Abstract
Sirtuins participate in sensing nutrient availability and directing metabolic activity to match energy needs with energy production and consumption. However, the pivotal targets for sirtuins in cancer are mainly unknown. In this study, we identify the M2 isoform of pyruvate kinase (PKM2) as a critical target of the sirtuin SIRT2 implicated in cancer. PKM2 directs the synthesis of pyruvate and acetyl-CoA, the latter of which is transported to mitochondria for use in the Krebs cycle to generate ATP. Enabled by a shotgun mass spectrometry analysis founded on tissue culture models, we identified a candidate SIRT2 deacetylation target at PKM2 lysine 305 (K305). Biochemical experiments including site-direct mutants that mimicked constitutive acetylation suggested that acetylation reduced PKM2 activity by preventing tetramerization to the active enzymatic form. Notably, ectopic overexpression of a deacetylated PKM2 mutant in Sirt2-deficient mammary tumor cells altered glucose metabolism and inhibited malignant growth. Taken together, our results argued that loss of SIRT2 function in cancer cells reprograms their glycolytic metabolism via PKM2 regulation, partially explaining the tumor-permissive phenotype of mice lacking Sirt2.
Keywords: Sirtuins, SIRT2, PKM2, Acetylation, Acetylome, Warburg effect, Glycolysis, Aging, Signaling
INTRODUCTION
Sirtuins are the human and murine homologs of the S. cerevisiae Sir2 deacetylase that regulate replicative and overall lifespan in several species (1–3). While these proteins determine longevity in mammals, they do appear to direct acetylome signaling networks in response to caloric restriction (CR) (4, 5). Furthermore, murine models lacking one of the seven sirtuin genes develop illnesses that mimic those commonly observed in older humans (6–9). Sirtuins are deacetylases that share a common catalytic domain and are localized to the nucleus (SIRT1, SIRT6, and SIRT7), mitochondria (SIRT3, SIRT4, and SIRT5), and cytoplasm (SIRT2) (10). One function of the sirtuin gene family is to sense an organism’s nutrient requirements and availability and optimize cellular pathways so that energy production matches consumption (11–15). Thus, sirtuins function as fidelity proteins that direct signaling networks via post-translational modifications involving lysine deacetylation (16, 17) in response to conditions of cellular stress including decreased nutrient availability.
Pyruvate kinase (PK) catalyzes the transfer of phosphate from phosphoenolpyruvate (PEP) to ADP to yield pyruvate and ATP (18, 19) and determines the production of ATP via oxidative phosphorylation or glycolytic intermediates (20–22). The human genome encodes two PK genes, PKLR and PKM2, which express four PK isoforms: L, R, M1, and M2 (23). When PKM2 is in its active tetramer state, glucose is used for ATP production via oxidative phosphorylation (24, 25). In contrast, when PKM2 dissociates a decrease in enzymatic activity is observed that blocks pyruvate production. This leads to the accumulation of upstream intermediates (24, 25) that are used for macromolecule synthesis, including lipids, nucleic acids, and amino acids, in newly dividing cells (26, 27).
PKM2’s function is dependent on post-translational modifications such as acetylation, oxidation, phosphorylation, prolyl hydroxylation, and sumoylation (28, 29). Acetylation of PKM2 at K433 by p300 acetyltransferase inhibits its activity leading to cell proliferation and tumorigenesis (30). Acetylation of K305 decreased the enzymatic activity of PKM2 and promoted its lysosomal-dependent degradation via chaperone-mediated autophagy, which ultimately stimulated the Warburg effect and tumor growth (30, 31). However, the mechanism by which these lysines are acetylated remains undiscovered.
SIRT2 plays an important role in tumor suppression (32, 33) and mice lacking Sirt2 develop liver, gastrointestinal, and mammary tumors (32, 33) and it has been suggested that a loss of metabolic and an increase in glucose metabolism, generally referred to as the Warburg Effect, may play a role in this phenotype (13, 34, 35). It has been shown that the dys-regulation of PKM2 favors both carcinogenesis and the Warburg Effect (24, 26, 31). As such, we hypothesized that the loss of SIRT2 activity, which may occur with increasing age, plays a role in solid tumors incidence and one mechanism may involve the dys-regulation of energy metabolism.
Materials and Methods
Cell culture
HEK-293T, HeLa, H1299, MCF-7, MDA-MB-231 were obtained from ATCC in 2012, authenticated using CellCheck 9 Plus by IDEXX Bioresearch, and tested for mycoplasma using PlasmoTest™ - Mycoplasma Detection Kit (InvivoGen, Inc). Sirt2−/−-MMT were cultured from Sirt2−/− mammary tumors and verified by PCR and western blotting. Cell lines were cultured in DMEM (Gibco) supplemented with 10% fetal bovine serum (Sigma). Genetically altered cell lines were constructed by infected established lines with lentivirus expressing PKM2, the PKM2 mutants, shPKM2, or other genes. For lentiviral infections 5 MOI were used.
Plasmids, short hairpin RNA constructs, and site-directed mutagenesis
pLKO.1 human SIRT2 shRNA was generated as AAGTAGTGACAGATGGTTGGC. pLKO.1 human and mouse PKM2 shRNA were generated as CATCTACCACTTGCAATTA and GTGCACTCCACTTCTGTCACT, respectively. pCMV5-Flag-SIRT2 was generated by standard PCR amplification of SIRT2 followed by cloning into the pCMV5-Flag (Sigma). The human SIRT2 and PKM2 genes were cloned into PCDH-CMV vectors (Lentiviral vector, System Biosciences; CD513B-1). Plasmids encoding Flag (DYKDDDDK)-tagged PKM2 (Open Biosystems) were purified and subjected to site-directed mutagenesis (BioInnovatise). WT amino acids (K62 and K305) were either converted to arginine (R) or glutamine (Q). Functionally inactive SIRT2 was created by conversion of H187 (Addgene) to tyrosine (Y).
Western blotting and immunoprecipitation
Cells were lysed for 30 min on ice in IP buffer (12) with protease inhibitors and TSA (Sigma). Samples were quantified with BCA assays and analyzed by western blotting. For IP, cell lysates were incubated with protein-specific antibodies or normal rabbit IgG (Santa Cruz Biotechnology, sc-2027) overnight at 4 °C, followed by incubation with protein A/G agarose beads (Millipore; IP10) for 3 h and washed in lysis buffer supplemented with protease inhibitors. Antibodies for western blotting were: Flag (Sigma; F3165), HA (Sigma; H9658), actin (Sigma; A5316), tubulin (Sigma; T6074), Acetyl-tubulin (Sigma; T7451), SIRT2 (Sigma; S8447), PKM2 (Cell Signaling; 3198), PKM2-Ac-K305 (Dr. Qun-Ying Lei; Fudan University, Shanghai, China), PCAF (Santa Cruz Biotechnology; sc-8999), Tip60 (Santa Cruz Biotechnology; sc-25378), Acetyl-lysine (Immunechem; ICP0380), and GAPDH (Millipore; MAB374).
Immunofluorescence
HeLa cells ( 1 × 104 cells/slide) were fixed with paraformaldehyde, permeabilized with 0.2% Triton X-100 at 4°C with antibodies to: rabbit anti-SIRT2 (Sigma; S8447), mouse anti-PKM2 (Santa Cruz; sc-365684); rabbit anti-Flag (Sigma; F7425), and mouse anti-HA (Sigma; H9658). The FITC- and Cys3-conjugated secondary antibody mixture (BD Pharmingen) was incubated for 1 h with Vectashield mounting medium with DAPI (Vector Laboratories) and visualized by fluorescence microscope (Olympus, FV1000).
