SUMMARY
Tamoxifen may require metabolic activation to endoxifen for efficacy in treating hormone receptor breast cancer. Dose escalation in patients with low endoxifen concentrations could enhance treatment efficacy. This approach is clinically feasible, and successfully increases endoxifen concentrations; however, it is unknown whether patients benefit from individualized tamoxifen dose escalation.
In this issue of Clinical Cancer Research, Fox and colleagues report a prospective tamoxifen dose-escalation study in patients with low endoxifen concentrations during treatment (1). Tamoxifen is highly effective for preventing recurrence of hormone receptor (HR+) breast cancer; however, approximately 15% of patients will recur within 5 years despite adjuvant treatment (2). Tamoxifen is believed to be a pro-drug with part of the anti-estrogen activity attributed to endoxifen, which is produced primarily via CYP2D6-catalyzed metabolism. There is substantial variability in endoxifen concentration in patients on tamoxifen treatment, and there is some evidence that patients with low endoxifen concentrations during tamoxifen treatment are at increased risk of cancer recurrence (3, 4). Based on this putative association between endoxifen concentration and tamoxifen treatment efficacy, several studies have prospectively dose escalated patients who have low activity CYP2D6 genotype and predicted, or in some cases confirmed, low endoxifen concentration (5–9). Consistently these studies of tamoxifen dose escalation show increased endoxifen concentrations in patients with low-activity genotypes, confirming the feasibility of genotype-guided tamoxifen treatment. None of these studies have detected meaningful increases in toxicity or diminished quality of life, further supporting the clinical feasibility of individualized dose escalation to improve treatment outcomes.
The major distinction between this study and those previously reported is the exclusive use of actual endoxifen concentration on 20 mg/day tamoxifen to select patients for dose escalation, ignoring the genetic surrogate that is predictive of low endoxifen. Using this adapted approach, Fox et al. confirmed that individualized tamoxifen dose escalation increases the proportion of patients who surpass a given threshold of endoxifen concentration without any notable increase in toxicity, specifically hot flashes.
Confirmation that individualized tamoxifen dose escalation is feasible and safe is a necessary first step toward clinical translation, but is insufficient on its own to warrant actual implementation. The association between endoxifen concentration and tamoxifen treatment effectiveness has only been identified in two retrospective analyses (3, 4), both of which are hypothesis generating, not confirmatory. The initial study by Madlensky et al., while interesting, reported an arbitrary cut point that may predict worse recurrence. The “replication” by Saladores et al. is similarly limited by its retrospective design and multiplicative a posteriori searches for factors that predict recurrence risk (CYP2D6 genotype, predicted CYP2D6 activity phenotype, endoxifen levels, metabolite ratios, etc.). To our knowledge only Love et al. used an a priori defined analysis plan to investigate the putative association between recurrence and endoxifen concentration using a case-control approach (10). This study actually found the opposite effect of that hypothesized; patients whose tumors had recurred had greater mean endoxifen concentrations than those whose tumors had not recurred. Given the discrepant findings in a small number of retrospective analyses there is insufficient evidence that endoxifen concentration is a meaningful surrogate of treatment efficacy, i.e. the association lacks clinical validity.
It is imperative that valid predictors of tamoxifen (in)effectiveness are discovered for clinical translation. As previously mentioned, in the adjuvant setting, patients with HR+ breast cancer the 5-year recurrence rate and cancer-related mortality rates are estimated to be 15% and 8.6%, respectively(2). Due to the high annual incidence of breast cancer, the majority of which is HR+, these relatively low rates equate to tens of thousands of recurrences and deaths. Subtherapeutic endoxifen concentration may be a useful predictor of treatment ineffectiveness, but the association requires validation in independent patient cohorts, either in retrospective analyses of prospective clinical studies (retrospective-prospective analyses), or directly in prospective clinical trials such as the ongoing ECOG-ACRIN E3018 study (NCT01124695; ClinicalTrials.gov). Confirmation of clinical validity is the necessary first step toward translation of this putative association into clinical practice.
After confirmation of clinical validity, a critical next step would be to develop an approach to individualized treatment. Previous studies have used a variety of primarily genotype-informed dose escalation approaches. The novel element of this study by Fox et al. was the exclusive use of actual endoxifen concentration to select patients for dose escalation. The principal advantage of this approach, illustrated by the secondary correlative analyses described within their report, is that CYP2D6 genetics is not the sole factor responsible for low endoxifen concentrations. CYP2D6 genetics and concomitant CYP2D6 inhibitor administration explains a relatively small amount of the variability in endoxifen concentration. The investigators report that baseline endoxifen concentration, and not CYP2D6 genetics, is the most useful predictor of endoxifen concentration after dose escalation. Their conclusion is that"the best method for determining endoxifen exposure is to measure endoxifen trough level rather than implying a particular level using genetic or clinical measures.” This is undoubtedly true; however, measuring endoxifen trough levels has some notable disadvantages. Trough endoxifen measurement requires the patient to return for sample collection and analysis after at least a month of treatment, based on the known half-life of approximately 1 week. Furthermore, endoxifen concentration analysis has some added cost and is not widely available in CLIA approved laboratories, which would be a requirement if these results were to be used to inform therapeutic decisions.
