SUMMARY
DNA and histone modifications, together with constraints imposed by nuclear architecture, contribute to the transcriptional regulatory landscape of the nervous system. Here, we provide select examples showing how these regulatory layers, often referred to as epigenetic, contribute to neuronal differentiation and function. We describe the interplay between DNA methylation and Polycomb-mediated repression during neuronal differentiation, the role of DNA methylation and long-range enhancer–promoter interactions in Protocadherin promoter choice, and the contribution of heterochromatic silencing and nuclear organization in singular olfactory receptor expression. Finally, we explain how the activity-dependent expression of a histone variant determines the longevity of olfactory sensory neurons.
Epigenetic mechanisms play important roles in neurogenesis. For example, DNA methylation regulates the differential expression of Protocadherins, cell-surface receptors required for neuronal identity.
OVERVIEW
The nervous system of higher organisms is characterized by an enormous diversity of cell types that function in concert to perform a myriad of neuronal functions. In mammals, there are an estimated 1011 neurons that form 1015 synapses. Differences in connectivity, and subsequent physiology of the connected neurons, are the result of differences in transcriptional programs. The extraordinary complexity of the nervous system requires a complex regulatory system. Neurons must interpret subtle fluctuations in spatiotemporal cues during development in order to commit to specific differentiation programs, but at the same time, they must retain a level of synaptic and transcriptional plasticity. It is well established that transcription factor combinations and the organization of cis-regulatory sequences control commitment to differentiation programs and preserve a nuclear plasticity required for neuronal functions. An additional level of regulation is provided by epigenetic regulatory mechanisms that allow stochastic, mutually exclusive transcriptional choices during neuronal differentiation. This affords long-lasting transcriptional changes in response to internal stimuli and external experiences. In fact, the evolution of epigenetic mechanisms that regulates the nervous system may have been a driving force in the expansion and evolution of the large numbers of chromatin- and DNA-modifying enzymes, capable of producing a highly complex, yet plastic nervous system in higher eukaryotes.
This article focuses primarily on epigenetic mechanisms involved in neurogenesis, the specification of neuronal fates, and the development of neural circuitry in the brain. We do not address epigenetic studies of neuronal plasticity, neurodegeneration, and psychiatric conditions, such as drug addiction (Fass et al. 2014; Nestler 2014; Rudenko and Tsai 2014). Changes in chromatin and DNA modifications occur during the very earliest stages of neuronal development (i.e., during neurogenesis), and they involve the interplay between DNA methylation and the Polycomb and Trithorax complexes. These changes coincide with the gene expression patterns that govern the developmental decision to adopt neuronal or nonneuronal (glia) cell identity. On commitment to the neuronal fate, developing neurons must make choices between similar, but functionally distinct transcription programs. Because these neurons are postmitotic, inheritance of histone and DNA posttranslational modifications cannot be tested. Thus, we use the word “epigenetic” in neural processes to describe posttranslational modifications of DNA that are associated with changes in gene expression. We describe two examples of this regulation, both of which are examples of the specification of neuronal identity during neural circuitry development. The first is the generation of enormous cell-surface diversity required for neuronal identity by the stochastic expression of the clustered protocadherin (Pcdh) genes. The second is the control of olfactory receptor (OR) gene expression in olfactory neurons, which govern the sense of smell. Both of these examples contribute to the establishment of complex neuronal architectures, as well as functional single-cell diversity, through mechanisms that involve distinct examples of monoallelic expression.
Monoallelic gene expression is an unusual form of gene regulation in which only one of two alleles of a gene is expressed. The challenge to the regulatory system is to discern and differentially express two alleles in the same nucleus. Recent studies have revealed an important role for chromatin modifications in this process, and similar mechanisms have also been identified in the immune system in which enormous single-cell diversity of immunoglobulin and T-cell receptors is generated (see Busslinger and Tarakhovsky 2014). It is not surprising that similar epigenetic mechanisms have evolved to generate cellular diversity in the nervous system. Generating panneuronal single-cell diversity in the central and peripheral nervous system occurs via mechanisms that ensure the stochastic combinatorial expression of cadherin-like Pcdh proteins from a cluster of genes. In the case of the OR genes, all but one of more than 1000 possible receptor isoforms are stochastically silenced. This stochastic process contrasts with genomic imprinting, which is an epigenetic process ensuring the parent of origin determined monoallelic expression of certain genes. These “classic” examples of monoallelic gene expression, however, do bear some similarities to genomic imprinting in that only one of two identical copies of a gene or genomic region are silenced in a single-cell nucleus.
