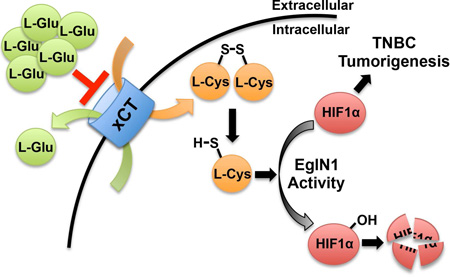
Summary
The HIF transcription factor promotes adaptation to hypoxia and stimulates the growth of certain cancers, including triple-negative breast cancer (TNBC). The HIFα subunit is usually prolyl-hydroxylated by EglN family members under normoxic conditions, causing its rapid degradation. We confirmed that TNBC cells secrete glutamate, which we found is both necessary and sufficient for the paracrine induction of HIF1α in such cells under normoxic conditions. Glutamate inhibits the xCT glutamate-cystine antiporter, leading to intracellular cysteine depletion. EglN1, the main HIFα prolyl hydroxylase, undergoes oxidative self-inactivation in the absence of cysteine both in biochemical assays and in cells, resulting in HIF1α accumulation. Therefore, EglN1 senses both oxygen and cysteine.
Graphical Abstract
eTOC Blurb
Increased glutamate secretion by triple negative breast cancer cells affects cellular cysteine levels and drives activation of HIF through disabling EglN1 under normoxic conditions. Thus. EglN1, which is known to sense oxygen, also senses cysteine.
Introduction
Breast cancers expressing the estrogen receptor, especially when they also express the progesterone receptor, are usually treated with hormonal manipulations, whereas those expressing the HER2 receptor tyrosine kinase are often treated with HER2 antagonists. New therapies and biomarkers are needed for triple-negative breast cancers (TNBCs), which do not express these receptors and are highly lethal.
Hypoxia-Inducible Factor (HIF) is a master transcriptional regulator of genes that support adaptation to hypoxia, including genes that promote angiogenesis, erythropoiesis, glycolysis, autophagy and energy conservation (Kaelin and Ratcliffe, 2008). HIF can promote or suppress tumor growth in a context-dependent manner (Blouw et al., 2003; Keith et al., 2012). In mouse models HIF promotes the growth, invasion, and metastasis of breast cancer cells, including TNBC cells (Chaturvedi et al., 2013; Chen et al., 2014; Montagner et al., 2012; Regan Anderson et al., 2013; Schito et al., 2012; Semenza, 2012; Wong et al., 2012; Zhang et al., 2012).
HIF consists of an unstable alpha subunit and a stable beta subunit. Under normoxic conditions HIFα is prolyl-hydroxylated by the EglN (also called PHD) 2-oxoglutarate (2-OG)-dependent dioxygenases, with EglN1 (PHD2) being the main HIFα prolyl hydroxylase (Kaelin and Ratcliffe, 2008). The pVHL ubiquitin ligase complex recognizes prolyl-hydroxylated HIFα and marks it for proteasomal degradation (Kaelin and Ratcliffe, 2008). The EglN oxygen KM values allow them to act as oxygen sensors, with their hydroxylase activity falling when oxygenation is inadequate (Kaelin and Ratcliffe, 2008). In some models, hypoxia generates reactive oxygen species (ROS) that can inhibit EglN activity although how, mechanistically, ROS inhibits EglN remains unclear (Kaelin, 2005). Therefore, HIF accumulates in hypoxic tissues.
HIF activation is common in solid tumors because they typically contain regions that are hypoxic due to poor blood flow. Additionally, many cancer-relevant mutations induce HIF irrespective of hypoxia. Cells lacking pVHL, such as most clear cell renal carcinoma cells (ccRCCs), accumulate high HIF levels due to impaired HIFα degradation. HIFα degradation is also decreased in Succinate Dehydrogenase and Fumarate Hydratase mutant tumors because they accumulate succinate and fumarate, respectively, which inhibit EglN activity (Kaelin and Ratcliffe, 2008). Cancers with activating mTOR mutations accumulate high HIF1 levels due to increased HIF1A transcription and translation (Kaelin and Ratcliffe, 2008). Therefore HIF activation in solid tumors can reflect bona fide hypoxia or the effects of oncogenic mutations on HIF synthesis and turnover.
Results
HIF Is Upregulated in TNBC
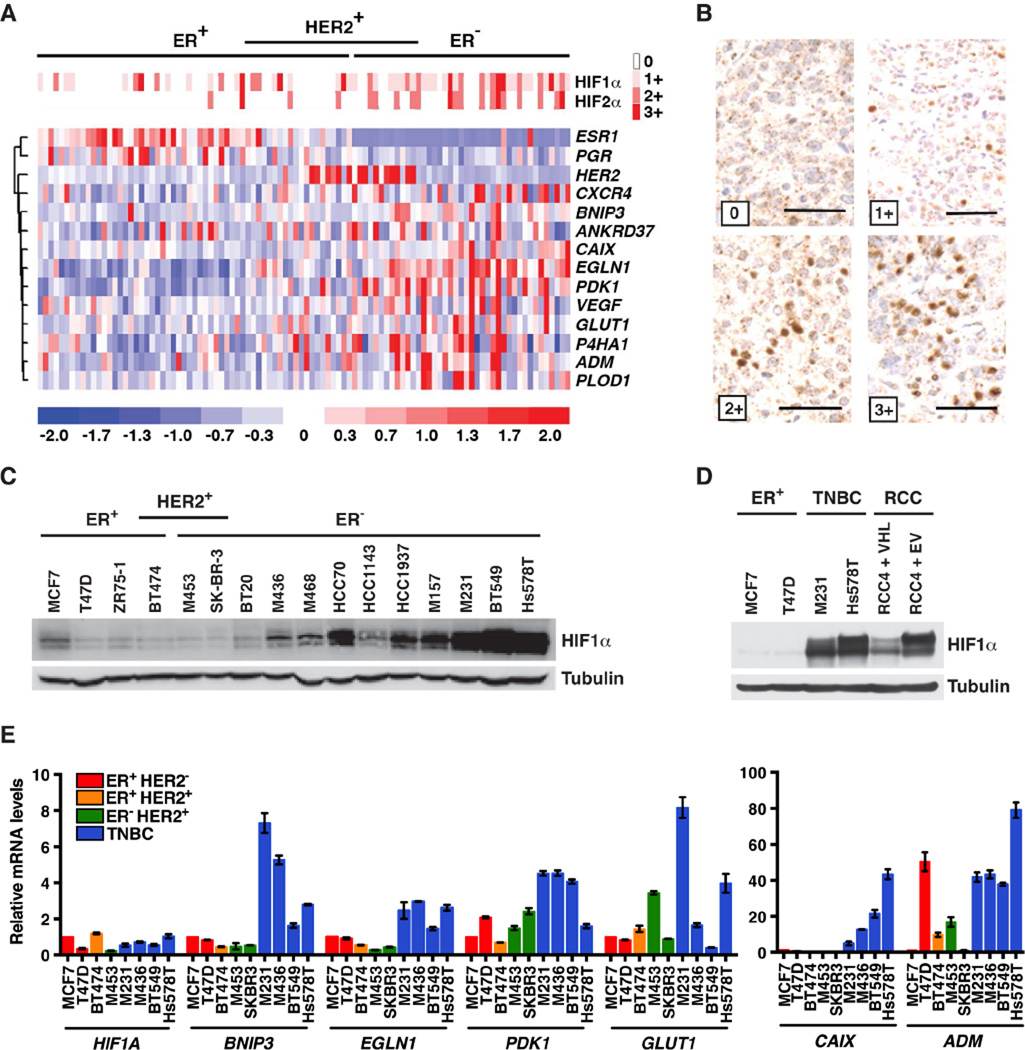
We performed immunohistochemical studies of breast cancer tissue microarrays using antibodies against HIF1α and HIF2α. In keeping with earlier reports, both proteins were readily detected in a subset of breast tumors, especially in TNBCs (Figures 1A and 1B) (Bos et al., 2002; Laurinavicius et al., 2012; Talks et al., 2000; Zhong et al., 1999). Consistent with these findings, various HIF-responsive mRNAs, such as the CXCR4, BNIP3, CAIX, EGLN1, PDK1, VEGF, GLUT1, and ADM mRNAs, were increased in TNBCs relative to other breast cancer subtypes (Figure 1A).
Figure 1. HIF Is Upregulated in Triple-Negative Breast Cancer.
(A) Heat maps depicting relative abundance of HIFα protein levels (top) and selected mRNAs (bottom), in a series of breast tumor specimens. Samples are arranged into subsets according to immunohistochemical staining for ER (Estrogen Receptor) and HER2, and each column refers to one specimen.
(B) Representative HIF1α immunohistochemistry from (A). Scale bar = 50 µm. (C–E) Immunoblot (C–D) and real-time PCR analysis (E) of the indicated breast cancer lines. RCC4 VHL−/− renal carcinoma cells infected to produce wild-type pVHL (VHL) or with the empty vector (EV) were included in (D) for comparison. M453, MDA-MB-453; M436, MDA-MB-436; M468, MDA-MB-468; M157, MDA-MB-157; M231, MDA-MB-231; RCC, renal cell carcinoma. In (E) transcript levels were normalized to ACTB, and then to the corresponding value in MCF7 cells. Data are represented as mean ± SEM.
See also Figure S1.
Increased HIFα protein levels in TNBC could reflect intratumoral hypoxia in vivo. Increased HIF1α protein levels were also noted, however, in subconfluent TNBC cell lines grown under well-oxygenated conditions (Figures 1C and S1A) and, in some cases, approached the HIF1α levels in VHL−/− ccRCC lines (Figure 1D). We did not reproducibly detect HIF2α in TNBC cell lines, and therefore focused on HIF1α in the experiments below.
Increased HIF1α protein levels in the TNBC cell lines correlated with increased levels of HIF1α–responsive mRNAs (Figure 1E). VEGF mRNA induction in TNBC lines was more variable, however (data not shown), possibly because it is mainly regulated by the HIF1α C-terminal transactivation domain that, unlike the HIF1α N-terminal transactivation domain, is suppressed by Factor Inhibiting HIF1 (FIH1) under normoxic conditions (Kaelin and Ratcliffe, 2008).
