Abstract
The impact of somatic missense mutation on cancer etiology and progression is often difficult to interpret. One common approach for assessing the contribution of missense mutations in carcinogenesis is to identify genes mutated with statistically nonrandom frequencies. Even given the large number of sequenced cancer samples currently available, this approach remains underpowered to detect drivers, particularly in less studied cancer types. Alternative statistical and bioinformatic approaches are needed. One approach to increase power is to focus on localized regions of increased missense mutation density or hotspot regions, rather than a whole gene or protein domain. Detecting missense mutation hotspot regions in three dimensional protein structure may also be beneficial, because linear sequence alone does not fully describe the biologically relevant organization of codons. Here, we present a novel and statistically rigorous algorithm for detecting missense mutation hotspot regions in 3D protein structures. We analyze ~3×105 mutations from The Cancer Genome Atlas (TCGA) and identify 216 tumor-type-specific hotspot regions. In addition to experimentally determined protein structures we consider high-quality structural models, which increases genomic coverage from ~5,000 to more than 15,000 genes. We provide new evidence that 3D mutation analysis has unique advantages. It enables discovery of hotspot regions in many more genes than previously shown and increases sensitivity to hotspot regions in tumor suppressor genes. While hotspot regions have long been known to exist in both tumor suppressor genes and oncogenes, we provide the first report that they have different characteristic properties in the two types of driver genes. We show how cancer researchers can use our results to link 3D protein structure and the biological functions of missense mutations in cancer, and to generate testable hypotheses about driver mechanisms. Our results are included in a new interactive website for visualizing protein structures with TCGA mutations and associated hotspot regions. Users can submit new sequence data, facilitating the visualization of mutations in a biologically relevant context.
Keywords: Molecular modeling, hotspot mutation regions, missense mutations
Major Findings
We use TCGA mutation data and identify 3D clusters of cancer mutations (“hotspot regions”) at amino-acid-residue resolution, in 91 genes of which 56 are known cancer-associated genes. The hotspot regions identified by our method are smaller than a protein domain or protein-protein interface and in many cases can be linked precisely with functional features such as binding sites, active sites, and sites of experimentally characterized mutations. The hotspot regions are shown to be biologically relevant to cancer, and we discover that there are characteristic differences between regions in the two types of driver genes: oncogenes and tumor suppressor genes. These differences include region size, mutational diversity, evolutionary conservation, and amino acid residue physiochemistry. For the first time, we quantify why the great majority of well-known hotspot regions occur in oncogenes. Because hotspot regions in tumor suppressor genes are larger, more heterogeneous than those in oncogenes, they are more difficult to detect using protein sequence alone and are likely to be underreported. Our results indicate that protein structure-based 3D mutation clustering increases power to find hotspot regions, particularly in tumor suppressor genes.
Quick Guide
An experimentally-determined or theoretically modeled protein structure consists of a set of atoms, each with a unique coordinate in three-dimensional Euclidean space. Each amino acid residue consists of many atoms and may harbor zero, one or multiple missense mutations in a cohort of sequenced cancer samples. Two key mathematical concepts in our study are the density of local missense mutations in 3D space, which underlies our statistical measure to define missense mutation hotspot regions, and mutational diversity of a hotspot region. Local missense mutation density
is defined for each amino acid residue r and each protein structure k. It considers the sum of the count of missense mutations that occurred at r and those that occurred at residues proximal to r i.e., in its “neighborhood”. Proximity is measured in 3D space and the neighborhood is limited to residues up to 1 nm away from r, where 1 nm was chosen because it is the order of magnitude of an amino acid side chain. The term is the missense mutation count for the nth residue neighbor of r. The observed value of is compared to simulations of its value under an empirical null distribution, where the total number of missense mutations observed in k remains the same, but they are distributed uniformly in 3D. Residue r has significantly increased if its adjusted P-value ≤ 0.01 after multiple testing correction. A 3D hotspot region is a grouping of residues with significantly increased that are linked as connected components in a neighbor graph. Our algorithm can find 3D hotspot regions directly on protein complexes, enabling detection of hotspot regions that occur on both sides of a protein-protein interface. It also handles complexes with multiple chains originating from a single gene product (e.g., a homodimer) by running identical simulations simultaneously.
Mutational diversity is computed for each hotspot region (where i indexes the region and g indexes the gene) based on the Shannon entropy of the joint probability of a missense mutation occurring at a specific residue r and having a specific mutant amino acid m
Because the maximum possible Shannon entropy grows with the number of residues in a hotspot region, the score is normalized so hotspot regions of different sizes can be compared.
N is the number of mutations in the hotspot region, R is the number of residues, and A is the number of possible alternate amino acids per residue. In this work, mutational diversity is found to be significantly different between hotspot regions that occur in oncogenes vs. those that occur in tumor suppressor genes.
Major assumptions of the model:
In the absence of selection for drivers, somatic missense mutations in cancers are equally likely to appear at any amino acid residue position in a protein structure of interest.
Many driving missense mutations have significantly increased local mutation density.
Residues with significantly increased mutation density and proximal to each other in three dimensions are likely to be subject to similar selective pressures and can be grouped together into hotspot regions.
The most parsimonious number of hotspot regions in a protein structure is preferred.
Carefully filtered theoretical protein structure models are accurate enough to capture local missense mutation densities and groupings of proximal residues with significantly increased densities.
Introduction
Missense mutations are perhaps the most difficult mutation type to interpret in human cancers. Truncating loss-of-function mutations and structural rearrangements generate major changes in the protein product of a gene, but a single missense mutation yields only a small change in protein chemistry. The impact of missense mutation on protein function, cellular behavior, cancer etiology and progression may be negligible or profound, for reasons that are not yet well understood. Missense mutations are frequent in most cancer types, accounting for ~85% of the somatic mutations observed in solid human tumors (1), and the cancer genomics community has prioritized the task of identifying important missense mutations discovered in sequencing studies. Whole-exome sequencing (WES) studies of cancer have created new opportunities to better understand the importance of missense mutations. This enormous collection of data now allows detection of patterns with power that was unheard of a few years ago.
The first approaches to identify cancer drivers from WES mutations looked for significantly mutated genes (SMGs), harboring a larger number of somatic mutations than expected by chance (2–5). Metrics to call SMGs now appear to be underpowered given the size of current cohorts in the Cancer Genome Atlas (TCGA) and International Cancer Genome Consortium (ICGC). A recent study suggested that ~1500 cases of endometrial cancer would need to be sequenced in order to attain 90% power to detect mutations in 90% of genes with a mutation frequency of 2% with the SMG approach (5). The recognition of the limitations of the SMG paradigm has motivated interest in orthogonal analysis techniques to detect mutational patterns associated with drivers (1,6–9).
Recurrence of somatic missense mutations in cancers at the same amino acid residue position is well known to be a characteristic feature of both oncogenes (OGs) and tumor suppressor genes (TSGs) (10). The observation that somatic mutations also frequently occur in positions proximal in protein sequence to the most highly recurrent positions has suggested that positional clustering of somatic missense mutations might be used to identify drivers (7). These clusters, known as “hotspots”, are regions where somatic missense mutations occur closer together in protein sequence than would be expected by chance. Hotspot regions can be rationalized as areas in a protein under positive selection in the cancer environment; missense mutations occurring in these regions are selected for because they alter protein function in a manner advantageous to the cancer cell. Several algorithms have been developed to identify protein functional domains and genes in which these regions are enriched (8,11) and to identify specific missense mutations in hotspots (7,12). These algorithms consider mutations in a coordinate system based on one-dimensional (1D) protein sequence. Whole-exome sequencing studies of cancer cohorts are increasingly incorporating missense “hotspot” detection as a routine analysis step in the search for new drivers.
Finding missense mutation hotspot regions in 1D is limited by the fact that functional proteins tend to fold into three-dimensional (3D) structures (with the exception of intrinsically disordered regions). Thus, positional clustering done in 1D will likely miss many hotspots that are present in 3D after folding. Gene- and protein domain-level testing may indicate the possibility of a 3D hotspot but cannot identify the specific positions in the hotspot. An algorithm that leverages 3D protein structure information, but still performs clustering in 1D through a dimensionality reduction step, has shown utility in detecting OGs (9). A recent study of an aggregated collection of TCGA cancer mutations from 21 tumor types presented an algorithm to identify cancer genes based on 3D clustering of somatic missense mutations, yielding ten such genes. They reported low correlation between 3D and 1D hotspot regions (13).
Here we present HotMAPS (Hotspot Missense mutation Areas in Protein Structure), a new, sensitive algorithm and a web-based community resource for high-throughput analysis of cancer missense mutation 3D hotspot regions. HotMAPS finds clusters of amino acid residues with significantly increased local mutation density in 3D protein space, compared to an empirical null distribution. The statistical model is designed to handle higher-order protein complexes and can capture regions that span protein-protein interfaces. We apply HotMAPS to missense mutations from 23 tumor types sequenced by TCGA. By careful use of both experimentally-derived protein biological assemblies in the Protein Data Bank (PDB) and theoretical protein structure models, we substantially increase the number of amino acids that can be mapped into 3D protein space and the number of detectable hotspot regions (13).
