Abstract
Purpose
The purpose of this study was to apply next-generation sequencing (NGS) technology to identify chromosomally normal embryos for transfer in preimplantation genetic diagnosis (PGD) cycles for translocations.
Methods
A total of 21 translocation couples with a history of infertility and repeated miscarriage presented at our PGD clinic for 24-chromosome embryo testing using copy number variation sequencing (CNV-Seq).
Results
Testing of 98 embryo samples identified 68 aneuploid (69.4 %) and 30 (30.6 %) euploid embryos. Among the aneuploid embryos, the most common abnormalities were segmental translocation imbalances, followed by whole autosomal trisomies and monosomies, segmental imbalances of non-translocation chromosomes, and mosaicism. In all unbalanced embryos resulting from reciprocal translocations, CNV-Seq precisely identified both segmental imbalances, extending from the predicted breakpoints to the chromosome termini. From the 21 PGD cycles, eight patients had all abnormal embryos and 13 patients had at least one normal/balanced and euploid embryo available for transfer. In nine intrauterine transfer cycles, seven healthy babies have been born. In four of the seven children tested at 18 weeks gestation, the karyotypes matched with the original PGD results.
Conclusion
In clinical PGD translocation cycles, CNV-Seq displayed the hallmarks of a comprehensive diagnostic technology for high-resolution 24-chromosome testing of embryos, capable of identifying true euploid embryos for transfer.
Electronic supplementary material
The online version of this article (doi:10.1007/s10815-016-0724-2) contains supplementary material, which is available to authorized users.
Keywords: Aneuploidy, Copy number variation, Next-generation sequencing, Preimplantation genetic diagnosis, Whole genome amplification
Introduction
Balanced translocations are abnormal chromosome derivatives with a carrier frequency of 0.1–0.2 % [1]. There are two types, comprising Robertsonian translocations formed by abnormal breakage and joining of two acrocentric chromosomes and reciprocal translocations formed by the exchange of terminal segments between two chromosomes. In most cases, these rearrangements do not lead to any significant loss of chromosomal material, and therefore, the vast majority of carriers do not exhibit any abnormal phenotype [2]. Balanced translocations can proceed through mitosis and replicate faithfully in somatic cells. However, in the germ line, the process of meiosis can lead to a variety of unbalanced translocation derivatives with duplication and deletion of terminal sequences on either side of the breakpoint [3]. Thus, recurrent miscarriage is a common reproductive outcome for couples who carry a translocation [4].
Preimplantation genetic diagnosis (PGD) is an alternative reproductive option to prenatal diagnosis for translocation carrier couples [5] which aims to identify balanced euploid embryos for intrauterine transplantation and subsequent development to a healthy term baby. In this assisted reproductive procedure, the embryos produced are biopsied at either the 8-cell cleavage stage to remove a single blastomere [6, 7] or at the blastocyst stage to remove 5–10 trophectoderm cells [8, 9]. The embryonic DNA is amplified by whole genome amplification and then tested by chromosome microarray analysis to distinguish balanced from unbalanced embryos. Successful application of these diagnostic technologies on a large number of patient embryos has resulted in increased implantation, pregnancy and live birth rates, and importantly, lower miscarriage rates [10–13].
Despite these successes, PGD for translocations remains challenging. Firstly, over half of the embryos may be abnormal due to unbalanced segregation of the two involved chromosomes [2, 14]. Further, in those embryos that are diagnosed as balanced, there is the possibility of additional incidental aneuploidies, including whole or partial chromosomal gains and losses, mosaicism, and segmental imbalances in other chromosomes that are known to commonly arise by either non-disjunction errors [15] or breakage-fusion-bridge cycles [16]. So far, the technology developed for PGD of translocations has primarily focused on the detection of unbalanced chromosome derivatives and incidental whole or partial chromosomal aneuploidies using 24-chromosome testing either by chromosome microarray analysis [11, 12] or next-generation sequencing (NGS) [13, 17–20]. We speculated that a higher-resolution technology with the capacity to additionally detect more subtle chromosomal abnormalities might further benefit patients undertaking PGD cycles for translocations.
Recently, we have developed a novel quantitative NGS method termed copy number variation sequencing (CNV-Seq) for fine analysis of embryonic chromosomes [19]. In a series of validation studies using cell lines with terminal deletions associated with chromosome disease syndromes [21], as well as whole genome amplification (WGA) products with known segmental translocation imbalances defined by array comparative genomic hybridization (CGH) [19, 22], we demonstrated that all the small CNVs tested in the range of 1–5 Mb could be reliably, accurately, and reproducibly detected. In addition, we have further validated CNV-Seq for the measurement of mosaicism in trophectoderm biopsy samples [23]. During the course of these validation studies, a small group of 21 translocation patients with either long-standing infertility or repeated miscarriage presented at our clinic for PGD. In this study, we applied CNV-Seq as the sole diagnostic technology for this group of PGD patients, with the aim of providing a more comprehensive assessment of their embryos for chromosomal abnormalities and improving reproductive outcomes.
