Summary
Can a heterotrophic organism be evolved to synthesize biomass from CO2 directly? So far, non-native carbon fixation in which biomass precursors are synthesized solely from CO2 has remained an elusive grand challenge. Here, we demonstrate how a combination of rational metabolic rewiring, recombinant expression, and laboratory evolution has led to the biosynthesis of sugars and other major biomass constituents by a fully functional Calvin-Benson-Bassham (CBB) cycle in E. coli. In the evolved bacteria, carbon fixation is performed via a non-native CBB cycle, while reducing power and energy are obtained by oxidizing a supplied organic compound (e.g., pyruvate). Genome sequencing reveals that mutations in flux branchpoints, connecting the non-native CBB cycle to biosynthetic pathways, are essential for this phenotype. The successful evolution of a non-native carbon fixation pathway, though not yet resulting in net carbon gain, strikingly demonstrates the capacity for rapid trophic-mode evolution of metabolism applicable to biotechnology.
PaperClip
Graphical Abstract
Highlights
-
•
Non-native Calvin-Benson cycle allows for sugar synthesis from CO2 in E. coli
-
•
Metabolic cutoff allows for the decoupling of energy harvesting from biomass synthesis
-
•
Chemostat-based directed evolution led to the emergence of sugar synthesis from CO2
-
•
Mutations in flux branchpoints are essential for the CBB cycle stable operation
Metabolic rewiring and directed evolution lead to a fully functional, non-native carbon fixation cycle, which synthesizes sugars and other major biomass components in E. coli.
Introduction
Whether CO2 can or cannot be transformed into sugar and biomass by carbon fixation is arguably the most basic distinction we make in defining the metabolism of an organism. Carbon fixation by autotrophs is the biochemical gateway to the organic world, as obligate heterotrophs are dependent on this supply of organic carbon. How difficult is it to evolve from one trophic mode of growth to another? Specifically, can the ability to synthesize biomass from CO2 be introduced into a heterotrophic organism? Exploring the process in which a heterotrophic bacterium, such as Escherichia coli, is evolved to synthesize sugars from CO2 can serve as a model system to tackle these questions.
The utilization of one-carbon compounds, such as CO2, methanol, and methane, has been recently drawing attention as a low-cost, abundant feedstock option for biochemical production (Li et al., 2012, Müller et al., 2015, Siegel et al., 2015). Recent studies have demonstrated that a wide variety of non-native metabolic pathways can be integrated into model microorganisms, allowing the synthesis of value-added chemicals using sugar as a feedstock (Galanie et al., 2015, Yim et al., 2011). However, efforts to synthesize sugar from inorganic CO2 by introducing a non-native carbon fixation cycle have never been successful. Carbohydrate biosynthesis through carbon fixation in E. coli would not only open exciting avenues to directly utilize CO2 for chemical production, but could also serve as a platform to rapidly optimize carbon fixation enzymes and pathways for subsequent implementation in agricultural crops (Lin et al., 2014, Mueller-Cajar and Whitney, 2008, Parikh et al., 2006, Shih et al., 2014). Furthermore, this experimental approach could shed light on cellular adaptations associated with horizontal-gene-transfer events, on the plasticity of metabolic networks, and on the evolutionary emergence of a biological novelty.
The achievement of novel biological phenotypes on laboratory timescales is at the heart of efforts such as the long-term evolutionary experiment, which studied how E. coli developed the ability to utilize citrate throughout several tens of thousands of generations (Blount et al., 2008, Maddamsetti et al., 2015, Wiser et al., 2013). These studies show the intricate dynamics of potentiating, actualizing, and refining steps during the evolutionary process (Quandt et al., 2015) and shed light on the interplay between selection, historical contingency, and epistatic effects. In parallel to lab evolution, which forces the selective conditions, synthetic biology efforts manipulate the genetic makeup with the aim of rationally designing desired phenotypes (Church et al., 2014, Galanie et al., 2015). In spite of significant progress, nothing as extreme as expressing a fully functional pathway that changes the trophic mode of an organism has been ever shown to be achievable.
The Calvin-Benson-Bassham cycle (Bassham et al., 1954) is, by far, the most dominant carbon fixation pathway in the biosphere out of all six known natural alternatives (Fuchs, 2011, Bar-Even et al., 2012). Previous efforts to show functional expression of the CBB cycle carboxylating enzyme, ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO), in E. coli have relied on the supply of a glycolytic carbon source (e.g., glucose) to replenish RuBisCO’s substrate, ribulose-bisphosphate (RuBP) (Durão et al., 2015, Gong et al., 2015, Mueller-Cajar and Whitney, 2008, Parikh et al., 2006, Zhuang and Li, 2013). These important studies therefore did not achieve the defining function of the CBB cycle of autocatalytic sugar synthesis from inorganic carbon. While this approach led to the elegant construction of a RuBisCO-dependent E. coli as a platform for directed evolution of RuBisCO activity (Durão et al., 2015, Mueller-Cajar and Whitney, 2008, Parikh et al., 2006), evolving a complete carbon fixation cycle capable of autocatalytically generating biomass from CO2 has remained an open challenge.
Here, we report achieving a fully functional and autocatalytic carbon fixation cycle in E. coli, capable of hexose, pentose and triose sugar synthesis with no input of organic carbon into the cycle. Energy and reducing power are supplied by the oxidation of an organic acid (pyruvate) in an isolated metabolic module and thus no net carbon gain is achieved at this point. This is the first example of a non-native carbon assimilation cycle in which all of the pathway intermediates and products are solely synthesized from CO2 and the required co-factors.
Results
Metabolic Cutoff in Gluconeogenesis Allows for the Decoupling of Energy Harvesting from Biomass Synthesis in E. coli
Out of the dozen of reactions utilized by autotrophs in the CBB cycle for the biosynthesis of sugars from CO2, only two enzymatic activities are absent in E. coli: phosphoribulokinase (prk) and RuBisCO. The native enzymes of gluconeogenesis and the pentose phosphate pathway in E. coli can catalyze all other reactions (Figure S1). Therefore, heterologous expression of RuBisCO and prk could, in principle, equip E. coli with the enzymatic machinery needed to execute all CBB cycle reactions and achieve the synthesis of sugar, as well as other major biomass constituents, from CO2.
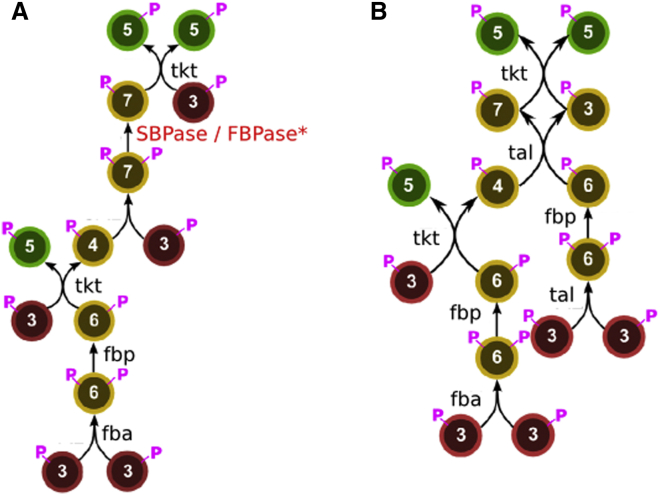
Figure S1.
Endogenous Pentose Phosphate Pathway Enzymes of E. coli Can Regenerate Pentoses in the Non-native CBB Cycle, Related to Figure 1
(A) Pentose regeneration in the CBB cycle is achieved in most photosynthetic organisms by utilizing a designated sedoheptulose-bisphosphate phosphatase (SBPase) to dephosphorylate sedoheptulose-bisphosphate (SBP). Alternatively, pentose regeneration can occur using a bifunctional FBPase as was found to be the case in cyanobacteria.
(B) While the E.coli genome does not contain bona fide SBPase, pentose-phosphates regeneration can be accomplished in an SBP-independent manner using the native enzymes of the pentose phosphate pathway (PPP). In Figure 1 we depict option (B) but E. coli can also operate option (A) using promiscuous activity of the endogenous FBPase.
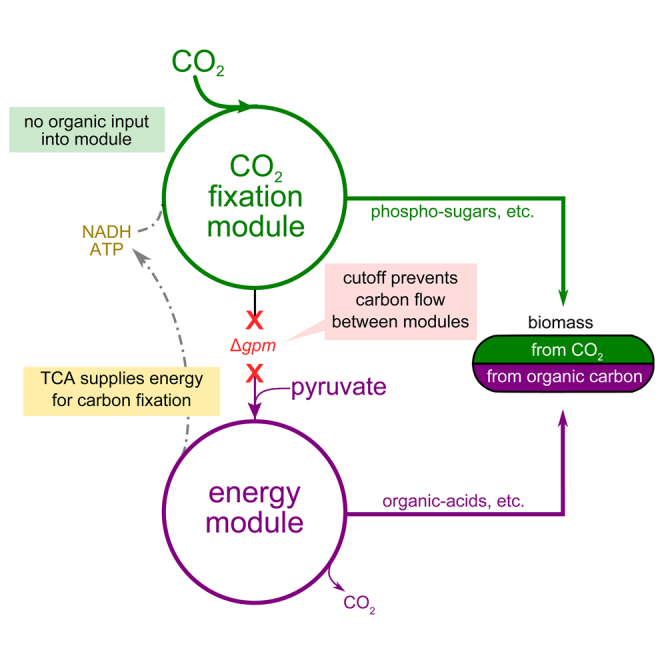
Inspired by the powerful tool of genetic screens, in which genomic perturbations and controlled growth conditions are combined to identify new phenotypes, we have used a computational framework based on flux balance analysis to analyze hundreds of gene-deletion combinations and identify perturbations in central carbon metabolism that, together with RuBisCO and prk expression, couple the rate of carbon fixation to cellular fitness (Figures S2 and S3; Experimental Procedures). Our analysis pointed to a yet unexplored scenario in which targeted severing of gluconeogenesis would decouple energy harvesting and biomass production into two independent metabolic networks (Figure 1). This rewired metabolism could drive the synthesis of sugars and other essential biomass components from CO2 as the sole carbon source, while production of cellular energy would be obtained from the catabolism of an organic carbon in the energy module. This scenario goes beyond previously described RuBisCO-dependent strains (Gong et al., 2015, Mueller-Cajar and Whitney, 2008, Parikh et al., 2006, Zhuang and Li, 2013) by employing a fully functional autocatalytic CBB cycle in which, as in autotrophic organisms, CO2, ATP, and reducing power are the only inputs for the synthesis of sugars. We termed this mode of growth, not known to occur in nature, hemiautotrophic growth (i.e., half-autotrophic, as it cuts metabolism into two parts, only one of which is autotrophic) and aimed to explore it in the lab. Our design for decoupling energy harvesting and CO2 fixation does not result in a net gain of carbon when energy is obtained from the oxidation of an organic carbon source (e.g., pyruvate). However, it creates a malleable platform to test the functionality of synthetic carbon fixation pathways and offers powerful modularity in the choice of energy module for carbon fixation.
