Abstract
Variation in the human leukocyte antigen (HLA) genes accounts for one-half of the genetic risk in type 1 diabetes (T1D). Amino acid changes in the HLA-DR and HLA-DQ molecules mediate most of the risk, but extensive linkage disequilibrium complicates the localization of independent effects. Using 18,832 case-control samples, we localized the signal to 3 amino acid positions in HLA-DQ and HLA-DR. HLA-DQβ1 position 57 (previously known; P = 1 × 10−1,355) by itself explained 15.2% of the total phenotypic variance. Independent effects at HLA-DRβ1 positions 13 (P = 1 × 10−721) and 71 (P = 1 × 10−95) increased the proportion of variance explained to 26.9%. The three positions together explained 90% of the phenotypic variance in the HLA-DRB1–HLA-DQA1–HLA-DQB1 locus. Additionally, we observed significant interactions for 11 of 21 pairs of common HLA-DRB1–HLA-DQA1–HLA-DQB1 haplotypes (P = 1.6 × 10−64). HLA-DRβ1 positions 13 and 71 implicate the P4 pocket in the antigen-binding groove, thus pointing to another critical protein structure for T1D risk, in addition to the HLA-DQ P9 pocket.
T1D is a highly heritable autoimmune disease that results from T cell–mediated destruction of insulin-producing pancreatic β cells. The worldwide incidence of T1D ranges from 0.1 per 100,000 persons in China to >36 per 100,000 persons in parts of Europe and has been steadily increasing1. Many autoimmune diseases, including T1D, rheumatoid arthritis, celiac disease and multiple sclerosis, have more genetic risk attributed to variants in the HLA genes within the major histocompatibility complex (MHC) region2–4 located at 6p21.3 than any other locus. HLA genes encode cell surface proteins that display antigenic peptides to effector immune cells to regulate self-tolerance and downstream immune responses. The risk of autoimmunity conferred by HLA molecules is likely the result of variation in amino acid residues at specific positions within the antigen-binding grooves, which may alter the repertoire of presented peptides5–8. In T1D, the largest allelic associations are in the HLA-DRB1–HLA-DQA1–HLA-DQB1 region, a three-gene ‘superlocus’ that encodes HLA-DR and HLA-DQ proteins9,10; additional associations have been identified in the genes encoding HLA-A, HLA-B, HLA-C and HLA-DP11–14.
Todd et al.15 initially identified strong T1D risk conferred by non-aspartate residues at position 57 of HLA-DQβ1. However, this amino acid position alone does not fully explain the HLA-mediated risk of T1D. Subsequently, many amino acid positions in HLA-DQβ1 and HLA-DRβ1 have been hypothesized to modify risk16, but extensive linkage disequilibrium (LD) spanning the 4-Mb MHC region makes it challenging to pinpoint the specific risk-associated variants. In addition, certain heterozygous genotypes confer the greatest disease risk13,17–19, consistent with synergistic interactions between classical HLA alleles. Despite evidence of non-additive effects within the MHC region on autoimmune disease risk, interactions have not been comprehensively examined in T1D. If risk-conferring amino acid positions and their interactions were understood, mechanistic investigation of how autoantigens interact with HLA proteins could become feasible. In this study, we used recently established accurate genotype imputation methods to examine a large case-control sample and rigorously identified independent amino acid positions, as well as interactions within the HLA region, that account for T1D risk (see Supplementary Fig. 1 for a schematic of the analyses).
RESULTS
HLA imputation and association testing
We fine mapped the MHC region in a collection of 8,095 T1D cases and 10,737 controls genotyped with the Immunochip array, provided by the Type 1 Diabetes Genetics Consortium (T1DGC)20–22. The data set included (i) case-control samples collected in the UK and (ii) a pseudocase-control set derived from European families (Online Methods and Supplementary Table 1). Using a set of 5,225 individuals with classical HLA typing as a reference22, we accurately imputed 8,617 binary markers (with minor allele frequency > 0.05%) from ~29 Mb to ~33 Mb on chromosome 6p21.3 (the 4-Mb classical MHC region) with SNP2HLA software21. The resulting data included 7,242 SNPs, 260 2- and 4-digit classical alleles, and amino acid residues at 399 positions for 8 HLA genes (HLA-A, HLA-B, HLA-C, HLA-DRB1, HLA-DQA1, HLA-DQB1, HLA-DPA1 and HLA-DPB1) with high imputation quality (INFO score >0.96; see Supplementary Table 2 for the list of variants and imputation quality). We have previously independently benchmarked the imputation strategy employed in this study for accuracy using a set of 918 samples with gold-standard HLA typing data. Starting with SNPs from the Immunochip genotyping platform and using the T1DGC reference panel, SNP2HLA obtained accuracies of 98.4%, 96.7% and 99.3% for all 2-digit alleles, 4-digit alleles and amino acid polymorphisms, respectively21.
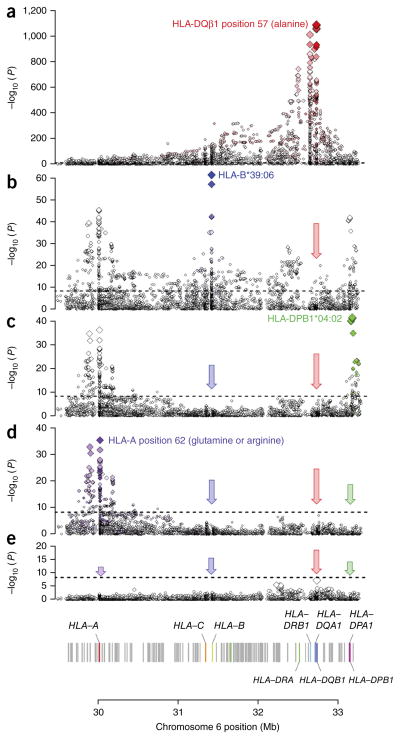
To test for T1D association with a given variant, we used a logistic regression model, assuming the log odds of disease to be proportional to the allelic dosage of the variant. We also included covariates to adjust for sex and region of origin (Supplementary Fig. 2 and Supplementary Note). As expected, the strongest associations with T1D were within the HLA-DRB1–HLA-DQA1–HLA-DQB1 locus. We confirmed that the leading risk variant was the presence of alanine at HLA-DQβ1 position 57 (P = 1 × 10−1,090; odds ratio (OR) = 5.17; Fig. 1a and Supplementary Table 2). In contrast, the single most significantly associated classical allele was HLA-DQB1*03:02 (P = 1 × 10−840), which encodes an alanine at HLA-DQβ1 position 57, although the classical allele was much more weakly associated than the amino acid residue itself. Common classical alleles tagged by each residue at key amino acid positions are listed in Table 1.
Figure 1.
HLA loci independently associated with T1D. Each binary marker was tested for T1D association, using the imputed allelic dosage (between 0 and 2). In each panel, the horizontal dashed line marks P = 5 × 10−8. The color gradient of the diamonds indicates LD (r2) with the most strongly associated variant; the darkest shade represents r2 = 1. (a) The strongest associations were located in the HLA-DRB1–HLA-DQA1–HLA-DQB1 locus. The single strongest risk variant was alanine at HLA-DQβ1 position 57 (OR = 5.17; P = 1 × 10−1,090). See Supplementary Table 2 for unadjusted associations for all markers. (b) Adjusting for all HLA-DRB1, HLA-DQA1 and HLA-DQB1 four-digit classical alleles, the strongest independent signals were in HLA-B. The strongest association was with HLA-B*39:06 (OR = 6.64; P = 1 × 10−75). (c) Adjusting for HLA-DRB1–HLA-DQA1–HLA-DQB1 and HLA-B, the next strongly associated variant was HLA-DPB1*04:02 (OR = 0.48; P = 1 × 10−55). (d) The final independent association was in HLA-A, led by glutamine at HLA-A position 62 (OR = 0.70; P = 1 × 10−25). (e) We found no residual independent association in the HLA-C or HLA-DPA1 genes.
