Abstract
MicroRNA-122 (miR-122) is the foremost liver-related micro-RNA, but its role in the hepatocyte is not fully understood. To evaluate whether circulating levels of miR-122 are elevated in chronic-HCV for a reason other than hepatic injury, we compared serum level in patients with chronic hepatitis C to other forms of liver injury including patients with acute liver failure and healthy controls. MiR-122 was quantitated using sera from 35 acute liver failure patients (20 acetaminophen-induced, 15 other etiologies), 39 chronic-HCV patients and 12 controls. In parallel, human genomic DNA (hgDNA) levels were measured to reflect quantitatively the extent of hepatic necrosis. Additionally, six HIV–HCV co-infected patients, who achieved viral clearance after undergoing therapy with interferon and ribavirin, had serial sera miR-122 and hgDNA levels measured before and throughout treatment. Serum miR-122 levels were elevated approximately 100-fold in both acute liver failure and chronic-HCV sera as compared to controls (P<0.001), whereas hgDNA levels were only elevated in acute liver failure patients as compared to both chronic-HCV and controls (P<0.001). Subgroup analysis showed that chronic-HCV sera with normal aminotransferase levels showed elevated miR-122 despite low levels of hepatocyte necrosis. All successfully treated HCV patients showed a significant Log10 decrease in miR-122 levels ranging from 0.16 to 1.46, after sustained viral response. Chronic-HCV patients have very elevated serum miR-122 levels in the range of most patients with severe hepatic injury leading to acute liver failure. Eradication of HCV was associated with decreased miR-122 but not hgDNA. An additional mechanism besides hepatic injury may be active in chronic-HCV to explain the exaggerated circulating levels of miR-122 observed.
Keywords: HCV, acetaminophen, biomarker, liver injury, micro-RNA
INTRODUCTION
Micro-RNAs (miRs) are increasingly under investigation as both targets for direct antiviral agents against the hepatitis C virus (HCV) [Jopling et al., 2005; Lanford et al., 2010; Machlin et al., 2012; Janssen et al., 2013] and as biomarkers for cancer and acute liver injury [Mitchell et al., 2008; Wang et al., 2009; Okada et al., 2010; Zhang et al., 2010; Bihrer et al., 2011; Antoine et al., 2013]. They are highly conserved, small, single stranded RNA molecules of either human or viral origin, and are created in the nucleus for future regulation of messenger RNAs (mRNA) [Kim, 2005; Grassmann and Jeang, 2008; Kumar, 2011]. They function by binding to complementary regions of corresponding mRNA strands and forming complexes for degradation, thereby inhibiting translation of protein products, limiting any number of cellular pathways [Kim, 2005; Okada et al., 2010].
Making miRs particularly useful are their tissue specific properties. For the hepatocyte, evidence suggests that microRNA-122 (miR-122) is necessary for HCV replication and hepatocyte differentiation and homeostasis [Jopling et al., 2005; Coulouarn et al., 2009; Kumar, 2011; Kambara et al., 2012; Conrad et al., 2013; Szabo and Bala, 2013]. Knock out of the miR-122 gene is associated with the loss of the hepatic phenotype and progression to cancer [Coulouarn et al., 2009; Hsu et al., 2012]. Though no association between HCV viral load and miR-122 has been observed [Sarasin-Filipowicz et al., 2009; Bihrer et al., 2011], lower cellular miR-122 levels at baseline may be predictive of treatment failure because of decreased cellular response to interferon [Coulouarn et al., 2009; Su et al., 2013].
Few studies have provided quantitation of miR-122 in the circulation in relation to disease type and severity. From first principles, the amount of cellular material released during hepatic injury should correlate in a general way with the degree of necrosis. In this light, circulating human genomic-DNA (hgDNA) was measured as a control, since release of cellular DNA should provide another “passive” serum marker for the quantity of hepatocyte necrosis in each patient [Wong et al., 2004].
MiR-122 sera levels in subjects with acute liver failure, chronic hepatitis C, healthy controls, and from chronic hepatitis C patients undergoing treatment were measured and compared with two other means of quantification of liver injury, alanine aminotransferase (ALT), and hgDNA [Benichou, 1990; Navarro and Senior, 2006].
METHODS
Sample Collection
As part of a pilot study of miR-122, sera were obtained from 35 acute liver failure subjects (20 acetaminophen induced, 6 autoimmune hepatitis, 4 drug induced, 2 hepatitis A virus, 1 hepatitis B virus, 1 Epstein Barr virus, and 1 Herpes Simplex virus). Acute liver failure sera were obtained from the Acute Liver Failure Study Group (ALFSG) National Institute of Diabetes and Digestive and Kidney (NIDDK) bio-sample repository.
The ALFSG registry database contains more than 2,100 patients with acute liver failure admitted to 1 of 23 sites between January 1, 1998 and December 31, 2012. Eligible acute liver failure patients had symptoms of jaundice or illness of less than 26 weeks prior to admission and mental status changes with coagulopathy defined as an international normalized ratio >1.5 without known underlying chronic liver disease. The ALFSG protocol was reviewed and approved by the Institutional Review Board at all of the participating sites and written informed consent was obtained from the patient’s next of kin.
Thirty-nine chronic hepatitis C samples were obtained from the clinics at UT Southwestern Medical Center and Parkland Health and Hospital Systems (22 HCV mono-infected and 17 human immunodeficiency virus [HIV] co-infected). Chronic hepatitis C samples were collected prospectively and stored either as part of a database of chronic hepatitis C patients consenting to future research use of their samples, or were banked during HCV treatment as part of a viral kinetics study in HCV–HIV co-infection conducted at UT Southwestern Medical Center. Current hepatitis C infection was confirmed via a positive RNA polymerase viral load.