We also describe another “epigenetic peculiarity” in olfactory neurons involving the expression of a histone H2b isoform (or variant) named H2be. This histone variant, which differs by only five amino acids from the canonical H2b protein, appears to be a gauge of the external olfactory sensory environment by being exclusively expressed from understimulated olfactory neurons, signaling the shortening of their life span.
1. EPIGENETIC REGULATION OF NEUROGENESIS
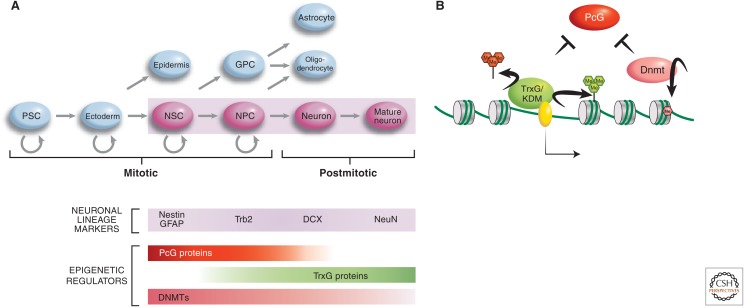
Multipotent neuronal stem and progenitor cells give rise to both neuronal and nonneuronal lineages of the nervous system (Fig. 1A) (reviewed in Olynik and Rastegar 2012). Epigenetic processes, some of which are described in this section, are involved in generating neuronal cells from nonneuronal multipotent precursor cells during neurogenesis. There are two main sites of adult neurogenesis: the subventricular (SVZ) zones in the forebrain and the dentate gyrus of the hippocampus (Alvarez-Buylla and Garcia-Verdugo 2002; Alvarez-Buylla and Lim 2004). These regions, which contain neuronal stem cells (NSCs), give rise to the majority of the neurons present throughout the lives of mice and humans (Doetsch et al. 1999). NSCs can be isolated, cultured, and differentiated into various types of neurons in vitro, thus, making it possible to study epigenetic regulation during neurogenesis in vitro, as well as in vivo. Studies thus far have revealed that DNA- and chromatin-modifying enzymes (particularly, the DNA methyltransferase, Trithorax group, and Polycomb group repressive enzymes) play key roles in this differentiation process (Fig. 1B) (Hirabayashi and Gotoh 2010).
Figure 1.
Epigenetic regulation of neurogenesis. (A) The temporal involvement of various major epigenetic regulator molecules, acting at key fate-defining genes, such as Dlx2, are indicated alongside the neurogenic differentiation pathway (i.e., cells shaded in pink). PcG, Polycomb group; TrxG, Trithorax group; DNMT, DNA methyltransferase; PSC, pluripotent stem cell; NSC, neuronal stem cell; NPC, neuronal precursor cell; GPC, glial progenitor cell. (B) The interplay and modes of action of the three types of epigenetic-regulating molecules involved in the neuronal differentiation process are illustrated. Dnmt3a methylates the gene bodies and upstream regulatory sequences of neurogenic genes, such as Dlx2. This nonpromoter methylation protects these genes from Polycomb-mediated silencing. MLL1, through the trimethylation of H3K4 and demethylation of H3K27me3 (by association of KDM6), also antagonizes Polycomb-mediated silencing of Dlx2 to promote neurogenesis.
DNA methylation is unequivocally implicated in NSC differentiation, as deletion of the de novo DNA methyltransferase, Dnmt3a, shows an approximately 10-fold reduction in the rate of differentiation of postmitotic cells expressing neuronal markers (Wu et al. 2010). Studies on the role of DNA methylation during neurogenesis (an epigenetic mark traditionally associated with repression; discussed in Li and Zhang 2014) have shown that, atypically, this type of DNA modification contributes to transcriptional activation, as well as repression of neurogenic genes (Wu et al. 2010). A closer look at the genome-wide binding of Dnmt3a revealed a widespread distribution of binding sites across the euchromatic genome with significant enrichment within genes and intergenic regions (Fig. 2). Many of these Dnmt3a-rich regions localize to transcriptionally active genes in the NSCs in which the chromatin is marked by high levels of H3K4me3 on promoters (Wu et al. 2010). A statistically significant number of these Dnmt3a target genes, such as Dlx2 and Sp8, are involved in neurogenesis or neuronal functions. In contrast, in transcriptionally inactive genes, the peak of Dnmt3a-dependent methylation maps to transcription start sites (TSS). Thus, it appeared that the role of DNA methylation in transcriptional activity may depend on the genomic context.