Table 1.
Haplotypes defined by HlA-DQβ1 position 57, HlA-DRβ1 position 13 and HlA-DRβ1 position 71 (control frequency > 0.1%)
| Haplotype | OR | Control freq. | Case freq. | Classical HLA-DQB1 alleles | Classical HLA-DRB1 alleles |
|---|---|---|---|---|---|
| A-H-K | 2.13 | 0.050 | 0.248 | 0201, 0202, 0302, 0304, 0305 | 0401, 0409 |
| A-H-E | 1.33 | 0.005 | 0.016 | 0201, 0202, 0302, 0304, 0305 | 0402, 0437 |
| A-S-K (ref) | 1.00 | 0.145 | 0.332 | 0201, 0202, 0302, 0304, 0305 | 0301, 0302, 0304, 1303 |
| A-H-R | 0.89 | 0.054 | 0.107 | 0201, 0202, 0302, 0304, 0305 | 0403, 0404, 0405, 0406, 0407, 0408, 0410, 0411 |
| A-S-E | 0.53 | 0.001 | 0.001 | 0201, 0202, 0302, 0304, 0305 | 1102, 1103, 1301, 1302, 1304 |
| D-F-R | 0.48 | 0.012 | 0.014 | 0301, 0303, 0401, 0402, 0503, 0601, 0602, 0603 | 0101, 0102, 0901, 1001 |
| A-F-R | 0.43 | 0.001 | 0.001 | 0201, 0202, 0302, 0304, 0305 | 0101, 0102, 0901, 1001 |
| S-R-R | 0.37 | 0.008 | 0.007 | 0502, 0504 | 1601, 1602 |
| V-F-R | 0.35 | 0.106 | 0.085 | 0501, 0604, 0609 | 0101, 0102, 0901, 1001 |
| V-S-E | 0.34 | 0.040 | 0.030 | 0501, 0604, 0609 | 1102, 1103, 1301, 1302, 1304 |
| D-G-R | 0.32 | 0.039 | 0.029 | 0301, 0303, 0401, 0402, 0503, 0601, 0602, 0603 | 0801–0806, 1201, 1202, 1404, 1415 |
| D-H-K | 0.27 | 0.068 | 0.042 | 0301, 0303, 0401, 0402, 0503, 0601, 0602, 0603 | 0401, 0409 |
| V-F-E | 0.24 | 0.013 | 0.006 | 0501, 0604, 0609 | 0103 |
| A-Y-R | 0.18 | 0.103 | 0.043 | 0201, 0202, 0302, 0304, 0305 | 0701 |
| D-S-E | 0.11 | 0.058 | 0.015 | 0301, 0303, 0401, 0402, 0503, 0601, 0602, 0603 | 1102, 1103, 1301, 1302, 1304 |
| D-F-E | 0.08 | 0.004 | 0.001 | 0301, 0303, 0401, 0402, 0503, 0601, 0602, 0603 | 0103 |
| D-H-R | 0.06 | 0.017 | 0.002 | 0301, 0303, 0401, 0402, 0503, 0601, 0602, 0603 | 0403–0408, 0410, 0411 |
| D-S-K | 0.06 | 0.010 | 0.001 | 0301, 0303, 0401, 0402, 0503, 0601, 0602, 0603 | 0301, 0302, 0304, 1303 |
| D-S-R | 0.05 | 0.083 | 0.010 | 0301, 0303, 0401, 0402, 0503, 0601, 0602, 0603 | 1101, 1104, 1106, 1108, 1305, 1401, 1402, 1405, 1406, 1407 |
| D-Y-R | 0.03 | 0.041 | 0.003 | 0301, 0303, 0401, 0402, 0503, 0601, 0602, 0603 | 0701 |
| D-R-A | 0.02 | 0.140 | 0.005 | 0301, 0303, 0401, 0402, 0503, 0601, 0602, 0603 | 1501 |
The 3 amino acid positions define 21 common haplotypes. We list their multivariate ORs and frequencies in controls and cases, as well as the classical four-digit alleles tagged by each haplotype. See Supplementary Table 5b for multivariate ORs and P values for all 31 haplotypes formed by HLA-DQβ1 position 57, HLA-DRβ1 position 13 and HLA-DRβ1 position 71. Freq., frequency.
Three amino acid positions independently drive T1D risk
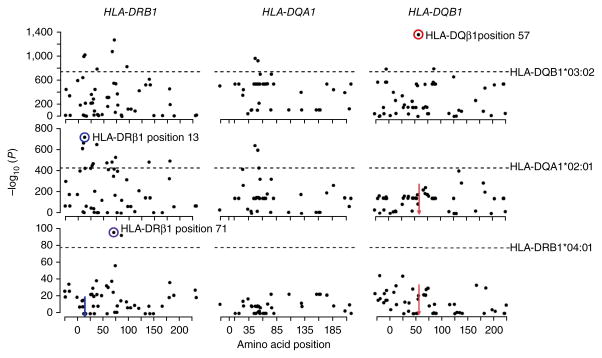
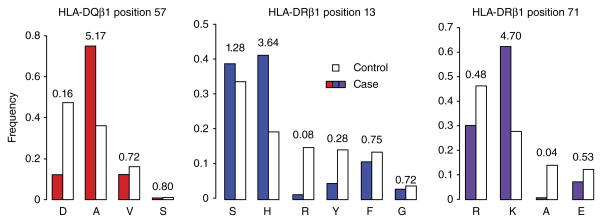
Given the strength and complexity of the association within HLA-DRB1–HLA-DQB1–HLA-DQA1, we aimed to first identify independent effects in this locus before examining the rest of the MHC region. We assessed the significance of multiallelic amino acid positions using conditional analysis by forward search (Online Methods). Unsurprisingly, the position most strongly associated with T1D was HLA-DQβ1 residue 57 (omnibus P = 1 × 10−1,355; Fig. 2 and Supplementary Tables 3 and 4a). At this position, alanine conferred the strongest risk (OR = 5.17; Fig. 3), whereas the most common residue in controls, aspartic acid, was the most protective (OR = 0.16). Conditioning on HLA-DQβ1 position 57, the second independent association was at HLA-DRβ1 position 13 (omnibus P = 1 × 10−721; Fig. 2). At this position, histidine (OR = 3.64) and serine (OR = 1.28) conferred the strongest risk, whereas arginine (OR = 0.08) and tyrosine (OR = 0.28) were protective (Fig. 3 and Supplementary Table 4a). The HLA-DRβ1 residue at position 71 was the third independently associated signal (omnibus P = 1 × 10−95; Fig. 2); lysine conferred strong risk (OR = 4.70), and alanine was strongly protective (OR = 0.04; Fig. 3 and Supplementary Table 4a). We note that, at these positions, the risk-conferring amino acid residues indeed tagged the HLA-DR3 and HLA-DR4 haplotypes, which confer the strongest risk among haplotypes. Histidine at position 13 tagged HLA-DRB1*04:01 and HLA-DRB1*04:04, whereas serine at this position tagged HLA-DRB1*03:01. Lysine at position 71 tagged both HLA-DRB1*03:01 and HLA-DRB1*04:01. The classical alleles tagged by residues at each key amino acid position and multivariate OR estimates for the haplotypes defined by these positions are listed in Table 1 and Supplementary Table 6.