HotMAPS systematically delineates 3D hotspot regions on the level of amino acid positions, and we provide a detailed catalog of 216 tumor-type-specific regions. We show how the catalog can be used as a discovery tool so that the links between 3D protein structure and the biological functions of missense mutations in cancer can be better utilized by the community. The catalog provides comprehensive identification of hotspot regions that overlap with many key biological features of proteins available in the literature (e.g., residue positions at active sites, small-molecule and metal-binding sites, protein interfaces, positions with published experimental mutagenesis results). This information can potentially provide a researcher with more fine-grained mechanistic understanding of missense mutation cancer relevance than is possible by 1D clustering or domain and gene enrichment approaches. Using the catalog, we are able for the first time to systematically analyze characteristic properties of 3D hotspot regions and differences between 3D hotspot regions in OGs and TSGs.
Materials and Methods
TCGA mutation collection
TCGA mutation annotation format (MAF) file data for 23 tumor types was downloaded from Xena data store (https://genome-cancer.soe.ucsc.edu/proj/site/xena/hub/) using their API.
3D protein structure and theoretical model collection and processing
PDB structures were obtained from the Worldwide Protein Data Bank (PDB) (10/17/2015). Only structures solved by x-ray crystallography and containing at least one human protein chain were used. Single-domain, theoretical protein structure models constructed based on homology to non-human proteins were included to increase coverage over a greater proportion of genes. Theoretical models were obtained from the ModPipe human 2013 dataset (ftp://salilab.org/databases/modbase/projects/genomes/H_sapiens/2013/), built with Modeller 9.11 (14). In addition to criteria required by ModPipe, we filtered the theoretical models to increase the quality of structures used in our assessment based on minimum length, target-template sequence identity, loop content and radius of gyration (Supplementary Materials and Methods).
Models were assessed by comparing 3D hotspot regions identified by HotMAPS in experimental structures with those identified in theoretical models of the same protein. First, we found all pairs of experimental structures and theoretical models of the same protein, in which there was overlap of the same amino acid residues. The agreement of a structure/model pair was the overlap of their hotspot region mutated residues. A false positive error was called when a model had a mutated residue in a hotspot region that was not in a hotspot region for any protein structure that it had been paired with. A false negative error was called when a structure had a mutated residue in a hotspot region that was not in a hotspot region for any of the models it had been paired with.
HotMAPS algorithm
HotMAPS identifies residue positions with higher local mutation density in each protein structure or model than expected from an empirical null distribution, based on simulations of a discrete uniform distribution. Residues are considered significant for increased local mutation density at FDR threshold of 0.01, after correction for multiple testing (Benjamini Hochberg). 3D missense mutation hotspots are identified as groupings of significant residues according to the principle of maximum parsimony, based on connected components in a neighbor graph. Construction of the neighbor graph and connected components are illustrated in Fig S1. HotMAPS is designed to run on both single-chain protein structures and biological assemblies with multiple chains originating from the same gene. Mathematical details are provided in Supplementary Materials and Methods.
Results
3D missense mutation hotspot regions identified in TCGA whole-exome sequencing
Mutation hotspot regions detectable in 3D
Applying HotMAPS to 19,368 PDB protein structures (PDB bioassemblies in which in vivo protein structure is represented) and 46,004 theoretical models, we identified 107 unique 3D mutation hotspot regions (aggregated across tumor types), of which 30 were only detectable by clustering in 3D (Table S1). When stratified by tumor type, 216 3D missense mutation hotspot regions were found in 19 out of the 23 TCGA tumor types, with none in Adrenocortical carcinoma (ACC), Kidney Renal Papillary Cell Carcinoma (KIRP), Liver hepatocellular carcinoma (LIHC) or Kidney chromophobe (KICH) (Table S2). KICH is known to be driven by alterations other than point mutation, such as structural breakpoints in the TERT promoter. Among all 23 tumor types, sample and mutation count is lower for these four tumor types (p=0.02 for sample count, p=0.04 for mutation count; Wilcoxon Rank Sum test), suggesting that at least for tumor types driven by missense mutations, larger sample size might increase our power to find more 3D regions. Our approach enabled us to consider the three-dimensional protein environment of a much higher fraction of TCGA mutations than has been described previously. We were able to map and analyze ~53% of the missense mutations in 23 TCGA tumor types (Table S3). Of these missense mutations, ~10% could be mapped to PDB protein structures and an additional 42% mapped to theoretical models, in the absence of PDB structure. Using hotspot regions identified in the PDB structures as a control, we estimate that the hotspots called in the models have a false positive rate of 0.058 and a false negative rate of 0.138. Therefore, very few hotspot regions found in the models are the result of modeling errors, justifying the increase in mutation coverage obtained.
Genes that harbor 3D mutation hotspot regions
When hotspot regions were stratified by tumor type, 91 genes contained at least one hotspot region in at least one tumor type (q=0.01) and 40 of these genes had regions that were only discoverable by consideration in 3D. Of the 91, 19 genes were previously annotated as OGs and 11 genes as TSGs with the 20/20 rule, a ratiometric method based on the proportions of different mutation consequence types observed in a gene (1). Twenty-five of the genes are listed in the Cancer Gene Census (CGC) (15) (Table 1). Of the remaining 58 genes, five (KL5, SMARCA2, RASA1, TGBFR2, KEAP1) have been identified as candidate TSGs in the literature (16–18) (19–24), and six (de-acetylated KL5, MAPK1, FSIP2, RANBP2, MTOR, CHEK2) as candidate OGs (16,25–30). Three of the genes are current or potential drug targets (SMARCA2, HDAC4, PARG, HLA-A) (18, 31–33). Two genes (ERCC2, CHEK2) are involved in hereditary cancer susceptibility when mutated in the germline (1). GTF2I is a prognostic biomarker in thymic epithelial tumors (34) (genes with literature support in Table 2, other genes in Table S4).
Table 1.
Cancer genes with 3D HotMAPS regions identified in TCGA tumor types and in Landscapes Benchmark or Cancer Gene Census.
| Gene | Landscapes Benchmark | Cancer Gene Census (CGC) | TCGA Tumor Type(s) |
|---|---|---|---|
| FGFR3 | OG | Dom | BLCA |
| SF3B1 | OG | Dom | BRCA, BLCA |
| FGFR2 | OG | Dom | BRCA, UCEC |
| KRAS | OG | Dom | CESC, UCS, PAAD, STAD, BLCA, UCEC, LUAD, BRCA |
| PIK3CA | OG | Dom | ESCA, CESC, UCS, LUSC, GBM, STAD, LGG*, BLCA, UCEC, PRAD, LUAD, KIRC, BRCA, HNSC |
| NFE2L2 | OG | Dom | ESCA, HNSC, BLCA, UCEC, LUSC |
| IDH1 | OG | Dom | GBM, LGG, SKCM |
| IDH2 | OG | Dom | LGG |
| PTPN11 | OG | Dom | LGG |
| MAP2K1 | OG | Dom | LUAD*, SKCM |
| GNAS | OG | Dom | PAAD |
| BRAF | OG | Dom | THCA, GBM, LUAD, SKCM, PRAD* |
| HRAS | OG | Dom | THCA, PCPG, BLCA, HNSC, LUSC* |
| NRAS | OG | Dom | THCA, SKCM |
| PPP2R1A | OG | Dom? | UCS, UCEC |
| SPOP | OG | Rec | PRAD |
| ERBB2 | OG | ESCA*, BRCA, BLCA | |
| EGFR | OG | GBM, LGG, LUAD | |
| RET | OG | PCPG | |
| PIK3R1 | TSG | Rec | BRCA*, GBM, UCEC*, LGG* |
| FBXW7 | TSG | Rec | CESC*, UCS, LUSC*, STAD, BLCA, UCEC, HNSC |
| TP53 | TSG | Rec | ESCA, UCS, PAAD, LUSC, GBM, STAD, LGG, BLCA, UCEC, PRAD, LUAD, OV, BRCA, HNSC |
| CIC | TSG | Rec | LGG |
| SMARCA4 | TSG | Rec | LGG* |
| BCOR | TSG | Rec | UCEC |
| PTEN | TSG | BRCA, GBM*, UCEC | |
| CDKN2A | TSG | ESCA* | |
| VHL | TSG | KIRC* | |
| NOTCH1 | TSG | LGG* | |
| SMAD4 | TSG | STAD* | |
| RHOA | Dom | BLCA*, HNSC, STAD | |
| RAC1 | Dom | HNSC, SKCM | |
| ERBB3 | Dom | STAD |
OG=oncogene. TSG=tumor suppressor gene (Landscapes benchmark). Cancer Gene Census Dom=dominant. Rec=recessive. Dom?=probably dominant. TCGA tumor types = tumor types in which the gene had a significant 3D mutation hotspot region (q=0.01). ACC=Adrenocortical carcinoma, BLCA=Bladder Urothelial Carcinoma, BRCA=Breast Invasive Carcinoma, CESC=Cervical squamous cell carcinoma and endocervical adenocarcinoma, ESCA=Esophagael Carcinoma, GBM=Glioblastoma Multiforme, HNSC=Head and Neck squamous cell carcinoma, KICH=Kidney chromophobe, KIRC=Kidney renal clear cell carcinoma, KIRP= Kidney Renal Papillary Cell Carcinoma, LGG=low-grade glima, LIHC=Liver hepatocellular carcinoma, LUAD=Lung adenocarcinoma, LUSC=Lung squamous cell carcinoma, OV=Ovarian serous cystadenocarcinom, PAAD=Pancreatic adenocarcinoma, PCPG=Pheochromocytoma and Paraganglioma, PRAD=Prostate adenocarcinoma, SKCM=Skin Cutaneous Melanoma, STAD=Stomach adenocarcinoma, THCA=Thyroid carcinoma, UCEC=Uterine Corpus Endometrial Carcinoma and UCS=Uterine Carcinosarcoma.