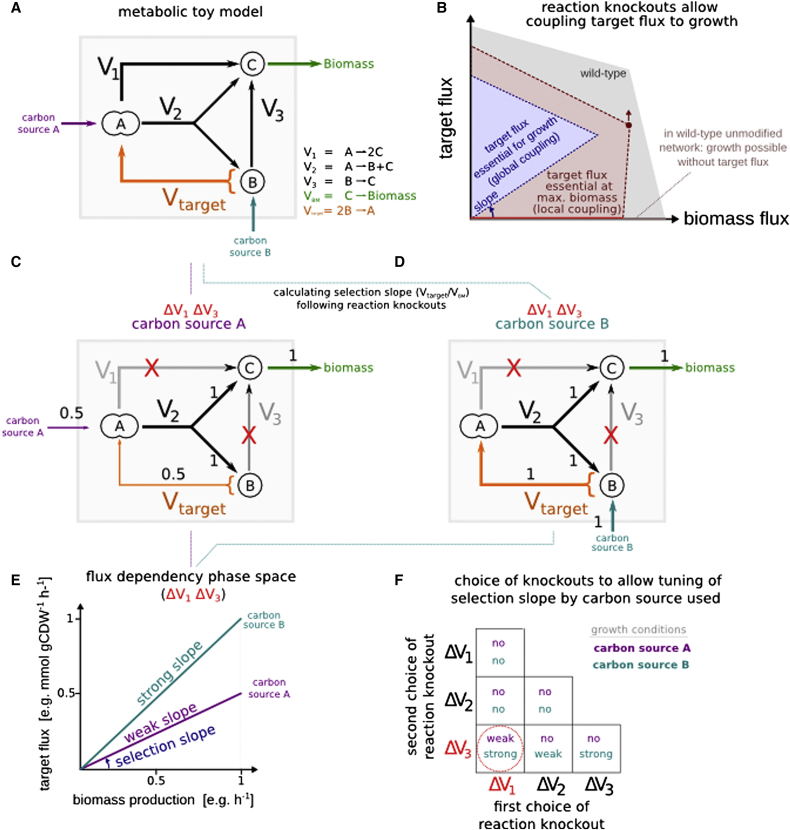
Figure S2.
Systematic Identification of Metabolic Configurations in Which Pathway Activity Is Coupled to Cellular Growth, Related to Figure 1
(A) Metabolic toy model containing three endogenous reactions (V1-V3) and one target recombinant reaction (Vtarget). Flux through the target reaction is not essential for biomass generation on either of two considered carbon sources (turquoise, purple).
(B) Computational frameworks for identifying gene deletions leading to the overproduction of a pre-specified metabolite (such as OptKnock) ensure maximal flux through the target pathway at the point of maximal biomass production (red background). However, maximal biomass production may not be the prevailing growth condition so global coupling between target flux and biomass production (red line) is not ensured. We performed an exhaustive search for combinations of reaction knockouts in which Vtarget is globally essential for growth (blue background). For example, eliminating reactions V1 and V3 results in making Vtarget essential for growth on either of the two carbon sources.
(C) While metabolizing carbon source A, the selection slope, defined as Vtarget/VBM at the origin, is 0.5.
(D) When carbon source B is metabolized, the selection slope increases, now requiring 1 unit of target flux per biomass flux, and thus a tighter coupling.
(E) Flux dependency space depicting the feasible fluxes for each of the metabolic configurations previously described. While the unmodified network allows biomass to be generated independently from the target flux (gray line), by eliminating reactions V1 and V3 the flux space is reduced only to flux modes in which biomass generation is coupled to the target flux; either with weak or strong coupling slope (purple and turquoise lines, respectively).
(F) If several alternatives lead to coupling, choosing a combination of knockouts that yields variable selection slopes on different carbon sources (red circle) is preferred, thus giving the experimentalist options to increase the selection stringency in steps. The software implementation is detailed in the Supplemental Experimental Procedures.
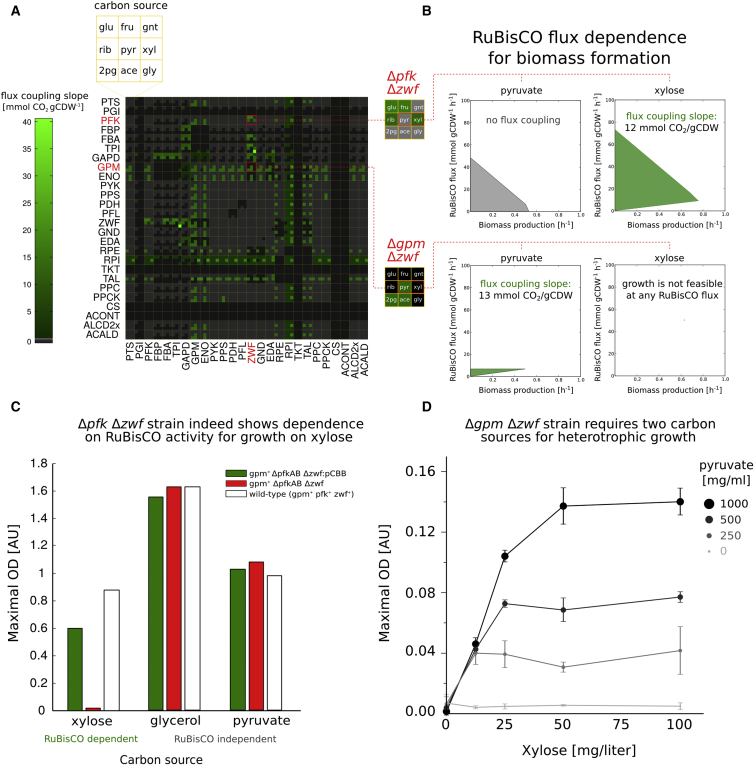
Figure S3.
Systematic Approach for Identifying Metabolic Configurations in Which Carbon Fixation Activity Is Essential for Cell Growth, Related to Figure 1
(A) Each block (see 3x3 yellow grid, top left) refers to a combination of two knockouts in metabolic reactions within central carbon metabolism of E. coli (knockout genes marked on axes). We computationally analyzed the coupling of growth (biomass formation) to RuBisCO target flux in each combination of reaction knockouts for nine different carbon sources (Supplemental Experimental Procedures). The coupling between target flux and biomass production is shown using a color code. Feasible fluxes are calculated for the perturbed metabolic network (Figure S2; Supplemental Experimental Procedures) when grown on each of the nine carbon sources as inputs (in addition to CO2). Metabolic configurations (a specific combination of knockout mutations and carbon source) in which carbon fixation activity is essential for cell growth are highlighted in shades of green. The coupling slope is defined as the minimal carbon fixation flux, required per unit of biomass produced. Gray cells refer to configurations in which carbon fixation is not essential for growth, while black cells are configurations where growth is not possible.
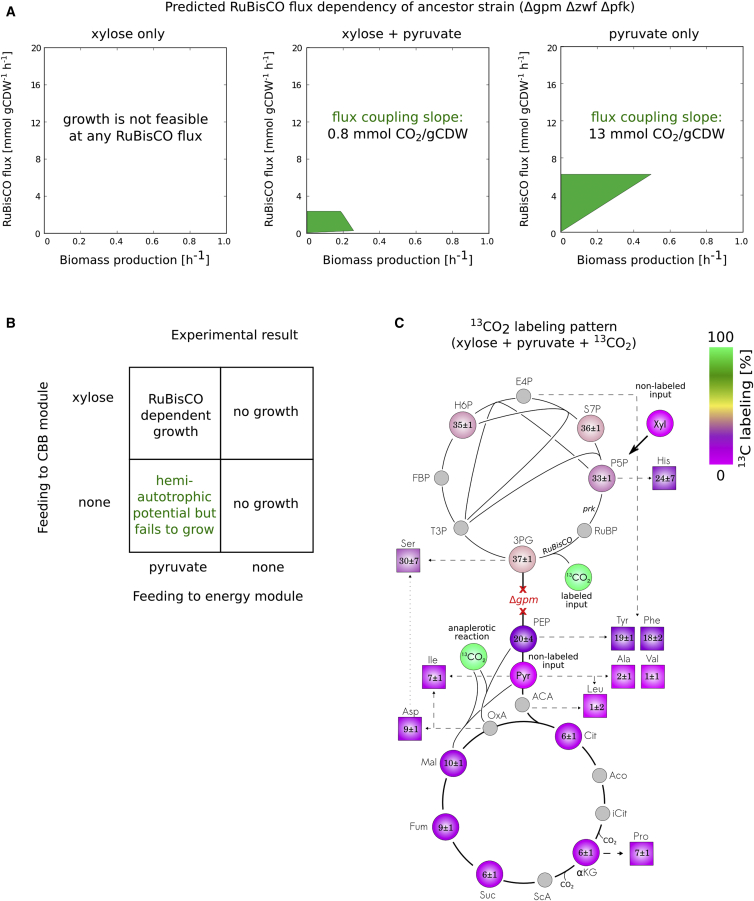
(B) The flux requirements for biomass formation of four metabolic configurations. A mutant strain which lacks the enzymatic activities of phosphofructokinase (pfkA and pfkB, denoted for simplicity as Δpfk) and glucose-6-phosphate dehydrogenase (Δzwf) is predicted to grow on pyruvate even without RuBisCO flux, and it thus contains points along the x axis and is colored gray. In contrast, the same set of knockouts when using pentoses (e.g., xylose) requires flux through RuBisCO (and prk). This result stems from the stoichiometry of the pentose phosphate pathway which converts three pentose sugars into two molecules of fructose-6-P (F6P) and a glyceraldehyde 3-phosphate triose. Downstream glycolysis of F6P requires pfk activity to generate fructose-1,6-P and feed the glycolytic pathway. Therefore, pfk deficient mutant is predicted not to grow on pentose sugars as a single carbon source without carbon fixation. Recombinant expression of RuBisCO and prk creates a pfk independent route from pentoses to trioses through the RuBisCO-dependent carboxylation of RuBP to 3-phosphoglycerate, hence rescuing growth. In contrast to pentose metabolism, three-carbon inputs to glycolysis (e.g., pyruvate) do not require pfk activity for catabolism, nor for the gluconeogenic biosynthesis of hexose phosphates which takes place via fructose 1,6-bisphosphatase (fbp). An interesting scenario is the combined knockout of zwf and phosphoglycerate mutase (gpmA and gpmM, denoted for simplicity as Δgpm). In this strain, expression of CBB enzymes allows growth on carbon sources which enter central carbon metabolism through lower glycolysis (e.g., pyruvate, bottom left). The metabolic cutoff prevents gluconeogenic carbon flow and therefore sugar synthesis from the organic carbon source. Since sugars (e.g., glucose-6-phosphate, ribose-5-phosphate) are essential components for biomass synthesis, heterotrophic growth is not feasible. However, pyruvate can be readily metabolized in the TCA cycle to generate ATP and reducing power, hence supplying the energy to allow carbon fixation using a non-native CBB cycle. We term such growth hemiautotrophic. The RuBisCO-dependent assimilation of CO2 into biomass, supplies the demand for biomass precursors, such as sugars, which cannot be synthesized from organic carbon in this Δgpm Δzwf strain. Uptake rate was set to 10 mmol gCDW-1 h-1 for xylose, and 16.7 for pyruvate.
(C) The RuBisCO dependency of a Δpfk Δzwf strain was experimentally validated. Gene deletions were performed using iterative P1 transductions (Supplemental Methods) and growth was tested on permissive (i.e., glycerol or pyruvate) versus restrictive carbon sources (i.e., xylose) under elevated CO2 conditions (pCO2 = 0.1 atm). As predicted from the stoichiometric model, growth on xylose was observed only in pCBB transformants, expressing RuBisCO and prk.
(D) E. coli BW25133, with an unperturbed metabolic network, can grow heterotrophically on minimal media supplied with a single carbon source. In contrast, a metabolically perturbed Δgpm Δzwf strain requires a minimum of two carbon sources for heterotrophic growth, each feeding into one of the two disconnected sub networks formed by the metabolic cutoff. When two carbon sources are co-supplied (e.g., xylose and pyruvate), lack of either one would limit growth, even in excess of the other. As the experimental results show, per given concentration of supplied pyruvate (feeding the energy module), maximal biomass formation is initially proportional to the amount of supplied xylose. An excess of xylose beyond the point in which the ratio of carbon sources matches the consumption uptake ratio, does not contribute to the formation of additional biomass since growth becomes limited by the availability of pyruvate. Knowledge of these different domains can be used to ensure a feeding ratio in the chemostat that will ensure xylose limited conditions with excess pyruvate ready to be used for making energy for carbon fixation. In a hemiautotrophic growth mode (in the presence of a fully functional CBB cycle), growth can be obtained using only pyruvate and CO2.