Figure 2.
Amino acid residues at HLA-DQβ1 position 57, HLA-DRβ1 position 13 and HLA-DRβ1 position 71 independently drive T1D risk associated with the HLA-DRB1–HLA-DQA1–HLA-DQB1 locus. To identify each independently associated position, we used conditional haplotypic analysis by forward search, using phased best-guess genotypes. In each panel, the dots mark amino acid positions along the gene (x axis) and their association P values (log10; y axis). The horizontal dashed lines mark the log10 (P value) of the most strongly associated classical allele for each gene. The most strongly associated signals are circled. The colored arrows indicate positions that have been conditioned on. The most strongly associated position was HLA-DQβ1 position 57 (P = 1 × 10−1,355). Conditioning on this site, HLA-DRβ1 position 13 was the next independently associated position (P = 1 × 10−721), followed by HLA-DRβ1 position 71 (P = 1 × 10−95). Each position was much more strongly associated than the best classical allele (HLA-DQB1*03:02, HLA-DQA1*02:01 and HLA-DRB1*04:01, respectively).
Figure 3.
Effect sizes for amino acid residues. Case (colored bars) and control (unfilled bars) frequencies, as well as unadjusted univariate OR estimates, are shown for each residue at HLA-DQβ1 position 57, HLA-DRβ1 position 13 and HLA-DRβ1 position 71.
Given the reported deviation from log-scale additivity for T1D risk effects in the HLA region19,23, we wanted to confirm that the contribution of these effects did not alter the risk-driving amino acid positions. By repeating the forward-search analysis while including non-additive terms in the regression model, we confirmed that HLA-DQβ1 position 57, HLA-DRβ1 position 13 and HLA-DRβ1 position 71 were the top three independent signals under the non-additive model as well as the additive model (Supplementary Fig. 3 and Supplementary Note).
We exhaustively tested all possible combinations of two, three and four amino acid positions for HLA-DRB1–HLA-DQA1–HLA-DQB1 and confirmed that HLA-DQβ1 position 57, HLA-DRβ1 position 13 and HLA-DRβ1 position 71 were the most strongly associated of all 457,450 combinations of 3 amino acids (P = 1 × 10−2,161; Supplementary Table 7). When conditioning on these 3 positions, more than 80 other positions and classical alleles remained highly significant (P < 1 × 10−8; Supplementary Table 4b,c), suggesting the presence of other independent associations. HLA-DQβ1 position −18 (located within the signal peptide) emerged as the fourth most significant association (P = 1 × 10−40) through the forward search; however, in the exhaustive test, many other combinations of four amino acid positions exceeded the goodness-of-fit for HLA-DQβ1 position 57, HLA-DRβ1 position 13, HLA-DRβ1 position 71 and HLA-DQβ1 position −18 (Supplementary Table 7). Therefore, we do not report subsequent positions that emerged through conditional analysis, as we could not confidently claim additional positions as independent drivers of T1D risk.
We wanted to confirm that the top three amino acid positions were not simply tagging the effects of specific haplotypes. To this end, we performed a permutation analysis in which we randomly reassigned amino acid sequences corresponding to each HLA-DRB1, HLA-DQB1 and HLA-DQA1 classical allele and retested for the most strongly associated amino acid positions (Online Methods). This approach preserved haplotypic associations; thus, if certain amino acids were tagging associated haplotypes, equally significant amino acid associations would be found in the permuted data. After 10,000 permutations, no combination of permuted amino acids resulted in a model that equaled or exceeded the goodness-of-fit for HLA-DQβ1 position 57, HLA-DRβ1 position 13 and HLA-DRβ1 position 71 in our data, as measured by either deviance or P value (Supplementary Fig. 4).
Finally, to ensure that the observed effects were not the result of heterogeneity between the UK and European subsets, we repeated the association analysis separately in the two subsets. The two sets yielded highly correlated effect sizes for all binary markers (Pearson r = 0.952; Supplementary Fig. 5a), as well as for all haplotypes formed by residues at HLA-DQβ1 position 57, HLA-DRβ1 position 13 and HLA-DRβ1 position 71 (Pearson r = 0.989; Supplementary Fig. 5b).
Key amino acids are located in the peptide-binding grooves
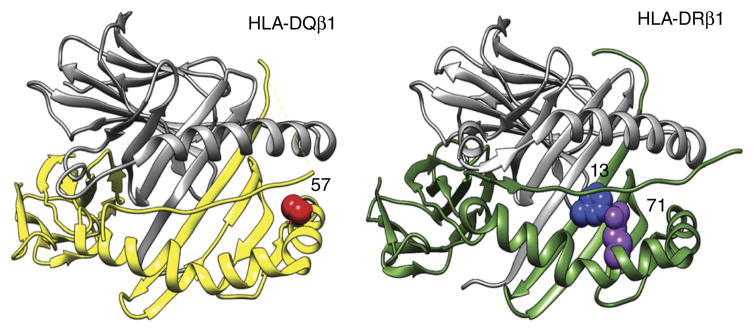
HLA-DQβ1 position 57, HLA-DRβ1 position 13 and HLA-DRβ1 position 71 are each located in the peptide-binding groove of the respective HLA molecule (Fig. 4). HLA-DRβ1 positions 13 and 71 line the P4 pocket of HLA-DR, which has previously been implicated in seropositive2 and seronegative24 rheumatoid arthritis and follicular lymphoma25. Although HLA-DRβ1 positions 13 and 71 are both involved in T1D and rheumatoid arthritis, the effects of individual residues at each position were discordant between the diseases (P < 1 × 10−232; Online Methods and Supplementary Fig. 6).
Figure 4.
HLA-DQβ1 position 57, HLA-DRβ1 position 13 and HLA-DRβ1 position 71 are each located in the respective molecule’s peptide-binding groove. HLA-DRβ1 positions 13 and 71 line the P4 pocket of the HLA-DR molecule.
Variance explained by the three amino acid positions in HLA-DRB1–HLA-DQA1–HLA-DQB1
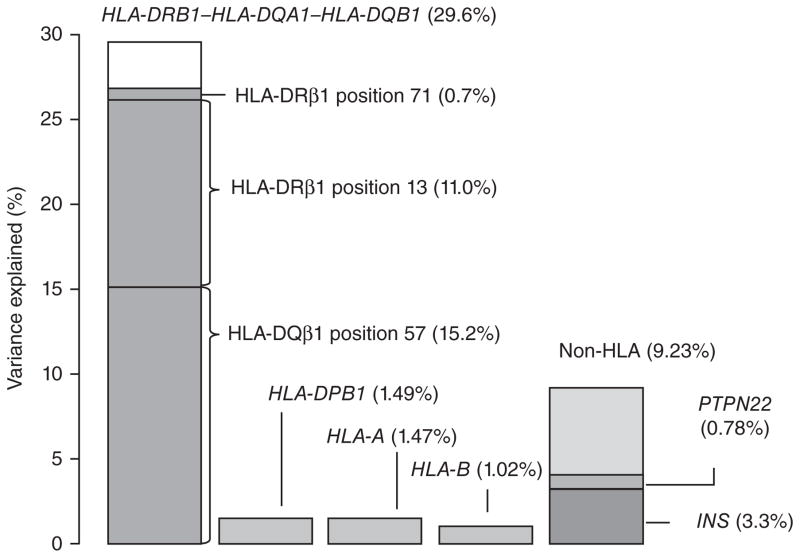
We quantified the proportion of phenotypic variance captured by the three amino acid positions using the liability threshold model26 (Supplementary Note). Assuming a T1D prevalence of 0.4% (ref. 27), the additive effects of all 67 haplotypes for HLA-DRB1–HLA-DQA1–HLA-DQB1 explained 29.6% of the total phenotypic variance (Supplementary Table 6). HLA-DQβ1 position 57 alone explained 15.2% of the total variance, and the addition of HLA-DRβ1 position 13 and HLA-DRβ1 position 71 increased the proportion explained by 11.7%. Therefore, these three amino acid positions together capture 26.9% of the total variance, accounting for over 90% of the T1D-HLA association in this locus (Fig. 5).
