Abstract
Cancer genome characterization efforts now provide an initial view of the somatic alterations in primary tumors. However, most point mutations occur at low frequency. and the function of these alleles remain undefined. We have developed a scalable systematic approach to interrogate the function of cancer-associated gene variants. We subjected 474 mutant alleles curated from 5,338 tumors to pooled in vivo tumor formation assays and gene expression profiling. We identified 12 transforming alleles including two in genes (PIK3CB, POT1) that have not been shown to be tumorigenic. One rare KRAS allele, D33E, displayed tumorigenicity and constitutive activation of known RAS effector pathways. By comparing gene expression changes induced upon expression of wild type and mutant alleles, we inferred the activity of specific alleles. Since alleles found to be mutated only once in 5,338 tumors rendered cells tumorigenic, these observations underscore the value of integrating genomic information with functional studies.
Keywords: rare variant, tumorigenesis, genetic screen, expression profiling, cancer
INTRODUCTION
Describing the complete list of genes altered in cancer genomes has been a major goal of cancer research, with an expectation that identifying mutated cancer genes would elucidate the molecular basis of cancer and nominate potential therapeutic targets (1). Advancements in sequencing technologies have facilitated the initial description of the mutational landscape in many types of cancers (2, 3). Although these efforts have identified some new classes of oncogenes and tumor suppressor genes that occur at high frequency, the majority of somatically altered alleles are found at low frequency, making it difficult to differentiate functionally relevant alleles from neutral, passenger mutations (2). Computational approaches to predict the functional consequences of these low incidence point mutants are informative but require experimental and clinical validation (4).
Increasing numbers of cancers are now being sequenced in clinical settings, and in some cases this information directs therapeutic decisions (5–8). Although such efforts will facilitate recruitment to clinical trials of molecularly targeted agents, it is already clear that such efforts identify many somatically altered but unstudied alleles in known oncogenes and tumor suppressor genes as well as genes not previously implicated in cancer initiation or progression (6, 9). At present, such alleles are either classified as variants of unknown significance (VUS) or are not reported (10, 11).
Although the in-depth study of single genes will eventually provide functional information for these cancer-associated alleles, it is now possible to systematically study the consequences of expressing mutant alleles at scale. To determine whether the systematic characterization of cancer alleles provides functional insights, we generated a large number of alleles identified in cancer genome sequencing studies and assessed the consequences of expressing these alleles on tumor formation and gene expression (Fig. 1A). This approach provides a scalable method to characterize and assign function to a large number of alleles identified by cancer genome sequencing efforts.
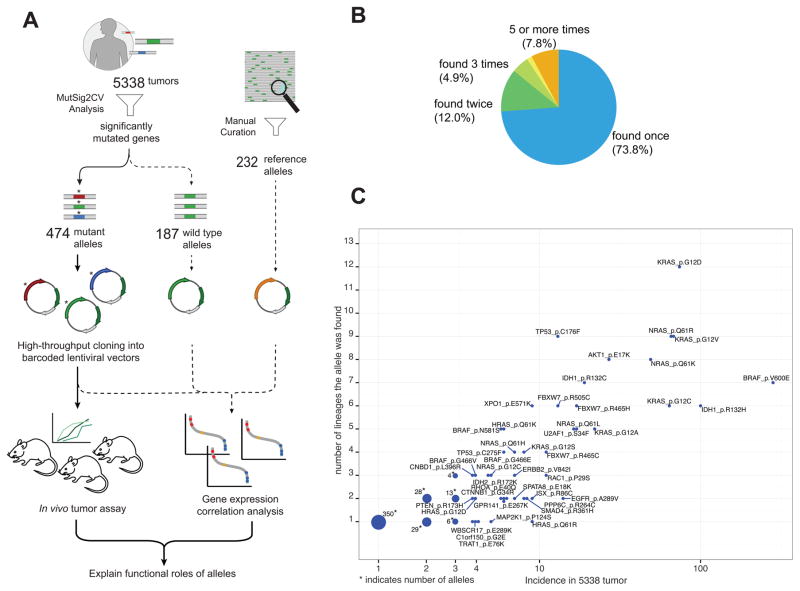
Figure 1. Project pipeline and summary of alleles included in this study.
(A) Project pipeline.
(B) Distribution of incidence of the alleles included in the project. 73.8% of the 474 alleles included in this study were found to be mutated only once.
(C) Alleles mutated frequently were also found to be mutated in larger number of lineages. The size of dots corresponds to the number of overlapping dots.
RESULTS
Creation of a Pan-Cancer candidate cancer allele panel
To create a panel of cancer alleles, we first identified candidate cancer genes by running MutSig2CV (12, 13) on a collection of 5,338 tumors representing 27 cancers that had been subjected to whole exome or whole genome sequencing. Specifically, we prioritized genes by their p-value calculated from their individualized background mutation rate, which was determined by considering covariates such as gene expression level and DNA replication timing (12). These analyses identified 381 genes, 220 of which (58%) templates were present in the hORFeome 8.1 collection of cDNA clones (14) (Supplementary Table S1). We selected 696 mutant alleles for reagent generation by considering local mutational density and evolutionary conservation (described in Methods). Of the 220 alleles for which we had templates, we generated 187 wild type alleles and 474 of the 696 nominated mutated alleles (68%, 178 genes). In addition, we constructed and included a set of 232 ORFs with known functions as well as 24 control ORFs. These alleles were introduced into uniquely barcoded lentiviral vectors. In total, this collection included 1163 ORFs (Methods; Supplementary Table S2).
The majority of the 474 mutant alleles were infrequently mutated in human cancers. Specifically, 350 (73.8%) of the mutant alleles were found only once, and 12.0%, and 4.9% of the alleles were found twice and three times, respectively (Fig. 1B). We noted that as the frequency of an allele increased, that allele was more likely to be found in multiple lineages (Fig. 1C). These observations suggest that testing these alleles in a single cell context may provide generalizable information.
High-throughput identification of transforming alleles in vivo
The assessment of tumor formation potential in mice is a widely used method to assess transforming function of specific alleles. We created a high-throughput platform to determine whether specific cancer-associated alleles induce tumor formation. For these studies, we used the genetically defined, immortalized human embryonic kidney cell line, HA1E (15), and a HA1E variant expressing an activated MEK1DD allele (HA1E-M) as model systems. HA1E-M cells are primed for cell transformation and have been previously used to identify genes involved in cell transformation (16, 17). We expressed each of the 474 alleles in HA1E-M cells and then used an in vivo pooled strategy to assess the tumorigenic potential of each allele (Fig. 2A).
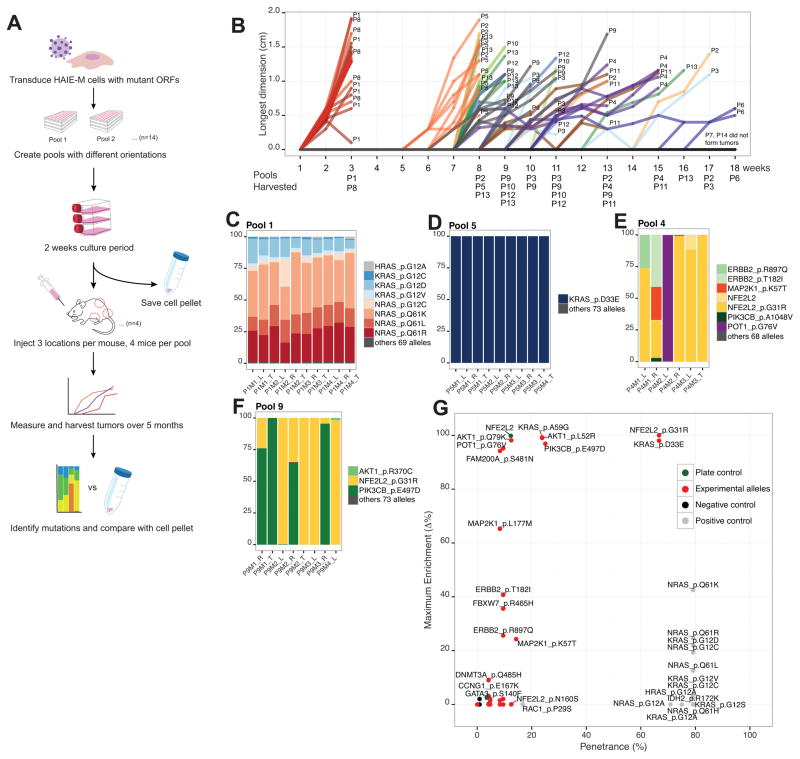
Figure 2. Pooled in vivo screen identifies novel transforming alleles.
