Abstract
Background
The IL-6/STAT3 axis promotes inflammation, angiogenesis, and cancer. The effect of genetic variants within this pathway on benefit from anti-angiogenic cancer therapy is unknown. We tested whether single nucleotide polymorphisms (SNPs) in genes involved in IL-6/STAT3 signaling can predict efficacy of bevacizumab-based chemotherapy in metastatic colorectal cancer (mCRC) patients
Methods
Associations between potentially functional IL-6 (rs2069837, rs1800795) and STAT3 (rs744166, rs4796793) SNPs and clinical outcomes (progression-free survival, overall survival and tumor response rate) were evaluated in mCRC patients receiving first-line FOLFIRI plus bevacizumab in two randomized phase III trials: TRIBE (n=223, training cohort) and FIRE-3 (n=288, validation cohort). Patients receiving FOLFIRI plus cetuximab in FIRE-3 (n=264) served as a control cohort. The interaction between genotype and primary tumor location with clinical outcomes was examined. Genomic DNA isolated from whole blood or tumor tissue was analyzed by PCR-based direct sequencing.
Results
Patients with an IL-6 rs2069837 G allele treated with FOLFIRI plus bevacizumab had an inferior PFS than those with the A/A genotype in TRIBE (9.4 v. 11.1 months, HR=1.53, 95% CI 1.12, 2.10, P=0.004) and FIRE-3 (8.8 v. 10.9 months, HR=1.40, 95% CI 1.06, 1.85, P=0.015). These associations were confirmed in multivariable analyses, and were not seen in the control cohort. In subgroup analysis, the effect of IL-6 rs2069837 on PFS was present only in patients with left-sided cancers, but the test for interaction was not significant.
Conclusion
IL-6 rs2069837 genotype is a clinically relevant prognostic factor in mCRC patients treated with first-line bevacizumab-based chemotherapy.
Keywords: bevacizumab, cetuximab, colorectal cancer, IL-6, single-nucleotide polymorphism
INTRODUCTION
Interleukin-6 (IL-6) is a pleiotropic, angiogenic cytokine with far-reaching effects across vascular disease, immunity, and cancer1,2. As initiators of pathologic inflammation, IL-6 and its downstream effector, STAT3 (signal transducer and activator of transcription-3), promote colorectal cancer (CRC) development3,4, invasion and metastasis5–7, by imposing genetic alterations in tumor cells3,8 and facilitating immune tolerance within the microenvironment6,9,10. In colitis-associated cancer in vivo models, IL-6 enhances tumor cell proliferation and protects normal and malignant intestinal epithelial cells from apoptosis in a STAT3 dependent fashion.
Another critical consequence of IL-6/STAT3 signaling is angiogenesis. Under hypoxic states, IL-6 promotes HIF-1α and STAT3 transcription which stimulates vascular endothelial growth factor (VEGF) expression, blood vessel formation and tumor growth11–13. In addition to tumor cells, stromal fibroblasts are an important source of IL-6 and provide a fertile environment for angiogenesis to occur14. Evidence from tumor xenografts suggests that IL-6 may trigger defective angiogenesis with reduced pericyte coverage, independent of VEGF signaling15, and may therefore serve as a resistance mechanism to anti-angiogenic therapy.
Previous data suggest that IL-6/STAT3 single nucleotide polymorphisms (SNPs) may have prognostic utility in CRC. Though the evidence has been mixed16–21, IL-6 SNPs have been shown to correlate with serum and intratumoral IL-6 levels22–24, as well as the susceptibility16–19,21,23,25 and survival20 of multiple cancers, including CRC. Similarly, STAT3 common genetic variants have been associated with cancer risk26. However, the potential of IL-6/STAT3 SNPs to predict efficacy of anti-angiogenic therapy in CRC has not been tested.
The primary objective of this study was to investigate the predictive and prognostic impact of functionally significant IL-6 and STAT3 polymorphisms in mCRC patients receiving cytotoxic and anti-angiogenic therapy. We determined associations between IL-6 (rs2069837, rs1800795) and STAT3 (rs744166, rs4796793) SNPs and outcomes in a phase III mCRC trial of first-line bevacizumab-based chemotherapy. We then validated our findings in an independent patient cohort from another large phase III study, and examined associations in a control cohort of patients receiving cetuximab-based treatment. Additionally, we assessed the influence of primary tumor site on the observed associations to identify potentially clinically relevant profiles.
METHODS
Study Design and Patient Population
A total of 775 patients were included in the current study. Patients treated with first-line FOLFIRI plus bevacizumab in the randomized, open-label, phase III TRIBE trial24 served as a training cohort, while patients treated with first-line FOLFIRI plus bevacizumab in the randomized, open-label, phase III FIRE-3 trial27 served as the validation cohort, and patients treated with first-line FOLFIRI plus cetuximab in FIRE-3 served as a control cohort. Patients without sufficient blood for analysis were excluded. Analysis of the effects of IL-6 (rs2069837, rs1800795) and STAT3 (rs744166, rs4796793) SNPs on clinical outcomes was conducted in 223 mCRC patients treated with first-line FOLFIRI plus bevacizumab in TRIBE24 with sufficient blood for analysis (87% of 256 enrolled patients). Relevant SNPs significantly associated with outcomes were then examined in the validation and control cohorts. The validation cohort consisted of 288 patients with sufficient tissue (98% enrolled of 295 patients) treated with first-line FOLFIRI plus bevacizumab in FIRE-327. The control set consisted of 264 patients with sufficient tissue (89% of 297 enrolled patients) treated with first-line FOLFIRI plus cetuximab in FIRE-3.
Patients in the training, validation and control cohorts had histologically confirmed stage IV colorectal adenocarcinoma, with measurable disease per Response Evaluation Criteria in Solid Tumors (RECIST) 1.0 and no prior treatment for metastatic disease, nor exposure to irinotecan, bevacizumab, or cetuximab. Standard inclusion and exclusion criteria were applied. Prior adjuvant oxaliplatin treatment was allowed in both TRIBE and FIRE-3 if more than 12 months had elapsed between the end of treatment and date of study enrollment. Patients received bevacizumab 5 mg/kg or cetuximab 400 mg/kg, followed by irinotecan 180 mg/m2 administered with folinic acid (leucovorin) 200 mg/m2, 5-fluorouracil 400 mg/m2 bolus infusion and 5-fluorouracil 2400 mg/m2 as a 48-hour continuous infusion. The regimen was repeated at two-week intervals. In the TRIBE study, patients received 12 cycles of FOLFIRI plus bevacizumab, followed by fluorouracil and bevacizumab maintenance therapy. Treatment was administered in all patients until the time of disease progression, intolerable toxicities, or patient withdrawal. Responses were measured by intravenous contrast-enhanced computed tomography (CT) scans every 8 weeks according to RECIST v1.0.
Study protocols were approved by the Institutional Review Boards of each participating center and conducted in accordance with the Declaration of Helsinki and Good Clinical Practice Guidelines. All patients signed informed consent for the analysis of molecular correlates.
Candidate Polymorphisms
Genes were chosen according to previous published literature and databases. Polymorphisms were chosen for investigation if the minor allele frequency exceeded 10% in Caucasians according to the ENSEMBL database (http://www.ensembl.org/index.html) and if the functional or predicted functional relevance of gene transcription or protein expression was previously described. Functional significance was predicted based on information provided by the National Institute of Environmental Health Science SNP Function Prediction, Queen’s University F-SNP, and the location of the SNP in the protein-coding region of the gene (http://snpinfo.niehs.nih.gov/snpinfo/snptag.htm).