Figure 2.
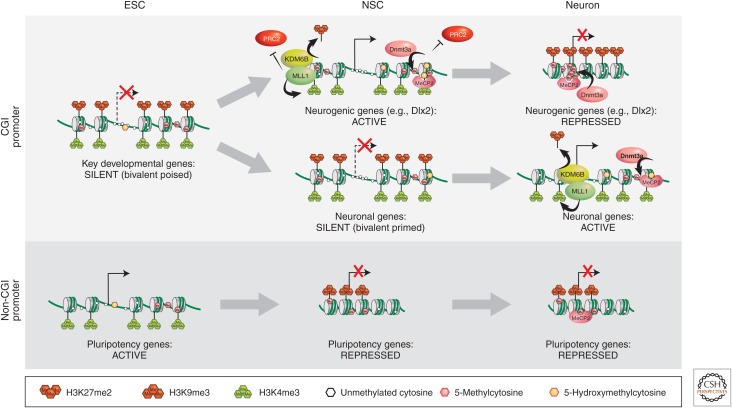
Epigenetic mechanisms involved in the regulation of gene expression during neurogenesis. Typically, CpG island–containing genes are mostly housekeeping genes and, consequently, are ubiquitously active. In early development, just postimplantation, these genes may not yet be active, but are marked by a bivalent chromatin signature, consisting of H3K4me3 and H3K27me3. Most of these genes will be activated during development via H3K27 demethylation (top line). A subset of these genes that are tissue-specific neurogenic genes may, subsequently, become silenced in nonneuronal or fully differentiated neuronal cells, through H3K9 methylated heterochromatin formation (top right). Other neuronal-specific genes may delay activation, but remain bivalently primed for expression until neuronal differentiation ensues (middle line). Pluripotency genes, largely, do not contain CpG islands in their promoters. They are active during early development and become repressed following differentiation by conventional H3K9 methylation and DNA methylation mechanisms. ESC, embryonic stem cell; NSC, neuronal stem cell; CGI, CpG island.
An explanation for this finding may be provided by the discovery that 5-hydroxymethylcytosine (5hmC) is present in the mammalian genome, and is approximately 10-fold more abundant in neurons than in peripheral tissues or embryonic stem (ES) cells (Kriaucionis and Heintz 2009; Munzel et al. 2010; Szulwach et al. 2011). Until recently, it was not possible to distinguish between 5mC and 5hmC. We now know that the distribution of 5hmC varies between distinct regions of the brain and between the brain and ES cells (Szulwach et al. 2011). For example, 5hmC is enriched in gene bodies and depleted from TSS in neurons, whereas the opposite is the case in ES cells. More recent studies revealed that the relationship between the distribution of 5hmC, 5mC, and gene expression is brain cell specific, and the methyl-CpG-binding protein 2 (MeCP2) binds to 5hmC in the brain (Mellen et al. 2012). Additional studies led to the proposal that when MeCP2 binds to 5hmC, it facilitates transcription in neural cell types, but can act as a repressor when bound to 5mC-containing DNA (Fig. 2). This proposal was based on the observation that 5hmC is enriched in euchromatin, whereas 5mC is enriched in heterochromatin (Mellen et al. 2012). Remarkably, MeCP2 proteins bearing a Rett syndrome mutation R133C display altered binding to 5hmC DNA, suggesting that MeCP2 mutations alter the chromatin distribution of the proteins. The investigators of this work speculated that neuronal cell-specific dynamic gene regulation is the consequence of the three-dimensional (3D) chromatin structure, which is determined by the levels of 5mC, 5hmC, and MeCP2 (Mellen et al. 2012).