In keeping with prior studies (Chaturvedi et al., 2013; Regan Anderson et al., 2013; Schito et al., 2012; Semenza, 2012; Wong et al., 2012; Zhang et al., 2012), two different HIF1α shRNAs suppressed orthotopic tumor formation by a TNBC line, suggesting that HIF1α deregulation promotes TNBC growth (Figures S1B–E). We therefore sought the mechanism responsible for normoxic accumulation of HIF1α in TNBC.
TNBC Cells Secrete a Small Molecule HIF1α Stabilizer
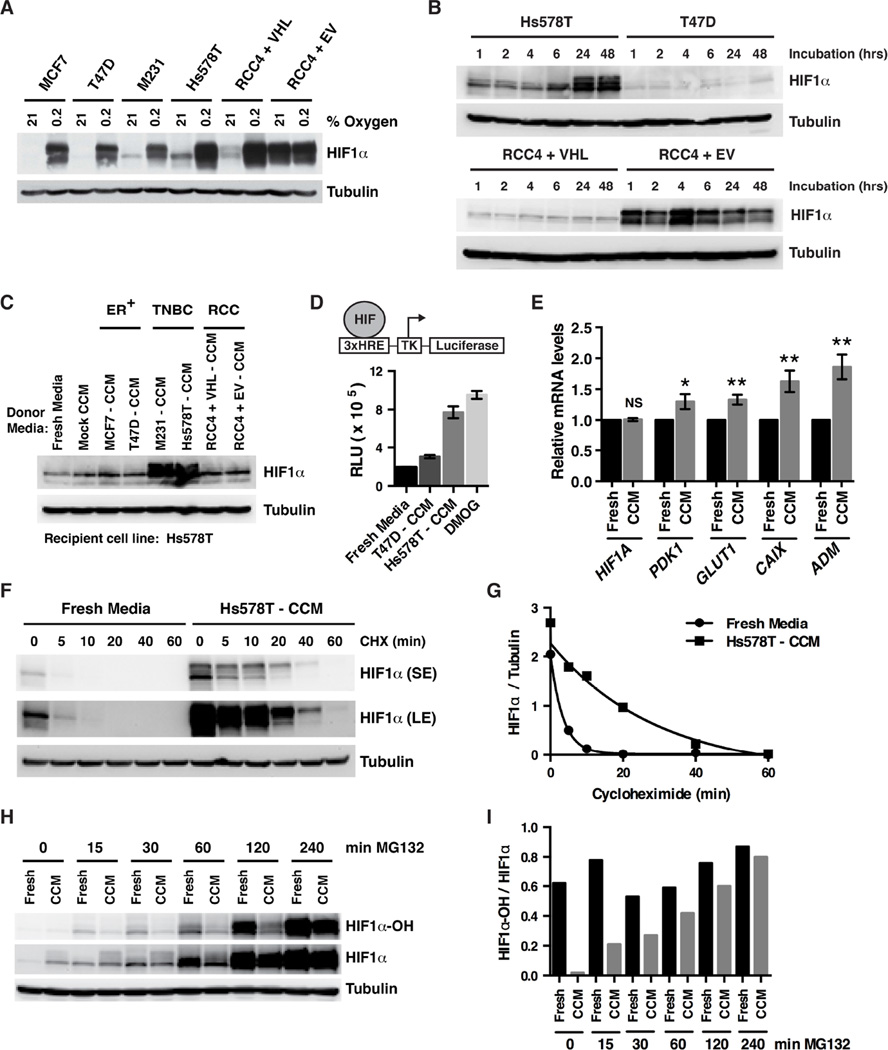
HIF1α accumulation in TNBC lines is posttranscriptional because HIF1A mRNA levels are not increased in TNBC cell lines compared to other breast cancer subtypes (Figure 1E). Of note, the protein levels of the EglNs that mark HIFα for destruction are not decreased in TNBC lines compared to hormone-responsive and HER2-positive breast cancer lines (Figure S1F). Moreover, the EglN-pVHL axis appeared to be functionally intact because freshly plated breast cancer lines, including the TNBC lines MDA-MB-231 and Hs578T, had low HIF1α levels that were robustly induced by hypoxia (Figure 2A). HIF1α accumulated in the TNBC lines to levels comparable to VHL−/− RCC4 ccRCCs, however, after 24–48 hours in culture under normoxic conditions (Figures 1D and 2B). In contrast, no such accumulation was noted in T47D hormone-responsive breast cancer cells (Figure 2B).
Figure 2. TNBC Cells Secrete a Small Molecule HIF1α Stabilizer.
(A) Immunoblot analysis of breast [MCF7, T47D, MDA-MB-231 (M231) and Hs578T] and renal (RCC4) cancer cell lines cultured in the presence of 21% oxygen or 0.2% oxygen for 6 hrs.
(B) Immunoblot analysis of breast (Hs578T and T47D) and renal (RCC4) cancer cell lines at the indicated timepoints after being plated in fresh media.
(C) Immunoblot analysis of Hs578T cells exposed for 6 hours to fresh media or media either incubated for 48 hours in the absence of cells (Mock) or conditioned by the indicated breast [MCF7, T47D, MDA-MB-231 (M231), Hs578T] or renal (RCC4) cell lines. CCM = cell-conditioned media.
(D) Luciferase activity of Hs578T cells expressing firefly luciferase (FLuc) under the control of a HIF-responsive promoter (3xHRE) after 6 hours exposure to fresh media, media conditioned by the indicated cell lines or fresh media supplemented with 1 mM DMOG. Data are represented as mean ± SEM.
(E) Real-time PCR analysis of Hs578T cells cultured in either HS578T-conditioned media or fresh media for 6 hours. Transcript levels were normalized to ACTB, and then to the corresponding value for “Fresh Media.” Data are represented as mean ± SEM. NS refers to not statistically significant, * refers to p-value < 0.05, ** refers to p-value < 0.01.
(F and H) Immunoblot analysis of Hs578T cells grown in fresh media or Hs578T-CCM. 100 µg/mL Cycloheximide (F) or 10 µM MG132 (H) was added for the indicated duration before cell lysis.
(G and I) Quantification of band intensities from (F and H).
See also Figure S2.
These observations suggested that a factor secreted by TNBC cells induces HIF1α. In support of this idea, the conditioned media from MDA-MB-231 and Hs578T cells induced HIF1α when added to freshly plated Hs578T cells (Figure 2C). This effect was specific, because HIF1α was not induced by conditioned media from hormone-responsive MCF7 and T47D breast cancer cells, or by conditioned media from RCC4 cells (Figure 2C). Moreover, the conditioned media from Hs578T cells did not induce HIF1α when added to freshly plated hormone-responsive breast cancer cells (Figure S2A).
To study this further, we made an Hs578T breast cancer subclone harboring a firefly luciferase (FLuc) reporter driven by a HIF-responsive (3xHRE) promoter. Conditioned media from parental Hs578T cells, but not T47D cells, increased FLuc activity in this subclone (Figure 2D). The prolyl hydroxylase inhibitor dimethyloxalylglycine (DMOG) and fresh media served as positive and negative controls, respectively. Consistent with these observations, Hs578T-conditioned media also induced endogenous HIF target gene expression when added to freshly plated Hs578T cells (Figure 2E). Hs578T-conditioned media, but not fresh media, stabilized HIF1α when added to freshly plated Hs578T cells, as evidenced by the slower disappearance of HIF1α after adding cycloheximide (Figures 2F and 2G). Collectively, these results suggest that TNBC cells can both secrete and respond to a factor that stabilizes HIF1α under normoxic conditions.
We next treated freshly plated Hs578T cells with fresh media or Hs578T-conditioned media and measured prolyl-hydroxylated HIF1α at varying timepoints after proteasomal blockade with MG132 using an antibody specific for HIF1α hydroxylated at proline 564. The total amount of HIF1α in the presence of fresh media and conditioned media was comparable after 6 hours of MG132 treatment, which is again consistent with conditioned media acting primarily at the level of HIF1α stability (Figures 2H and 2I). Notably, prolyl-hydroxylated HIF1α (both absolute and relative to total HIF1α) was decreased at each of the earlier timepoints in cells treated with Hs578T-conditioned media compared to cells fed fresh media (Figures 2H and 2I), suggesting that TNBC cells secrete a factor that suppresses HIF1α prolyl hydroxylation and thereby stabilizes HIF1α.
L-Glutamate Is Sufficient to Induce HIF1α
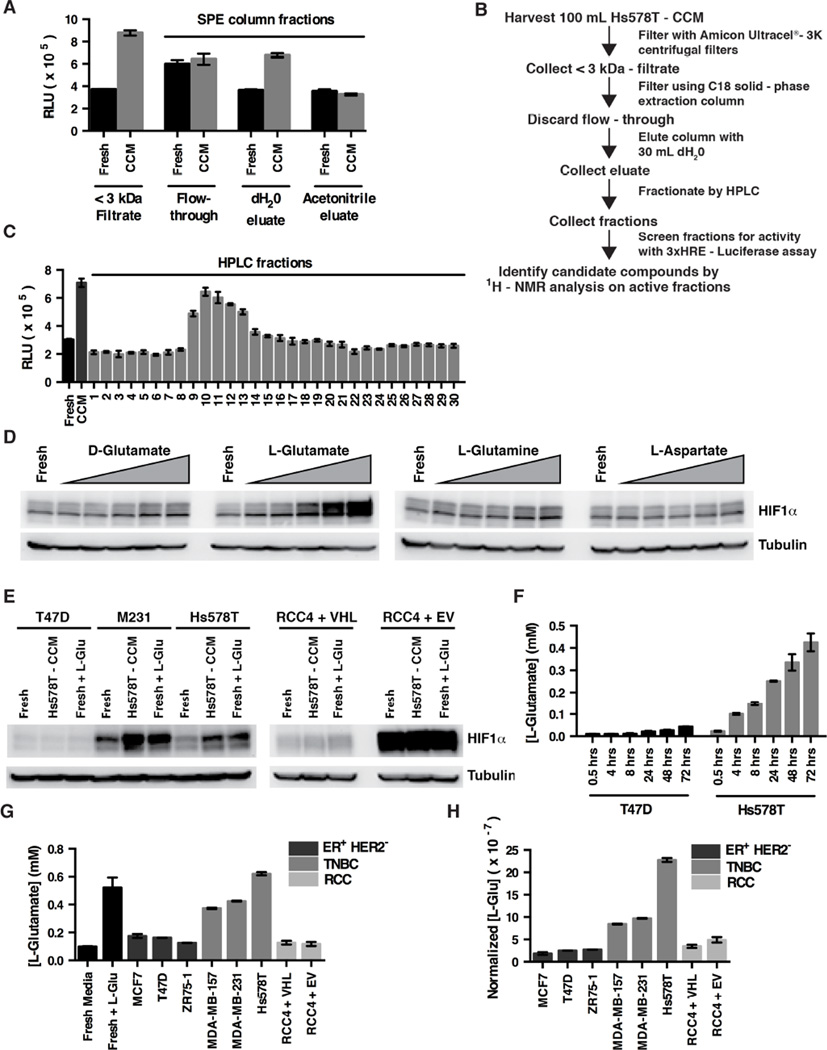
This factor was not inactivated by boiling or multiple freeze/thaw cycles, was insensitive to RNAse, DNAse and Proteinase K treatment, was less than 3 kDa in size, and was eluted by water from C18 solid phase extraction (SPE) columns, as measured by its ability to induce FLuc in the Hs578T 3xHRE-FLuc cells (Figure 3A and data not shown). The C18 water eluate was then further fractionated by HPLC (Figures 3B and 3C). NMR spectroscopy of the HPLC fraction containing the maximal HIF-inducing activity revealed the presence of glutamate, as well as several other metabolites including lysine, serine, malate, 2,3-diaminopropionate and inositol.