Genotyping
Genomic DNA was extracted from the peripheral whole blood of patients in the training cohort; and from formalin-fixed paraffin-embedded (FFPE) tissues of patients from the validation and control cohorts using the QIAmp Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s protocol (www.qiagen.com). PCR-based direct DNA sequence analysis using ABI 3100A Capillary Genetic Analyzer and Sequencing Scanner v1.0 (Applied Biosystems, Waltham, MA, USA) was performed for genotyping the SNPs. The extracted DNA was amplified using the primer sets shown in Supplementary Table 1, and analyzed by PCR-based direct DNA sequencing.
For quality control purposes, a random selection of 10% of the samples were re-examined for each polymorphism, and the genotype concordance rate was 100%. The investigator analyzed the sequencing data using the ABI Sequencing Scanner v1.0 (Applied Biosystems, Life Technologies, Grand Island, NY, USA) and was blinded to the clinical data set.
Statistical Analysis
The primary outcome measure was progression-free survival (PFS), defined as time from randomization to first documented disease progression or death from any cause. If progression or death was not observed, PFS was censored on the day of the last CT scan. Secondary endpoints were response rate (RR) and overall survival (OS). Patients were dichotomized into responders (including complete or partial response) and non-responders (including stable or progressive disease) as defined by RECIST. OS was defined as the period from randomization to the date of death or censored on the date of last contact if alive. Allelic distribution of polymorphisms by ethnicity was tested for deviation from Hardy–Weinberg equilibrium (HWE) using the exact test.
The differences between baseline characteristics for the three cohorts were compared by using the Chi-square test or the Kruskal-Wallis test whenever appropriate. Associations between SNPs and PFS, OS and RR were examined using Kaplan–Meier curves, log-rank test, and Fisher’s exact test, respectively. A Cox proportional hazards regression model with stratification factors was fitted to re-evaluate the association between SNPs and PFS and OS considering imbalances in the distributions of baseline characteristics among the cohorts. Codominant, dominant and additive models for each SNP were analyzed. For the additive model, a trend test was calculated assigning a linear score to each genotype (0 for homozygous common allele, 1 for heterozygous common allele, and 2 for homozygous rare allele. The baseline demographic and clinical characteristics that remained significantly associated with endpoints in the multivariable analysis (P < 0.10) were included in the final model. These findings were then evaluated in the independent validation and control cohorts. Subgroup analyses by tumor location were also investigated. Interaction terms of SNPs and tumor location were included, and likelihood ratio tests were performed in the multivariable Cox proportional hazards regression models. Case-wide deletion for missing polymorphisms was applied in univariate and multivariable analysis.
The training cohort consisted of 223 patients (171 PFS events); therefore, there was 80% power to detect an association between a SNP and PFS with a minimum hazard ratio (HR) from 1.54 to 2.07 using a two-sided 0.05-level log-rank test. We assumed that the minor allele frequency varied from 0.05 to 0.4, and the dominant model was used. In the validation cohort, there was greater than 88% power using the same test to detect the same HRs with the same allele frequencies under the dominant model (n = 288, 245 PFS events). In the control cohort (n = 264, 229 PFS events), there was 95% power to demonstrate no significant association between a SNP and PFS, with a HR equal to 1.00 under the alternative hypothesis against the null hypothesis (HR 1.54) for a SNP with a minor allele frequency of 0.3, using a dominant model and a one-sided 0.05-level test.
SAS 9.4 (SAS Institute, Cary, NC, USA) was used to perform all analyses. All tests were two-sided at a significance level of 0.05.
RESULTS
Patient and Tumor Characteristics
Clinicopathologic characteristics for the training, validation, and control cohorts are presented in Table 1. The median follow-up was 4.1 years in the training cohort, 3.4 years in the validation cohort, and 3.4 years in the control cohort. The median PFS and OS in each cohort were as follows: 8.2 months and 26.1 months (TRIBE training cohort) 10.1 months and 24.2 months (FIRE-3 validation cohort), and 9.6 months and 28.0 months (FIRE-3 control cohort).
Table 1.
Baseline patient and tumor characteristics
| Training Cohort (TRIBE) (n=223) |
Validation Cohort (FIRE-3) (n=288) |
Control Cohort (FIRE-3) (n=264) |
P value† | ||||
|---|---|---|---|---|---|---|---|
| n | % | n | % | n | % | ||
| Gender | 0.53 | ||||||
| Male | 136 | 61 | 189 | 66 | 171 | 65 | |
| Female | 87 | 39 | 99 | 34 | 93 | 35 | |
| Age (years) | |||||||
| Median (range) | 60 (29 – 75) | 65 (31 – 76) | 65 (38 – 79) | < 0.001 | |||
| ≤ 65 | 160 | 72 | 151 | 52 | 140 | 53 | < 0.001 |
| > 65 | 63 | 28 | 137 | 48 | 124 | 47 | |
| Performance status | < 0.001 | ||||||
| ECOG 0 | 183 | 82 | 159 | 55 | 135 | 51 | |
| ECOG ≥1 | 39 | 17 | 129 | 45 | 129 | 49 | |
| Unknown* | 1 | 1 | |||||
| Primary tumor site | 0.038 | ||||||
| Right-sided colon | 57 | 26 | 63 | 22 | 38 | 14 | |
| Left-sided colon | 151 | 68 | 173 | 60 | 174 | 66 | |
| Unknown* | 15 | 7 | 52 | 18 | 52 | 20 | |
| Liver only metastases | 0.94 | ||||||
| Yes | 71 | 32 | 95 | 33 | 88 | 33 | |
| No | 152 | 68 | 193 | 67 | 176 | 67 | |
| Number of metastases | 0.97 | ||||||
| < 2 | 95 | 43 | 105 | 36 | 92 | 35 | |
| ≥2 | 128 | 57 | 137 | 48 | 125 | 47 | |
| Unknown* | 46 | 16 | 47 | 18 | |||
| Time to metastasis | 0.15 | ||||||
| Synchronous | 184 | 83 | 183 | 64 | 163 | 62 | |
| Metachronous | 39 | 17 | 58 | 20 | 52 | 20 | |
| Unknown* | 46 | 16 | 47 | 18 | |||
| Primary tumor resected | < 0.001 | ||||||
| Yes | 142 | 64 | 249 | 86 | 219 | 83 | |
| No | 81 | 36 | 39 | 14 | 44 | 17 | |
| Adjuvant chemotherapy | 0.027 | ||||||
| Yes | 27 | 12 | 53 | 18 | 56 | 21 | |
| No | 196 | 88 | 235 | 82 | 207 | 79 | |
| High LDH | 0.66 | ||||||
| Yes | 104 | 47 | 71 | 25 | 64 | 24 | |
| No | 87 | 39 | 70 | 24 | 64 | 24 | |
| Unknown* | 32 | 14 | 147 | 51 | 136 | 52 | |
| KRAS status | < 0.001 | ||||||
| Wildtype | 95 | 43 | 242 | 84 | 217 | 82 | |
| Mutant | 90 | 40 | 46 | 16 | 47 | 18 | |
| Unknown* | 38 | 17 | |||||
Based on Chi-square test, or Kruskal-Wallis test whenever appropriate.