Figure 5.
HLA-DQβ1 position 57, HLA-DRβ1 position 13 and HLA-DRβ1 position 71 explain over 90% of the phenotypic variance from the HLA-DRB1–HLA-DQA1–HLA-DQB1 locus. Assuming the liability threshold model and a global T1D prevalence of 0.4%, all haplotypes in HLA-DRB1–HLA-DQA1–HLA-DQB1 together explain 29.6% of total phenotypic variance. HLA-DQβ1 position 57 alone explains 15.2% of the variance; the addition of HLA-DRβ1 position 13 and HLA-DRβ1 position 71 increases the explained proportion to 26.9%. Therefore, these three amino acid positions together capture over 90% of the signal within HLA-DRB1–HLA-DQA1–HLA-DQB1. In contrast, variation in HLA-A, HLA-B and HLA-DPB1 together explain approximately 4% of total variance. Genome-wide independently associated SNPs outside the HLA together explain about 9% of variance; rs678 (in INS) and rs2476601 (in PTPN22) explain 3.3% and 0.78%, respectively.
Independent HLA associations in HLA-B, HLA-DPB1 and HLA-A
We then sought to identify HLA associations with T1D independent of those in the HLA-DRB1–HLA-DQA1–HLA-DQB1 locus. We conservatively conditioned on all HLA-DRB1, HLA-DQA1 and HLA-DQB1 four-digit classical alleles to eliminate all effects at these loci. We observed the next strongest association across the MHC region in HLA-B, where the classical allele HLA-B*39:06 was the most significant signal (OR = 6.64; P = 1 × 10−75; Fig. 1b and Supplementary Table 5a)11. After adjusting for HLA-B*39:06, other classical alleles and amino acid positions in HLA-B remained significantly associated, including HLA-B*18:01 and HLA-B*50:01. Upon additionally adjusting for all HLA-B alleles, HLA-DPB1*04:02 was the next strongest independent signal (OR = 0.47; P < 1 × 10−55; Fig. 1c), which is nearly perfectly tagged by methionine at amino acid position 178 of HLA-DPβ1. Conditioning on HLA-DPB1*04:02, additional associations were present for HLA-DPB1, including amino acid position 65 and HLA-DPB1*01:01 (Supplementary Table 5b). After conditioning on HLA-DPB1 alleles as well, we observed independent effects in HLA-A led by amino acid position 62 (P = 1 × 10−45; Fig. 1d); additional signals included HLA-A*03 and HLA-A*24:02 (Supplementary Table 5c). We observed no independent association with T1D in HLA-C or HLA-DPA1 (Fig. 1e). The independent effects of all haplotypes in HLA-B, HLA-DPB1 and HLA-A together explained ~4% of the total phenotypic variance. The total T1D risk variance explained by additive effects in the eight HLA genes was ~34%, consistent with the estimates by Speed et al.28.
HLA haplotypic interaction effects are common in T1D
The previously observed excess risk of T1D in HLA-DR3/HLA-DR4 heterozygotes (HLA-DRB1*03:01–HLA-DQA1*05:01–HLA-DQB1*02:01/HLA-DRB1*04:XX–HLA-DQA1*03:01–HLA-DQB1*03:02) might represent a synergistic interaction between two distinct alleles23. Here we conducted an unbiased search for interactions among all haplotypes within the HLA-DRB1–HLA-DQA1–HLA-DQB1 locus (Online Methods). As interactions cannot be observed reliably with rare genotypes, we focused this analysis on the seven HLA-DRB1–HLA-DQA1–HLA-DQB1 haplotypes with frequencies >5%; all of these haplotypes had very high imputation accuracies (INFO score > 0.94; Supplementary Table 8 and Supplementary Note).
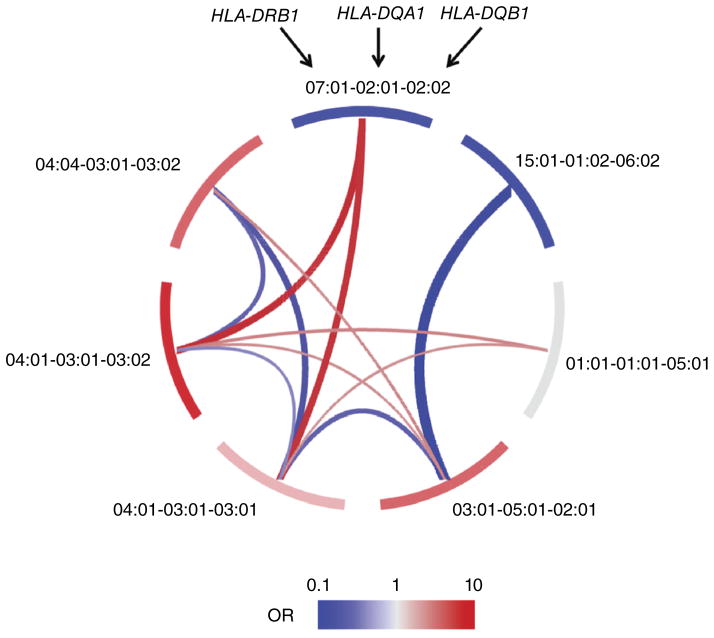
We tested for interactions between all possible pairs of haplotypes using a global multivariate regression model that included 21 interaction terms as well as 7 additive terms. The inclusion of interactions in the model resulted in a statistically significant improvement in fit over the additive model (P = 1.6 × 10−64). Of the 21 potential interactions, 11 were significant after correcting for the 21 tests (P < 0.05/21 = 2.4 × 10−3; Fig. 6, Table 2 and Supplementary Table 9). Consistent with previous reports9,19, we observed a significant interaction between the HLA-DR3 haplotype (HLA-DRB1*03:01–HLA-DQA1*05:01–HLA-DQB1*02:01) and the HLA-DR4 haplotype (HLA-DRB1*04:01–HLA-DQA1*03:01–HLA-DQB1*03:02) (P = 1.2 × 10−5). This interaction resulted in an OR of 30.42, in comparison to an expected OR of 15.51 due to only additive contributions. Likewise, we confirmed an independent interaction between HLA-DRB1*03:01–HLA-DQA1*05:01–HLA-DQB1*02:01 and HLA-DRB1*04:04–HLA-DQA1*03:01–HLA-DQB1*03:02 (P = 1.9 × 10−4).
Figure 6.