In vitro/tissue culture deacetylation assay
For the in vitro deacetylation assay of PKM2, HEK-293T cells were co-transfected with Flag-PKM2-WT or Flag-PKM2 mutants and HATs (PCAF and Tip60), and treated with 1 μM TSA and 20 mM nicotinamide for 12 h, as described (36). For purification of SIRT2, pCMV-Flag-SIRT2-WT or -H187Y was transfected into HEK-293T cells and acetylated PKM2 proteins were resuspended in 20 μl deacetylation reaction buffer (36), and analyzed by western blotting with anti-acetyl-lysine antibodies (Millipore). For the tissue culture deacetylation assay, HeLa cells were co-transfected with pcDNA3.1-Flag-PKM2, HATs, and HA-SIRT2-WT or -H150Y, IPed with anti-Flag agarose beads (Sigma; A2220), and blotted with anti-acetyl lysine antibody.
Protein purification
The SIRT2 plasmid (pcDNA3-Flag-hSIRT2-WT, -H187Y) or HATs (PCAF, Tip60) and PKM2 plasmid (pcDNA3.1-Flag-PKM2-WT, -K62R, -K305R, K62R/K305R were transiently transfected into HEK-293T cells using polyethylenimine (Polysciences, 24885). Cells were lysed as described (9), resolved by SDS-PAGE, and stained with Coomassie blue.
Pyruvate kinase assay
Cells were washed twice with ice-cold PBS and lysed with pyruvate kinase assay buffer, collected, and PK activity was determined using a PK activity assay kit according to the manufacturer’s instructions (BioVision; K709). Total protein was adjusted to 3 μg.
Detection of protein oligomerization using SDS and native gel electrophoresis
Eluted proteins were cross-linked by 0.05% glutaraldehyde on ice for 10 min. The reaction was stopped by Laemmli buffer and heating for 5 min at 95 °C, separated by SDS-PAGE (Invitrogen). Assays in the absence of SDS were harvested and lysed on ice in Native PAGE sample buffer (Invitrogen) supplemented with 0.5% n-dodecyl-β-maltoside and protease inhibitors. Samples were separated followed by western blotting with anti-PKM2 antibody.
Mass spectrometry sample preparation and mass spectrometry-based proteomics
HATs (p300/CBP, PCAF, GCN5, Tip60) plasmids were co-transfected into HeLa cells, TSA (1 μM) of nicotinamide (20 mM) for 12 h, lysed in IP buffer, and followed by an in vitro deacetylation assay. The samples were digested by 12.5 ng/μl proteomics-grade trypsin (Sigma; T6567) at a ratio of 1:40 enzyme to protein, re-IPed with acetyl-lysine-conjugated beads (Immunchem; ICP0388), and peptides were then desalted by solid-phase extraction (Sep-pak C18 cartridges, Waters Corporation, Milford, MA) (37). LC-MS-MS analysis of the peptides was performed using a LTQ-Orbitrap mass spectrometer (Thermo Scientific) with a nanospray source and an Eksigent NanoLC 1D Plus and AS1 Autosampler, as previously described (11).
Cell proliferation and soft agar assays
For the cell proliferation assay cells were infected with PKM2-WT and mutants and 50 or 100 cells per well were seeded onto a 6-well plate. After 14 days, the colonies were stained with crystal violet. For the soft-agar colony formation assay, 1×103 cells were plated on 0.3% agar and after 14 days, colonies were stained with methylene blue and counted (Microtec Nition).
Human breast cancer tissue array immunohistochemistry
Breast cancer tissue arrays (Cat No. BC081120) from US Biomax, Inc and sections were deparaffinized and rehydrated in ethanol in series and processed as previously published (8) using an anti-SIRT2 antibody (1:500, Sigma; S8447), rabbit anti-PKM2 (1:500, Cell Signaling; 3198), or rabbit anti-PKM2-Ac-K305 (1:500). Visualization was performed using a DAB kit (SK-4100, Vector Laboratories). Bright-field images were captured using a Tissuegnostics microscope and analyzed by Tissuerequest software (Vienna, Austria).
ATP production and glucose uptake assays
The level of intracellular ATP was determined by ATP colorimetric assay kit (Bio vision, K354). Glucose uptake was measured by fluorimetric cell-based glucose uptake assay kit (Bioassay systems, #EFGU-100). All the measurements were normalized to cell numbers and protein concentration.
Statistical Methods
For comparison of the data presented statistics were done using a two group, unpaired Student’s t-test while for the comparison of three groups, one-way ANOVA with post-hoc analyses were performed via GraphPad Prism software.
RESULTS
Glycolytic metabolism signaling proteins contain reversible acetyl lysines
Sirtuins sense cellular energy requirements as well as availability and direct metabolic pathways to assure that energy production matches consumption. As such, it seemed reasonable to attempt to identify SIRT2 downstream targets involved in glucose metabolism by proteomic analysis. To address this either Flag-SIRT2 or Flag-overexpressing HeLa cells were co-transfected with histone acetyltransferases (HATs), including p300/CBP, PCAF, GCN5, and Tip60. HeLa cells were used since we have significant previous experience using them for sirtuin target discovery experiments and HATs were included to decrease the possibility of false negatives. Proteins were immunoprecipitated (IPed) using Flag-beads and eluted peptides were then IPed again using acetyl-lysine-conjugated beads to enriching the samples for acetylated proteins. LC-MS-MS analysis of the final eluted SIRT2 trapped proteins was performed using an LTQ-Orbitrap mass spectrometer. These experiments identified SIRT2 downstream deacetylation target proteins, including those involved in the glycolysis pathway (Table 1).
Table 1.