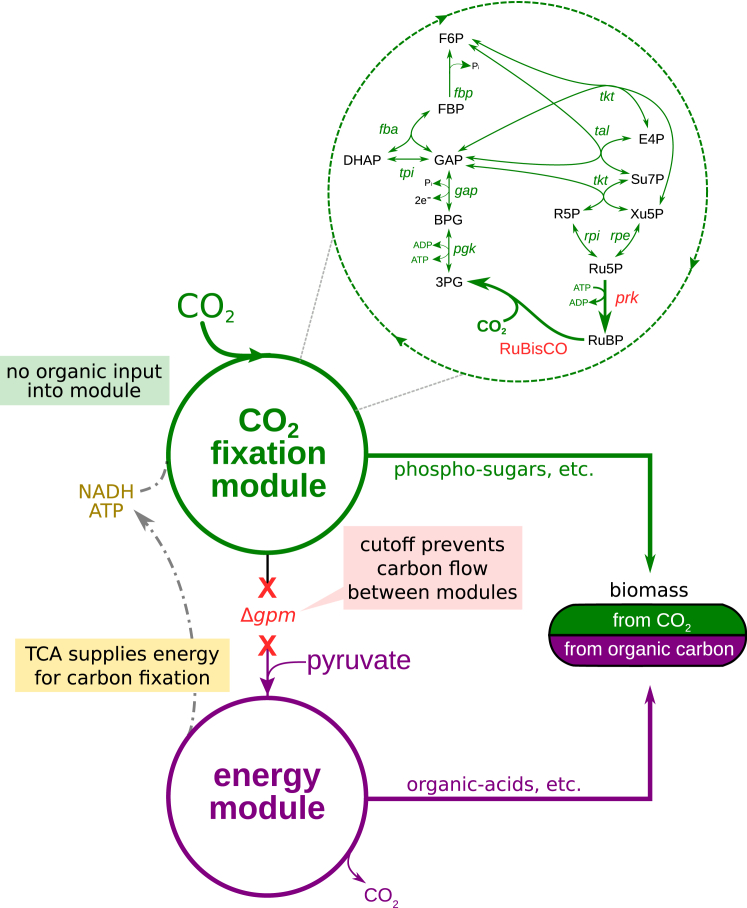
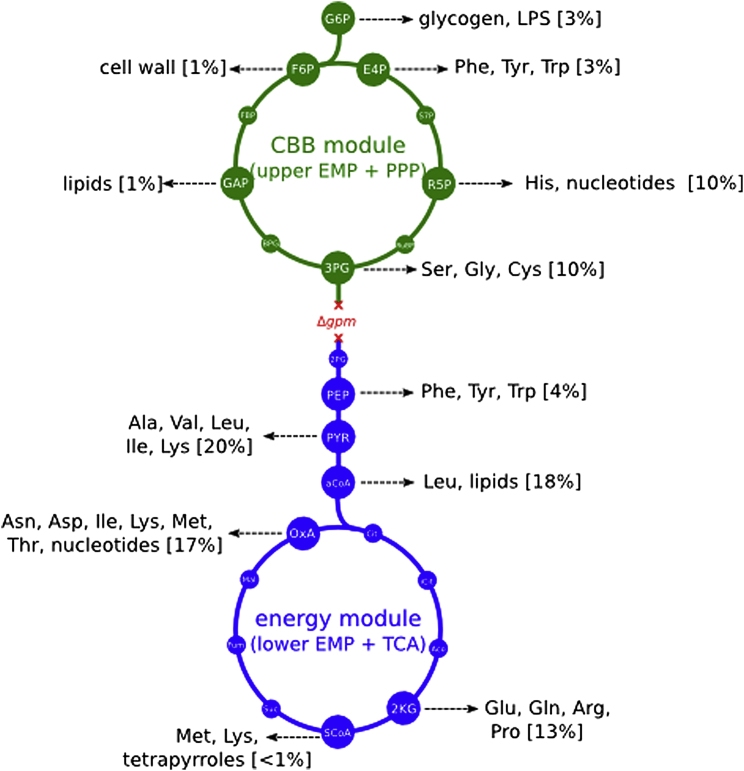
Figure 1.
Decoupling Energy Production and Carbon Fixation in E. coli to Achieve Hemiautotrophic Growth
Two recombinant enzymes are needed to complete a carbon fixation cycle in E. coli: RuBisCO, the carboxylating enzyme, and the kinase prk. The remaining reactions required for the reduction and substrate regeneration phases of the cycle are endogenous to the metabolic network of the host, as part of gluconeogenesis and the pentose phosphate pathway. Deletion of the phosphoglycerate mutase genes (ΔgpmA and ΔgpmM) disrupts carbon flow in the glycolytic/gluconeogenic backbone and generates two disconnected sub-networks: (1) a carbon fixation module containing upper glycolysis, the pentose phosphate pathway, and the two foreign CBB enzymes and (2) an energy module, containing lower glycolysis and the TCA cycle, supplying reducing power and ATP. In a scenario in which an organic carbon source (e.g., pyruvate) is utilized by the energy module to supply the energetic demands of the carbon fixation cycle, the cellular building blocks derived from carbon fixation module metabolites (e.g., phospho sugars, such as ribose-P; see Figure S4 for details) can be synthesized from inorganic carbon using the non-native CBB cycle. The remainder of the biomass building blocks (those emanating from the energy module metabolites, e.g., organic acids for many of the amino acids), as well as the energy requirements of the cell, are supplied directly via the catabolism of the organic carbon source. In such a hemiautotrophic growth mode, CO2 and energy carriers are the sole inputs for the production of biomass precursors in a carbon fixation cycle.
See also Figures S1, S2, S3, and S4.
Heterologous Expression of CBB Cycle Components Is Not Sufficient to Sustain Hemiautotrophic Growth
We have experimentally followed this scenario by severing gluconeogenesis through the deletion of the phosphoglycerate mutase genes gpmA and gpmM. These deletions separate central metabolism into two disconnected sub-networks (Figure 1): (1) a carbon-fixing CBB module containing upper glycolysis, the pentose phosphate pathway, and the recombinant CBB enzymes and (2) an energy module containing lower glycolysis and the TCA cycle, which supplies ATP and reducing equivalents to power carbon fixation by the first module. Notably, all of the six cellular phospho-sugars (G6P, F6P, R5P, E4P, GAP, and 3PG) that are starting points for biosynthetic pathways diverging out of central carbon metabolism have to be synthesized completely from CO2 by the CBB module (Figure S4). An organic-acid feeding the energy module, such as pyruvate, could provide reducing power and ATP for carbon fixation and thereby allow the synthesis of biomass precursors in the CBB module from CO2. Importantly, our metabolic design couples the biosynthesis of sugars and other essential biomass constituents to the activity of the complete carbon fixation cycle, and not to individual enzymatic steps or sub-modules in the pathway.
Figure S4.
Schematic Representation of the Main Sinks for Biomass Precursor Metabolites in Central Carbon Metabolism, Related to Figure 1
12 precursor metabolites in glycolysis, the PPP and the TCA cycle serve as metabolic branchpoints, in which flux branches out from central carbon metabolism to build the components of the cell’s biomass. We detail the percentage (by mass) contributed to the total of cellular carbon derived from each of these key metabolites (as calculated based on Neidhardt, 1987). This serves as a basis for predicting the expected total labeled carbon fraction in the hemiautotrophic strain. In mutant E. coli lacking the phosphoglycerate mutase activity (gpmA, gpmM) central carbon metabolism is divided into two sub-networks disconnected in terms of carbon flow between them. Due to the cutoff, heterotrophic growth requires a minimum of two carbon sources: one feeding lower glycolysis and the TCA cycle (energy module, blue), while the other feeding upper glycolysis and the pentose phosphate pathway (CBB module, green). The relative percentage of the biomass emanating from precursor metabolites in the CBB module sums to approximately 30% of total biomass in the cell (or almost equivalently, carbon biomass), while the metabolites derived from the energy module contribute the remaining 70%. In evolved cells showing a hemiautotrophic phenotype, CO2 is the sole carbon input into the CBB module, hence about 30% of the cell’s carbon is expected to be supplied from RuBisCO-dependent assimilation of inorganic carbon.
In contrast to our computational prediction, a gpm double-deletion mutant expressing recombinant RuBisCO, prk, and carbonic anhydrase, provided pyruvate as a single organic carbon source, failed to grow in an elevated CO2 atmosphere. This strain grew only when a second carbon source, a pentose sugar directly feeding the CBB module, was also provided (e.g., xylose), thus allowing biomass precursors to be synthesized from the externally supplied sugar (Figures 2A and S3D). Since our model does not account for regulatory and kinetic effects, we hypothesized that the organism failed to grow hemiautotrophically because a finely tuned flux distribution might be required, for example, to balance the rate of sugar uptake by biosynthetic pathways branching from the CBB cycle intermediates with the rate of sugar biosynthesis from CO2 fixation.
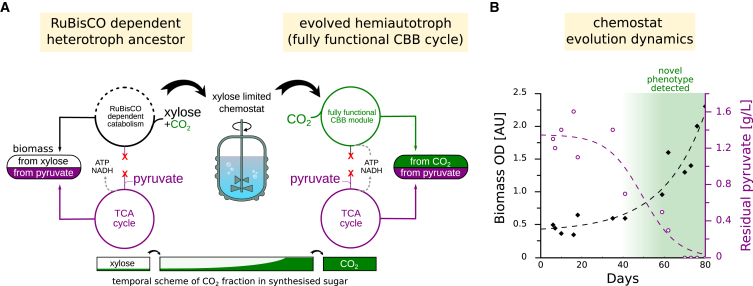
Figure 2.
Chemostat Evolution Leads to a Hemiautotrophic Phenotype
(A) The ancestor strain (left) containing gpmA and gpmM deletions was evolved in a xylose-limited chemostat supplied with an excess of pyruvate and CO2. Additional deletions of pfkA, pfkB, and zwf resulted in RuBisCO-dependent (but not hemiautotrophic) catabolism of xylose in the initial heterotrophic growth (Figure S5). Propagation in a chemostat ensured xylose-limited growth, resulting in strong selective pressure toward increased carbon fixation flux. When mutations arise that create a fully functioning CBB cycle, they enable CO2 to be the sole carbon input for the required biomass precursors. If carbon fixation by the CBB cycle could meet cellular demand for phospho-sugars, growth dependency on xylose would be alleviated and the hemiautotrophic strain (right) could take over the population.
(B) Around day 50 of chemostat evolution, an increase in optical density (OD) (black diamonds) and a decrease in pyruvate concentration (purple circles) were observed, indicating a takeover by an evolved clone with a metabolically distinct phenotype. Culture samples from day ≈50 onward (shaded green) were able to grow in minimal media with pyruvate as the only organic carbon source in an elevated CO2 atmosphere, both on liquid media and agar plates. Dashed lines are sigmoidal fits.
See also Figure S5.
Continuous Evolution in a Chemostat of RuBisCO-Expressing Cells Leads to Sugar Synthesis from CO2
While rational design is limited in predicting necessary changes in regulatory or kinetic parameters, directed evolution is a powerful alternative to explore the multivariate fine-tuning required for operating non-native pathways (Sauer, 2001). Our metabolic design, which couples the biosynthesis of sugars and derived biomass components to the functionality of the non-native CBB cycle, allowed us to harness natural selection by using a chemostat-based evolution regimen that continuously maintained selective conditions for a hemiautotrophic phenotype. The ancestral strain of this evolution experiment was a double-knockout gpm strain expressing recombinant RuBisCO, prk, and carbonic anhydrase with the additional deletions of phosphofructokinase (pfkA and pfkB) and glucose-6-phosphate-1-dehydrogenase (zwf). Collectively, these metabolic manipulations made RuBisCO carboxylation essential for xylose catabolism during heterotrophic growth and eliminated oxidation of hexose sugars through the oxidative pentose phosphate pathway (Figure S5).
Figure S5.
