Abstract
The MDMX protein was identified as a p53-interacting protein with a strong similarity to MDM2. Like Mdm2, Mdmx expression is essential for curbing p53 activity during embryonic development, indicating nonredundant functions of Mdmx and Mdm2. There is now a large body of evidence indicating that cancers frequently up-regulate MDMX expression as a means to dampen p53 tumor-suppressor function. Importantly, MDMX also shows p53-independent oncogenic functions. These data make MDMX an attractive therapeutic target for cancer therapy. Here, we summarize the mechanisms used by cancer cells to increase MDMX expression and promising pharmacological strategies to target MDMX in cancer—in particular, the recent findings that antisense oligonucleotides (ASOs) can be used to efficiently modulate MDMX messenger RNA (mRNA) splicing.
MDMX up-regulation contributes to p53 inactivation in tumors that retain wild-type p53. The reactivation of p53 by targeting MDMX or aspects of its production (e.g., mRNA splicing) is, therefore, an attractive therapeutic option.
The identification of MDMX (also known as MDM4) was first reported almost 20 years ago. It was discovered as a p53-binding protein and showed high structural similarity to MDM2, hence its name. A few years later, MDMX was independently identified in an unbiased screen for MDM2-interacting proteins placing MDMX in the center of the p53–MDM2 network. Although overexpression studies established the ability of MDMX to inhibit the p53 transcriptional activation function, the p53 community remained skeptical about its biological importance. “Why do we need MDMX when MDM2 can do the job,” appeared to be the consensus at the time.
The key genetic finding that Mdmx, like Mdm2, is essential to keep p53 activity in check during embryonic development changed this sentiment. This observation clearly indicated that Mdmx and Mdm2 have nonredundant functions in regulation of p53 activity. Since then, MDMX has attracted more attention in the p53 community and the MDM proteins earned their own international workshop.
Much more insight into the regulation of MDMX abundance and function under physiological and pathological conditions has been obtained. It has especially become clear that MDMX is highly expressed in great numbers of rather rare (retinoblastoma, ocular melanoma) but also more common tumor types like cutaneous melanoma and breast cancer. There is also increasing recognition that MDMX is a promising and rather safe therapeutic target for p53 reactivation therapy. Although most efforts aimed at pharmacological reactivation of p53 in tumors harboring wild-type p53 focused on the MDM2–p53 interaction, recent data argue that targeting specifically MDMX as a means to liberate p53 is likely to be more effective, at least in some specific cases, and less toxic for the patients.
In this work, we will primarily review what we have learned about the mechanisms that drive high MDMX expression in cancer and the opportunities that this creates for developing new, promising anticancer therapeutic strategies.
FROM DISCOVERY TO STRUCTURE AND BIOCHEMICAL FUNCTIONS
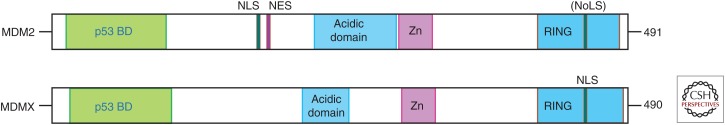
The Mdmx protein was discovered as a p53-binding protein and shows a strong similarity to MDM2 in its primary amino acid sequence (Fig. 1) (Shvarts et al. 1996).
Figure 1.
Schematic representation of the MDM2 and MDMX proteins. The main conserved domains are shown. BD, Binding domain; NLS, nuclear localization signal; NES, nuclear export signal; NoLS, nucleolar localization signal.
Four main conserved regions can be identified: the amino-terminal hydrophobic region important for interaction with p53 and p73, the carboxy-terminal RING domain involved in heterodimerization with MDM2, and possibly other RING domain–containing proteins, the zinc-finger domain and the acidic domain (AD).
The Amino-Terminal p53-Associating Domain
The amino-terminal hydrophobic region or “pocket” is best conserved between MDM2 and MDMX. In vitro studies have indicated similar affinities of the amino-terminal domains of MDMX and MDM2 for the p53 transactivation domain (TAD) (Bottger et al. 1999). However, it should be emphasized that the affinity of the MDMX and MDM2 hydrophobic pockets for p53-TAD in vitro cannot be directly extrapolated to the affinity in living cells. The interaction is affected by other domains within MDM2/MDMX and will be affected by additional factors. First, it has been shown that the very amino-terminal “lid” of MDM2 is involved in the interaction of the hydrophobic pocket with p53-TAD; because the lid of MDM2 appears to be much more flexible than the MDMX lid and it can be affected by modification, this distinction between MDM2 and MDMX will affect their affinity to the p53-TAD (McCoy et al. 2003; Worrall et al. 2009, 2010). In addition, and very interestingly, MDMX contains an internal motif, which is structurally analogous to the interaction domain within p53, leading to an intramolecular interaction within the MDMX protein, strongly diminishing the binding of MDMX to p53. Interaction with other proteins or posttranslational modifications within this intramolecular motif, as has been so clearly shown for CK1α, will modulate the affinity of MDMX for p53 and the functional inhibition (Chen et al. 2005b, 2015; Wu et al. 2012; Bista et al. 2013). Even so, it has been shown that swapping the MDM2 and MDMX amino-terminal domains does not affect the efficiency of p53 ubiquitination in transfection experiments (Kawai et al. 2003b; Meulmeester et al. 2003), indicating a functional similarity.
The amino-terminal pocket of MDM2 not only interacts well with p53-TAD, but also with p73-TAD and affects its function, although the literature is not consistent regarding the biochemical consequences of the MDM2–p73-TAD interaction (i.e., whether MDM2 leads to ubiquitination of p73 or not) (Balint et al. 1999; Dobbelstein et al. 1999; Zeng et al. 1999; Kubo et al. 2010; Wu and Leng 2015). So far, only transfection or in vitro studies have been published showing the interaction between MDMX and p73-TAD (Ongkeko et al. 1999; Wang et al. 2001; Zdzalik et al. 2010). We have recently been able to show endogenous interaction between MDMX and TA-p73α in cells highly expressing MDMX, which could be disrupted by Nutlin-3 treatment. Indeed, the Nutlin-3-induced growth arrest in these cells appeared to be partly determined by up-regulation of TA-p73α activity (J Burger, K Ahmed, and AG Jochemsen, unpubl.).
Although the literature is more ambiguous regarding putative interactions with p63-TAD and a specific interaction with the amino-terminal pockets of MDM2 and MDMX in vitro with very low affinity has been shown, it is highly questionable whether physiological conditions will ever be encountered in which such an interaction can occur (Zdzalik et al. 2010).
In conclusion, the amino-terminal hydrophobic pocket of MDMX interacts with p53-TAD and p73-TAD, and thereby has the potential to inhibit the tumor-suppressor functions of these proteins.
The RING Domain
The second-best conserved domain between MDM2 and MDMX is the carboxy-terminal RING domain. MDM2 functions as an ubiquitin ligase targeting p53 for ubiquitination and degradation. The RING-finger domain is essential for this activity through its ability to interact with the E2 ubiquitin-conjugating enzyme. Despite the conservation, the RING domain of MDMX has no detectable ubiquitin-ligase activity itself (Kawai et al. 2003b; Meulmeester et al. 2003). There is, however, biochemical evidence indicating that the MDMX RING domain can stimulate the ubiquitin-ligase activity of MDM2 through heterodimerization, thereby providing an extended interaction domain for the E2 ubiquitin-conjugating enzyme (Poyurovsky et al. 2007; Linke et al. 2008). In agreement, the Mdm2–Mdmx complex formation appears to be essential to keep p53’s activity under control during mouse embryonic development (Huang et al. 2011; Pant et al. 2011). However, interpretation of these data is complicated by the fact that disruption/mutation of the RING domain of Mdmx most likely will have more profound consequences than just preventing its interaction with Mdm2. In addition, in contrast to Mdm2-null embryonic tissues, the steady-state p53 levels are only very moderately elevated in tissues from Mdmx-null embryos (Francoz et al. 2006). This observation indicates that Mdm2 is perfectly capable of keeping p53 protein at low levels even in the absence of Mdmx.