Interactions between common HLA-DRB1–HLA-DQA1–HLA-DQB1 haplotypes lead to observed non-additive effects. We exhaustively tested the seven common haplotypes for pairwise interaction. Of the 21 possible pairs, 11 showed significant interaction effects. Along the perimeter of the diagram, each segment represents one haplotype; red or blue color indicates a risk-conferring or protective additive effect for each haplotype, respectively. Each arc connecting two haplotypes represents a significant interaction. Red indicates additional risk due to the interaction beyond the additive effects, whereas blue indicates reduced risk (protection) due to the interaction beyond the additive effects. The thickness of each arc represents the effect size of the interaction (a thicker red arc means a larger risk effect, whereas a thicker blue arc means a more protective effect). See Table 2 and Supplementary Table 9 for P values and effect sizes for all pairwise haplotypic interactions.
Table 2.
Pairwise haplotypic interactions in HLA-DRB1–HLA-DQA1–HLA-DQB1
| HLA-DRB1 | 15:01 | 07:01 | 04:04 | 04:01 | 04:01 | 03:01 | 01:01 | ||
|---|---|---|---|---|---|---|---|---|---|
| HLA-DQA1 | 01:02 | 02:01 | 03:01 | 03:01 | 03:01 | 05:01 | 01:01 | ||
| HLA-DQB1 | 06:02 | 02:02 | 03:02 | 03:02 | 03:01 | 02:01 | 05:01 | ||
|
|
|||||||||
| Amino acids | D-R-A | A-Y-R | A-H-R | A-H-K | D-H-K | A-S-K | V-F-R | ||
| HLA-DRB1 | Amino acids | Additive OR | 0.16 | 0.19 | 2.77 | 5.49 | 1.40 | 2.83 | 1.00 (ref) |
| HLA-DQA1 | |||||||||
| HLA-DQB1 | |||||||||
| 01:01 | V-F-R | 1.00 (ref) | 0.14 | 2.32 | 0.71 | 2.16 | 1.95 | 1.04 | |
| 01:01 | (0.004) | (0.04) | (0.19) | (1.2 × 10−4) | (7.7 × 10−4) | (0.77) | |||
| 05:01 | |||||||||
| 03:01 | A-S-K | 2.83 | 0.09 | 2.24 | 2.12a | 1.96a | 0.32 | ||
| 05:01 | (1.2 × 10−5) | (0.03) | (1.9 × 10−4)a | (1.2 × 10−5)a | (9.2 × 10−11) | ||||
| 02:01 | |||||||||
| 04:01 | D-H-K | 1.40 | 0.26 | 4.78 | 0.23 | 0.48 | |||
| 03:01 | (0.03) | (1.1 × 10−4) | (2.4 × 10−6) | (1.1 × 10−3) | |||||
| 03:01 | |||||||||
| 04:01 | A-H-K | 5.49 | 0.36 | 5.09 | 0.33 | ||||
| 03:01 | (0.03) | (4.2 × 10−5) | (3.5 × 10−5) | ||||||
| 03:02 | |||||||||
| 04:04 | A-H-R | 2.77 | 0.62 | 2.16 | |||||
| 03:01 | (0.03) | (0.08) | |||||||
| 03:02 | |||||||||
| 07:01 | A-Y-R | 0.19 | 0.76 | ||||||
| 02:01 | (0.74) | ||||||||
| 02:02 | |||||||||
| 15:01 | D-R-A | 0.16 | |||||||
| 01:02 | |||||||||
| 06:02 | |||||||||
The table shows, for each given pair of haplotypes, the fold change in OR (from additive effect only) due to interaction. The amino acids column and row denote the residues at HLA-DQβ1 position 57, HLA-DRβ1 position 13 and HLA-DRβ1 position 71 corresponding to each haplotype. For each haplotype pair, the P value of the interaction term is shown in parentheses. Cells in bold indicate interactions that are significant after Bonferroni correction (P < 0.05/21 = 0.0024). The OR of a given diploid genotype is calculated as additive effecthaplotype 1 × additive effecthaplotype 2 × interactionhaplotype 1, haplotype 2 (Supplementary Table 9).
The known HLA-DR3/HLA-DR4 heterozygote effect.
We observed many other significant haplotypic interactions beyond the well-studied HLA-DR3/HLA-DR4 heterozygote effect (Table 2 and Supplementary Table 9). Most interactions increased T1D risk. For example, the combination of HLA-DRB1*04:01–HLA-DQA1*03:01–HLA-DQB1*03:02 and HLA-DRB1*07:01–HLA-DQA1*02:01–HLA-DQB1*02:02 dramatically increased risk by 5.09-fold (beyond the risk predicted by the additive model). Other pairs significantly reduced risk. Notably, whereas HLA-DRB1*04:01–HLA-DQA1*03:01–HLA-DQB1*03:02 and HLA-DRB1*04:04–HLA-DQA1*03:01–HLA-DQB1*03:02 each conferred risk, the heterozygous combination elicited a threefold reduction relative to the expected risk. Because we restricted our analysis to haplotypes with an allele frequency of at least 5%, other interaction effects are likely present but unobserved19.
Interaction effects are mediated by HLA-DQβ1 57 and HLA-DRβ1 13
The HLA-DQ αβ trans heterodimer formed by the proteins encoded by HLA-DQA1*05:01 and HLA-DQB1*03:02 may confer a particularly high risk for individuals with the HLA-DR3/HLA-DR4 genotype owing to its unique antigen-binding properties29. To identify the possible drivers of this haplotypic interaction, we tested pairwise interactions among the HLA-DRB1, HLA-DQA1 and HLA-DQB1 four-digit alleles. We observed a significant interaction between HLA-DQA1*05:01 and HLA-DQB1*03:02 (P = 1.71 × 10−25). However, because of high LD across the locus, several pairs of classical alleles (including HLA-DQB1*02:01/HLA-DQB1*03:02 and HLA-DRB1*03:01/HLA-DQB1*03:02; Supplementary Table 10) achieved similarly significant P values. Therefore, although our model is consistent with a risk-conferring interaction between HLA-DQA1*05:01 and HLA-DQB1*03:02, we cannot eliminate the possibility that interactions between other alleles within the two haplotypes are driving this specific interaction.
We next assessed whether these haplotypic interactions could be explained by amino acid positions. We exhaustively tested for all pairwise interactions among amino acid residues for HLA-DRB1–HLA-DQA1–HLA-DQB1, again limiting the analysis to residues with a frequency of at least 5%. Of the 3,773 pairs of amino acid positions tested, we observed that interactions between HLA-DQβ1 position 57 and HLA-DRβ1 position 13 yielded the largest improvement over the additive model (Supplementary Table 11). We note that two other pairs of amino acid positions achieved similarly significant P values. These analyses suggest that the same amino acid positions that explain the greatest proportion of the additive risk may also be the positions that mediate interaction effects within this locus.
DISCUSSION
Fine mapping the MHC locus in T1D demonstrates that amino acid polymorphisms at HLA-DQβ1 position 57, HLA-DRβ1 position 13 and HLA-DRβ1 position 71 independently modulate T1D risk and capture over 90% of the phenotypic variance explained by the HLA-DRB1–HLA-DQA1–HLA-DQB1 locus (and 80% of the variance explained by the entire MHC region). Previous studies have suggested that other amino acid positions within the HLA class II molecules confer T1D risk (for example, HLA-DRβ1 position 86, HLA-DRβ1 position 74 and HLA-DRβ1 position 57 in the P1, P4 and P9 pockets, respectively)16. Although our analysis highlights the top three amino acid positions as the main contributors of T1D risk, there is also evidence of other allelic effects within the HLA-DRB1–HLA-DQA1–HLA-DQB1 locus; however, the corresponding relative effect sizes were very modest in comparison to those for the three leading positions identified. We note that our results are derived from cases and controls from a relatively homogeneous population (from the UK), and our ability to interrogate rare alleles in this population may be limited. For instance, HLA-DRB1*04:03, a common protective allele in the Sardinian population highlighted by Cucca et al.16, is rare in this data set, with an allele frequency of 0.3%. As such, the observed effects of amino acid positions that best define this allele (HLA-DRβ1 positions 74 and 86) may have been less pronounced than what might be observed in a more diverse data set. Additional variants may be conclusively identified in the future with increased sample size. Finally, although coding variants contribute to the majority of the phenotypic variance in T1D, there is the possibility that there are other mechanisms, such as gene and protein expression, that further modulate susceptibility30,31.