Twelve normal human serum samples were purchased commercially to serve as the healthy controls.
Separately, serial sera from six HCV–HIV co-infected patients, who achieved a sustained viral response after treatment with pegylated interferon and ribavirin, had banked serum samples measured for miR-122 and hgDNA at baseline, week 1, week 4, end of treatment (week 48), and at sustained viral response (measured at 12 or 24 weeks after therapy completion).
Corresponding ALT levels for all acute liver failure and chronic hepatitis C patients were obtained from case report forms or patient charts from the date of initial serum collection. ALT levels were not available for controls. Fibrosis stages for chronic hepatitis C patients were retrospectively extracted from biopsy (Batts–Ludwig) or ultrasound reports in patient records from time of sera collection, when available.
Serum miR-122 levels have previously been correlated with plasma levels and hepatocyte expression of miR-122, qualifying its use in this study [Su et al., 2013].
Preparation and Measurement of Serum miR-122 Levels
Total small RNA was extracted from serum with mirVana™ PARIS™ Kit according to the instructions from the manufacturer (Ambion, AM1556, Austin, TX). The eluate containing the microRNA was stored at −20°C, until use. Five microliters of the eluate was used for the reverse transcription reaction. For generation of standard curves, chemically synthesized miR-122 was dissolved in distilled H2O and diluted to varying concentrations (102–106 copies/µl). Input RNA was reverse transcribed using the TaqMan miRNA Reverse Transcription Kit and miRNA-specific stem-loop primers (Applied BioSystems, Carlsbad, CA) in the reverse transcription reaction. Measurement of serum miR-122 was performed using real-time PCR on an Applied BioSystems 7900HT thermocycler and data computed with SDS Absolute Quantification Software Version 2.2.2 (Applied BioSystems).
Preparation and Measurement of Serum Human Genomic DNA Levels
Total human genomic DNA (hgDNA) was extracted from serum by QIAamp DNA Blood Mini kit. The single copy gene beta-actin was chosen to represent hgDNA and was measured with a Taqman real-time PCR assay. Known 10 ng/µl concentrations of hgDNA were purchased from Applied BioSystems and used at varying dilutions in the real-time PCR reaction (3030, 303, 30.3, 3, 1.0 copies/µl) to generate a standard curve for estimating serum hgDNA levels. Measurement of serum hgDNA was performed using real-time PCR on an Applied BioSystems 7900HT thermocycler and data computed with SDS Absolute Quantification Software Version 2.2.2 (Applied BioSystems).
Statistics
The three main study categories were acute liver failure, chronic hepatitis C, and controls. Subgroups were then created for acute liver failure and chronic hepatitis C. Acute liver failure was subdivided into acetaminophen-induced and other causes. To further evaluate the impact of hepatic necrosis and inflammation on miR-122, chronic hepatitis C patients were stratified into two arms by ALT level: Elevated and Normal (>40 and ≤40 IU/L, respectively).
Groups were compared on demographic and clinical features using the non-parametric Kruskal–Wallis test. When the Kruskal–Wallis test was significant, a Tukey type multiple comparisons test was used to determine which groups were significantly different [Dunn, 1964; Elliott and Hynan, 2011]. Spearman rank order correlation (rho) was used to examine the association between ALT, liver fibrosis stage, miR-122, and hgDNA. In the case of both analytes, quantitative results were analyzed as copies/µl to determine relative copy number across different etiologies.
Among the serial sustained viral response samples, change from baseline (copies/µl) was calculated as Log10(Baseline) − Log10(time), where time was week 1, week 4, end of treatment, and sustained viral response; a mixed model analysis of variance (ANOVA) was used to examine change across time for these samples. If the ANOVA was significant, post hoc, pairwise comparisons were used to examine change across the four time points.
IBM© SPSS Statistics V20 (SPSS, Inc., IBM© SPSS Statistics V20, Chicago, IL, 2011) and SAS V9.3 (SAS Institute, Inc., SAS V9.3, Cary, NC, 2010) were used to analyze these data. Statistical significance was set at P<0.05. Figures were created using SPSS Statistics V20, SPSS Statistics V21, or Prism V6 (GraphPad Software Incorporated, San Diego, CA). All figures are presented in a Log10 scale.