SIRT2 downstream deacetylation target proteins in the glycolysis pathway
| Gene | amino acid | sequence | spectral counts | |
|---|---|---|---|---|
| GFP | SIRT2 | |||
| PKM2 | 62 | SVETLKEMIK | 11 | 0 |
| PKM2 | 305 | GDLGIEIPAEKVFLAQK | 8 | 0 |
|
| ||||
| ENO1 | 60 | YMGKGVSK | 1 | 0 |
| ENO1 | 126 | AGAVEKGVPLYR | 3 | 0 |
| ENO1 | 221 | EGGFAPNILENKEGLELLK | 1 | 0 |
| ENO1 | 199 | NVIKEKYGK | 2 | 0 |
| ENO1 | 256 | SGKYDLDFKSPDDPSR16 | 0 | |
| ENO1 | 422 | AKFAGR | 1 | 0 |
|
| ||||
| LDHB | 7 | EKLIAPVAEEEATVPNNK | 5 | 1 |
| LDHB | 82 | IVADKDYSVTANSK | 1 | 0 |
|
| ||||
| ALDOA | 42 | GILAADESTGSIAKR | 5 | 0 |
| ALDOA | 147 | DGADFAKWR | 2 | 0 |
| ALDOA | 330 | AAQEEYVKR | 1 | 0 |
|
| ||||
| PGK1 | 11 | LTLDKLDVK | 5 | 0 |
| PGK1 | 106 | GPEVEKAANPAAGSVILLEN | 1 | 0 |
| PGK1 | 146 | AEPAKIEAFR | 2 | 0 |
| PGK1 | 291 | ITLPVDFVTADKFDENAK | 6 | 0 |
| PGK1 | 361 | ALMDEVVKATSR | 5 | 0 |
|
| ||||
| GAPDH | 194 | TVDGPSGKLWR | 2 | 0 |
| GAPDH | 254 | LEKPAKYDDIKK | 14 | 0 |
| GAPDH | 259 | LEKPAKYDDIKK | 7 | 0 |
| GAPDH | 334 | VVDLMAHMASKE | 4 | 1 |
|
| ||||
| PCDHB | 198 | TATPQQAQEVHEKLR | 2 | 0 |
|
| ||||
| TRFP | 124 | SNTPILVDGKDVMPEVNKVLDK | 1 | 0 |
SIRT2 knockdown or genetic deletion alters glycolytic metabolism and PK activity
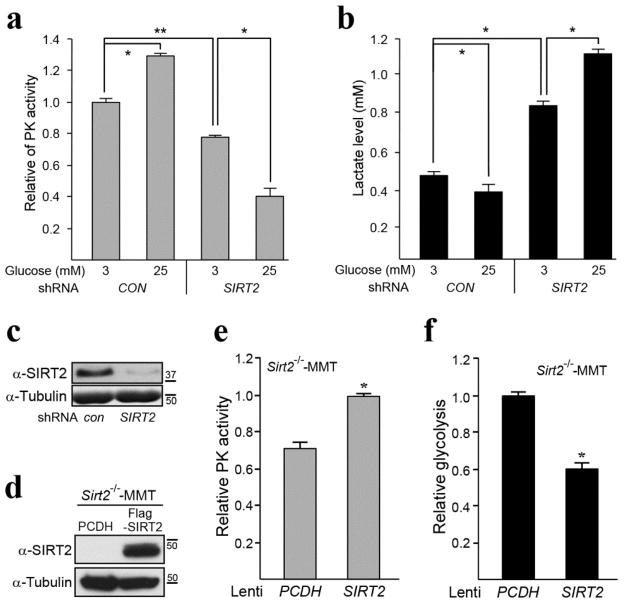
A careful analysis of the mass spectrometry data identified PKM2 as a potential reversible acetyl protein that directs glucose metabolism. As such, SIRT2 was knocked down by transfecting shSIRT2 RNA into H1299 cells to create a tissue culture model that contains decreased SIRT2 deacetylation activity. These genetically altered cells exhibited a decrease in PKM2 activity (Fig. 1a) as well as an increase in lactate production (Fig. 1b) suggesting a dys-regulation of PKM2 activity and glucose metabolism. This was observed both under low (3 mM) and high glucose (25 mM) conditions, though the difference was more dramatic at high glucose. The effectiveness of the shSIRT2 was validated through western blotting (Fig. 1c). These experiments were repeated in MCF-7, MDA-MB-231, and HeLa cell lines and a very similar change in PK activity (Fig. S1a) and glycolysis (Fig. S1b) was observed. Finally, similar outcomes were assessed using a tumor cell line isolated from a Sirt2−/− mouse mammary tumor (referred to as Sirt2−/−-MMT). Sirt2−/−-MMT cells transfected with wild-type (WT) SIRT2 exhibited an increase in PK activity, as well as a decrease in lactate production (Fig. 1d).
Figure 1. SIRT2 knockdown alters pyruvate kinase activity and glycolytic metabolism.
(a–b) H1299 cells were infected with a control vector or shSIRT2 and cultured in low (3 mM) and high glucose (25 mM) media. Pyruvate kinase activity (a) and lactate production (b) assays were performed according to the manufacturer’s procedures. Lactate production was used as a measure of the glycolytic rate. (c) SIRT2 was knocked down in H1299 cells and SIRT2 and tubulin levels were determined by western analysis using anti-SIRT2 and tubulin antibodies. (d) Sirt2−/−-MMT cells were isolated from a mammary tumor in mice lacking Sirt2 and subsequently infected with either a control virus (PCDH) or lenti-SIRT2. Cellular extracts were isolated and immunoblotted with an anti-SIRT2 or tubulin antibody. (e–f) Sirt2−/−-MMT cells infected with PCDH or lenti-SIRT2 were used to measure (e) pyruvate kinase activity and (f) glycolysis. All experiments were done in triplicate. Representative images are shown. Error bars represent one standard deviation from the mean. * indicates P < 0.05 and ** indicates P < 0.01.
SIRT2 physically interacts with PKM2
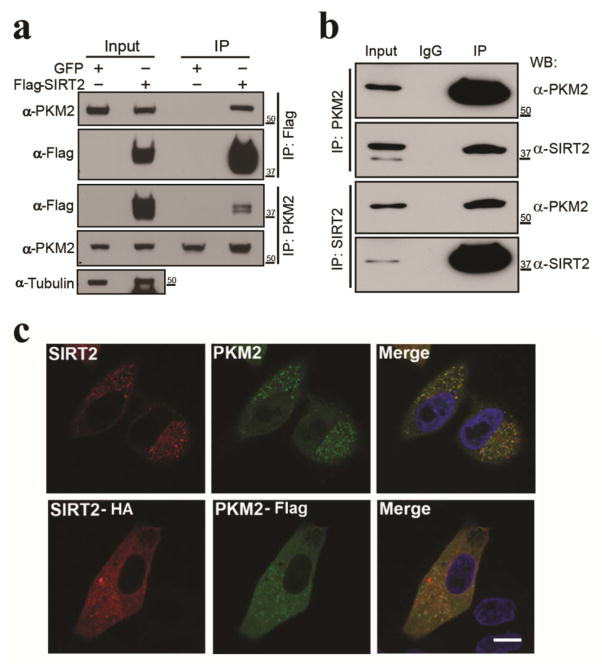
Since mass spectrometry data identified PKM2 as a potential reversible acetyl protein (Table 1) and PKM2 activity is dys-regulated in cells lacking or exhibiting a decrease in SIRT2 activity (Fig. 1a–d). As such, HEK-293T cells were transiently transfected with Flag-SIRT2 or pGFP and total protein extracts were IPed using anti-Flag or PKM2 antibodies, respectively. The results from these experiments showed an interaction between PKM2 and SIRT2 (Fig. 2a). To further confirm this interaction, a reverse co-immunoprecipitation (co-IP) was performed after PKM2 transfection and demonstrated that endogenous SIRT2 binds to PKM2 (Fig. 2b). Immunofluorescence staining showed that SIRT2 and PKM2 co-localized in the cytoplasm (Fig. 2c, upper panels) and similar results were obtained when HA-SIRT2 and Flag-PKM2 were co-transfected into HeLa cells (Fig. 1c, lower panels). We also performed in vitro acetylation assays to identify histone acetyltransferases that acetylate PKM2. For this experiment p300, CBP, GCN5, PCAF, Tip60 as well as Flag-PKM2 were co-transfected into HEK-293T cells, IPed by anti-Flag and anti-PKM2 antibodies, followed by immunoblotting with anti-pan-acetyl-lysine antibody. These results indicate that PCAF (P300/CBP-associated factor) and Tip60, supporting previous data (30), are PKM2 acetyltransferases (Fig. S2a–b). In addition, PKM2 is not deacetylated by SIRT1, SIRT6, or HDAC6 proteins (Fig. S2c). Finally, knocking down PKM2 expression did not change PKM1 protein levels (Fig. S2d).
Figure 2. SIRT2 physically interacts with PKM2.