Materials and methods
In vitro fertilization and preimplantation genetic diagnosis
In vitro fertilization (IVF) and embryo biopsy procedures were performed for 21 patients at the Department of Obstetrics and Gynecology, Chinese PLA General Hospital, Beijing, using previously published procedures [19]. In brief, fertilization of MII oocytes was achieved by intracytoplasmic sperm injection of a single sperm. Fertilized embryos were either cultured for 48 h to the cleavage stage or transferred to G2 medium and incubated 48–72 h for development to the blastocyst stage. Five patients had cleavage-stage biopsy, using a finely pulled glass pipette to remove a single blastomere. During the study, the embryology shifted PGD practice to blastocyst biopsy. The remaining 16 patients had blastocyst biopsy, by firstly creating a hole in the zona pellicuda with a non-contact laser at the cleavage stage and then, secondly, removing 5–10 trophectoderm cells using a rubbing dissection method.
Biopsied cell(s) were washed in phosphate-buffered saline (PBS), transferred to a polymerase chain reaction (PCR) tube containing 2 μl of sterile PBS, and then frozen at −20 °C prior to WGA. All embryos biopsied either at day 3 or day 5 were cryopreserved by vitrification. All solutions used during vitrification and thawing were prepared according to the manufacturer’s protocol (Kitazato Supply). Frozen embryos diagnosed as normal/balanced and euploid following CNV-Seq analysis were thawed, and then one to two embryos were placed in an embryo catheter and transferred to the women’s uterus for implantation. Serum β-HCG was detected 14 days after transfer to confirm the biochemical pregnancy. Ultrasound examination was performed to confirm the clinical pregnancy 28 days after transfer.
Next-generation sequencing
Single blastomeres or five to ten trophectoderm cells transferred to lysis buffer were subjected to WGA using the single-cell SurePlex amplification kit (SurePlex, Rubicon). Embryonic WGA products were purified using DNA Clean and Concentrator columns (Zymo Research, USA) and quantitated using the Quant-iT Assay kit with the Qubit fluorometer (Invitrogen, USA). The quality control criteria for judging successful WGA included a concentration of >40 ng/μl (20 μl final volume after column purification) and a size range of 100–500 bp. A total of 50 ng of WGA product was used as a template for CNV-Seq [24, 25]. WGA products were fragmented to an average size of 300 bp and end-ligated with 9-bp barcoded sequencing adaptors [26]. Tagged fragments were then amplified with PCR primers for 15 cycles to generate sequencing libraries. Massively parallel sequencing of embryo libraries was performed on the HiSeq2500 platform (Illumina, USA).
Sequence data analysis and derivation of 24 chromosome profiles
Approximately 5 million 45-bp reads, comprising the 9-bp barcode and 36 bp of genomic sequence, were generated from each embryo WGA product. After applying the Burrows-Wheeler transform algorithm [27] to filter out low-quality reads, a total of 2.8–3.2 million reads were perfectly and uniquely aligned to the hg19 reference genome. The filtered sequencing data was then allocated to 20-kb sequencing bins with an average number of 30–35 reads per bin. To calculate the mean CNV variation, binned read counts at each genomic location were internally compared across a minimum of 15 sample data sets using the fused lasso smoothing algorithm [28]. Plots of log2 mean CNV (Y-axis) versus 20-kb bins (X-axis) were generated for each of the 24 chromosomes. Color markings superimposed on the plots included a blue line to indicate mean CNV, red lines to indicate repetitive regions, and a black box to mark the centromere. Dashed lines at log2 [1.5] and log2 [0.5] were used to indicate the expected CNV for duplications (three copies) and deletions (one copy), respectively. Gain or loss of chromosomal regions was called using stringent cutoff copy number values of >2.8 for duplications and <1.2 for deletions over the entire length of the CNV interval.
Criteria for selecting embryos for transfer
Embryos diagnosed as normal/balanced and euploid were available for transfer, whereas embryos diagnosed as either aneuploid, unbalanced, or unbalanced and aneuploid were excluded for transfer. In regard to embryos diagnosed as diploid-aneuploid mosaics involving a single chromosome (exceptions: chromosomes 21, 18, and 13), if the level of mosaicism was 40 % or less, the embryos were considered for transfer in situations where no other normal/balanced and euploid embryos were available. All embryos with multiple mosaicism, regardless of the level, were excluded for transfer.
Results
PGD patients
A total of 21 couples with either long-standing infertility (n = 10) or a history of recurrent miscarriage (n = 11) presented at the General PLA Hospital IVF clinic for PGD treatment (Table 1). In all couples, prior karyotyping revealed the presence of either a Robertsonian (n = 4) or a reciprocal (n = 17) balanced translocation in one of the partners (Table 2). The mean maternal age was 32.5 years (25–39 years), indicating a moderate risk of incidental aneuploidies. All 21 couples consented to IVF combined with PGD, with five undertaking cleavage-stage biopsy and 16 blastocyst biopsy. In their first IVF and PGD cycle, all couples produced a cohort of embryos for PGD (Supplementary Table 1). Biopsy samples were amplified by WGA and embryonic DNA products subjected to NGS using CNV-Seq technology [19].