RESULTS
Serum miR-122 and hg DNA Levels in Acute Liver Failure and Chronic Hepatitis C
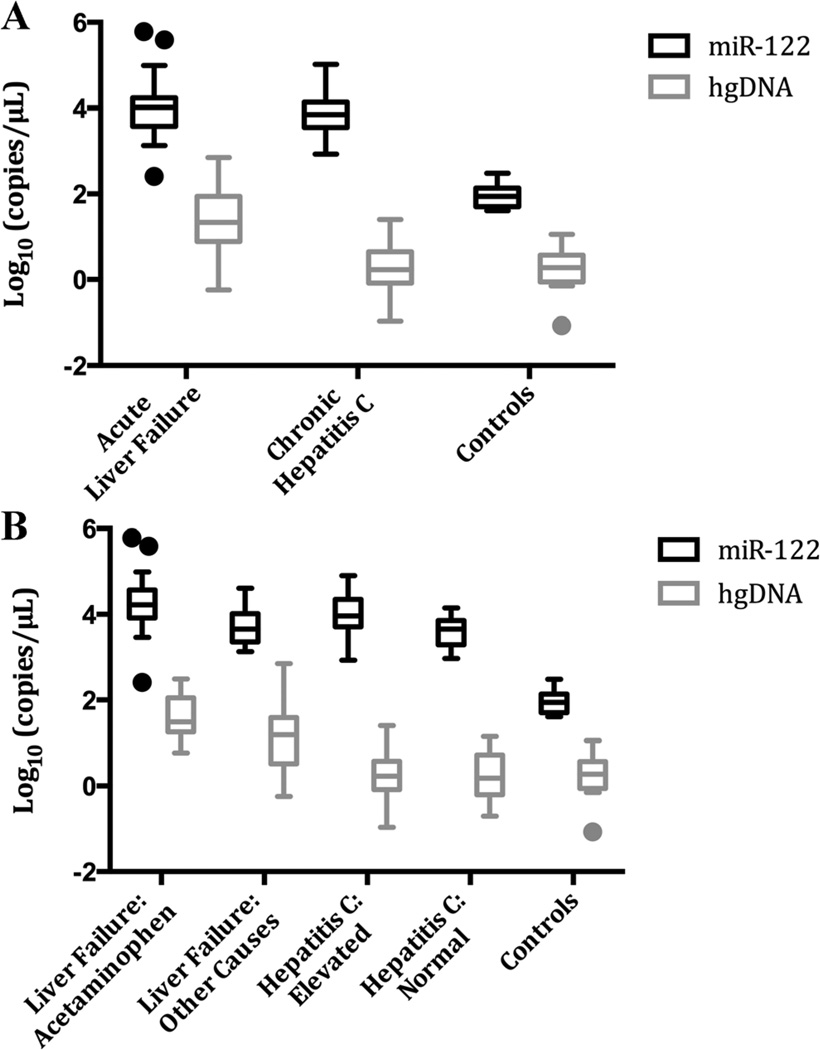
The median serum miR-122 level and range was 9,806 (257.3–597,300) copies/µl in acute liver failure, 7,072 (855.2–103,782) copies/µl in chronic hepatitis C, and 88.7 (40.4–298.7) copies/µl in controls (Fig. 1). Median and range values for subgroups are presented in Table I.
Fig. 1.
Serum miR-122 and hgDNA levels across groups and subgroups. Values have been transformed using Log10. A: Three main study groups. B: Five subgroups. Plots presented using Tukey method. hgDNA, human genomic DNA; ALT, alanine aminotransferase; miR-122, microRNA-122.
TABLE I.
Median and Range miR-122, hgDNA, and ALT Levels Across Subgroups
| Group | Statistic | miR-122 (copy/µl) |
Log10 miR-122 (copy/µl) |
hgDNA (copy/µl) |
Log10 hgDNA (copy/µl) |
ALT (IU/L) |
|---|---|---|---|---|---|---|
| Acetaminophen induced | N | 20 | 20 | 19 | 19 | 20 |
| Median | 16,703 | 4.2 | 31.2 | 1.5 | 3,377 | |
| Min | 257.3 | 2.4 | 5.8 | 0.8 | 175.0 | |
| Max | 597,300 | 5.8 | 308.9 | 2.5 | 12,695 | |
| Other cause | N | 15 | 15 | 15 | 15 | 15 |
| Median | 4,608 | 3.7 | 15.5 | 1.2 | 586.0 | |
| Min | 1,354 | 3.1 | 0.6 | −0.2 | 100.0 | |
| Max | 41,054 | 4.6 | 714.0 | 2.9 | 1,982 | |
| Elevated chronic hepatitis C | N | 22 | 22 | 22 | 22 | 22 |
| Median | 9,404 | 4.0 | 1.8 | 0.3 | 82.0 | |
| Min | 855.2 | 2.9 | 0.1 | −1.0 | 41.0 | |
| Max | 79,922 | 4.9 | 25.3 | 1.4 | 423.0 | |
| Normal chronic hepatitis C | N | 14 | 14 | 14 | 14 | 14 |
| Median | 4,721 | 3.7 | 1.4 | 0.2 | 31.5 | |
| Min | 932.2 | 3.0 | 0.2 | −0.7 | 19.0 | |
| Max | 14,271 | 4.2 | 14.2 | 1.2 | 39.0 | |
| Control | N | 12 | 12 | 12 | 12 | |
| Median | 88.7 | 2.0 | 0.7 | 0.3 | ||
| Min | 40.4 | 1.6 | 0.1 | −1.1 | ||
| Max | 298.7 | 2.5 | 3.6 | 1.1 | ||
| Kruskal–Wallis | P-value | <0.001 | <0.001 | <0.001 |
Sample size, median, and range for miR-122, hgDNA, and ALT levels across the five subgroups. ALT levels were not measured in controls.
The Kruskal–Wallis tests for miR-122 comparing both the three and five sub-groups were significant (Fig. 1). Post hoc comparisons for the three groups showed similar levels in acute liver failure and chronic hepatitis C, with both acute liver failure and chronic hepatitis C markedly elevated as compared to controls. Post hoc pairwise comparison for the five sub-groups showed that all subgroups had greater miR-122 levels than healthy controls. In addition, acetaminophen induced acute liver failure samples were found to be significantly higher than Normal-chronic hepatitis C samples, but not higher than the Elevated-chronic hepatitis C group. No other pairwise comparisons were significant (Table II).
TABLE II.