(A) Pooled in vivo screen design.
(B) Tumor formation over an 18 week timeframe per pool.
(C) Pool 1, a positive control pool, showed consistent tumor composition across tumors. Each tumor is represented as a bar. The compositions of tumors were shown as different colors.
(D) KRASD33E induced tumor formation in pool 5.
(E) NFE2L2G31R and POT1G76V induced tumor formation in pool 4.
(F) NFE2L2G31R and PIK3CBE497D induced tumor formation in pool 9.
(G) Summary of the in vivo pooled screen. X-axis shows penetrance, which was calculated to be (times each allele was more than 0.01% of tumor reads) / (number of sites the allele was implanted). Since tumor size cannot exceed 2cm in the longest dimension, not all sites were observed for the full length of time. Y-axis shows maximum enrichment, which was calculated to be (maximum percentage of allele in any tumor) – (percentage of the allele in pre-injection cell pellet). Positive controls (colored in grey) had penetrance of around 80%, and low maximum enrichment due to competition against each other.
Based on optimization experiments, we placed all 474 alleles into seven different pools (Pool 1–7) and segregated known oncogenic alleles into Pool 1, to reduce the possibility that known transforming alleles would dominate tumor formation and mask weaker oncogenic alleles. Pool 8 is a biological replicate of Pool 1. We scrambled alleles in Pool 2–7 into Pool 9–14 to create an additional set of pools, to give each allele two different sets of pool neighbors to increase sensitivity. The pool composition is described in Supplementary Table S3. We transduced each of the alleles into HA1E-M cells in an arrayed format, then pooled and expanded cells for tumorigenicity studies (Fig. 2A; Methods). Barcode sequencing of ORFs confirmed that nearly all of the alleles were represented upon implantation, although we noted that the representation of the alleles was not equal, likely due to the differences in viral titer because of differences in the length of each ORF and nucleotide composition (Supplementary Fig. S1A–S1D).
Pools consisting of known cancer alleles (Pool 1 and 8), formed tumors within 1–2 weeks (Fig. 2B), and all eight mice in these pools were sacrificed by week 3. Pool 7 and 14, experimental pools with a total of 110 unique alleles, failed to form any tumors after 18 weeks, confirming previous works showing that the background rate of tumor formation is low in this experimental model (16, 17) (Fig. 2B). We harvested 69 tumors from 168 implantation sites and quantified the barcodes associated with each ORF by PCR amplification and sequencing (Methods; Supplementary Table S4A–S4N).
We observed that tumors derived from pools 1 and 8, which were composed of known oncogenic alleles, repeatedly demonstrated a similar pattern of allele representation, mainly composed of NRAS and KRAS alleles (Fig. 2C). In contrast, we found that tumors derived from other experimental pools showed a wide diversity of allele representation. Some pools contained a single dominant oncogenic allele while others included several oncogenic alleles (Fig. 2D, E, F). Certain alleles, such as KRASD33E, were found enriched in all tumors in which they were assessed; we labeled these alleles as highly penetrant (Fig. 2G). Other alleles such as POT1G76V were less penetrant but they were highly enriched in a few tumors (Fig. 2E, G). We considered alleles that were found at more than 1% in at least two tumors or more than 90% in at least one tumor to have scored. KRASA59G, AKT1L52R, AKT1Q79K, NFE2L2G31R, NFE2L2WT, PIK3CBE497D, FAM200AS481N alleles also scored in the pooled screen (Fig. 2G; Supplementary Fig. S2A–S2H).
The pooled nature of the screen forces competition among alleles in the same pool. For example, Pool 1, only eight alleles out of 77 were represented at 1% or higher in tumors and when lower threshold of 0.01% was applied, 24 alleles met the cutoff (Supplementary Table S4C). Known oncogenic alleles such as AKT1E17K failed to score due to competition, even though this allele is known to transform in this cell context (17). Nevertheless, these observations allowed us to identify a subset of somatically altered alleles that induce tumor formation in this context.
Gene expression correlation analysis differentiates allele function
In parallel to testing the tumorigenic potential of each allele in vivo, we created expression signatures for each of these alleles by expressing the 1163 constructs in HA1E cells (15). We selected this cell line since established cancer cell lines harbor many genetic alterations, which could confound the interpretation of expressing each allele. We decided to use HA1E cells, and not HA1E-M cells, which was used in the in vivo screen, to eliminate the contribution of an overexpressed MEKDD allele. We measured transcript levels of 978 landmark genes using the L1000 Luminex bead-based gene expression assay (18) (Methods). Using the normalized gene expression change induced by each overexpressed allele, we calculated the pairwise Spearman correlation coefficient of all the alleles included in the study (Fig. 3A). We excluded alleles with low infection efficiency (less than 40%), allowing us to assess 1036 perturbations (Methods; Supplementary Table S5).
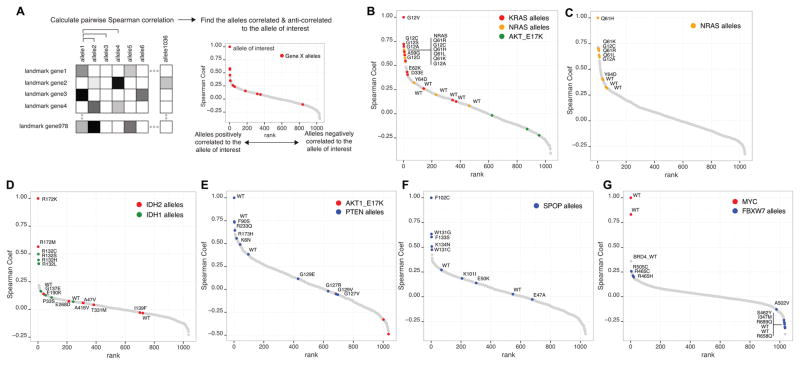
Figure 3. Gene expression profiling differentiates functional variants.
(A) Expression signatures were analyzed by pairwise Spearman correlation to identify similar or dissimilar alleles to the allele of interest.
(B) KRASG12V induces similar gene expression changes as other known activating alleles of KRAS and NRAS.
(C) NRASQ61H induces similar gene expression changes as other known activating alleles of NRAS. However, the signature from the novel Y64D allele had a lower correlation, similar to wild type.
(D) IDH1/2 alleles were correlated to known activating mutant IDH2R172K. Other known activating alleles of IDH1/2 are highly correlated to IDH2R172K.
(E) When correlated to the PTEN wild type, F90S, R233Q, K6N, R173H correlated strongly with the wild type PTEN. The known loss-of-function, dominant negative allele G129E showed a lower correlation. G127R, G129V, G127V also showed low correlation to the wild type.
(F) When alleles were correlated against SPOPF102C, a loss-of-function, dominant negative SPOP allele, other known loss-of-function, dominant negative alleles W131G, F133S, K134N, and W131C were highly correlated. On the other hand, E50K, K101I, E47A had lower correlation to F102C.