Dnmt3a-dependent de novo DNA methylation has been shown to inhibit the binding of the Polycomb repressive complex PRC2 (Wu et al. 2010). The neurogenic genes that require Dnmt3a are known targets of PRC2 in nonneuronal lineages (Hirabayashi et al. 2009). For this reason, it is thought that Dnmt3a-dependent DNA methylation prevents the binding of the inhibitor, in this case, PRC2 (Fig. 2; see the active NSC locus). Indeed, deletion of Dnmt3a results in an increase in PRC2 binding to these neurogenic genes in NSCs and, thus, enrichment of H3K27me3 on their promoters. Conversely, partial rescue of the neurogenic Dnmt3a deficit and reexpression of neuronal markers occurs following knockdown of PRC2 components in these Dnmt3a knockout NSCs.
Dnmt3a-dependent DNA methylation is not the only factor involved in the control of Polycomb-mediated silencing in neurogenesis. The Trithorax (trxG) protein complex is the bona fide inhibitor of the Polycomb group, as shown by complementary genetic and biochemical studies first performed in Drosophila and, subsequently, extended to other model organisms (described in Grossniklaus and Paro 2014; Kingston and Tamkun 2014). It is important to note that genes subject to PcG regulation typically contain CpG islands at their promoters and are initially bivalently marked by H3K4me3 and H3K27me3 in pluripotent stem cells (Fig. 2). A member of the trx group, Mll1 (mixed lineage leukemia 1), is required for neurogenesis in the postnatal brain, as is Dnmt3a (Fig. 1) (Lim et al. 2009). Mll1 trimethylates H3K4 (H3K4me3), an epigenetic mark that is associated with transcriptional competence. Mll1 also mediates H3K27 demethylation by recruiting the histone demethylase KDM6 (Fig. 1B) (Burgold et al. 2008). Conditional deletion of MLL1 in NSCs results in postnatal atrophy of brain regions, which involves postnatal neurogenesis, ataxia, and death between P25 and P30 (Lim et al. 2009). Expression analysis of the MLL1 knockout NSCs revealed significant down-regulation of neurogenic genes, such as Dlx2. Further analysis of the SVZ showed that, in wild-type mice, nucleosomes in the Dlx2 promoter are marked only by H3K4me3, but in Mll1 knockout mice, it is bivalently marked by H3K4me3 and H3K27me3. The simultaneous presence of these histone marks is typically found in ES cells at bivalent loci that remain poised until cell differentiation signals cause transcriptional activation and H3K27me3 demethylation of certain neural genes (Bernstein et al. 2006). Therefore, a possible role of Mll1 in neurogenesis is to protect Dlx2 from Polycomb-mediated silencing by reversing H3K27me3. Indeed, the neurogenesis deficits observed in the Mll1 knockout mice were partially rescued by Dlx2 overexpression. Thus, the intersection of the three epigenetic pathways on the Dlx2 gene, which encodes a transcription factor with a pivotal role in neurogenesis, determines whether a multipotent cell commits to the neuronal lineage. When Dnmt3a and Mll1 prevent Polycomb-mediated repression of the Dlx2 promoter, the cell differentiates into a neuron (top line of Fig. 2). When an NSC fails to activate Dlx2, then a glial fate ensues (see Fig. 1A). For nonneuronal cells, Dnmt3a and Mll1 are not activated at neuronal-specific loci and, thus, PRC2 acts to trimethylate H3K27, resulting in gene repression.
There are, however, numerous open questions regarding the epigenetic control of early neurogenesis and, without a doubt, Dnmt3a, Mll1, and PRC2 are only a few of the regulatory factors involved in this developmental process. For example, as mentioned earlier, it is not yet clear if further modification of 5-methylcytocine to 5-hydroxymethylcytocine in gene bodies and intergenic regions contributes to the competition between Polycomb and Trithorax, and whether additional DNA- or chromatin-modifying complexes and transcription factors are involved in determining the final balance between neurogenesis or gliogenesis (Riccio 2010). Research is also needed to understand the cross talk between the various epigenetic mechanisms (Jobe et al. 2012). Significant advances are being made into understanding the role of nucleosome remodelers (elaborated in Becker and Workman 2013), such as microRNAs, histone variants, and histone deacetylases (see Seto and Yoshida 2014) in the process of neurogenesis (reviewed in Hirabayashi and Gotoh 2010; Ma et al. 2010; Tyssowski et al. 2014).