Not included in the Chi-square test.
Associations between baseline characteristics and clinical outcomes were examined using the log-rank test in univariate analysis. In the training cohort, ECOG performance status, primary tumor resection and BRAF status were significantly associated with PFS and OS. In addition, age, primary tumor site, number of metastases, time to metastasis, and adjuvant chemotherapy were also significantly correlated with OS (Supplementary Table 1). In the validation cohort, Eastern Cooperative Oncology Group (ECOG) performance status was significantly associated with PFS and OS. Additionally, primary tumor site, liver only disease, number of metastases and primary tumor resection were also significantly associated with OS (Supplementary Table 2). In the control cohort, primary tumor site was significantly associated with PFS and OS. Additionally, sex was significantly correlated with PFS; lung metastasis, liver metastasis, number of metastases, primary tumor resection, time to metastasis, high LDH and KRAS status were also significantly associated with OS (Supplementary Table 3).
Clinical Outcomes by IL-6 and STAT3 Genetic Variants in Patients Receiving FOLFIRI with Bevacizumab: TRIBE and FIRE-3
All genotype frequencies for candidate polymorphisms analyzed were within HWE (P > 0.05) for each ethnic/race group in each cohort.
Genotyping for the IL-6 rs2069837 candidate SNPs was successful in 223 patients and 270 (94%) patients in the training and validation cohorts, respectively. In 18 patients of the validation cohort, genotyping was not successful due to limited quantity or quality of extracted genomic DNA.
Associations between IL-6 rs2069837 and outcomes were analyzed (Table 2). Among evaluable patients in the training (n = 223) and validation (n = 270) cohorts, 58.3% and 32.2% carried a G allele in IL-6 rs2069837, respectively.
Table 2.
Association between IL-6 rs2069837 with clinical outcomes
| Tumor Response
|
Progression-Free Survival
|
Overall Survival
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | CR+PR | SD+PD | P value * | Median, months (95%CI) |
HR (95%CI) † | P value* | HR (95%CI) ‡ | P value* | Median, months (95%CI) |
HR (95%CI) † | P value* | HR (95%CI) ‡ | P value* | |
| Training Cohort | 0.11 | 0.001 | 0.017 | 0.34 | 0.70 | |||||||||
| A/A | 93 | 57 (63%) | 34 (37%) | 11.1 (9.4, 12.6) | 1 (Reference) | 1 (Reference) | 26.8 (23.9, 35.8) | 1 (Reference) | 1 (Reference) | |||||
| A/G | 116 | 58 (52%) | 53 (48%) | 9.2 (8.0, 9.7) | 1.68 (1.22, 2.33) | 1.67 (1.14, 2.46) | 22.5 (19.4, 27.9) | 1.24 (0.91, 1.69) | 1.17 (0.81, 1.70) | |||||
| G/G | 14 | 11 (79%) | 3 (21%) | 12.7 (8.2, 14.2) | 0.81 (0.41, 1.62) | 0.73 (0.29, 1.84) | 31.9 (16.3, 44.1) | 0.97 (0.52, 1.83) | 1.05 (0.45, 2.44) | |||||
| 0.33 | 0.004 | 0.033 | 0.22 | 0.43 | ||||||||||
| A/A | 93 | 57 (63%) | 34 (37%) | 11.1 (9.4, 12.6) | 1 (Reference) | 1 (Reference) | 26.8 (23.9, 35.8) | 1 (Reference) | 1 (Reference) | |||||
| Any G | 130 | 69 (55%) | 56 (45%) | 9.4 (8.6, 10.3) | 1.53 (1.12, 2.10) | 1.50 (1.03, 2.17) | 22.9 (19.8, 27.9) | 1.21 (0.89, 1.63) | 1.16 (0.81, 1.66) | |||||
| Additive Model: HR per G | 1.21 (0.96, 1.52) | 0.10 | 1.20 (0.90, 1.59) | 0.21 | 1.10 (0.87, 1.40) | 0.42 | 1.10 (0.82, 1.48) | 0.51 | ||||||
|
| ||||||||||||||
| Validation Cohort | 0.040 | 0.024 | 0.072 | 0.15 | 0.59 | |||||||||
| A/A | 183 | 113 (67%) | 56 (33%) | 10.9 (9.9, 12.4) | 1 (Reference) | 1 (Reference) | 26.1 (23.1, 28.4) | 1 (Reference) | 1 (Reference) | |||||
| A/G | 58 | 25 (47%) | 28 (53%) | 7.8 (6.5, 9.3) | 1.54 (1.12, 2.11) | 1.47 (1.06, 2.03) | 22.7 (14.7, 25.9) | 1.21 (0.85, 1.73) | 1.11 (0.78, 1.59) | |||||
| G/G | 29 | 16 (62%) | 10 (38%) | 10.2 (8.2, 12.7) | 1.17 (0.75, 1.82) | 1.12 (0.71, 1.77) | 19.5 (15.4, 29.1) | 1.51 (0.96, 2.38) | 1.26 (0.78, 2.03) | |||||
| 0.026 | 0.015 | 0.047 | 0.084 | 0.36 | ||||||||||
| A/A | 183 | 113 (67%) | 56 (33%) | 10.9 (9.9, 12.4) | 1 (Reference) | 1 (Reference) | 26.1 (23.1, 28.4) | 1 (Reference) | 1 (Reference) | |||||
| Any G | 87 | 41 (52%) | 38 (48%) | 8.8 (7.2, 10.2) | 1.40 (1.06, 1.85) | 1.34 (1.00, 1.79) | 22.1 (16.7, 25.9) | 1.30 (0.96, 1.76) | 1.16 (0.85, 1.58) | |||||
| Additive Model: HR per G | 0.18 (0.98, 1.42) | 0.076 | 1.15 (0.94, 1.39) | 0.17 | 1.23 (1.00, 1.51) | 0.053 | 1.12 (0.90, 1.39) | 0.31 | ||||||
|
| ||||||||||||||
| Control Cohort | 0.042 | 0.22 | 0.24 | 0.66 | 0.43 | |||||||||
| A/A | 179 | 108 (70%) | 46 (30%) | 9.5 (8.3, 10.4) | 1 (Reference) | 1 (Reference) | 28.7 (23.5, 36.4) | 1 (Reference) | 1 (Reference) | |||||
| A/G | 71 | 43 (70%) | 18 (30%) | 9.7 (7.9, 12.2) | 1.04 (0.78, 1.39) | 0.98 (0.73, 1.31) | 28.5 (20.3, 38.3) | 1.09 (0.77, 1.55) | 1.10 (0.77, 1.56) | |||||
| G/G | 14 | 4 (33%) | 8 (67%) | 6.0 (1.9, 10.9) | 1.64 (0.93, 2.91) | 1.61 (0.91, 2.85) | 20.2 (10.0, 36.6) | 1.33 (0.67, 2.63) | 1.56 (0.78, 3.13) | |||||
| 0.36 | 0.46 | 0.77 | 0.48 | 0.41 | ||||||||||
| A/A | 179 | 108 (70%) | 46 (30%) | 9.5 (8.3, 10.4) | 1 (Reference) | 1 (Reference) | 28.7 (23.5, 36.4) | 1 (Reference) | 1 (Reference) | |||||
| Any G | 85 | 47 (64%) | 26 (36%) | 9.6 (7.8, 10.9) | 1.11 (0.84, 1.45) | 1.04 (0.79, 1.37) | 25.2 (19.9, 37.0) | 1.13 (0.81, 1.57) | 1.15 (0.82, 1.61) | |||||
| Additive Model: HR per G | 1.15 (0.92, 1.44) | 0.23 | 1.10 (0.88, 1.39) | 0.41 | 1.12 (0.86, 1.47) | 0.39 | 1.17 (0.89, 1.54) | 0.26 | ||||||
CI, confidence interval; HR, hazard ratio
P value was based on Fisher’s exact test for tumor response, log-rank test for PFS and OS in the univariate analysis (†) and Wald test for PFS and OS in the multivariable Cox regression model adjusted for age, performance status, primary tumor site, primary tumor resection, BRAF mutation status and adjuvant chemotherapy in the training cohort; adjusted for sex, performance status, liver only disease, primary tumor resection in the validation cohort and control cohort (‡).