(G) FBXW7 wild type, R658Q, I347M, S462Y, and R689Q, were strongly anti-correlated to MYC. Known dominant negative alleles (R505C, R465C, R465H) no longer were anti-correlated to MYC. BRD4 wild type was the most closely correlated to MYC.
Using the pairwise Spearman correlation coefficient between every pair of alleles included in the study, we first examined whether we could detect known relationships. For instance, we found that the expression relationship of KRASG12V, a well-known gain-of-function mutant of KRAS, correlated highly with other known oncogenic KRAS and NRAS mutants (Fig. 3B). Other known oncogenic alleles such as AKTE17K did not correlate with the KRAS signature, demonstrating that this correlation was not simply the consequence of a pro-survival signal induced by an oncogenic allele. Novel alleles of KRAS, D33E and E62K correlated less strongly to known KRAS activating mutants but were clearly differentiated from the wild type alleles, suggesting they may be activating mutants (Fig. 3B). In addition, when we examined NRASQ61H, a known activating mutant of NRAS, we found that this allele was highly correlated with other oncogenic NRAS mutants, but that the novel Y64D allele was more similar to the wild type NRAS allele suggesting that this allele is likely to be a passenger allele (Fig. 3C). Indeed, Y64D did not score in the pooled in vivo screen.
In addition to oncogenic RAS alleles, we also found a correlation among activating mutations for other known oncogenes such as IDH1/2 (Fig. 3D). Specifically, we found that other known gain-of-function mutants IDH2R172M, IDH1R132C, IDH1R132S, IDH1R132H and IDH1R132L were highly correlated to the known gain-of-function mutant IDH2R172K (19). On the other hand, the IDH1 E190K and P33S alleles and the IDH2 G137E, E268D, A416V, A47V, T331M, and I138F alleles failed to correlate to known activating mutants, suggesting these alleles were more similar to the WT allele (Fig. 3D).
Next, we investigated PTEN, a commonly mutated tumor suppressor gene, whose loss of function leads to constitutive activation of the phosphatidylinositol-3-kinase (PI3K) signaling pathway (20). Among the eight PTEN alleles included in this study, F90S, R233Q, K6N, and R173H correlated with the signature induced by overexpressing wild type PTEN, suggesting that these alleles did not completely inactivate PTEN function (Fig. 3E). F90S mutant was recently shown to retain lipid phosphatase activity but was unable to translocate to the plasma membrane (21). R233Q may also affect localization (22). The R173H variant was previously reported to lack phosphoinositide phosphatase activity (23), but its effect was later reported to be less severe (24). Our data supports the notion that R173H retains residual PTEN function. In contrast, a known loss-of-function, dominant interfering allele (G129E) (25, 26) failed to correlate with the wild type allele. We also found that signatures from the G129V, G127V and G127R alleles were clearly distinct from the wild type allele and moderately correlated to G129E (Supplementary Fig. S3A, S3B), suggesting that these alleles are also likely to be loss-of-function variants. Other alleles that activate PI3K signaling (AKT1E17K) were anti-correlated with wild type PTEN (Fig. 3E).
We used a similar approach to differentiate several alleles of SPOP, a gene mutated in prostate and endometrial cancers (27, 28) (Fig. 3F). Specifically, we found that the W131G, F133S, K134N, and W131C alleles strongly correlated with F102C, a known loss-of-function, dominant negative variant (29, 30), but that the WT, K101I, E50K, and E47A did not correlate with the F102C allele. Codons F102, W131, F133 and K134 are mutated mostly in prostate cancers and E47 and E50 are altered in endometrial cancers (27, 28, 31). Recently, SPOP was shown to induce ubiquitination and degradation of androgen receptor and ERG in prostate cancer and estrogen receptor-α in endometrial cancer, but the SPOP mutants were impaired in this ubiquitination activity (29, 30, 32, 33). When we looked for alleles correlated to the E50K, loss-of-function allele in endometrial cancer (32), E47A was highly correlated, suggesting that this allele may also be a loss-of-function allele (Supplementary Fig. S3C). Gene expression signatures of E47 and E50 variants clustered with that of wild type but were distinct from F102, W131, F133 and K134 variants (Supplementary Fig. S3D). These findings suggest that gene expression analysis may allow nuanced interpretation of loss-of-function alleles that are associated with specific context. Since missense mutations in tumor suppressor genes tend to occur throughout their coding sequences, it is often difficult to differentiate functional from nonfunctional mutations by inspecting of the mutations or their frequency. Examining gene expression changes induced by these mutations may facilitate the classification of missense mutant alleles.
We also examined which of the included alleles correlated with the proto-oncogene MYC, a commonly amplified oncogenic transcription factor (34). The most positively correlated allele in our dataset was wild type BRD4, which is a transcriptional activator of MYC (Fig. 3G) (35). BRD4 has been shown to regulate MYC transcription, and pharmacologic modulation of BRD4 inhibited proliferation in MYC-dependent cancers (35). We found that the FBXW7 wild type, R658Q, I347M, R689Q, and S462Y alleles were anti-correlated to wild type MYC (Fig. 3G). FBXW7 is the substrate recognition component of the SCF ubiquitin ligase targeting MYC (36), suggesting that these four alleles do not affect FBXW7 function. In contrast, we found that the known dominant interfering alleles, FBXW7 R505C, R465C, and R465H (37, 38), were anti-correlated to wild type FBXW7, in consonance with the interpretation that these alleles inhibit endogenous wild type FBXW7 (Supplementary Fig. S3E).
Validation of rare oncogenic alleles
To validate the tumor formation of rare alleles, we performed individual tumorigenicity experiments with the candidate oncogenic alleles and their allelic series (Fig. 4A–D; Supplementary Fig. S4A–S4C). We defined tumorigenic allele in as an allele that formed any tumor larger than 500 mm3 by 130 days. We validated that AKT1L52R, NFE2L2G31R, POT1G76V, KRASD33E, and KRASA59G were tumorigenic. In addition, some alleles that did not score in pooled screen formed tumors in individual experiment including KRASE62K, PIK3CBA1048V, NFE2L2G31A, NFE2L2G31V, NFE2L2N160S, AKT1E267G and AKT1R370C.
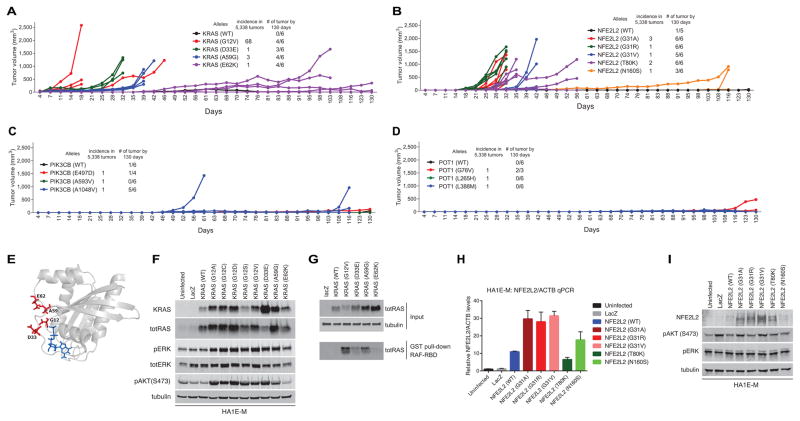
Figure 4. Validation of rare oncogenic alleles.
(A) Individual tumor validation of KRAS alleles. The KRASD33E and KRASA59G alleles formed tumors robustly. E62K did not form tumors in the pooled assay but formed tumors in individual assays, at a later time point.