2. EPIGENETIC REGULATION OF Pcdh PROMOTER CHOICE
Gene regulation involving DNA and chromatin modifications enables the fine-tuning of the nervous system. This is illustrated by two examples discussed in the following sections: (1) the control of the expression of clustered Pcdh genes in this section, and (2) the OR gene choice described in Section 3. A common feature of the two gene families is that their regulation generates vast diversity among neurons, which allows them to adopt specialized functional identities. However, this is accomplished by distinct mechanisms. In the case of the Pcdh genes, stochastic promoter choice occurs by multiple independent, random choice events across three clusters of genes on two chromosomes. This usually results in monoallelic expression as a result of simple probability, but there is not a mechanism that enforces monoallelism. Promoter choice in the OR system is truly “monoallelic” in that it relies on random activation of a single OR gene coupled with strict monoallelic expression (Chess 2013).
2.1. Clustered Pcdh Genes
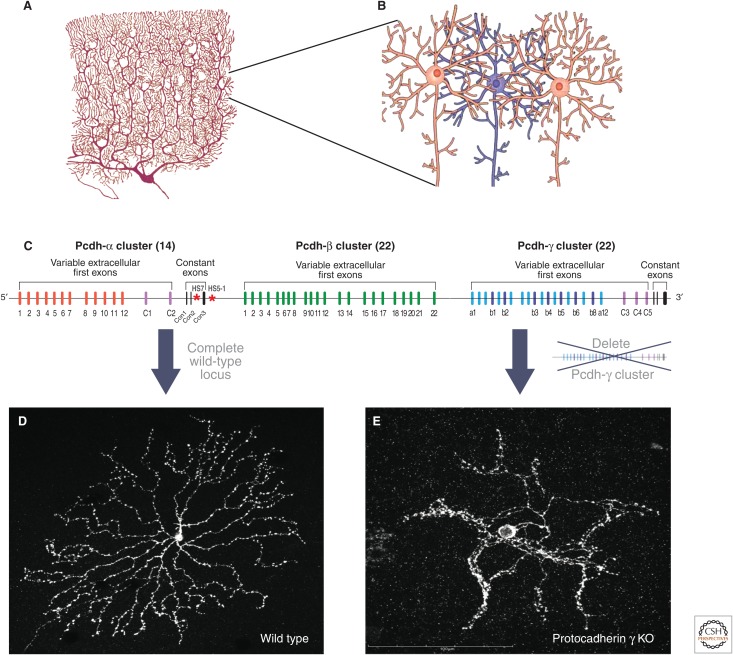
Individual neurons engage in multiple and precise synaptic connections with other neurons through an elaborate network of dendrites and axons, which collectively form a neural arbor (see Fig. 3A). During development, a growing neurite (i.e., axons and dendrites) must precisely navigate through a sea of hundreds, if not millions, of other developing neurites and find the appropriate target to connect with, while maintaining the ability to distinguish and minimize interactions with neurites emerging from its own cell (i.e., sister neurites; see Fig. 3B). Thus, prevention of synaptic interactions between sister neurites is essential for proper neural connectivity, signal transmission, and computation of neuronal signals. In both vertebrates and higher invertebrates, the solution to this problem is to provide a unique extracellular identity tag to every neuron, similar to a “barcode,” in which neurites containing the same barcode recognize and repel each other (Cameron and Rao 2010; Grueber and Sagasti 2010). This mechanism is called self-avoidance, a process that ensures that neuronal processes from the same neuron repel each other during arborization and axon branching to avoid clumping. Self-avoidance also prevents extensive overlapping in the arborization pattern and facilitates the coverage of the neuronal processes across different regions of the nervous system during development.
Figure 3.
Neurons require a mechanism for self-recognition. (A) Drawing of a Purkinje cerebellar neuron emphasizes the elaborate dendritic arborization. (B) Arbors of the blue neuron must make connections with the neighboring orange neurons while avoiding connectivity with other blue arbors. This image emphasizes the need for a “barcoding” mechanism for self-avoidance. (C) The genomic organization of clustered Pcdh genes. Pcdh-α, -β, and -γ clusters span a genomic region of ∼1 Mb. The α cluster has 12 monoallelically variable exons and two biallelically expressed variable exons (C1 and C2). All variable exons splice to the three constant exons that code for the intracellular region. Similarly, the Pcdh-γ cluster has 19 monoallelically expressed variable exons and three variable exons (C3, C4, and C5) that are biallelically expressed. All variable exons splice to the constant exons except at the β cluster. (D) Wild-type retinal starburst amacrine cells. (E) Deletion of the 22 variable exons of the Pcdh-γ cluster abolishes self-avoidance of retinal starburst amacrine cells resulting in dendritic collapse and luck of arborization.