Beyond the previously described HLA-DR3/HLA-DR4 interactions, we find nine additional pairwise interactions between HLA haplotypes that contribute to T1D risk, suggesting that non-additive effects are common within this locus. Notably, we showed that HLA-DQβ1 position 57 and HLA-DRβ1 position 13 are the strongest contributors to both additive and interactive risk effects. Interestingly, the two strongest interacting amino acid positions are in separate HLA molecules (HLA-DQ and HLA-DR, respectively). HLA-DQA1, which is in strong LD with HLA-DRB1 and HLA-DQB1, appears to have a minimal role in modulating T1D risk. This finding suggests that the interaction effects are possibly due to alteration of the antigen presentation repertoire created by the combination of different HLA molecules, rather than the consequence of specific HLA-DQ αβ heterodimers with particular structural features that confer extreme binding affinities.
The HLA amino acid variants identified in our study may mediate recognition of one or more autoantigens and cause autoimmunity through different mechanisms. In particular, our findings implicate the HLA-DR P4 pocket in T1D in addition to the known role of the HLA-DQ P9 pocket; this is the first instance, to our knowledge, where the HLA-DR P4 pocket has an important but secondary role to a different locus (HLA-DQβ1 position 57). The HLA-DR P4 pocket has been shown to have primary roles in other autoimmune diseases. For example, in rheumatoid arthritis, the risk-conferring amino acid residues in P4 likely facilitate the binding of citrullinated peptides7. In T1D, the anti-islet autoantibody reactivity in sera from patients is largely accounted for by four autoantigens (preproinsulin, glutamate decarboxylase (GAD), islet antigen 2 (IA-2) and ZnT8), although the identification of specific peptides that affect autoreactivity is still work in progress8,32–37. Cucca et al. implicated the signal peptide sequences of preproinsulin as potentially important in T1D, by modeling the associations of HLA class II alleles and their polymorphic amino acid positions with structural features of the peptide-binding pockets16. The discovery of critical variants that drive T1D risk enables future functional investigations. Synthesis of HLA molecules containing single-residue alterations at risk-modulating positions may demonstrate the effects of these positions on the physicochemical properties of the antigen-binding pockets. Furthermore, the use of peptide display or small molecule libraries may directly identify and characterize peptides that differentially bind to HLA molecules that differ at risk-modulating positions, thereby uncovering the essential pathogenic peptides and the mechanisms through which they evoke autoimmunity.
ONLINE METHODS
Sample collection
The data set was provided by T1DGC20 and consisted of (i) a UK case-control data set and (ii) a European family-based data set. All samples were collected after obtaining informed consent. The UK case-control data set consisted of a total of 16,086 samples (6,670 cases and 9,416 controls) from 3 collections: (i) cases from the UK-GRID, (ii) shared controls from the British 1958 Birth Cohort and (iii) shared controls from Blood Services controls (data release 4 February 2012; hg18). The UK samples were collected from 13 regions (listed in Supplementary Table 1). The European family-based data set consisted of 10,791 samples (5,571 affected children and 5,220 controls) from 2,699 European-ancestry families (data release 30 January 2013; hg18). All samples were genotyped on the Immunochip array. After quality control, 6,223 and 6,608 markers, respectively, were genotyped in the MHC region between 29 Mb and 45 Mb on chromosome 6 in the 2 data sets. Using the family data, we constructed 1,662 pairs of pseudocase and pseudocontrol samples (Supplementary Note).
HLA imputation
We used SNP2HLA (with default input parameters) to impute SNPs, amino acid residues, indels, and two- and four-digit classical alleles for eight HLA genes in the MHC region from 29,602,876 to 33,268, 403 bp on chromosome 6. We used the reference panel provided by T1DGC, which included 5,225 European samples with classical typing for HLA-A, HLA-B, HLA-C, HLA-DRB1, HLA-DQA1, HLA-DQB1, HLA-DPB1 and HLA-DPA1 4-digit alleles21,22. The imputed genotype data set included 8,961 binary markers before frequency thresholding. For each marker and each individual, two types of output were produced: a phased best-guess genotype (for example, AA/AT/TT) and a dosage, which accounted for imputation uncertainty and could be continuous between 0 (0 copies of the alternative allele) and 2 (2 copies of the alternative allele).
We imputed the UK case-control data set and the European family data set independently; within each set, cases and controls were imputed together to avoid disparity in imputation quality. We used 4,604 and 5,125 SNPs in the MHC region for imputation in the UK and European data sets, respectively. After combining the UK and European data sets, we excluded a total of 344 binary markers because of allele missingness or rareness (allele frequency < 0.05%); we then removed individuals who carried the missing or rare alleles. The final data set after quality control consisted of 18,832 samples, comprising 8,095 cases (including 1,662 pseudocases) and 10,737 controls (including 1,662 pseudocontrols).
Statistical framework
We tested a given variant’s association with disease status using the logistic regression model:
where variant xi may be an imputed dosage or the best-guess genotype for a SNP, classical allele, amino acid or haplotype. β0 is the logistic regression intercept and β1,j is the additive effects of allele j of variant xi. The number of alleles at each variant is m; for a binary variant (presence or absence of xi), m equals 2. The covariate yi,k denotes each region of sample collection (n = 14). We included sex as covariate z. β2 and β3 are the effect sizes of the region and sex covariates, respectively.
To account for population stratification, we included region codes as covariates (Supplementary Note). Samples from the European data set were considered as the fourteenth region. To assess the statistical significance of a tested variant, we calculated the improvement of fit for the model containing the test variant over the null model (with only region and sex as covariates). We calculated the model improvement as deviance, defined by Δdeviancealt – null = −2ln(likelihoodalt/likelihoodnull), which follows a χ2 distribution with m − 1 degrees of freedom, from which we calculated the P value. We considered P = 5 × 10−8 to be the significance threshold.
Analysis of amino acid positions
To test amino acid effects within HLA-DRB1–HLA-DQA1–HLA-DQB1, we applied conditional haplotypic analysis. We tested each single amino acid position by first identifying the m amino acid residues occurring at that position and partitioning all samples into m groups with identical residues at that position. We estimated the effect of each of the m groups using the logistic regression model (including covariates as above) and assessed the significance of model improvement by Δdeviance in comparison to the null model, with m − 1 degrees of freedom. This approach is equivalent to testing a single multiallelic locus for association with m alleles. To test the effect of a second amino acid position while conditioning on the first, we further updated the model to include all unique haplotypes created by residues at both positions. We then tested whether the updated model improved on the previous model by calculating Δdeviance, taking into consideration the increased number of degrees of freedom.