The RING–RING interaction between MDM2 and MDMX is also necessary for the regulation of MDMX ubiquitination on DNA damage, in that way diminishing the levels of MDMX leading to full activation of p53 (Kawai et al. 2003a; Chen et al. 2005a; Okamoto et al. 2005; Pereg et al. 2005). Intriguingly, 5S rRNA binds to the Mdmx RING domain and inhibits the MDM2-mediated degradation of MDMX; the mechanism of action has not yet been revealed (Li and Gu 2011). The RING of MDMX is also involved in an intramolecular interaction with the acidic middle domain, in that way affecting the subcellular localization. The MDMX RING domain contains a cryptic nuclear localization signal (NLS) that is hidden by the intramolecular interaction. DNA damage leads to phosphorylation of MDMX on several residues, leading to 14-3-3 interaction, disruption of the intramolecular RING–AD interaction, and nuclear localization and degradation (Okamoto et al. 2005; Pereg et al. 2005, 2006; LeBron et al. 2006; Chen et al. 2015).
The Acidic Domain and Zinc-Finger Domain
MDM2 and MDMX also contain a conserved zinc-finger domain, of which less is known regarding functionality. The MDM2 zinc-finger domain is involved in the interaction with and regulation of the retinoblastoma protein and in its regulation by at least some of the ribosomal proteins (Hsieh et al. 1999; Liu et al. 2014). Although MDMX also has been reported to interact with the retinoblastoma protein, the domain involved in this interaction and the putative functional consequences have not been unequivocally elucidated (Uchida et al. 2006; Zhang et al. 2015).
Although not conserved as primary amino acid sequences, both MDM2 and MDMX contain an AD in the middle of the protein. In MDMX, this domain is involved in the intramolecular interactions with the p53-binding domain and the RING domain mentioned above. In MDM2, the AD is essential for ubiquitination of p53, a function that cannot be fulfilled by the analogous domain of MDMX (Kawai et al. 2003b; Meulmeester et al. 2003). The AD in MDM2 is also involved in the interaction with ribosomal proteins and with p14ARF; none of these proteins were found to interact strongly with MDMX. However, both p14ARF and some ribosomal proteins affect the stability of MDMX through their interaction with MDM2 (Gilkes et al. 2006; Li et al. 2012; Daftuar et al. 2013).
Lessons from Mice
Loss-of-function studies in mice confirmed that Mdmx acts as an essential negative regulator of p53, at least during embryonic development. The functional relationship between Mdmx and p53 was clearly genetically established by the observation that inactivation of p53 rescued the embryonic developmental defects in Mdmx-deficient mice (Parant et al. 2001; Finch et al. 2002; Migliorini et al. 2002).
Early embryonic lethality associated with Mdm2 and Mdmx-null mutations has made it difficult to assess the physiological contributions of Mdm2 and Mdmx to the regulation of p53 levels and activity. This was overcome by the generation of Mdmx- and Mdm2-conditional knockout (KO) alleles (Grier et al. 2002, 2006; Steinman and Jones 2002; Mendrysa et al. 2003). For example, both Mdm2 and Mdmx were conditionally inactivated in neuronal progenitors (Xiong et al. 2006). Similarly, conditional expression of p53 was restored specifically in neuronal progenitor cells or in postmitotic cells of mice lacking Mdm2 and/or Mdmx (Francoz et al. 2006). Loss of Mdmx or Mdm2 led to distinct phenotypes but, importantly, all phenotypes disappear in the absence of p53. These observations showed that both Mdm2 and Mdmx are required to inhibit p53 activity in the same cell type and confirmed that Mdm2 cannot compensate for Mdmx loss in vivo, at least in these cell types.
As discussed above, the precise role of Mdmx in the control of p53 stability remains somewhat unclear. p53 levels were below the detection limit in progenitor and in postmitotic neuronal cells lacking Mdmx (Francoz et al. 2006). Similarly, p53 could not be detected at E10.5 in Mdmx-KO neural progenitor cells. In contrast, p53 is clearly detectable in Mdm2-deficient cells at the same stage of development (Xiong et al. 2006). Loss of both Mdm2 and Mdmx does not lead to any further increase in p53 levels compared with loss of Mdm2 alone, suggesting that Mdmx does not participate in the regulation of p53 stability independently of Mdm2 (Francoz et al. 2006). However, whether it does so in an Mdm2-dependent manner remains a matter of debate.
Important insights into Mdmx function also came from the analysis of the hypomorphic p53ΔP mouse model, in which Mdmx-deficiency can be fully rescued (Toledo et al. 2006). In this model, the consequences of Mdmx loss were studied in a compromised p53 context. Together with the genetic models described above, these studies provided the first genetic evidence that Mdmx can inhibit p53 transcriptional activity independent of Mdm2. This is further illustrated by the observation that loss of Mdmx led to an increase in p53 activity in cultured MEFs even in the absence of Mdm2 (Francoz et al. 2006).
Although it is established that direct physical interaction is required, the molecular mechanisms underlying attenuation of p53 transcriptional activity by Mdmx remain unclear. However, there is evidence that Mdmx binding to p53 interferes with the recruitment and/or activity of the p53 cofactor p300 (Sabbatini and McCormick 2002; Danovi et al. 2004).
Importantly, the genetic model systems described above provide clear evidence that Mdmx and Mdm2 cooperate to antagonize p53, at least in some specific embryonic cell types. For instance, ablation of Mdm2 and Mdmx specifically in the central nervous system (CNS) led to a phenotype that was more severe and appeared earlier than the phenotype seen on loss of Mdm2 alone (Xiong et al. 2006). Similarly, the extent of p53-mediated apoptosis was significantly greater in the neuroepithelium of mice lacking both Mdm2 and Mdmx than in mice lacking Mdm2 alone (Francoz et al. 2006). These data indicate that to achieve full p53 reactivation in cells expressing both Mdm2 and Mdmx one needs to target both Mdm proteins.
Another key observation originating from these genetic studies is that, in contrast to Mdm2, inhibition of p53 by Mdmx is only required in a restricted number of cell types and/or under certain physiological conditions. For example, in contrast to Mdm2, conditional inactivation of Mdmx in cardiomyocytes (Grier et al. 2006) and smooth muscle cells of the gastrointestinal (GI) tract (Boesten et al. 2006) led to only minor defects, if any, in histogenesis and tissue homeostasis. Similarly, although transient restoration of the activity of a p53 variant (p53-ER [TAM]) in mice deficient for Mdm2 is ineluctably lethal (Ringshausen et al. 2006), the same genetic manipulation was well tolerated on the Mdmx-deficient background (Garcia et al. 2011).
Consistently, Mdmx protein, in contrast to Mdm2, is preferentially expressed in brain, and to a lesser extent in spleen and thymus (De Clercq et al. 2010). Its expression was extremely low to undetectable in all other adult tissues analyzed. One reasonable interpretation of these data is that, whereas Mdm2 functions in both proliferating and terminally differentiated cells, Mdmx assists Mdm2 in keeping p53 in check only in highly proliferating cells, such as embryonic tissues or in the proliferative compartment of the intestinal epithelium (Valentin-Vega et al. 2009).
These observations have important implications when it comes to p53 reactivation therapeutic strategies. Indeed, whereas systemic exposure to potent MDM2-inhibitors is predicted to be toxic in patients, MDMX inhibition is likely to be well-tolerated.
MDMX UP-REGULATION IN CANCER
Cancers that retain wild-type p53 often find alternative ways to subvert p53 function, through either deregulation of upstream modulators and/or inactivation of downstream effectors (Vogelstein et al. 2000). An increasing body of evidence indicates that p53 inactivation in these cancers is often a consequence of up-regulation of MDMX protein levels.