In the TRIBE training cohort, patients carrying an IL-6 rs2069837 G allele had an inferior median PFS of 9.4 months compared with those with the A/A genotype who had a median PFS of 11.1 months (HR, 1.53; 95% CI, 1.12 – 2.10; P = 0.004) (Table 2, Figure 1A). This association remained significant in multivariable analysis (HR, 1.50, adjusted = 0.033) (Table 3). Additive models showed no evidence of significant effect for each G allele. There was no evidence for an association between IL-6 rs2069837 genotype and RR, nor any significant relationships between IL-6 rs1800795 and STAT3 (rs744166, rs4796793) genotype and outcomes in the training cohort (Supplementary Table 4).
Figure 1.
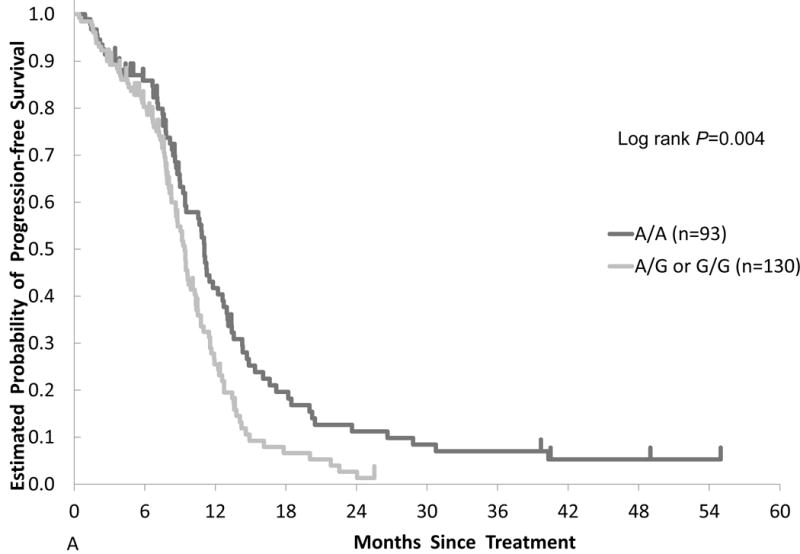
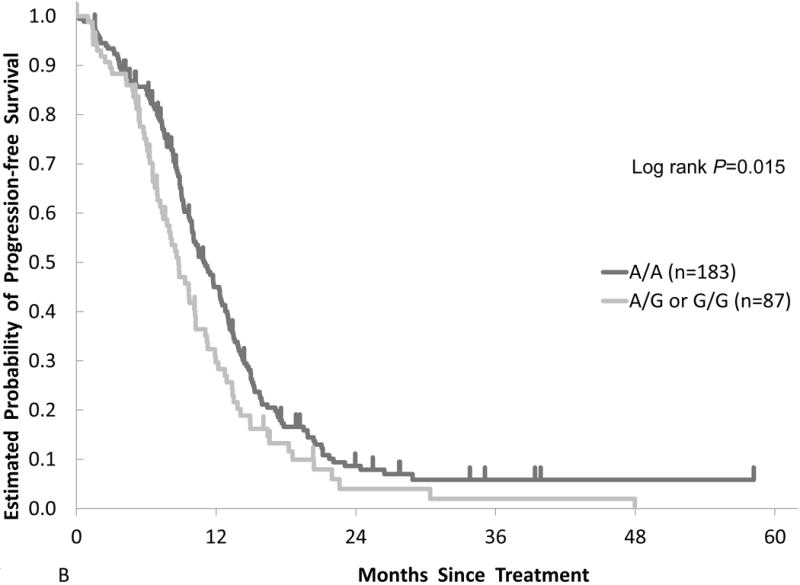
Comparison of clinical outcomes by IL-6 gene polymorphism in two cohorts. Kaplan-Meier cumulative progression-free survival probability curves stratified by IL-6 rs2069837 genotype in the (A) training cohort and (B) validation cohort.
Table 3.
Association between IL-6 rs2069837 and clinical outcomes stratified by primary tumor location
| Tumor Response
|
Progression-Free Survival
|
Overall Survival
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | CR+PR | SD+PD | P value * | Median, months (95%CI) |
HR (95%CI) † | P value* | HR (95%CI) ‡ | P value * | Median, months (95%CI) |
HR (95%CI) † | P value* | HR (95%CI) ‡ | P value* | |
| Left-Sided | ||||||||||||||
| Training Cohort | 0.19 | 0.031 | 0.058 | 0.88 | 0.87 | |||||||||
| A/A | 65 | 39 (61%) | 25 (39%) | 11.1 (9.4, 12.7) | 1 (Reference) | 1 (Reference) | 33.1 (24.8, 39.7) | 1 (Reference) | 1 (Reference) | |||||
| A/G | 76 | 38 (51%) | 36 (49%) | 9.6 (8.3, 10.8) | 1.57 (1.05, 2.36) | 1.68 (1.05, 2.69) | 27.9 (20.5, 34.4) | 1.10 (0.75, 1.62) | 1.13 (0.72, 1.78) | |||||
| G/G | 10 | 8 (80%) | 2 (20%) | 12.7 (4.5, 24.0) | 0.86 (0.39, 1.89) | 0.74 (0.26, 2.09) | 34.5 (5.2, 55.5+) | 1.02 (0.48, 2.15) | 1.02 (0.40, 2.61) | |||||
| 0.50 | 0.050 | 0.088 | 0.65 | 0.62 | ||||||||||
| A/A | 65 | 39 (61%) | 25 (39%) | 11.1 (9.4, 12.7) | 1 (Reference) | 1 (Reference) | 33.1 (24.8, 39.7) | 1 (Reference) | 1 (Reference) | |||||
| Any G | 86 | 46 (55%) | 38 (45%) | 9.7 (8.6, 11.0) | 1.43 (0.97, 2.12) | 1.48 (0.94, 2.32) | 28.3 (21.1, 34.4) | 1.09 (0.75, 1.58) | 1.12 (0.72, 1.73) | |||||
| Additive Model: HR per G | 1.20 (0.75, 1.93) | 0.45 | 1.17 (0.84, 1.63) | 0.36 | 1.05 (0.78, 1.42) | 0.74 | 1.06 (0.75, 1.51) | 0.73 | ||||||
| Validation Cohort | 0.30 | 0.13 | 0.23 | 0.14 | 0.75 | |||||||||
| A/A | 122 | 76 (68%) | 35 (32%) | 11.7 (9.9, 13.0) | 1 (Reference) | 1 (Reference) | 27.5 (24.8, 31.9) | 1 (Reference) | 1 (Reference) | |||||
| A/G | 28 | 13 (52%) | 12 (48%) | 10.1 (6.5, 12.2) | 1.53 (0.97, 2.44) | 1.47 (0.92, 2.34) | 30.3 (16.7, 42.5) | 1.01 (0.59, 1.73) | 0.95 (0.55, 1.63) | |||||
| G/G | 14 | 9 (64%) | 5 (36%) | 10.2 (7.4, 12.9) | 1.35 (0.75, 2.43) | 1.32 (0.72, 2.43) | 18.9 (15.1, 29.6) | 1.81 (0.98, 3.35) | 1.27 (0.64, 2.51) | |||||
| 0.18 | 0.047 | 0.090 | 0.32 | 0.85 | ||||||||||
| A/A | 122 | 76 (68%) | 35 (32%) | 11.7 (9.9, 13.0) | 1 (Reference) | 1 (Reference) | 27.5 (24.8, 31.9) | 1 (Reference) | 1 (Reference) | |||||
| Any G | 42 | 22 (56%) | 17 (44%) | 10.2 (7.4, 12.0) | 1.46 (0.99, 2.16) | 1.41 (0.95, 2.11) | 21.3 (17.3, 34.6) | 1.25 (0.80, 1.93) | 1.04 (0.66, 1.65) | |||||
| Additive Model: HR per G | 1.24 (0.96, 1.60) | 0.093 | 1.22 (0.93, 1.59) | 0.15 | 1.26 (0.94, 1.69) | 0.12 | 1.08 (0.79, 1.48) | 0.64 | ||||||
|
| ||||||||||||||
| Right-Sided | ||||||||||||||
| Training Cohort | 0.80 | 0.14 | 0.43 | 0.073 | 0.19 | |||||||||
| A/A | 22 | 14 (67%) | 7 (33%) | 9.3 (6.7, 13.4) | 1 (Reference) | 1 (Reference) | 24.3 (12.5, 33.9) | 1 (Reference) | 1 (Reference) | |||||
| A/G | 32 | 16 (55%) | 13 (45%) | 8.8 (6.2, 9.9) | 1.53 (0.83, 2.81) | 1.38 (0.63, 3.03) | 18.3 (14.1, 20.3) | 1.68 (0.92, 3.08) | 1.69 (0.78, 3.69) | |||||
| G/G | 3 | 2 (67%) | 1 (33%) | |||||||||||
| Additive Model: HR per G | 1.29 (0.74, 2.24) | 0.38 | 1.11 (0.61, 2.00) | 0.73 | 1.80 (0.99, 3.30) | 0.056 | 1.55 (0.82, 2.93) | 0.17 | ||||||