Metabolic Perturbations Allow Selections for RuBisCO Activity during Chemostat Evolution toward Hemiautotrophic Growth, Related to Figure 2
(A) In contrast to our stoichiometric model prediction, a Δgpm Δzwf strain expressing the pCBB plasmid failed to grow hemiautotrophically in an elevated CO2 atmosphere while provided with pyruvate as a single organic carbon source. To harness natural selection toward establishing hemiautotrophic growth we analyzed the effect of further deleting phosphofructokinase (pfkA and pfkB) in the context of a xylose limited (pyruvate and CO2 enriched) chemostat evolution experiment. As indicated in the phenotypic phase space resulting from the computational analysis (Supplemental Methods), pfk deletion does not affect the feasibility of hemiautotrophic growth (using solely pyruvate as an organic carbon source), as phosphofructokinase is not required to operate the CBB cycle. However, this additional deletion couples carbon fixation by RuBisCO to cellular growth even when a second organic carbon source feeding directly to the CBB module (e.g., xylose) is supplied in addition to pyruvate (Figure S3), as occurs during xylose limited chemostat evolution. Usefully for the adaptation, when xylose is supplied (in addition to pyruvate and CO2), the minimal RuBisCO activity required to support growth is significantly lower than the activity required for hemiautotrophic growth. Furthermore, under these conditions, while xylose utilization is dependent on the activity of RuBisCO, the CBB cycle is not required to operate in an autocatalytic manner, as required in the hemiautotrophic growth mode (Figure 1). Uptake rate was set to 10 mmol gCDW-1 h-1 for xylose, 16.7 for pyruvate and 6.2 mmol gCDW-1 h-1 for each of the two carbon sources when combined.
(B) We constructed the Δgpm Δzwf Δpfk strain using iterative P1 transductions and transformed it with the pCBB plasmid (Experimental Procedures). In contrast to the predicted feasibility of hemiautotrophic growth for this strain, growth on minimal media was observed only when xylose (0.1 g/L) was supplemented in addition to pyruvate (5 g/l) and CO2 (pCO2 = 0.1 atm), and not on pyruvate and CO2 alone.
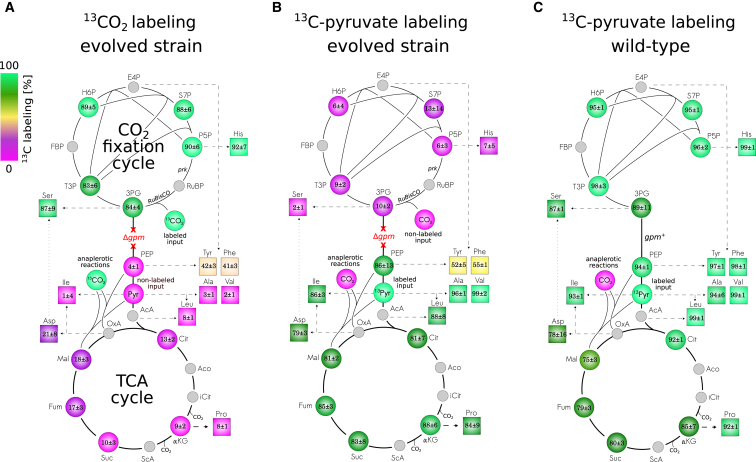
(C) As shown in a 13CO2 labeling experiment, expression of RuBisCO and prk in a Δgpm Δzwf Δpfk strain supported growth on minimal media supplemented with pyruvate, xylose and isotopically labeled CO2 (p13CO2 = 0.1 atm). Labeling of intermediates of upper glycolysis and the pentose phosphate pathway is due to the flux from pentose-phosphates to 3PG through RuBisCO, in which 13CO2 is fixed. Intermediates of lower glycolysis and the TCA cycle are synthesized from pyruvate and therefore are mostly unlabeled. Low levels of labeling in TCA intermediates results from inorganic carbon fixation in anaplerotic reactions. The mean percentage (±SD) of labeled carbon in intracellular metabolites and hydrolyzed amino acids was calculated according to the mass isotopologues distributions obtained using LC-MS (n = 3).
The ancestral strain was propagated in a xylose-limited chemostat (dilution rate of 0.08 h−1, doubling time of ≈9 hr). The surrogate sugar (i.e., xylose) was supplied to initiate the evolutionary process, as shown in Figure 2A. We set the composition of the feed media such that the CBB module surrogate sugar ([xylose] = 0.1 g/l) became strongly limiting, and the energy module substrate was supplied in excess ([pyruvate] = 5 g/l). The scarcity of xylose in the chemostat imposes a strong and continuous selective pressure on cells to utilize the CBB cycle for sugar biosynthesis directly from the abundant inorganic carbon (i.e., CO2) in order to alleviate the dependency on xylose. The inherent dynamics of a chemostat forces cell growth to equal the dilution rate and thus ensures that the concentration of xylose in the chemostat is extremely low and decreases even further as cells adapt. We hypothesized that this selection process will favor mutations that lead to a fully functional CBB cycle that bypasses the need for xylose, thus enabling a hemiautotrophic strain to take over the population. High levels of CO2 (pCO2 = 0.25 atm) were maintained throughout the experiment to maximize the RuBisCO carboxylation rate and prevent competing oxygenation reactions.
After inoculation, xylose concentration quickly fell below the detection limit (<1 mg/l) as expected for a sugar-limited chemostat regimen, while the concentration of pyruvate remained in considerable excess (≈2 g/l), as shown in Figure 2B. Due to severing gluconeogenesis by gpm deletion, carbon from pyruvate could not be used for sugar biosynthesis to compensate for the xylose limitation. However, excess pyruvate could potentially serve as a source of energy and reducing power to be utilized by the CBB module as it evolves to function as a xylose-independent CO2 fixation cycle. During the first 40 days of growth (≈100 chemostat generations), we observed no significant change in cell density and nutrient concentrations in the effluent media. Over the following 20 days, we noticed a gradual increase in cell density, accompanied by a steady decrease in pyruvate concentration. Finally, around day 60 (≈150 chemostat generations; Figure 2B), the concentration of pyruvate dropped to an undetectable level (<1 mg/l), suggesting that growth was no longer limited by xylose availability and that pyruvate became fully utilized. Importantly, in contrast to the ancestral strain, culture samples from day 50 onward (Figure 2B) were able to grow in minimal media when supplied with only pyruvate and elevated CO2 (doubling time of ≈6 hr; Figure 3). In ambient CO2, no growth was detected in either liquid media or agar plates.
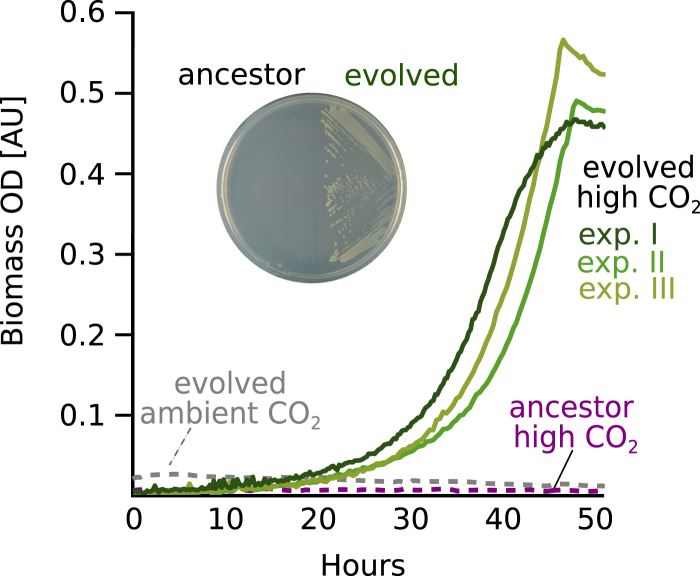
Figure 3.
Growth without Xylose Is Dependent on CO2 Availability
In contrast to the ancestral strain, evolved clones isolated from all three chemostat experiments were able to grow in minimal media, supplemented solely with pyruvate (doubling time of ≈6 hr). In all cases, growth required elevated CO2 conditions (pCO2 = 0.1 atm) and no growth was detected under ambient atmosphere. Similarly, evolved clones, but not the ancestral strain, were able to form colonies on minimal media agar plates when supplemented with pyruvate under a high CO2 atmosphere (inset).
See also Figure S6.
Plasmid curing from an evolved clone isolated from the chemostat resulted in the loss of the newly acquired phenotype. Cured cells lacking the RuBisCO- and prk-encoding plasmid did not grow in minimal media when only supplemented with CO2 and pyruvate (Figures S6A and S6B; Experimental Procedures). We retransformed the cured strain with a modified plasmid, in which RuBisCO, prk, and carbonic anhydrase were placed under the control of two separate inducible promoters. Regaining the hemiautotrophic phenotype was only observed on co-induction of both enzymes (Figure S6C). This result indicates that the novel growth mode is indeed dependent on the co-expression of RuBisCO and prk, as expected of CBB-cycle-mediated synthesis of sugars from CO2. We observed the evolution of clones able to grow solely on CO2 and pyruvate in two additional independent chemostat experiments (appearing on days 60 and 130; Figures S7A–S7C; Experimental Procedures).
Figure S6.
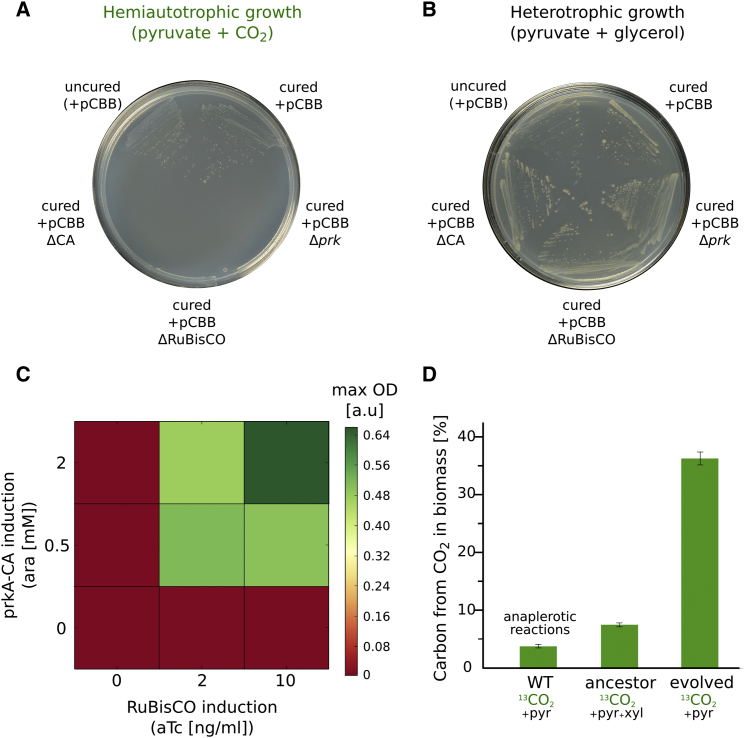
Hemiautotrophic Growth Is Dependent on Co-expression of RuBisCO and prk, Related to Figure 3
(A) Evolved hemiautotrophic cells that were cured from their pCBB plasmid could not grow on M9 minimal media agar plate supplemented only with pyruvate and elevated CO2 (pCO2 = 0.1 atm). Growth was restored only upon re-transformation with a plasmid containing all three genes (cbbM coding for RuBisCO, prk and CA). Cured cells re-transformed with modified plasmids, in which either cbbM, prkA or CA was deleted, did not grow hemiautotrophically. The reason for the apparent lack of growth with pCBB without CA awaits further research.
(B) In a control experiment, agar plates supplemented with glycerol (0.2%) in addition to pyruvate support growth which is independent of CBB activity (as glycerol can be used as carbon source for sugar synthesis; Figure S3). As expected, with glycerol, plasmid cured cells formed colonies independently from the expression of cbbM, prk or CA.
(C) Re-transformation of cured cells with a modified plasmid in which RuBisCO and a bicistronic operon of prk and CA were placed under the control of inducible promoters (Ptet and Para promoters, respectively) restores the ability to grow hemiautotrophically only when both inducers (aTc and arabinose) were added. Cells were incubated for ∼96 hr in elevated CO2 atmosphere (pCO2 = 0.1 atm) at 37°C. Notably, the BW 25113 strain cannot utilize arabinose as a carbon source due to ara operon deletion.