The overexpression of MDMX affects a wide range of tumor types, including sarcomas, carcinomas, melanomas, and even leukemias.
MDMX up-regulation in cancer can be driven by different mechanisms, such as gene amplification, increased transcription, and protein stabilization.
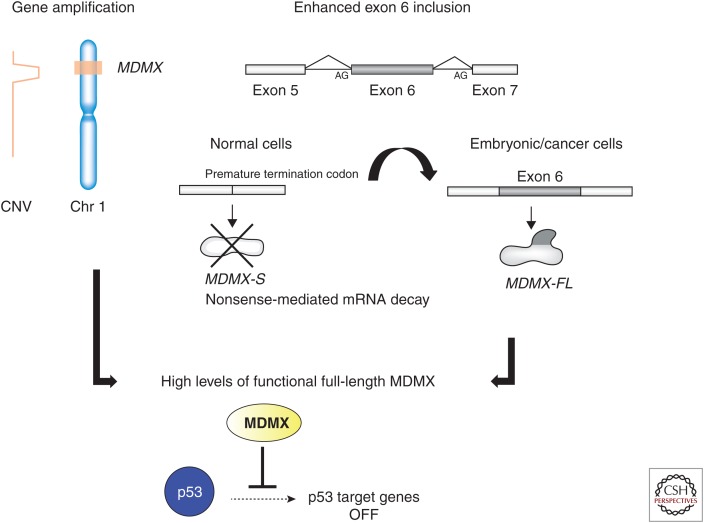
Riemenschneider and colleagues were the first to report aberrant MDMX expression in tumors; they reported MDMX gene amplification correlating with high MDMX messenger RNA (mRNA) levels in ∼4% of wild-type p53 malignant gliomas (Riemenschneider et al. 1999). A few years later, we showed by in situ hybridization that MDMX mRNA levels are increased in ∼20% of breast, lung, and colon cancer samples (Danovi et al. 2004). In breast carcinomas, the overexpression could be explained by gene amplification in 5% of the cases. Together these studies identified gene amplification as one key mechanism that contributes to MDMX up-regulation in human cancers (Fig. 2).
Figure 2.
Gene amplification and enhanced exon 6 inclusion are two important mechanisms that drive MDMX overexpression in human cancers. CNV, Copy number variation; mRNA, messenger RNA; FL, full length.
Amplification of the MDMX gene was reported subsequently in a subset of soft-tissue sarcomas (17%). Interestingly, amplification of MDMX correlated with poor prognosis (Bartel et al. 2005). MDMX mRNA was detected in all tumor samples, but whether mRNA levels correlated with gene copy number was not determined.
Increased MDMX copy number was also found in ∼65% of retinoblastomas. MDMX protein was found highly expressed in retinoblastoma samples and to be essential for retinoblastoma tumor cell proliferation and survival (Laurie et al. 2006). MDMX mRNA was also found highly expressed in all investigated tumors. Surprisingly, however, total mRNA levels did not correlate with MDMX gene copy numbers and total mRNA levels were not significantly higher in retinoblastomas compared with fetal retina (McEvoy et al. 2012). These results strongly indicate that posttranscriptional and/or posttranslational mechanisms contribute to the regulation of MDMX protein levels in tumors.
It actually remains unclear whether enhanced MDMX promoter activity/transcription per se can contribute, at least to some extent, to increased expression in tumors. Jiandong Chen and colleagues show that the MDMX promoter contains, among others, Ets-family transcription-factor binding sites and that transcription of MDMX could be increased by mitogenic signaling activating the mitogen-activated protein kinase (MAPK) pathway (e.g., by insulin-like growth factor [IGF]-I stimulation of serum-starved cells) (Gilkes et al. 2008). Their data also indicate that transient BRAF-V600E expression into H1299 cells increases endogenous Mdmx levels. However, the relevance of these transfection data is not clear as we found no correlation among Mdmx protein levels in the BRAF or NRAS status in melanoma, and inhibition of BRAF-V600E did not cause any significant decrease in MDMX mRNA or protein levels (Marine Laboratory, unpubl.). In the same study, Chen and colleagues reported that ∼50% of colon cancer samples express high MDMX protein levels, correlating with high phospho-ERK staining. Unfortunately, mRNA levels were not determined in these samples. It is, therefore, still unclear whether high ERK activity is causatively involved in up-regulation of MDMX transcription. High MDMX protein or mRNA levels were also reported in some types of leukemias (Han et al. 2007; Bo et al. 2010; Li et al. 2014). However, because expression levels in healthy control tissues were partly missing and protein data were not always available, interpretation of these data is complicated.
Recently, we showed that the MDMX protein is highly expressed in a very substantial fraction (>65%) of cutaneous melanoma (Gembarska et al. 2012). Strikingly, there was no correlation between MDMX protein levels and total MDMX mRNA levels. This observation is again in favor of a model in which up-regulation of MDMX expression in these samples relies on posttranscriptional mechanisms.
Alternative splicing (AS) is one posttranscriptional mechanism that modulates gene expression by adding or removing protein domains, affecting protein activity, or altering the stability of the mRNA transcripts (Kornblihtt et al. 2013; Perez-Ortin et al. 2013). Interestingly, both in normal tissues and tumor cells, the MDMX gene produces two main isoforms, the protein-coding full-length MDMX transcript and a shorter transcript referred to as MDMX-S, an abbreviation for MDMX-Short. The MDMX-S isoform lacks exon 6, resulting in a frameshift and a premature stop codon, thereby directing the potential synthesis of a carboxy-terminal truncated protein, which essentially only retains its p53-binding domain (Rallapalli et al. 1999). Overexpression in cancer cell lines initially identified the MDMX-S protein as a more potent inhibitor of p53 than full-length MDMX. In contrast to MDMX, ectopically expressed MDMX-S protein primarily localized to the nucleus and showed higher affinity for p53 (Rallapalli et al. 1999, 2003). However, to date, there is no conclusive evidence supporting detectable expression of endogenous MDMX-S protein in any normal or cancer cell lines. Concomitant to p53 activation, several types of DNA damage induce a clear splicing switch from MDMX-FL (FL [full length]) to MDMX-S mRNA (Lenos and Jochemsen 2011). Yet, even in these experimental conditions, only the full-length MDMX protein could be detected. Similarly, in tumor cell lines expressing higher levels of MDMX-S mRNA compared with MDMX-FL mRNA, only the full-length protein was detected. We conclude from these data that endogenous MDMX-S is possibly a very unstable protein and that the switch from MDMX-FL to MDMX-S is one mechanism that cells undergoing stress use to lower the amount of MDMX-FL protein, and thereby activate p53 (Lenos and Jochemsen 2011). In addition, there is recent evidence that exon 6 functions as an “nonsense-mediated decay (NMD) switch” exon (Bezzi et al. 2013; Boutz et al. 2015). The in-frame insertion of the premature termination codon in exon 7, which is located far more than 50 nucleotides away from the poly(A) signal, creates a recognition site for the NMD machinery (for review, see Popp and Maquat 2013). Consequently, exon 6 skipping might result in a relatively unstable MDMX-S transcript. Consistent with all of the above, heterozygous mice engineered for an obligatory Mdmx exon 6 skipping showed a decrease in Mdmx-FL protein expression and a concomitant increase in p53 activity (Bardot et al. 2015). Importantly, the MDMX-FL/MDMX-S ratio correlated with overall levels of full-length MDMX protein in panels of several tumor cell lines, including breast cancer cells, osteosarcomas, and uveal melanoma (Lenos and Jochemsen 2011; de Lange et al. 2012; Bezzi et al. 2013). Together, these data raise the possibility that high levels of MDMX protein expression in cancer cells may be a result, at least in part, of their ability to affect the balance between skipping and inclusion of exon 6 (Fig. 2).