Exhaustive test
To ensure that the independently associated amino acids were not identified only as a result of the forward-search approach, which might possibly converge on local minima, we exhaustively tested all possible combinations of one, two, three and four amino acid positions for HLA-DRB1, HLA-DQA1 and HLA-DQB1. For each number of amino acid positions being combined, we selected the best model according to Δdeviance from the null (with only sex and region as covariates).
Haplotype–amino acid permutation analysis
Given the polymorphic nature of the HLA genes and the strong effect sizes in the HLA-DRB1–HLA-DQA1–HLA-DQB1 locus, we wanted to assess whether the observed associations at HLA-DQβ1 position 57, HLA-DQβ1 position 13 and HLA-DQβ1 position 71 could emerge by chance, owing to the ability of these positions to tag classical alleles with different effects on risk. To eliminate this possibility, we conducted a permutation test. In each permutation, for each of the three genes (for example, HLA-DQB1, HLA-DRB1 and HLA-DQA1), we preserved the sample’s case or control status and the sex and region covariates. To maintain allelic associations, we preserved groups of samples with the same amino acid sequence (four-digit classical allele) for each gene. We then randomly reassigned the amino acid sequence corresponding to each classical allele in each permutation and repeated the forward-search analysis. We repeated this permutation 10,000 times, each time selecting the combinations of two, three and four amino acid positions that produced the best model (as measured by deviance). If the amino acids were merely tagging the effects of certain haplotypes, the effects we observed in the real data would not be more significant than those generated from permutations. To obtain the permutation-based P value, we calculated the proportion of permutated models that exceeded the goodness-of-fit of the best model in the unpermuted data.
Testing for non-additivity and interactions
We defined haplotypes across the HLA-DRB1–HLA-DQA1–HLA-DQB1 locus on the basis of unique combinations of amino acid residues encoded across the three genes. As non-additive effects can be observed only when sufficient numbers of homozygous individuals are present, we limited the interaction analysis to a subset of common haplotypes or classical alleles with frequencies greater than 5%. We excluded all individuals with one or more haplotypes that fell below this threshold.
We constructed an interaction model, which included additive terms for each common haplotype and interaction terms for all possible pairs of common haplotypes:
where ϕ is the interaction effect size. We determined the improvement in fit with each successive model by calculating the change in deviance and used a significance threshold of P = 0.05/h, where h is the total number of interaction parameters added to the original additive model.
HLA-DR3/HLA-DR4 classical allele interactions
To characterize the HLA-DR3/HLA-DR4 interaction, we defined 12 interaction terms, where each term represented a potential interaction between a classical allele on the HLA-DR3 haplotype (HLA-DRB1*03:01, HLA-DQA1*05:01 and HLA-DQB1*02:01) and a classical allele on the HLA-DR4 haplotype (HLA-DRB1*04:01 or HLA-DRB1*04:04, HLA-DQA1*03:01 and HLA-DQB1*03:02). We only looked at trans interactions, as haplotype analyses already account for classical alleles that occur together in cis. We began with a null model that included additive effects for all haplotypes. We then tested each of the 12 interaction terms individually by adding each term to the null model separately. Once again, we used the change in deviance to assess the improvement in fit, using P = 0.05/21 = 2.4 × 10−3 as the threshold for significance.
Amino acid interaction analysis
To determine whether amino acid positions could explain haplotypic interactions, we defined haplotypes across the HLA-DRB1–HLA-DQA1–HLA-DQB1 locus on the basis of the 141 amino acid positions imputed for this locus. To ensure that a sufficiently large number of homozygous individuals were present, we excluded all amino acid residues with a frequency less than 5% before creating the haplotypes. We also excluded any individual who had one or more amino acids that fell below this threshold.
We began with a null model that included additive effects for each amino acid haplotype. Then, for each pair of amino acid positions {q, r}, we added a set of nq × nr interaction terms, where each term specifies a trans interaction between one variant at each position and np represents the total number of variants at position p. Each pair of amino acids was tested in a separate model, and we calculated the change in deviance to determine the improvement in fit. Monomorphic amino acid positions were excluded from this analysis, as they were constant across all individuals.
Supplementary Material
Acknowledgments
This research makes use of resources provided by the Type 1 Diabetes Genetics Consortium (T1DGC), a collaborative clinical study sponsored by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDKD), the National Institute of Allergy and Infectious Diseases (NIAID), the National Human Genome Research Institute (NHGRI), the National Institute of Child Health and Human Development (NICHHD) and Juvenile Diabetes Research Foundation International (JDRFI) and supported by grant U01DK062418. This work is supported in part by funding from the US National Institutes of Health (5R01AR062886-02 (P.I.W.d.B.), 1R01AR063759 (S.R.), 5U01GM092691-05 (S.R.), 1UH2AR067677-01 (S.R.) and R01AR065183 (P.I.W.d.B.)), a Doris Duke Clinical Scientist Development Award (S.R.), the Wellcome Trust (J.A.T.) and the UK National Institute for Health Research (NIHR; J.A.T. and J.M.M.H.) and by a Vernieuwingsimpuls VIDI Award (016.126.354) from the Netherlands Organization for Scientific Research (P.I.W.d.B.). T.L.L. was supported by the German Research Foundation (LE 2593/1-1 and LE 2593/2-1).
Footnotes
AUTHOR CONTRIBUTIONS
X.H. and S.R. conceived the study. X.H., A.J.D., T.L.L., S.R., B.H., P.I.W.d.B. and S.S.R. contributed to the study design and analysis strategy. X.H., A.J.D., T.L.L. and S.R. conducted all analyses. X.H. and A.J.D. wrote the initial manuscript. B.H. contributed critical analytical methods. S.O.-G., W.-M.C. and S.S.R. organized and contributed subject samples and provided SNP genotype data. J.M.M.H., J.A.T., P.I.W.d.B., S.S.R. and S.R. contributed critical writing and review of the manuscript. All authors contributed to the final manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Maahs DM, West NA, Lawrence JM, Mayer-Davis EJ. Epidemiology of type 1 diabetes. Endocrinol Metab Clin North Am. 2010;39:481–497. doi: 10.1016/j.ecl.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Raychaudhuri S, et al. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat Genet. 2012;44:291–296. doi: 10.1038/ng.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trynka G, Wijmenga C, van Heel DA. A genetic perspective on coeliac disease. Trends Mol Med. 2010;16:537–550. doi: 10.1016/j.molmed.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 4.Gourraud PA, Harbo HF, Hauser SL, Baranzini SE. The genetics of multiple sclerosis: an up-to-date review. Immunol Rev. 2012;248:87–103. doi: 10.1111/j.1600-065X.2012.01134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee KH, Wucherpfennig KW, Wiley DC. Structure of a human insulin peptide–HLA-DQ8 complex and susceptibility to type 1 diabetes. Nat Immunol. 2001;2:501–507. doi: 10.1038/88694. [DOI] [PubMed] [Google Scholar]