(B) Individual tumor validation of NFE2L2 alleles. In the pooled assay, only G31R scored in multiple tumors. In the individual assay, G31V, G31A, T80K formed tumors as well. N160S formed tumors at a later time point. NFE2L2 wild type formed one small tumor by the end of the experiment.
(C) Individual tumor validation of PIK3CB alleles. E497D and the wild type formed tumors after long latency. PIK3CBA1048V formed tumors with shorter latency at the majority of injection sites.
(D) Individual tumor validation of POT1 alleles. The G76V allele formed tumor at a later time point. One of the POT1G76V mice died of unknown cause.
(E) The structure of KRAS (PDB: 4EPV) shows that all four of the mutants are in close spatial proximity. Mutated residues are shown in red, GDP bound to the substrate pocket is shown in blue.
(F) Immunoblot of KRAS alleles (including other positive control alleles) shows increased phospho-ERK and phospho-AKT1 levels in KRASD33E, and KRASA59G overexpressed cells.
(G) RAF binding domain pull down assay shows increased GTP bound KRAS in D33E and A59G mutants.
(H) Quantitative PCR of NFE2L2 mRNA expression shows all alleles were expressed.
(I) Immunoblot of NFE2L2 alleles show increased NFE2L2 protein level in G31A, G31R, G31V and T80K overexpressed cells. There was no change in phospho-ERK or phospho-AKT1 levels.
We found that the KRASD33E and KRASA59G alleles were potently tumorigenic, while the KRASE62K allele induced tumor formation at much longer latencies (Fig. 4A). When we mapped the KRASD33E, KRASE62K, and KRASA59G on the KRAS structure (39), we found that these mutations occur in close proximity with known transforming alleles (Fig. 4E). Cells expressing KRASD33E and KRASA59G showed increased activation of the MAP kinase and PI3K pathways as assessed by phosphorylation of specific effectors and a RAF binding domain pull down assay (Fig. 4F, G). These observations suggest that these rare KRAS alleles are indeed oncogenic.
When we examined the NFE2L2 allelic series, we found that the G31R, G31V, G31A, and T80K alleles robustly formed tumors (Fig. 4B), while the N160S allele formed small tumors at a much later time point. We note that expression of wild type NFE2L2 induced the formation of a single tumor formation at long latency. Tumor formation by NFE2L2 wild type overexpression was also observed in the pooled screen (Supplementary Fig. S2C, S2G). In consonance with these observations, we found that tumorigenic NFE2L2 mutants were expressed a higher levels, likely due to defective degradation by endogenous KEAP1 (Fig. 4H, I). Gene expression analysis of NFE2L2 mutants showed a similar gene expression pattern to that of wild type, presumably because overexpression of the wild type allele induced similar gene expression changes as did the overexpression of gain-of-function mutants in the short term gene expression assay (Supplementary Fig. S5). These observations demonstrate that the in vivo tumorigenicity assay differentiates gain-of-function mutants even when we were unable to detect a difference in short term in vitro gene expression assays.
In individual tumor assays, PIK3CBE497D showed delayed tumor formation, similar to what we observed when we expressed the wild type PIK3CB (Fig. 4C), implying that E497D is a passenger mutation. Wild type PIK3CB was previously shown to induce foci in a foci formation assay (40). PIK3CBA1048V, on the other hand, induced tumors in the majority of replicates with shorter latency, demonstrating that PIK3CBA1048V is a transforming gain-of-function mutant. In the POT1 allelic series, we noted that only POT1G76V formed tumors in individual tumor experiments after long latency (Fig. 4D). POT1 was recently shown to be mutated in familial melanoma (41, 42), chronic lymphocytic leukemia (43), familial glioma (44), and cardiac angiosarcoma (45). In particular, the Y89C, Q94E, R273L, Y223C, and S270N alleles were previously shown to be loss-of-function alleles, resulting in elongated telomeres and increased genomic instability (41, 42). These observations suggest that POT1G76V may also contribute to cell transformation through a similar mechanism.
Although some of the alleles that we found induced tumor formation were recurrently observed in particular human cancer types, we noted that many of the alleles that we found were able to induce tumor formation, including KRASD33E, KRASE62K, NFE2L2G31R, NFE2L2G31V, NFE2L2N160S, POT1G76V and PIK3CBA1048V, were found to be mutated only once in our set of 5,338 tumors. These observations demonstrate that rare alleles may be functionally important in tumorigenesis.
To investigate whether high throughput functional phenotyping complements in silico predictions, we compared our observations pertaining to 71 alleles analyzed herein to four different in silico methods, Polyphen2 (46), Mutation Assessor (47), CHASM (48), and VEST (49). Each of these methods makes predictions about whether a mutation is likely to affect protein function but does not attempt to predict whether the mutation induces gain or loss of function. To compare these approaches, we used the term “functional variant” to denote both gain-of-function and loss-of-function alleles (50) and “neutral variant” for all other alleles. The concordance rates between each of these methods and our approach ranged from 66% to 77% (Methods; Supplementary Table S6; Supplementary Fig. S6A, S6B), suggesting that gene expression comparisons provided additional information about gene function. For example, Polyphen2 and CHASM predicted that SPOPK134N was likely to be a functional variant while Mutation Assessor and VEST assessed this to be a neutral variant. We found that the gene expression of SPOPK134N correlated with that of SPOPF102C, providing evidence that this allele is a functional variant. Together, these observations suggest that the experimental characterization of alleles complements in silico methods.
DISCUSSION
Cancer genome sequencing projects have already identified thousands of variants of unknown significance, and this number is likely to increase rapidly as more tumors are sequenced. Here we report a pilot study to facilitate functional characterization of these alleles by creating a large number of cancer-associated variants and testing them in two phenotypic assays. Using an in vivo tumorigenesis assay and gene expression profiling, we identified a subset of these variants that exhibit tumorigenic phenotypes and induced changes in gene expression different from that of wild type or known gain-of-function counterparts. This study provides proof of principle evidence that large-scale mutant characterization is both tractable and provides new information about the functional relevance of many alleles.
We recognize that these studies are not exhaustive. For example, we performed all experiments using immortalized kidney epithelial cells, thus limiting those genes that are potentially transforming in a specific tissue context. In addition, the tumorigenesis assay we used here does not assess all tumor-essential phenotypes, and this experimental design does not permit the discovery of loss-of-function tumor suppressor alleles. For example, alleles involved in metastasis, angiogenesis, immune response, and splicing changes may not score in this assay. Weaker transforming alleles may be masked by stronger oncogenic alleles in the pooled format used in these experiments, and it is possible that there are both productive and inhibitory interactions between cells harboring different alleles. Furthermore, alleles that affect pathways that were already perturbed in our engineered system, which include inhibition of TP53 and RB as well as hTERT and MEKDD overexpression, are not likely to be discovered in this context. Also, in cases where presumable mechanisms involve stochastic accumulation of mutations over long time periods, as in the case of genes involved in genomic instability such as POT1, these genes may not reliably score in this context. However, considering the very low background tumor formation rate in this assay, even a single instance of tumor formation lends support for future studies. As such, this approach provides a powerful paradigm to discover functionally relevant rare alleles that may otherwise not be considered for functional studies due to their rarity. Further studies such as those described herein using similar approaches in other genetic and lineage contexts will facilitate the comprehensive discovery of transforming alleles.
Using gene expression signatures generated by expressing wild type or mutant alleles, we found that some PTEN, FBXW7, NRAS, IDH1/2, and SPOP alleles resembled the wild type alleles or known functional variants, suggesting that these alleles are functionally similar to those alleles. On the other hand, in oncogenes such as NFE2L2, we found that gain-of-function mutants induced similar gene expression signatures as the wild type allele. This observation suggests that some truly transforming alleles may not score in the short term in vitro gene expression assay. Furthermore, for genes whose mechanism of action involves longer-term processes such as DNA repair, the acute effect of overexpressing alleles may not be reflected in gene expression changes. Combining expression profiling with tumor formation or other phenotypic experiments may provide complementary information in these cases.