Consistent with this possibility, we have recently shown that mouse Mdmx mRNA is unproductively spliced in most normal adult tissues (Dewaele et al. 2015). As discussed above, whereas Mdmx protein is detectable in embryonic tissues, it is undetectable in most adult tissues, except brain and thymus. Remarkably, we observed a striking correlation between Mdmx protein levels and the ratio between Mdmx-FL and Mdmx-S mRNA. Importantly, we also obtained evidence that enhanced exon 6 inclusion correlated with high MDMX protein expression levels in the vast majority (>90%) of melanoma clinical samples and cell lines analyzed (n = 30). These data indicate that MDMX protein expression is primarily controlled by a splicing switch and that the MDMX-FL/MDMX-S ratio is a reliable predictor of MDMX protein abundance in melanoma (Fig. 2) (Dewaele et al. 2015).
Interestingly, we identified the splicing enhancer SRSF3 as necessary, but not sufficient, to promote inclusion of exon 6 in MDMX mRNA in melanoma cells. This observation is consistent with the previously recognized oncogenic role of SRSF3 (Corbo et al. 2013). It is also consistent with previous findings that inhibition of SR protein kinase CLKs, by the small molecule TG003, resulted in a similar decrease in MDMX-FL/MDMX-S ratio and protein abundance, both in neural stem/progenitors and several other cancer cell lines (Bezzi et al. 2013). Indeed, TG003 led to similar effects in all melanoma cell lines tested and reduced their survival rates (Dewaele et al. 2015).
Note, however, that the correlation between MDMX-FL/MDMX-S ratio and MDMX protein abundance is not strict as we identified a few (<10%) melanoma samples in which this correlation could not be established. This indicates that other mechanisms—possibly posttranslational—may also account for MDMX up-regulation in a small proportion of melanoma. Although, in contrast to MDM2, there is so far no evidence for enhanced translation of MDMX mRNA in tumors (Landers et al. 1994, 1997), a number of reports indicate that posttranslational events may contribute to MDMX up-regulation by protein stabilization in human cancers.
Phosphorylation by AKT was found to induce 14-3-3 binding and stabilization of both MDMX and MDM2 proteins and inhibition of p53 activity (Lopez-Pajares et al. 2008). These data are surprising, however, because the identified AKT phosphorylation site, Ser367, is the same site that is phosphorylated by Chk2 to trigger MDMX degradation in response to DNA damage (Chen et al. 2005a; LeBron et al. 2006; Pereg et al. 2006). It may mean that regulation of MDMX stability is more complex than we previously anticipated. One possible explanation for this apparent conundrum is that MDMX levels are also under the control of deubiquitination enzymes (DUBs), which, depending on the cell context, might (or not) counteract the ubiquitin ligase activity of Mdm2. We have shown that USP7 is essential to maintain normal MDMX levels in cells (Meulmeester et al. 2005). High USP7 expression in several tumor types has been reported, and, recently, a small molecule inhibitor of USP7 was found to inhibit the growth of multiple myeloma cells, partly through MDM2 regulation (Chauhan et al. 2012). It is well possible that such a USP7 inhibitor also affects the growth of tumor cells with high levels of MDMX by reducing MDMX levels. Another DUB, USP22, is highly expressed in many tumor types and in general correlates with poor prognosis in patients with lung or hepatocellular carcinomas (Hu et al. 2015; Tang et al. 2015). USP22 is also highly expressed in a high proportion of non-small-cell-lung cancer samples and cell lines (Ding et al. 2015). Targeting USP22 by shRNA inhibits tumor growth and induces apoptosis by activating p53, correlating with down-regulation of MDMX protein levels. Indeed, the investigators show that the effect of USP22 down-regulation could be rescued by ectopic MDMX expression. It will be interesting to further determine MDMX protein abundance in tumors with high USP22 levels to determine the generality of these observations.
Stabilization of MDMX by USP2a has also been reported. Down-regulation of USP2a in N-TERA2 and MCF-7 cells, both expressing high levels of MDMX protein, resulted in decreased MDMX levels and growth inhibition (Allende-Vega et al. 2010). Interestingly, more recently, the stabilization of MDMX by USP2a was also described in glioblastoma (Wang et al. 2014). Unexpectedly, MDMX overexpression correlated with good prognosis in this particular case. The investigators found higher levels of MDMX and Ser46-phosphorylated p53 at the mitochondria of MDMX overexpressing cells, which sensitizes the cells to ultraviolet (UV)-induced apoptosis. These data are in line with data from Moretti’s group, which reported that high levels of MDMX sensitize tumor types for UV- and adriamycin-induced apoptosis (Mancini et al. 2004, 2009, 2015). Although most studies so far indicate that reducing MDMX levels in tumors with wild-type p53 sensitizes these cells for treatments, these provocative results should be kept in mind when developing MDMX-targeting strategies.
MDMX AS A THERAPEUTIC TARGET
Several studies established a causative link between MDMX overexpression and cancer formation in vivo and, importantly, underlined the addiction of cancer cells to high levels of MDMX. Depletion of MDMX decreased the growth of a series of p53 wild-type cancer cell lines, including breast cancer cells, retinoblastoma, uveal, and cutaneous melanoma, in a largely p53-dependent manner (Danovi et al. 2004; Laurie et al. 2006; de Lange et al. 2012; Gembarska et al. 2012). Moreover, as discussed above, mouse genetic data strongly argue that MDMX inhibition is likely to be a less hazardous strategy than MDM2 inhibition for restoring p53 function in tumors. All of these data designate MDMX as a promising target for p53 reactivation therapy.
Unfortunately, potent and specific small molecule inhibitors of the MDMX–p53 interaction with clinical perspective are still lacking. A specific inhibitor of the MDMX–p53 interaction has been reported, but the very complex mechanism of action prevents the further development into a clinically relevant compound (Reed et al. 2010; Bista et al. 2012).
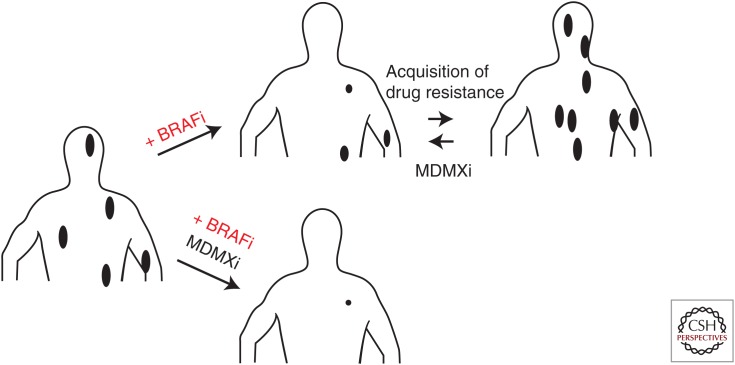
As an alternative, Bernal and coworkers reported the design, synthesis, and evaluation of stabilized α-helical (SAH) peptides (Bernal et al. 2010). One of these compounds, SAH-p53-8, binds Mdmx with high affinity and disrupts MDMX–p53 complexes in vitro and in vivo (Bernal et al. 2010). Consistently, SAH-p53-8 was sufficient to induce p53-dependent apoptosis in cultured melanoma cells (Gembarska et al. 2012). Importantly, SAH-p53-8 sensitized melanoma cells to conventional chemotherapeutics and to inhibition of BRAF-V600E by vemurafenib and inhibited growth of BRAF-V600E-mutant melanoma cells that acquired resistance to BRAF-V600E inhibitors (Fig. 3). These data, therefore, established MDMX as a promising target for antimelanoma combination therapy. Even so, although stapled peptides targeting both MDM2 and MDMX have recently been introduced into the clinic (phase II), it is questionable whether these types of therapeutics can make it as blockbuster-selling drugs. These peptides are expensive to produce and have to be delivered through intravenous injections by trained professionals, which very significantly increases the logistic burden associated with their use in the clinic.
Figure 3.