Table 1.
Patient demographics and PGD outcomes
| Clinical parameter | IVF/PGD outcome |
|---|---|
| Number of patients (first PGD cycle) | 21 |
| Mean maternal age (25–39 years) | 32.5 |
| Mean number of miscarriages (0–6) | 2.8 |
| Mean number of MII oocytes (2–23) | 11 |
| Number of patients having blastomere biopsy | 5 (23.8 %) |
| Number of patients having blastocyst biopsy | 16 (76.2 %) |
| Mean number of cleavage-stage embryos biopsied (2–9) | 5.4 |
| Mean number of blastocysts biopsied (1–16) | 5.7 |
| Number of patients with all abnormal embryosa | 8 (38.1 %) |
| Number of patients with normal embryos for transferb | 13 (61.9 %) |
| Number of patients with transferred embryos | 9 (42.9 %) |
| Number of transferred embryos | 10 |
| Mean number of transferred embryos (1 or 2) | 1.1 |
| Implantation rate | 8 (80.0 %) |
| Clinical pregnancy rate at 20 weeks gestation (one twin) | 6 (60.0 %) |
| Miscarriage rate (two biochemical pregnancies) | 2 (20.0 %) |
| Number of fetuses karyotyped | 4/7 (57.2 %) |
| Fetuses confirmed with a normal/balanced karyotype | 4 (100 %) |
| Live birth rate | 7 (70.0 %) |
Implantation and live birth rates were calculated per embryos transferred
aAbnormal embryos defined as either aneuploid, unbalanced, or unbalanced + aneuploid
bNormal embryos defined as normal/balanced + euploid
Table 2.
Chromosomal abnormalities in individual PGD embryo
| Robertsonian or reciprocal translocation | MA | No. of embryos tested | CNV-Seq diagnosis | |||
|---|---|---|---|---|---|---|
| Unbalanced | Balanced | |||||
| Incidental CNVs (%) | No CNVs (%) | Incidental CNVs (%) | No CNVs (%) | |||
| Blastomere biopsy | ||||||
| 46,XX,t(3;14)(q29;q23) | 27 | 7 | – | E2, 4, 9, 12 | E1, 3 | E8 |
| 46,XX,t(2;6)(p25;q14) | 39 | 5 | E2, 3, 5 | – | E1, 4 | – |
| 46,XX,t(2;9)(p23;q34) | 31 | 9 | – | E3, 9, 14, 17 | E1, 2, 7 | E12, 18 |
| 46,XX,t(8;14)(p12;q11) | 37 | 1 | E1 | – | – | – |
| 46,XY,t(5;13)(q33;q14) | 39 | 2 | – | – | E1, 3 | – |
| Translocations (n = 5) | 24 | 4 (16.7) | 8 (33.3) | 9 (37.5) | 3 (12.5) | |
| Trophectoderm biopsy | ||||||
| 45,XY,der(14;22)(q10;q10) | 36 | 1 | – | – | E3 | – |
| 45,XX,der(13;14)(q10;q10) | 31 | 10 | E7 | E11 | E5, 13 | E3, 4, 10, 12, 15, 16 |
| 45,XY,der(14;21)(q10;q10) | 28 | 8 | – | E2, 11 | E5 | E3, 4, 9, 10, 12 |
| 45,XX,der(14;15)(q10;q10) | 26 | 5 | E3, 4 | – | E2 | E5, 6 |
| 46,XY,t(2;18)(q31;q21.1) | 34 | 3 | – | E1, 2 | – | E3 |
| 46,XY,t(11;16)(q13;q12) | 25 | 4 | – | E1 | – | E2, 3, 4 |
| 46,XX,t(4;9)(q11.3;q22) | 36 | 3 | – | E3, 8 | – | E6 |
| 46,XY,t(7;14)(q22;q24.3) | 30 | 9 | E1 | E4, 6, 7, 8 | – | E2, 3, 5, 10 |
| 46,XY,t(1;2)(q32.3;q23) | 33 | 5 | E1 | E9, 10 | E8 | E6 |
| 46,XY,t(1;4)(p32;q31.3) | 33 | 11 | E1, 6, 8 | E9, 10, 14, 19 | E4, 5, 22 | E11 |
| 46,XY,t(5;8)(q34;q23) | 30 | 4 | – | E9 | E5, 14 | E4 |
| 46,XX,t(8;13)(q24.1;q12) | 29 | 1 | – | E6 | – | – |
| 46,XX,t(7;11)(p22;q23) | 37 | 4 | – | E1, 3 | – | E2, 4 |
| 46,XX,t(7;13)(q11.2;q14) | 37 | 2 | E1 | E2 | – | – |
| 46,XY,t(7;10)(q32;q23) | 31 | 1 | – | E2 | – | – |
| 46,XY,t(10;12)(q22;q14) | 34 | 3 | E1, 3 | E4 | – | – |
| Translocations (n = 16) | 74 | 11 (14.9) | 25 (33.8) | 11 (14.9) | 27 (36.4) | |
| All translocations (n = 21) | 98 | 15 (15.3) | 33 (33.7) | 20 (20.4) | 30 (30.6) | |
MA maternal age
Chromosomal analysis of PGD embryos
Interpretable CNV-Seq embryo profiles were obtained from 98 of 113 (87 %) successfully amplified embryo biopsy samples (Table 2, Supplementary Table 1). Results from the remaining 15 embryos could not be obtained due to failed QC caused by suboptimal WGA. From the 98 CNV-Seq results, embryos were initially classified based on the primary translocation abnormality as either balanced or unbalanced, with or without the presence of incidental CNVs (Table 2). Within the balanced group of 50 embryos, there were 20 (40 %) with incidental aneuploidies and 30 (60 %) without any CNVs. Similarly, in the unbalanced group of 48 embryos, there were 15 (31.3 %) with incidental aneuploidies and 33 (68.7 %) without any CNVs. Thus, of the 98 embryos analyzed, only 30 (30.6 %) were suitable for transfer.