Using the in vivo tumorigenesis assay, we identified rare mutants with transforming function, such as KRASD33E. As this variant was identified only once in the cohort of 5,338 tumors, a large number of tumors would need to be sequenced before the frequency of this allele reached statistical significance. As KRAS mutational status is already used in directing therapeutic decisions (51), this observation demonstrates the importance of studying rare alleles for accurate patient stratification. PIK3CBA1048V and POT1G76V were also rare alleles that were found only once in our cohort. PIK3CB was recently shown to be mutated in prostate cancer (52), and computational analysis using network mutation burden nominated PIK3CB to be a significantly mutated gene (53). Although further studies are required to elucidate the mechanisms by which PIK3CBA1048V and POT1G76V contribute to malignant transformation, this study provides evidence that these alleles are indeed transforming alleles.
In this study, we focused on alleles that have been identified in cancer genome sequencing efforts. An alternative approach would be to create a set of alleles where each amino acid is substituted to prospectively identify alleles that alter wild type gene function and to interrogate the relationship among evolutionary conservation, gene function and prevalence of mutations in tumors. Although this type of study is not yet feasible at the scale presented here, our studies suggest that expanding the number of alleles in genes will provide useful information. We acknowledge that arbitrarily limiting the number of alleles per gene, especially in known cancer genes, excluded some well-studied alleles. Including additional criteria, such as 3D spatial clustering (54), may increase the sensitivity of discovering functional alleles. Expanding the number of alleles in genes, especially those already used in clinical decision-making, is also desirable. Furthermore, high throughput adaptation of other functional assays, such as experiments that quantify morphologic changes as well as proteomic and epigenetic differences will expand our knowledge of the functional consequences of mutant alleles.
In summary, these studies demonstrate that systematically performing functional assays complements the structural information gathered from the sequencing efforts to accelerate the interpretation of cancer associated variants. We anticipate that as additional tumors are characterized in both research and clinical settings, additional cancer associated genes and alleles will be identified, and the approach described here can be useful to ascertain the function of these alleles. Using diverse cellular backgrounds and different phenotypic assays will also increase the power to detect functional variants and reduce false negatives. As more functional data become available, we may also be able to gain insights on empirically improving the accuracy of mutation impact calling algorithms by incorporating information from high confidence functional data. This iterative process between functional and structural genomics will synergistically facilitate the complete description of cancer-associated mutations.
Methods
Mutated gene curation
271 mutated genes were called from the analysis of 5,338 tumor normal pairs by running MutSig2CV and setting the q-value cutoff at 0.1. The algorithm was described previously (13). 13 genes were manually added (PIK3C2G, PIK3R2, PIK3CG, PIK3C2B, PIK3CB, PIK3C2A, PIK3R4, BCL2, BCL3, BCL6, BCL9, BCOR, ISX). 49 likely false positive genes (genes with high background mutation rate) and 48 randomly chosen, likely neutral genes, were added. Total of 381 genes were selected for the project. 220 of these genes had matching template in the hORFeome 8.1 collection and these were used for subsequent steps (Supplementary Table S1).
Selection of alleles from significantly mutated genes
For each missense mutation, “priority” was calculated, which was defined as “density” (local concentration of mutations) multiplied by conservation.
Mutation density was calculated by counting the number of mutations in 20 bp window, with the allele of interest at the center of the window. Conservation was calculated by using phyloP (55), which scores evolutionary conservation from sequence alignment of 46 vertebrates. Conservation values were scaled linearly to range from 0 to 100.
We chose an allele by taking the highest-priority mutated allele. The same procedure was repeated until we selected as many alleles as desired. The number of alleles selected for each gene was decided by the number of times the gene was mutated in patients.
If a gene was mutated in 120 patients or more, then 8 alleles were chosen.
If a gene was mutated in 100 patients or more, then 7 alleles were chosen.
If a gene was mutated in 80 patients or more, then 6 alleles were chosen.
If a gene was mutated in 70 patients or more, then 5 alleles were chosen.
If a gene was mutated in 60 patients or more, then 4 alleles were chosen.
If a gene was mutated in 50 patients or more, then 3 alleles were chosen.
If a gene was mutated in 30 patients or more, then 2 alleles were chosen.
Otherwise, one allele per gene was chosen.
For HRAS, SPOP, MAP2K1, B2M, AKT1, RHOA, IDH1, and IDH2, 8 alleles were chosen.
For genes with one or two alleles selected, we considered all the mutations as ‘experimental’ alleles. For genes with three or more alleles selected, we selected one allele that we predicted to be less likely to be functional as a ‘control’ allele. The other alleles were considered ‘experimental’ alleles. The ‘control’ allele was chosen as an internal control that is less likely than the ‘experimental’ alleles to be functional. The ‘control’ alleles were chosen by the following criteria.
Remove any positions that were chosen above.
Remove any mutations with conservation above a threshold of 60.
For the remaining mutations, define controlpriority = (100 - conservation) / (# of times that exact mutation occurs)^2.
Add a bonus for mutations that are close to the first or second mutations chosen above. If distance between first or second experimental allele and the control allele was less than one fifth of the total gene length, bonus of 20 was given. If distance between first or second experimental allele and the control allele was less than one third of the total gene length, bonus of 10 was given.
Choose the mutated allele with the highest controlpriority + bonus.
All selected alleles are shown in Supplemental Table S1.
Barcoded mutant allele generation in lentiviral vectors
We used a previously published method to perform high-throughput mutagenesis (56). Briefly, each ORF was PCR amplified by using primers that contain mutated sequence incorporated. These fragments were transferred to pDONR223 vector (Invitrogen) through BP cloning (Invitrogen) and the constructs were transformed into competent cells. The discontinuity at the mutation introduction site was repaired by endogenous bacterial repair mechanism. The mutated ORF was transferred to the barcoded destination vector by LR reaction (Invitrogen).
Lentivirus generation
Virus were prepared according to the RNAi Consortium (TRC) virus protocol.
Cell lines
HA1E-M and HA1E cell lines were established in our laboratory and were authenticated by using Fluidigm-enable genotyping assay that queries a set of 96 single nucleotide polymorphism (SNP) markers. Both cell lines were cultured in MEM-alpha (Invitrogen) with 10% FBS (Sigma-Aldrich) and 1% penicillin/streptomycin (Gibco) supplementation. Both cell lines tested negative for mycoplasma.
Multiplexed in vivo screening
All animal experiments were approved by the Institutional Animal Care and Use Committee at the Dana-Farber Cancer Institute. To determine whether the number of cells transduced with a particular allele in a pool of ~80 alleles was sufficient to form tumors, we performed serial dilution and subcutaneous injection with activating KRAS allele, G12V, and found that cells diluted to a representation of 1/96 dilution (approximately 20,000 cells) formed tumors in all injection sites. For the screen, 2,500 HA1E-M cells were plated in 100 ul of media per well in a 96-well plate on day 1. On day 2, polybrene was added to a final concentration of 4 ug/ml and 12 ul of arrayed viral supernatant was added to the target cell plates. Plates were spun at 2,250 rpm for 30 min at room temperature. After 4 hours, media was changed. After 18 hours, puromycin was added to a final concentration of 2ug/ml. After 48 hours of puromycin selection, cells were trypsinized and pooled. 96 wells were combined into one pool per pool composition (Supplementary Table S3). Cell pellets were taken immediately after pooling (called “pre-expansion”), and also on day 15 to use as a reference points for future analysis. Transduced HA1E-M cells were propagated for 15 days to obtain at least 60 million cells per pool. More than 90% of the ORFs in each pool were represented at 0.01% of the injected cell population (Supplementary Fig. S1C, S1D). We note that alleles with even lower representation, such as NFE2L2G31R, which represented 0.0089% of the cells in pre-injection cell pellet of Pool4, formed multiple tumors.
