figure 5C) [112].
Structural and Mechanistic Studies of Deoxyribozymes
Finally, the First Deoxyribozyme Crystal Structure
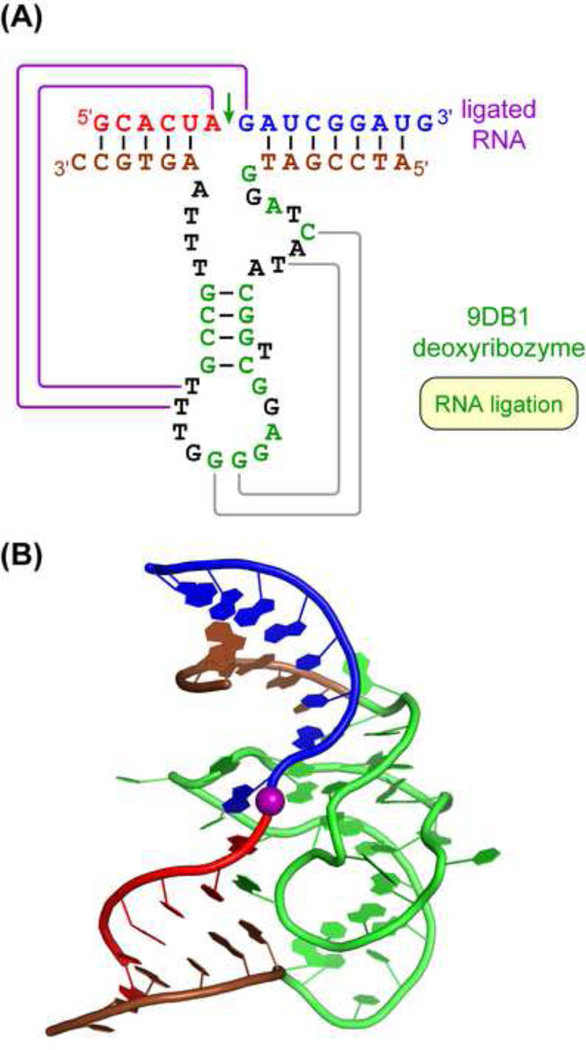
Although deoxyribozymes have been known for over 20 years, until very recently no high-resolution structure was available [113]. In early 2016, a research team led by Höbartner and Pena reported the first deoxyribozyme X-ray crystal structure [114], that of the RNA-ligating 9DB1 deoxyribozyme [115]. This structure reveals several important facets of catalysis by DNA, including the fundamental confirmation that deoxyribozymes can have well-defined tertiary structures that are responsible for their catalysis. The intricate three-dimensional structure of 9DB1 complexed with its RNA ligation product is a double pseudoknot, in which two adjacent T residues interact with the A-G RNA nucleotides directly at the ligation junction (Figure 6). Recognizing these interactions, which were not evident from biochemical experiments, enabled reengineering of the deoxyribozyme to broaden its RNA substrate scope by straightforward Watson-Crick covariation. The structure also revealed contacts that explain why the newly formed phosphodiester bond is 3′–5′ (native) rather than 2′–5′. This outcome was demanded by the selection strategy that led to the identification of 9DB1, but the underlying structural explanation was unknown.
Figure 6. The First Deoxyribozyme Crystal Structure [114].
(A) Double pseudoknot secondary structure of the minimized 9DB1 deoxyribozyme that ligates RNA, bound to its ligation product. Red and blue denote the two halves of the RNA product, corresponding to the two RNA substrates. The deoxyribozyme binding arms are brown; black and green nucleotides are nonconserved and conserved core nucleotides, respectively. Pseudoknot interactions are purple and grey lines. Two adjacent T residues of the deoxyribozyme interact with the A and G nucleotides in the RNA product at the ligation junction (purple lines), enabling general RNA ligation by covariation of these nucleotides. (B) Three-dimensional structure, with same colors as panel A except the entire core of the deoxyribozyme is green. The ligation site is marked with a purple sphere. Nucleobases are shown explicitly; the remainder of each nucleotide is shown in ribbon form. Image courtesy of C. Höbartner (PDB 5ckk).
Initially all ribozymes were thought to require divalent metal ions as catalytic cofactors [116], but both natural and synthetic ribozymes have been found that lack this requirement [117,118]. The 9DB1 deoxyribozyme requires either Mg2+ or Mn2+ for its catalysis [115], but its crystal structure reveals no divalent metal ion at the active site. Instead, an internucleotide phosphodiester oxygen appears to make a critical contact. Therefore, this first deoxyribozyme crystal structure supports the conclusion that DNA enzymes are not obligatory metalloenzymes with regard to the catalytic role of divalent metal ions [89,104,119,120].