| Validation Cohort | 0.15 | 0.22 | 0.28 | 0.35 | 0.20 | |||||||||
| A/A | 29 | 19 (70%) | 8 (30%) | 10.5 (9.0, 13.0) | 1 (Reference) | 1 (Reference) | 21.0 (16.5, 24.2) | 1 (Reference) | 1 (Reference) | |||||
| A/G | 19 | 7 (41%) | 10 (59%) | 6.6 (5.7, 8.8) | 1.40 (0.78, 2.52) | 1.41 (0.76, 2.62) | 15.9 (9.3, 23.7) | 1.33 (0.72, 2.46) | 1.55 (0.79, 3.02) | |||||
| G/G | 10 | 5 (71%) | 2 (29%) | |||||||||||
| Additive Model: HR per G | 1.16 (0.78, 1.72) | 0.47 | 1.14 (0.75, 1.74) | 0.54 | 1.26 (0.74, 1.71) | 0.58 | 1.19 (0.76, 1.86) | 0.45 | ||||||
CI, confidence interval; HR, hazard ratio
P value was based on Fisher’s exact test for tumor response, log-rank test for PFS and OS in the univariate analysis (†) and Wald test for PFS and OS in the multivariable Cox regression model adjusted for age, performance status, primary tumor site, primary tumor resection, BRAF mutation status and adjuvant chemotherapy in the training cohort; adjusted for sex, performance status, liver only disease, primary tumor resection in the validation cohort and control cohort (‡).
In the FIRE-3 validation cohort, IL-6 rs2069837 was significantly associated with PFS in univariate analysis. Patients harboring any G allele had a shorter median PFS (8.8 months) compared with those with an A/A genotype (10.9 months, HR, 1.40; 95% CI, 1.06 – 1.85; P = 0.015) (Table 2, Figure 1B). In multivariable analysis, IL-6 rs2069837 genotype remained significantly associated with PFS (HR, 1.34, adjusted P = 0.047). Additionally, those with an A/A genotype had a significantly higher tumor response rate compared to those with any G allele (67% v. 52%, Fisher’s exact P = 0.026).
There was no evidence for an association between IL-6 rs2069837 with OS in either the training or validation cohorts.
Clinical Outcomes by IL-6 Genetic Variants in Patients Receiving FOLFIRI with Cetuximab: FIRE-3
In the FIRE-3 control cohort, genotyping for the IL-6 rs2069837 was successful in 264 patients (89%). Genotyping was not successful in 33 patients because of a limited quantity and quality of extracted genomic DNA. Patients with the G/G genotype had a significantly lower RR (33%) than those with the A/G or A/A variants (70% for both, Fisher’s exact P = 0.042, Table 3). There was no evidence for an association between the IL-6 rs2069837 genotype with PFS or OS in univariate or multivariate analyses (Table 3).
Effect of IL-6 Genetic Variants on Clinical Outcomes by Primary Tumor Location in Patients Receiving FOLFIRI with Bevacizumab: TRIBE and FIRE-3
In a prior study, we showed that the impact of genetic variants involved in pericyte maturation on outcomes was influenced by tumor subsite28. Here, we explored whether associations between IL-6 rs2069837 and outcomes were affected by primary tumor location.
In patients with left-sided colon cancer, IL-6 rs2069837 genotype was significantly associated with PFS in the training and validation cohorts. In the TRIBE training cohort, patients with any G allele had a significantly inferior PFS (9.7 months) compared to those with an A/A genotype in univariate analysis (11.1 months; HR, 1.43; 95% CI, 0.97 – 2.12; P=0.050) (Table 4). Similarly, in FIRE-3 patients receiving bevacizumab-based therapy, patients with any G allele had significantly shorter PFS (10.2 months) compared to those with an A/A genotype in univariate analysis (11.7 months; HR, 1.46; 95% CI, 0.99 – 2.16; P=0.047) (Table 4). In multivariable analysis, there was a trend towards inferior PFS among IL-6 rs2069837 G allele carriers in both cohorts but this did not reach statistical significance (adjusted P = 0.088 and 0.090 in the training and validation cohorts, respectively) (Table 4).
There was no evidence for an association between IL-6 rs2069837 genotype and outcomes in patients with right-sided tumors in either the training or validation cohorts. However, the interaction term of IL-6 rs2069837 and tumor location showed no significance in both cohorts.