Figure 3. L-Glutamate Secreted by TNBC is Sufficient to Induce HIF1α.
(A and C) FLuc activity of Hs578T cells expressing FLuc under the control of the 3xHRE promoter after 6 hours exposure to the indicated partially purified fractions derived from Hs578T-CCM. Panel (A) shows data from the SPE-column elutions. Panel (C) shows data from the HPLC fractions. Data are represented as mean ± SEM. (B) Schema for purifying HIF1α-inducing factor from Hs578T-CCM.
(D) Immunoblot analysis of Hs578T cells grown for 4 hours in fresh standard culture media (containing 4 mM L-glutamine and 0.2 mM L-cystine) supplemented with increasing concentrations of the indicated amino acids. Triangle indicates 0.075, 0.15, 0.3, 0.6, and 1.2 mM.
(E) Immunoblot analysis of breast [T47D, MDA-MB-231 (M231), Hs578T] and renal (RCC4) cancer cell lines cultured in fresh media, Hs578T-CCM, or fresh media supplemented with 0.6 mM L-glutamate.
(F) L-glutamate concentrations in cell culture media conditioned by T47D cells or Hs578T cells for the indicated timepoints. Data are represented as mean ± SEM.
(G and H) L-glutamate concentrations in cell culture media conditioned by the indicated cell lines for 72 hours. Fresh media (Fresh) or fresh media supplemented with 0.6 mM L-glutamate (+L-Glu) serve as controls. Cells were plated in order to achieve similar confluence at 72 hours. In (H) data were normalized to cell number. Data are represented as mean ± SEM.
See also Figure S2.
Of these potential mediators, only L-glutamate acutely induced HIF1α in TNBC cells when added to fresh media (Figures 3D and 3E, and data not shown). This effect was specific because it was not seen with D-glutamate, L-glutamine, or L-aspartate (Figure 3D). Moreover, L-glutamate, like TNBC-conditioned media, did not induce HIF1α when added to T47D breast cancer cells, or RCC4 cells (Figure 3E), but did induce HIF1α in PC-9 lung cancer cells, M34 melanoma cells, immortalized astrocytes, and a subset of glioma lines, indicating that HIF1α induction by L-glutamate is not unique to TNBC (Figure S2B–G). Importantly, these L-glutamate effects were seen at concentrations comparable to the L-glutamate concentrations achieved in TNBC conditioned media, and the ability of conditioned media from T47D cells, MDA-MB-231 cells, and Hs578T cells to induce HIF1α correlated with their L-glutamate concentrations (Figures 2C and 3F–3H).
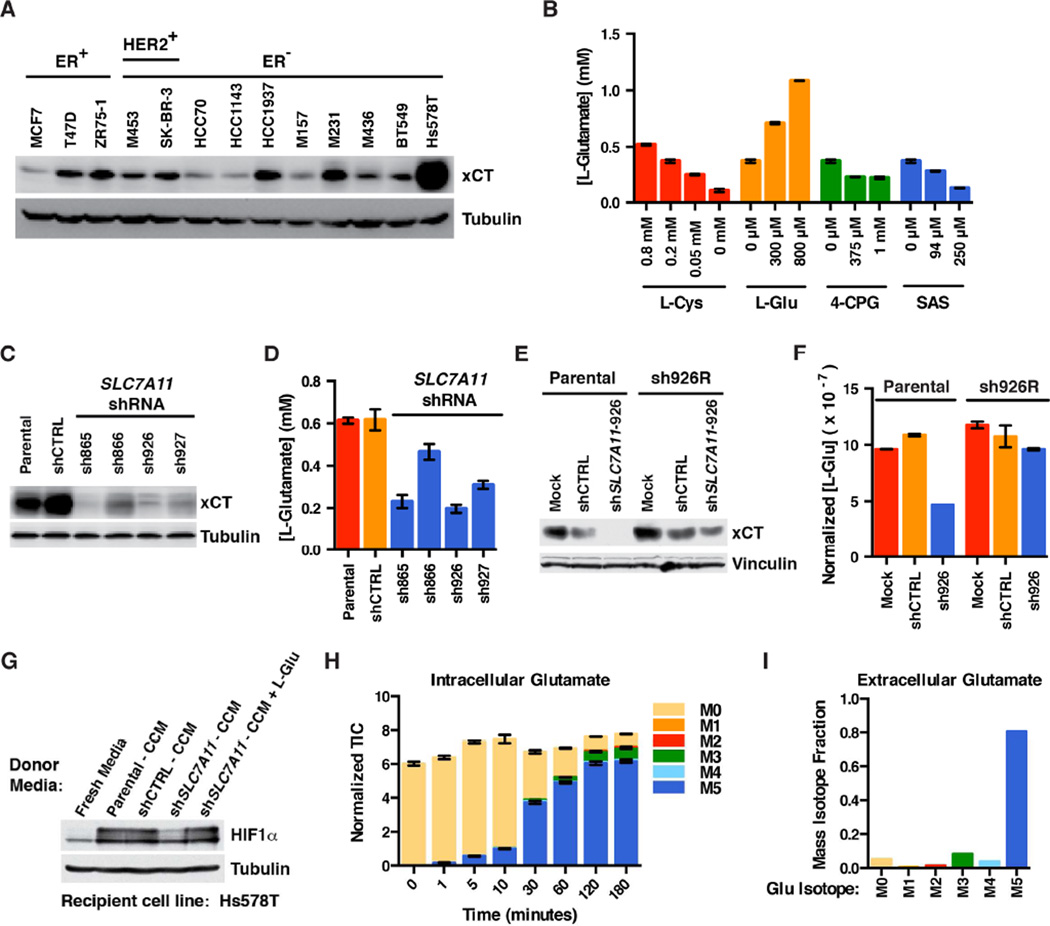
xCT Is Required for L-Glutamate Secretion by TNBC
We reasoned that glutamate secretion by TNBC cells might be mediated by the xCT cystine-glutamate antiporter (encoded by SLC7A11), which has been linked to glutamate secretion by cancer cells and implicated as a potential therapeutic target in TNBC (Narang et al., 2003, 2007; Timmerman et al., 2013; Yang and Yee, 2014). In TNBC, high xCT protein levels are an independent predictor of poor prognosis (Figure S3). In keeping with earlier reports, we found that the xCT protein is detectable in all of the breast cancer lines we tested, but to varying degrees (Figure 4A). Eliminating xCT with either shRNA or CRISPR-based gene editing modestly inhibited Hs578T cell proliferation under standard cell culture conditions (Figure S4A–C).
Figure 4. xCT Is Required for Glutamate Secretion by TNBC.
(A) Immunoblot analysis of the indicated breast cancer lines.
(B) L-glutamate concentration in cell culture media conditioned by Hs578T cells treated with the indicated concentrations of L-cystine (L-Cys), L-glutamate (L-Glu), or the xCT inhibitors (S)4-carboxyphenylglycine (4-CPG) or sulfasalazine (SAS) for 24 hours. All media contained 0.2 mM L-Cys except where indicated. Data are represented as mean ± SEM.
(C and E) Immunoblot of Hs578T cells infected with lentiviruses encoding the indicated SLC7A11 shRNAs or a control (CTRL) shRNA. In (E) either parental Hs578T cells or cells stably expressing an exogenous SLC7A11 cDNA resistant to SLC7A11 shRNA-926 (sh926R) were infected with lentiviruses encoding SLC7A11 shRNA-926 (sh926) or a control shRNA (shCTRL), or underwent a mock infection in the absence of lentivirus (Mock).
(D and F) L-glutamate concentrations in cell culture media conditioned by the cell lines used in (C) and (E), respectively, for 48 hours. In (F) L-glutamate quantifications were normalized to total cell number. Data are represented as mean ± SEM.
(G) Immunoblot analysis of Hs578T treated with conditioned media from cells from (C) and (D). Where indicated exogenous L-glutamate was added to the CCM to a final concentration of 0.6 mM.
(H and I) 13C-labeled intracellular (H) and extracellular (I) glutamate detected after growing Hs578T cells either for the indicated timepoints (H), or 24 hours (I) in media containing uniformly labeled (M5) 13C-glutamine. Ion counts in for each mass isotopomer in (H) were normalized to an internal standard. Data are represented as mean ± SEM (H), or representative of two independent experiments (I).
See also Figure S3.
Pharmacologically inhibiting xCT with (S)4-carboxyphenylglycine [(S)4-CPG], sulfasalazine (SAS), or growth in L-cystine-free media, decreased glutamate accumulation in Hs578T-conditioned media (Figure 4B). Of note, neither (S)4-CPG nor SAS affected Hs578T proliferation at these concentrations. (S)4-CPG and SAS did, however, inhibit Hs578T proliferation at concentrations above 1 mM and 500 µM, respectively, in a way that is likely off-target as revealed by the sensitivity of Hs578T cells lacking xCT to these two agents (Figure S4D–H).