MDMX targeting sensitizes cells to BRAF-V600E inhibitors and reduces cell viability/growth of melanoma cells that have acquired resistance to BRAF-V600E inhibitors.
One additional limitation of targeting the MDMX–p53 interaction is that this will not counteract p53-independent oncogenic activity of MDMX. There is indeed an increasing body of evidence that MDMX possesses p53-independent oncogenic functions (De Clercq et al. 2010; de Lange et al. 2012; Carrillo et al. 2015). Consistently, knocking down MDMX in most melanoma cells caused cell-cycle arrest, which could not be rescued on concomitant inactivation of p53 (Gembarska et al. 2012). Inhibition of melanoma growth on MDMX depletion could also be observed in some melanoma cells with mutant p53 (Marine Laboratory, unpubl.). These data point to p53-independent oncogenic functions of MDMX, in addition to its known ability to suppress p53. As an alternative approach, which could overcome this limitation, methods that target MDMX protein abundance should be explored.
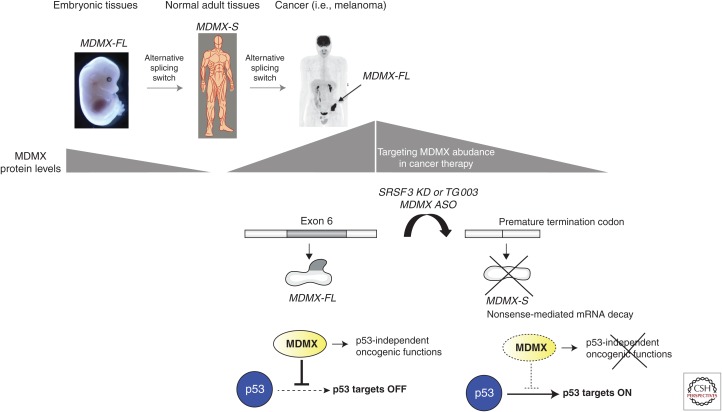
The identification of the above-described alternative splicing-dependent mechanism responsible for the high MDMX protein levels in a large fraction of melanoma has offered novel therapeutic opportunities. Accordingly, splice-switching antisense oligonucleotides (ASOs) flanking the exon–intron boundaries of exon 6 have been designed. Exposure of melanoma cells to these agents leads to efficient exon 6 skipping and subsequent decrease in MDMX protein abundance, accompanied by an increase in p53 activity (Fig. 4). Importantly, ASO-mediated exon 6 skipping decreased the ability of p53 wild-type melanoma cultures to grow in vitro and in patient-derived xenograft (PDX) mouse models and enhanced their sensitivity to MAPK-targeting therapeutics (Dewaele et al. 2015). In addition, we showed that targeting MDMX splicing with ASOs is also applicable to other cancers that overexpress MDMX (Dewaele et al. 2015). Moreover, because the approach targets MDMX protein abundance rather than its interaction with p53, it is expected to target the p53-independent oncogenic activities of MDMX and is applicable to an even broader spectrum of human tumors including those harboring p53 mutations (Fig. 4).
Figure 4.
Although MDMX is unproductively spliced in most normal adult tissues, the MDMX protein is highly expressed in embryonic tissues and in cancers as a result of enhanced exon 6 inclusion. Inducing MDMX exon 6 skipping via antisense oligonucleotides (ASOs) is a very specific, efficient, and clinically compatible approach to inhibit MDMX oncogenic functions.
Importantly, >100 different antisense drugs have been enrolled into various phase II and III clinical trials, and three of them (including an oral ASO) have recently been approved by the Food and Drug Administration (FDA) (Raal et al. 2010; Hair et al. 2013; Monteleone et al. 2015). Given the recent surge of optimism about ASO-based therapeutics, the targeting of MDMX exon 6 splicing may be rapidly applicable to the clinic.
To increase the chance of success of such clinical trials, it will be essential to stratify patients into MDMX-high and MDMX-low expressers. Because no correlation exists between MDMX mRNA and protein levels, screening of patient samples should be ideally performed at the protein level. In this context, the development of new molecular diagnostic tools, such as the proximity ligation assay, may become an important priority. However, the implementation of such assays in diagnostic laboratories is often a challenge. Alternatively, because recent data have indicated that the MDMX-FL/MDMX-S ratio is a reliable predictor of MDMX protein levels, this analysis, relatively easy to implement from a low amount of starting materials, could be used as a guide.
CONCLUDING REMARKS
Over the last 20 years, MDMX evolved from the status of the little brother of its well-studied and important “older” relative MDM2 to another key negative regulator of p53 in embryonic and, perhaps most importantly, cancer cells. A large body of evidence indicates that up-regulation of MDMX is a main mechanism that contributes to p53 inactivation in tumors that retain wild-type p53. MDMX has also become a very attractive therapeutic target for p53 reactivation therapy and to sensitize cancer cells to other therapeutic strategies. While awaiting the development of potent and specific small-molecule inhibitors of the MDMX–p53 interaction, the recent identification of one key mechanism that controls MDMX abundance—namely, alternative splicing—has offered alternative routes for its targeting. We sincerely hope that these findings will help in translating >30 years dedicated to p53 research into clinical benefits for the patients.
Footnotes
Editors: Guillermina Lozano and Arnold J. Levine
Additional Perspectives on The p53 Protein available at www.perspectivesinmedicine.org
REFERENCES
- Allende-Vega N, Sparks A, Lane DP, Saville MK. 2010. MdmX is a substrate for the deubiquitinating enzyme USP2a. Oncogene 29: 432–441. [DOI] [PubMed] [Google Scholar]
- Balint E, Bates S, Vousden KH. 1999. Mdm2 binds p73α without targeting degradation. Oncogene 18: 3923–3929. [DOI] [PubMed] [Google Scholar]
- Bardot B, Bouarich-Bourimi R, Leemput J, Lejour V, Hamon A, Plancke L, Jochemsen AG, Simeonova I, Fang M, Toledo F. 2015. Mice engineered for an obligatory Mdm4 exon skipping express higher levels of the Mdm4-S isoform but exhibit increased p53 activity. Oncogene 34: 2943–2948. [DOI] [PubMed] [Google Scholar]
- Bartel F, Schulz J, Bohnke A, Blumke K, Kappler M, Bache M, Schmidt H, Wurl P, Taubert H, Hauptmann S. 2005. Significance of HDMX-S (or MDM4) mRNA splice variant overexpression and HDMX gene amplification on primary soft tissue sarcoma prognosis. Int J Cancer 117: 469–475. [DOI] [PubMed] [Google Scholar]
- Bernal F, Wade M, Godes M, Davis TN, Whitehead DG, Kung AL, Wahl GM, Walensky LD. 2010. A stapled p53 helix overcomes HDMX-mediated suppression of p53. Cancer Cell 18: 411–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezzi M, Teo SX, Muller J, Mok WC, Sahu SK, Vardy LA, Bonday ZQ, Guccione E. 2013. Regulation of constitutive and alternative splicing by PRMT5 reveals a role for Mdm4 pre-mRNA in sensing defects in the spliceosomal machinery. Genes Dev 27: 1903–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bista M, Smithson D, Pecak A, Salinas G, Pustelny K, Min J, Pirog A, Finch K, Zdzalik M, Waddell B, et al. 2012. On the mechanism of action of SJ-172550 in inhibiting the interaction of MDM4 and p53. PLoS ONE 7: e37518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bista M, Petrovich M, Fersht AR. 2013. MDMX contains an autoinhibitory sequence element. Proc Natl Acad Sci 110: 17814–17819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bo MD, Secchiero P, Degan M, Marconi D, Bomben R, Pozzato G, Gaidano G, Del PG, Forconi F, Zauli G, et al. 2010. MDM4 (MDMX) is overexpressed in chronic lymphocytic leukaemia (CLL) and marks a subset of p53wild-type CLL with a poor cytotoxic response to Nutlin-3. Br J Haematol 150: 237–239. [DOI] [PubMed] [Google Scholar]