(a) Flag-SIRT2 or pGFP plasmid were transiently transfected into HEK-293T cells. Total protein extracts were IPed with either anti-PKM2 or Flag antibodies and subsequently separated by SDS-PAGE followed by western blotting with an anti-PKM2, Flag, or tubulin antibody. (b) Endogenous PKM2 or SIRT2 were IPed from HeLa cell lysates and separated via SDS-PAGE followed by western blotting with anti-PKM2 and SIRT2 antibodies. (c) Endogenous SIRT2 and PKM2 were stained by anti-SIRT2 and -PKM2 antibodies in HeLa cells (upper panel). HA-SIRT2 and Flag-PKM2 were co-transfected into HeLa cells (lower panel), stained with anti-HA and FLAG antibodies, and subsequently visualized by confocal microscopy. Scale bar: 30 μm. Representative images are shown. All experiments were done in triplicate.
PKM2 lysines 305 is a direct SIRT2 deacetylation targets
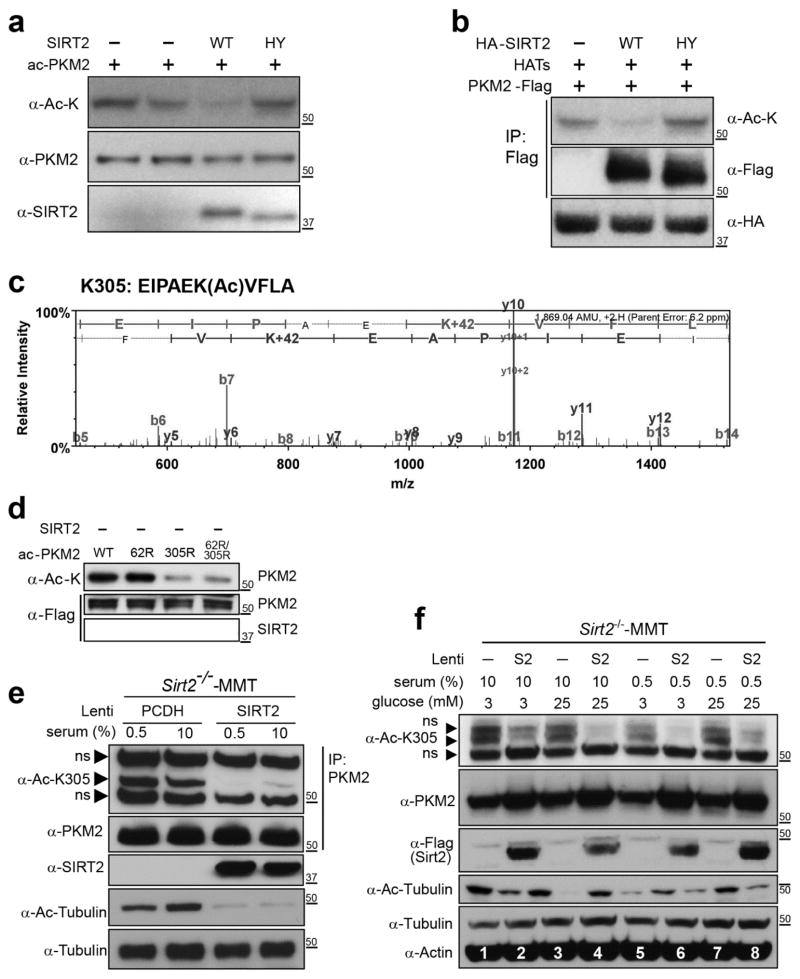
To determine the mechanisms through which SIRT2 directs PKM2, an in vitro deacetylation assay was done. HEK-293T cells were transfected with Flag-PKM2 in the presence of HATs. This is done as a tissue culture model system where proteins are relatively acetylated as might be expected in a nutrient rich cellular environment. Cell lysate was IPed with an anti-Flag antibody and mixed with either purified WT (SIRT2-WT) or enzymatically inactive (SIRT2-H187Y) SIRT2. Samples were next immunoblotted with a pan anti-acetyl-lysine, anti-PKM2, or anti-SIRT2 antibody. These experiments showed a decrease in PKM2 total protein acetylation when mixed with WT SIRT2, but not in control samples or those mixed with the deacetylation-null SIRT2-H187Y (Fig. 3a, lane 1, 2, and 4 versus 3). A tissue culture deacetylation assay was also performed by transiently transfecting HATs and Flag-PKM2 with either HA-SIRT2-WT or HA-SIRT2-H187Y into HeLa cells. These experiments also showed that cells transfected with WT SIRT2 exhibited significantly lower PKM2 acetylation, as compared to either control cells (Fig. 3b, lane 2 versus 1) or cells transfected with SIRT2-H187Y (lane 2 versus 3). Finally, these samples were subject to LC-MS-MS analysis to detect acetylated PKM2 peptides. Such analysis showed that lysines K62 (Fig. S3a) and K305 (Fig. 3c) were acetylated. A genomic analysis was done demonstrating that K62 and K305 are highly conserved among different species throughout evolution (Fig. S3b–c). Overall, these results support the idea that PKM2 is a direct SIRT2 deacetylation target.
Figure 3. PKM2 lysine 305 is a direct SIRT2 deacetylation target.
(a) For in vitro deacetylation assay, HEK-293T cells were treated with 1 μM of TSA for 12 h and transfected with HATs (p300/CBP, PCAF, GCN5, Tip60) as well as Flag-PKM2. Cells were harvested and PKM2 was IPed with an anti-Flag antibody. PKM2 was subsequently mixed with WT or deacetylation-null isolated SIRT2 in the presence of NAD+, and the reaction mixtures were separated by SDS-PAGE and immunoblotted with anti-pan-acetyl-lysine, PKM2, and SIRT2 antibodies. (b) For tissue culture deacetylation assay, HeLa cells were transfected with HATs, Flag-PKM2, and with either HA-SIRT2-WT or HA-SIRT2-H187Y followed by a Flag-IP. Extracts were separated and immunoblotted with anti-pan-acetyl-lysine, Flag, and HA antibodies. (c) LC-MS-MS of PKM2 showing the acetylation of PKM2 K305. HeLa cells were treated with 1 μM of TSA and transfected with HATs (p300/CBP, PCAF, GCN5, Tip60) along with either a SIRT2 or GFP expression vector. Cellular protein extracts were sent for LC-MS-MS analysis of the acetylation intensity of PKM2 peptides. (d) HEK-293T cells were transfected with HA-PKM2-WT or deacetylation mimetic mutants, PKM2K62R, PKM2K305R, or PKM2K6R2/K305R, as well as HATs and TSA. PKM2 was IPed, mixed with WT isolated SIRT2 with NAD+ (and NAM as a control) and reaction mixtures were separated and immunoblotted with anti-pan-acetyl-lysine, and flag antibodies. (e) Sirt2−/−-MMT cells isolated from a mammary tumor in mice lacking Sirt2 were stably infected with PCDH or Flag-SIRT2-WT, cultured in 0.5% or 10% serum and 3 mM (low) or 25 mM (high) glucose, IPed with an anti-PKM2 antibody, and immunoblotted with either anti-PKM2-Ac-K305, PKM2, or SIRT2, Ac-tubulin, and tubulin antibodies. (f) Sirt2−/−-MMT cells were stably infected with PCDH or Flag-SIRT2-WT, were cultured in 0.5% or 10% serum and 3 mM (low) or 25 mM (high) glucose. Cell lysates were immunoblotted by anti-PKM2-Ac-K305, PKM2, Flag, Ac-tubulin, tubulin, and actin antibodies. ns = non-specific band. Representative images are shown. All experiments were done in triplicate.