The mechanisms by which a highly diverse barcode is generated in vertebrates and flies are different. Diversity in Drosophila neurons is generated by one of the most complex examples of alternative pre-mRNA splicing known, whereas, in mammals, diversity is achieved through a combination of higher-order chromatin organization, stochastic promoter choice, and alternative splicing. Drosophila neurons express the Dscam1 gene, which encodes a transmembrane protein with extracellular immunoglobulin-like domains. Through alternative splicing of a large number of variable exons, an individual Drosophila neuron expresses approximately 10 to 15 isoforms, out of about 38,000 available combinations, of which 19,008 have a different extracellular domain (Schmucker et al. 2000). The Dscam1 immunoglobulin domains on the cell surface of one dendrite interact exclusively (i.e., “homophilically” with its counterpart on the opposing dendrite). These homophilic interactions ultimately lead to repulsion of the dendrites through a process requiring the intracellular domains of the Dscam1 proteins.
Single-cell identify of mammalian neurons is thought to be provided by the clustered Pcdhs rather than the mammalian Dscam homologs, as the latter have a simple organization compared with their Drosophila counterparts, and therefore cannot generate high levels of cell-surface diversity (Schmucker and Chen 2009). The clustered Pcdh gene family consists of approximately 70 family members in the mouse and 58 in humans (Wu and Maniatis 1999; Wu et al. 2001). The mechanism by which Pcdh diversity is generated is distinct from that which generates Dscam diversity. The organization of the Pcdh gene clusters is, however, remarkably similar to that of the immunoglobulin and T-cell receptor gene clusters discussed in Busslinger and Tarakhovsky (2014) and Wu and Maniatis (1999). The extracellular domains of Pcdhs are encoded by “variable” exons, whereas the intracellular domains are encoded by three “constant” exons (see Fig. 3C). The variable extracellular domains of Pcdh proteins function in self-recognition through homophilic interactions (Schreiner and Weiner 2010; Chen and Maniatis 2013; Zipursky and Grueber 2013; Thu et al. 2014), whereas the constant intracellular domain appears to be involved in cell signaling (Wu and Maniatis 2000; Han et al. 2010; Schalm et al. 2010; Suo et al. 2012). The intracellular domain can be released from the membrane by the combined activities of a metalloproteinase and γ-secretase in a process that requires endocytosis (Haas et al. 2005; Hambsch et al. 2005; Reiss et al. 2006; Bonn et al. 2007; Buchanan et al. 2010). The intracellular domain is then thought to translocate to the nucleus, but neither the regulation of this process nor the cytoplasmic or nuclear function of the intracellular domain is understood.
The extracellular domain of Pcdh proteins, like Dscams, appears to mediate homotypic repulsion (Schreiner and Weiner 2010; Zipursky and Sanes 2010; Lefebvre et al. 2012; Yagi 2012, 2013; Chen and Maniatis 2013; Zipursky and Grueber 2013; Thu et al. 2014). This has been shown by genetic manipulation of the Pcdh gene clusters; deletion of the 22 Pcdh-γ genes (Fig. 3C) results in the disruption of dendritic self-avoidance in retinal starbust amacrine cells (a type of retinal interneuron) (see Fig. 3E) and cerebellar Purkinje cells (inhibitory neurons involved in motor coordination) (Lefebvre et al. 2012). Considering the large difference in the number of distinct Dscam and Pcdh protein isoforms (19,000 compared with 58), it is surprising that the Pcdhs could generate the cell-surface diversity sufficient for single-cell identity. However, cell aggregation studies provided evidence that Pcdhs engage in homophilic interactions as multimeric complexes, thus dramatically increasing the potential single-cell diversity provided by the Pcdhs (Schreiner and Weiner 2010; Thu et al. 2014). In fact, theoretical calculations suggest that comparable levels of single-cell diversity can be generated by the invertebrate Dscam gene and vertebrate clustered Pcdhs (Yagi 2012; Thu et al. 2014). Recent studies have shown that all but one of the members of the Pcdh-α, -β, and -γ gene clusters are capable of individually engaging in homophilic interactions as monomers, and that multimers display highly specific interactions. Examination of homophilic interactions between specific combinations of multiple Pcdh isoforms revealed that Pcdh combinatorial recognition specificities depend on the identity of all of the expressed isoforms. However, the nature of the multimer (dimer, trimer, or higher-order multimers) and complexes on the cell surface have yet to be established (Thu et al. 2014).