- 6.Astill TP, Ellis RJ, Arif S, Tree TI, Peakman M. Promiscuous binding of proinsulin peptides to Type 1 diabetes–permissive and –protective HLA class II molecules. Diabetologia. 2003;46:496–503. doi: 10.1007/s00125-003-1070-3. [DOI] [PubMed] [Google Scholar]
- 7.Scally SW, et al. A molecular basis for the association of the HLA-DRB1 locus, citrullination, and rheumatoid arthritis. J Exp Med. 2013;210:2569–2582. doi: 10.1084/jem.20131241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Lummel M, et al. Posttranslational modification of HLA-DQ binding islet autoantigens in type 1 diabetes. Diabetes. 2014;63:237–247. doi: 10.2337/db12-1214. [DOI] [PubMed] [Google Scholar]
- 9.Erlich H, et al. HLA DR-DQ haplotypes and genotypes and type 1 diabetes risk: analysis of the Type 1 Diabetes Genetics Consortium families. Diabetes. 2008;57:1084–1092. doi: 10.2337/db07-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Noble JA, et al. The role of HLA class II genes in insulin-dependent diabetes mellitus: molecular analysis of 180 Caucasian, multiplex families. Am J Hum Genet. 1996;59:1134–1148. [PMC free article] [PubMed] [Google Scholar]
- 11.Nejentsev S, et al. Localization of type 1 diabetes susceptibility to the MHC class I genes HLA-B and HLA-A. Nature. 2007;450:887–892. doi: 10.1038/nature06406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cucca F, et al. The HLA-DPB1–associated component of the IDDM1 and its relationship to the major loci HLA-DQB1, -DQA1, and -DRB1. Diabetes. 2001;50:1200–1205. doi: 10.2337/diabetes.50.5.1200. [DOI] [PubMed] [Google Scholar]
- 13.Deschamps I, et al. HLA genotype studies in juvenile insulin-dependent diabetes. Diabetologia. 1980;19:189–193. doi: 10.1007/BF00275267. [DOI] [PubMed] [Google Scholar]
- 14.Howson JM, Walker NM, Clayton D, Todd JA Type 1 Diabetes Genetics Consortium. Confirmation of HLA class II independent type 1 diabetes associations in the major histocompatibility complex including HLA-B and HLA-A. Diabetes Obes Metab. 2009;11(suppl 1):31–45. doi: 10.1111/j.1463-1326.2008.01001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Todd JA, Bell JI, McDevitt HO. HLA-DQβ gene contributes to susceptibility and resistance to insulin-dependent diabetes mellitus. Nature. 1987;329:599–604. doi: 10.1038/329599a0. [DOI] [PubMed] [Google Scholar]
- 16.Cucca F, et al. A correlation between the relative predisposition of MHC class II alleles to type 1 diabetes and the structure of their proteins. Hum Mol Genet. 2001;10:2025–2037. doi: 10.1093/hmg/10.19.2025. [DOI] [PubMed] [Google Scholar]
- 17.Thomson G, et al. Genetic heterogeneity, modes of inheritance, and risk estimates for a joint study of Caucasians with insulin-dependent diabetes mellitus. Am J Hum Genet. 1988;43:799–816. [PMC free article] [PubMed] [Google Scholar]
- 18.Svejgaard A, Ryder LP. HLA genotype distribution and genetic models of insulin-dependent diabetes mellitus. Ann Hum Genet. 1981;45:293–298. doi: 10.1111/j.1469-1809.1981.tb00340.x. [DOI] [PubMed] [Google Scholar]
- 19.Koeleman BP, et al. Genotype effects and epistasis in type 1 diabetes and HLA-DQ trans dimer associations with disease. Genes Immun. 2004;5:381–388. doi: 10.1038/sj.gene.6364106. [DOI] [PubMed] [Google Scholar]
- 20.Onengut-Gumuscu S, et al. Fine mapping of type 1 diabetes susceptibility loci and evidence for colocalization of causal variants with lymphoid gene enhancers. Nat Genet. 2015;47:381–386. doi: 10.1038/ng.3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jia X, et al. Imputing amino acid polymorphisms in human leukocyte antigens. PLoS ONE. 2013;8:e64683. doi: 10.1371/journal.pone.0064683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown WM, et al. Overview of the MHC fine mapping data. Diabetes Obes Metab. 2009;11(suppl 1):2–7. doi: 10.1111/j.1463-1326.2008.00997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noble JA, Erlich HA. Genetics of type 1 diabetes. Cold Spring Harb Perspect Med. 2012;2:a007732. doi: 10.1101/cshperspect.a007732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han B, et al. Fine mapping seronegative and seropositive rheumatoid arthritis to shared and distinct HLA alleles by adjusting for the effects of heterogeneity. Am J Hum Genet. 2014;94:522–532. doi: 10.1016/j.ajhg.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Foo JN, et al. Coding variants at hexa-allelic amino acid 13 of HLA-DRB1 explain independent SNP associations with follicular lymphoma risk. Am J Hum Genet. 2013;93:167–172. doi: 10.1016/j.ajhg.2013.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Witte JS, Visscher PM, Wray NR. The contribution of genetic variants to disease depends on the ruler. Nat Rev Genet. 2014;15:765–776. doi: 10.1038/nrg3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sivertsen B, Petrie KJ, Wilhelmsen-Langeland A, Hysing M. Mental health in adolescents with Type 1 diabetes: results from a large population-based study. BMC Endocr Disord. 2014;14:83. doi: 10.1186/1472-6823-14-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Speed D, Hemani G, Johnson MR, Balding DJ. Improved heritability estimation from genome-wide SNPs. Am J Hum Genet. 2012;91:1011–1021. doi: 10.1016/j.ajhg.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reichstetter S, Kwok WW, Nepom GT. Impaired binding of a DQ2 and DQ8-binding HSV VP16 peptide to a DQA1*0501/DQB1*0302 trans class II heterodimer. Tissue Antigens. 1999;53:101–105. doi: 10.1034/j.1399-0039.1999.530111.x. [DOI] [PubMed] [Google Scholar]
- 30.Ettinger RA, Liu AW, Nepom GT, Kwok WW. Exceptional stability of the HLA-DQA1*0102/DQB1*0602 αβ protein dimer, the class II MHC molecule associated with protection from insulin-dependent diabetes mellitus. J Immunol. 1998;161:6439–6445. [PubMed] [Google Scholar]
- 31.Miyadera H, Ohashi J, Lernmark A, Kitamura T, Tokunaga K. Cell-surface MHC density profiling reveals instability of autoimmunity-associated HLA. J Clin Invest. 2015;125:275–291. doi: 10.1172/JCI74961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knight RR, et al. A distinct immunogenic region of glutamic acid decarboxylase 65 is naturally processed and presented by human islet cells to cytotoxic CD8 T cells. Clin Exp Immunol. 2015;179:100–107. doi: 10.1111/cei.12436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kronenberg D, et al. Circulating preproinsulin signal peptide–specific CD8 T cells restricted by the susceptibility molecule HLA-A24 are expanded at onset of type 1 diabetes and kill β-cells. Diabetes. 2012;61:1752–1759. doi: 10.2337/db11-1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakayama M, et al. Prime role for an insulin epitope in the development of type 1 diabetes in NOD mice. Nature. 2005;435:220–223. doi: 10.1038/nature03523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baekkeskov S, et al. Identification of the 64K autoantigen in insulin-dependent diabetes as the GABA-synthesizing enzyme glutamic acid decarboxylase. Nature. 1990;347:151–156. doi: 10.1038/347151a0. [DOI] [PubMed] [Google Scholar]
- 36.Karlsen AE, et al. Recombinant glutamic acid decarboxylase (representing the single isoform expressed in human islets) detects IDDM-associated 64,000-Mr autoantibodies. Diabetes. 1992;41:1355–1359. doi: 10.2337/diab.41.10.1355. [DOI] [PubMed] [Google Scholar]
- 37.Roep BO, Peakman M. Antigen targets of type 1 diabetes autoimmunity. Cold Spring Harb Perspect Med. 2012;2:a007781. doi: 10.1101/cshperspect.a007781. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.