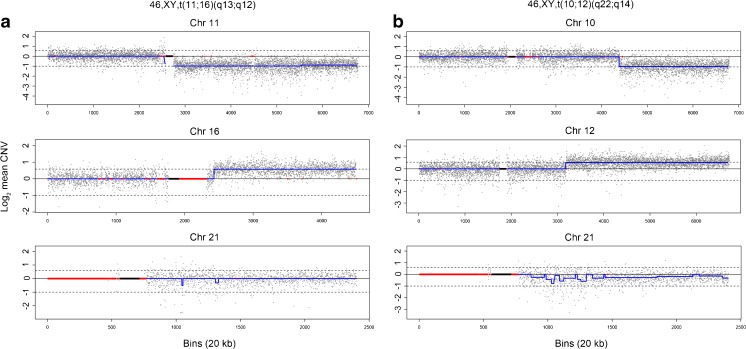
In the unbalanced group comprising 48 embryos, CNV-Seq detected the single trisomy/monosomy derivatives of the unbalanced Roberstsonian translocations and the two terminal duplication/deletion derivatives of each reciprocal translocation (Supplementary Table 1). The terminal CNVs detected mapped precisely on either side of the known breakpoints for each reciprocal translocation and the copy number along the length of the CNV interval paralleled closely with theoretical values. Two examples of the segmental imbalances resulting from t(11;16)(q13;q12) and t(10;12)(q22;q14) derivatives are shown in Fig. 1a, b, respectively. In the couple with a t(2;9)(p23;q34) balanced translocation (case 3, embryos 3, 14, and 17), CNV-Seq precisely identified the 0.8-Mb imbalance at the terminal end of 9q and the 26.48 Mb imbalance at the terminal end of 2p (Supplementary Table 1).
Fig. 1.
CNV-Seq chromosome plots of unbalanced embryos. a Unbalanced derivatives of t(11;16)(q13;q12). The two terminal imbalances were detected as an 80.1-Mb 11q deletion and a 41.5-Mb 16q duplication, with mean copy numbers of 1.0 and 3.0, respectively. The control chromosome 21 has a mean copy number of 2.0. b Unbalanced derivatives of t(10;12)(q22;q14). The two terminal imbalances were detected as an 87.5-Mb 10p deletion and a 70.2-Mb 12q duplication, with mean copy numbers of 1.0 and 3.0, respectively. An incidental aneuploidy involving mosaic chromosome 21 with a mean copy number of 1.8 was detected in the same embryo
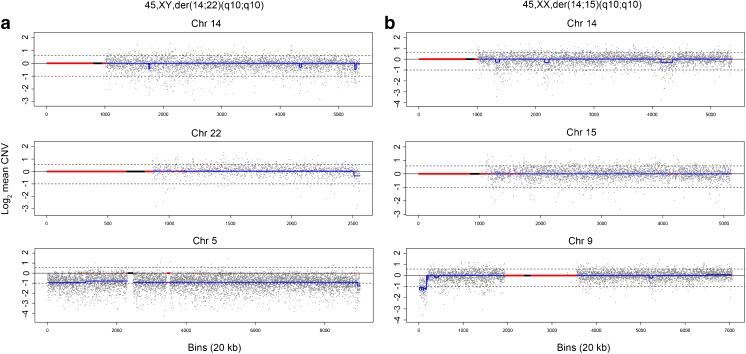
The incidental aneuploidies found in the 20 balanced and 15 unbalanced embryos were extremely diverse (Table 3, Supplementary Table 1). Of the 35 embryos, 21 (60 %) had single aneuploidies, four double aneuploidies (11.4 %), and 10 (28.6 %) complex aneuploidies associated with three or more chromosomes. The 21 embryos with single aneuploidies comprised 13 with either monosomies or trisomies (example, Fig. 2a), six with a non-translocation chromosome segmental imbalance (example, Fig. 2b), and two with mosaicism (example, Fig. 1b). The four embryos with double aneuploidies only involved trisomies and monosomies. In all 10 embryos with complex abnormalities, seven exhibited combined autosomal trisomies/monosomies and segmental imbalances of non-translocation chromosomes and three had additional mosaicism.