To determine if PKM2 K62 and K305 are direct SIRT2 deacetylation targets, we generated a series of PKM2 K62 and K305 mutants, including Flag-PKM2, Flag-PKM2K62R, Flag-PKM2K305R, and Flag-PKM2K62R/K305R. In these mutants, the substitution of a lysine with an arginine mimics constitutive deacetylation (9). These vectors were subsequently transfected into HEK-293T cells, and an in vitro deacetylation assay was performed. These results showed that PKM2-K305R and PKM2-K62/K305R exhibited lower acetylation levels as compared to PKM2-K62R (Fig. 3d, lanes 3 and 4 versus lane 2), suggesting that lysine 305, but not 62, is deacetylated by SIRT2. Thus, when K305 is mutated, only K62 can be acetylated, and as such, the greater acetylation in the presence of K62R means that K305 was the primary contributor to the acetylation, as has been shown by other (31), whereas the lower acetylation in the presence of K305R means that K62 was not acetylated.
A tissue culture deacetylation assay was performed using Sirt2−/−-MMT that stably overexpress WT SIRT2 or PCDH (a vector only negative control). Cells in low and high serum were isolated and exogenous PKM2 was IPed using an anti-Flag antibody and extracts were subsequently immunoblotted with a previously described anti-PKM2-Ac-K305 antibody (30). These experiments showed a decrease in PKM2 K305 acetylation in Sirt2−/−-MMT cells expressing WT SIRT2 in low (Fig. 3e, lane 1 versus 3) and high serum (lane 2 versus 4) conditions. The deacetylation of a-tubulin is a positive control (bottom two panels). Interestingly, the decrease was greatest in cells cultured in low serum suggesting that high tissue culture nutrient conditions may slightly decrease SIRT2 activity resulting in a small increase in PKM2 acetylation (lane 3 versus 4). These tissue culture deacetylation assays using the anti-PKM2-Ac-K305 antibody were repeated with both high and low serum and high and low glucose.
To further address the idea that nutrient conditions may direct SIRT2 deacetylation activity these experiments were repeated in high and low serum as well as low glucose levels. These tissue culture deacetylation assays using the anti-PKM2-Ac-K305 antibody showed that co-transfection of SIRT2 decreased PKM2 acetylation levels (Fig. 3f, lanes 1, 3, 5, and 7 versus lanes 2, 4, 6, and 8). In addition, the acetylation of PKM2 was lowest in cells co-transfection of SIRT2 that were cultured in both low serum and glucose (Fig. 3f, lane 6) as compared to cells cultured in high serum and low glucose (lane 2) or high glucose and low serum (lane 8). These results suggest that SIRT2 is less active in cells maintained in a nutrient-rich environment, whereas in low serum or low glucose levels, SIRT2 deacetylation activity is increased, resulting in a decrease in PKM2 K305 acetylation. Overall, these results strongly support the idea that PKM2 lysine 305 is a reversibly acetylated lysine and a direct SIRT2 deacetylation target.
Acetylation status of K305 directs PKM2 activity
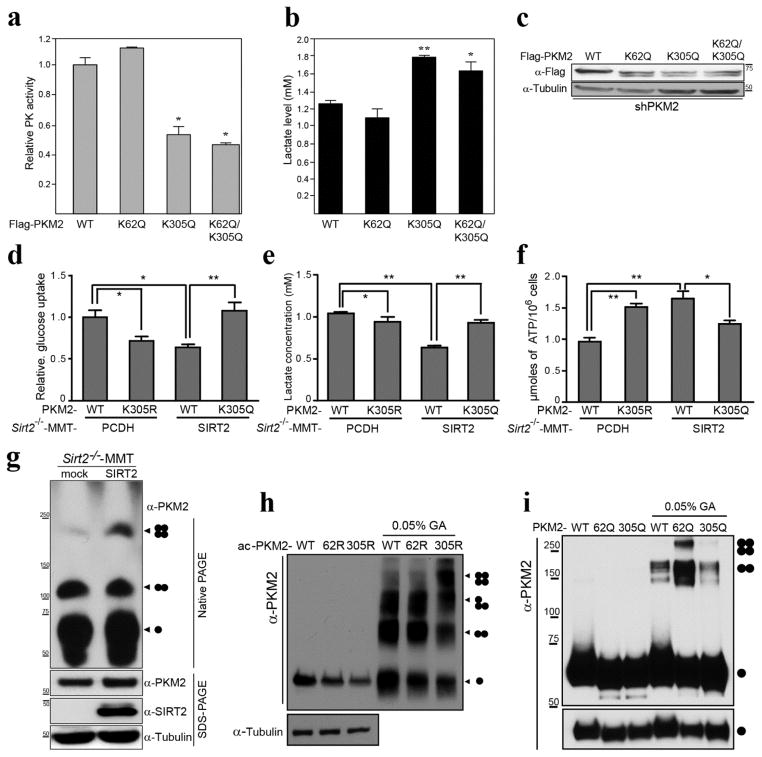
To determine if the acetylation status of PKM2 directs enzymatic activity an in vitro an PK activity assay was performed in HeLa cells infected with shPKM2 to decrease WT PKM2 levels. These HeLa- PKM2-knockdown cells were subsequently transfected with Flag-PKM2-WT and the expression vectors for several PKM2 mutants (FlagPKM2K62Q, FlagPKM2K305Q, and FlagPKM2K62Q/K305Q). In these mutants, the substitution of a lysine with a glutamine mimics constitutive acetylation (8, 9, 38). When lysine 305 was substituted with glutamine (K305Q or K62Q/K305Q), PK activity was significantly lower (Fig. 4a) and lactate production was higher, as compared to WT- or K62Q-transfected control cells (Fig. 4b) although PKM2 expression levels were similar (Fig. 4c). In contrast, when K62 was substituted with glutamine (K62Q), there was no significant change in PK activity or lactate levels compared to those of WT control cells (Fig. 4a, b).
Figure 4. Acetylation status of K305 directs PKM2 activity.
(a–c) HeLa cells were infected with shPKM2 and were subsequently transiently transfected with Flag-PKM2 and the PKM2 mutant acetylated mimic expression vectors Flag-PKM2K62Q, Flag-PKM2K305Q, Flag-PKM2K62Q/K305Q). (a) Pyruvate kinase activity and (b) lactate production assay were performed using eluted PKM2-WT protein as well as protein from cells transfected with the PKM2 mutants. * indicates P < 0.05 and ** indicates P < 0.01. (c) Extracts from the shPKM2 cells transfected with these expression vectors were immunoblotted with anti-Flag and tubulin antibodies. (d–f) PKM2 was knocked down in Sirt2−/−-MMT cells were subsequently transfected with Flag-PKM2WT, Flag-PKM2K305R, and Flag-PKM2K305Q. (d) Glucose uptake, (e) lactate production and (f) ATP production assays were performed. (g) Sirt2−/−-MMT cells were infected with lenti-SIRT-WT, and control and SIRT2-expressing stable cells were lysed in native page sample buffer, separated by native-PAGE or SDS-PAGE, and immunoblotted with anti-PKM2, SIRT2, and tubulin antibodies. (h–i) HeLa cells were transfected with HATs (PCAF/Tip60) with either Flag-PKM2 or the PKM2 deacetylation mimic mutants, Flag-PKM2K62R and Flag-PKM2K305R (h). HeLa cells were transfected Flag-PKM2 or Flag-PKM2 acetylation mimic mutants, Flag-PKM2K62Q and Flag-PKM2K305Q (i). Cells then were lysed and acetylated PKM2 proteins were eluted by Flag peptide. Eluted proteins were cross-linked by 0.05% glutaraldehyde on ice. The samples were separated by SDS-PAGE, and immunoblotted with anti-PKM2 and tubulin antibodies.