Kruskal–Wallis Analyses and Post Hoc Comparisons
| P-value | |
|---|---|
| 3 Groups miR-122 | <0.001 |
| Acute liver failure vs. chronic hepatitis C | <0.999 |
| Acute liver failure vs. controls | <0.001 |
| Chronic hepatitis C vs. controls | <0.001 |
| 3 Groups hgDNA | <0.001 |
| Acute liver failure vs. chronic hepatitis C | <0.001 |
| Acute liver failure vs. controls | <0.001 |
| Chronic hepatitis C vs. controls | 0.197 |
| 5 Subgroups miR-122 | <0.001 |
| Acetaminophen induced vs. controls | <0.001 |
| Other causes vs. controls | 0.004 |
| Elevated chronic hepatitis C vs. controls | <0.001 |
| Normal chronic hepatitis C vs. controls | 0.040 |
| Acetaminophen induced vs. normal chronic hepatitis C |
0.021 |
| 5 Subgroups hgDNA | <0.001 |
| Acetaminophen induced vs. elevated chronic hepatitis C |
<0.001 |
| Acetaminophen induced vs. normal chronic hepatitis C |
<0.001 |
| Acetaminophen induced vs. controls | <0.001 |
| Other cause vs. normal chronic hepatitis C | 0.042 |
| Other causes vs. controls | <0.001 |
Kruskal–Wallis and post hoc pairwise results comparison for miR-122 and hgDNA across three main study groups and five subgroups. All pairwise results presented for main groups comparison. Only significant pairwise comparisons presented for subgroup analysis.
The median serum hgDNA level and range was 21.9 (0.6–714.0) copies/µl in acute liver failure, 1.7 (0.1–25.4) copies/µl in chronic hepatitis C, and 0.7 (0.1–3.6) copies/µl in controls (Fig. 1). Median and range values for all five subgroups are presented in Table I.
The Kruskal–Wallis tests for hgDNA comparing both the three and five sub-groups were significant. For the three sub-group post hoc comparisons, the chronic hepatitis C and controls were similar, whereas acute liver failure hgDNA levels were significantly greater than either chronic hepatitis C or controls. Post hoc pairwise comparisons of the five subgroups demonstrated that acetaminophen induced-acute liver failure levels for hgDNA were significantly higher than Elevated-chronic hepatitis C, Normal-chronic hepatitis C, and controls. Other-acute liver failure hgDNA levels were also significantly higher than levels in Normal-chronic hepatitis C patients and controls. No other pairwise comparisons were significant (Table II).
Correlations between ALT, Liver Fibrosis Stage, miR-122, and hgDNA
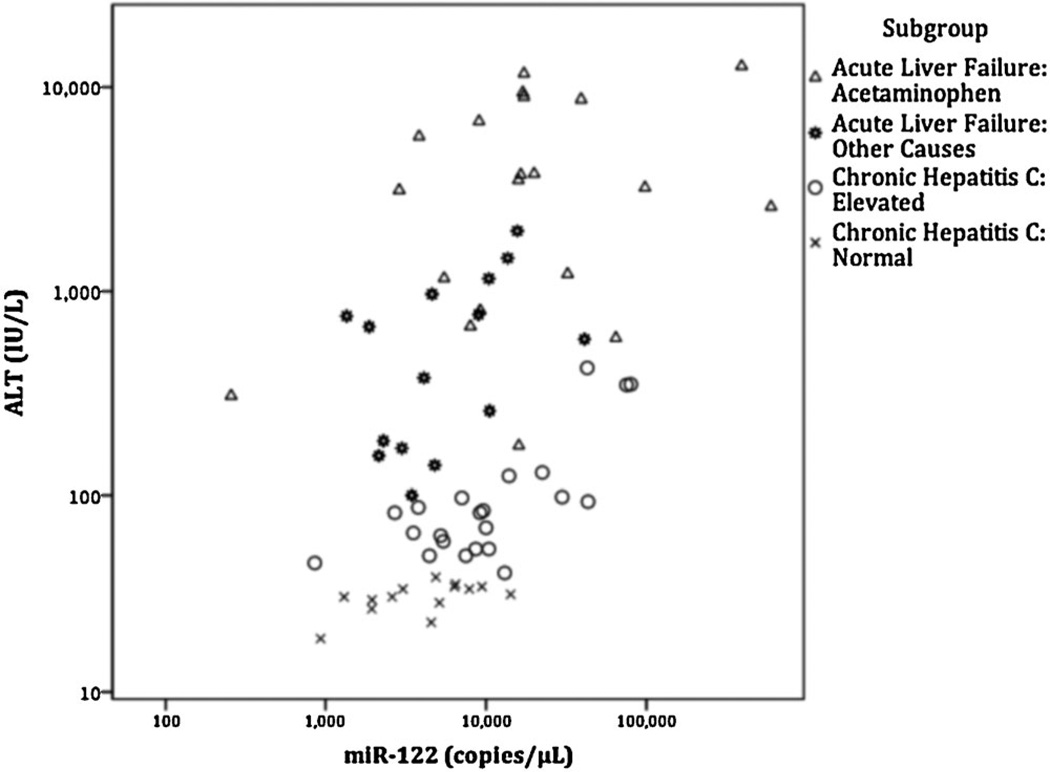
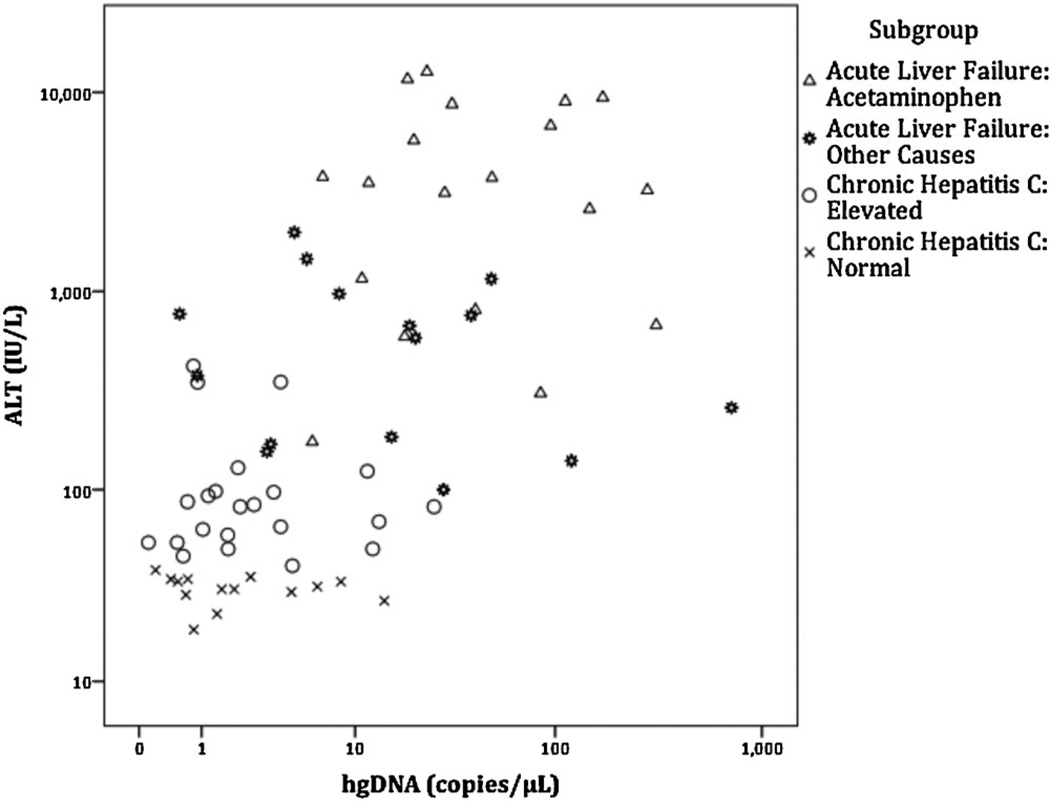
Serum miR-122 (Fig. 2) and hgDNA (Fig. 3) levels were significantly correlated with serum ALT levels across all patient groups. However, sub-group correlations for miR-122 with ALT levels were only significant for the two chronic hepatitis C subgroups, and no hgDNA subgroups were significantly correlated with ALT levels (Table III).