At least one 3D hotspot region in the gene/tumor type was not detected with the 1D-only version of the algorithm.
Table 2. Genes with HotMAPS regions identified in TCGA tumor types.
Genes that are candidate OGs and TSGs, hereditary cancer genes, associated with cancer phenotypes and drug targets.
| Gene | TCGA Tumor Type(s) | Gene Details |
|---|---|---|
| AP2B1 | ESCA | Involved in FGFR signaling. Knockdown promotes the formation of matrix degrading invadopodia, adhesion structures linked to invasive migration in cancer cells (Pignatelli 2012). |
| CAND1 | BLCA* | Component of many protein complexes involved in proteasome-dependent protein degredation via ubiquitination and neddylation. CAND1 binding to the complexes inactivates ubiquitin ligase activity and may block adaptor and NEDD8 conjugation sites. (Bosu 2008). May play a role in PLK4-mediated centriole overduplication and Disrupted in prostate cancer (Korzeniewski 2012). |
| CHEK2 | ESCA, LGG, BLCA, HNSC, PRAD, LUAD, PCPG, KIRC | Checkpoint kinase involved in DNA damage response signaling. Significantly mutated gene and candidate OG in papillary thyroid carcinoma (PTC) cohort of 296 patients (TCGA 2014 #85). Breast cancer susceptibility gene (inherited germline variants) (Vogelstein 2013) |
| CUL1 | BLCA | Candidate TSG. SCF complex E3 ubiquitin ligase scaffold protein. Suppressor of centriole multiplication through regulation of PLK4 level (Korzeniewski 2009) |
| ERCC2 | BLCA, LGG* | DNA-repair (Nucleotide excision repair) protein. Significantly mutated in cisplatin-responders vs. non-responders in cohort of 50 patients with muscle-invasive urothelial carcinoma (MIUC). ERCC2 mutation status may inform cisplatin-containing regimen usage in MIUC (Van Allen 2014). Recurrently mutated in cohort of 17 patients with urothelial bladder cancer (UBC) (Balbas-Martinez 2013). Xeroderma pigmentosum susceptibility gene (inherited germline variants) (Vogelstein 2013) |
| FSIP2 | ESCA* | Candidate OG. Recurrently amplified in testicular germ cell tumors (TGCTs)(Litchfield 2015). |
| GNA13 | BLCA | Significantly mutated in cohort of 55 patients with diffuse large B-cell lymphoma (DLBCL) (Lohr 2012) |
| GTF2I | UCEC | Highly recurrent missense mutation in Thymic epithelial tumors and associated with increased patient survival (Petrini 2014). |
| HDAC4 | ESCA | Histone de-aceytlation enzyme. Drug target. Overexpression shown to promote growth of colon cancer cells via p21 repression. Regulator of colon cell proliferation. (Wilson 2008). May regulate cancer cell response to hypoxia via its regulates HIF1a acetylation and stability (Geng 2011) |
| HLA-A | BLCA, HNSC, LGG, PRAD | Immune system. Encodes MHC-Class 1A protein, which presents antigens for T cell recognition. Somatic mutations previously suggested to contribute to tumor immune escape (Shukla 2015). |
| KEAP1 | LUAD* | Candidate TSG. Inhibits NRF2 (aka NFE2L2). In cohort of 76 non-small cell lung cancer (NSCLC) patients, KEAP1 found mutated in 2 patients with advanced adenocarcinoma and smoking history. KEAP1 mutation was mutually exclusive of EGFR, Kas, ERBB2 and NFE2L2 mutation in the cohort and KEAP1 mutation status proposed as marker for personalized therapy selection. (Sasaki 2013) Proposed TSG in lung squamous cell carcinomas (Hast 2014) Proposed as therapeutic target for thyroid-transcription-factor-1 (TTF1)-negative lung adenocarcinoma (LUAD) (Cardnell 2015). |
| KLF5 | BLCA* | Transcription factor that promotes breast cancer cell proliferation, survival, migration and tumour growth. Upregulates TNFAIP2, which interacts with the two small GTPases Rac1 and Cdc42, thereby increasing their activities to change actin cytoskeleton and cell morphology (Jia 2015). Proposed as playing dual role as both TSG when acetylated and OG when de-acetylated in prostate cancer (Atala 2015). Recurrently mutated in mucinous ovarian carcinoma (Ryland 2015) |
| MAPK1 | CESC*, HNSC | Kinase involved in cell proliferation, differentiation, transcription regulation, and development; key signaling component of the toll-like receptor pathway. Candidate OG in pancreatic cancer (Furukawa 2006), laryngeal squamous cell carcinoma cell lines (Kostrzewska-Poczekaj 2010). Significantly mutated in cohort of 91 chronic lymphocytic leukemia CLL patients.(Wang 2011). |
| MSN | ESCA* | Protein homolog of TSG NF2 (Merlin) (Golovnina 2005). Member of the Ezrin-Radixin-Moesin (ERM) protein family. Links membrane and cytoskeleton involved in contact-dependent regulation of EGFR (Chiasson-MacKenzie 2015). Regulates the motility of oral cancer cells via MT1-MMP and E-cadherin/p120-catenin adhesion complex. Cytoplasmic expression of MSN correlates with nodal metastasis and poor prognosis of oral squamous cell carcinomas (OSCCs), may be potential candidate for targeted gene therapy for OSCCs (Li 2015). |
| MTOR | KIRC | Candidate OG. Serine/threonine protein kinase regulates cell growth, proliferation and survival. Frequently activated in human cancer and a major therapeutic target. Randomly selected mutants in HEAT repeats and kinase domain induced transformation in NIH3T3 cells and rapid tumor growth in nude mice (Mueugan 2013) |
| NBPF10 | BLCA* | Somatic missense mutation reported in prostate cancer cohort of 141 patients (Manson-Bahr 2015). In gene family with numerous tandem repeats and pseudogenes, possible read alignment and mutation calling errors. |
| PARG | GBM, LGG, BLCA, HNSC, PRAD, LUAD, PCPG, KIRC* | Involved in DNA damage repair (with PARP1). Cells deficient in these proteins are sensitive to lethal effects of ionizing radiation and alkylating agents (17). Potential Drug target for BRCA2-deficient cancers (Fathers 2012). |
| RANBP2 | ESCA | Candidate OG (Gylfe 2013). A large multimodular and pleiotropic protein with SUMO E3 ligase function. (Zhu 2015) Interacts with mTOR (to regulate cell growth and proliferation via cellular anabolic processes) (Kazyken 2014). Hot spot mutation previously found in MSI colorectal cancer (CRC). Hot spot suggested as useful for personalized tumor profling and therapy in CRC. (Gylfe 2013) |
| RASA1 | HNSC* | Identified as TSG in another squamous cell cancer, cutaneous squamous cell skin cancer (cSCC) (Pickering 2014) |
| RGPD3 | BLCA*, UCEC, PAAD | Component of ubiqutin E3 ligase complex. Named for similarity to RANBP2. |
| SIRPB1 | HNSC, PRAD | Ig-like cell-surface receptor. Negatively regulates RTK processes. Related to FGFR signaling. |
| SMARCA2 | BLCA* | Actin-dependent regulator of chromatin. Its ATPase domain named as Drug target in SWI/SNF mutant cancers (e.g., lung, synovial sarcoma, leukemia, and rhabdoid tumors) (Vangamudi 2015). Proposed TSG, and synthetic lethal target in SMARCA4 (aka BRG1)-deficient cancers.(Hoffman 2014) |
| TGFBR2 | HNSC | TSG in HNSC (Rothenberg,2012) MSI CRC (Biswas 2008), epithelial transformation and invasive squamous cell carcinoma in the mouse forestomach (Yang 2014). |
At least one 3D hotspot region in the gene/tumor type was not detected with the 1D-only version of the algorithm.