On day 15, cells were trypsinized, washed, and counted (called “pre-injection”). Five million cells were prepped in 200ul of PBS per injection site, except pools 2 and 11, for which 4 million cells were prepped per site. Three sites—inter-scapular area, right and left flanks—were injected in each mouse and four mice were injected per pool (12 sites per pool). Mice were monitored for tumor formation and the longest dimension of each tumor was measured. Tumors were harvested when they reached around 2cm. The tumor tissue was finely minced and subjected to genomic DNA extraction with Qiagen DNeasy blood and tissue kit. 1ug of genomic DNA was subjected to PCR amplification for barcode de-multiplexing by sequencing. To amplify the barcodes with Illumina sequencing primer integrated, following primers were used (different sequence components are demarcated by “<>”):
P5 ORF primer:
<P5 flow cell attachment sequence><Illumina sequencing primer><Vector primer binding>
<AATGATACGGCGACCACCGAGATCT><ACACTCTTTCCCTACACGACGCTCTTCCGATCT[s]><TCTT GTGGAAAGGACGA>
P7 ORF primer:
<P7 flow cell attachment sequence><Barcode><Illumina sequencing primer><Vector primer binding>
<CAAGCAGAAGACGGCATACGAGAT><NNNNNNNN><GTGACTGGAGTTCAGACGTGTGCTCTTCCG ATCT><TAAAGCAGCGTATCCACATAGCGT>
Upon amplification, the PCR products were purified with AMPure beads and subjected to Illumina sequencing. On average, 1.6 million reads were obtained per tumor.
In vivo screening analysis
The barcode reads were de-multiplexed by custom scripts. Less than 1% of contaminating reads (barcode reads that do not belong to the specific pool) were found and removed. The rest of the reads were normalized by dividing the number of reads by the total number of reads from the tumor. Penetrance was calculated by (number of times in which specific allele was represented at more than 0.01%) / (number of times that allele was injected). Since the mouse needs to be sacrificed when the biggest tumor reaches certain diameter per protocol, not all three sites per mouse were observed for full 18 weeks. Maximum enrichment was calculated by (maximum percentage of tumor reads each allele accounted for) – (percentage of that allele in pre-injection cell pellet). We defined “scoring allele” as an allele that was found at more than 1% in at least two tumors or more than 90% in at least one tumor.
Expression profiling using L1000
L1000 is a high-throughput, bead-based gene expression assay in which mRNA is extracted from cultured human cells treated with various chemical or genomic perturbagens (small molecules, gene knockdowns, or gene over-expression constructs). HA1E cells were plated at 400 cells per well in 384 well plates. The next day cells were transduced with 3 μl of lentiviral supernatant by spin infection. Infections were performed in 5 replicates, 2 of which were used to assess infection efficiency and the remaining 3 for gene expression profiling. Following 24-h incubation, media and virus were removed and replaced with complete growth media or media containing antibiotics (for infection efficiency calculation). Cell plates used for gene expression analysis were not selected to reduce the effect of antibiotics on the gene expression. 96 hours after infection, cells were lysed with addition of TCL buffer (Qiagen) and incubated for 30 minutes at room temperature. mRNA is reverse-transcribed into first-strand cDNA. Gene specific probes containing barcodes and universal primer sites are annealed to the first strand cDNA. The probes are ligated to form a template for PCR. The template is PCR amplified with biotinylated universal primers. The end products are biotinylated, fixed length, barcoded amplicons. The amplicons are then mixed with Luminex beads that contain complementary barcodes to those encoded in each of the 978 amplified landmark genes. These beads are then stained with fluorescent streptavidin-phycoerythrin (SAPE) and detected in 384 well plate format on a Luminex FlexMap flow cytometry-based scanner. The resulting readout is a measure of mean fluorescent intensity (MFI) for each landmark gene. The raw expression data are log2-scaled, quantile normalized, and z-scored, such that a differential expression value is achieved for each gene in each well. These differential expression values are collapsed across replicate wells using a weighted average to yield a differential expression signature for each perturbagen. Each replicate is weighted according to its correlation with the others. These signatures were used for subsequent analysis. Detailed protocol is available at LINCS website.
Gene expression correlation analysis
Each normalized gene expression data was filtered by infection efficiency, which was calculated by dividing cell viability after antibiotic selection with cell viability without antibiotic selection by CellTiter-Glo Luminescent Cell Viability Assay (Promega). Viability was assessed 96h post-infection. 40% infection efficiency was used as cutoff to filter inadequately transduced alleles. 1036 gene expression signatures were Spearman correlated with gene expression signature of all other ORFs. “cor(method=”spearman”)” function in R was used for Spearman correlation coefficient calculation (57). Negative controls (BFP, eGFP, HcRed, LacZ, Luciferase), L1000 expression plate controls (NFE2L2, RHEB, NFKB1A, DNMT3A) were also included. After pairwise Spearman correlation, alleles at the extreme ends of the spectrum were manually curated to find alleles that are consistent with previously known relationship.
Stable cell line generation for validation
For individual validation experiments, the same vector used for the pooled screen was used to generate lentiviruses. 80,000 293T cells were plated in one well of 6-well plates. Delta8.9 (900ng), vsv-g (100ng), the ORF vectors (1ug) were transfected in 3ul of TransIT-LT1 Transfection Reagent (Mirus Bio). The viral supernatant was collected after 48 hrs and was frozen at −80C until use. HA1E-M cells were plated in 6-well plate at 100,000 cells per well. HA1E-M cells were transduced with 300ul of viral supernatant in 8ul/ml polybrene and were spin-infected at 2250rpm for 30 minutes. The next day, the media was changed to selection media (puromycin 2ug/ml). After 48 hrs of selection, cells were cultured in puromycin free MEM-alpha complete media (Invitrogen).
Screen validation
Six-week old male homozygous NCR-Nu mice (Taconic) were used for xenograft experiments. HA1E-M cell lines stably expressing individual candidate alleles were injected at two million cells per site, except for NFE2L2 alleles, which were injected at one million cells per site. Each stable cell line was injected at three sites per animal, and into two animals, with the total of six sites per cell line. Tumor formation was monitored using calipers twice weekly for 130 days (or 106 days for ERBB2 alleles). Tumor volume was calculated as ((tumor length)*(tumor width)^2))/2. We defined scoring allele in validation experiment as an allele that formed any tumor larger than 500 mm3 by 130 days.
KRAS structure analysis
KRAS mutations of interest were overlaid onto the structure of the protein product (PDB: 4EPV) and visualized the structure using PyMOL (The PyMOL Molecular Graphics System, Version 1.7.4 Schrödinger, LLC.).
Immunoblots
Protein lysates were resolved on 7.5, 4–12, or 8–16% polyacrylamide SDS gels (Bio-Rad), transferred onto nitrocellulose membranes (Bio-Rad) using standard wet-transfer procedures, and incubated with primary antibodies as indicated. All immunoblot assays were visualized using a LI-COR Odyssey infrared imager. The following antibodies were used: KRAS (Proteintech Group 12063-1-AP), RAS (CST 3965), RAS (clone 10, EMD Millipore 05-516), pERK (CST 4370), ERK (CST 9102), pAKT (S473, CST 4060), a-tubulin (Sigma Aldrich, clone DM1A, T9026), NRF2 (CST 12721), and NRF2 (R&D Systems AF3925) (CST: Cell Signaling Technologies). Secondary anti-rabbit and anti-mouse IRDye antibodies were from LI-COR Biosciences.