Another notable finding from the new structure is an important insight into the relationship between DNA and RNA enzymes. The lack of 2′-hydroxyl groups in DNA is expected to reduce its functional capacity relative to RNA, all other things equal, but the magnitude of this effect is unknown. DNA and RNA aptamers in general have very similar distributions of binding constants (Kd values) [10], and a deoxyribozyme for a Diels-Alder reaction was found to have similar catalytic parameters as an independently obtained ribozyme [76]. The new structure reveals that the 9DB1 deoxyribozyme has a much broader range of sugar-phosphate backbone conformations than is found in natural and synthetic ribozymes. Therefore, the missing 2′-hydroxyl groups of DNA, while removing one source of functional interactions, in parallel expand the conformational variability of DNA relative to RNA. These counteracting effects appear to leave DNA at least as catalytically competent as RNA. Indeed, the greater backbone freedom for single-stranded DNA as observed in the 9DB1 crystal structure may enable DNA catalysis in ways not possible for RNA.
What does the future hold for high-resolution structural studies of deoxyribozymes? Now that the first crystal structure has been reported [114], surely others will follow. Höbartner’s elegant combinatorial minimization methods, which were applied to 9DB1 before its crystallization [121], will likely be invaluable for enabling crystallization of unrelated deoxyribozymes. In parallel, NMR studies of deoxyribozymes are surely possible, although none have yet been reported.
Mechanistic Studies of Deoxyribozymes
Investigations of biochemical mechanisms are often intimately linked to structural studies. For ribozymes, meaningful mechanistic insights have derived from biochemical experiments that were enabled by high-resolution structures (e.g., [122]). Considering the dearth of deoxyribozyme structures, it is unsurprising that few substantial mechanistic reports on DNA enzymes have yet appeared. For example, many studies have addressed functional roles of metal ions and investigated kinetics, but not to the extent where we truly understand mechanisms of DNA catalysis. With the first deoxyribozyme crystal structure now reported [114] and hopefully more on the way, the prospects are strong for understanding the detailed mechanisms by which DNA enzymes catalyze many reactions. A reasonable expectation is that deoxyribozymes and ribozymes will share common contributions to catalysis, such as acid-base chemistry involving the nucleobases [123].
Concluding Remarks
This Trends Review has described the many conceptual and practical advantages of DNA as a catalyst relative to proteins and RNA. From the relatively modest first report in 1994, deoxyribozymes have grown to encompass a broad range of catalytic abilities, including with the assistance of modified DNA nucleotide components. The first experiments to understand key aspects of deoxyribozyme structure and mechanism are currently emerging. Many questions are still unanswered about the capabilities and applications of deoxyribozymes (see Outstanding Questions). The advantages of DNA as a synthetic catalyst provide strong encouragement for the continued development of deoxyribozymes.
Outstanding Questions.
What are the limits on the scope of reaction catalysis by DNA?
How important are modified nucleotides or other nonstandard features to enable catalysis by DNA?
For what practical applications (established or new) will deoxyribozymes have the greatest utility?
What are the structures of deoxyribozymes? In this regard, how are deoxyribozymes similar to, or different from, ribozymes and protein enzymes?
What are the mechanisms by which deoxyribozymes perform catalysis?
Trends.
As a synthetic catalyst identified by in vitro selection, DNA has many advantages over proteins and RNA. The growing reaction scope of deoxyribozymes is increasing our fundamental understanding of biocatalysis and enabling applications in chemistry and biology.
Including modified DNA nucleotides enhances the catalytic ability of deoxyribozymes. Several approaches for this purpose are being evaluated.
Investigators are pursuing practical applications for deoxyribozymes, such as in vivo mRNA cleavage and sensing of metal ions and small molecules.
High-resolution structural analysis of deoxyribozymes is a new field; the first crystal structure was recently reported. This structure provides an understanding of how DNA can be as catalytically competent as RNA. Structural information will enable mechanistic studies of deoxyribozymes.
Acknowledgments
I apologize to investigators whose work was not mentioned, cited, or described in greater detail due to space constraints, and I regret that certain topics were addressed or cited only briefly or not at all. Studies on deoxyribozymes in the Silverman laboratory are supported by the U.S. National Institutes of Health.
Glossary
- Aptamer
A DNA or RNA (or other oligonucleotide variant) sequence that binds to a molecular target. Aptamers are identified by in vitro selection, which for aptamers is often called SELEX (Systematic Evolution of Ligands by EXponential enrichment), noting that the aptamer is the “ligand” for the molecular target.
- Deoxyribozyme
A particular DNA sequence that has catalytic activity, in analogy to a protein enzyme as a catalytic sequence of amino acids. Synonyms for “deoxyribozyme” include DNA enzyme, DNAzyme, DNA catalyst, and catalytic DNA. While deoxyribozymes are unknown in nature, synthetic catalytic DNA sequences can be identified by in vitro selection.