Fig. 2.
Relationship of serum miR-122 levels with serum ALT levels. ALT versus miR-122 levels using Log10 scale. Spearman rank order correlation for all cases was significant (rho = 0.481, P<0.001). Only the subgroups Elevated-chronic hepatitis C (rho = 0.621, P = 0.002) and Normal-chronic hepatitis C (rho = 0.620, P = 0.018) were significantly correlated with ALT. ALT levels were not available for healthy controls.
Fig. 3.
Relationship of serum hgDNA levels with serum ALT levels. ALT versus hgDNA levels using Log10 scale. Spearman rank order correlation for all cases was significant (rho = 0.622, P<0.001). No subgroups were significantly correlated with ALT levels. ALT levels were not available for healthy controls.
TABLE III.
Spearman’s Rank Order Correlations (rho) Between ALT Levels and miR-122 or hgDNA
| Rho | P-value | N | |
|---|---|---|---|
| miR-122 all cases | 0.481 | <0.001 | 71 |
| Acetaminophen induced | 0.310 | 0.184 | 20 |
| Other causes | 0.439 | 0.101 | 15 |
| Elevated chronic hepatitis C | 0.621 | 0.002 | 22 |
| Normal chronic hepatitis C | 0.620 | 0.018 | 14 |
| hgDNA all cases | 0.622 | <0.001 | 70 |
| Acetaminophen induced | 0.104 | 0.673 | 19 |
| Other causes | −0.171 | 0.541 | 15 |
| Elevated chronic hepatitis C | 0.020 | 0.928 | 22 |
| Normal chronic hepatitis C | −0.315 | 0.272 | 14 |
hgDNA, human genomic DNA; ALT, alanine aminotransferase; miR-122, microRNA-122.
Spearman rank order correlations (rho) between ALT levels and either miR-122 or hgDNA for all cases were significant, but significant subgroup correlations were only found for miR-122 and ALT levels in the chronic hepatitis C subgroups.
Twenty-four chronic hepatitis C patients had fibrosis staging documented around time of serum collection. Of these, liver fibrosis stage was not significantly correlated with miR-122 (rho = 0.188) or hgDNA (rho = −0.273).
Serial miR-122 and hgDNA Measurements
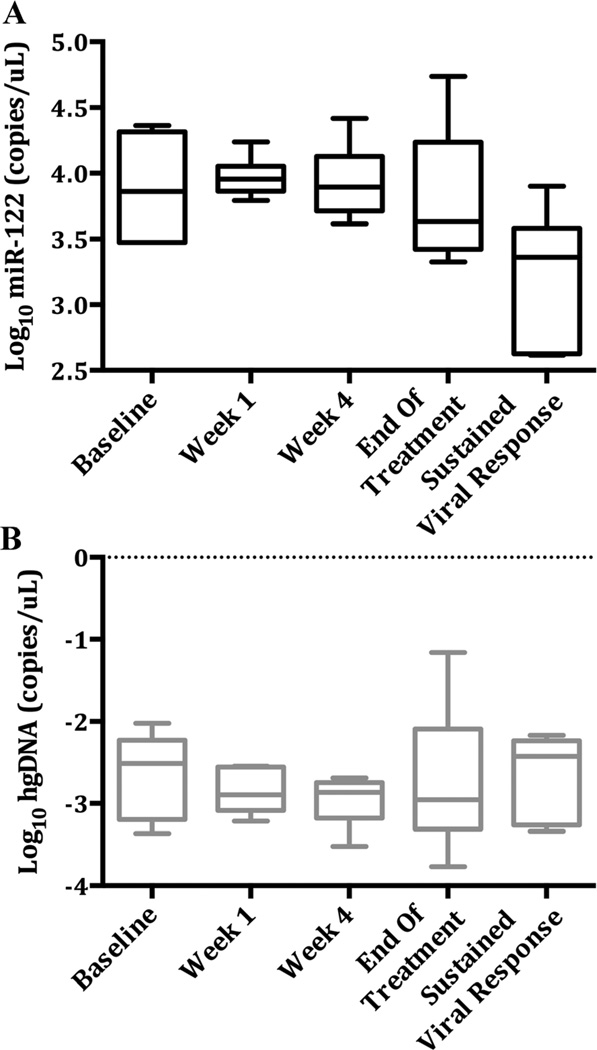
From the sustained viral response serial samples, baseline miR-122 levels ranged from 2,971 to 43,109 copies/µl (median: 16,040). Median miR-122 values decreased at all time points measured, and from Week 1 on after Log10 transformation (Fig. 4). All subjects had lower miR-122 values at sustained viral response from baseline, with a Log10 decrease ranging from 0.16 to 1.46. The mixed model ANOVA was significant (F(3,14) = 4.39, P = 0.023); the significant post hoc, pairwise comparisons were: Week 1 versus sustained viral response (P = 0.006), Week 4 versus sustained viral response (P = 0.009), and end of treatment versus sustained viral response (P = 0.046).