RAS activation assay
Ras activation assays were performed according to the manufacturer’s protocol (Millipore 17–218). In brief, cells were cultured on 6-well dishes and harvested for lysates. A sample of each lysate was saved for input (total Ras load) and the remaining lysate was rocked with glutathione-sepharose 1:1 Raf-RBD slurry in lysis buffer for 1 hour at 4°C. The beads were then washed three times with ice-cold lysis buffer, followed by addition of Laemmli/SDS buffer to elute the bound proteins. The Ras-GTP pull-down samples were loaded and resolved on 12% polyacrylamide SDS gels (Bio-Rad).
Quantitative real-time PCR (qPCR)
RNeasy kit (Qiagen) was used to purify total RNA from cells and cDNA was generated using Superscript III Vilo (Life Technologies). Quantitative real-time PCR was performed using SYBR reagents (Life Technologies) on an ABI-7300 instrument following a two-step cycling protocol with the following primers:
NFE2L2_FWD: CACATCCAGTCAGAAACCAGTGG
NFE2L2_REV: GGAATGTCTGCGCCAAAAGCTG
ACTB FWD: CACCATTGGCAATGAGCGGTTC
ACTB REV: AGGTCTTTGCGGATGTCCACGT
Relative expression was calculated using the ΔΔCt method with ACTB for normalization between samples.
Comparison to the in silico methods
We compared our observations to four different in silico methods, Polyphen2 (46), Mutation Assessor (47), CHASM (48), and VEST (49). We used the term “functional variant,” to denote both gain and loss of function alleles (50), and “neutral variant” otherwise. For PolyPhen2, “possibly damaging” and “probably damaging” categories were considered functional variants. We used the HumDiv-trained version of Polyphen2. For Mutation Assessor, “high” and “medium” were considered functional variants. For CHASM and VEST, alleles with FDR <0.05 were considered functional variants. Default parameters were used for PolyPhen2 and Mutation Assessor and “cancer type: other” was chosen for CHASM analysis. The Venn diagram was drawn with Venny (58)
Supplementary Material
SIGNIFICANCE.
Experimentally inferring the functional status of cancer-associated mutations facilitates the interpretation of genomic information in cancer. Pooled in vivo screen and gene expression profiling identified functional variants and demonstrated that expression of rare variants induced tumorigenesis. Variant phenotyping through functional studies will facilitate defining key somatic events in cancer.
Acknowledgments
Financial support: Samsung Scholarship (to E.K.), Susan G. Komen Postdoctoral Fellowship PDF12230602 (to N.I.), Long-term postdoctoral fellowship by the European Molecular Biology Laboratory (to A.K.), Conquer Cancer Foundation of ASCO Young Investigator Award (to S.M.C), U.S. NCI grant, U01 CA176058 (to W.C.H.). This work was conducted as part of the Slim Initiative for Genomic Medicine, a project funded by the Carlos Slim Foundation in Mexico.
We thank I. Fung and B. Wong for assistance with illustrations.
Footnotes
Contributions
E.K., N.I., Y.S, G.G., J.S.B. and W.C.H. conceived the study.
M.S.L., and A.K. led cancer genomics analysis for variant selection.
Y.S., C.Z., X.Y., R.L., N.T., C.N. and D.E.R. generated cDNA reagents.
E.K., N.I., Y.S., C.R.C. and J.G.D. performed tumorigenesis experiments
F.P., M.B., X.W, Y.S. and D E.R. performed gene expression experiments.
E.K., L.Z, L.H., T.N. and S.M.C. performed gene expression analyses.
H.H., K.L., P.T. and S.M.C. advised on analysis strategy.
E.K., N.I., A.K., Y.S. and C.R.C. performed validation experiments.
E.K., J.S.B. and W.C.H. wrote the manuscript.
All authors participated on discussion and commented on the manuscript.
Conflicts of interest: W.C.H. is a consultant and receives grant support from Novartis.
References
- 1.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–24. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garraway Levi A, Lander Eric S. Lessons from the Cancer Genome. Cell. 2013;153:17–37. doi: 10.1016/j.cell.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 3.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer Genome Landscapes. Science. 2013;339:1546–58. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cooper GM, Shendure J. Needles in stacks of needles: finding disease-causal variants in a wealth of genomic data. Nature reviews Genetics. 2011;12:628–40. doi: 10.1038/nrg3046. [DOI] [PubMed] [Google Scholar]
- 5.Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. The New England journal of medicine. 2010;362:2380–8. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 6.Van Allen EM, Wagle N, Levy MA. Clinical analysis and interpretation of cancer genome data. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2013;31:1825–33. doi: 10.1200/JCO.2013.48.7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 8.Douillard JY, Oliner KS, Siena S, Tabernero J, Burkes R, Barugel M, et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. The New England journal of medicine. 2013;369:1023–34. doi: 10.1056/NEJMoa1305275. [DOI] [PubMed] [Google Scholar]
- 9.Garraway LA. Genomics-Driven Oncology: Framework for an Emerging Paradigm. Journal of Clinical Oncology. 2013;31:1806–14. doi: 10.1200/JCO.2012.46.8934. [DOI] [PubMed] [Google Scholar]
- 10.Domchek SM, Greenberg RA. Breast cancer gene variants: separating the harmful from the harmless. The Journal of clinical investigation. 2009;119:2895–7. doi: 10.1172/JCI40577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genetics in medicine : official journal of the American College of Medical Genetics. 2015;17:405–24. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–8. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang X, Boehm JS, Yang X, Salehi-Ashtiani K, Hao T, Shen Y, et al. A public genome-scale lentiviral expression library of human ORFs. Nat Methods. 2011;8:659–61. doi: 10.1038/nmeth.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hahn WC, Counter CM, Lundberg AS, Beijersbergen RL, Brooks MW, Weinberg RA. Creation of human tumour cells with defined genetic elements. Nature. 1999;400:464–8. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- 16.Boehm JS, Zhao JJ, Yao J, Kim SY, Firestein R, Dunn IF, et al. Integrative genomic approaches identify IKBKE as a breast cancer oncogene. Cell. 2007;129:1065–79. doi: 10.1016/j.cell.2007.03.052. [DOI] [PubMed] [Google Scholar]
- 17.Dunn GP, Cheung HW, Agarwalla PK, Thomas S, Zektser Y, Karst AM, et al. In vivo multiplexed interrogation of amplified genes identifies GAB2 as an ovarian cancer oncogene. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:1102–7. doi: 10.1073/pnas.1311909111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peck D, Crawford ED, Ross KN, Stegmaier K, Golub TR, Lamb J. A method for high-throughput gene expression signature analysis. Genome biology. 2006;7:R61. doi: 10.1186/gb-2006-7-7-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, et al. The Common Feature of Leukemia-Associated IDH1 and IDH2 Mutations Is a Neomorphic Enzyme Activity Converting alpha-Ketoglutarate to 2-Hydroxyglutarate. Cancer Cell. 2010;17:225–34. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salmena L, Carracedo A, Pandolfi PP. Tenets of PTEN tumor suppression. Cell. 2008;133:403–14. doi: 10.1016/j.cell.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 21.Nguyen HN, Yang JM, Jr, Rahdar M, Keniry M, Swaney KF, Parsons R, et al. A new class of cancer-associated PTEN mutations defined by membrane translocation defects. Oncogene. 2015;34:3737–43. doi: 10.1038/onc.2014.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tilot AK, Gaugler MK, Yu Q, Romigh T, Yu W, Miller RH, et al. Germline disruption of Pten localization causes enhanced sex-dependent social motivation and increased glial production. Human molecular genetics. 2014;23:3212–27. doi: 10.1093/hmg/ddu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han SY, Kato H, Kato S, Suzuki T, Shibata H, Ishii S, et al. Functional evaluation of PTEN missense mutations using in vitro phosphoinositide phosphatase assay. Cancer Res. 2000;60:3147–51. [PubMed] [Google Scholar]