(D) Cells were propagated in M9 minimal media supplemented with non-labeled pyruvate (5 g/L) and isotopically labeled 13CO2 atmosphere (p13CO2 = 0.1 atm). Growth of the ancestor strain required the addition of xylose (non-labeled, 0.2 g/L). Isotopic carbon composition in total cell biomass was measured (mean ± SD) by analyzing lyophilized cells pellet using an elemental mass analyzer (n = 3; Supplemental Experimental Procedures). In the evolved hemiautotrophic strain over one third of cellular carbon originated from fixed inorganic carbon. This value is in line with the expected fraction of biomass precursors originating from the CBB module in the literature (Figure S4). Labeled carbon in the ancestor strain results from the RuBisCO dependent catabolism of xylose (due to the pfkA, pfkB, and zwf deletions, as detailed in Figures S3 and S5) and from the assimilation of inorganic carbon via anaplerotic reactions. Similarly, labeling in the metabolically unperturbed E. coli BW25113 (lacking the RuBisCO and prk encoding plasmid) is due to anaplerotic reactions.
Figure S7.
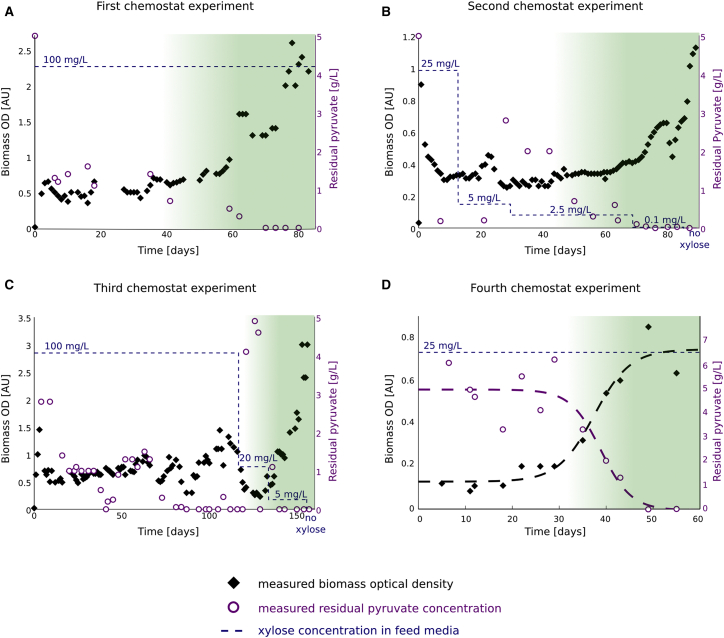
Hemiautotrophic Growth Evolved in Three Independent Chemostat Experiments and in a Fourth Experiment in Which prs and ppsR Mutations Were Introduced to the Ancestor, Related to Figure 5
(A) Ancestral strain was cultured in xylose limited chemostat with a dilution rate of 0.08 h-1. Xylose concentration in the feed media was kept constant at 100 mg/L. Pyruvate was provided in excess and its concentration in the feed media was 5 g/L. Residual concentrations and cell density inside the chemostat were measured by routine sampling. Once steady-state was reached, residual xylose concentration fell below detection limit while cell density remained relatively constant for the next 40 days. From this point onward, cell density increased while the residual pyruvate levels decreased. In addition, culture samples from day 50 onward were able to grow on liquid media, as well as on agar plates, in elevated CO2 conditions while supplemented solely with pyruvate. The number of colonies formed increased from day 50, reaching saturation at approximately day 70. Green shading denotes the emergence of the hemiautotrophic phenotype on pyruvate plates.
(B) For the second experiment we further modified the ancestor strain by knocking out a component of the mismatch repair system (mutS) known to increase the mutation rate. In addition, we slowed the dilution rate to 0.035 h-1 and decreased the xylose concentration in the feed to 25 mg/L. We further decreased the xylose concentration in the feed media whenever residual pyruvate concentration in the culture fell below detection limit, thus avoiding the possibility that pyruvate becomes the limiting nutrient instead of xylose. Clones displaying the hemiautotrophic phenotype were isolated from day 55 onward.
(C) In the third experiment, the ancestor strain was propagated under the same conditions as in the first experiment but the xylose concentration in the feed media was reduced whenever pyruvate concentration approached the detection limit as in described for the second experiment. Clones with hemiautotrophic phenotype were first isolated on day 130. Xylose was completely omitted from the feed media from day 150 onward, validating the ability of cells to grow in the complete absence of an organic carbon source feeding into the CBB module.
(D) Based on the genomic changes observed in the first three experiments, the ancestral strain in the fourth xylose limited chemostat evolution experiment contained a mutation in the prs gene (R105_A110dup) in addition to a knockout of the ppsR gene encoding the PEP synthase regulatory protein. The dilution rate was set to 0.035 h-1. The xylose concentration in the feed media was kept constant at 25 mg/L. Pyruvate was provided in excess and its concentration in the feed media was 5 g/L. Random mutagenesis was not performed prior to the inoculation into the chemostat. As early as day 30 onward an increase in biomass optical density was observed along with a decrease in the residual concentration of pyruvate. Culture samples from day 50 onward (shaded green) were able to grow in minimal media with pyruvate as the only organic carbon source in an elevated CO2 incubator, both on liquid media and agar plates. Sigmoid fits are added as guides to the eye. Note that the novel phenotype appears on a faster time-scale than in previous experiments.
Mass Spectrometry Reveals that CO2 Is the Sole Source of Carbon for Sugar Biosynthesis in the Evolved Cells
The ability of evolved bacteria to grow in the absence of xylose suggested that the CBB cycle evolved to operate in the absence of an organic input into the CBB module. To test if the intracellular pool of phospho-sugars and other CBB-cycle-derived biomass components are indeed synthesized solely from CO2 fixation, we propagated the evolved strain in minimal media, supplemented with isotopically labeled 13CO2 (p13CO2 = 0.1 atm) and non-labeled pyruvate. Isotopic labeling analysis using liquid chromatography-tandem mass spectrometry (LC-MS/MS) showed that sugars and biomass building blocks derived from the CBB module were almost completely 13C-labeled, while those derived from the energy module were almost totally unlabeled as shown in Figure 4A (Experimental Procedures). This serves as evidence of a fully functional CBB module operating in E. coli, capable of synthesizing sugars directly from CO2. The labeling pattern was further verified in a reciprocal experiment in which we propagated the bacteria in the presence of uniformly labeled 13C-pyruvate and unlabeled CO2 (Figure 4B). Small deviations from homogeneous labeling of CBB produced molecules were expected due to the refixation of CO2 released from the TCA cycle. Similarly, low levels of labeling in TCA cycle intermediates resulting from inorganic carbon usage in anaplerotic reactions can be seen in control experiments (Figures 4C, S5C, and S6D). Finally, isotopic analysis of the overall biomass content revealed that ≈35% of cellular carbon originated from CO2 fixation (Figure S6D), matching the known fraction of biomass produced from CBB module metabolites (Neidhardt, 1987) (Figure S4).
Figure 4.
Isotopic Labeling Experiments Demonstrate the Biosynthesis of Sugars and Other Biomass Components from CO2
(A) Isotopic labeling analysis, in which evolved clones were grown with isotopically labeled 13CO2 as an inorganic carbon source and non-labeled pyruvate as an energy source, showed almost full labeling of all CBB intermediates and the derived biomass building blocks. In marked contrast, TCA intermediates and biomass components originating from the energy module show low levels of labeling, as predicted for a hemiautotrophic growth mode. This indicates that the evolved strain synthesizes CBB module biomass precursors from CO2 using the non-native CBB cycle, while the biomass precursors originating from the energy module are synthesized from the supplemented pyruvate.
(B) In a reciprocal setup in which non-labeled CO2 was supplied in addition to uniformly labeled pyruvate, the labeling pattern was again in agreement with hemiautotrophic growth expectations: CBB intermediates and the derived amino acids were mostly non-labeled (as the supplied CO2), while metabolites in the energy module were mostly labeled.
(C) Metabolically unperturbed BW25113 E. coli in which RuBisCO and prk were not recombinantly expressed. As expected when lacking the gpm metabolic cutoff and RuBisCO, when uniformly labeled 13C-pyruvate was supplied as a carbon source, the intermediates of glycolysis and the pentose-phosphate pathway were fully labeled. TCA cycle intermediates and the derived biomass components were not fully labeled due to the usage of non-labeled bicarbonate in anaplerotic reactions (denoted CO2 for simplicity). The mean percentage (±SD) of three replicates is shown.
The Only Gene Mutated in All Three Lab Evolution Experiments Is at the Main Flux Branchpoint of the CBB Cycle
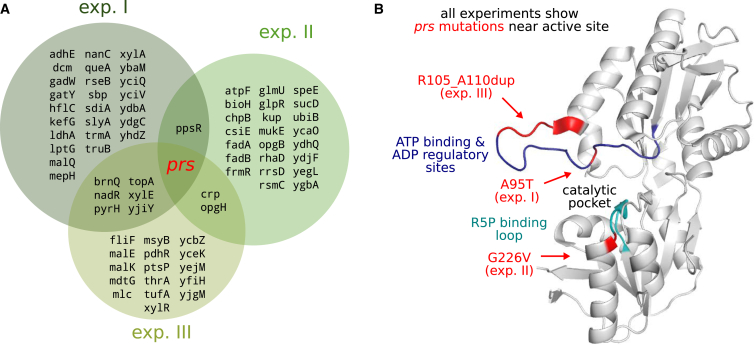
To identify the mutations responsible for the emerging metabolic phenotype, we performed whole-genome sequencing of clones displaying hemiautotrophic growth, isolated from three independent evolutionary chemostat experiments (Figures S7A–S7C; Experimental Procedures). ≈80 mutational events were identified in our analysis, none of them in RuBisCO or prk (Tables S1, S2, and S3). Ten genes were independently mutated in more than one clone (Figure 5A), and only a single gene was mutated in all three experiments. This gene, ribose-phosphate diphosphokinase (prs), encodes for an enzyme that phosphorylates ribose-5-phosphate and is the main flux branchpoint of the CBB module in which a pentose sugar is diverted toward biomass production. All three mutations were within the coding sequence: two of which were missense mutations (A95T and G226V), and the third was an 18-bp tandem duplication, leading to in-frame insertion of six amino acids (R105_A110dup). Structural analysis showed that all prs mutations arose within loops participating in substrate binding and catalytic activity (Figure 5B) (Eriksen et al., 2000). When the prs wild-type sequence was reintroduced to the evolved genetic background (Experimental Procedures), replacing the mutated prs sequence, the phenotype was lost: growth was only observed when xylose was supplemented in addition to pyruvate and CO2, as in the ancestral strain. Given that prs is the main branchpoint in which a pentose sugar is shunted from the CBB cycle toward biomass, we hypothesized that a change in one or more of the enzymatic parameters of prs was required for CBB cycle operation as discussed in the next section.
Figure 5.
The Genetic Basis Underlying the Hemiautotrophic Phenotype
(A) Venn diagram summarizing the intersection among mutations accumulated in hemiautotrophic evolved strains isolated from three distinct chemostat experiments. A detailed description of the mutations can be found in Tables S1, S2, and S3. In the ancestral strain for the second chemostat experiment (exp. II) a ΔmutS deletion that induces hyper-mutability was introduced (only mutations occurring between chemostat inoculation and hemiautotrophic phenotype emergence are depicted).
(B) Ribose-phosphate diphosphokinase (prs), the main flux branching enzyme of the CBB module by which ribose-phosphate is diverted toward biomass production, was the only gene in which mutations appeared in all of the chemostat evolution experiments. Structural analysis (based on the most closely available crystal structure, that of Bacillus subtilis showing ≈70% sequence similarity to the E.coli homolog) indicates that the mutations are located in catalytically active regions of the enzyme, either on the ribose-5-phosphate substrate binding loop (turquoise) or on a second loop containing the ATP binding site and an allosteric regulatory site (dark blue).