- Directed evolution
The process by which a known enzyme sequence is improved with regard to some property, such as rate, yield, or substrate scope. The enzyme may be protein, DNA, or RNA. For protein enzymes, practical considerations dictate that only a very small subset of amino acids within the sequence may be varied; these residues are chosen on the basis of structural or mechanistic information. Alternatively, more randomly targeted approaches such as gene shuffling may be used. For DNA and RNA enzymes, by contrast, the entire nucleotide sequence is typically included in the directed evolution process.
- In vitro selection
The experimental process by which random sequences, such as those of DNA or RNA nucleotides, are examined in parallel (not one at a time) to identify those particular sequences that have a desired function such as binding or catalysis. This term is defined in analogy to natural selection, which is an undirected process. In vitro selection is conceptually distinct from screening, which is a one-at-a-time process.
- Ribozyme
A particular RNA sequence that has catalytic activity (cf. deoxyribozyme). Both natural and synthetic ribozymes are known; the latter are identified by in vitro selection.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kruger K, et al. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell. 1982;31:147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- 2.Guerrier-Takada C, et al. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- 3.Higgs PG, Lehman N. The RNA World: molecular cooperation at the origins of life. Nat. Rev. Genet. 2015;16:7–17. doi: 10.1038/nrg3841. [DOI] [PubMed] [Google Scholar]
- 4.Pressman A, et al. The RNA World as a Model System to Study the Origin of Life. Curr. Biol. 2015;25:R953–R963. doi: 10.1016/j.cub.2015.06.016. [DOI] [PubMed] [Google Scholar]
- 5.Jimenez RM, et al. Chemistry and Biology of Self-Cleaving Ribozymes. Trends Biochem. Sci. 2015;40:648–661. doi: 10.1016/j.tibs.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 7.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 8.Robertson DL, Joyce GF. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature. 1990;344:467–468. doi: 10.1038/344467a0. [DOI] [PubMed] [Google Scholar]
- 9.Joyce GF. Forty Years of In Vitro Evolution. Angew. Chem. Int. Ed. 2007;46:6420–6436. doi: 10.1002/anie.200701369. [DOI] [PubMed] [Google Scholar]
- 10.McKeague M, et al. Analysis of In Vitro Aptamer Selection Parameters. J. Mol. Evol. 2015;81:150–161. doi: 10.1007/s00239-015-9708-6. [DOI] [PubMed] [Google Scholar]
- 11.Henkin TM. The T box riboswitch: A novel regulatory RNA that utilizes tRNA as its ligand. Biochim. Biophys. Acta. 2014;1839:959–963. doi: 10.1016/j.bbagrm.2014.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mandal M, et al. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell. 2003;113:577–586. doi: 10.1016/s0092-8674(03)00391-x. [DOI] [PubMed] [Google Scholar]
- 13.Serganov A, Nudler E. A decade of riboswitches. Cell. 2013;152:17–24. doi: 10.1016/j.cell.2012.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Breaker RR, Joyce GF. A DNA enzyme that cleaves RNA. Chem. Biol. 1994;1:223–229. doi: 10.1016/1074-5521(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 15.Cuchillo CM, et al. Bovine pancreatic ribonuclease: fifty years of the first enzymatic reaction mechanism. Biochemistry. 2011;50:7835–7841. doi: 10.1021/bi201075b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Silverman SK. Catalytic DNA (deoxyribozymes) for synthetic applications—current abilities and future prospects. Chem. Commun. 2008:3467–3485. doi: 10.1039/b807292m. [DOI] [PubMed] [Google Scholar]