3D mutation hotspot regions are important in cancer
3D Hotspot regions are enriched in well-known cancer genes
Amongst the set of genes with available protein structure or models (n=15,697), the genes harboring a 3D hotspot region are enriched for OGs and TSGs (p=6.1E-30 for OGs and p=2.4E-13 for TSGs; one-tailed Fisher’s Exact Test). They are also enriched for genes in the CGC list (p=1.4E-30; one-tailed Fisher’s Exact Test). The subset of these genes harboring only a 3D hotspot region not detectable in 1D is also significantly enriched (p=4.3E-09 for OGs, p=7.9E-12 for TSGs, p=8.0E-11 for CGC genes; one-tailed Fisher’s Exact Test). An additional 23 genes that are proposed OGs, TSGs and/or drug targets or hereditary cancer genes contained at least one 3D hotspot region. This enrichment of known and candidate driver genes supports our claim that many of the regions are biologically relevant and not simply artifacts. While regions were detected in only ~18% of established cancer genes, we expect that many of these genes harbor drivers other than missense mutations, some are drivers in tumor types not represented in our study and many lack structural coverage.
Mutations in 3D Hotspot regions are different from other somatic mutations in cancers
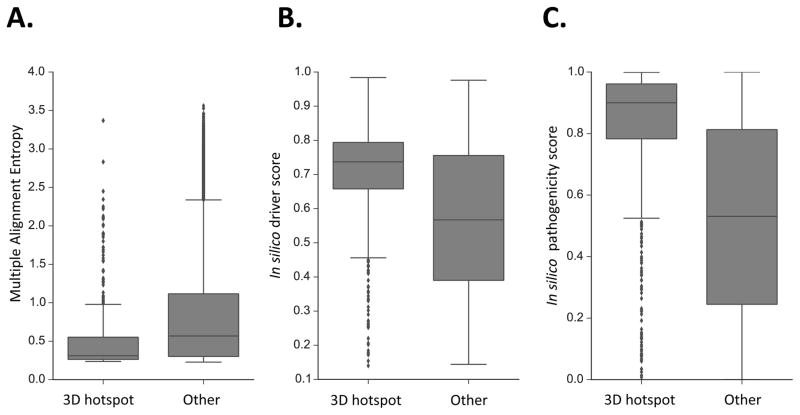
We examined whether the amino acid residue positions and the missense mutations in the 3D hotspot regions had distinctive features suggestive of a special biological importance, when compared to the remaining mutations in our study. Four candidate distinguishing features were tested: 1) vertebrate evolutionary conservation; 2) occurrence at a protein-protein interface, which increases the potential for a missense mutation to disrupt protein-protein interactions; 3) in silico cancer driver scores generated with the CHASM algorithm (6) and 4) in silico pathogenicity scores generated with the VEST algorithm (35), which are predictors of increased missense mutation impact (Fig 1). In comparison to mutated residues not in 3D hotspot regions, vertebrate evolutionary conservation was higher and protein-protein interface occurrence was higher in the 3D hotspot regions (conservation p=2.9E-29, Mann-Whitney U test; protein interface p=5.2E-13, one-tailed Fisher’s Exact test). In silico driver scores and pathogenicity scores were higher for missense mutations in 3D hotspot regions (driver score p=3.0E-47, pathogenicity score p=3.0E-16; Mann-Whitney U-test) than for the remaining mutations (Fig 1).
Fig. 1. 3D Hotspot regions are different from other mutated protein residues.
Three distinguishing features of HotMAPS regions. A. HotMAPS mutated residues are more conserved in vertebrate evolution than mutated residues not in hotspot regions, as shown by lower Multiple Alignment Entropy (p=1.2E-29; Mann-Whitney U test). Multiple Alignment Entropy is calculated as the Shannon entropy of protein-translated 46-way vertebrate genome alignments from UCSC Genome Browser, which is lowest for the most conserved residues. B. HotMAPS missense mutations have higher in silico cancer driver scores from the CHASM algorithm (p=5.3E-47; Mann-Whitney U test) than those mutations not in hotspot regions, and C. higher in silico pathogenicity scores from the VEST algorithm (p=7.0E-162; Mann-Whitney U-test). Finally, HotMAPS mutated residues occur more frequently at protein-protein interfaces (p=1.3E-11; one-tailed Fisher’s Exact test) (Table S8).
3D Hotspot regions are different in OGs and TSGs
The catalog contains 37 regions stratified by tumor type in bonafide TSGs and 77 in bonafide OGs (114 regions in 30 genes), using as a benchmark the classifications of Vogelstein et al. (1) (Landscapes benchmark). We used this data to explore possible differences between TSG and OG regions at amino acid resolution. We found that in TSGs, 3D hotspot regions were larger than in OGs (region size p=9.6E-06; Mann-Whitney U test). They were also more mutationally diverse (mutational diversity p=2.1E-07; Mann-Whitney U test). Additionally, OG 3D hotspot regions were more conserved in vertebrate evolution than TSGs and more solvent accessible in protein structure, meaning that they tend to occur at the protein surface (evolution p=4.7E-07, solvent accessible p=1.5E-06; Mann-Whitney U test). TSG hotspot regions harbored increased mutation net change in hydrophobicity (p=3.3E-07; Mann-Whitney U test) and mutation net change in volume (p=2.2E-07; Mann-Whitney U test), suggesting that their impact on protein function could be due to decreased stability. The in silico missense mutation cancer driver scores were higher for OG regions (p=0.003; Mann-Whitney U test). We also tested differences between in silico pathogenicity scores and occurrence at protein-protein interfaces between OG and TSG regions, but these were not significant (pathogenicity scores p=0.37, protein interface p=0.34; Mann-Whitney U test).
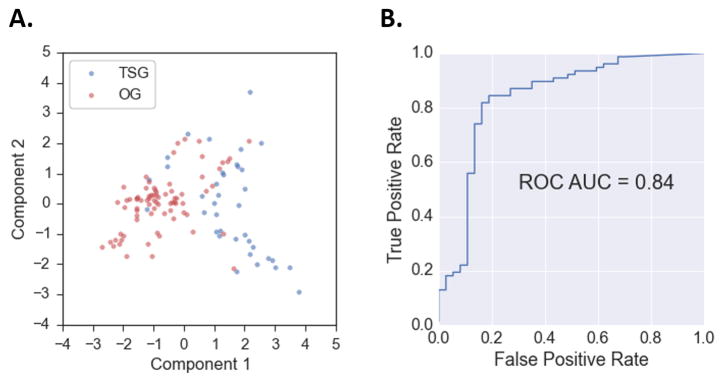
The fact that these differences between OG and TSG regions were statistically significant suggested that they might have predictive value. Principal components analysis (PCA) of the six significant features indicated some separation (Fig 2A). Next, we trained a Naive Bayes machine learning classifier to discriminate between OG and TSG hotspot regions, using region size, mutational diversity, vertebrate conservation, residue solvent accessibility, mutation net hydrophobicity change and residue volume change as features. A rigorous gene-level holdout protocol was used to avoid overfitting (Supplementary Materials and Methods). A Naive Bayes score closer to 1.0 indicates that the hotspot region is likely in an OG while a score closer to 0.0 indicates that it is in a TSG. Area under receiver operating characteristic (ROC) curve or AUC, a standard measure of classifier performance, was 0.84 out of 1.0, a result that supports our claim that 3D hotspot regions in OGs and TSGs have distinctive characteristics (Fig 2B). AUC of a classifier with random performance is 0.5. Performance did not improve when the other features were included in the classifier. Table 3 lists the 30 genes and the median Naive Bayes score, across all regions in each gene. The median values for each predictive feature are also shown.
Fig. 2. HotMAPS regions have different characteristic features in oncogenes (OGs) and tumor suppressor genes (TSGs).
A. Principal components analysis (PCA) plot shows a clustering pattern in hotspot regions identified in OGs (red) and TSGs (blue). Each point is a region represented by six numeric features, projected into two dimensions. The features are region size, mutational diversity, vertebrate evolutionary conservation, residue relative solvent accessibility, mutation net change in hydropobicity and mutation net change in residue volume. B. OG and TSG HotMAPS regions can be discriminated with machine learning, based on four features. A Gaussian Naive Bayes classifier trained on provides a reasonable separation between the two classes with AUC=0.84 out of 1.0. Performance of a random classifier is AUC=0.5. ROC=Receiver Operating Characteristic (ROC), AUC = area under the ROC curve.
Table 3. Median scores and feature values for oncogene and tumor suppressor gene hotspot regions.