Consistent with our results with pharmacological xCT inhibitors, downregulating SLC7A11 with four independent shRNAs decreased L-glutamate secretion (Figures 4C and 4D). This effect was on-target because it was reversed by expressing an shRNA-resistant SLC7A11 cDNA (Figures 4E and 4F). As expected, the conditioned media from cells expressing an effective SLC7A11 shRNA did not induce HIF1α unless supplemented with exogenous L-glutamate (Figure 4G). Together with the data in Figure 3, these results indicate that L-glutamate is secreted by xCT and is both sufficient and necessary for TNBC-conditioned media to induce HIF.
HIF1 reportedly can regulate SLC7A11 in some cells, including breast cancer cells (Lu et al., 2015; Sims et al., 2012). However, xCT levels did not correlate with HIF1α levels in our breast cancer cell line panel (compare Figure 1C with Figure 4A) and, unlike the well-studied HIF1α target BNIP3, was unaffected by effective HIF1A shRNAs or DMOG (Figure S1B and C). Perhaps some of the previously described effects of hypoxia on SLC7A11 were caused by redox stress and induction of the SLC7A11 regulator NRF2 instead of HIF.
The highest xCT levels we detected were in Hs578T cells (Figure 4A), which secrete very high amounts of L-glutamate (Figures 3G and 3H). Nonetheless, differences in xCT levels do not fully explain the differences in glutamate secretion between TNBC cells and hormone-responsive cells such as T47D and ZR75-1 (compare Figure 4A to Figures 3G and 3H). Conceivably other differences, such as differences in glutamate metabolism, play a role. Growing Hs578T cells in media containing 13C-glutamine confirmed that the glutamate they secrete is derived from glutamine (Figure 4H and 4I), consistent with reports that TNBC cells avidly consume glutamine and are glutamine auxotrophs (Gross et al., 2014; Timmerman et al., 2013).
Inhibition of xCT by Glutamate Depletes Intracellular Cysteine and Induces HIF1α
How does glutamate induce HIF1α? We reasoned that glutamate might act through a glutamate receptor, such as metabotropic glutamate receptor 1 (GRM1), which is expressed by some breast cancers (Mehta et al., 2013; Speyer et al., 2012). However, HIF1α induction by glutamate in TNBC lines was not blocked by glutamate receptor antagonists such as the GRM1 antagonists, BAY 36-7620 and JNJ 16259685; or the GRM2/3 antagonist, LY 341495 (data not shown). Nor was HIF1α induced by glutamate receptor agonists such as the GRM1/5 agonist, (S)-3,5-DHPG; the AMPA and GRM1/5 agonist, L-quisqualic acid; the GRM2/3 agonist, DCG IV; or the GRM4, 6–8 agonist, L-AP4 (data not shown).
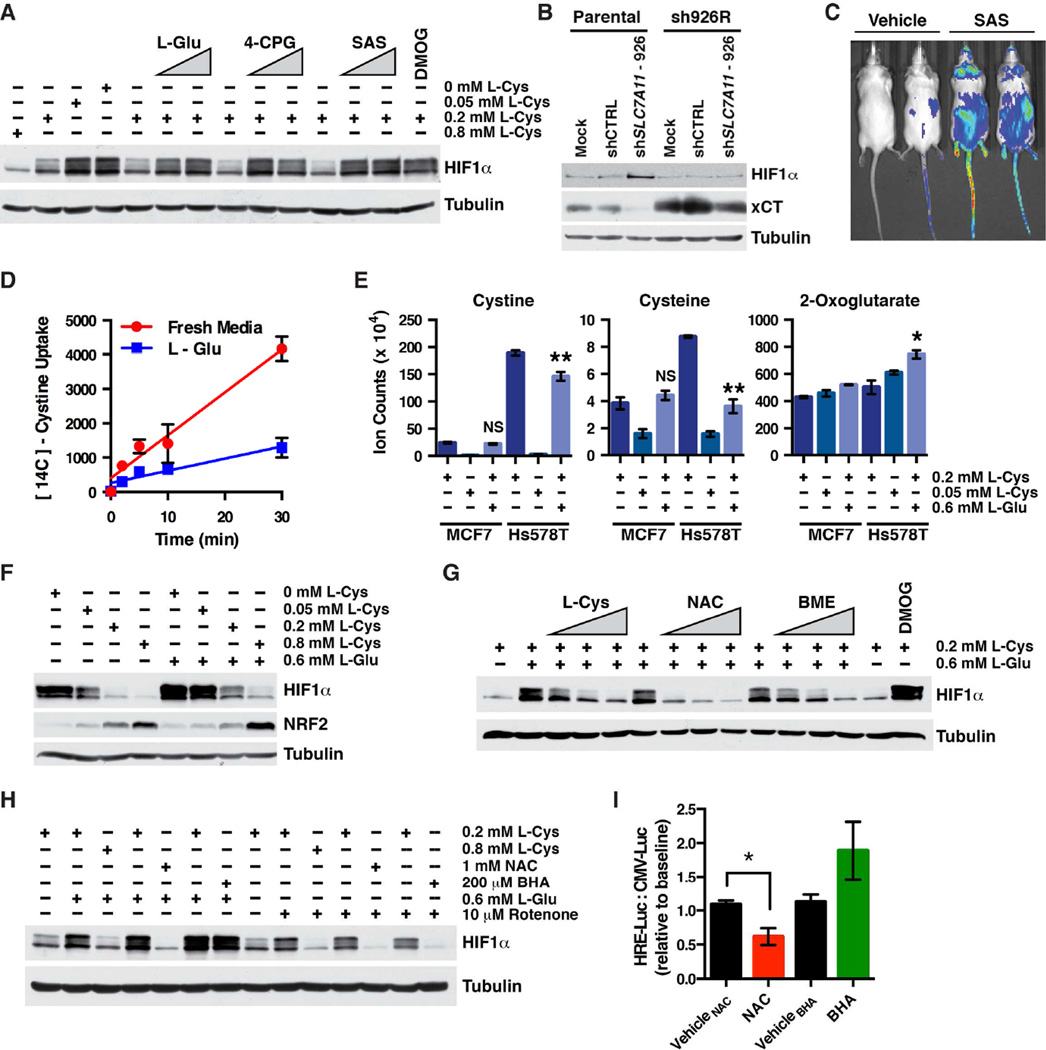
In the course of our xCT inhibition experiments, however, we noted that HIF1α was induced in TNBC cells in which xCT activity was pharmacologically or genetically suppressed (Figures 5A, 5B and S4A). The xCT inhibitor SAS also inhibited EglN activity in vivo, and stabilized HIF1α, as determined by bioluminescent imaging of reporter mice that ubiquitously express a HIF1α-FLuc fusion protein (Figures 5C, S5A and S5B). Moreover, extracellular glutamate inhibits xCT activity with a Ki of ~150 µM (Makowske and Christensen, 1982), which is below the glutamate concentration we measured in TNBC-conditioned media (see Figures 3F and 3G). We confirmed that 600 µM L-glutamate inhibited xCT, as shown by decreased cystine uptake, decreased intracellular cystine and cysteine, and increased intracellular glutamate (Figures 5D, 5E, S5C and S5D), without affecting cellular proliferation (data not shown). Collectively these observations suggested that extracellular glutamate induces HIF by inhibiting xCT and decreasing cystine uptake.
Figure 5. Inhibition of xCT Depletes Cysteine and Induces HIF.
(A) Immunoblot of Hs578T cells grown in fresh media supplemented with the indicated concentrations of L-cystine (L-Cys) and increasing concentrations of either L-glutamate (L-Glu; 300 and 800 µM), the xCT inhibitors (S)4-carboxyphenylglycine (4-CPG; 0.375 and 1 mM) or sulfasalazine (SAS; 94 and 250 µM), or the EglN inhibitor, DMOG (0.1 mM) for 4 hours.
(B) Immunoblot analysis of cells prepared as in Figure 4E.
(C) Representative dorsal bioluminescent images (BLI) of HIF1α-luciferase mice after two doses of 250 mg/kg sulfasalazine (SAS) or vehicle. Images were obtained 18 hours after the first dose.
(D) Cystine uptake in Hs578T cells grown in fresh media or media supplemented with 0.6 mM L-glutamate. Data are represented as mean ± SEM.
(E) Intracellular levels of the indicated metabolites, as determined by LC-MS, for MCF7 and Hs578T cells cultured in the presence of the indicated concentrations of L-cystine and L-glutamate. Data are represented as mean ± SEM, * refers to p-value < 0.05, and ** refers to p-value < 0.01.
(F) Immunoblot of Hs578T grown in media containing the indicated concentrations of L-cystine (L-Cys) and L-glutamate (L-Glu).
(G and H) Immunoblot analysis of Hs578T cells grown for 4 hours in media containing additives at the indicated concentrations, and for (G), increasing concentrations of L-cystine (L-Cys; to a final concentration of 0.4, 0.6 and 0.8 mM), N-acetylcysteine (NAC; 0.2, 0.5, and 1.0 mM) and β-mercaptoethanol (BME; 20, 50 and 100 µM).
(I) Change in FLuc activity, as determined by BLI, of orthotopic Hs578T cell tumors expressing FLuc driven by the 3xHRE promoter after treating mice with NAC, BHA, or their respective vehicles. For each mouse the values were first normalized to the BLI signal from a contralateral orthotopic Hs578T cell tumor expressing FLuc driven by a constitutive promoter (CMV) and then divided by the corresponding pretreatment ratio for that mouse.
See also Figures S4 and S5.
Consistent with this idea, HIF1α was induced in Hs578T cells grown in cystine-poor media (0 or 0.05 mM cystine compared to standard culture media, which contains 0.2 mM cystine), while high cystine levels reversed the effects of exogenous L-glutamate (Figures 5A and 5F). Notably, these manipulations did not affect HIF1α in MCF7 cells, which were likewise insensitive to TNBC-conditioned media (Figures S2A, S5E and S5F). Treating Hs578T cells with β-mercaptoethanol (BME), which promotes cystine uptake in an xCT-independent manner (Ishii et al., 1981; Janjic and Wollheim, 1992), also reversed the effects of glutamate on HIF1α (Figure 5G).