Table 3.
Chromosomal characteristics of embryo biopsies
| Type of abnormality | Blastomeres | TE cells | All cells (%) |
|---|---|---|---|
| Embryo | |||
| Total embryos analyzed | 24 | 74 | 98 |
| Balanced + euploidy | 3 | 27 | 30 (30.6) |
| Balanced + aneuploidy | 9 | 11 | 20 (20.4) |
| Single aneuploidy | 6 | 7 | 13 (65.0) |
| Double aneuploidy | 2 | 2 | 4 (20.0) |
| Complex aneuploidya | 1 | 2 | 3 (15.0) |
| Unbalanced | 8 | 25 | 33 (33.7) |
| Unbalanced + aneuploidy | 4 | 11 | 15 (15.3) |
| Single aneuploidy | 0 | 8 | 8 (53.3) |
| Double aneuploidy | 0 | 0 | 0 (0) |
| Complex aneuploidya | 4 | 3 | 7 (46.7) |
| Chromosomal | |||
| Total chromosomes analyzed | 576 | 1776 | 2352 |
| Euploid | 508 | 1670 | 2178 (92.6) |
| Aneuploid | 68 | 106 | 174 (7.4) |
| Trisomic | 15 | 9 | 24 (13.7) |
| Monosomic | 14 | 11 | 25 (14.4) |
| Mosaic | 8 | 10 | 18 (10.4) |
| Segmental imbalances | 31 | 76 | 107 (61.5) |
| Translocation | 17 | 62 | 79 (73.8) |
| Non-translocation | 14 | 14 | 28 (26.2) |
aComplex aneuploidy = three or more aneuploidies
Fig. 2.
CNV-Seq chromosome plots of balanced embryos. a Balanced chromosomes of der(14;22)(q10;q10). Chromosomes 14 and 22 have a copy number of 2.0. An incidental monosomy 5 with a copy number of 1.0 was detected in the same embryo. b Balanced chromosomes of der(14;15)(q10;q10). Chromosomes 14 and 15 have a copy number of 2.0. An incidental 3.27-Mb 7p terminal deletion with a copy number of 1.0 was detected in the same embryo
At the individual chromosome level, from the 2352 chromosomes analyzed in the 98 embryos, 174 (7.4 %) were associated with a range of different types of structural abnormalities (Table 3, Supplementary Table 1). The most common aneuploidies involved 107 segmental imbalances (61.5 %), comprising 79 (73.8 %) related to the translocation chromosomes and 28 (26.2 %) related to non-translocation chromosomes. There were 49 whole chromosome aneuploidies (28.1 %) consisting of an approximately equal proportion of autosomal trisomies and monosomies, including three instances of monosomy X. The remaining 18 aneuploidies involved mosaic chromosome gains and losses at levels of 20–40 %.
Clinical PGD outcomes
Following CNV-Seq diagnoses of the 21 cohorts of embryos, eight patients (38 %) had all abnormal embryos and 13 patients (62 %) had at least one euploid embryo available for transfer (Table 1). Nine of the 13 couples proceeded to a frozen embryo transfer. Overall, the implantation rate was 80 % and ongoing pregnancy rate at 20 weeks gestation was 60 %. Two patients had a biochemical pregnancy (no fetal heartbeat) and subsequently miscarried. Four of the six couples with successful pregnancies (cases 3, 13, 14, and 15) consented to fetal karyotyping at 18 weeks gestation (Supplementary Table 1). The remaining two couples, one with a twin pregnancy (case 7) and the other with a single pregnancy (case 12), refused karyotyping due to the risk of miscarriage. Of the four fetuses tested, three had normal karyotypes and one had a balanced karyotype, consistent with the original PGD results. All six pregnancies developed to term, resulting in five singleton births and one twin birth.
Discussion
We report the first clinical experience using the high-resolution NGS technology CNV-Seq as the sole diagnostic method without array CGH support for assessing the chromosomal constitution of embryos produced in PGD translocation cycles. In this study of 21 patients, interpretable 24 chromosome profiles were obtained in all 98 embryo biopsy samples that produced a high-quality WGA product. All chromosomal CNVs identified by CNV-Seq showed the correct quantitative copy number change for both loses (deletions) and gains (duplications). In addition, the segmental imbalances resulting from unbalanced translocation derivatives could be mapped precisely to the predicted cytogenetic breakpoints for the 17 different reciprocal translocations analyzed. In addition, CNV-Seq elucidated a variety of incidental chromosomal abnormalities including trisomies, monosomies, non-translocation segmental imbalances, and mosaicism. On this basis, PGD embryos could be clearly separated into those that were either normal/balanced and euploid, aneuploid, unbalanced, or unbalanced and aneuploid, enabling selection of true euploid embryos for intrauterine transfer.