Thirty genes classified as OGs or TSGs in Landscapes benchmark. The genes contain a total of 114 tumor-type-specific hotspot regions. Each row in the table shows the median Naive Bayes classification score for the regions in the gene. Scores close to 1.0 predict that a region is in an OG and scores close to 0.0 predict that region is in a TSG. Genes are ranked in decreasing order by the Naive Bayes scores. Ranking generally agrees with the Landscapes benchmark. Also shown for each gene is the median value of the six features used to train the Naive Bayes classifier. The features are region size (number of residues), mutational diversity, vertebrate evolutionary conservation (Shannon entropy of alignment position, where lower entropy=higher conservation), mutation net hydrophobicity change, residue solvent accessibility and mutation net volume change.
| Gene | Landscapes Benchmark | Naïve Bayes score | Region Size | Mutational diversity | Conservation | Change in Hydrophobicity | Residue Solvent Accessibility | Change in Volume |
|---|---|---|---|---|---|---|---|---|
| FGFR3 | OG | 1.00 | 1.00 | 0.00 | 0.29 | −3.00 | 0.61 | −1.80 |
| KRAS | OG | 1.00 | 2.50 | 0.58 | 0.26 | 4.49 | 0.73 | −1.90 |
| NRAS | OG | 1.00 | 2.50 | 0.37 | 0.25 | 6.24 | 0.49 | −0.98 |
| BCOR | TSG | 1.00 | 1.00 | 0.00 | 0.32 | −5.40 | 0.55 | 0.84 |
| PIK3R1 | TSG | 1.00 | 1.00 | 0.00 | 0.26 | 4.20 | 0.08 | −0.70 |
| BRAF | OG | 1.00 | 4.00 | 0.25 | 0.25 | 8.70 | 0.42 | −0.07 |
| HRAS | OG | 1.00 | 2.00 | 0.62 | 0.32 | 4.72 | 0.43 | −1.27 |
| PTPN11 | OG | 1.00 | 1.00 | 0.00 | 0.41 | 10.80 | 0.25 | −0.75 |
| FGFR2 | OG | 1.00 | 1.50 | 0.13 | 0.41 | 8.91 | 0.34 | −1.81 |
| PPP2R1A | OG | 0.99 | 3.00 | 0.43 | 0.29 | 4.82 | 0.21 | −1.84 |
| PIK3CA | OG | 0.99 | 2.00 | 0.35 | 0.26 | 0.63 | 0.32 | −0.81 |
| MAP2K1 | OG | 0.97 | 2.00 | 0.71 | 0.25 | −1.02 | 0.29 | −0.49 |
| NFE2L2 | OG | 0.97 | 3.00 | 0.69 | 0.27 | −1.49 | 0.30 | −0.34 |
| SF3B1 | OG | 0.96 | 3.00 | 0.30 | 0.25 | 0.01 | 0.23 | 0.29 |
| ERBB2 | OG | 0.93 | 1.50 | 0.41 | 0.31 | −0.95 | 0.20 | −1.27 |
| RET | OG | 0.92 | 1.00 | 0.00 | 0.26 | 2.20 | 0.10 | 1.56 |
| GNAS | OG | 0.84 | 1.00 | 0.33 | 0.29 | −10.73 | 0.38 | 1.10 |
| CIC | TSG | 0.79 | 6.00 | 0.55 | 0.41 | −8.41 | 0.39 | −0.68 |
| SMAD4 | TSG | 0.69 | 5.00 | 0.92 | 0.30 | −5.15 | 0.16 | −0.34 |
| FBXW7 | TSG | 0.61 | 5.00 | 0.72 | 0.28 | −10.28 | 0.27 | 1.96 |
| PTEN | TSG | 0.61 | 6.00 | 1.00 | 0.26 | −1.33 | 0.07 | 0.80 |
| IDH2 | OG | 0.60 | 1.00 | 0.67 | 0.28 | −7.85 | 0.10 | 0.49 |
| SMARCA4 | TSG | 0.11 | 1.00 | 0.00 | 0.30 | −13.30 | 0.08 | 3.78 |
| NOTCH1 | TSG | 0.08 | 1.00 | 0.00 | 0.55 | −0.80 | 0.17 | 0.79 |
| IDH1 | OG | 0.07 | 1.00 | 0.22 | 0.41 | −9.83 | 0.16 | 1.09 |
| SPOP | OG | 0.06 | 7.00 | 0.73 | 0.25 | 1.37 | 0.09 | 2.46 |
| CDKN2A | TSG | 0.05 | 2.00 | 0.58 | 0.60 | −4.27 | 0.35 | −0.57 |
| VHL | TSG | 0.04 | 8.00 | 0.89 | 0.40 | 0.92 | 0.02 | 0.87 |
| EGFR | OG | 0.01 | 8.00 | 0.53 | 0.34 | −1.01 | 0.30 | −0.76 |
| TP53 | TSG | 0.00 | 30.50 | 0.81 | 0.44 | −4.11 | 0.19 | −0.09 |
The ROC performance and PCA plot support our claim that characteristic differences between OG and TSG hotspots can be quantified. However, some hotspot regions remain misclassified, according to their labels in the Landscapes benchmark (Discussion).
What is gained by 3D hotspot region detection vs. 1D?
The larger size and mutational diversity of hotspot regions in TSGs vs. OGs suggests that they could be more difficult to detect and perhaps they have been underreported by 1D approaches. OG hotspot regions consisting of recurrent missense mutations at one or two residues can be seen by eye with lollipop plots and are straightforward to detect computationally based on 1D primary sequence. We hypothesized that detection of many TSG hotspot regions might require a 3D algorithm. To maximize the interpretability of this analysis, regions that occurred in multiple tumor types were merged so that each region was represented only once in each gene (Materials and Methods).
For a well-controlled comparison of 3D and 1D hotspot region detection, we applied a 1D version of our method to the protein chain sequences of the same set of PDB protein bioassemblies and theoretical protein structure models to detect non-uniform clustering patterns on primary protein sequence (Supplementary Materials and Methods, Table S5 and Table S6). 72% of hotspot regions identified in 3D were identifiable in 1D.
Next, we compared the number of hotspot regions identified in OGs and TSGs. We considered regions identified in 3D only, in both 3D and 1D, and in 1D only. Using the bona fide OGs and TSGs (Table 1), there were significantly more OG regions that TSG regions identified by the 1D algorithm (p=0.03; one-sided Fisher’s Exact Test). The 1D-only version of the algorithm detected 5 OG and 2 TSG regions; 1D further detected an additional 25 OG and 7 TSG regions that were also identified by the 3D algorithm. The 3D algorithm identified an additional 4 OG and 6 TSG regions. To increase our power, we repeated this test again using the bona fide OGs and TSGs plus additional regions in 5 candidate OGs and TSGs reported in the literature (OGs were FSIP2, MTOR, RANBP2, CHEK2, and MAPK1; TSGs were RASA1, SMARCA2, KEAP1, CUL1, TGFBR2) (all are listed and cited in Table 2), yielding increased statistical significance (p=0.009, one-sided Fisher’s Exact Test). The results suggest that 1D detection methods may be better suited to detecting regions in OGs rather than TSGs.
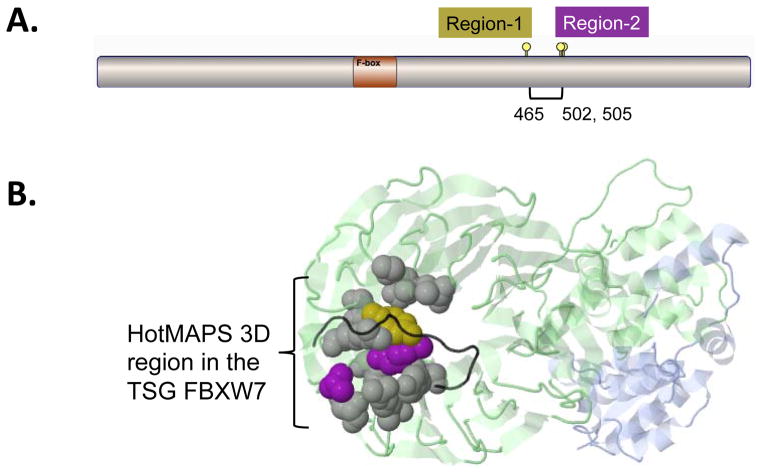
A further problem with sequence-based 1D hotspot region detection is that larger regions detectable in 3D may be only partially characterized and/or split into multiple pieces. Fig 3 shows an example of a TSG hotspot region in FBXW7 found in 3D by HotMAPS that has been split into two pieces by the 1D algorithm. In 1D protein sequence, residue 465 is not close enough to residues 502 and 505 to be identified in one hotspot region. On the 3D protein structure of FBXW7 (PDB code 2OVQ), the three residues are spatially close and a single hotspot region is detected.
Fig. 3. Comparison of hot spot detection in the TSG FBXW7 in 1D and 3D.