Fig. 4.
Serial miR-122 and hgDNA values across treatment time points for patients undergoing hepatitis C treatment. A: Log miR-122 levels across treatment time points. Median values decrease following Week 1, and are significantly lower at sustained viral response. B: Serial Log10 hgDNA levels across treatment time points. No significant differences were observed in hgDNA levels. Plots presented using Tukey method.
There was one significant outlier identified at end of treatment in the treated chronic hepatitis C group. Examination of patient records identified a simultaneous mild ALT elevation of 72 IU/L for this patient. No additional cause was determined for this persistent elevation in miR-122 despite viral clearance, except to note the ALT elevation, not associated with viral breakthrough. Removing the outlier at end of treatment, the mixed model ANOVA [F(3,13) = 3.78, P = 0.038] and post hoc pairwise comparisons for Week 1 versus sustained viral response and Week 4 versus sustained viral response remained significant, although there was no longer a significant difference between end of treatment and the subsequent sustained viral response time point. No significant changes were observed for serial hgDNA levels over time in the chronic hepatitis C patients.
DISCUSSION
This study demonstrated that serum levels of miR-122 were very elevated to statistically similar amounts in acute liver failure and in chronic hepatitis C, despite a more than 20-fold discrepancy in median ALT levels between the two groups. Both chronic hepatitis C as a whole and the subgroups within chronic hepatitis C showed elevated miR-122 as compared to healthy controls. At the same time, no difference in hgDNA levels were observed between chronic hepatitis C and controls, presumably because of the relatively small amount of necrosis in chronic hepatitis C compared to acute liver failure. Thus, the high miR-122 levels in hepatitis C infection appear not to solely reflect hepatic necrosis, but an additional mechanism, as has been suggested by Coulouarn et al. [2009]. Levels of hgDNA as well as the striking difference in aminotransferase levels confirmed the large differences in cellular necrosis between acute liver failure and chronic hepatitis C patient groups. For the acute liver failure patients, the hgDNA values were 30-fold higher than those observed in controls, while the chronic hepatitis C levels of hgDNA were much lower than acute liver failure sera, only 2-fold elevated compared to controls; this is consonant with the minimal ALT elevations observed in chronic hepatitis C as opposed to the massive elevations in acute liver failure (Table I and Fig. 1).
The serial analysis of the treated hepatitis C group provides evidence that the high miR-122 is associated with the miR-122 over-expression in chronic hepatitis C; after sustained viral response, the elevated miR-122 levels resolved to normal values.
Cellular miRs both inhibit and promote replication of viruses, and cells infected with other viruses have been observed to increase expression of miRs [Jopling et al., 2005; Lecellier et al., 2005; Yeung et al., 2005; Grassmann and Jeang, 2008; Wang et al., 2008]. Thus, it is conceivable that HCV employs similar mechanisms to increase miR-122 production. Since much of the hepatic injury associated with chronic hepatitis C appears to be immune regulated [Rosen, 2011], and miRs are known to have immune-regulatory roles [Xiao and Rajewsky, 2009], it is also possible the observed miR-122 elevation is an intrinsic but non-specific hepatocyte response to chronic infection. This might explain the observed delay in reduction of miR-122 levels until sustained viral response in the serial data, well after cessation of treatment.
The finding that miR-122 levels in chronic hepatitis C are equivalent to acute liver failure levels identifies a problem regarding its use as a biomarker for severity of liver injury in chronic hepatitis C (Fig. 4). Physicians treating chronic hepatitis C patients will not be able to substitute miR-122 for aminotransferase measurements to assess hepatic injury, as highly elevated miR-122 levels appear in all chronic hepatitis C patients, regardless of the degree of ongoing hepatic necrosis in the absence of viral clearance.
This study was limited by the small sample size used in the analyses. Despite these limitations, significant differences across subject groups were identified, suggesting a possible large effect size. The sample size in the serial analysis was too small for conclusive proof of this point, but in each instance serial samples allowed the patient to serve as his own control. Given the established relationship of ALT with liver injury [Benichou, 1990; Navarro and Senior, 2006], and the data’s significant positive correlations of ALT with both miR-122 and hgDNA, the authors acknowledge that increased serum miR-122 in chronic hepatitis C remains partly associated with hepatic injury. Thus, these data support the view that profound upregulation of miR-122 occurs in HCV-infected hepatocytes, augmenting the role of hepatic injury in determining the quantity of miR in the systemic circulation.