The rate of mutation accumulation identified in the evolved hemiautotrophic strains exceeds those reported in previous adaptive evolution studies (Barrick et al., 2009, Foster et al., 2015). This can be the result of several contributing factors, such as the known increased mutation rate under carbon starvation (Notley-McRobb et al., 2003, Novick and Szilard, 1950). Similarly, the tailored selective conditions we have used give ample ground for beneficial mutations that can (partially) restore fitness and shift the evolutionary dynamics toward a “strong selection, strong mutation” regime (Desai and Fisher, 2007) in which successive mutational sweeps and mutational cohorts are known to lead to relatively fast fixation of mutations. Interestingly, we note a bias in the spectrum of fixed mutations in some of the strains isolated from distinct experiments (Tables S1, S2, and S3). The underlying basis of this observation is currently unclear. In terms of copy number of the plasmid harboring RuBisCO and prk, we did not observe any consistent variations in the evolved strains versus the ancestor and the plasmid sequence did not show any mutations (Experimental Procedures).
Modeling the Autocatalytic Carbon Fixation Cycle Suggests that Fine-Tuning of prs Catalytic Rate Is Essential for Metabolic Stability
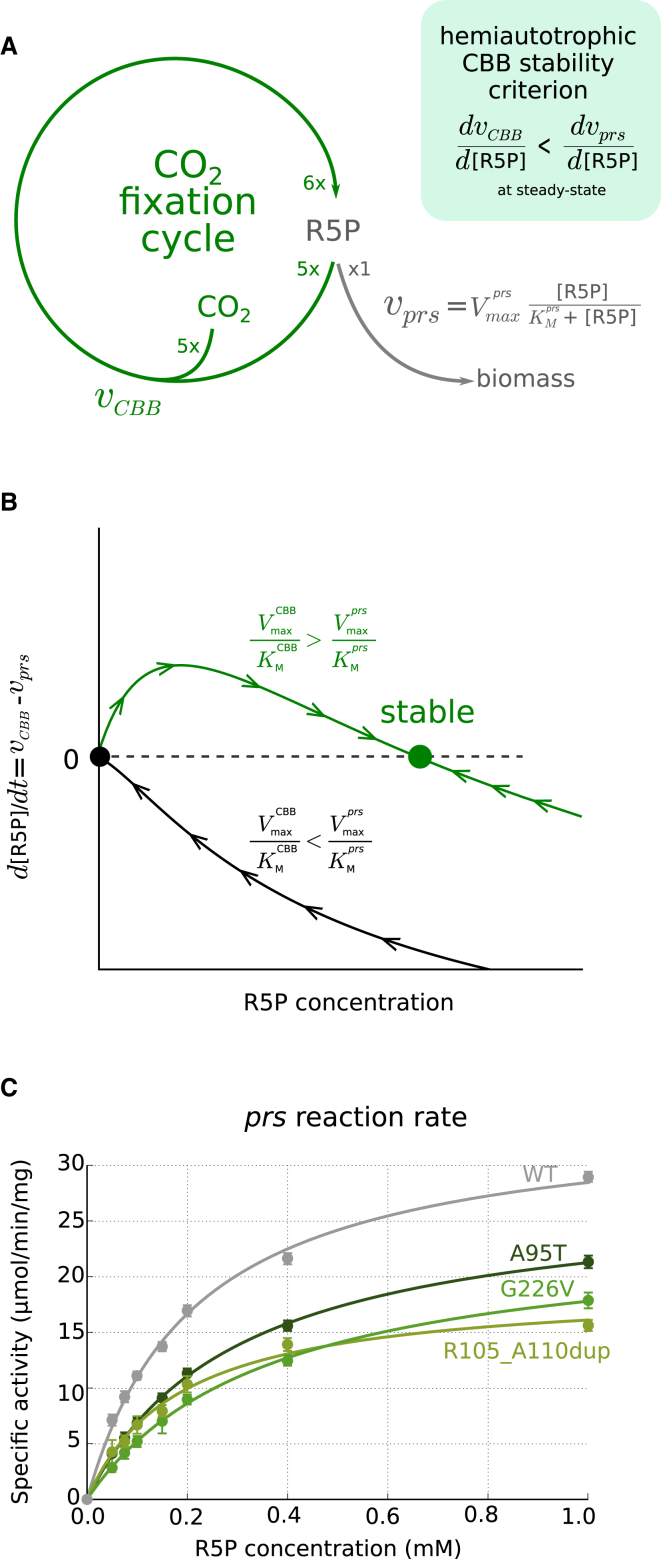
In contrast to linear metabolic pathways in which stable metabolic steady-state can be achieved, regardless of the specific kinetic properties, the stability of autocatalytic cycles (consisting of a set of metabolites which regenerate more of themselves with each turn of the cycle) is not guaranteed. The stability depends on the topology of the cycle and the kinetic parameters of the enzymes involved (Reznik and Segrè, 2010). This is demonstrated in the simplified model shown in Figure 6A, consisting of an autocatalytic effective carbon fixation reaction and a biomass generating reaction from which assimilated carbon is shunted toward biosynthesis. The stability of the steady state in this metabolic network depends on the kinetic properties of the enzymes, as these parameters govern the response to a perturbation in the intracellular concentration of the metabolite at the flux branchpoint. Explicitly, for a stable steady state to exist in the carbon fixation cycle, the Michaelis constant of prs must satisfy: (where and and are effective parameters that are a function of all the reactions composing the CBB cycle). This relation ensures that enough flux remains in the cycle to ensure autocatalysis (Figure 6B; Supplemental Experimental Procedures).
Figure 6.
Stability Analysis of Autocatalytic Carbon Fixation Cycles
(A) Simplified model for an autocatalytic carbon fixation cycle. We consider a two-reaction pathway in which a generalized carbon fixation reaction autocatalytically produces a metabolite (R5P) from an external supply of CO2 (effectively performing five carboxylations, ). A second reaction consumes R5P for the production of biomass. For clarity, we assume irreversible Michaelis-Menten rate laws. Model parameters for each reaction are the Michaelis constant (KM) and the maximal reaction rate (, where [E] is the enzyme concentration and is the turnover number). For a steady-state concentration to be stable, the derivative of with respect to R5P concentration has to be higher than the derivative of at the steady-state point. This relation will imply that if R5P concentration deviates from its steady state, the flux through the reaction branching to biomass synthesis would stabilize the R5P concentration. In terms of metabolic control analysis, this is equivalent to requiring that the elasticity of the prs reaction is greater than the elasticity of the CBB reaction at the steady-state point.
(B) Schematic of the stability analysis in the phase space of R5P concentration showing the net flux at the branchpoint. The steady-state concentration of R5P (inferred from setting ) and the stability of the steady state are determined by values of the parameters. Assuming the maximal rate of carbon fixation is lower than biomass synthesis , a non-zero stable steady state exists if the enzymatic parameters of prs satisfy the relation > .
(C) Experimental in vitro reaction rate measurements of purified prs enzymes, with either wild-type or mutated sequences. All of the mutated prs enzymes from the evolved strains show ≈2-fold decrease in their values, as predicted by the stability analysis. The measured values in respect to the wild-type enzyme were 57% ± 2%, 65% ± 6%, and 38% ± 3% for the A95T, G226V, and R105_A110dup prs mutations, respectively. The mean percentage (±SD) of three replicates is shown.
In order to test the model prediction, we measured in vitro the reaction rates of mutated prs toward its substrate ribose-5-phosphate. In all cases, as shown in Figure 6C, we found an approximately 2-fold decrease in following the evolutionary process, consistent with the stability analysis requirement (Experimental Procedures; no changes were observed in the Prs protein expression levels). This observation of convergent evolution strengthens the hypothesis that, in addition to the expression of recombinant CBB cycle enzymes, changes in the kinetic properties of endogenous components, which interact with the non-native pathway, are essential for a stably operating carbon fixation cycle and hence for the hemiautotrophic phenotype to emerge. We found that a mutated prs was not sufficient to allow hemiautotrophic growth when introduced on the ancestral strain background (Experimental Procedures), demonstrating that while fine-tuning of fluxes at the flux branchpoint via the prs mutation is essential, this mutation alone is not sufficient to fully reconstruct the phenotype.
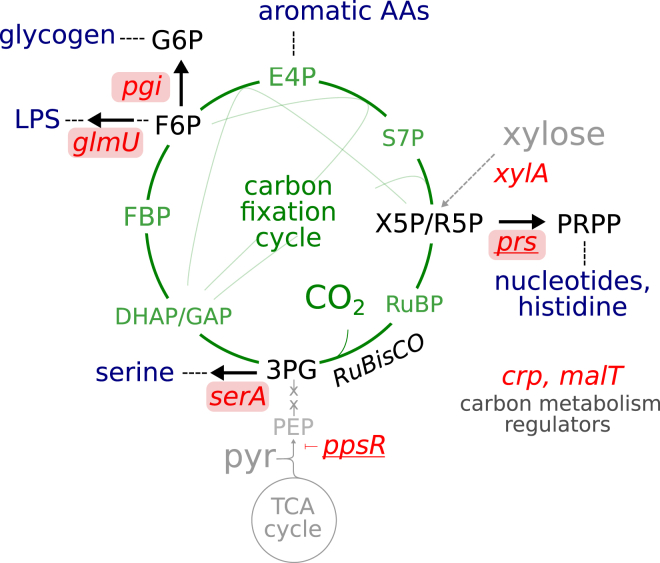
To further identify endogenous metabolic components that require fine-tuning to achieve stable operation of the carbon fixation pathway, we conducted a fourth chemostat evolution experiment (Figure S7D), in which one of the observed prs mutations (R105_A110dup) was introduced to the genetic background of the ancestral strain. In addition, we knocked out the PEP synthase regulatory protein (ppsR), as this gene is the only shared mutation between experimental repeats I and II beyond prs (Experimental Procedures). While the introduction of these two mutations to the ancestral background was not sufficient to achieve hemiautotrophic growth, xylose-limited chemostat evolution (dilution rate of 0.035 h−1) rapidly led to the emergence of a hemiautotrophic phenotype in <100 chemostat generations (Figure S7D). Whole-genome sequencing of hemiautotrophic clones isolated from this chemostat experiment revealed that six mutations were fixed in the evolutionary process (Figure 7; Table S4). Strikingly, almost all of the newly acquired mutations appeared in either flux branching points toward biomass from the CBB cycle (serA, pgi, and glmU) or global regulators of carbon metabolism (crp and malT). Four out of these six loci were mutated in at least one previous experiment (crp, glmU, xylA, and the mal operon). These results demonstrate that the functionality of the non-native CBB cycle depends not only on the introduction of heterologous enzymes required to construct the pathway but also on the fine-tuning of endogenous components interacting with it, mostly biosynthetic enzymes feeding from the carbon pool of the cycle (Figure 7).
Figure 7.
Acquired Mutations Focus on Flux Branchpoints and Carbon Metabolism Regulators
A fourth evolution experiment was initialed with a modified ancestral strain, containing additional mutations in prs (R105_A110dup, underlined) and ppsR (knockout, underlined). Emergence of the hemiautotrophic phenotype was detected in <100 chemostat generations. Whole-genome sequencing of isolated clones revealed that six mutations, in addition to prs and ppsR, were acquired in the evolutionary transition toward hemiautotrophic growth (red). Out of a total of eight mutations appearing in this hemiautotrophic strain, four are found in flux branchpoints, shunting cycle intermediates toward biomass (serA, pgi, and glmU in addition to prs), an additional three are in regulators of carbon metabolism (crp, malT, and ppsR), and one (xylA) is in the catabolism of the surrogate xylose sugar not used by the final evolved phenotype. A detailed description of the mutations can be found in Table S4.
See also Table S4.