Neither cystine depletion nor treatment with L-glutamate lowered 2-OG levels, making loss of this EglN cofactor an unlikely explanation for their ability to induce HIF1α (Figure 5E). Cystine depletion was associated, however, with decreased intracellular cysteine levels and the effects of L-glutamate on HIF1α were reversed by N-acetylcysteine (NAC)(Figures 5E and 5G).
Cysteine is used to make the ROS scavenger, reduced glutathione (GSH). Increased ROS can inhibit EglN in some cellular contexts (Kaelin, 2005). Glutathione (reduced/oxidized) was not, however, decreased in TNBC cells deprived of cystine or treated with L-glutamate under conditions sufficient to induce HIF1α (Figure S5H). As a control, we did detect decreased glutathione levels in cells treated with the glutathione synthase inhibitor, buthionine sulfoximine (BSO)(Figure S5H). Moreover, ROS was not increased in cells treated with L-glutamate (Figure S5I).
As an alternative way to monitor ROS, we exploited the fact that ROS inactivates the KEAP1 ubiquitin ligase and thereby stabilizes NRF2 (Sporn and Liby, 2012). Cystine depletion and L-glutamate treatment did not increase NRF2 (Figure 5F) and did not activate an exogenous NRF2-FLuc fusion protein we made as an ROS reporter (Figure S5J). The ROS-inducing agent diethyl maleate (DEM), which depletes cells of reduced glutathione, served as a positive control (Figures S5H and S5J). Additionally, the ROS scavenger butylated hydroxyanisole (BHA), which is structurally unrelated to NAC, blocked the accumulation of HIF1α caused by the ROS-inducing agent rotenone, but not by L-glutamate (Figure 5H). Finally, lowering glutathione levels in Hs578T with BSO or DEM did not induce HIF1α (Figure S5K). Collectively these results implied that HIF1α induction by L-glutamate was not caused by cellular ROS.
To study this further, we grew Hs578T cells expressing FLuc driven by the 3xHRE promoter or a constitutive promoter in opposing mammary fat pads in immunocompromised mice. Treating such mice with SAS minimally induced the 3xHRE-FLuc signal (data not shown), possibly because xCT was already inhibited by glutamate. Consistent with this idea, and consistent with the cell culture results depicted in Figure 5H, the 3xHRE-FLuc signal was inhibited by NAC, but not BHA (Figure 5I).
2-OG-dependent dioxygenases, such as EglN1, are prone to self-catalyzed inactivation, thought to reflect oxidation of their ferrous irons or reactive cysteine residues (Flashman et al., 2010; Hirsila et al., 2005; Myllyla et al., 1978). For this reason, in vitro assays with such enzymes often contain reducing agents such as ascorbate. We noted that HIF1α induction in TNBC cells by glutamate was reversed by ascorbate and, to a much lesser extent, ferrous iron, consistent with glutamate causing EglN oxidation (Figures S6A and S6B). Importantly, baseline ascorbate levels are higher in Hs578T cells than in hormone-responsive MCF7 cells (Figures S5C and S6C), indicating that the sensitivity of Hs578T cells to glutamate, relative to MCF7 cells, is not caused by ascorbate deficiency.
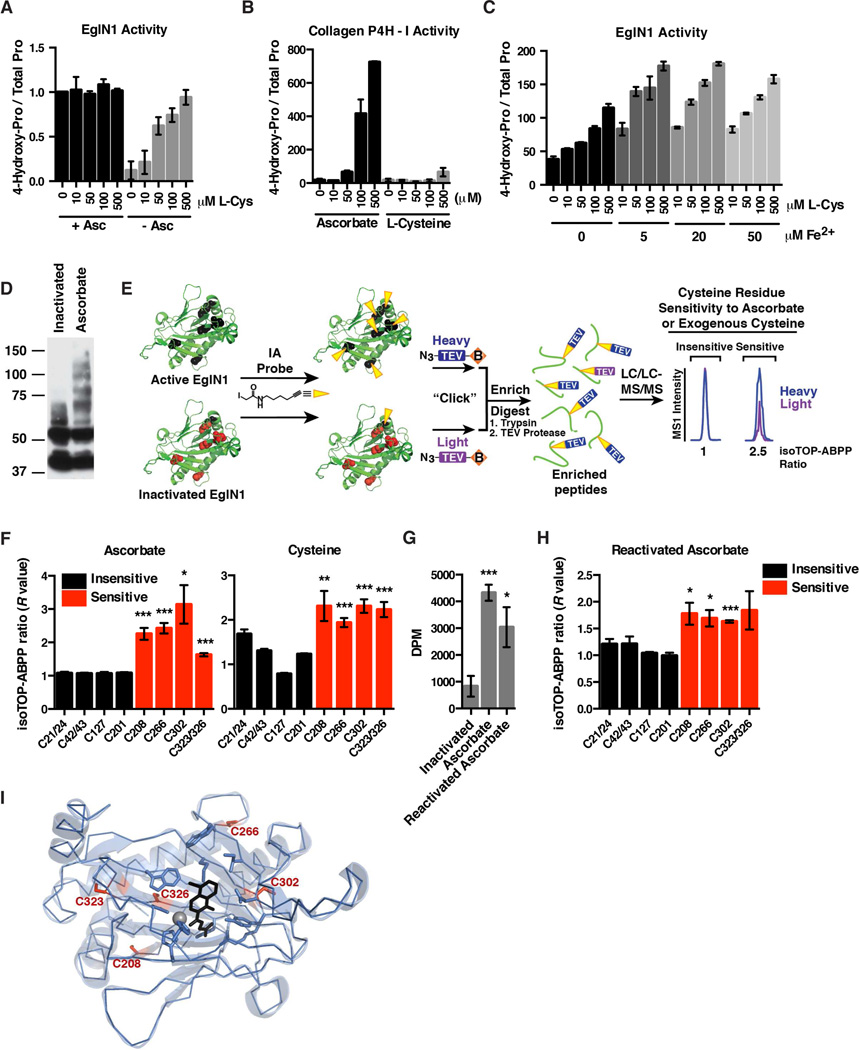
Free Cysteine Maintains EglN1 Activity by Preventing Oxidation of Specific EglN1 Cysteine Residues
In vitro assays done with recombinant EglN1 in the presence of ascorbate were insensitive to the addition of L-cysteine (Figure 6A). Remarkably, however, ascorbate-free reactions were stimulated by L-cysteine in the same concentration range as in our cellular studies (Figure 6A). In contrast, L-cysteine could not substitute for ascorbate in collagen 4-prolyl hydroxylase assays (Figure 6B). These findings suggest that EglN1 senses both oxygen and free cysteine.
Figure 6. Cysteine Maintains EglN1 Activity by Preventing Oxidation of Specific Intramolecular Cysteine Residues.
(A and B) In vitro EglN1 activity (A) or collagen-4-prolyl hydroxylase-1 activity (B) in the presence or absence of ascorbate, and increasing L-cysteine concentrations. Data shown are represented as mean ± SEM of four independent experiments (A), or two independent experiments (B). Data in (A) were first normalized to the value for “0 µM L-Cys + Asc.”
(C) In vitro activity of EglN1 supplemented with increasing concentrations of Lcysteine (L-Cys) and ferrous iron. Data shown are representative of two independent experiments.
(D) Immunoblot of self-inactivated or active recombinant EglN1 after covalent attachment of PEG-maleimide followed by SDS-PAGE. EglN1 activity was maintained using ascorbate.
(E) EglN1 isoTOP-ABPP schematic. Active or inactivated recombinant EglN1 samples are treated with cysteine-directed iodoacetamide-alkyne probe (IA) (reduced cysteine residues represented in black, oxidized cysteine residues represented in red). The samples are then conjugated to isotopically differentiated TEV protease-cleavable biotin tags [light (purple) and heavy (blue)] by click chemistry and mixed. The IA-labeled proteins are enriched using streptavidin-conjugated beads, and digested stepwise on-bead with trypsin and TEV to yield IA-labeled peptides for MS analysis. Competition ratios or R values are measured by dividing the MS1 ion peaks for IA-labeled peptides in active (ascorbate-treated) (heavy, or blue) versus inactivated (light, or purple) samples.
(F and H) Iodoacetamide alkyne-labeling of cysteine residues in full-length, recombinant EglN1. Residues sensitive to ascorbate or exogenous cysteine are indicated by an increase in the isoTOP-ABPP ratio (red bars). Data are represented as mean ± SEM. * refers to p-value < 0.05, ** refers to p-value < 0.01, *** refers to p-value < 0.001.
(G) In vitro activity of recombinant EglN1 in the absence of ascorbate (Inactivated), the presence of ascorbate, or activity of previously inactivated EglN1 following reactivation with ascorbate (Reactivated Ascorbate). Data shown are represented as mean ± SEM of four independent experiments. * refers to p-value < 0.05, *** refers to p-value < 0.001.
(I) Crystal structure of EglN1 catalytic domain (PDB ID: 2G19) showing location of the reactive cysteine residues, iron (gray sphere), and a 2-OG-competitive EglN inhibitor, N-[(4-hydroxy-8-iodoisoquinolin-3-YL)carbonyl]glycine (black).
See also Figure S6.
EglN1 activity in ascorbate-free reactions was also sustained in the presence of dithiothreitol (DTT) or high concentrations of reduced glutathione, but not in the presence of BHA, BME or L-cystine (Figure S6D). The ability of DTT and glutathione to substitute for ascorbate in such assays has been reported before (Flashman et al., 2010). EglN1 activation by L-cysteine occurred over a wide range of ferrous iron concentrations and in both the presence and absence of catalase, suggesting that the effects of L-cysteine did not involve altered iron oxidation or protection from hydrogen peroxide (Figures 6C and S6E). A caveat regarding the former is that iron is tightly bound by EglN1 and might not rapidly exchange with free iron in solution. We did not, however, detect a change in the oxidation status of EglN1-bound iron by electron paramagnetic resonance (EPR) spectroscopy under conditions that promote EglN1 self-inactivation (Figure S6F).