- 17.Schlosser K, Li Y. Biologically inspired synthetic enzymes made from DNA. Chem. Biol. 2009;16:311–322. doi: 10.1016/j.chembiol.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 18.Silverman SK. Deoxyribozymes: Selection Design and Serendipity in the Development of DNA Catalysts. Acc. Chem. Res. 2009;42:1521–1531. doi: 10.1021/ar900052y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silverman SK. DNA as a Versatile Chemical Component for Catalysis, Encoding, and Stereocontrol. Angew. Chem. Int. Ed. 2010;49:7180–7201. doi: 10.1002/anie.200906345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu J, et al. Functional nucleic acid sensors. Chem. Rev. 2009;109:1948–1998. doi: 10.1021/cr030183i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang XB, et al. Metal ion sensors based on DNAzymes and related DNA molecules. Annu. Rev. Anal. Chem. 2011;4:105–128. doi: 10.1146/annurev.anchem.111808.073617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Silverman SK. Pursuing DNA Catalysts for Protein Modification. Acc. Chem. Res. 2015;48:1369–1379. doi: 10.1021/acs.accounts.5b00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hollenstein M. DNA Catalysis: The Chemical Repertoire of DNAzymes. Molecules. 2015;20:20777–20804. doi: 10.3390/molecules201119730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Renata H, et al. Expanding the Enzyme Universe: Accessing Non-Natural Reactions by Mechanism-Guided Directed Evolution. Angew. Chem. Int. Ed. 2015;54:3351–3367. doi: 10.1002/anie.201409470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roiban GD, Reetz MT. Expanding the toolbox of organic chemists: directed evolution of P450 monooxygenases as catalysts in regio- and stereoselective oxidative hydroxylation. Chem. Commun. 2015;51:2208–2224. doi: 10.1039/c4cc09218j. [DOI] [PubMed] [Google Scholar]
- 26.Blackwell HE, Grubbs RH. Highly Efficient Synthesis of Covalently Cross-Linked Peptide Helices by Ring-Closing Metathesis. Angew. Chem. Int. Ed. 1998;37:3281–3284. doi: 10.1002/(SICI)1521-3773(19981217)37:23<3281::AID-ANIE3281>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 27.Tinoco I, Jr, Bustamante C. How RNA Folds. J. Mol. Biol. 1999;293:271–281. doi: 10.1006/jmbi.1999.3001. [DOI] [PubMed] [Google Scholar]
- 28.Kiss G, et al. Computational enzyme design. Angew. Chem. Int. Ed. 2013;52:5700–5725. doi: 10.1002/anie.201204077. [DOI] [PubMed] [Google Scholar]
- 29.Giger L, et al. Evolution of a designed retro-aldolase leads to complete active site remodeling. Nat. Chem. Biol. 2013;9:494–498. doi: 10.1038/nchembio.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keefe AD, Szostak JW. Functional proteins from a random-sequence library. Nature. 2001;410:715–718. doi: 10.1038/35070613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tawfik DS, Griffiths AD. Man-made cell-like compartments for molecular evolution. Nat. Biotechnol. 1998;16:652–656. doi: 10.1038/nbt0798-652. [DOI] [PubMed] [Google Scholar]
- 32.Miller OJ, et al. Directed evolution by in vitro compartmentalization. Nat. Methods. 2006;3:561–570. doi: 10.1038/nmeth897. [DOI] [PubMed] [Google Scholar]
- 33.Agresti JJ, et al. Selection of ribozymes that catalyse multiple-turnover Diels-Alder cycloadditions by using in vitro compartmentalization. Proc. Natl. Acad. Sci. USA. 2005;102:16170–16175. doi: 10.1073/pnas.0503733102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zaher HS, Unrau PJ. Selection of an improved RNA polymerase ribozyme with superior extension and fidelity. RNA. 2007;13:1017–1026. doi: 10.1261/rna.548807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wochner A, et al. Ribozyme-catalyzed transcription of an active ribozyme. Science. 2011;332:209–212. doi: 10.1126/science.1200752. [DOI] [PubMed] [Google Scholar]
- 36.Ryckelynck M, et al. Using droplet-based microfluidics to improve the catalytic properties of RNA under multiple-turnover conditions. RNA. 2015;21:458–469. doi: 10.1261/rna.048033.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Santoro SW, Joyce GF. A general purpose RNA-cleaving DNA enzyme. Proc. Natl. Acad. Sci. USA. 1997;94:4262–4266. doi: 10.1073/pnas.94.9.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Faulhammer D, Famulok M. The Ca2+ Ion as a Cofactor for a Novel RNA-Cleaving Deoxyribozyme. Angew. Chem. Int. Ed. Engl. 1996;35:2837–2841. [Google Scholar]
- 39.Bruesehoff PJ, et al. Improving metal ion specificity during in vitro selection of catalytic DNA. Comb. Chem. High Throughput Screen. 2002;5:327–335. doi: 10.2174/1386207023330264. [DOI] [PubMed] [Google Scholar]
- 40.Carrigan MA, et al. Quantitative analysis of a RNA-cleaving DNA catalyst obtained via in vitro selection. Biochemistry. 2004;43:11446–11459. doi: 10.1021/bi049898l. [DOI] [PubMed] [Google Scholar]