In summary, elevated serum miR-122 levels will require further understanding of the role of liver injury and an as yet undetermined additional mechanism of active synthesis of miR-122 that is most evident during chronic HCV infection, and accounts for the high levels of circulating miR-122 observed. Further understanding of miR function may indicate whether induced levels are secondary to an intrinsic hepatocyte response or represent a mechanism unique to HCV. Understanding miR-122 elevations in the hepatitis C setting might lead to new antiviral strategies.
Acknowledgments
Members and institutions participating in the Acute Liver Failure Study Group 1998–2011 are as follows: W.M. Lee, M.D. (Principal Investigator); Anne M. Larson, M.D., Iris Liou, M.D., University of Washington, Seattle, WA; Timothy Davern, M.D., University of California, San Francisco, CA (current address: California Pacific Medical Center, San Francisco, CA), Oren Fix, M.D., University of California, San Francisco; Michael Schilsky, M.D., Mount Sinai School of Medicine, New York, NY (current address: Yale University, New Haven, CT); Timothy McCashland, M.D., University of Nebraska, Omaha, NE; J. Eileen Hay, M.B.B.S., Mayo Clinic, Rochester, MN; Natalie Murray, M.D., Baylor University Medical Center, Dallas, TX; A. Obaid S. Shaikh, M.D., University of Pittsburgh, Pittsburgh, PA; Andres Blei, M.D., Northwestern University, Chicago, IL (deceased), Daniel Ganger, M.D., Northwestern University, Chicago, IL; Atif Zaman, M.D., University of Oregon, Portland, OR; Steven H.B. Han, M.D., University of California, Los Angeles, CA; Robert Fontana, M.D., University of Michigan, Ann Arbor, MI; Brendan McGuire, M.D., University of Alabama, Birmingham, AL; Raymond T. Chung, M.D., Massachusetts General Hospital, Boston, MA; Alastair Smith, M.B., Ch.B., Duke University Medical Center, Durham, NC; Robert Brown, M.D., Cornell/Columbia University, New York, NY; Jeffrey Crippin, M.D., Washington University, St Louis, MO; Edwin Harrison, Mayo Clinic, Scottsdale, AZ; Adrian Reuben, M.B.B.S., Medical University of South Carolina, Charleston, SC; Santiago Munoz, M.D., Albert Einstein Medical Center, Philadelphia, PA; Rajender Reddy, M.D., University of Pennsylvania, Philadelphia, PA; R. Todd Stravitz, M.D., Virginia Commonwealth University, Richmond, VA; Lorenzo Rossaro, M.D., University of California Davis, Sacramento, CA; Raj Satyanarayana, M.D., Mayo Clinic, Jacksonville, FL; and Tarek Hassanein, M.D., University of California, San Diego, CA. The University of Texas Southwestern Administrative Group included Grace Samuel, Ezmina Lalani, Carla Pezzia, and Corron Sanders, Ph.D., Nahid Attar, Linda S. Hynan, Ph.D., and the Medical University of South Carolina Data Coordination Unit included Valerie Durkalski, Ph.D., Wenle Zhao, Ph.D., Catherine Dillon, Holly Battenhouse, and Tomoko Goddard. Serial samples collection during HCV treatment was supported by career development award to Mamta K. Jain K23-AI-065630.
Grant sponsor: National Institute of Diabetes and Digestive and Kidney Diseases (to The Acute Liver Failure Study Group) U-01-DK58369-014; Grant sponsor: Mary Ella and Rollin King Fund of the Southwestern Medical Foundation (additional funding); Grant sponsor: Doris Duke Charitable Foundation (to Clinical Research Fellow Perry H. Dubin)
Conflict of Interest: Mamta K. Jain receives research funding from Vertex Pharmaceuticals, Gilead Sciences, Bristol-Myers Squibb, Janssen Pharmaceuticals, and Boehringer Ingelheim. William M. Lee receives consulting fees from Eli Lilly and Novartis, and has unrestricted grants/contracts from Merck, BMS, BI, Novartis, Vertex, Anadys, Siemens, and Gilead.