- Boesten LS, Zadelaar SM, De CS, Francoz S, van NA, Biessen EA, Hofmann F, Feil S, Feil R, Jochemsen AG, et al. 2006. Mdm2, but not Mdm4, protects terminally differentiated smooth muscle cells from p53-mediated caspase-3-independent cell death. Cell Death Differ 13: 2089–2098. [DOI] [PubMed] [Google Scholar]
- Bottger V, Bottger A, Garcia-Echeverria C, Ramos YF, van der Eb AJ, Jochemsen AG, Lane DP. 1999. Comparative study of the p53-mdm2 and p53-MDMX interfaces. Oncogene 18: 189–199. [DOI] [PubMed] [Google Scholar]
- Boutz PL, Bhutkar A, Sharp PA. 2015. Detained introns are a novel, widespread class of post-transcriptionally spliced introns. Genes Dev 29: 63–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo AM, Bouska A, Arrate MP, Eischen CM. 2015. Mdmx promotes genomic instability independent of p53 and Mdm2. Oncogene 34: 846–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan D, Tian Z, Nicholson B, Kumar KG, Zhou B, Carrasco R, McDermott JL, Leach CA, Fulcinniti M, Kodrasov MP, et al. 2012. A small molecule inhibitor of ubiquitin-specific protease-7 induces apoptosis in multiple myeloma cells and overcomes bortezomib resistance. Cancer Cell 22: 345–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Gilkes DM, Pan Y, Lane WS, Chen J. 2005a. ATM and Chk2-dependent phosphorylation of MDMX contribute to p53 activation after DNA damage. EMBO J 24: 3411–3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Li C, Pan Y, Chen J. 2005b. Regulation of p53-MDMX interaction by casein kinase 1 α. Mol Cell Biol 25: 6509–6520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Borcherds W, Wu S, Becker A, Schonbrunn E, Daughdrill GW, Chen J. 2015. Autoinhibition of MDMX by intramolecular p53 mimicry. Proc Natl Acad Sci 112: 4624–4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbo C, Orru S, Salvatore F. 2013. SRp20: An overview of its role in human diseases. Biochem Biophys Res Commun 436: 1–5. [DOI] [PubMed] [Google Scholar]
- Daftuar L, Zhu Y, Jacq X, Prives C. 2013. Ribosomal proteins RPL37, RPS15 and RPS20 regulate the Mdm2-p53-MdmX network. PLoS ONE 8: e68667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danovi D, Meulmeester E, Pasini D, Migliorini D, Capra M, Frenk R, de GP, Francoz S, Gasparini P, Gobbi A, et al. 2004. Amplification of Mdmx (or Mdm4) directly contributes to tumor formation by inhibiting p53 tumor suppressor activity. Mol Cell Biol 24: 5835–5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clercq S, Gembarska A, Denecker G, Maetens M, Naessens M, Haigh K, Haigh JJ, Marine JC. 2010. Widespread overexpression of epitope-tagged Mdm4 does not accelerate tumor formation in vivo. Mol Cell Biol 30: 5394–5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange J, Teunisse AF, Vries MV, Lodder K, Lam S, Luyten GP, Bernal F, Jager MJ, Jochemsen AG. 2012. High levels of Hdmx promote cell growth in a subset of uveal melanomas. Am J Cancer Res 2: 492–507. [PMC free article] [PubMed] [Google Scholar]
- Dewaele M, Tabaglio T, Willekens K, Bezzi M, Teo SX, Low DH, Koh CM, Rambow F, Fiers M, Rogiers A, et al. 2015. Antisense oligonucleotide-mediated MDM4 exon 6 skipping impairs tumor growth. J Clin Invest 10.1172/JCI82534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding F, Bao C, Tian Y, Xiao H, Wang M, Xie X, Hu F, Mei J. 2015. USP22 promotes NSCLC tumorigenesis via MDMX up-regulation and subsequent p53 inhibition. Int J Mol Sci 16: 307–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbelstein M, Wienzek S, Konig C, Roth J. 1999. Inactivation of the p53-homologue p73 by the mdm2-oncoprotein. Oncogene 18: 2101–2106. [DOI] [PubMed] [Google Scholar]
- Finch RA, Donoviel DB, Potter D, Shi M, Fan A, Freed DD, Wang CY, Zambrowicz BP, Ramirez-Solis R, Sands AT, et al. 2002. mdmx is a negative regulator of p53 activity in vivo. Cancer Res 62: 3221–3225. [PubMed] [Google Scholar]
- Francoz S, Froment P, Bogaerts S, De CS, Maetens M, Doumont G, Bellefroid E, Marine JC. 2006. Mdm4 and Mdm2 cooperate to inhibit p53 activity in proliferating and quiescent cells in vivo. Proc Natl Acad Sci 103: 3232–3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia D, Warr MR, Martins CP, Brown SL, Passegue E, Evan GI. 2011. Validation of MdmX as a therapeutic target for reactivating p53 in tumors. Genes Dev 25: 1746–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gembarska A, Luciani F, Fedele C, Russell EA, Dewaele M, Villar S, Zwolinska A, Haupt S, de LJ, Yip D, et al. 2012. MDM4 is a key therapeutic target in cutaneous melanoma. Nat Med 18: 1239–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilkes DM, Chen L, Chen J. 2006. MDMX regulation of p53 response to ribosomal stress. EMBO J 25: 5614–5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilkes DM, Pan Y, Coppola D, Yeatman T, Reuther GW, Chen J. 2008. Regulation of MDMX expression by mitogenic signaling. Mol Cell Biol 28: 1999–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grier JD, Yan W, Lozano G. 2002. Conditional allele of mdm2 which encodes a p53 inhibitor. Genesis 32: 145–147. [DOI] [PubMed] [Google Scholar]
- Grier JD, Xiong S, Elizondo-Fraire AC, Parant JM, Lozano G. 2006. Tissue-specific differences of p53 inhibition by Mdm2 and Mdm4. Mol Cell Biol 26: 192–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hair P, Cameron F, McKeage K. 2013. Mipomersen sodium: First global approval. Drugs 73: 487–493. [DOI] [PubMed] [Google Scholar]
- Han X, Garcia-Manero G, McDonnell TJ, Lozano G, Medeiros LJ, Xiao L, Rosner G, Nguyen M, Fernandez M, Valentin-Vega YA, et al. 2007. HDM4 (HDMX) is widely expressed in adult pre-B acute lymphoblastic leukemia and is a potential therapeutic target. Mod Pathol 20: 54–62. [DOI] [PubMed] [Google Scholar]
- Hsieh JK, Chan FS, O’Connor DJ, Mittnacht S, Zhong S, Lu X. 1999. RB regulates the stability and the apoptotic function of p53 via MDM2. Mol Cell 3: 181–193. [DOI] [PubMed] [Google Scholar]
- Hu J, Yang D, Zhang H, Liu W, Zhao Y, Lu H, Meng Q, Pang H, Chen X, Liu Y, et al. 2015. USP22 promotes tumor progression and induces epithelial–mesenchymal transition in lung adenocarcinoma. Lung Cancer 88: 239–245. [DOI] [PubMed] [Google Scholar]
- Huang L, Yan Z, Liao X, Li Y, Yang J, Wang ZG, Zuo Y, Kawai H, Shadfan M, Ganapathy S, et al. 2011. The p53 inhibitors MDM2/MDMX complex is required for control of p53 activity in vivo. Proc Natl Acad Sci 108: 12001–12006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai H, Wiederschain D, Kitao H, Stuart J, Tsai KK, Yuan ZM. 2003a. DNA damage-induced MDMX degradation is mediated by MDM2. J Biol Chem 278: 45946–45953. [DOI] [PubMed] [Google Scholar]