- 41.Schlosser K, Li Y. A versatile endoribonuclease mimic made of DNA: characteristics and applications of the 8–17 RNA-cleaving DNAzyme. ChemBioChem. 2010;11:866–879. doi: 10.1002/cbic.200900786. [DOI] [PubMed] [Google Scholar]
- 42.Cuenoud B, Szostak JW. A DNA metalloenzyme with DNA ligase activity. Nature. 1995;375:611–614. doi: 10.1038/375611a0. [DOI] [PubMed] [Google Scholar]
- 43.Flynn-Charlebois A, et al. Deoxyribozymes with 2'–5' RNA Ligase Activity. J. Am. Chem. Soc. 2003;125:2444–2454. doi: 10.1021/ja028774y. [DOI] [PubMed] [Google Scholar]
- 44.Baum DA, Silverman SK. Deoxyribozyme-Catalyzed Labeling of RNA. Angew. Chem. Int. Ed. 2007;46:3502–3504. doi: 10.1002/anie.200700357. [DOI] [PubMed] [Google Scholar]
- 45.Büttner L, et al. Site-specific labeling of RNA at internal ribose hydroxyl groups: terbium-assisted deoxyribozymes at work. J. Am. Chem. Soc. 2014;136:8131–8137. doi: 10.1021/ja503864v. [DOI] [PubMed] [Google Scholar]
- 46.Carmi N, et al. In vitro selection of self-cleaving DNAs. Chem. Biol. 1996;3:1039–1046. doi: 10.1016/s1074-5521(96)90170-2. [DOI] [PubMed] [Google Scholar]
- 47.Carmi N, et al. Cleaving DNA with DNA. Proc. Natl. Acad. Sci. USA. 1998;95:2233–2237. doi: 10.1073/pnas.95.5.2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carmi N, Breaker RR. Characterization of a DNA-cleaving deoxyribozyme. Bioorg. Med. Chem. 2001;9:2589–2600. doi: 10.1016/s0968-0896(01)00035-9. [DOI] [PubMed] [Google Scholar]
- 49.Wang M, et al. In vitro selection of DNA-cleaving deoxyribozyme with site-specific thymidine excision activity. Nucleic Acids Res. 2014;42:9262–9269. doi: 10.1093/nar/gku592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang M-Q, et al. Characterization of deoxyribozymes with site-specific oxidative cleavage activity against DNA obtained by in vitro selection. Org. Biomol. Chem. 2016;14:2347–2351. doi: 10.1039/c6ob00148c. [DOI] [PubMed] [Google Scholar]
- 51.Chandra M, et al. DNA-catalyzed sequence-specific hydrolysis of DNA. Nat. Chem. Biol. 2009;5:718–720. doi: 10.1038/nchembio.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gu H, et al. Small, highly active DNAs that hydrolyze DNA. J. Am. Chem. Soc. 2013;135:9121–9129. doi: 10.1021/ja403585e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parker DJ, et al. DNA catalysis of a normally disfavored RNA hydrolysis reaction. J. Am. Chem. Soc. 2013;135:8472–8475. doi: 10.1021/ja4032488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sheppard TL, et al. A DNA enzyme with N-glycosylase activity. Proc. Natl. Acad. Sci. USA. 2000;97:7802–7807. doi: 10.1073/pnas.97.14.7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Höbartner C, et al. Site-selective depurination by a periodate-dependent deoxyribozyme. Chem. Commun. 2007:2255–2257. doi: 10.1039/b704507g. [DOI] [PubMed] [Google Scholar]
- 56.Dokukin V, Silverman SK. Lanthanide Ions as Required Cofactors for DNA Catalysts. Chem. Sci. 2012;3:1707–1714. doi: 10.1039/C2SC01067D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li Y, Breaker RR. Phosphorylating DNA with DNA. Proc. Natl. Acad. Sci. USA. 1999;96:2746–2751. doi: 10.1073/pnas.96.6.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li Y, et al. Capping DNA with DNA. Biochemistry. 2000;39:3106–3114. doi: 10.1021/bi992710r. [DOI] [PubMed] [Google Scholar]
- 59.Sreedhara A, et al. Ligating DNA with DNA. J. Am. Chem. Soc. 2004;126:3454–3460. doi: 10.1021/ja039713i. [DOI] [PubMed] [Google Scholar]
- 60.Chinnapen DJ, Sen D. A deoxyribozyme that harnesses light to repair thymine dimers in DNA. Proc. Natl. Acad. Sci. USA. 2004;101:65–69. doi: 10.1073/pnas.0305943101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sekhon GS, Sen D. Unusual DNA-DNA cross-links between a photolyase deoxyribozyme, UV1C, and its bound oligonucleotide substrate. Biochemistry. 2009;48:6335–6347. doi: 10.1021/bi900531z. [DOI] [PubMed] [Google Scholar]
- 62.Chinnapen DJ, Sen D. Towards elucidation of the mechanism of UV1C, a deoxyribozyme with photolyase activity. J. Mol. Biol. 2007;365:1326–1336. doi: 10.1016/j.jmb.2006.10.062. [DOI] [PubMed] [Google Scholar]