2.2. Pcdh Gene Regulation
This section describes the genomic organization of the three Pcdh gene clusters, and Section 2.3 describes the chromatin and epigenetic mechanisms that underlie their stochastic expression in individual neurons. Remarkably, the pattern of Pcdh expression in mouse and human cell lines is maintained indefinitely through many rounds of cell division. The timing of promoter choice during neuronal differentiation in vivo and its stability during cell division in vivo (if it occurs) remains to be shown. Nevertheless, the stability of promoter choice in neuroblastoma cell lines suggests the existence of mechanisms that faithfully maintain expression patterns during DNA replication. Such mechanisms could allow a stable pattern of Pcdh expression through the life of an individual neuron, thereby maintaining neuronal self-identity. Continued studies on the mechanisms of Pcdh promoter choice during development, and the epigenetic mechanisms involved in determining whether and how the choice is maintained, should provide important insights into the role of Pcdh diversity in the assembly of neural circuits.
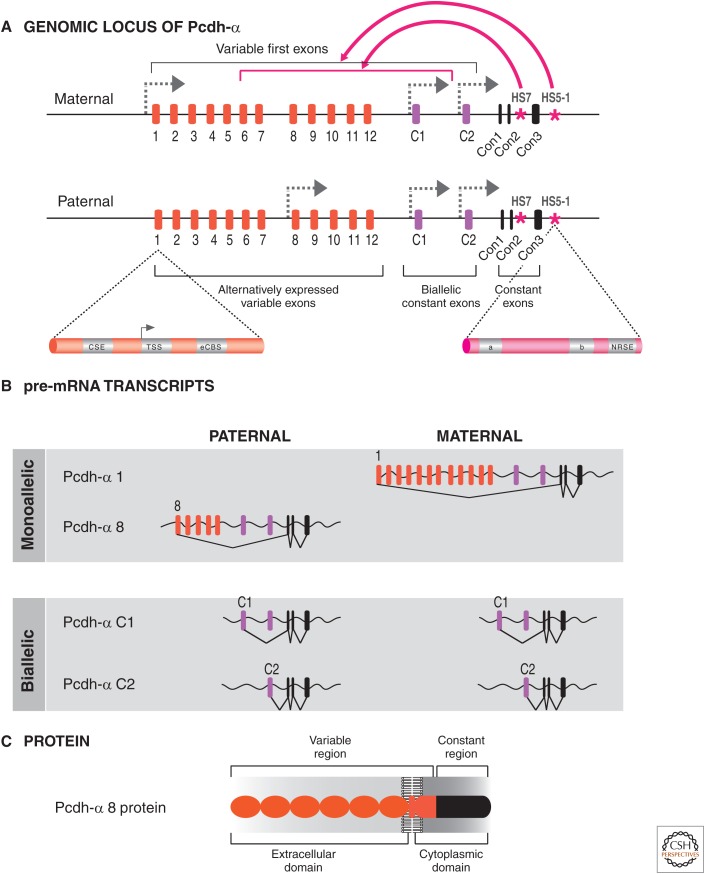
To understand the epigenetic regulation of this gene cluster, it is necessary to describe its genomic organization and expression. The extracellular domains of the mouse Pcdh-α and -γ genes are encoded by 14 and 22 variable exons, respectively (Fig. 3C). Each of the variable exons is transcribed from its own promoter. Transcripts initiated from these promoters read through the downstream variable exons (i.e., C1 and C2 in Fig. 4A). The 5′ splice site nearest the start site of transcription is then cis-spliced to the first constant exon (Con1 in Fig. 4B). Two types of variable exons have been identified: the alternately expressed exons (i.e., Pcdh-α 1-12, and Pcdh-γ 1–22) and the biallelic ubiquitously expressed exons (Pcdh-α C1 and C2, and Pcdh-γ C3, C4, C5) (Esumi et al. 2005).
Figure 4.