REFERENCES
- Antoine DJ, Dear JW, Starkey-Lewis P, Platt V, Coyle J, Masson M, Thanacoody RH, Gray AJ, Webb DJ, Moggs JG, Bateman DN, Goldring CE, Park BK. Mechanistic biomarkers provide early and sensitive detection of acetaminophen-induced acute liver injury at first presentation to hospital. Hepatology. 2013;58:777–787. doi: 10.1002/hep.26294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benichou C. Criteria of drug-induced liver disorders. Report of an international consensus meeting. J Hepatol. 1990;11:272–276. doi: 10.1016/0168-8278(90)90124-a. [DOI] [PubMed] [Google Scholar]
- Bihrer V, Friedrich-Rust M, Kronenberger B, Forestier N, Haupenthal J, Shi Y, Peveling-Oberhag J, Radeke HH, Sarrazin C, Herrmann E, Zeuzem S, Waidmann O, Piiper A. Serum miR-122 as a biomarker of necroinflammation in patients with chronic hepatitis C virus infection. Am J Gastroenterol. 2011;106:1663–1669. doi: 10.1038/ajg.2011.161. [DOI] [PubMed] [Google Scholar]
- Conrad KD, Giering F, Erfurth C, Neumann A, Fehr C, Meister G, Niepmann M. MicroRNA-122 dependent binding of Ago2 protein to hepatitis C virus RNA is associated with enhanced RNA stability and translation stimulation. PLoS ONE. 2013;8:e56272. doi: 10.1371/journal.pone.0056272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulouarn C, Factor VM, Andersen JB, Durkin ME, Thorgeirsson SS. Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. Oncogene. 2009;28:3526–3536. doi: 10.1038/onc.2009.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn OJ. Multiple contrasts using rank sums. Technometrics. 1964;6:241–252. [Google Scholar]
- Elliott AC, Hynan LS. A SAS((R)) macro implementation of a multiple comparison post hoc test for a Kruskal–Wallis analysis. Comput Methods Programs Biomed. 2011;102:75–80. doi: 10.1016/j.cmpb.2010.11.002. [DOI] [PubMed] [Google Scholar]
- Grassmann R, Jeang KT. The roles of microRNAs in mammalian virus infection. Biochim Biophys Acta. 2008;1779:706–711. doi: 10.1016/j.bbagrm.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu SH, Wang B, Kota J, Yu J, Costinean S, Kutay H, Yu L, Bai S, La Perle K, Chivukula RR, Mao H, Wei M, Clark KR, Mendell JR, Caligiuri MA, Jacob ST, Mendell JT, Ghoshal K. Essential metabolic, anti-inflammatory, and antitumorigenic functions of miR-122 in liver. J Clin Invest. 2012;122:2871–2883. doi: 10.1172/JCI63539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, van der Meer AJ, Patick AK, Chen A, Zhou Y, Persson R, King BD, Kauppinen S, Levin AA, Hodges MR. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368:1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- Kambara H, Fukuhara T, Shiokawa M, Ono C, Ohara Y, Kamitani W, Matsuura Y. Establishment of a novel permissive cell line for the propagation of hepatitis C virus by expression of microRNA miR122. J Virol. 2012;86:1382–1393. doi: 10.1128/JVI.06242-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN. MicroRNA biogenesis: Coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- Kumar A. MicroRNA in HCV infection and liver cancer. Biochim Biophys Acta. 2011;1809:694–699. doi: 10.1016/j.bbagrm.2011.07.010. [DOI] [PubMed] [Google Scholar]
- Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, Orum H. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecellier CH, Dunoyer P, Arar K, Lehmann-Che J, Eyquem S, Himber C, Saib A, Voinnet O. A cellular microRNA mediates antiviral defense in human cells. Science. 2005;308:557–560. doi: 10.1126/science.1108784. [DOI] [PubMed] [Google Scholar]
- Machlin ES, Sarnow P, Sagan SM. Combating hepatitis C virus by targeting microRNA-122 using locked nucleic acids. Curr Gene Ther. 2012;12:301–306. doi: 10.2174/156652312802083558. [DOI] [PubMed] [Google Scholar]
- Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro VJ, Senior JR. Drug-related hepatotoxicity. N Engl J Med. 2006;354:731–739. doi: 10.1056/NEJMra052270. [DOI] [PubMed] [Google Scholar]
- Okada H, Kohanbash G, Lotze MT. MicroRNAs in immune regulation–opportunities for cancer immunotherapy. Int J Biochem Cell Biol. 2010;42:1256–1261. doi: 10.1016/j.biocel.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen HR. Clinical practice. Chronic hepatitis C infection. N Engl J Med. 2011;364:2429–2438. doi: 10.1056/NEJMcp1006613. [DOI] [PubMed] [Google Scholar]
- Sarasin-Filipowicz M, Krol J, Markiewicz I, Heim MH, Filipowicz W. Decreased levels of microRNA miR-122 in individuals with hepatitis C responding poorly to interferon therapy. Nat Med. 2009;15:31–33. doi: 10.1038/nm.1902. [DOI] [PubMed] [Google Scholar]
- Su TH, Liu CH, Liu CJ, Chen CL, Ting TT, Tseng TC, Chen PJ, Kao JH, Chen DS. Serum microRNA-122 level correlates with virologic responses to pegylated interferon therapy in chronic hepatitis C. Proc Natl Acad Sci USA. 2013;110:7844–7849. doi: 10.1073/pnas.1306138110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo G, Bala S. MicroRNAs in liver disease. Nat Rev Gastroenterol Hepatol. 2013;10:542–552. doi: 10.1038/nrgastro.2013.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang FZ, Weber F, Croce C, Liu CG, Liao X, Pellett PE. Human cytomegalovirus infection alters the expression of cellular microRNA species that affect its replication. J Virol. 2008;82:9065–9074. doi: 10.1128/JVI.00961-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Zhang S, Marzolf B, Troisch P, Brightman A, Hu Z, Hood LE, Galas DJ. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc Natl Acad Sci USA. 2009;106:4402–4407. doi: 10.1073/pnas.0813371106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong DK, Yuen MF, Tse E, Yuan H, Sum SS, Hui CK, Lai CL. Detection of intrahepatic hepatitis B virus DNA and correlation with hepatic necroinflammation and fibrosis. J Clin Microbiol. 2004;42:3920–3924. doi: 10.1128/JCM.42.9.3920-3924.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C, Rajewsky K. MicroRNA control in the immune system: Basic principles. Cell. 2009;136:26–36. doi: 10.1016/j.cell.2008.12.027. [DOI] [PubMed] [Google Scholar]
- Yeung ML, Bennasser Y, Myers TG, Jiang G, Benkirane M, Jeang KT. Changes in microRNA expression profiles in HIV-1-transfected human cells. Retrovirology. 2005;2:81. doi: 10.1186/1742-4690-2-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Jia Y, Zheng R, Guo Y, Wang Y, Guo H, Fei M, Sun S. Plasma microRNA-122 as a biomarker for viral-, alcohol-, and chemical-related hepatic diseases. Clin Chem. 2010;56:1830–1838. doi: 10.1373/clinchem.2010.147850. [DOI] [PubMed] [Google Scholar]