The alternative to measuring trough endoxifen concentration is to use available information to predict the concentration and select an appropriate starting dose. Previous studies have been limited to only CYP2D6 genetic information; however, there are several other genetic and clinical factors that are known to predict endoxifen concentration. Teft et al. recently published an endoxifen prediction algorithm that integrates CYP2D6 genotype with other genetic (CYP3A4*22) and clinical (age, weight, concomitant inhibitors, season) information (11). Unlike endoxifen concentration, this information can be obtained prior to treatment initiation and most of it is freely available. The notable exception is patient genetic information, which someday is likely to be available for the majority of cancer patients due to the proliferation of tumor genetic analyses (12) or prior genotyping for personalized medicine. Refinement of this algorithm to include additional clinical and genetic predictors to more accurately predict endoxifen concentration, and even more usefully, to predict the necessary tamoxifen dose for a patient to achieve a target endoxifen concentration, would be a critical next step toward clinical translation of tamoxifen dose individualization.
Establishing the clinical validity of endoxifen as a surrogate of treatment efficacy is the first step toward clinical implementation of individualized tamoxifen treatment. A reasonable next step would be to develop predictive tools that can accurately estimate the patient’s ideal starting dose. Endoxifen concentration can then be measured after a month of personalized dosing to inform further dose optimization in an integrated personalized treatment strategy (Figure 1). Depending on the strength of the evidence that endoxifen concentration is a useful surrogate of treatment efficacy, this integrated approach may need to be tested prospectively to demonstrate that it improves efficacy, and perhaps that it does so in a cost-effective manner. This prospective demonstration of clinical utility will incite uniform uptake of personalized tamoxifen treatment to maximize efficacy of HR+ breast cancer therapy.
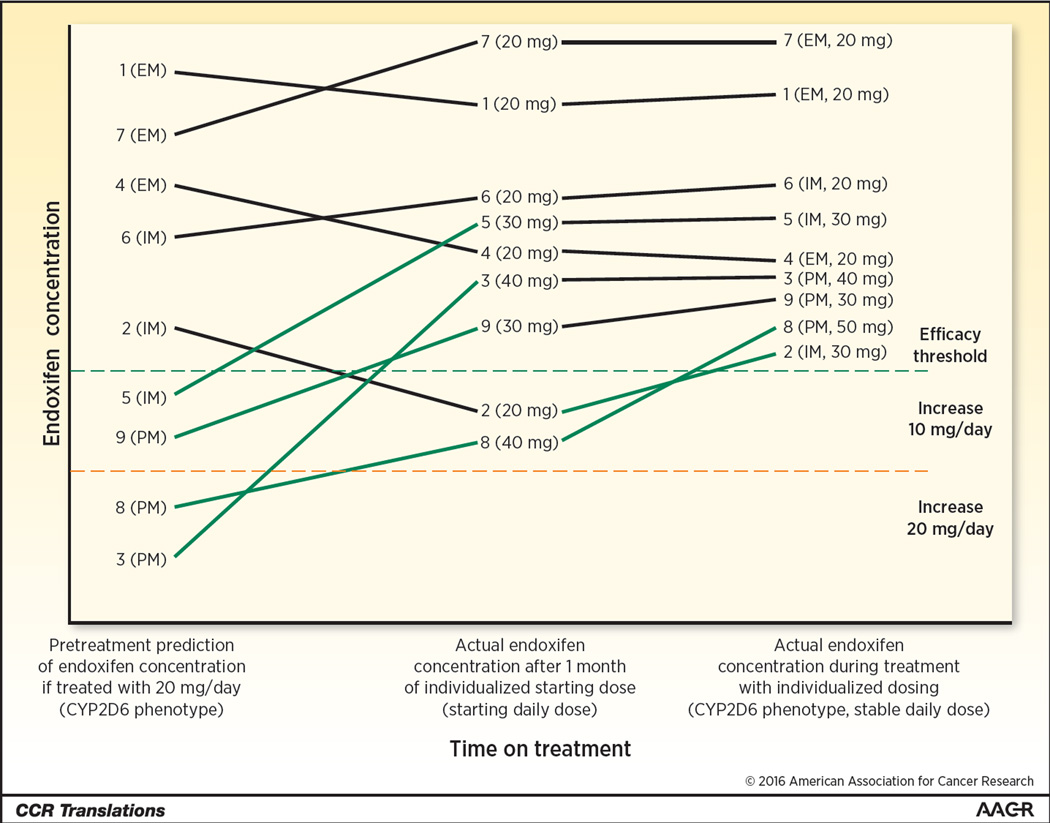
Figure 1. Illustration of 9 patients matriculating through an integrated personalized treatment strategy.