A. A simplified 1D version of HotMAPS found two regions in FBXW7. The 3D version of HotMAPS found a single larger region, encompassing both regions. Diagram shows protein sequence of FBXW7, which contains a single F-box functional domain. Region-1 = residue 465 (left lollipop), Region-2 = residues 502 and 505 (right lollipops). B. HotMAPS identifies a single 3D hotspot region in FBXW7. Structure of SCFFbw7 ubiquitin ligase complex (PDB 2OVQ), containing FBXW7 (Green), SKP1 (Blue) and CCNE1 fragment (degron peptide) (Black). Residue coloring: 1D Region-1 (Gold), 1D Region-2 (Purple). Residues missed by 1D detection but included in HotMAPS 3D=Gray. Although the 1D regions are far in the primary protein sequence, residues 505 and 465 spatially contact at the interface with CCNE1. Protein structure figures are generated by JSMol in MuPIT (http://mupit.us).
3D Hotspot regions may increase interpretability of driver mechanisms
Three-dimensional consideration of hotspot regions in protein structure can potentially provide researchers with a rich source of hypothesis generation about driver mechanisms. While gene- or domain-level mutation enrichment analysis can point to potential protein functions, interactions, biological processes and pathways important for cancer etiology and progression, more detailed information may be available once a specific set of mutated amino acid residues has been identified as significant.
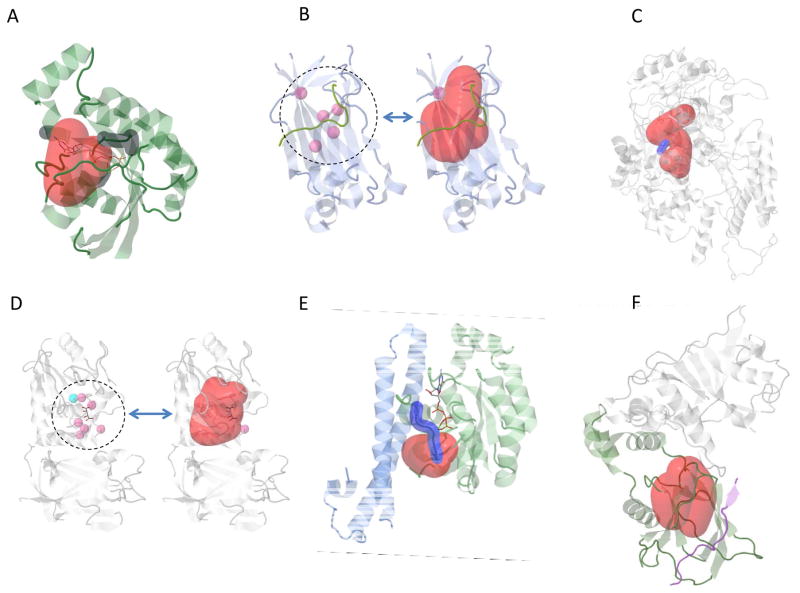
For many of the 3D hotspot regions found by HotMAPS, the literature contains evidence that they are in direct contact with or proximal to amino acid residues of known functional importance. Fig 4 shows six cancer-associated proteins in which the hotspot region is either overlapping or proximal to important functional sites.
Fig. 4. HotMAPS hotspot regions overlap and are proximal to important functional sites.
A. HNSC hotspot region (red) in RAC1 (green) and GTP/GDP binding residues (dark gray) (PDB 2FJU). B. PRAD hotspot region (red) in SPOP-substrate complex (PDB 3HGH) with SPOP (blue) and H2AFY substrate (green). Left shows 5 residues (pink) that when mutated show strongly reduced affinity for substrate. C. BLCA Hotspot region (red) in ERCC2 (gray) shown on theoretical model of ERCC2 helicase ATP-binding domain. The hotspot is proximal to the DEAH box (blue), a highly conserved motif containing residues that interact with Mg2+ and are critical for ATP binding and helicase activity. D. UCEC hotspot region (red) in PTEN (PDB 1D5R) with active site phosphocysteine residue (blue), residues when mutated annotated to reduce phosphatase activity (pink). E. STAD hotspot region (red) in RHOA with a GTP analog bound (sticks) (PDB 1CXZ). GTP binding residues and effector region (dark blue). F. KIRC hotspot region (red) in VHL-TCEB1-TCEB2 complex, bound to HIF1A peptide (PDB 4AJY). Proximity to the interaction site of VHL (Green) and HIF1A (Blue), suggests possible decreased ubiquitination of HIF1A, resulting in increased protein expression of HIF1A. TCEB1 and TCEB2 (Gray). HNSC= Head and Neck Squamous Cell Carcinoma, PRAD= Prostate Adenocarcinoma BLCA= Bladder Urothelial Carcinoma, UCEC= Uterine Corpus Endometrial Carcinoma, STAD= Stomach Adenocarcinoma. KIRC = Kidney Renal Clear Cell Carcinoma.
RAC1 hotspot in squamous head and neck cancer (HNSCC)
RAC1 is a Rho GTPase important in signaling systems that regulate the organization of actin cytoskeleton and cell motility. The hotspot overlaps the GTP/GDP binding site and could impact regulation of normal RAC1 cycling between GTP- and GDP-bound states (Fig 4A). It contains a previously identified recurrent mutation in melanoma (P29S) which dysregulates RAC1 by a fast cycling mechanism (36).
SPOP hotspot in prostate cancer (PRAD)
SPOP is the substrate recognition component of a cullin3-based E3 ubiquitin-protein ligase complex, which targets multiple substrates for proteasomal degradation. The hotspot overlaps with a binding groove harboring five residue positions (pink) where mutagenesis has strongly reduced affinity for the substrate (annotated in the UniProtKB).
ERCC2 hotspot in bladder cancer (BLCA)
ERCC2 is an ATP-dependent helicase that is part of the protein complex TFIIH involved in RNA polymerase II transcription and nucleotide excision repair (NER). We identified a hotspot region, proximal to the DEAH box, a highly conserved motif containing residues that interact with Mg2+ and are critical for ATP binding and helicase activity (Fig 4C). This proximity suggests that the hotspot mutations could disrupt ATPase activity and yield defective NER (37).
PTEN hotspot
PTEN is a phosphatase for both proteins and phosphoinositides, and it removes a phosphate from PIP3, critical for signaling to AKT. The hotspot region identified in endometrial cancer (UCEC) spans two functionally important loops in the protein (P and WPD loops) at the boundary of the active site pocket. Residues in these loops are critical for catalysis (blue dot) and are important for the P-loop’s conformation. Mutagenesis of residues in the WPD loop reduces phosphatase activity and increases colony formation in cell culture(38). Pink dots show residues that impact phosphatase activity.
RHOA hotspots
RHOA is a small GTPase oncogene, and like RAC1 is a member of the Ras superfamily (39). We identified hotspot regions in bladder cancer (BLCA), squamous head and neck cancer (HNSC) and stomach adenocarcinoma (STAD). The hotspot regions overlap with the RHOA effector region, a highly conserved motif that is involved in Ras superfamily signaling with downstream effector proteins. The regions are immediately proximal to a magnesium ion, which has been implicated in regulating the kinetics of Rho family GTPases (40).
VHL hotspot (KIRC)
VHL is a component of an E3 ubiquitin protein ligase complex, and it ubiquitinates the OG transcription factor HIF1A, targeting it for proteasomal degradation (41). One impact of VHL loss of function with failure to ubiquitinate HIF1A is increased protein expression of HIF1A. The hotspot region is proximal to its interaction site with HIF1A and could potentially have an impact on this interaction (Fig 4F). The TCGA kidney cancer (KIRC) samples were stratified based on their missense mutation status: VHL hotspot, non-hotspot, or no missense (WT). HIF1A protein expression was not significantly different between VHL non-hotspot and VHL WT groups (p=0.5; Mann-Whitney U-test), but was significantly higher between VHL hotspot and VHL WT groups (p=0.03; Mann-Whitney U-test). This result is consistent with a special role for VHL hotspot missense mutations in regulating HIF1A protein expression. However, increased HIF1A expression in these KIRC samples is likely impacted by additional genetic and other factors. We might see a substantially lower p value If VHL hotspot mutations were the only cause of the observed increase. Also, there are many VHL missense mutations outside of the hotspot region, and it is likely that several of these also have a functional impact. In particular, several of them are at the interface of VHL and the elongins B and C in the complex and could impact VHL/elongin binding.
Discussion
Catalog of TCGA 3D missense mutation hotspot regions at amino acid resolution
The large-scale whole-exome sequencing and mutation calling efforts of the TCGA have identified hundreds of thousands of somatic missense mutations in human cancers. While some of these mutations are private, many are shared across multiple patients and multiple tumor types. The biological and therapeutic relevance of these shared mutations is of great interest to the cancer research community. For example, patients can be stratified for clinical trials and treatment protocols selected based on missense mutation status in several key driver genes, including BRAF, KRAS, EGFR. A special type of shared missense mutations are those which occur recurrently not only at the same genomic codon, but at neighboring codons in translated protein sequence and more generally, neighboring amino acid residues in protein 3D structure. These clusters of neighboring missense mutations are known as missense mutation hotspot regions. They have been proposed to have particular relevance to oncogenic processes (12), since the increased frequency of missense mutation at a highly localized region in a protein may be a signature of positive selection (42). Missense hotspot regions may be informative in detecting driver genes (7). A number of groups have developed algorithms to detect enrichment of these regions on the gene- and domain-level (7–9,11–13), but until now, there have been not been systematically characterized on a large number of protein structures and models at the resolution of individual amino acid residues.