Sequencing read counting NGS-based technologies for PGD are evolving at a rapid pace [29]. These methods rely on PCR-based amplification of embryonic DNA [30] to provide a suitable template for library construction and massively parallel sequencing. While there are inherent differences in the genomic read length, the number of reads generated, and the algorithms used to precisely map the chromosomal position of sequencing reads, in principle, all methods generate a set of mapped reads for data analysis and allow direct comparisons to reference for identifying copy number changes. In validation studies against gold standard array comparative genomic hybridization [31, 32], these methods have been proven to be very reliable and accurate for the detection of whole and partial chromosome aneuploidies. More recently, an NGS method based on a semiconductor ion torrent platform was developed and validated for PGD of translocations, with a reported resolution of 5 Mb for segmental imbalances [20].
The main advantage offered by CNV-Seq over other NGS methods is higher resolution and quantitation of chromosomal aneuploidy. This was achieved by incorporating two key modifications into the standard sequencing pipeline [25]. Firstly, we generated 2.8–3.2 million mapped sequencing reads of 36 bp in length, which is approximately threefold higher than that used by other published NGS methods developed for embryo chromosome analysis [20, 31]. Secondly, sequencing reads are binned at 20-kb intervals (30–35 reads per bin), in contrast to 1-Mb chromosomal intervals used in the data analysis of other NGS methods. Thirdly, samples are run in the same flow cell and the mapped binned data internally compared between 15 or more data sets from the same sequencing batch. Thus, the increased data points generated provide greater statistical power, allowing quantitation of copy number change and precise delineation of CNV. For example, to define a copy number change across a 1-Mb interval, 50 sequential data points can be utilized to calculate the mean CNV and provide a more confident call even if there is local WGA bias in this region of the genome. Thus, this resolution will enable more accurate diagnosis of unbalanced translocations where the breakpoints lie close to the terminal p and q arms. As highlighted in this study, we successfully diagnosed one translocation imbalance involving duplication and deletion of a 0.8-Mb region at the terminal end of 9q. In addition, we also demonstrated detection of mosaicism down to 20 %, which is equivalent to one aneuploid cell in a five-cell blastocyst biopsy. However, while CNV-Seq offers high-resolution chromosome analysis of embryos, the major disadvantage at present is the high depth of sequencing (2.8–3.2 mapped reads) which adds extra cost to the PGD test.
At the clinical level, comprehensive chromosomal analysis of the patient’s embryos using CNV-Seq allowed for clear clinical decision-making in each PGD cycle. In eight of the 21 patient embryo cohorts, all embryos were abnormal as a result of aneuploidy, unbalanced translocations, or both. This important information was rapidly conveyed to these eight patients, and following discussion with their treating doctor, all decided to undertake a second PGD cycle. In the remaining 13 patient embryo cohorts, at least one euploid embryo was identified for transfer. Of note, there were four embryos within these cohorts whereby CNV-Seq identified small pathogenic segmental imbalances that may have been missed by other lower-resolution NGS technologies. Thus, by using CNV-Seq, patients avoided the transfer of any of these embryos and potentially prevented the possibility of an adverse reproductive outcome. In nine transfer cycles with euploid embryos, we achieved seven live healthy births. Importantly, the follow-up fetal karyotypes undertaken matched with the PGD results, validating the accuracy of CNV-Seq at the clinical level.
In conclusion, translocation patients are at a high risk of producing unbalanced embryos with a further risk of incidental aneuploidies. As demonstrated in this pilot clinical study, CNV-Seq proved a reliable and accurate NGS technology for clearly distinguishing between abnormal embryos with deleterious chromosomal constitutions and those that were truly normal/balanced and euploid. The application of a higher-resolution NGS technology like CNV-Seq should further benefit patients undertaking PGD cycles for translocations by avoiding unproductive transfers with embryos harboring more subtle pathogenic chromosomal abnormalities.
Electronic supplementary material
Below is the link to the electronic supplementary material.
(DOCX 37 kb)
Acknowledgments
The study was supported by a grant awarded to Yuanqing Yao by the Key Program of the “Twelfth Five-Year Plan” of the People’s Liberation Army (BWS11J058) and the National High Technology Research and Development Program (SS2015AA020402). Li Wang was supported by a grant from the China Postdoctoral Science Foundation.
Abbreviations
- IVF
In vitro fertilization
- NGS
Next-generation sequencing
- PGD
Preimplantation genetic diagnosis
- t
Translocation
- WGA
Whole genome amplification
Compliance with ethical standards
Ethical approval
Clinical PGD was conducted at the Department of Obstetrics and Gynecology, Chinese PLA General Hospital, Beijing, with approval from the Ethics Committee of the Chinese PLA General Hospital (S2013-092-02). All patients provided written informed consent for IVF and PGD.
Footnotes
Capsule
Next-generation sequencing offers increased chromosome resolution for comprehensive preimplantation genetic diagnosis of translocations.
Wenke Zhang, Ying Liu and Li Wang contributed equally to this work.