- Kawai H, Wiederschain D, Yuan ZM. 2003b. Critical contribution of the MDM2 acidic domain to p53 ubiquitination. Mol Cell Biol 23: 4939–4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblihtt AR, Schor IE, Allo M, Dujardin G, Petrillo E, Munoz MJ. 2013. Alternative splicing: A pivotal step between eukaryotic transcription and translation. Nat Rev Mol Cell Biol 14: 153–165. [DOI] [PubMed] [Google Scholar]
- Kubo N, Okoshi R, Nakashima K, Shimozato O, Nakagawara A, Ozaki T. 2010. MDM2 promotes the proteasomal degradation of p73 through the interaction with Itch in HeLa cells. Biochem Biophys Res Commun 403: 405–411. [DOI] [PubMed] [Google Scholar]
- Landers JE, Haines DS, Strauss JF III, George DL. 1994. Enhanced translation: A novel mechanism of mdm2 oncogene overexpression identified in human tumor cells. Oncogene 9: 2745–2750. [PubMed] [Google Scholar]
- Landers JE, Cassel SL, George DL. 1997. Translational enhancement of mdm2 oncogene expression in human tumor cells containing a stabilized wild-type p53 protein. Cancer Res 57: 3562–3568. [PubMed] [Google Scholar]
- Laurie NA, Donovan SL, Shih CS, Zhang J, Mills N, Fuller C, Teunisse A, Lam S, Ramos Y, Mohan A, et al. 2006. Inactivation of the p53 pathway in retinoblastoma. Nature 444: 61–66. [DOI] [PubMed] [Google Scholar]
- LeBron C, Chen L, Gilkes DM, Chen J. 2006. Regulation of MDMX nuclear import and degradation by Chk2 and 14-3-3. EMBO J 25: 1196–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenos K, Jochemsen AG. 2011. Functions of MDMX in the modulation of the p53-response. J Biomed Biotechnol 2011: 876173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Gu W. 2011. A critical role for noncoding 5S rRNA in regulating Mdmx stability. Mol Cell 43: 1023–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Gilkes D, Li B, Cheng Q, Pernazza D, Lawrence H, Lawrence N, Chen J. 2012. Abnormal MDMX degradation in tumor cells due to ARF deficiency. Oncogene 31: 3721–3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Tan Y, Chen X, Xu Z, Yang S, Ren F, Guo H, Wang X, Chen Y, Li G, et al. 2014. MDM4 overexpressed in acute myeloid leukemia patients with complex karyotype and wild-type TP53. PLoS ONE 9: e113088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linke K, Mace PD, Smith CA, Vaux DL, Silke J, Day CL. 2008. Structure of the MDM2/MDMX RING domain heterodimer reveals dimerization is required for their ubiquitylation in trans. Cell Death Differ 15: 841–848. [DOI] [PubMed] [Google Scholar]
- Liu Y, He Y, Jin A, Tikunov AP, Zhou L, Tollini LA, Leslie P, Kim TH, Li LO, Coleman RA, et al. 2014. Ribosomal protein-Mdm2-p53 pathway coordinates nutrient stress with lipid metabolism by regulating MCD and promoting fatty acid oxidation. Proc Natl Acad Sci 111: E2414–E2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Pajares V, Kim MM, Yuan ZM. 2008. Phosphorylation of MDMX mediated by Akt leads to stabilization and induces 14-3-3 binding. J Biol Chem 283: 13707–13713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini F, Gentiletti F, D’Angelo M, Giglio S, Nanni S, D’Angelo C, Farsetti A, Citro G, Sacchi A, Pontecorvi A, et al. 2004. MDM4 (MDMX) overexpression enhances stabilization of stress-induced p53 and promotes apoptosis. J Biol Chem 279: 8169–8180. [DOI] [PubMed] [Google Scholar]
- Mancini F, Di CG, Pellegrino M, Rinaldo C, Prodosmo A, Giglio S, D’Agnano I, Florenzano F, Felicioni L, Buttitta F, et al. 2009. MDM4 (MDMX) localizes at the mitochondria and facilitates the p53-mediated intrinsic-apoptotic pathway. EMBO J 28: 1926–1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini F, Pieroni L, Monteleone V, Lucà R, Fici L, Luca E, Urbani A, Xiong S, Soddu S, Masetti R, et al. 2015. MDM4/HIPK2/p53 cytoplasmic assembly uncovers coordinated repression of molecules with anti-apoptotic activity during early DNA damage response. Oncogene 10.1038/onc.2015.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy MA, Gesell JJ, Senior MM, Wyss DF. 2003. Flexible lid to the p53-binding domain of human Mdm2: Implications for p53 regulation. Proc Natl Acad Sci 100: 1645–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEvoy J, Ulyanov A, Brennan R, Wu G, Pounds S, Zhang J, Dyer MA. 2012. Analysis of MDM2 and MDM4 single nucleotide polymorphisms, mRNA splicing and protein expression in retinoblastoma. PLoS ONE 7: e42739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendrysa SM, McElwee MK, Michalowski J, O’Leary KA, Young KM, Perry ME. 2003. mdm2 is critical for inhibition of p53 during lymphopoiesis and the response to ionizing irradiation. Mol Cell Biol 23: 462–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulmeester E, Frenk R, Stad R, de GP, Marine JC, Vousden KH, Jochemsen AG. 2003. Critical role for a central part of Mdm2 in the ubiquitylation of p53. Mol Cell Biol 23: 4929–4938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulmeester E, Maurice MM, Boutell C, Teunisse AF, Ovaa H, Abraham TE, Dirks RW, Jochemsen AG. 2005. Loss of HAUSP-mediated deubiquitination contributes to DNA damage-induced destabilization of Hdmx and Hdm2. Mol Cell 18: 565–576. [DOI] [PubMed] [Google Scholar]
- Migliorini D, Lazzerini DE, Danovi D, Jochemsen A, Capillo M, Gobbi A, Helin K, Pelicci PG, Marine JC. 2002. Mdm4 (Mdmx) regulates p53-induced growth arrest and neuronal cell death during early embryonic mouse development. Mol Cell Biol 22: 5527–5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteleone G, Neurath MF, Ardizzone S, Di SA, Fantini MC, Castiglione F, Scribano ML, Armuzzi A, Caprioli F, Sturniolo GC, et al. 2015. Mongersen, an oral SMAD7 antisense oligonucleotide, and Crohn’s disease. N Engl J Med 372: 1104–1113. [DOI] [PubMed] [Google Scholar]
- Okamoto K, Kashima K, Pereg Y, Ishida M, Yamazaki S, Nota A, Teunisse A, Migliorini D, Kitabayashi I, Marine JC, et al. 2005. DNA damage-induced phosphorylation of MdmX at serine 367 activates p53 by targeting MdmX for Mdm2-dependent degradation. Mol Cell Biol 25: 9608–9620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongkeko WM, Wang XQ, Siu WY, Lau AW, Yamashita K, Harris AL, Cox LS, Poon RY. 1999. MDM2 and MDMX bind and stabilize the p53-related protein p73. Curr Biol 9: 829–832. [DOI] [PubMed] [Google Scholar]
- Pant V, Xiong S, Iwakuma T, Quintas-Cardama A, Lozano G. 2011. Heterodimerization of Mdm2 and Mdm4 is critical for regulating p53 activity during embryogenesis but dispensable for p53 and Mdm2 stability. Proc Natl Acad Sci 108: 11995–12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parant J, Chavez-Reyes A, Little NA, Yan W, Reinke V, Jochemsen AG, Lozano G. 2001. Rescue of embryonic lethality in Mdm4-null mice by loss of Trp53 suggests a nonoverlapping pathway with MDM2 to regulate p53. Nat Genet 29: 92–95. [DOI] [PubMed] [Google Scholar]