DISCUSSION
IL-6/STAT3 signaling is crucial for angiogenesis29,30. As activators of VEGF expression and tumor vessel development, IL-6 and STAT3 may serve as important potential biomarkers of anti-angiogenic therapy, yet the clinical relevance of IL-6/STAT3 genetic variants has previously not been reported. Here, we examined the predictive and prognostic impact of IL-6 and STAT3 SNPs in patients receiving cytotoxic and targeted agents. To our knowledge, we are the first to show that IL-6 rs2069837 genotype is significantly associated with PFS in mCRC patients receiving first-line chemotherapy with bevacizumab.
IL-6 maps to chromosome 7p21 and encodes a phosphorylated, glycoprotein with 212 amino acids, consisting of four introns and five exons, spanning ~4.9 kb of the genomic DNA31. Binding of IL-6 to the signal transducer, gp130, along with either membrane-bound or soluble IL-6 receptor (IL-6R), leads to IL-6 classical or trans-signaling, respectively. Trans-signaling is implicated in T-cell activation, stromal tissue inflammation as well as cardiovascular disorders, inflammatory bowel disease, and colitis-associated cancers1. Knockout IL6 mice models demonstrate deficiencies in wound healing32 and impaired recovery from colitis33. Clinically, elevated serum IL-6 has been associated with inferior survival in CRC34, and in a phase II study of rectal cancer patients treated with neoadjuvant bevacizumab and chemoradiation, change in plasma IL-6 correlated with degree of tumor regression35. Data regarding STAT3 have been less conclusive, with studies supporting both positive36–38 and negative39,40 prognostic roles for intratumoral STAT3 expression.
Evidence relating IL-6 SNPs to CRC outcomes has been inconsistent. While most studies have shown a decreased CRC risk with the IL-6 rs1800795 C allele18,19,21, particularly in those taking NSAIDs15,18, Landi et al. reported an increased susceptibility17, and others have been unable to demonstrate any significant association16,20. A study by Wilkening and colleagues showed a heterozygote survival advantage over those with the IL-6 rs1800795 CC genotype20. Our study was unable to demonstrate any significant association between this SNP and outcomes. IL-6 rs2069837 is purported to be at a transcription factor binding site and as a tag SNP, may exert functional effects through linked polymorphisms at other loci. There are no reports linking IL-6 rs2069837 with cancer survival, but the G allele has been associated with increased susceptibility to cervical23 and liver25 cancers, in addition to increased IL-6 tumor protein expression23. In our study, patients with a G allele had a significantly shorter median PFS compared to those with the A/A variant. Though we were not able to measure IL-6 protein expression, the IL-6 rs2069837 G allele may lead to increased IL-6 expression and serve as a surrogate for resistance towards anti-VEGF therapy in mCRC patients.
Notably, our data showed that the association between IL-6 rs2069837 and outcomes may differ by primary tumor location. We previously demonstrated that genetic variants in angiogenesis pathways, specifically pericyte maturation, variably affect outcomes in mCRC by tumor subsite28. In the present study, the effect of IL-6 rs2069837 on PFS remained significant in patients with distal but not proximal tumors. However, the test for interaction between IL-6 genotype and primary tumor site was not significant, and these findings warrant further exploration. The mechanisms remain to be elucidated, though prior studies have suggested a prominent role IL-6 plays in colitis-associated cancers, which are predominantly left-sided41. On the other hand, Dejea et al42 demonstrated that bacterial biofilms induce IL-6 expression and STAT3 activation and were particularly associated with the development of right-sided colorectal cancers. Taken together, this suggests distinct environmental and inherited mechanisms for IL-6 mediated carcinogenesis in right and left-sided colorectal cancers, respectively.
In addition to carcinogenesis, IL-6 contributes to hemostasis43,44 and vasculopathy45,46, and IL-6 genetic variants have been implicated in atherosclerosis43,47, hypertension47, and cancer-associated thromboembolism48. Accordingly, we explored whether IL-6 SNPs could predict bevacizumab toxicity (i.e. hypertension, bleeding, and thromboembolic events) but found no significant associations in our cohort (data not shown).
Our study is limited by its retrospective design and sample size. Importantly, we were not able to correlate IL-6 polymorphisms with serum or intratumoral IL-6 expression levels which may lend insight on mechanisms of bevacizumab resistance. Nor were we able to determine interactions between microsatellite instability8,49, NSAID use, or inflammatory bowel disease, all of which may be affect the influence of IL-6 polymorphisms on outcomes. Functional studies are certainly needed to determine the significance of each examined genetic variant. Whether the observed associations can be generalized to patients receiving oxaliplatin-containing regimens should also be tested.
Comprehensive molecular profiling has advanced the understanding of several cancers, but the potential of patient genetic profiling to influence therapeutic decisions in mCRC has yet to be realized. Here, we present the first evidence that IL-6 common genetic variants are associated with PFS in mCRC patients treated with first-line bevacizumab-based chemotherapy. Our findings further suggest that the effect of IL-6 rs2069837 may rely upon primary tumor site. If confirmed in larger prospective studies, IL-6 genotype may serve as a promising predictive and prognostic biomarker in mCRC patients receiving bevacizumab-based chemotherapy.
Supplementary Material
STATEMENT OF TRANSLATIONAL RELEVANCE.
The IL-6/STAT3 signaling pathway promotes inflammation, angiogenesis, and cancer. There are currently no validated markers of benefit from anti-angiogenic cancer therapy, and understanding the prognostic impact of genetic variants within the IL-6/STAT3 axis has the potential to inform therapeutic decisions. We analyzed associations between IL-6 and STAT3 polymorphisms and outcomes in metastatic colorectal cancer (mCRC) patients who underwent targeted and cytotoxic therapy in two randomized phase III studies. Our data show for the first time that IL-6 rs2069837 genotype is significantly associated with progression-free survival in mCRC patients treated with first-line bevacizumab-based chemotherapy, but not in those receiving cetuximab-based treatment. Furthermore, our results suggest that the effect of IL-6 polymorphisms on outcomes may depend on primary tumor location. Therefore, IL-6 genetic variants may serve as a promising biomarker in patients with mCRC treated with bevacizumab-based chemotherapy.
Acknowledgments
Satoshi Matsusaka is a recipient of Takashi Tsuruo Memorial Fund. Martin D. Berger received a grant from the Swiss Cancer League (BIL KLS-3334-02-2014).
The project described was supported in part by award number P30CA014089 from the National Cancer Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health. This project was also supported in part by the Wunderglo Project and Care to Cure Research Fund.
Footnotes
Disclosures: The authors do not have any potential conflicts of interest to declare.