Contributor Information
David S. Cram, Phone: +86 10 5325 9188 ext 5073, FAX: +86 10 8430 6824, Email: david.cram@berrygenomics.com
Yuanqing Yao, Phone: +86 10 6693 9258, FAX: +86 10 6693 9258, Email: yqyao_ghpla@163.com.
References
- 1.Ogilvie CM, Braude P, Scriven PN. Successful pregnancy outcomes after preimplantation genetic diagnosis (PGD) for carriers of chromosome translocations. Hum Fertil (Camb) 2001;4:168–71. doi: 10.1080/1464727012000199252. [DOI] [PubMed] [Google Scholar]
- 2.Scriven PN, Handyside AH, Ogilvie CM. Chromosome translocations: segregation modes and strategies for preimplantation genetic diagnosis. Prenat Diagn. 1998;18:1437–49. doi: 10.1002/(SICI)1097-0223(199812)18:13<1437::AID-PD497>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 3.Munné S. Analysis of chromosome segregation during preimplantation genetic diagnosis in both male and female translocation heterozygotes. Cytogenet. Genome Res. 2005;111:305–9. doi: 10.1159/000086904. [DOI] [PubMed] [Google Scholar]
- 4.Suzumori N, Sugiura-Ogasawara M. Genetic factors as a cause of miscarriage. Curr Med Chem. 2010;17:3431–7. doi: 10.2174/092986710793176302. [DOI] [PubMed] [Google Scholar]
- 5.Munné S, Sandalinas M, Escudero T, Fung J, Gianaroli L, Cohen J. Outcome of preimplantation genetic diagnosis of translocations. Fertil Steril. 2000;73:1209–18. doi: 10.1016/S0015-0282(00)00495-7. [DOI] [PubMed] [Google Scholar]
- 6.Hardy K, Handyside AH. Biopsy of cleavage stage human embryos and diagnosis of single gene defects by DNA amplification. Arch Pathol Lab Med. 1992;116:388–92. [PubMed] [Google Scholar]
- 7.De Vos A, Van Steirteghem A. Aspects of biopsy procedures prior to preimplantation genetic diagnosis. Prenat Diagn. 2001;21:767–80. doi: 10.1002/pd.172. [DOI] [PubMed] [Google Scholar]
- 8.Kokkali G, Vrettou C, Traeger-Synodinos J, Jones GM, Cram DS, Stavrou D, et al. Birth of a healthy infant following trophectoderm biopsy from blastocysts for PGD of beta-thalassaemia major. Hum Reprod. 2005;20:1855–9. doi: 10.1093/humrep/deh893. [DOI] [PubMed] [Google Scholar]
- 9.McArthur SJ, Leigh D, Marshall JT, de Boer KA, Jansen RP. Pregnancies and live births after trophectoderm biopsy and preimplantation genetic testing of human blastocysts. Fertil Steril. 2005;84:1628–36. doi: 10.1016/j.fertnstert.2005.05.063. [DOI] [PubMed] [Google Scholar]
- 10.Fischer J, Colls P, Escudero T, Munné S. Preimplantation genetic diagnosis (PGD) improves pregnancy outcome for translocation carriers with a history of recurrent losses. Fertil Steril. 2010;94:283–9. doi: 10.1016/j.fertnstert.2009.02.060. [DOI] [PubMed] [Google Scholar]
- 11.Fiorentino F, Spizzichino L, Bono S, Biricik A, Kokkali G, Rienzi L, et al. PGD for reciprocal and Robertsonian translocations using array comparative genomic hybridization. Hum Reprod. 2011;26:1925–35. doi: 10.1093/humrep/der082. [DOI] [PubMed] [Google Scholar]
- 12.Treff NR, Northrop LE, Kasabwala K, Su J, Levy B, Scott RT., Jr Single nucleotide polymorphism microarray-based concurrent screening of 24-chromosome aneuploidy and unbalanced translocations in preimplantation human embryos. Fertil Steril. 2011;95:1606–12.e1-2. doi: 10.1016/j.fertnstert.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 13.Tan Y, Yin X, Zhang S, Jiang H, Tan K, Li J, et al. Clinical outcome of preimplantation genetic diagnosis and screening using next generation sequencing. Gigascience. 2014;3:30. doi: 10.1186/2047-217X-3-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Munné S. Preimplantation genetic diagnosis for aneuploidy and translocations using array comparative genomic hybridization. Curr. Genomics. 2012;13:463–70. doi: 10.2174/138920212802510457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vanneste E, Voet T, Le Caignec C, Ampe M, Konings P, Melotte C, et al. Chromosome instability is common in human cleavage-stage embryos. Nat Med. 2009;15:577–83. doi: 10.1038/nm.1924. [DOI] [PubMed] [Google Scholar]
- 16.Voet T, Vanneste E, Van der Aa N, Melotte C, Jackmaert S, Vandendael T, et al. Breakage-fusion-bridge cycles leading to inv dup del occur in human cleavage stage embryos. Hum Mutat. 2011;32:783–93. doi: 10.1002/humu.21502. [DOI] [PubMed] [Google Scholar]