Baseline information including CYP2D6 phenotype (poor (PM), intermediate (IM), or extensive (EM) metabolizer) and clinical information (body mass index, drug-drug interactions, etc.) are used to predict endoxifen concentration during treatment with the typical dose of 20 mg/day. Patients predicted to be above the efficacy threshold (hatched green horizontal line) are started at the typical dose. Patients predicted to be below the threshold are pre-emptively dose escalated by 10 or 20 mg/day. After 1 month of treatment, endoxifen concentration is measured to determine whether further dose personalization is necessary. Some patients who were not expected to require escalation (i.e. #2) have sub-therapeutic concentration and require a dose increase. Other patients who had a dose escalation (i.e. #8) require further escalation to reach therapeutic endoxifen levels. Ultimately, all patients achieve therapeutic endoxifen concentrations, maximizing tamoxifen treatment effectiveness.
Acknowledgments
Grant Support: This work was supported by the Breast Cancer Research Foundation (BCRF) (N003173; to J.M. Rae) and the National Institute of General Medical Sciences (GM099143; to J.M. Rae).
Footnotes
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest were disclosed.
Contributor Information
Daniel L. Hertz, Email: DLHertz@med.umich.edu.
James M. Rae, Email: jimmyrae@med.umich.edu.
References
- 1.Fox P, Balleine RL, Lee C, Gao B, Balakrishnar B, Menzies AM, et al. Dose escalation of tamoxifen in patients with low endoxifen level: evidence for therapeutic drug monitoring—the TADE Study. Clin Cancer Res. 2016 Feb 4; doi: 10.1158/1078-0432.CCR-15-1470. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 2.Davies C, Godwin J, Gray R, Clarke M, Cutter D, et al. Early Breast Cancer Trialists' Collaborative Group (EBCTCG) Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: patient-level meta-analysis of randomised trials. Lancet. 2011;378:771–784. doi: 10.1016/S0140-6736(11)60993-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saladores P, Murdter T, Eccles D, Chowbay B, Zgheib NK, Winter S, et al. Tamoxifen metabolism predicts drug concentrations and outcome in premenopausal patients with early breast cancer. Pharmacogenomics J. 2015;15:84–94. doi: 10.1038/tpj.2014.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Madlensky L, Natarajan L, Tchu S, Pu M, Mortimer J, Flatt SW, et al. Tamoxifen metabolite concentrations, CYP2D6 genotype, and breast cancer outcomes. Clin Pharmacol Ther. 2011;89:718–725. doi: 10.1038/clpt.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dezentje VO, Opdam FL, Gelderblom H, Hartigh den J, Van der Straaten T, Vree R, et al. CYP2D6 genotype- and endoxifen-guided tamoxifen dose escalation increases endoxifen serum concentrations without increasing side effects. Breast Cancer Res Treat. 2015;153:583–590. doi: 10.1007/s10549-015-3562-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Irvin WJ, Walko CM, Weck KE, Ibrahim JG, Chiu WK, Dees EC, et al. Genotype-guided tamoxifen dosing increases active metabolite exposure in women with reduced CYP2D6 metabolism: a multicenter study. J Clin Oncol. 2011;29:3232–3239. doi: 10.1200/JCO.2010.31.4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barginear MF, Jaremko M, Peter I, Yu C, Kasai Y, Kemeny M, et al. Increasing tamoxifen dose in breast cancer patients based on CYP2D6 genotypes and endoxifen levels: effect on active metabolite isomers and the antiestrogenic activity score. Clin Pharmacol Ther. 2011;90:605–611. doi: 10.1038/clpt.2011.153. [DOI] [PubMed] [Google Scholar]
- 8.Kiyotani K, Mushiroda T, Imamura CK, Tanigawara Y, Hosono N, Kubo M, et al. Dose-adjustment study of tamoxifen based on CYP2D6 genotypes in Japanese breast cancer patients. Breast Cancer Res Treat. 2012;131:137–145. doi: 10.1007/s10549-011-1777-7. [DOI] [PubMed] [Google Scholar]
- 9.Martinez de Duenas E, Ochoa Aranda E, Blancas Lopez-Barajas I, Ferrer Magdalena T, Bandres Moya F, Chicharro Garcia LM, et al. Adjusting the dose of tamoxifen in patients with early breast cancer and CYP2D6 poor metabolizer phenotype. Breast. 2014 Aug;23:400–406. doi: 10.1016/j.breast.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 10.Love RR, Desta Z, Flockhart D, Skaar T, Ogburn ET, Ramamoorthy A, et al. CYP2D6 genotypes, endoxifen levels, and disease recurrence in 224 Filipino and Vietnamese women receiving adjuvant tamoxifen for operable breast cancer. Springerplus. 2013;2:52. doi: 10.1186/2193-1801-2-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teft WA, Gong IY, Dingle B, Potvin K, Younus J, Vandenberg TA, et al. CYP3A4 and seasonal variation in vitamin D status in addition to CYP2D6 contribute to therapeutic endoxifen level during tamoxifen therapy. Breast Cancer Res Treat. 2013 May;139:95–105. doi: 10.1007/s10549-013-2511-4. [DOI] [PubMed] [Google Scholar]
- 12.Hertz DL, McLeod HL. Integrated patient and tumor genetic testing for individualized cancer therapy. Clin Pharmacol Ther. 2016;99:143–146. doi: 10.1002/cpt.294. [DOI] [PubMed] [Google Scholar]