Discussion
While several synthetic metabolic pathways have been successfully integrated into model microorganisms, such as E. coli and S. cerevisiae, the introduction of an autocatalytic carbon fixation pathway into a heterotrophic host has remained a standing challenge (Müller et al., 2015, Siegel et al., 2015). In this study, we demonstrate that the combination of rational design and laboratory evolution can rapidly evolve a strain to convert CO2 into sugars and other biomass components via non-native carbon fixation machinery. The striking ability of E. coli to rapidly change its growth mode demonstrates that central carbon metabolism can be extremely adaptive when subjected to selective conditions and extends the possibilities for achieving novel metabolic phenotypes. Successful introduction of carbon fixation into a heterotrophic model organism suggests that closely related radical modulations of C1 central carbon metabolism, such as the desired synthetic methylotrophy (Whitaker et al., 2015), synthetic mixotrophy (Fast et al., 2015), and synthetic electrotrophy (Lovley, 2011), can be well within reach with a similar approach. From an industrial perspective, even though not translated to immediate industrial applications, a fully functional non-native CO2 fixation cycle is a critical milestone toward synthetic lithoautotrophy. Our strategy to decouple energy and carbon fixation metabolism enables powerful modularity in the choice of energy module used to energize the synthetic cycle. The methodology we present of selection-based evolutionary transitions can be directly extended to establish a synthetic methylotrophic strain, utilizing industrially relevant energy sources, such as methanol, to drive CO2 fixation. Furthermore, previous studies described the “paper biochemistry” of several fully synthetic carbon fixation cycles, not known to occur in nature, with promising predicted kinetic properties (Bar-Even et al., 2010). Our results suggest an experimental route to implement these pathways in a fast growing, genetically malleable model organism, such as E. coli.
In contrast to a traditional design scheme, in which individual components or sub-modules are tested and then integrated, our metabolic design focused on the coupling of cellular fitness to the desired functionality. While the initial rational design, arising from our in silico analysis, was not sufficient to achieve the desired phenotype, it formed the genetic basis and suggested the selective conditions under which a complex metabolic transition evolved. The ability to create synergy between rational metabolic design and laboratory evolution by tailoring the appropriate experimental setup is essential for harnessing natural selection to fine-tuning the metabolic network. Quantitative a priori prediction of the interactions between endogenous and newly introduced metabolic functions is often challenging due to the lack of sufficient information regarding the kinetics, energetics, and regulation of the relevant components. A design that couples cellular fitness to the activity of a non-native pathway is therefore useful. Such coupling allows the harnessing of natural selection in a controlled environment to obtain the necessary fine-tuning between native and non-native metabolic functions to achieve the desired phenotype. Since chemostats inherently implement a feedback mechanism that continuously increases the selection stringency in response to improvements in the desired pathway activity, they can serve as an ideal platform for evolving pathways that utilize non-native substrates, such as inorganic carbon. A dynamic selection stringency, as in the case of chemostats, enables growth during the early phases of laboratory evolution by supplying limiting amounts of a surrogate sugar (i.e., xylose), which compensates for the lack of full pathway activity, even if the carbon fixation pathway performs suboptimally (Kleeb et al., 2007).
The hybrid approach demonstrated here of rational design combined with laboratory evolution has allowed us, for the first time, to achieve a fully functional carbon fixation cycle in a heterologous host. Our results make evident the remarkable plasticity of metabolism and provide a malleable platform for deciphering the biochemistry and evolution of carbon fixation. The rapid emergence of a novel metabolic phenotype in laboratory timescales suggests a route for synthetic biology efforts to optimize pathways of biotechnological importance that can drive transformative advances in our ability to tackle the grand challenge of resource sustainability in years to come.
Experimental Procedures
Strains and Genomic Modifications
An E. coli BW25113 strain (Grenier et al., 2014), referred to in the text as “wild-type,” was used as the parental strain for further genomic modifications. The genome sequence of the parent used for constructing the ancestor strain differed from the E. coli strain BW25113 at four loci: ptsI (D464N), fabR (V42G), btuB (G162A), and fhuA (frameshift). These mutations were acquired during early handling of the strain prior to chemostat inoculation. The strain inoculated in the chemostat for the evolutionary experiment, referred to in the text as “ancestor,” contained further genomic modifications: we deleted phosphoglycerate mutase genes (gpmA and gpmM), phosphofructokinase genes (pfkA and pfkB), and 6-phosphate-1-dehydrogenase (zwf). In addition, we further deleted the aceBAK operon encoding for the enzymes of the glyoxylate shunt, thus ensuring that a bypass using the tartronate semialdehyde pathway cannot be used to grow on pyruvate as a sole organic carbon source in Δgpm mutant. A detailed description of the methods used for performing genomic modifications can be found in the Supplemental Experimental Procedures. The ancestral strain inoculated in the second chemostat experiment contained a ΔmutS mutation that induces hyper-mutability. Detailed information regarding the genotype of clones showing hemiautotrophic phenotype, referred to in the text as “evolved isolated clones,” appears in Tables S1, S2, S3, and S4.
Recombinant Expression of RuBisCO, prk, and Carbonic Anhydrase
For recombinant expression of the CBB cycle components, we constructed a synthetic operon encoding the His-tagged type II RuBisCO (cbbM) from Rhodospirillum rubrum ATCC 11170 (courtesy of Prof. Michal Shapira), His-tagged phosphoribulokinase (prkA) from Synechococcus elongatus PCC 7942 (courtesy of Prof. Ichiro Matsumura), and carbonic anhydrase (Rru_A2056) from Rhodospirillum rubrum (CA). Synthetic ribosomal binding sites were used to achieve different levels of expression, as previously described (Zelcbuch et al., 2013). The synthetic operon was cloned into a pZA11 expression vector (Expressys). Detailed information regarding the construction and sequence of the plasmid containing the synthetic operon (pCBB), plasmid curing, and the reintroduction of modified plasmids can be found in the Supplemental Experimental Procedures.
Growth Conditions
Cells were grown on M9 minimal media, supplemented with 34 mg/l chloramphenicol and suitable carbon sources, as specified in the text. Agar plates were prepared using ultrapure agarose (Hispanagar). Growth curves were obtained by culturing cells in 96-well plates incubated at 37°C in a gas-controlled shaking incubator (Infinite M200, Tecan). In experiments in which frozen bacterial stock was used to inoculate the culture (e.g., 13CO2 labeling experiments), cells were first streaked on solid minimal media supplemented with pyruvate (0.5%) and xylose (0.1%) to facilitate initial growth. Several colonies were then streaked on solid minimal media supplemented with pyruvate. Once growth was observed, liquid minimal medium supplemented only with pyruvate was inoculated. We found that, in order to achieve the transition between xylose containing medium and pyruvate alone, it was required to sample several colonies to ensure hemiautotrophic growth initiation. All growth was performed in enriched CO2 atmosphere (pCO2 = 0.1 atm).
Computational Analysis of RuBisCO-Dependent Strains
To computationally identify candidate mutants in which cell growth is coupled to the flux through non-native CBB enzymes, we implemented an algorithm based on the principles of flux balance analysis (FBA) (Orth et al., 2010). We started by considering combinations of up to three enzymatic reaction knockouts in central metabolism and filtered out all those combinations that allow growth without any flux in RuBisCO. For those that cannot produce any biomass without RuBisCO, we calculated a slope defined as the biomass production rate achieved by allowing a unit of flux in RuBisCO. All other constraints were the same as in standard FBA, where the rate of biomass production is maximized. Details regarding software implementation can be found in the Supplemental Experimental Procedures.
Chemostat Evolution
Chemostat-based laboratory evolution was conducted in four independent experiments. Gas inflow composition was 25% CO2, 5% O2, and 70% N2 and temperature was maintained at 37°C. We used Bioflo 110 chemostats (New Brunswick Scientific) with a working volume of 0.7 l except in the second experiment, which used a DASBox bacterial fermentation system (DASGIP) with a working volume of 0.1 l. In the first and third experiments, the dilution rate was set to 0.08 h−1 (equivalent to 9 hours of doubling time) with a feed input of M9 minimal media supplemented with 5 g/l sodium pyruvate, 100 mg/l xylose, and 34 mg/l chloramphenicol. In the second and fourth experiments, the dilution rate was set to 0.035 h−1 (equivalent to 20 hours of doubling time) with a feed input of M9 minimal media supplemented with 5 g/l sodium pyruvate, 25 mg/l xylose, and 34 mg/l chloramphenicol.
Isotopic Labeling Experiments
For mass isotopologues distribution analysis, cells were cultured in M9 minimal media, either in the presence of a uniformly labeled 13C-pyruvate and unlabeled CO2 or in a reciprocal setup with isotopically labeled 13CO2 (Cambridge Isotope Laboratories) and a non-labeled pyruvate. For experiments in which gaseous 13CO2 was used, culture tubes were placed in a transparent air-tight container and flushed with five volumes of isotopically labeled gas mixture (10% 13CO2, 10% O2, and 80% N2). Cells were incubated in a shaking incubator at 37°C and harvested during exponential growth. For the extraction of intracellular metabolites, we used cold (−20°C) acetonitrile:methanol:water (40:40:20) extraction solution. A ZIC-pHILIC column (4.6 × 150 mm, guard column 4.6 × 10 mm; Merck) was used for liquid chromatography separation using gradient elution with a solution of 20 mM ammonium carbonate, with 0.1% ammonium hydroxide and acetonitrile at 0.1 ml/min. Detection of metabolites was performed using a Thermo Scientific Exactive high-resolution mass spectrometer with electrospray ionization, examining metabolites in a polarity switching mode over the mass range of 75–1,000 m/z. Compound identities were verified by matching masses and retention times to authenticated standards library. In addition we analyzed the labeling of proteinogenic amino acids obtained from the acid hydrolysis of protein biomass as described in the Supplemental Experimental Procedures. Data analysis was performed using TargetLynx (Waters) and the Maven software suite (Melamud et al., 2010). Total carbon labeling was calculated according to the formula:
Further details and the description for the measurement of isotopic carbon ratio in whole-biomass samples can be found in the Supplemental Experimental Procedures.
Whole-Genome Sequencing and Analysis
DNA was extracted from sampled cultures using a DNeasy Blood and Tissue Kit (QIAGEN), and sequencing was performed using an Illumina Hiseq 2500 platform. Detailed information regarding sample preparation can be found in the Supplemental Experimental Procedures. A reference genome was constructed based on the E. coli strain BW25113 (Grenier et al., 2014) and the sequence of the CBB enzymes encoding plasmid as a second contig. Sequence alignment and variant calling was achieved by use of the breseq pipeline (Deatherage and Barrick, 2014). The breseq program was used to identify genomic variants, including SNPs and insertion-deletion polymorphisms (INDELs). We note that our analysis does not capture certain types of mutations, such as copy number variations.
Author Contributions
A.B.-E., N.A., E.N., and R.M. designed the project; N.A., L.Z., E.H., T.B., and S.A. constructed the plasmids and performed genome engineering; E.N., U.B., N.T., N.A., A.W., D.D., and A.B.-E. developed the computational framework; N.A., S.G., Y.Z., E.H., S.A., S.M., G.J., T.B., I.S., and D.G.W. performed the experiments; N.A., S.G., E.N., and S.M. performed and analyzed LC-MS/MS experiments; S.G. and Y.Z. generated the sequencing data; N.A., S.G., Y.B., and U.B. analyzed the sequencing data; and N.A., S.G., A.B.-E., and R.M. wrote the paper.