The results above measure total lactate production and as such, it seems reasonable to determine if the increase in glucose is a result of maintaining a constant flux split ratio from increased lactate production and mitochondrial respiration. Thus, PKM2 was knocked down in Sirt2−/−-MMT cells (i.e., loss of Sirt2 and PKM2 cells) and these cells were subsequently transfected with FlagPKM2WT, FlagPKM2K305R, and FlagPKMK305Q (Fig. S4). These experiments showed an increase in glucose uptake (Fig. 4d) and lactate production (Fig. 4e) in cells transfected with the PKM2 K-Q acetylated mutant. In contrast, ATP production was increased in cells transfected with the PKM2 K-R deacetylated mutant (Fig. 4f, bar 1 vs. 2) and decreased with the PKM2 K-Q acetylated mutant (bar 3 vs. 4).
It is known that the active form of PKM2 is a tetramer, and it is proposed that post-translational modifications disrupt tetramer formation and, thereby, PK activity. As such, Sirt2−/−-MMT and Sirt2−/−-MMT cells stably expressing exogenous SIRT2 were harvested and prepared for native-PAGE analysis. SIRT2 re-expression in Sirt2−/−-MMT produced higher levels of PKM2 tetramer formation, as compared to Sirt2−/−-MMT cells (Fig. 4g). In addition, similar experiments were done using the PKM2 deacetylation mimic mutants (K62R and K305R) and the PKM2 acetylation mimic mutants (K62Q and K305Q), these experiments showed that K305 favors the formation of the active, tetrameric form of PKM2 (Fig. 4h, i, and Fig. S5). These results suggest, for the first time, that the acetylation status of PKM2, as directed by SIRT2, determines enzymatic activity via a mechanism involving the complex formation that forms the PKM2 tetramer.
Acetylated PKM2 contributes to tumor cell proliferation
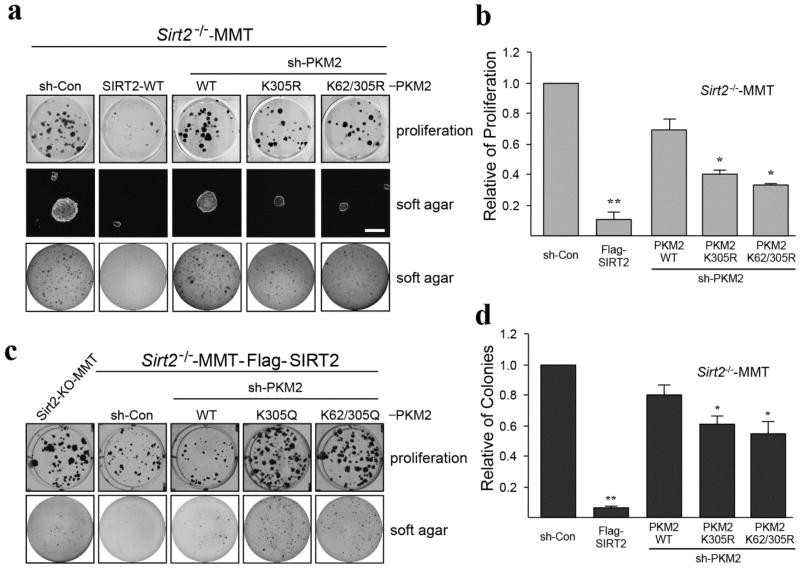
It is well known that aberrant glycolysis is one of the major hallmarks of carcinogenesis and that increased glucose metabolism is related to tumor cell proliferation. Therefore, to assess the contribution of the SIRT2/PKM2 axis to these events, PKM2 was knocked down in Sirt2−/−-MMT cells by shPKM2, and cells were subsequently infected with Flag-PKM2-WT, FlagPKM2K305R, and FlagPKM2K62R/K305R to stably overexpress these genes. Protein levels in these cell lines were confirmed by western blotting (Fig. S4). These experiments clearly showed that cells expressing the deacetylated PKM2 protein exhibited a decrease in tumor cell proliferation (Fig. 5a top panel, b) and growth in soft agar (Fig. 5a middle, low panel, c), consistent with a less aggressive phenotype. In addition, similar experiments were done using the PKM2 acetylation mimic mutants (FlagPKM2K305Q, and FlagPKM2K62R/K305Q) in re-overexpressed SIRT2 of Sirt2−/−-MMT cells showed that cells expressing the acetylated K305-PKM2 protein exhibited an increase in tumor cell proliferation (Fig. 5d top panel) and growth in soft agar (Fig. 5d low panel).
Figure 5. The PKM2 deacetylation mimetic mutants inhibit tumor growth.
(a–c) Sirt2−/−-MMT cells were infected with shPKM2 and re-infected with Flag-SIRT2-WT, Flag-PKM2-WT, Flag-PKM2K305R, or Flag-PKM2K62R/K305R. These cell lines were subsequently used for (a, top panel and b) representative images of cell proliferation assays (doubling time) and (a, middle, low panel and c) growth in soft agar assays, as measured by colony formation (invasion assay). Scale bar = 100 μm. *p < 0.05, **p < 0.01. (d) Sirt2−/−-MMT cells were infected with shPKM2 and re-infected with Flag-SIRT2-WT, Flag-PKM2-WT, Flag-PKM2K305Q, or Flag-PKM2K62R/K305Q. These cells were used for cell proliferation assays (top panel) and soft agar assays (low panel). Experiments were repeated three times and data expressed as mean ± SEM. Representative images are shown.
PKM2 K305 acetylation status correlates with recurrence in human breast cancer samples
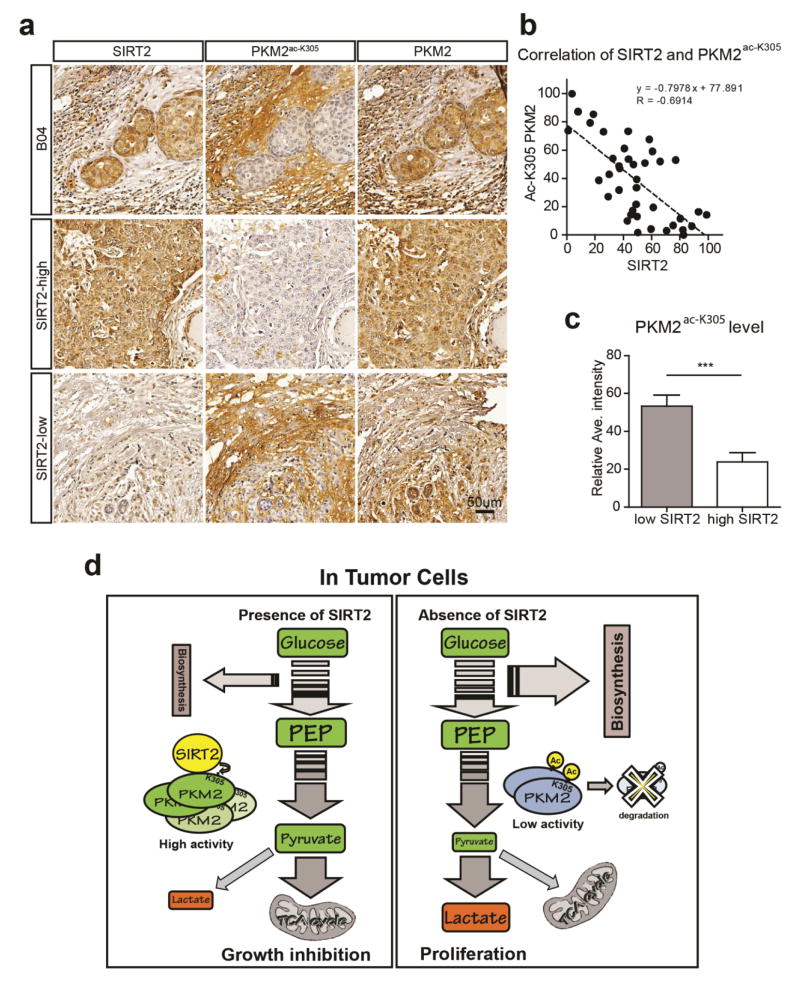
Based on the results above it seemed reasonable to determine a potential relationship between SIRT2 and PKM2 acetylation in breast cancer tumor clinical samples. Thus, we compared the immunohistochemical (IHC) staining intensities of SIRT2 and PKM2 acetylated at K305 (PKM2-Ac-K305) in human breast cancer samples (Fig. 6a, b, c). A total of 40 breast tumors were analyzed by IHC assay and blindly analyzed by two expert investigators. Interestingly, a negative correlation between the levels of SIRT2 and PKM2-Ac-K305 was found (Pearson’s correlation coefficient R=−0.6914, p<0.0001, n=40 patients; Fig. 6b). We further separated the tumor samples into high and low SIRT2 expression groups, according to their staining intensities. Tumors with high SIRT2 expression showed low PKM2-Ac-K305 expression, and vice versa (p<0.001, n=20; Fig. 6a, c). Taken together, these results suggest that a negative correlation exists between SIRT2 and PKM2 acetylation in breast cancer, and thus PKM2 K305 acetylation status might be used to predict tumor progression in human breast cancer.