We next asked if EglN1 self-inactivation might reflect oxidation of specific EglN1 cysteine residues, which would decrease their reactivity with electrophilic probes such as maleimide and iodoacetamide. In support of this idea, active EglN1 was more readily labeled than self-inactivated EglN1 with PEG-maleimide, as seen by its retarded electrophoretic mobility (Figure 6D). To map the responsible cysteine residues, we used a quantitative chemical proteomic method termed isoTOP-ABPP (Weerapana et al., 2010), wherein active and inactive EglN1 were reacted with an iodoacetamide-alkyne (IA) probe, which was then conjugated to an isotopically heavy or light, respectively, TEV protease-cleavable biotin tag using click chemistry (Figure 6E). The two EglN1 samples were then mixed and the labeled EglN1 was recovered with streptavidin agarose and digested with trypsin. Bound peptides containing IA-labeled cysteine residues were released using TEV protease and analyzed by liquid chromatography-MS/MS, using the heavy/light (isoTOP-ABPP) ratio to infer changes in cysteine reactivity (ratio of ~1 = ~ no change, ratio > 1 = more reactive in active EglN1 than in self-inactivated EglN1). Previous isoTOP-ABPP studies showed that the IA-alkyne probe, in general, reacts strongly with redox-regulated cysteines in the native human proteome (Weerapana et al., 2010). We therefore used native EglN1 in our isoTOP-ABPP experiments in an attempt to minimize denaturation-induced artifactual changes in cysteine oxidation states. These experiments reproducibly quantified the relative reactivity of 11 of 15 total cysteines in EglN1.
We reproducibly observed increased IA-labeling of specific EglN1 cysteine residues within the EglN1 catalytic domain (Cys 208, Cys 266, Cys 302, and Cys 323/326; note that cysteines on the same tryptic peptide cannot be differentiated) in active EglN1 compared to self-inactivated EglN1, consistent with these residues becoming oxidized during self-inactivation (Figures 6F and 6I). These same residues regained IA-reactivity after reactivating self-inactivated EglN1 with ascorbate (Figures 6G and 6H). These changes were specific because multiple other EglN1 cysteine residues (Cys 21/24, Cys 42/43, Cys 127, and Cys 201) were not differentially labeled comparing active and inactive EglN1 (Figure 6F and 6H). These results suggest that free cysteine protects EglN1 from reversible auto-oxidation of specific intramolecular cysteine residues.
Discussion
We discovered that TNBC cells induce HIF in a paracrine fashion by secreting glutamate. High extracellular glutamate levels inhibit the glutamate-cystine antiporter, xCT, and thereby interfere with cystine uptake and lower intracellular cysteine levels. Decreased intracellular cysteine levels inhibit the EglN prolyl hydroxylases and thereby stabilize HIF1α.
xCT is required for the secretion of glutamate by TNBC cells and thus appears to play both an efferent and afferent role in intercellular signaling by glutamate with respect to HIF. Glutamate secretion by cancer cells via xCT has been reported previously (Seidlitz et al., 2009; Sharma et al., 2010). HIF induction by glutamate does not exclude other roles of glutamate secretion in cancer. Glutamate can also alter cancer cell behavior by modulating ionotropic glutamate receptors, such as the AMPA receptor and NMDA receptors, and metabotropic glutamate receptors (Stepulak et al., 2014; Willard and Koochekpour, 2013). Indeed, a role for the NMDA receptor and metabotropic glutamate receptor-1 in breast cancer has been previously suggested (Mehta et al., 2013; North et al., 2010; Speyer et al., 2012). Secretion of glutamate by glioblastoma cells promotes invasion and tumorigenesis at least partly by causing neuroexcitatory death of surrounding normal brain cells and by activating AMPA receptors (Stepulak et al., 2014; Willard and Koochekpour, 2013). Glutamate can also, via AMPA and NMDA receptors, promote blood vessel leakiness, which could facilitate metastasis (Andras et al., 2007). Therefore, glutamate might have multiple downstream targets in cancer.
Our data suggest that some cancers induce HIF in response to glutamate and others do not. In this regard, we noted that ER-positive breast cancer lines and HER2-positive breast cancer lines do not secrete high levels of glutamate and do not increase HIF in response to exogenous glutamate despite the presence of xCT. This differential sensitivity presumably reflects, at least in part, differences in intracellular metabolism such as differences in glutamine uptake (and conversion to glutamate), intracellular cysteine turnover, and basal EglN activity.
TNBCs have particularly high xCT levels relative to other cancers (Timmerman et al., 2013; Yang and Yee, 2014) and we found that TNBCs with high xCT levels are highly lethal, conceivably because they can secrete and respond to extracellular glutamate. Clearly it is dangerous to infer causality from a prognostic biomarker. For example, the association of xCT with poor clinical outcomes might simply reflect the fact that aggressive tumors experience redox stress, which can induce NRF2 and hence xCT.
Our work, however, suggests the intriguing possibility that TNBCs with high xCT have a poor prognosis because they accumulate extracellular glutamate, which inhibits xCT function and induces HIF. If intratumoral glutamate levels were to fall the inhibition of xCT would be relieved, leading to more glutamate secretion and reestablishment of the xCT-inhibited steady state.
On the other hand pharmacological and shRNA-mediated disruption of xCT function inhibits many cancers, including TNBC, in preclinical models (Dixon et al., 2012; Lu et al., 2015; Narang et al., 2003, 2007; Timmerman et al., 2013; Yang et al., 2014; Yang and Yee, 2014). Can this be reconciled with our model that inhibition of xCT by glutamate stimulates tumor growth? We confirmed that loss of xCT causes a modest proliferation defect under standard culture conditions. Nonetheless, we readily created TNBC cells with undetectable xCT levels using CRISPR-based gene editing. Of note, these cells remained sensitive to (S)4-CPG and SAS, suggesting that off-target effects confounded their previously published anticancer activities. More importantly, glutamate did not inhibit TNBC proliferation at concentrations that were more than sufficient (up to 25 mM; data not shown) to inhibit xCT and induce HIF, suggesting there are quantitative or qualitative differences between inhibiting xCT with glutamate compared to genetically eliminating xCT or inhibiting its function with drugs such as (S)4-CPG and SAS.
Among the natural amino acids, L-cysteine and L-histidine are uniquely capable of increasing EglN activity in cells (Lu et al., 2005). L-cysteine can be converted to the endogenous ROS scavenger, reduced glutathione. We therefore initially hypothesized that the induction of HIF1α observed after xCT blockade was caused by ROS-mediated EglN inactivation resulting from glutathione depletion. We did not, however, detect decreased glutathione (reduced/oxidized) or increased ROS in TNBC cells grown in cystine-free media or in cells in which xCT was blocked by extracellular glutamate. In addition, we observed decreased, rather than increased, levels of the ROS-inducible NRF2 transcription factor in cystine-starved cells. Conversely, NRF2 was induced in cells in which HIF1 was suppressed by exogenous cystine. Finally, ROS scavengers structurally unrelated to cysteine prevented HIF induction by ROS, but not by glutamate. These findings strongly suggest that the effects of cysteine loss on HIF1α in TNBC cells are not caused by decreased glutathione and increased cellular ROS.
Instead, we found that cysteine directly stimulates recombinant EglN1 in cell-free assays in the absence of other reducing agents and does so at concentrations that modulate EglN1 activity in cells. This finding is consistent with earlier work in which EglN1 activity assays were performed with crude cell extracts (Lu et al., 2005). Ascorbate is used to support EglN1 activity in vitro but is dispensable for EglN1 activity in vivo (Nytko et al., 2011). While EglN1 acts as a key oxygen sensor, these data indicate that it can also directly sense intracellular cysteine, and that cysteine is sufficient to protect EglN1 from self-inactivation.
Although both cysteine and histidine can alter iron redox status, we did not detect a change in EglN1 iron oxidation status in self-inactivated EglN1. Instead, our data suggest that free cysteine prevents the oxidation of specific cysteine residues within EglN1 that are critical for its function. There is growing appreciation that oxidative cysteine posttranslational modifications play important roles in cell signaling (Chung et al., 2013; Green and Paget, 2004; Jacob, 2011). Indeed, cysteine appears to be an evolutionarily ancient sensor of oxygen and oxidative stress (Green and Paget, 2004; Mecinovic et al., 2009).
EglN1 has an N-terminal zinc finger domain with 7 cysteine residues, a catalytic domain (residues 181–426) with 7 highly conserved cysteines, and an intervening spacer region with one cysteine. Earlier work showed that cysteine residues 127, 201 and 208 are particularly prone to oxidation (Chowdhury et al., 2011; Mecinovic et al., 2009; Nytko et al., 2011). We found that self-inactivation of EglN1 is linked to oxidation of cysteine residues 208, 266, 302, and either 323 or 326 (or both). Of note, it was recently reported that EglN1 cysteine residues 302 and 326 mediate EglN1 dimer formation when EglN1 is oxidatively inactivated (Lee et al., 2016). Clearly, additional studies will be required to determine how, mechanistically, oxidation of the intramolecular cysteine residues identified here regulates EglN1 activity and to understand why this enzyme evolved to sense both oxygen and cysteine. It will also be important to ask whether other 2-OG-dependent dioxygenases, including chromatin regulatory enzymes, share this property.
Experimental Procedures
Full details are provided in the Extended Experimental Procedures.
Identification of L-Glutamate by Fractionation and 1NMR
In pilot experiments, the < 3 kDa media fractions from either fresh media or media conditioned by Hs578T for 72 hours (Hs578T-CCM) were isolated using Ultracel 3K centrifugal filters (Amicon), loaded onto Sep-Pak 1 g C18 6 cc Vac Cartridges (Waters WAT036905), and the flow-through collected. The column was eluted with 1 mL dH20, followed by acetonitrile. The eluates were lyophilized and resuspended in dH20 (flow-through and dH20 eluate) or DMSO (acetonitrile eluate) to test for activity in the 3xHRE-FLuc assay.
To isolate and identify the HIF-inducing factor, the fractionation schema used for the pilot experiments was scaled up proportionally. 30 mL media was loaded onto a 10 g C18-SPE column and the flow-through collected. The column was then eluted twice with 10 mL dH2O, once with 1:1 methanol:dH2O and once with methanol. The eluates were collected and lyophilized, and an aliquot of each was resuspended in water prior to the 3xHRE-FLuc assay.