- 63.Barlev A, Sen D. Catalytic DNAs that harness violet light to repair thymine dimers in a DNA substrate. J. Am. Chem. Soc. 2013;135:2596–2603. doi: 10.1021/ja309638j. [DOI] [PubMed] [Google Scholar]
- 64.Thorne RE, et al. A deoxyribozyme, Sero1C, uses light and serotonin to repair diverse pyrimidine dimers in DNA. J. Mol. Biol. 2009;388:21–29. doi: 10.1016/j.jmb.2009.02.064. [DOI] [PubMed] [Google Scholar]
- 65.Li Y, Sen D. A catalytic DNA for porphyrin metallation. Nat. Struct. Biol. 1996;3:743–747. doi: 10.1038/nsb0996-743. [DOI] [PubMed] [Google Scholar]
- 66.Travascio P, et al. DNA-enhanced peroxidase activity of a DNA-aptamer-hemin complex. Chem. Biol. 1998;5:505–517. doi: 10.1016/s1074-5521(98)90006-0. [DOI] [PubMed] [Google Scholar]
- 67.Cheng X, et al. General peroxidase activity of G-quadruplex-hemin complexes and its application in ligand screening. Biochemistry. 2009;48:7817–7823. doi: 10.1021/bi9006786. [DOI] [PubMed] [Google Scholar]
- 68.Pradeepkumar PI, et al. DNA-Catalyzed Formation of Nucleopeptide Linkages. Angew. Chem. Int. Ed. 2008;47:1753–1757. doi: 10.1002/anie.200703676. [DOI] [PubMed] [Google Scholar]
- 69.Wong O, et al. DNA-Catalyzed Covalent Modification of Amino Acid Side Chains in Tethered and Free Peptide Substrates. Biochemistry. 2011;50:4741–4749. doi: 10.1021/bi200585n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chu C, et al. A Generalizable DNA-Catalyzed Approach to Peptide-Nucleic Acid Conjugation. ChemBioChem. 2014;15:1905–1910. doi: 10.1002/cbic.201402255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Walsh SM, et al. DNA Catalysts with Tyrosine Kinase Activity. J. Am. Chem. Soc. 2013;135:14928–14931. doi: 10.1021/ja407586u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Walsh SM, et al. Identification of Sequence-Selective Tyrosine Kinase Deoxyribozymes. J. Mol. Evol. 2015;81:218–224. doi: 10.1007/s00239-015-9699-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chandrasekar J, Silverman SK. Catalytic DNA with Phosphatase Activity. Proc. Natl. Acad. Sci. USA. 2013;110:5315–5320. doi: 10.1073/pnas.1221946110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chandrasekar J, et al. Phosphoserine Lyase Deoxyribozymes: DNA-Catalyzed Formation of Dehydroalanine Residues in Peptides. J. Am. Chem. Soc. 2015;137:9575–9578. doi: 10.1021/jacs.5b06308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brandsen BM, et al. DNA-Catalyzed Lysine Side Chain Modification. Angew. Chem. Int. Ed. 2014;53:9045–9050. doi: 10.1002/anie.201404622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chandra M, Silverman SK. DNA and RNA Can Be Equally Efficient Catalysts for Carbon-Carbon Bond Formation. J. Am. Chem. Soc. 2008;130:2936–2937. doi: 10.1021/ja7111965. [DOI] [PubMed] [Google Scholar]
- 77.Dokukin V, Silverman SK. A modular tyrosine kinase deoxyribozyme with discrete aptamer and catalyst domains. Chem. Commun. 2014;50:9317–9320. doi: 10.1039/c4cc04253k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Golub E, et al. Nucleoapzymes: Hemin/G-Quadruplex DNAzyme-Aptamer Binding Site Conjugates with Superior Enzyme-like Catalytic Functions. J. Am. Chem. Soc. 2016;138:164–172. doi: 10.1021/jacs.5b09457. [DOI] [PubMed] [Google Scholar]
- 79.Bauke Albada H, et al. Rational design of supramolecular hemin/G-quadruplex-dopamine aptamer nucleoapzyme systems with superior catalytic performance. Chem. Sci. 2016;7:3092–3101. doi: 10.1039/c5sc04832j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lermer L, et al. Toward an RNaseA mimic: A DNAzyme with imidazoles and cationic amines. J. Am. Chem. Soc. 2002;124:9960–9961. doi: 10.1021/ja0205075. [DOI] [PubMed] [Google Scholar]
- 81.Santoro SW, et al. RNA cleavage by a DNA enzyme with extended chemical functionality. J. Am. Chem. Soc. 2000;122:2433–2439. doi: 10.1021/ja993688s. [DOI] [PubMed] [Google Scholar]