Figure 6. PKM2 K305 acetylation status correlates with recurrence risk in human breast cancer samples.
(a) Representative bright-field images showing SIRT2, PKM2-Ac-K305, and PKM2 staining (brown) in human breast cancer sections. Nuclei (blue) were marked by hematoxylin staining. Scale bar: 50 μm. (b) Dot plot correlates the level of PKM2-Ac-K305 (y-axis) and SIRT2 (x-axis) expression in 40 human breast cancer cases. The dotted line shows the negative correlation of SIRT2 and PKM2-Ac-K305. Pearson r=−0.6914, p<0.0001 (correlation analysis by Prism 6). (c) The bar graph shows the average intensity of PKM2-Ac-K305 signals in 20 high SIRT2 expression cases and 20 low SIRT2 expression cases. ***p < 0.001. Experiments were repeated three times and data expressed as mean ± SEM. Representative images are shown. (d) Schematic diagram summarizing the role of SIRT2 in directing PKM2 acetylation (PMK2-K305) in tumor cell growth.
DISCUSSION
Tumor cells exhibit increased rates of glycolysis even in the presence of oxygen and this physiological reprogramming is referred to as the Warburg effect (22, 34, 39). PKM2 is a critical regulatory node in directing glucose metabolism, and its dysregulation via one of several potentially aberrant processes is associated with a phenotype similar to the Warburg effect (18, 20, 25, 40). PKM2 is a unique metabolic enzyme that shows a cancer-specific switch of expression from PKM1 to PKM2 (30, 31). It has previously been shown that PKM2 activity is directed by changes in acetylation of lysines 305 (31) and 433 (30); however, the mechanism that directing its post-translational acetylation remains unknown.
This work suggest that this is one mechanism by which acetylation directs PKM2 enzymatic activity is due to the deacetylation of K305, by SIRT2, that directs activity via a mechanism that favors a tetrameric structure. In addition, using anti-SIRT2 and PKM2-Ac-K305 antibodies, we have shown that human breast cancer samples that exhibit high SIRT2 expression showed low PKM2-Ac-K305 levels. We also showed vesicular-like cytoplasm punctate staining cytoplasm that might result from its co-localization with microtubules. Since we have previously shown that female mice lacking Sirt2 developed mammary tumors, and we have now extended these results to identify a subgroup of breast cancer malignancies that exhibit a loss-of-SIRT2/PMK2 signature. This information may be used in the future in personalized therapy to predict risk for tumorigenesis and tumor recurrence.
Thus, under conditions of nutrient excess, SIRT2 activity is decreased which increase PKM2 acetylation as well as enzymatic activity. This favors lactate production while simultaneously decreasing the accumulation of pyruvate creating a metabolic state similar to the Warburg Effect (Fig. 6d). It is also proposed that a decrease in PKM2 activity results in a tumor proliferation permissive resistance phenotype. In contrast, under conditions lacking sufficient nutrients it would be proposed that SIRT2, and other sirtuins, would be active resulting in the deacetylation of multiple downstream targets including PKM2. This would activate PKM2 and favor the accumulation of pyruvate that would feed substrates used by Krebs cycle and oxidative phosphorylation (Fig. 6d).
One important and un-addressed question in regards to SIRT2 biology is: what are the dys-regulated downstream targets that create a tumor permissive phenotype. For example, loss of Sirt2 has been shown to dys-regulate DNA repair (41), cell cycle checkpoints (32), energy metabolism and/or lipid biosynthesis (42), and inflammation (43) and all of these pathways may play a role, at least in some part, in a tumor permissive phenotype. This work adds to this list. Finally, it has been previously shown that lysine acetylation modifications can direct the complex formation of PKM2 and acetylation of K305 prevents tetramer formation that subsequently promotes a glycolytic as well as pro-growth tumor cell phenotype.
Supplementary Material
Acknowledgments
DG is supported by 2R01CA152601-06A1, 1R01CA152799-01A1, 1R01CA168292-01A1, and 1R01CA16383801A1. A.V. is supported by NCI-R01CA182506-01A1, the Lefkofsky Family Foundation/Liz and Eric Lefkofsky Innovation Research Award. Yueming Zhu is supported by a Robert H. Lurie Translation Bridge Fellowship Award. Melissa Stauffer, PhD, of Scientific Editing Solutions, provided editorial assistance.
Footnotes
The authors have no conflict of interest.