The active fraction (first dH20 eluate) was separated on a Hypercarb column (250 × 10 mm; 5 µ •; 2 mL/min; with water for 10 minutes, transitioned to 100% methanol in 10 minutes, and maintained on 100% methanol for an additional 10 minutes) using an Agilent HPLC instrument. 30 subfractions were collected (one subfraction per minute), lyophilized and tested in the 3xHRE-FLuc assay.
The 1H NMR spectrum of the active subfractions, collected from a 600 MHz Varian NMR spectrometer, revealed the presence of inositol, malic acid, lactic acid, serine, DAP, glutamic acid, lysine, and glycine. These compounds (both enantiomers, when applicable)(Sigma) were tested in the 3xHRE-FLuc assay.
In Vitro Prolyl Hydroxylation Assays
Recombinant EglN1 was produced in H5 insect cells and affinity purified with anti-Flag affinity column as described earlier (Hirsila et al., 2005). In vitro prolyl hydroxylation assays were done with 0.5 µM recombinant EglN1 and 0.2 µM L-[2,3,4,5-3H]proline-labeled HIF1α oxygen-dependent degradation domain (Koivunen et al., 2006) in hydroxylase buffer containing 50 mM Tris-HCl pH 7.8, 60 µg/ml catalase and 5 µM FeSO4 for 30 min at 37°C. Ascorbate, L-cysteine, L-cystine, GSH, DTT, BHA and BME were added as indicated. 4-hydroxy[3H]proline was measured by a specific radiochemical procedure (Juva and Prockop, 1966). Collagen prolyl-4-hydroxylase activity was determined with the same procedure using purified recombinant Col-P4H-I, [14C]proline-labeled type I protocollagen as a substrate and detecting 4-hydroxy[14C]proline.
Reactivation of self-inactivated EglN1 was assessed by measuring the hydroxylation-coupled decarboxylation of 2-oxo[1-14C]glutarate in a reaction that contained a synthetic peptide DLD19 (50 µM), representing the Pro564 hydroxylation site in HIF1α as a substrate (Hirsila et al., 2005) and 15 µM EglN1 in presence or absence of 0.5 mM ascorbate in hydroxylase buffer for 30 min at 37°C. Omitting ascorbate resulted in self-inactivation. Reactivation was induced by adding 0.5 mM ascorbate to the self-inactivated EglN1 and continuing the incubation for an additional 30 min at 37°C.
In Vitro Iodoacetamide Alkyne Labeling and Enrichment of Recombinant EglN1
Recombinant EglN1 (0.7 mg/mL) was used in in vitro prolyl hydroxylation assays either in the absence of reducing agent (Inactive), or in the presence of 0.5 mM ascorbate or 0.1 mM L-cysteine (Active). 10 µL of active and inactive EglN1 were labeled separately with 1 mM iodoacetamide alkyne (1 µL of 10 mM stock in DMSO). After 60 minutes incubation at RT, 90 µL of soluble MDA-MB-231 proteome (1 mg/mL) was added to each sample. Copper-mediated azide-alkyne cycloaddition (CuAAC or click) chemistry was done for 1h by adding 100 µM heavy-TEV-tag or light-TEV-tag to the active or inactive EglN1 preparations, respectively (2 µL of a 5 mM stock), 1 mM TCEP (50X solution in water), 100 µM ligand (17X solution in DMSO:t-butanol 1:4) and 1 mM CuSO4 (50X solution in water). The labeled samples were then combined, processed and analyzed according the published isoTOP-ABPP protocol (Weerapana et al., 2007)(Weerapana et al., 2007). Cys 33/36, Cys 58 and Cys 283 could not be detected reproducibly, so they were omitted from the final analysis.
Supplementary Material
Highlights.
HIF1 Promotes Triple-Negative Breast Carcinogenesis
Triple-Negative Breast Cancers Secrete Large Amounts of Glutamate
Extracellular Glutamate Inhibits Cystine Uptake by the xCT Antiporter
Intracellular Cysteine Depletion Directly Inhibits the HIF Prolyl Hydroxylases
Acknowledgments
We thank members of the Kaelin Laboratory for useful discussions, Gang Lu for multiple reagents and cloning advice, Jason Marineau for fractionation advice and reagents, Julie Losman for her FACS expertise, Amanda Silva for performing gavage, and Humayun Sharif and Michael Eck for generating Figure 6I. Supported by grants to WGK from the NIH and the Breast Cancer Research Foundation, and T32 and F32 NIH training grants to KJB. WGK is an HHMI Investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
K.J.B. conducted the experiments and, together with W.G.K., Jr., designed the experiments, analyzed data, and assembled and wrote the manuscript. P.K. purified the recombinant EglN1 and performed the in vitro prolyl hydroxylation assays. S.C. and J.C. performed the HPLC fractionation and the NMR analysis of Hs578T-conditioned media. K.M.B. and B.F.C. performed the iodoacetamide alkyne labeling experiments. B.A.O. performed the GC-MS methodology and analysis. H.P. and G.J.G. performed the EPR analysis. Q.Z. assisted with the orthotopic mouse model experiments. S.S. performed the HIF and xCT immunohistochemistry. A.R. analyzed the HIF immunohistochemistry and gene expression analysis. A.W. analyzed and performed statistical analysis on the xCT immunohistochemistry.
References
- Andras IE, Deli MA, Veszelka S, Hayashi K, Hennig B, Toborek M. The NMDA and AMPA/KA receptors are involved in glutamate-induced alterations of occludin expression and phosphorylation in brain endothelial cells. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 2007;27:1431–1443. doi: 10.1038/sj.jcbfm.9600445. [DOI] [PubMed] [Google Scholar]
- Blouw B, Song H, Tihan T, Bosze J, Ferrara N, Gerber HP, Johnson RS, Bergers G. The hypoxic response of tumors is dependent on their microenvironment. Cancer Cell. 2003;4:133–146. doi: 10.1016/s1535-6108(03)00194-6. [DOI] [PubMed] [Google Scholar]
- Bos R, van Der Hoeven JJ, van Der Wall E, van Der Groep P, van Diest PJ, Comans EF, Joshi U, Semenza GL, Hoekstra OS, Lammertsma AA, et al. Biologic correlates of (18)fluorodeoxyglucose uptake in human breast cancer measured by positron emission tomography. J Clin Oncol. 2002;20:379–387. doi: 10.1200/JCO.2002.20.2.379. [DOI] [PubMed] [Google Scholar]
- Chaturvedi P, Gilkes DM, Wong CC, Kshitiz, Luo W, Zhang H, Wei H, Takano N, Schito L, Levchenko A, et al. Hypoxia-inducible factor-dependent breast cancer-mesenchymal stem cell bidirectional signaling promotes metastasis. J Clin Invest. 2013;123:189–205. doi: 10.1172/JCI64993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Iliopoulos D, Zhang Q, Tang Q, Greenblatt MB, Hatziapostolou M, Lim E, Tam WL, Ni M, Chen Y, et al. XBP1 promotes triple-negative breast cancer by controlling the HIF1alpha pathway. Nature. 2014;508:103–107. doi: 10.1038/nature13119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury R, Flashman E, Mecinovic J, Kramer HB, Kessler BM, Frapart YM, Boucher JL, Clifton IJ, McDonough MA, Schofield CJ. Studies on the reaction of nitric oxide with the hypoxia-inducible factor prolyl hydroxylase domain 2 (EGLN1) J Mol Biol. 2011;410:268–279. doi: 10.1016/j.jmb.2011.04.075. [DOI] [PubMed] [Google Scholar]
- Chung HS, Wang SB, Venkatraman V, Murray CI, Van Eyk JE. Cysteine oxidative posttranslational modifications: emerging regulation in the cardiovascular system. Circulation research. 2013;112:382–392. doi: 10.1161/CIRCRESAHA.112.268680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, Patel DN, Bauer AJ, Cantley AM, Yang WS, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flashman E, Davies SL, Yeoh KK, Schofield CJ. Investigating the dependence of the hypoxia-inducible factor hydroxylases (factor inhibiting HIF and prolyl hydroxylase domain 2) on ascorbate and other reducing agents. Biochem J. 2010;427:135–142. doi: 10.1042/BJ20091609. [DOI] [PubMed] [Google Scholar]
- Green J, Paget MS. Bacterial redox sensors. Nature reviews Microbiology. 2004;2:954–966. doi: 10.1038/nrmicro1022. [DOI] [PubMed] [Google Scholar]
- Gross MI, Demo SD, Dennison JB, Chen L, Chernov-Rogan T, Goyal B, Janes JR, Laidig GJ, Lewis ER, Li J, et al. Antitumor Activity of the Glutaminase Inhibitor CB-839 in Triple-Negative Breast Cancer. Mol Cancer Ther. 2014;13:890–901. doi: 10.1158/1535-7163.MCT-13-0870. [DOI] [PubMed] [Google Scholar]
- Hirsila M, Koivunen P, Xu L, Seeley T, Kivirikko KI, Myllyharju J. Effect of desferrioxamine and metals on the hydroxylases in the oxygen sensing pathway. FASEB J. 2005;19:1308–1310. doi: 10.1096/fj.04-3399fje. [DOI] [PubMed] [Google Scholar]
- Ishii T, Hishinuma I, Bannai S, Sugita Y. Mechanism of growth promotion of mouse lymphoma L1210 cells in vitro by feeder layer or 2-mercaptoethanol. Journal of cellular physiology. 1981;107:283–293. doi: 10.1002/jcp.1041070215. [DOI] [PubMed] [Google Scholar]
- Jacob C. Redox signalling via the cellular thiolstat. Biochemical Society transactions. 2011;39:1247–1253. doi: 10.1042/BST0391247. [DOI] [PubMed] [Google Scholar]
- Janjic D, Wollheim CB. Effect of 2-mercaptoethanol on glutathione levels, cystine uptake and insulin secretion in insulin-secreting cells. European journal of biochemistry / FEBS. 1992;210:297–304. doi: 10.1111/j.1432-1033.1992.tb17421.x. [DOI] [PubMed] [Google Scholar]
- Juva K, Prockop DJ. Modified procedure for the assay of H-3-or C-14-labeled hydroxyproline. Analytical biochemistry. 1966;15:77–83. doi: 10.1016/0003-2697(66)90249-1. [DOI] [PubMed] [Google Scholar]