- 82.Sidorov AV, et al. Sequence-specific cleavage of RNA in the absence of divalent metal ions by a DNAzyme incorporating imidazolyl and amino functionalities. Nucleic Acids Res. 2004;32:1591–1601. doi: 10.1093/nar/gkh326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hollenstein M, et al. A self-cleaving DNA enzyme modified with amines, guanidines and imidazoles operates independently of divalent metal cations (M2+) Nucleic Acids Res. 2009;37:1638–1649. doi: 10.1093/nar/gkn1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hollenstein M, et al. A DNAzyme with three protein-like functional groups: enhancing catalytic efficiency of M2+-independent RNA cleavage. ChemBioChem. 2009;10:1988–1992. doi: 10.1002/cbic.200900314. [DOI] [PubMed] [Google Scholar]
- 85.Hollenstein M, et al. Toward the combinatorial selection of chemically modified DNAzyme RNase A mimics active against all-RNA substrates. ACS Comb. Sci. 2013;15:174–182. doi: 10.1021/co3001378. [DOI] [PubMed] [Google Scholar]
- 86.Zhou C, et al. J. Am. Chem. Soc. Vol. 138. ASAP; 2016. DNA-Catalyzed Amide Hydrolysis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Brandsen BM, et al. DNA-Catalyzed Hydrolysis of Esters and Aromatic Amides. J. Am. Chem. Soc. 2013;135:16014–16017. doi: 10.1021/ja4077233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Taylor AI, et al. Catalysts from synthetic genetic polymers. Nature. 2015;518:427–430. doi: 10.1038/nature13982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Roth A, Breaker RR. An amino acid as a cofactor for a catalytic polynucleotide. Proc. Natl. Acad. Sci. USA. 1998;95:6027–6031. doi: 10.1073/pnas.95.11.6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Emilsson GM, et al. Ribozyme speed limits. RNA. 2003;9:907–918. doi: 10.1261/rna.5680603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Breaker RR, et al. A common speed limit for RNA-cleaving ribozymes and deoxyribozymes. RNA. 2003;9:949–957. doi: 10.1261/rna.5670703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Brown CW, III, et al. Catalytic molecular logic devices by DNAzyme displacement. ChemBioChem. 2014;15:950–954. doi: 10.1002/cbic.201400047. [DOI] [PubMed] [Google Scholar]
- 93.Orbach R, et al. Catalytic nucleic acids (DNAzymes) as functional units for logic gates and computing circuits: from basic principles to practical applications. Chem. Commun. 2015;51:4144–4160. doi: 10.1039/c4cc09874a. [DOI] [PubMed] [Google Scholar]
- 94.Wang F, et al. DNA switches: from principles to applications. Angew. Chem. Int. Ed. 2015;54:1098–1129. doi: 10.1002/anie.201404652. [DOI] [PubMed] [Google Scholar]
- 95.Zhang M, et al. Investigation of a deoxyribozyme as a biofuel cell catalyst. J. Am. Chem. Soc. 2011;133:15890–15893. doi: 10.1021/ja206787h. [DOI] [PubMed] [Google Scholar]
- 96.Bobbin ML, Rossi JJ. RNA Interference (RNAi)-Based Therapeutics: Delivering on the Promise? Annu. Rev. Pharmacol. Toxicol. 2016;56:103–122. doi: 10.1146/annurev-pharmtox-010715-103633. [DOI] [PubMed] [Google Scholar]
- 97.Young DD, et al. Activation and deactivation of DNAzyme and antisense function with light for the photochemical regulation of gene expression in mammalian cells. J. Am. Chem. Soc. 2010;132:6183–6193. doi: 10.1021/ja100710j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cho EA, et al. Safety and tolerability of an intratumorally injected DNAzyme, Dz13, in patients with nodular basal-cell carcinoma: a phase 1 first-in-human trial (DISCOVER) Lancet. 2013;381:1835–1843. doi: 10.1016/S0140-6736(12)62166-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Krug N, et al. Allergen-induced asthmatic responses modified by a GATA3-specific DNAzyme. N. Engl. J. Med. 2015;372:1987–1995. doi: 10.1056/NEJMoa1411776. [DOI] [PubMed] [Google Scholar]
- 100.Fokina AA, et al. Targeting insulin-like growth factor I with 10–23 DNAzymes: 2'-O-methyl modifications in the catalytic core enhance mRNA cleavage. Biochemistry. 2012;51:2181–2191. doi: 10.1021/bi201532q. [DOI] [PubMed] [Google Scholar]