References
- 1.Wang RH, et al. Interplay among BRCA1, SIRT1, and Survivin during BRCA1-associated tumorigenesis. Molecular cell. 2008;32(1):11–20. doi: 10.1016/j.molcel.2008.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang RH, et al. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer cell. 2008;14(4):312–323. doi: 10.1016/j.ccr.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Finkel T, Deng CX, Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature. 2009;460:587–591. doi: 10.1038/nature08197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sinclair DA. Toward a unified theory of caloric restriction and longevity regulation. Mechanisms of ageing and development. 2005;126(9):987–1002. doi: 10.1016/j.mad.2005.03.019. [DOI] [PubMed] [Google Scholar]
- 5.Guarente L, Picard F. Calorie restriction--the SIR2 connection. Cell. 2005;120(4):473–482. doi: 10.1016/j.cell.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 6.Kim EJ, Kho JH, Kang MR, Um SJ. Active regulator of SIRT1 cooperates with SIRT1 and facilitates suppression of p53 activity. Molecular cell. 2007;28(2):277–290. doi: 10.1016/j.molcel.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 7.Wood JG, et al. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. 2004;430(7000):686–689. doi: 10.1038/nature02789. [DOI] [PubMed] [Google Scholar]
- 8.Kim HS, et al. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer cell. 2010;17(1):41–52. doi: 10.1016/j.ccr.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tao R, et al. Sirt3-mediated deacetylation of evolutionarily conserved lysine 122 regulates MnSOD activity in response to stress. Molecular cell. 2010;40(6):893–904. doi: 10.1016/j.molcel.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hallows WC, Albaugh BN, Denu JM. Where in the cell is SIRT3?--functional localization of an NAD+-dependent protein deacetylase. Biochem J. 2008;411(2):e11–13. doi: 10.1042/BJ20080336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vassilopoulos A, et al. SIRT3 Deacetylates ATP Synthase F Complex Proteins in Response to Nutrient- and Exercise-Induced Stress. Antioxid Redox Signal. 2014 doi: 10.1089/ars.2013.5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vassilopoulos A, et al. SIRT3 Deacetylates ATP Synthase F1 Complex Proteins in Response to Nutrient and Exercise-Induced Stress. Antioxid Redox Signal. 2013 doi: 10.1089/ars.2013.5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guarente L. The many faces of sirtuins: Sirtuins and the Warburg effect. Nature medicine. 2014;20(1):24–25. doi: 10.1038/nm.3438. [DOI] [PubMed] [Google Scholar]
- 14.Verdin E. The many faces of sirtuins: Coupling of NAD metabolism, sirtuins and lifespan. Nature medicine. 2014;20(1):25–27. doi: 10.1038/nm.3447. [DOI] [PubMed] [Google Scholar]
- 15.Baur JA, et al. Dietary restriction: standing up for sirtuins. Science. 2010;329(5995):1012–1013. doi: 10.1126/science.329.5995.1012. author reply 1013–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwer B, Verdin E. Conserved metabolic regulatory functions of sirtuins. Cell metabolism. 2008;7(2):104–112. doi: 10.1016/j.cmet.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 17.Saunders LR, Verdin E. Sirtuins: critical regulators at the crossroads between cancer and aging. Oncogene. 2007;26(37):5489–5504. doi: 10.1038/sj.onc.1210616. [DOI] [PubMed] [Google Scholar]
- 18.Ferguson EC, Rathmell JC. New roles for pyruvate kinase M2: working out the Warburg effect. Trends Biochem Sci. 2008;33(8):359–362. doi: 10.1016/j.tibs.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gius D, Spitz DR. Redox signaling in cancer biology. Antioxid Redox Signal. 2006;8(7–8):1249–1252. doi: 10.1089/ars.2006.8.1249. [DOI] [PubMed] [Google Scholar]
- 20.Bayley JP, Devilee P. The Warburg effect in 2012. Current opinion in oncology. 2012;24(1):62–67. doi: 10.1097/CCO.0b013e32834deb9e. [DOI] [PubMed] [Google Scholar]
- 21.Redel BK, et al. Glycolysis in preimplantation development is partially controlled by the Warburg Effect. Molecular reproduction and development. 2012;79(4):262–271. doi: 10.1002/mrd.22017. [DOI] [PubMed] [Google Scholar]
- 22.Meldrum NU, Tarr HL. The reduction of glutathione by the Warburg-Christian system. Biochem J. 1935;29(1):108–115. doi: 10.1042/bj0290108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mazurek S. Pyruvate kinase type M2: a key regulator of the metabolic budget system in tumor cells. Int J Biochem Cell Biol. 2011;43(7):969–980. doi: 10.1016/j.biocel.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 24.Hitosugi T, et al. Tyrosine phosphorylation inhibits PKM2 to promote the Warburg effect and tumor growth. Science signaling. 2009;2(97):ra73. doi: 10.1126/scisignal.2000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang W, Lu Z. Nuclear PKM2 regulates the Warburg effect. Cell cycle (Georgetown, Tex) 2013;12(19):3154–3158. doi: 10.4161/cc.26182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dang CV. PKM2 tyrosine phosphorylation and glutamine metabolism signal a different view of the Warburg effect. Science signaling. 2009;2(97):pe75. doi: 10.1126/scisignal.297pe75. [DOI] [PubMed] [Google Scholar]
- 27.Yang W, et al. ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nature cell biology. 2012;14(12):1295–1304. doi: 10.1038/ncb2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo W, Semenza GL. Emerging roles of PKM2 in cell metabolism and cancer progression. Trends in endocrinology and metabolism: TEM. 2012;23(11):560–566. doi: 10.1016/j.tem.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chaneton B, Gottlieb E. Rocking cell metabolism: revised functions of the key glycolytic regulator PKM2 in cancer. Trends Biochem Sci. 2012;37(8):309–316. doi: 10.1016/j.tibs.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 30.Lv L, et al. Mitogenic and oncogenic stimulation of K433 acetylation promotes PKM2 protein kinase activity and nuclear localization. Molecular cell. 2013;52(3):340–352. doi: 10.1016/j.molcel.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lv L, et al. Acetylation targets the M2 isoform of pyruvate kinase for degradation through chaperone-mediated autophagy and promotes tumor growth. Molecular cell. 2011;42(6):719–730. doi: 10.1016/j.molcel.2011.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim HS, et al. SIRT2 maintains genome integrity and suppresses tumorigenesis through regulating APC/C activity. Cancer cell. 2011;20(4):487–499. doi: 10.1016/j.ccr.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park SH, et al. SIRT2 is a tumor suppressor that connects aging, acetylome, cell cycle signaling, and carcinogenesis. Translational cancer research. 2012;1(1):15–21. [PMC free article] [PubMed] [Google Scholar]
- 34.Warburg O, Wind F, Negelein E. The Metabolism of Tumors in the Body. The Journal of general physiology. 1927;8(6):519–530. doi: 10.1085/jgp.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Warburg O. The Chemical Constitution of Respiration Ferment. Science. 1928;68(1767):437–443. doi: 10.1126/science.68.1767.437. [DOI] [PubMed] [Google Scholar]
- 36.Ozden O, et al. SIRT3 deacetylates and increases pyruvate dehydrogenase activity in cancer cells. Free Radic Biol Med. 2014;76:163–172. doi: 10.1016/j.freeradbiomed.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vassilopoulos A, et al. SIRT3 deacetylates ATP synthase F1 complex proteins in response to nutrient- and exercise-induced stress. Antioxid Redox Signal. 2014;21(4):551–564. doi: 10.1089/ars.2013.5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haigis MC, Deng CX, Finley LW, Kim HS, Gius D. SIRT3 Is a Mitochondrial Tumor Suppressor: A Scientific Tale That Connects Aberrant Cellular ROS, the Warburg Effect, and Carcinogenesis. Cancer Res. 2012;72(10):2468–2472. doi: 10.1158/0008-5472.CAN-11-3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Warburg O. Iron, the Oxygen-Carrier of Respiration-Ferment. Science. 1925;61(1588):575–582. doi: 10.1126/science.61.1588.575. [DOI] [PubMed] [Google Scholar]
- 40.Stoyanovsky DA, et al. Thioredoxin and lipoic acid catalyze the denitrosation of low molecular weight and protein S-nitrosothiols. J Am Chem Soc. 2005;127(45):15815–15823. doi: 10.1021/ja0529135. [DOI] [PubMed] [Google Scholar]
- 41.Zhang H, et al. SIRT2 directs the replication stress response through CDK9 deacetylation. Proc Natl Acad Sci U S A. 2013;110(33):13546–13551. doi: 10.1073/pnas.1301463110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin R, et al. Acetylation stabilizes ATP-citrate lyase to promote lipid biosynthesis and tumor growth. Molecular cell. 2013;51(4):506–518. doi: 10.1016/j.molcel.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rothgiesser KM, Erener S, Waibel S, Luscher B, Hottiger MO. SIRT2 regulates NF-kappaB dependent gene expression through deacetylation of p65 Lys310. Journal of cell science. 2010;123(Pt 24):4251–4258. doi: 10.1242/jcs.073783. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.