We systematically identify 3D missense hotspot regions using TCGA somatic mutation data from 6,594 samples in 23 tumor types. HotMAPS identified 107 unique, tumor-type-aggregated gene level regions and 216 unique tumor-type-specific gene level regions (Materials and Methods, Tables S1, S2). This catalog enables assessment of how the specific missense mutations in a hotspot contribute to cancer-associated molecular mechanisms.
TCGA 3D hotspot regions have functional importance
We compared features of residues in 3D hotspot regions to other missense mutated residues in the TCGA data. The hotspot regions have characteristic features that support their putative functional importance: high evolutionary conservation, high in silico-predicted missense mutation impacts and increased frequency of occurrence at protein interfaces. Genes containing the 3D hotspot regions appear to be particularly relevant to cancer. Landscapes benchmark TSG and OGs are overrepresented and the list includes many candidate TSGs, OGs, drug targets and hereditary cancer genes (Tables 1, 2). For several TSGs and OGs, the regions coincide with enzymatic active sites, positions that have been shown to alter protein function in experimental mutagenesis assays and sites of interaction with protein and nucleotide interaction partners (Fig 4, Table S7).
TCGA hotspot regions are different in OGs and TSGs
Although recurrent missense mutations have long been known to occur in both OGs and TSGs (10), they have been observed more frequently in OGs. We show that there are systematic differences in hotspot regions found in OGs and TSGs. OG regions are smaller, less mutationally diverse, more evolutionarily conserved, and more solvent accessible than TSG regions. TSG regions are more likely to harbor mutations that may impact protein stability through changes in hydrophobicity or volume. Potential explanations for these differences are that there are more ways to lose the function of a protein than to gain function (43). Loss-of-function tumor suppressor mutations can occur at many residue positions and involve many types of amino acid residue substitutions, while oncogene mutations will occur at a few functionally important positions and involve fewer substitution types.
A consequence of these differences is that TSG regions are harder to detect visually or by 1D clustering approaches than OG regions. Thus, they have been missed by 1D analysis methods. A major contribution of 3D analysis is enabling detection of hotspot regions in TSGs in addition to those in OGs. We suspect that novel hotspot regions in TSGs will continue to be discovered as more samples are sequenced in more tumor types.
We are able to leverage the characteristic differences to distinguish between hotspot regions in TSGs and OGs, with a simple machine learning method, achieving an area under the receiver operating characteristic curve (ROC AUC) of ~0.80. However, not all regions are correctly classified by this method. Interestingly, we find that some of these “undistinguishable” genes may act as both TSGs and OGs, depending on context or be atypical of their class. PIK3R1 has been described as an OG (44) and SPOP as a TSG (45), in agreement with our Naive Bayes scores, but not with the Landscapes benchmark. The OGs IDH1 and IDH2 both have high net hydrophobicity changes, which are protein destabilizing and characteristic of TSGs. IDH1/IDH2 hotspot mutation may cause a (TSG-like) partial loss of enzymatic function, yielding accumulation of 2 hydoxyglutarate (2HG), a carcinogenic catalytic intermediate (46). EGFR has two regions in GBM and LGG which are scored as TSG-like. In these tumor types, EGFR mutation patterns is atypical, because EGFR amplification is an early event. This amplification has been linked to increased mutation load in EGFR itself, including in aberrant extrachromosomal copies of EGFR (43). Fig S2 indicates the locations of these misclassified regions on the PCA plot.
HotMAPS has increased sensitivity and coverage than previous 3D hotspot detection algorithms
A disadvantage of working with experimentally-derived protein structures is that they are available for a limited number of human proteins (39%). For many of these genes, the structure data is incomplete, so that only a single protein domain or small fragment is represented in PDB. In this work, by careful use biological assemblies of PDB structures and also theoretical protein models, we mapped ~53% of unique residue positions harboring a TCGA missense mutation into 3D protein space. In a recent study of 21 TCGA tumor types that used a different algorithm and PDB structures only, 11.2% positions were mapped (13). We note that theoretical protein models are well suited for this kind of analysis. HotMAPS considers the center of geometry for each amino acid residue, a metric that is not highly sensitive to atomic-resolution errors common in theoretical protein models (47). The increased sensitivity and coverage of HotMAPS is supported by the number of tumor types in which 3D hotspot regions were detected (19 out of 23), the total number of regions detected (107 unique, tumor-type-aggregated gene level regions and 216 unique tumor-type-specific gene level regions), and the number of genes in which regions were detected (91). The only previous systematic attempt to find 3D hotspots in TCGA data detected statistically significant regions in 10 genes, based on 21 TCGA tumor types (13), and nine of these were also detected by HotMAPS.
We hope that some HotMAPS regions found by our algorithm point to novel driver genes, however functional studies are warranted to find out if they are discoveries or false positives
An interactive 3D protein viewer where users can submit their own mutations and compare to the HotMAPS catalog (48) is available at http://mupit.us
HotMAPS software is open source at https://github.com/karchinlab/HotMAPS
Additional material is available in Supplementary Materials and Methods: detailing mapping from genomic coordinates to protein structures (Fig S3), overview flow chart of HotMAPS (Fig S4), an example of single-residue hotspot region discovery in 1D vs. 3D (Fig S5) and a stratified analysis of HotMAPS properties by solvent accessibility (Fig S6), and in Supplementary Tables: a list of blacklisted residues (Table S9).
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health, National Cancer Institute fellowship F31CA200266 to CT and grant U01CA180956 to RK.
We thank Drs. Bert Vogelstein and Jing Zhu of the UCSC Xena team for valuable discussion on the manuscript.
Footnotes
No conflicts of interest
References
- 1.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339(6127):1546–58. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318(5853):1108–13. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 3.Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446(7132):153–8. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dees ND, Zhang Q, Kandoth C, Wendl MC, Schierding W, Koboldt DC, et al. MuSiC: identifying mutational significance in cancer genomes. Genome Res. 2012;22(8):1589–98. doi: 10.1101/gr.134635.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499(7457):214–8. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carter H, Chen S, Isik L, Tyekucheva S, Velculescu VE, Kinzler KW, et al. Cancer-specific high-throughput annotation of somatic mutations: computational prediction of driver missense mutations. Cancer Research. 2009;69(16):6660–67. doi: 10.1158/0008-5472.CAN-09-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gonzalez-Perez A, Perez-Llamas C, Deu-Pons J, Tamborero D, Schroeder MP, Jene-Sanz A, et al. IntOGen-mutations identifies cancer drivers across tumor types. Nature methods. 2013;10(11):1081–2. doi: 10.1038/nmeth.2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nehrt NL, Peterson TA, Park D, Kann MG. Domain landscapes of somatic mutations in cancer. BMC Genomics. 2012;13(Suppl 4):S9. doi: 10.1186/1471-2164-13-S4-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ryslik GA, Cheng Y, Cheung K-H, Modis Y, Zhao H. Utilizing protein structure to identify non-random somatic mutations. BMC bioinformatics. 2013;14(1):190. doi: 10.1186/1471-2105-14-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253(5015):49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 11.Tamborero D, Gonzalez-Perez A, Lopez-Bigas N. OncodriveCLUST: exploiting the positional clustering of somatic mutations to identify cancer genes. Bioinformatics. 2013;29(18):2238–44. doi: 10.1093/bioinformatics/btt395. [DOI] [PubMed] [Google Scholar]