- Kaelin WG., Jr ROS: really involved in oxygen sensing. Cell Metab. 2005;1:357–358. doi: 10.1016/j.cmet.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Kaelin WG, Jr, Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell. 2008;30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- Keith B, Johnson RS, Simon MC. HIF1alpha and HIF2alpha: sibling rivalry in hypoxic tumour growth and progression. Nat Rev Cancer. 2012;12:9–22. doi: 10.1038/nrc3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koivunen P, Hirsila M, Kivirikko KI, Myllyharju J. The length of peptide substrates has a marked effect on hydroxylation by the hypoxia-inducible factor prolyl 4-hydroxylases. J Biol Chem. 2006;281:28712–28720. doi: 10.1074/jbc.M604628200. [DOI] [PubMed] [Google Scholar]
- Laurinavicius A, Laurinaviciene A, Ostapenko V, Dasevicius D, Jarmalaite S, Lazutka J. Immunohistochemistry profiles of breast ductal carcinoma: factor analysis of digital image analysis data. Diagnostic pathology. 2012;7:27. doi: 10.1186/1746-1596-7-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G, Won HS, Lee YM, Choi JW, Oh TI, Jang JH, Choi DK, Lim BO, Kim YJ, Park JW, et al. Oxidative Dimerization of PHD2 is Responsible for its Inactivation and Contributes to Metabolic Reprogramming via HIF-1alpha Activation. Scientific reports. 2016;6:18928. doi: 10.1038/srep18928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Dalgard CL, Mohyeldin A, McFate T, Tait AS, Verma A. Reversible inactivation of HIF-1 prolyl hydroxylases allows cell metabolism to control basal HIF-1. J Biol Chem. 2005;280:41928–41939. doi: 10.1074/jbc.M508718200. [DOI] [PubMed] [Google Scholar]
- Lu H, Samanta D, Xiang L, Zhang H, Hu H, Chen I, Bullen JW, Semenza GL. Chemotherapy triggers HIF-1-dependent glutathione synthesis and copper chelation that induces the breast cancer stem cell phenotype. Proc Natl Acad Sci U S A. 2015;112:E4600–E4609. doi: 10.1073/pnas.1513433112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makowske M, Christensen HN. Contrasts in transport systems for anionic amino acids in hepatocytes and a hepatoma cell line HTC. J Biol Chem. 1982;257:5663–5670. [PubMed] [Google Scholar]
- Mecinovic J, Chowdhury R, Flashman E, Schofield CJ. Use of mass spectrometry to probe the nucleophilicity of cysteinyl residues of prolyl hydroxylase domain 2. Analytical biochemistry. 2009;393:215–221. doi: 10.1016/j.ab.2009.06.029. [DOI] [PubMed] [Google Scholar]
- Mehta MS, Dolfi SC, Bronfenbrener R, Bilal E, Chen C, Moore D, Lin Y, Rahim H, Aisner S, Kersellius RD, et al. Metabotropic glutamate receptor 1 expression and its polymorphic variants associate with breast cancer phenotypes. PLoS One. 2013;8:e69851. doi: 10.1371/journal.pone.0069851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagner M, Enzo E, Forcato M, Zanconato F, Parenti A, Rampazzo E, Basso G, Leo G, Rosato A, Bicciato S, et al. SHARP1 suppresses breast cancer metastasis by promoting degradation of hypoxia-inducible factors. Nature. 2012;487:380–384. doi: 10.1038/nature11207. [DOI] [PubMed] [Google Scholar]
- Myllyla R, Kuutti-Savolainen ER, Kivirikko KI. The role of ascorbate in the prolyl hydroxylase reaction. Biochem Biophys Res Commun. 1978;83:441–448. doi: 10.1016/0006-291x(78)91010-0. [DOI] [PubMed] [Google Scholar]
- Narang VS, Pauletti GM, Gout PW, Buckley DJ, Buckley AR. Suppression of cystine uptake by sulfasalazine inhibits proliferation of human mammary carcinoma cells. Anticancer Res. 2003;23:4571–4579. [PubMed] [Google Scholar]
- Narang VS, Pauletti GM, Gout PW, Buckley DJ, Buckley AR. Sulfasalazine-induced reduction of glutathione levels in breast cancer cells: enhancement of growth-inhibitory activity of Doxorubicin. Chemotherapy. 2007;53:210–217. doi: 10.1159/000100812. [DOI] [PubMed] [Google Scholar]
- North WG, Gao G, Memoli VA, Pang RH, Lynch L. Breast cancer expresses functional NMDA receptors. Breast Cancer Res Treat. 2010;122:307–314. doi: 10.1007/s10549-009-0556-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nytko KJ, Maeda N, Schlafli P, Spielmann P, Wenger RH, Stiehl DP. Vitamin C is dispensable for oxygen sensing in vivo. Blood. 2011;117:5485–5493. doi: 10.1182/blood-2010-09-307637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regan Anderson TM, Peacock DL, Daniel AR, Hubbard GK, Lofgren KA, Girard BJ, Schorg A, Hoogewijs D, Wenger RH, Seagroves TN, et al. Breast tumor kinase (Brk/PTK6) is a mediator of hypoxia-associated breast cancer progression. Cancer Res. 2013;73:5810–5820. doi: 10.1158/0008-5472.CAN-13-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schito L, Rey S, Tafani M, Zhang H, Wong CC, Russo A, Russo MA, Semenza GL. Hypoxia-inducible factor 1-dependent expression of platelet-derived growth factor B promotes lymphatic metastasis of hypoxic breast cancer cells. Proc Natl Acad Sci U S A. 2012;109:E2707–E2716. doi: 10.1073/pnas.1214019109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidlitz EP, Sharma MK, Saikali Z, Ghert M, Singh G. Cancer cell lines release glutamate into the extracellular environment. Clinical & experimental metastasis. 2009;26:781–787. doi: 10.1007/s10585-009-9277-4. [DOI] [PubMed] [Google Scholar]
- Semenza GL. Molecular mechanisms mediating metastasis of hypoxic breast cancer cells. Trends in molecular medicine. 2012;18:534–543. doi: 10.1016/j.molmed.2012.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma MK, Seidlitz EP, Singh G. Cancer cells release glutamate via the cystine/glutamate antiporter. Biochem Biophys Res Commun. 2010;391:91–95. doi: 10.1016/j.bbrc.2009.10.168. [DOI] [PubMed] [Google Scholar]
- Sims B, Clarke M, Francillion L, Kindred E, Hopkins ES, Sontheimer H. Hypoxic preconditioning involves system Xc- regulation in mouse neural stem cells. Stem cell research. 2012;8:285–291. doi: 10.1016/j.scr.2011.09.002. [DOI] [PubMed] [Google Scholar]
- Speyer CL, Smith JS, Banda M, DeVries JA, Mekani T, Gorski DH. Metabotropic glutamate receptor-1: a potential therapeutic target for the treatment of breast cancer. Breast Cancer Res Treat. 2012;132:565–573. doi: 10.1007/s10549-011-1624-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporn MB, Liby KT. NRF2 and cancer: the good, the bad and the importance of context. Nat Rev Cancer. 2012;12:564–571. doi: 10.1038/nrc3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepulak A, Rola R, Polberg K, Ikonomidou C. Glutamate and its receptors in cancer. Journal of neural transmission. 2014 doi: 10.1007/s00702-014-1182-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talks KL, Turley H, Gatter KC, Maxwell PH, Pugh CW, Ratcliffe PJ, Harris AL. The expression and distribution of the hypoxia-inducible factors HIF-1alpha and HIF-2alpha in normal human tissues, cancers, and tumor-associated macrophages. Am J Pathol. 2000;157:411–421. doi: 10.1016/s0002-9440(10)64554-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmerman LA, Holton T, Yuneva M, Louie RJ, Padro M, Daemen A, Hu M, Chan DA, Ethier SP, van 't Veer LJ, et al. Glutamine sensitivity analysis identifies the xCT antiporter as a common triple-negative breast tumor therapeutic target. Cancer Cell. 2013;24:450–465. doi: 10.1016/j.ccr.2013.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerapana E, Speers AE, Cravatt BF. Tandem orthogonal proteolysis-activity-based protein profiling (TOP-ABPP)--a general method for mapping sites of probe modification in proteomes. Nat Protoc. 2007;2:1414–1425. doi: 10.1038/nprot.2007.194. [DOI] [PubMed] [Google Scholar]
- Weerapana E, Wang C, Simon GM, Richter F, Khare S, Dillon MB, Bachovchin DA, Mowen K, Baker D, Cravatt BF. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature. 2010;468:790–795. doi: 10.1038/nature09472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willard SS, Koochekpour S. Glutamate signaling in benign and malignant disorders: current status, future perspectives, and therapeutic implications. International journal of biological sciences. 2013;9:728–742. doi: 10.7150/ijbs.6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CC, Zhang H, Gilkes DM, Chen J, Wei H, Chaturvedi P, Hubbi ME, Semenza GL. Inhibitors of hypoxia-inducible factor 1 block breast cancer metastatic niche formation and lung metastasis. Journal of molecular medicine. 2012;90:803–815. doi: 10.1007/s00109-011-0855-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WS, SriRamaratnam R, Welsch ME, Shimada K, Skouta R, Viswanathan VS, Cheah JH, Clemons PA, Shamji AF, Clish CB, et al. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156:317–331. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Yee D. IGF-I Regulates Redox Status in Breast Cancer Cells by Activating the Amino Acid Transport Molecule xC. Cancer Res. 2014;74:2295–2305. doi: 10.1158/0008-5472.CAN-13-1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Wong CC, Wei H, Gilkes DM, Korangath P, Chaturvedi P, Schito L, Chen J, Krishnamachary B, Winnard PT., Jr HIF-1-dependent expression of angiopoietin-like 4 and L1CAM mediates vascular metastasis of hypoxic breast cancer cells to the lungs. Oncogene. 2012;31:1757–1770. doi: 10.1038/onc.2011.365. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhong H, De Marzo A, Laughner E, Lim M, Hilton D, Zagzag D, Buechler P, Isaacs W, Semenza G, Simons J. Overexpression of hypoxia-inducible factor 1alpha in common human cancers and their metastases. Cancer Res. 1999;59:5830–5835. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.