Acknowledgments
We thank Ichiro Matsumura for kindly supplying the prkA plasmid and Michal Shapira for kindly supplying the R. rubrum culture. We also thank Amir Aharoni, Asaph Aharoni, Uri Alon, Gil Amitai, Abdussalam Azem, Naama Barkai, Sima Benjamin, Sonjia Billerbeck, Dvora Biran, Elad Chomski, Ahuva Cooperstein, Daniel Dar, Michal Dayagi, Shani Doron, Noa Dvir, Avigdor Eldar, David Fell, Avi Flamholz, Shay Fleishon, Idan Frumkin, Shlomit Gilad, Pierre Goloubinoff, Michael Gurevitz, Jacob Hanna, Tami Hayon, Lee-Or Herzog, Keren Kahil, Miriam Kaltenbach, Mechael Kanovsky, Aaron Kaplan, Leeat Keren, Roy Kishony, Hagar Kroytoro, Rob Last, Paola Laurino, Tali Lavy, Ronen Levi, Ayelet Levin, Yishai Levin, Avi Levy, Hannes Link, Adar Lopez, Yehuda Marcus, Barak Marcus, Adi Millman, Uri Moran, Oliver Mueller-Cajar, Yotam Nadav, Amnon Naziri, Ravit Netzer, Gal Ofir, Nigel Orme, Rob Phillips, Uri Pick, Noa Rippel, Orel Rivni, Liat Rockah, Ilana Rogachev, Eliora Ron, Dave Savage, Alon Savidor, Efrat Schwartz, Lior Shachar, Tali Shalit, Tomer Shlomi, Rotem Sorek, Zvika Tamari, Dan Tawfik, Noam Vardi, Adi Volpert, Alon Wellner, Sagit Yahav, Dan Yakir, Oren Yishai, and Yonatan Zegman for support and feedback on this research. This work was funded by the European Research Council (projects SYMPAC 260392 and NOVCARBFIX 646827), Dana and Yossie Hollander, the Helmsley Charitable Foundation, the Larson Charitable Foundation, the Estate of David Arthur Barton, the Anthony Stalbow Charitable Trust, and Stella Gelerman, Canada. R.M. is the Charles and Louise Gartner professional chair and an EMBO young investigator program member. D.G.W. is supported by the United States-Israel Education Foundation.
Published: June 23, 2016
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, seven figures, and four tables and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2016.05.064.
An audio PaperClip is available at http://dx.doi.org/10.1016/j.cell.2016.05.064#mmc7.
Supplemental Information
References
- Bar-Even A., Noor E., Lewis N.E., Milo R. Design and analysis of synthetic carbon fixation pathways. Proc. Natl. Acad. Sci. USA. 2010;107:8889–8894. doi: 10.1073/pnas.0907176107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Even A., Noor E., Milo R. A survey of carbon fixation pathways through a quantitative lens. J. Exp. Bot. 2012;63:2325–2342. doi: 10.1093/jxb/err417. [DOI] [PubMed] [Google Scholar]
- Barrick J.E., Yu D.S., Yoon S.H., Jeong H., Oh T.K., Schneider D., Lenski R.E., Kim J.F. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature. 2009;461:1243–1247. doi: 10.1038/nature08480. [DOI] [PubMed] [Google Scholar]
- Bassham J.A., Benson A.A., Kay L.D., Harris A.Z., Wilson A.T., Calvin M. The path of carbon in photosynthesis. XXI. The cyclic regeneration of carbon dioxide acceptor1. J. Am. Chem. Soc. 1954;76:1760–1770. [Google Scholar]
- Blount Z.D., Borland C.Z., Lenski R.E. Historical contingency and the evolution of a key innovation in an experimental population of Escherichia coli. Proc. Natl. Acad. Sci. USA. 2008;105:7899–7906. doi: 10.1073/pnas.0803151105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church G.M., Elowitz M.B., Smolke C.D., Voigt C.A., Weiss R. Realizing the potential of synthetic biology. Nat. Rev. Mol. Cell Biol. 2014;15:289–294. doi: 10.1038/nrm3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deatherage D.E., Barrick J.E. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol. Biol. 2014;1151:165–188. doi: 10.1007/978-1-4939-0554-6_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai M.M., Fisher D.S. Beneficial mutation selection balance and the effect of linkage on positive selection. Genetics. 2007;176:1759–1798. doi: 10.1534/genetics.106.067678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durão P., Aigner H., Nagy P., Mueller-Cajar O., Hartl F.U., Hayer-Hartl M. Opposing effects of folding and assembly chaperones on evolvability of Rubisco. Nat. Chem. Biol. 2015;11:148–155. doi: 10.1038/nchembio.1715. [DOI] [PubMed] [Google Scholar]
- Eriksen T.A., Kadziola A., Bentsen A.K., Harlow K.W., Larsen S. Structural basis for the function of Bacillus subtilis phosphoribosyl-pyrophosphate synthetase. Nat. Struct. Biol. 2000;7:303–308. doi: 10.1038/74069. [DOI] [PubMed] [Google Scholar]
- Fast A.G., Schmidt E.D., Jones S.W., Tracy B.P. Acetogenic mixotrophy: novel options for yield improvement in biofuels and biochemicals production. Curr. Opin. Biotechnol. 2015;33:60–72. doi: 10.1016/j.copbio.2014.11.014. [DOI] [PubMed] [Google Scholar]
- Foster P.L., Lee H., Popodi E., Townes J.P., Tang H. Determinants of spontaneous mutation in the bacterium Escherichia coli as revealed by whole-genome sequencing. Proc. Natl. Acad. Sci. USA. 2015;112:E5990–E5999. doi: 10.1073/pnas.1512136112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs G. Alternative pathways of carbon dioxide fixation: insights into the early evolution of life? Annu. Rev. Microbiol. 2011;65:631–658. doi: 10.1146/annurev-micro-090110-102801. [DOI] [PubMed] [Google Scholar]
- Galanie S., Thodey K., Trenchard I.J., Filsinger Interrante M., Smolke C.D. Complete biosynthesis of opioids in yeast. Science. 2015;349:1095–1100. doi: 10.1126/science.aac9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong F., Liu G., Zhai X., Zhou J., Cai Z., Li Y. Quantitative analysis of an engineered CO2-fixing Escherichia coli reveals great potential of heterotrophic CO2 fixation. Biotechnol. Biofuels. 2015;8:86. doi: 10.1186/s13068-015-0268-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenier F., Matteau D., Baby V., Rodrigue S. Complete genome sequence of Escherichia coli BW25113. Genome Announc. 2014;2 doi: 10.1128/genomeA.01038-14. e01038-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleeb A.C., Edalat M.H., Gamper M., Haugstetter J., Giger L., Neuenschwander M., Kast P., Hilvert D. Metabolic engineering of a genetic selection system with tunable stringency. Proc. Natl. Acad. Sci. USA. 2007;104:13907–13912. doi: 10.1073/pnas.0705379104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Opgenorth P.H., Wernick D.G., Rogers S., Wu T.-Y., Higashide W., Malati P., Huo Y.-X., Cho K.M., Liao J.C. Integrated electromicrobial conversion of CO2 to higher alcohols. Science. 2012;335:1596. doi: 10.1126/science.1217643. [DOI] [PubMed] [Google Scholar]
- Lin M.T., Occhialini A., Andralojc P.J., Parry M.A.J., Hanson M.R. A faster Rubisco with potential to increase photosynthesis in crops. Nature. 2014;513:547–550. doi: 10.1038/nature13776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovley D.R. Powering microbes with electricity: direct electron transfer from electrodes to microbes. Environ. Microbiol. Rep. 2011;3:27–35. doi: 10.1111/j.1758-2229.2010.00211.x. [DOI] [PubMed] [Google Scholar]
- Maddamsetti R., Lenski R.E., Barrick J.E. Adaptation, clonal interference, and frequency-dependent interactions in a long-term evolution experiment with Escherichia coli. Genetics. 2015;200:619–631. doi: 10.1534/genetics.115.176677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melamud E., Vastag L., Rabinowitz J.D. Metabolomic analysis and visualization engine for LC-MS data. Anal. Chem. 2010;82:9818–9826. doi: 10.1021/ac1021166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller-Cajar O., Whitney S.M. Evolving improved Synechococcus Rubisco functional expression in Escherichia coli. Biochem. J. 2008;414:205–214. doi: 10.1042/BJ20080668. [DOI] [PubMed] [Google Scholar]
- Müller J.E.N., Meyer F., Litsanov B., Kiefer P., Potthoff E., Heux S., Quax W.J., Wendisch V.F., Brautaset T., Portais J.-C., Vorholt J.A. Engineering Escherichia coli for methanol conversion. Metab. Eng. 2015;28:190–201. doi: 10.1016/j.ymben.2014.12.008. [DOI] [PubMed] [Google Scholar]
- Neidhardt F.C., editor. Escherichia coli and Salmonella typhimurium: cellular and molecular biology, volumes I and II. American Society for Microbiology; 1987. [Google Scholar]
- Notley-McRobb L., Seeto S., Ferenci T. The influence of cellular physiology on the initiation of mutational pathways in Escherichia coli populations. Proc. Biol. Sci. 2003;270:843–848. doi: 10.1098/rspb.2002.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick A., Szilard L. Experiments with the chemostat on spontaneous mutations of bacteria. Proc. Natl. Acad. Sci. USA. 1950;36:708–719. doi: 10.1073/pnas.36.12.708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth J.D., Thiele I., Palsson B.Ø. What is flux balance analysis? Nat. Biotechnol. 2010;28:245–248. doi: 10.1038/nbt.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh M.R., Greene D.N., Woods K.K., Matsumura I. Directed evolution of RuBisCO hypermorphs through genetic selection in engineered E.coli. Protein Eng. Des. Sel. 2006;19:113–119. doi: 10.1093/protein/gzj010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quandt E.M., Gollihar J., Blount Z.D., Ellington A.D., Georgiou G., Barrick J.E. Fine-tuning citrate synthase flux potentiates and refines metabolic innovation in the Lenski evolution experiment. eLife. 2015;4:e09696. doi: 10.7554/eLife.09696. pii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reznik E., Segrè D. On the stability of metabolic cycles. J. Theor. Biol. 2010;266:536–549. doi: 10.1016/j.jtbi.2010.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer U. Evolutionary engineering of industrially important microbial phenotypes. Adv. Biochem. Eng. Biotechnol. 2001;73:129–169. doi: 10.1007/3-540-45300-8_7. [DOI] [PubMed] [Google Scholar]
- Shih P.M., Zarzycki J., Niyogi K.K., Kerfeld C.A. Introduction of a synthetic CO2-fixing photorespiratory bypass into a cyanobacterium. J. Biol. Chem. 2014;289:9493–9500. doi: 10.1074/jbc.C113.543132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel J.B., Smith A.L., Poust S., Wargacki A.J., Bar-Even A., Louw C., Shen B.W., Eiben C.B., Tran H.M., Noor E. Computational protein design enables a novel one-carbon assimilation pathway. Proc. Natl. Acad. Sci. USA. 2015;112:3704–3709. doi: 10.1073/pnas.1500545112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitaker W.B., Sandoval N.R., Bennett R.K., Fast A.G., Papoutsakis E.T. Synthetic methylotrophy: engineering the production of biofuels and chemicals based on the biology of aerobic methanol utilization. Curr. Opin. Biotechnol. 2015;33:165–175. doi: 10.1016/j.copbio.2015.01.007. [DOI] [PubMed] [Google Scholar]
- Wiser M.J., Ribeck N., Lenski R.E. Long-term dynamics of adaptation in asexual populations. Science. 2013;342:1364–1367. doi: 10.1126/science.1243357. [DOI] [PubMed] [Google Scholar]
- Yim H., Haselbeck R., Niu W., Pujol-Baxley C., Burgard A., Boldt J., Khandurina J., Trawick J.D., Osterhout R.E., Stephen R. Metabolic engineering of Escherichia coli for direct production of 1,4-butanediol. Nat. Chem. Biol. 2011;7:445–452. doi: 10.1038/nchembio.580. [DOI] [PubMed] [Google Scholar]
- Zelcbuch L., Antonovsky N., Bar-Even A., Levin-Karp A., Barenholz U., Dayagi M., Liebermeister W., Flamholz A., Noor E., Amram S. Spanning high-dimensional expression space using ribosome-binding site combinatorics. Nucleic Acids Res. 2013;41:e98. doi: 10.1093/nar/gkt151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Z.-Y., Li S.-Y. Rubisco-based engineered Escherichia coli for in situ carbon dioxide recycling. Bioresour. Technol. 2013;150:79–88. doi: 10.1016/j.biortech.2013.09.116. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.