- 12.Ye J, Pavlicek A, Lunney EA, Rejto PA, Teng C-H. Statistical method on nonrandom clustering with application to somatic mutations in cancer. BMC bioinformatics. 2010;11(1):11. doi: 10.1186/1471-2105-11-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kamburov A, Lawrence MS, Polak P, Leshchiner I, Lage K, Golub TR, et al. Comprehensive assessment of cancer missense mutation clustering in protein structures. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(40):E5486–95. doi: 10.1073/pnas.1516373112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pieper U, Eswar N, Webb BM, Eramian D, Kelly L, Barkan DT, et al. MODBASE, a database of annotated comparative protein structure models and associated resources. Nucleic acids research. 2009;37(Database issue):D347–54. doi: 10.1093/nar/gkn791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, et al. A census of human cancer genes. Nature reviews Cancer. 2004;4(3):177–83. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Atala A. Re: Interruption of KLF5 Acetylation Converts its Function from Tumor Suppressor to Tumor Promoter in Prostate Cancer Cells. The Journal of urology. 2015;194(5):1505. doi: 10.1002/ijc.29028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Helming KC, Wang X, Roberts CW. Vulnerabilities of mutant SWI/SNF complexes in cancer. Cancer cell. 2014;26(3):309–17. doi: 10.1016/j.ccr.2014.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoffman GR, Rahal R, Buxton F, Xiang K, McAllister G, Frias E, et al. Functional epigenetics approach identifies BRM/SMARCA2 as a critical synthetic lethal target in BRG1-deficient cancers. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(8):3128–33. doi: 10.1073/pnas.1316793111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pickering CR, Zhou JH, Lee JJ, Drummond JA, Peng SA, Saade RE, et al. Mutational landscape of aggressive cutaneous squamous cell carcinoma. Clinical cancer research: an official journal of the American Association for Cancer Research. 2014;20(24):6582–92. doi: 10.1158/1078-0432.CCR-14-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rothenberg SM, Ellisen LW. The molecular pathogenesis of head and neck squamous cell carcinoma. The Journal of clinical investigation. 2012;122(6):1951–7. doi: 10.1172/JCI59889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biswas S, Trobridge P, Romero-Gallo J, Billheimer D, Myeroff LL, Willson JK, et al. Mutational inactivation of TGFBR2 in microsatellite unstable colon cancer arises from the cooperation of genomic instability and the clonal outgrowth of transforming growth factor beta resistant cells. Genes, chromosomes & cancer. 2008;47(2):95–106. doi: 10.1002/gcc.20511. [DOI] [PubMed] [Google Scholar]
- 22.Sasaki H, Suzuki A, Shitara M, Okuda K, Hikosaka Y, Moriyama S, et al. mutations in lung cancer patients. Oncology letters. 2013;6(3):719–21. doi: 10.3892/ol.2013.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Korzeniewski N, Zheng L, Cuevas R, Parry J, Chatterjee P, Anderton B, et al. Cullin 1 functions as a centrosomal suppressor of centriole multiplication by regulating polo-like kinase 4 protein levels. Cancer Res. 2009;69(16):6668–75. doi: 10.1158/0008-5472.CAN-09-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hast BE, Cloer EW, Goldfarb D, Li H, Siesser PF, Yan F, et al. Cancer-derived mutations in KEAP1 impair NRF2 degradation but not ubiquitination. Cancer Res. 2014;74(3):808–17. doi: 10.1158/0008-5472.CAN-13-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Furukawa T, Kanai N, Shiwaku HO, Soga N, Uehara A, Horii A. AURKA is one of the downstream targets of MAPK1/ERK2 in pancreatic cancer. Oncogene. 2006;25(35):4831–9. doi: 10.1038/sj.onc.1209494. [DOI] [PubMed] [Google Scholar]
- 26.Kostrzewska-Poczekaj M, Giefing M, Jarmuz M, Brauze D, Pelinska K, Grenman R, et al. Recurrent amplification in the 22q11 region in laryngeal squamous cell carcinoma results in overexpression of the CRKL but not the MAPK1 oncogene. Cancer biomarkers: section A of Disease markers. 2010;8(1):11–9. doi: 10.3233/DMA-2011-0814. [DOI] [PubMed] [Google Scholar]
- 27.Litchfield K, Summersgill B, Yost S, Sultana R, Labreche K, Dudakia D, et al. Whole-exome sequencing reveals the mutational spectrum of testicular germ cell tumours. Nature communications. 2015;6:5973. doi: 10.1038/ncomms6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gylfe AE, Kondelin J, Turunen M, Ristolainen H, Katainen R, Pitkanen E, et al. Identification of candidate oncogenes in human colorectal cancers with microsatellite instability. Gastroenterology. 2013;145(3):540–3. e22. doi: 10.1053/j.gastro.2013.05.015. [DOI] [PubMed] [Google Scholar]
- 29.Murugan AK, Alzahrani A, Xing M. Mutations in critical domains confer the human mTOR gene strong tumorigenicity. The Journal of biological chemistry. 2013;288(9):6511–21. doi: 10.1074/jbc.M112.399485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cancer Genome Atlas Research N. Integrated genomic characterization of papillary thyroid carcinoma. Cell. 2014;159(3):676–90. doi: 10.1016/j.cell.2014.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.West AC, Johnstone RW. New and emerging HDAC inhibitors for cancer treatment. The Journal of clinical investigation. 2014;124(1):30–9. doi: 10.1172/JCI69738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fathers C, Drayton RM, Solovieva S, Bryant HE. Inhibition of poly(ADP-ribose) glycohydrolase (PARG) specifically kills BRCA2-deficient tumor cells. Cell cycle. 2012;11(5):990–7. doi: 10.4161/cc.11.5.19482. [DOI] [PubMed] [Google Scholar]
- 33.Shukla SA, Rooney MS, Rajasagi M, Tiao G, Dixon PM, Lawrence MS, et al. Comprehensive analysis of cancer-associated somatic mutations in class I HLA genes. Nature biotechnology. 2015;33(11):1152–58. doi: 10.1038/nbt.3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petrini I, Meltzer PS, Kim IK, Lucchi M, Park KS, Fontanini G, et al. A specific missense mutation in GTF2I occurs at high frequency in thymic epithelial tumors. Nature genetics. 2014;46(8):844–9. doi: 10.1038/ng.3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Douville C, Carter H, Kim R, Niknafs N, Diekhans M, Stenson PD, et al. CRAVAT: cancer-related analysis of variants toolkit. Bioinformatics. 2013;29(5):647–48. doi: 10.1093/bioinformatics/btt017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davis MJ, Ha BH, Holman EC, Halaban R, Schlessinger J, Boggon TJ. RAC1P29S is a spontaneously activating cancer-associated GTPase. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(3):912–7. doi: 10.1073/pnas.1220895110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Winkler GS, Araujo SJ, Fiedler U, Vermeulen W, Coin F, Egly JM, et al. TFIIH with inactive XPD helicase functions in transcription initiation but is defective in DNA repair. The Journal of biological chemistry. 2000;275(6):4258–66. doi: 10.1074/jbc.275.6.4258. [DOI] [PubMed] [Google Scholar]
- 38.Lee JO, Yang H, Georgescu MM, Di Cristofano A, Maehama T, Shi Y, et al. Crystal structure of the PTEN tumor suppressor: implications for its phosphoinositide phosphatase activity and membrane association. Cell. 1999;99(3):323–34. doi: 10.1016/s0092-8674(00)81663-3. [DOI] [PubMed] [Google Scholar]
- 39.Rojas AM, Fuentes G, Rausell A, Valencia A. The Ras protein superfamily: evolutionary tree and role of conserved amino acids. The Journal of cell biology. 2012;196(2):189–201. doi: 10.1083/jcb.201103008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang B, Zhang Y, Wang Z, Zheng Y. The role of Mg2+ cofactor in the guanine nucleotide exchange and GTP hydrolysis reactions of Rho family GTP-binding proteins. The Journal of biological chemistry. 2000;275(33):25299–307. doi: 10.1074/jbc.M001027200. [DOI] [PubMed] [Google Scholar]
- 41.Gossage L, Eisen T, Maher ER. VHL, the story of a tumour suppressor gene. Nature reviews Cancer. 2015;15(1):55–64. doi: 10.1038/nrc3844. [DOI] [PubMed] [Google Scholar]
- 42.Wagner A. Rapid detection of positive selection in genes and genomes through variation clusters. Genetics. 2007;176(4):2451–63. doi: 10.1534/genetics.107.074732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nikolaev S, Santoni F, Garieri M, Makrythanasis P, Falconnet E, Guipponi M, et al. Extrachromosomal driver mutations in glioblastoma and low-grade glioma. Nature communications. 2014;5:5690. doi: 10.1038/ncomms6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Philp AJ, Campbell IG, Leet C, Vincan E, Rockman SP, Whitehead RH, et al. The phosphatidylinositol 3′-kinase p85alpha gene is an oncogene in human ovarian and colon tumors. Cancer Res. 2001;61(20):7426–9. [PubMed] [Google Scholar]
- 45.Li C, Ao J, Fu J, Lee DF, Xu J, Lonard D, et al. Tumor-suppressor role for the SPOP ubiquitin ligase in signal-dependent proteolysis of the oncogenic co-activator SRC-3/AIB1. Oncogene. 2011;30(42):4350–64. doi: 10.1038/onc.2011.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang H, Ye D, Guan KL, Xiong Y. IDH1 and IDH2 mutations in tumorigenesis: mechanistic insights and clinical perspectives. Clinical cancer research: an official journal of the American Association for Cancer Research. 2012;18(20):5562–71. doi: 10.1158/1078-0432.CCR-12-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baker D, Sali A. Protein structure prediction and structural genomics. Science. 2001;294(5540):93–6. doi: 10.1126/science.1065659. [DOI] [PubMed] [Google Scholar]
- 48.Niknafs N, Kim D, Kim R, Diekhans M, Ryan M, Stenson PD, et al. MuPIT interactive: webserver for mapping variant positions to annotated, interactive 3D structures. Human genetics. 2013;132(11):1235–43. doi: 10.1007/s00439-013-1325-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.