References
- 1.Hunter CA, Jones SA. IL-6 as a keystone cytokine in health and disease. Nat Immunol. 2015;16:448–57. doi: 10.1038/ni.3153. [DOI] [PubMed] [Google Scholar]
- 2.Middleton K, Jones J, Lwin Z, Coward JI. Interleukin-6: an angiogenic target in solid tumours. Crit Rev Oncol Hematol. 2014;89:129–39. doi: 10.1016/j.critrevonc.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 3.Lin L, Liu A, Peng Z, Lin HJ, Li PK, Li C, et al. STAT3 is necessary for proliferation and survival in colon cancer-initiating cells. Cancer Res. 2011;71:7226–37. doi: 10.1158/0008-5472.CAN-10-4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Waldner MJ, Foersch S, Neurath MF. Interleukin-6–a key regulator of colorectal cancer development. Int J Biol Sci. 2012;8:1248–53. doi: 10.7150/ijbs.4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu H, Ren G, Wang T, Chen Y, Gong C, Bai Y, et al. Aberrantly expressed Fra-1 by IL-6/STAT3 transactivation promotes colorectal cancer aggressiveness through epithelial-mesenchymal transition. Carcinogenesis. 2015;36:459–68. doi: 10.1093/carcin/bgv017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu H, Lai W, Zhang Y, Liu L, Luo X, Zeng Y, et al. Tumor-associated macrophage-derived IL-6 and IL-8 enhance invasive activity of LoVo cells induced by PRL-3 in a KCNN4 channel-dependent manner. BMC Cancer. 2014;14:330. doi: 10.1186/1471-2407-14-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou C, Tong Y, Wawrowsky K, Melmed S. PTTG acts as a STAT3 target gene for colorectal cancer cell growth and motility. Oncogene. 2014;33:851–61. doi: 10.1038/onc.2013.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tseng-Rogenski SS, Hamaya Y, Choi DY, Carethers JM. Interleukin 6 alters localization of hMSH3, leading to DNA mismatch repair defects in colorectal cancer cells. Gastroenterology. 2015;148:579–89. doi: 10.1053/j.gastro.2014.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patel SA, Gooderham NJ. IL6 mediates immune and colorectal cancer cell crosstalk via miR-21 and miR-29b. Mol Cancer Res. 2015;13:1502–8. doi: 10.1158/1541-7786.MCR-15-0147. [DOI] [PubMed] [Google Scholar]
- 10.Wang J, Cao Z, Zhang XM, Nakamura M, Sun M, Hartman J, et al. Novel mechanism of macrophage-mediated metastasis revealed in a zebrafish model of tumor development. Cancer Res. 2015;75:306–15. doi: 10.1158/0008-5472.CAN-14-2819. [DOI] [PubMed] [Google Scholar]
- 11.Niu G, Wright KL, Huang M, Song L, Haura E, Turkson J, et al. Constitutive Stat3 activity up-regulates VEGF expression and tumor angiogenesis. Oncogene. 2002;21:2000–8. doi: 10.1038/sj.onc.1205260. [DOI] [PubMed] [Google Scholar]
- 12.Wei LH, Kuo ML, Chen CA, Chou CH, Lai KB, Lee CN, et al. Interleukin-6 promotes cervical tumor growth by VEGF-dependent angiogenesis via a STAT3 pathway. Oncogene. 2003;22:1517–27. doi: 10.1038/sj.onc.1206226. [DOI] [PubMed] [Google Scholar]
- 13.Xu Q, Briggs J, Park S, Niu G, Kortylewski M, Zhang S, et al. Targeting Stat3 blocks both HIF-1 and VEGF expression induced by multiple oncogenic growth signaling pathways. Oncogene. 2005;24:5552–60. doi: 10.1038/sj.onc.1208719. [DOI] [PubMed] [Google Scholar]
- 14.Nagasaki T, Hara M, Nakanishi H, Takahashi H, Sato M, Takeyama H. Interleukin-6 released by colon cancer-associated fibroblasts is critical for tumour angiogenesis: anti-interleukin-6 receptor antibody suppressed angiogenesis and inhibited tumour-stroma interaction. Br J Cancer. 2014;110:469–78. doi: 10.1038/bjc.2013.748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gopinathan G, Milagre C, Pearce OM, Reynolds LE, Hodivala-Dilke K, Leinster DA, et al. Interleukin-6 Stimulates Defective Angiogenesis. Cancer Res. 2015;75:3098–107. doi: 10.1158/0008-5472.CAN-15-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abuli A, Fernandez-Rozadilla C, Alonso-Espinaco V, Muñoz J, Gonzalo V, Bessa X, et al. Case-control study for colorectal cancer genetic susceptibility in EPICOLON: previously identified variants and mucins. BMC Cancer. 2011;11:339. doi: 10.1186/1471-2407-11-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Landi S, Moreno V, Gioia-Patricola L, Guino E, Navarro M, de Oca J, et al. Association of common polymorphisms in inflammatory genes interleukin (IL)6, IL8, tumor necrosis factor alpha, NFKB1, and peroxisome proliferator-activated receptor gamma with colorectal cancer. Cancer Res. 2003;63:3560–6. [PubMed] [Google Scholar]
- 18.Slattery ML, Wolff RK, Herrick JS, Caan BJ, Potter JD. IL6 genotypes and colon and rectal cancer. Cancer Causes Control. 2007;18:1095–105. doi: 10.1007/s10552-007-9049-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Theodoropoulos G, Papaconstantinou I, Felekouras E, Nikiteas N, Karakitsos P, Panoussopoulos D, et al. Relation between common polymorphisms in genes related to inflammatory response and colorectal cancer. World J Gastroenterol. 2006;12:5037–43. doi: 10.3748/wjg.v12.i31.5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilkening S, Tavelin B, Canzian F, Enquist K, Palmqvist R, Altieri A, et al. Interleukin promoter polymorphisms and prognosis in colorectal cancer. Carcinogenesis. 2008;29:1202–6. doi: 10.1093/carcin/bgn101. [DOI] [PubMed] [Google Scholar]
- 21.Yu Y, Wang W, Zhai S, Dang S, Sun M. IL6 gene polymorphisms and susceptibility to colorectal cancer: a meta-analysis and review. Mol Biol Rep. 2012;39:8457–63. doi: 10.1007/s11033-012-1699-4. [DOI] [PubMed] [Google Scholar]
- 22.Belluco C, Olivieri F, Bonafe M, Giovagnetti S, Mammano E, Scalerta R, et al. −174 G>C polymorphism of interleukin 6 gene promoter affects interleukin 6 serum level in patients with colorectal cancer. Clin Cancer Res. 2003;9:2173–6. [PubMed] [Google Scholar]
- 23.Shi TY, Zhu ML, He J, Wang MY, Li QX, Zhou XY, et al. Polymorphisms of the Interleukin 6 gene contribute to cervical cancer susceptibility in Eastern Chinese women. Hum Genet. 2013;132:301–12. doi: 10.1007/s00439-012-1245-4. [DOI] [PubMed] [Google Scholar]
- 24.Loupakis F, Cremolini C, Masi G, Lonardi S, Zagonel V, Salvatore L, et al. Initial therapy with FOLFOXIRI and bevacizumab for metastatic colorectal cancer. N Engl J Med. 2014;371:1609–18. doi: 10.1056/NEJMoa1403108. [DOI] [PubMed] [Google Scholar]
- 25.Zheng X, Han C, Shan R, Zhang H, Zheng Z, Liu Y, et al. Association of interleukin-6 polymorphisms with susceptibility to hepatocellular carcinoma. Int J Clin Exp Med. 2015;8:6252–6. [PMC free article] [PubMed] [Google Scholar]