- 101.Xiang Y, Lu Y. DNA as sensors and imaging agents for metal ions. Inorg. Chem. 2014;53:1925–1942. doi: 10.1021/ic4019103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li J, et al. In vitro selection and characterization of a highly efficient Zn(II)-dependent RNA-cleaving deoxyribozyme. Nucleic Acids Res. 2000;28:481–488. doi: 10.1093/nar/28.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wu P, et al. A DNAzyme-gold nanoparticle probe for uranyl ion in living cells. J. Am. Chem. Soc. 2013;135:5254–5257. doi: 10.1021/ja400150v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Torabi S-F, et al. In vitro selection of a sodium-specific DNAzyme and its application in intracellular sensing. Proc. Natl. Acad. Sci. USA. 2015;112:5903–5908. doi: 10.1073/pnas.1420361112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hollenstein M, et al. A Highly Selective DNAzyme Sensor for Mercuric Ions. Angew. Chem. Int. Ed. 2008;47:4346–4350. doi: 10.1002/anie.200800960. [DOI] [PubMed] [Google Scholar]
- 106.Willner I, et al. DNAzymes for sensing, nanobiotechnology and logic gate applications. Chem. Soc. Rev. 2008;37:1153–1165. doi: 10.1039/b718428j. [DOI] [PubMed] [Google Scholar]
- 107.Xiang Y, Lu Y. An invasive DNA approach toward a general method for portable quantification of metal ions using a personal glucose meter. Chem. Commun. 2013;49:585–587. doi: 10.1039/c2cc37156a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wang F, et al. Amplified analysis of DNA by the autonomous assembly of polymers consisting of DNAzyme wires. J. Am. Chem. Soc. 2011;133:17149–17151. doi: 10.1021/ja2076789. [DOI] [PubMed] [Google Scholar]
- 109.Shen Z, et al. A Catalytic DNA Activated by a Specific Strain of Bacterial Pathogen. Angew. Chem. Int. Ed. 2016;55:2431–2434. doi: 10.1002/anie.201510125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Xiao Y, et al. Catalytic beacons for the detection of DNA and telomerase activity. J. Am. Chem. Soc. 2004;126:7430–7431. doi: 10.1021/ja031875r. [DOI] [PubMed] [Google Scholar]
- 111.Shimron S, et al. Amplified detection of DNA through the enzyme-free autonomous assembly of hemin/G-quadruplex DNAzyme nanowires. Anal. Chem. 2012;84:1042–1048. doi: 10.1021/ac202643y. [DOI] [PubMed] [Google Scholar]
- 112.Yin B-C, et al. An allosteric dual-DNAzyme unimolecular probe for colorimetric detection of copper(II) J. Am. Chem. Soc. 2009;131:14624–14625. doi: 10.1021/ja9062426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Nowakowski J, et al. Crystal structure of an 82-nucleotide RNA-DNA complex formed by the 10–23 DNA enzyme. Nat. Struct. Biol. 1999;6:151–156. doi: 10.1038/5839. [DOI] [PubMed] [Google Scholar]
- 114.Ponce-Salvatierra A, et al. Crystal structure of a DNA catalyst. Nature. 2016;529:231–234. doi: 10.1038/nature16471. [DOI] [PubMed] [Google Scholar]
- 115.Purtha WE, et al. General Deoxyribozyme-Catalyzed Synthesis of Native 3’–5’ RNA Linkages. J. Am. Chem. Soc. 2005;127:13124–13125. doi: 10.1021/ja0533702. [DOI] [PubMed] [Google Scholar]
- 116.Pyle AM. Ribozymes: a distinct class of metalloenzymes. Science. 1993;261:709–714. doi: 10.1126/science.7688142. [DOI] [PubMed] [Google Scholar]
- 117.Murray JB, et al. The hammerhead, hairpin and VS ribozymes are catalytically proficient in monovalent cations alone. Chem. Biol. 1998;5:587–595. doi: 10.1016/s1074-5521(98)90116-8. [DOI] [PubMed] [Google Scholar]
- 118.Jayasena VK, Gold L. In vitro selection of self-cleaving RNAs with a low pH optimum. Proc. Natl. Acad. Sci. USA. 1997;94:10612–10617. doi: 10.1073/pnas.94.20.10612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Geyer CR, Sen D. Evidence for the metal-cofactor independence of an RNA phosphodiester-cleaving DNA enzyme. Chem. Biol. 1997;4:579–593. doi: 10.1016/s1074-5521(97)90244-1. [DOI] [PubMed] [Google Scholar]
- 120.Mazumdar D, et al. Activity, Folding and Z-DNA Formation of the 8–17 DNAzyme in the Presence of Monovalent Ions. J. Am. Chem. Soc. 2009;131:5506–5515. doi: 10.1021/ja8082939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wachowius F, et al. Combinatorial mutation interference analysis reveals functional nucleotides required for DNA catalysis. Angew. Chem. Int. Ed. 2010;49:8504–8508. doi: 10.1002/anie.201003940. [DOI] [PubMed] [Google Scholar]
- 122.Das SR, Piccirilli JA. General acid catalysis by the hepatitis delta virus ribozyme. Nat. Chem. Biol. 2005;1:45–52. doi: 10.1038/nchembio703. [DOI] [PubMed] [Google Scholar]
- 123.Wilcox JL, et al. Charged nucleobases and their potential for RNA catalysis. Acc. Chem. Res. 2011;44:1270–1279. doi: 10.1021/ar2000452. [DOI] [PMC free article] [PubMed] [Google Scholar]