- 17.Yin X, Tan K, Vajta G, Jiang H, Tan Y, Zhang C, et al. Massively parallel sequencing for chromosomal abnormality testing in trophectoderm cells of human blastocysts. Biol Reprod. 2013;88:69. doi: 10.1095/biolreprod.112.106211. [DOI] [PubMed] [Google Scholar]
- 18.Huang J, Yan L, Fan W, Zhao N, Zhang Y, Tang F, et al. Validation of multiple annealing and looping-based amplification cycle sequencing for 24-chromosome aneuploidy screening of cleavage-stage embryos. Fertil Steril. 2014;102:1685–91. doi: 10.1016/j.fertnstert.2014.08.015. [DOI] [PubMed] [Google Scholar]
- 19.Wang L, Cram DS, Shen J, Wang X, Zhang J, Song Z, et al. Validation of copy number variation sequencing for detecting chromosome imbalances in human preimplantation embryos. Biol Reprod. 2014;91:37. doi: 10.1095/biolreprod.114.120576. [DOI] [PubMed] [Google Scholar]
- 20.Bono S, Biricik A, Spizzichino L, Nuccitelli A, Minasi MG, Greco E, et al. Validation of a semiconductor next-generation sequencing-based protocol for preimplantation genetic diagnosis of reciprocal translocations. Prenat Diagn. 2015;35:938–44. doi: 10.1002/pd.4665. [DOI] [PubMed] [Google Scholar]
- 21.Fan J, Wang L, Wang H, Ma M, Wang S, Liu Z, et al. The clinical utility of next-generation sequencing for identifying chromosome disease syndromes in human embryos. Reprod. Biomed. Online. 2015;31:62–70. doi: 10.1016/j.rbmo.2015.03.010. [DOI] [PubMed] [Google Scholar]
- 22.Wang H, Wang L, Ma M, Song Z, Zhang J, Xu G, et al. A PGD pregnancy achieved by embryo copy number variation sequencing with confirmation by non-invasive prenatal diagnosis. J Genet Genomics. 2014;41:453–6. doi: 10.1016/j.jgg.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 23.Ruttanajit T, Chanchamroen S, Cram DS, Sawakwongpra K, Suksalak W, Leng X, et al. Detection and quantitation of chromosomal mosaicism in human blastocysts using copy number variation sequencing. Prenat Diagn. 2015;36:154–62. doi: 10.1002/pd.4759. [DOI] [PubMed] [Google Scholar]
- 24.Liang D, Peng Y, Lv W, Deng L, Zhang Y, Li H, et al. Copy number variation sequencing for comprehensive diagnosis of chromosome disease syndromes. J Mol Diagn. 2014;16:519–26. doi: 10.1016/j.jmoldx.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y, Chen Y, Tian F, Zhang J, Song Z, Wu Y, et al. Maternal mosaicism is a significant contributor to discordant sex chromosomal aneuploidies associated with noninvasive prenatal testing. Clin Chem. 2014;60:251–9. doi: 10.1373/clinchem.2013.215145. [DOI] [PubMed] [Google Scholar]
- 26.Liang D, Lv W, Wang H, Xu L, Liu J, Li H, et al. Non-invasive prenatal testing of fetal whole chromosome aneuploidy by massively parallel sequencing. Prenat Diagn. 2013;33:409–15. doi: 10.1002/pd.4033. [DOI] [PubMed] [Google Scholar]
- 27.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–95. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tibshirani R, Wang P. Spatial smoothing and hot spot detection for CGH data using fused lasso. Biostatistics. 2008;9:18–29. doi: 10.1093/biostatistics/kxm013. [DOI] [PubMed] [Google Scholar]
- 29.Handyside AH. 24-chromosome copy number analysis: a comparison of available technologies. Fertil Steril. 2013;100:595–602. doi: 10.1016/j.fertnstert.2013.07.1965. [DOI] [PubMed] [Google Scholar]
- 30.Li N, Wang L, Wang H, Ma M, Wang X, Li Y, et al. The performance of whole genome amplification methods and next-generation sequencing for pre-implantation genetic diagnosis of chromosomal abnormalities. J. Genet. Genomics. 2015;42:151–9. doi: 10.1016/j.jgg.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 31.Fiorentino F, Biricik A, Bono S, Spizzichino L, Cotroneo E, Cottone G, et al. Development and validation of a next-generation sequencing-based protocol for 24-chromosome aneuploidy screening of embryos. Fertil Steril. 2014;101:1375–82. doi: 10.1016/j.fertnstert.2014.01.051. [DOI] [PubMed] [Google Scholar]
- 32.Wells D, Kaur K, Grifo J, Glassner M, Taylor JC, Fragouli E, et al. Clinical utilisation of a rapid low-pass whole genome sequencing technique for the diagnosis of aneuploidy in human embryos prior to implantation. J Med Genet. 2014;51:553–62. doi: 10.1136/jmedgenet-2014-102497. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 37 kb)