- 26.Yan R, Lin F, Hu C, Tong S. Association between STAT3 polymorphisms and cancer risk: a meta-analysis. Mol Genet Genomics. 2015;290:2261–70. doi: 10.1007/s00438-015-1074-y. [DOI] [PubMed] [Google Scholar]
- 27.Heinemann V, von Weikersthal LF, Decker T, Kiani A, Vehling-Kaiser U, Al-Batran SE, et al. FOLFIRI plus cetuximab versus FOLFIRI plus bevacizumab as first-line treatment for patients with metastatic colorectal cancer (FIRE-3): a randomised, open-label, phase 3 trial. Lancet Oncol. 2014;15:1065–75. doi: 10.1016/S1470-2045(14)70330-4. [DOI] [PubMed] [Google Scholar]
- 28.Volz NB, Stintzing S, Zhang W, Yang D, Ning Y, Wakatsuki T, et al. Genes involved in pericyte-driven tumor maturation predict treatment benefit of first-line FOLFIRI plus bevacizumab in patients with metastatic colorectal cancer. Pharmacogenomics J. 2015;15:69–76. doi: 10.1038/tpj.2014.40. [DOI] [PubMed] [Google Scholar]
- 29.Mar AC, Chu CH, Lee HJ, Chien CW, Cheng JJ, Yang SH, et al. Interleukin-1 Receptor Type 2 Acts with c-Fos to Enhance the Expression of IL-6 and VEGF-A in Colon Cancer Cells and Induce Angiogenesis. J Biol Chem. 2015;290:22212–24. doi: 10.1074/jbc.M115.644823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Motro B, Itin A, Sachs L, Keshet E. Pattern of interleukin 6 gene expression in vivo suggests a role for this cytokine in angiogenesis. Proc Natl Acad Sci U S A. 1990;87:3092–6. doi: 10.1073/pnas.87.8.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gruol DL, Nelson TE. Physiological and pathological roles of interleukin-6 in the central nervous system. Mol Neurobiol. 1997;15:307–39. doi: 10.1007/BF02740665. [DOI] [PubMed] [Google Scholar]
- 32.McFarland-Mancini MM, Funk HM, Paluch AM, Zhou M, Giridhar PV, Mercer CA, et al. Differences in wound healing in mice with deficiency of IL-6 versus IL-6 receptor. J Immunol. 2010;184:7219–28. doi: 10.4049/jimmunol.0901929. [DOI] [PubMed] [Google Scholar]
- 33.Sommer J, Engelowski E, Baran P, Garbers C, Floss DM, Scheller J, et al. Interleukin-6, but not the interleukin-6 receptor plays a role in recovery from dextran sodium sulfate-induced colitis. Int J Mol Med. 2014;34:651–60. doi: 10.3892/ijmm.2014.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim YW, Kim SK, Kim CS, Kim IY, Cho MY, Kim NK. Association of serum and intratumoral cytokine profiles with tumor stage and neutrophil lymphocyte ratio in colorectal cancer. Anticancer Res. 2014;34:3481–7. [PubMed] [Google Scholar]
- 35.Willett CG, Duda DG, di Tomaso E, Boucher Y, Ancukiewicz M, Sahani DV, et al. Efficacy, safety, and biomarkers of neoadjuvant bevacizumab, radiation therapy, and fluorouracil in rectal cancer: a multidisciplinary phase II study. J Clin Oncol. 2009;27:3020–6. doi: 10.1200/JCO.2008.21.1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin C, Wang A, Chen J, Liu X, Wang G. Relationship between expression and prognostic ability of PTEN, STAT3 and VEGF-C in colorectal cancer. Exp Ther Med. 2012;4:633–9. doi: 10.3892/etm.2012.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Monnien F, Zaki H, Borg C, Mougin C, Bosset JF, Mercier M, et al. Prognostic value of phosphorylated STAT3 in advanced rectal cancer: a study from 104 French patients included in the EORTC 22921 trial. J Clin Pathol. 2010;63:873–8. doi: 10.1136/jcp.2010.076414. [DOI] [PubMed] [Google Scholar]
- 38.Gordziel C, Bratsch J, Moriggl R, Knösel T, Friedrich K. Both STAT1 and STAT3 are favourable prognostic determinants in colorectal carcinoma. Br J Cancer. 2013;109:138–46. doi: 10.1038/bjc.2013.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hbibi AT, Lagorce C, Wind P, Spano JP, Des Guetz G, Milano G, et al. Identification of a functional EGF-R/p60c-src/STAT3 pathway in colorectal carcinoma: analysis of its long-term prognostic value. Cancer Biomark. 2008;4:83–91. doi: 10.3233/cbm-2008-4204. [DOI] [PubMed] [Google Scholar]
- 40.Klupp F, Diers J, Kahlert C, Neumann L, Halama N, Franz C, et al. Expressional STAT3/STAT5 Ratio is an Independent Prognostic Marker in Colon Carcinoma. Ann Surg Oncol. 2015;22:1548–55. doi: 10.1245/s10434-015-4485-4. [DOI] [PubMed] [Google Scholar]
- 41.Neurath MF. Cytokines in inflammatory bowel disease. Nat Rev Immunol. 2014;14:329–42. doi: 10.1038/nri3661. [DOI] [PubMed] [Google Scholar]
- 42.Dejea CM, Wick EC, Hechenbleikner EM, White JR, Mark Welch JL, Rossetti BJ, et al. Microbiota organization is a distinct feature of proximal colorectal cancers. Proc Natl Acad Sci U S A. 2014;111:18321–6. doi: 10.1073/pnas.1406199111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kerr R, Stirling D, Ludlam CA. Interleukin 6 and haemostasis. Br J Haematol. 2001;115:3–12. doi: 10.1046/j.1365-2141.2001.03061.x. [DOI] [PubMed] [Google Scholar]
- 44.Kaser A, Brandacher G, Steurer W, Kaser S, Offner FA, Zoller H, et al. Interleukin-6 stimulates thrombopoiesis through thrombopoietin: role in inflammatory thrombocytosis. Blood. 2001;98:2720–5. doi: 10.1182/blood.v98.9.2720. [DOI] [PubMed] [Google Scholar]
- 45.Zhang W, Wang W, Yu H, Zhang Y, Dai Y, Ning C, et al. Interleukin 6 underlies angiotensin II-induced hypertension and chronic renal damage. Hypertension. 2012;59:136–44. doi: 10.1161/HYPERTENSIONAHA.111.173328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reitter EM, Ay C, Kaider A, Pirker R, Zielinski C, Zlabinger G, et al. Interleukin levels and their potential association with venous thromboembolism and survival in cancer patients. Clin Exp Immunol. 2014;177:253–60. doi: 10.1111/cei.12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Humphries SE, Luong LA, Ogg MS, Hawe E, Miller GJ. The interleukin-6 −174 G/C promoter polymorphism is associated with risk of coronary heart disease and systolic blood pressure in healthy men. Eur Heart J. 2001;22:2243–52. doi: 10.1053/euhj.2001.2678. [DOI] [PubMed] [Google Scholar]
- 48.Malaponte G, Polesel J, Candido S, Sambataro D, Bevelacqua V, Anzaldi M, et al. IL-6 −174 G > C and MMP-9-1562 C > T polymorphisms are associated with increased risk of deep vein thrombosis in cancer patients. Cytokine. 2013;62:64–9. doi: 10.1016/j.cyto.2013.02.017. [DOI] [PubMed] [Google Scholar]
- 49.Slattery ML, Wolff RK, Curtin K, Fitzpatrick F, Herrick J, Potter JD, et al. Colon tumor mutations and epigenetic changes associated with genetic polymorphism: insight into disease pathways. Mutat Res. 2009;660:12–21. doi: 10.1016/j.mrfmmm.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