- Pereg Y, Shkedy D, de GP, Meulmeester E, Edelson-Averbukh M, Salek M, Biton S, Teunisse AF, Lehmann WD, et al. 2005. Phosphorylation of Hdmx mediates its Hdm2- and ATM-dependent degradation in response to DNA damage. Proc Natl Acad Sci 102: 5056–5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereg Y, Lam S, Teunisse A, Biton S, Meulmeester E, Mittelman L, Buscemi G, Okamoto K, Taya Y, Shiloh Y, et al. 2006. Differential roles of ATM- and Chk2-mediated phosphorylations of Hdmx in response to DNA damage. Mol Cell Biol 26: 6819–6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Ortin JE, Alepuz P, Chavez S, Choder M. 2013. Eukaryotic mRNA decay: Methodologies, pathways, and links to other stages of gene expression. J Mol Biol 425: 3750–3775. [DOI] [PubMed] [Google Scholar]
- Popp MW, Maquat LE. 2013. Organizing principles of mammalian nonsense-mediated mRNA decay. Annu Rev Genet 47: 139–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poyurovsky MV, Priest C, Kentsis A, Borden KL, Pan ZQ, Pavletich N, Prives C. 2007. The Mdm2 RING domain C-terminus is required for supramolecular assembly and ubiquitin ligase activity. EMBO J 26: 90–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raal FJ, Santos RD, Blom DJ, Marais AD, Charng MJ, Cromwell WC, Lachmann RH, Gaudet D, Tan JL, Chasan-Taber S, et al. 2010. Mipomersen, an apolipoprotein B synthesis inhibitor, for lowering of LDL cholesterol concentrations in patients with homozygous familial hypercholesterolaemia: A randomised, double-blind, placebo-controlled trial. Lancet 375: 998–1006. [DOI] [PubMed] [Google Scholar]
- Rallapalli R, Strachan G, Cho B, Mercer WE, Hall DJ. 1999. A novel MDMX transcript expressed in a variety of transformed cell lines encodes a truncated protein with potent p53 repressive activity. J Biol Chem 274: 8299–8308. [DOI] [PubMed] [Google Scholar]
- Rallapalli R, Strachan G, Tuan RS, Hall DJ. 2003. Identification of a domain within MDMX-S that is responsible for its high affinity interaction with p53 and high-level expression in mammalian cells. J Cell Biochem 89: 563–575. [DOI] [PubMed] [Google Scholar]
- Reed D, Shen Y, Shelat AA, Arnold LA, Ferreira AM, Zhu F, Mills N, Smithson DC, Regni CA, Bashford D, et al. 2010. Identification and characterization of the first small molecule inhibitor of MDMX. J Biol Chem 285: 10786–10796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riemenschneider MJ, Buschges R, Wolter M, Reifenberger J, Bostrom J, Kraus JA, Schlegel U, Reifenberger G. 1999. Amplification and overexpression of the MDM4 (MDMX) gene from 1q32 in a subset of malignant gliomas without TP53 mutation or MDM2 amplification. Cancer Res 59: 6091–6096. [PubMed] [Google Scholar]
- Ringshausen I, O’Shea CC, Finch AJ, Swigart LB, Evan GI. 2006. Mdm2 is critically and continuously required to suppress lethal p53 activity in vivo. Cancer Cell 10: 501–514. [DOI] [PubMed] [Google Scholar]
- Sabbatini P, McCormick F. 2002. MDMX inhibits the p300/CBP-mediated acetylation of p53. DNA Cell Biol 21: 519–525. [DOI] [PubMed] [Google Scholar]
- Shvarts A, Steegenga WT, Riteco N, van Laar T, Dekker P, Bazuine M, van Ham RC, van der Houven van Oordt W, Hateboer G, et al. 1996. MDMX: A novel p53-binding protein with some functional properties of MDM2. EMBO J 15: 5349–5357. [PMC free article] [PubMed] [Google Scholar]
- Steinman HA, Jones SN. 2002. Generation of an Mdm2 conditional allele in mice. Genesis 32: 142–144. [DOI] [PubMed] [Google Scholar]
- Tang B, Tang F, Li B, Yuan S, Xu Q, Tomlinson S, Jin J, Hu W, He S. 2015. High USP22 expression indicates poor prognosis in hepatocellular carcinoma. Oncotarget 6: 12654–12667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo F, Krummel KA, Lee CJ, Liu CW, Rodewald LW, Tang M, Wahl GM. 2006. A mouse p53 mutant lacking the proline-rich domain rescues Mdm4 deficiency and provides insight into the Mdm2-Mdm4-p53 regulatory network. Cancer Cell 9: 273–285. [DOI] [PubMed] [Google Scholar]
- Uchida C, Miwa S, Isobe T, Kitagawa K, Hattori T, Oda T, Yasuda H, Kitagawa M. 2006. Effects of MdmX on Mdm2-mediated downregulation of pRB. FEBS Lett 580: 1753–1758. [DOI] [PubMed] [Google Scholar]
- Valentin-Vega YA, Box N, Terzian T, Lozano G. 2009. Mdm4 loss in the intestinal epithelium leads to compartmentalized cell death but no tissue abnormalities. Differentiation 77: 442–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Lane D, Levine AJ. 2000. Surfing the p53 network. Nature 408: 307–310. [DOI] [PubMed] [Google Scholar]
- Wang X, Arooz T, Siu WY, Chiu CH, Lau A, Yamashita K, Poon RY. 2001. MDM2 and MDMX can interact differently with ARF and members of the p53 family. FEBS Lett 490: 202–208. [DOI] [PubMed] [Google Scholar]
- Wang CL, Wang JY, Liu ZY, Ma XM, Wang XW, Jin H, Zhang XP, Fu D, Hou LJ, Lu YC. 2014. Ubiquitin-specific protease 2a stabilizes MDM4 and facilitates the p53-mediated intrinsic apoptotic pathway in glioblastoma. Carcinogenesis 35: 1500–1509. [DOI] [PubMed] [Google Scholar]
- Worrall EG, Wawrzynow B, Worrall L, Walkinshaw M, Ball KL, Hupp TR. 2009. Regulation of the E3 ubiquitin ligase activity of MDM2 by an N-terminal pseudo-substrate motif. J Chem Biol 2: 113–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worrall EG, Worrall L, Blackburn E, Walkinshaw M, Hupp TR. 2010. The effects of phosphomimetic lid mutation on the thermostability of the N-terminal domain of MDM2. J Mol Biol 398: 414–428. [DOI] [PubMed] [Google Scholar]
- Wu H, Leng RP. 2015. MDM2 mediates p73 ubiquitination: A new molecular mechanism for suppression of p73 function. Oncotarget 28: 21479–21492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Chen L, Becker A, Schonbrunn E, Chen J. 2012. Casein kinase 1α regulates an MDMX intramolecular interaction to stimulate p53 binding. Mol Cell Biol 32: 4821–4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong S, Van Pelt CS, Elizondo-Fraire AC, Liu G, Lozano G. 2006. Synergistic roles of Mdm2 and Mdm4 for p53 inhibition in central nervous system development. Proc Natl Acad Sci 103: 3226–3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdzalik M, Pustelny K, Kedracka-Krok S, Huben K, Pecak A, Wladyka B, Jankowski S, Dubin A, Potempa J, Dubin G. 2010. Interaction of regulators Mdm2 and Mdmx with transcription factors p53, p63 and p73. Cell Cycle 9: 4584–4591. [DOI] [PubMed] [Google Scholar]
- Zeng X, Chen L, Jost CA, Maya R, Keller D, Wang X, Kaelin WG Jr, Oren M, Chen J, Lu H. 1999. MDM2 suppresses p73 function without promoting p73 degradation. Mol Cell Biol 19: 3257–3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Hu L, Qiu W, Deng T, Zhang Y, Bergholz J, Xiao ZX. 2015. MDMX exerts its oncogenic activity via suppression of retinoblastoma protein. Oncogene 10.1038/onc.2015.11. [DOI] [PubMed] [Google Scholar]