- 24.Xu J, Li Z, Wang J, Chen H, Fang J-Y. Combined PTEN Mutation and Protein Expression Associate with Overall and Disease-Free Survival of Glioblastoma Patients. Translational oncology. 2014;7:196–205. e1. doi: 10.1016/j.tranon.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Papa A, Wan L, Bonora M, Salmena L, Song MS, Hobbs RM, et al. Cancer-associated PTEN mutants act in a dominant-negative manner to suppress PTEN protein function. Cell. 2014;157:595–610. doi: 10.1016/j.cell.2014.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Myers MP, Pass I, Batty IH, Van der Kaay J, Stolarov JP, Hemmings BA, et al. The lipid phosphatase activity of PTEN is critical for its tumor supressor function. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:13513–8. doi: 10.1073/pnas.95.23.13513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44:685–9. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le Gallo M, O’Hara AJ, Rudd ML, Urick ME, Hansen NF, O’Neil NJ, et al. Exome sequencing of serous endometrial tumors identifies recurrent somatic mutations in chromatin-remodeling and ubiquitin ligase complex genes. Nat Genet. 2012;44:1310–5. doi: 10.1038/ng.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.An J, Ren S, Murphy SJ, Dalangood S, Chang C, Pang X, et al. Truncated ERG Oncoproteins from TMPRSS2-ERG Fusions Are Resistant to SPOP-Mediated Proteasome Degradation. Molecular cell. 2015;59:904–16. doi: 10.1016/j.molcel.2015.07.025. [DOI] [PubMed] [Google Scholar]
- 30.Gan W, Dai X, Lunardi A, Li Z, Inuzuka H, Liu P, et al. SPOP Promotes Ubiquitination and Degradation of the ERG Oncoprotein to Suppress Prostate Cancer Progression. Mol Cell. 2015;59:917–30. doi: 10.1016/j.molcel.2015.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cancer Genome Atlas Research N. Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang P, Gao K, Jin X, Ma J, Peng J, Wumaier R, et al. Endometrial cancer-associated mutants of SPOP are defective in regulating estrogen receptor-alpha protein turnover. Cell death & disease. 2015;6:e1687. doi: 10.1038/cddis.2015.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Geng C, Rajapakshe K, Shah SS, Shou J, Eedunuri VK, Foley C, et al. Androgen receptor is the key transcriptional mediator of the tumor suppressor SPOP in prostate cancer. Cancer Res. 2014;74:5631–43. doi: 10.1158/0008-5472.CAN-14-0476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–17. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davis RJ, Welcker M, Clurman BE. Tumor suppression by the Fbw7 ubiquitin ligase: mechanisms and opportunities. Cancer Cell. 2014;26:455–64. doi: 10.1016/j.ccell.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akhoondi S, Sun D, von der Lehr N, Apostolidou S, Klotz K, Maljukova A, et al. FBXW7/hCDC4 is a general tumor suppressor in human cancer. Cancer Res. 2007;67:9006–12. doi: 10.1158/0008-5472.CAN-07-1320. [DOI] [PubMed] [Google Scholar]
- 38.O’Neil J, Grim J, Strack P, Rao S, Tibbitts D, Winter C, et al. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to gamma-secretase inhibitors. The Journal of experimental medicine. 2007;204:1813–24. doi: 10.1084/jem.20070876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun Q, Burke JP, Phan J, Burns MC, Olejniczak ET, Waterson AG, et al. Discovery of small molecules that bind to K-Ras and inhibit Sos-mediated activation. Angewandte Chemie. 2012;51:6140–3. doi: 10.1002/anie.201201358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kang S, Denley A, Vanhaesebroeck B, Vogt PK. Oncogenic transformation induced by the p110beta, -gamma, and -delta isoforms of class I phosphoinositide 3-kinase. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:1289–94. doi: 10.1073/pnas.0510772103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robles-Espinoza CD, Harland M, Ramsay AJ, Aoude LG, Quesada V, Ding Z, et al. POT1 loss-of-function variants predispose to familial melanoma. Nat Genet. 2014;46:478–81. doi: 10.1038/ng.2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi J, Yang XR, Ballew B, Rotunno M, Calista D, Fargnoli MC, et al. Rare missense variants in POT1 predispose to familial cutaneous malignant melanoma. Nat Genet. 2014;46:482–6. doi: 10.1038/ng.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramsay AJ, Quesada V, Foronda M, Conde L, Martinez-Trillos A, Villamor N, et al. POT1 mutations cause telomere dysfunction in chronic lymphocytic leukemia. Nat Genet. 2013;45:526–30. doi: 10.1038/ng.2584. [DOI] [PubMed] [Google Scholar]
- 44.Bainbridge MN, Armstrong GN, Gramatges MM, Bertuch AA, Jhangiani SN, Doddapaneni H, et al. Germline mutations in shelterin complex genes are associated with familial glioma. J Natl Cancer Inst. 2015;107:384. doi: 10.1093/jnci/dju384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Calvete O, Martinez P, Garcia-Pavia P, Benitez-Buelga C, Paumard-Hernandez B, Fernandez V, et al. A mutation in the POT1 gene is responsible for cardiac angiosarcoma in TP53-negative Li-Fraumeni-like families. Nature communications. 2015;6:8383. doi: 10.1038/ncomms9383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39:e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carter H, Chen S, Isik L, Tyekucheva S, Velculescu VE, Kinzler KW, et al. Cancer-specific high-throughput annotation of somatic mutations: computational prediction of driver missense mutations. Cancer Res. 2009;69:6660–7. doi: 10.1158/0008-5472.CAN-09-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carter H, Douville C, Stenson PD, Cooper DN, Karchin R. Identifying Mendelian disease genes with the variant effect scoring tool. BMC genomics. 2013;14(Suppl 3):S3. doi: 10.1186/1471-2164-14-S3-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gonzalez-Perez A, Mustonen V, Reva B, Ritchie GR, Creixell P, Karchin R, et al. Computational approaches to identify functional genetic variants in cancer genomes. Nat Methods. 2013;10:723–9. doi: 10.1038/nmeth.2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stephen AG, Esposito D, Bagni RK, McCormick F. Dragging ras back in the ring. Cancer Cell. 2014;25:272–81. doi: 10.1016/j.ccr.2014.02.017. [DOI] [PubMed] [Google Scholar]
- 52.Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–28. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Horn H, Lawrence MS, Hu JX, Worstell E, Ilic N, Shrestha Y, et al. A comparative analysis of network mutation burdens across 21 tumor types augments discovery from cancer genomes. 2015 bioRxiv. [Google Scholar]
- 54.Kamburov A, Lawrence MS, Polak P, Leshchiner I, Lage K, Golub TR, et al. Comprehensive assessment of cancer missense mutation clustering in protein structures. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E5486–95. doi: 10.1073/pnas.1516373112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–21. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Suzuki Y, Kagawa N, Fujino T, Sumiya T, Andoh T, Ishikawa K, et al. A novel high-throughput (HTP) cloning strategy for site-directed designed chimeragenesis and mutation using the Gateway cloning system. Nucleic Acids Res. 2005;33:e109. doi: 10.1093/nar/gni103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2014. [Google Scholar]
- 58.Venny Oliveros JC. An interactive tool for comparing lists with Venn’s diagrams. [Internet] 2007–2015 Available from: http://bioinfogp.cnb.csic.es/tools/venny/index.html.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.