Alternative splicing produces variable Pcdh isoforms. (A) Hypothetical example showing the expression pattern of the Pcdh-α cluster in a single neuron, in which one of the alternative promoters is activated in a seemingly stochastic fashion on each chromosome. An enlargement of the exon 1 promoter region shows the two CCCTC-binding factor (CTCF) binding sites (conserved sequence element [CSE] and exonic CTCF binding site [eCBS]). The enlargement of the HS5-1 enhancer shows the two CTCF binding sites (a and b) and the neuron-restrictive silencer factor (NRSF) binding sequence, neuron-restrictive silencer element (NRSE). (B) The pre-mRNAs produced the maternal and paternal alleles, showing monoallelic expression of variable exons α1 and α8, but biallelic expression of the C-type exons. The variable pre-mRNAs contain all the variable exons located downstream from the selected promoter, which are subsequently spliced to the Con1–3 exons. Only the exon located immediately downstream from the selected promoter will be spliced to the exons encoding the intracellular constant region. (C) Each variable exon encodes for six ecto- (cadherin) domains involved in self-recognition, most probably through homotypic repulsion.
The expression of the alternately expressed exons is determined by random promoter choice. For example, in the case of the Pcdh-α cluster, promoter choice is stochastic and occurs independently on each chromosome in an individual neuron, based on single-cell analyses of Purkinje cells from F1 mouse hybrids (Esumi et al. 2005). Thus, different maternal and paternal alternate promoters are stochastically activated in each neuron. This choice is not strictly limited to a single alternate promoter; sometimes, two or more different alternate Pcdh isoforms from a cluster can be expressed on each chromosome. This mechanism ultimately results in monoallelic promoter choice (i.e., each chromosome expresses one or more unique isoforms) (Fig. 4B). This is different than the monoallelic expression of X-linked genes in females (see Brockdorff and Turner 2014) or immunoglobulin receptors (described in Busslinger and Tarakhovsky 2014); in these cases, a single copy is expressed and chosen at random from only one chromosome in each cell (see Chess 2013 for a comparison of the different monoallelic expression mechanisms). Another distinction from monoallelic immunoglobulin genes is that, in addition to the alternately expressed exons, five “C”-type exons are expressed ubiquitously and biallelically (C1–C5, of which two are expressed from the α cluster and three from the γ cluster) (Esumi et al. 2005; Kaneko et al. 2009). These C1–C5-type exons (Fig. 3B) are also spliced to the constant exons (Con1–3) (Fig. 4B). The Pcdh-β cluster differs in that the variable exons encode complete proteins with only short cytoplasmic domains and, thus, are unlikely to have signaling potential (at least on their own). Taken together, this stochastic, combinatorial (i.e., a combination of 1–2 Pcdh-α, 1–2 Pcdh-β, and 1–2 Pcdh-γ isoforms in a single cell), and partially monoallelic pattern of expression of the three Pcdh gene clusters can potentially generate extraordinary cell-surface diversity (see Yagi 2012 for discussion).
2.3. Epigenetic Control of Pcdh Expression
In this section, we will focus on what is known about the genetic and epigenetic mechanisms that regulate promoter choice in the Pcdh-α cluster as a paradigm, but it is virtually certain that similar mechanisms are used in the Pcdh-β and -γ gene clusters.
The promoters of every alternately expressed exon, and a subset of the biallelic exons, share a conserved sequence element (CSE) required for transcriptional activation (Wu et al. 2001; Tasic et al. 2002). Beyond this common promoter element, long-distance enhancers are required for maximum levels of Pcdh expression. For the Pcdh-α cluster, these include two cis-regulatory sequences, hypersensitive sites HS7 and HS5-1, identified by in vivo DNase I sensitivity assays, which function as nervous-system-specific enhancers based on transgenic reporter assays (Ribich et al. 2006).
Deletion of HS5-1 in mice results in a decrease in Pcdh-α gene expression throughout the brain (Kehayova et al. 2011). In particular, expression of exons proximal to HS5-1, which contain a CSE element (i.e., Pcdh-α6–12, in addition to Pcdhα-C1) is most affected by the deletion (Fig. 5A). In nonneuronal tissues, however, deletion of HS5-1 results in up-regulation of Pcdh-α gene expression. Examination of the DNA sequence of HS5-1 revealed that this enhancer contains a sequence element known to function as a silencer (Kehayova et al. 2011). This element, referred to as the neuron-restrictive silencer element (Fig. 4A) (Kehayova et al. 2011), is known to act as a repressor when bound by NRSF, a repressor expressed in nonneuronal tissues that inhibits neurogenic transcription by the recruitment of repressive protein complexes (Lunyak and Rosenfeld 2005).
Figure 5.
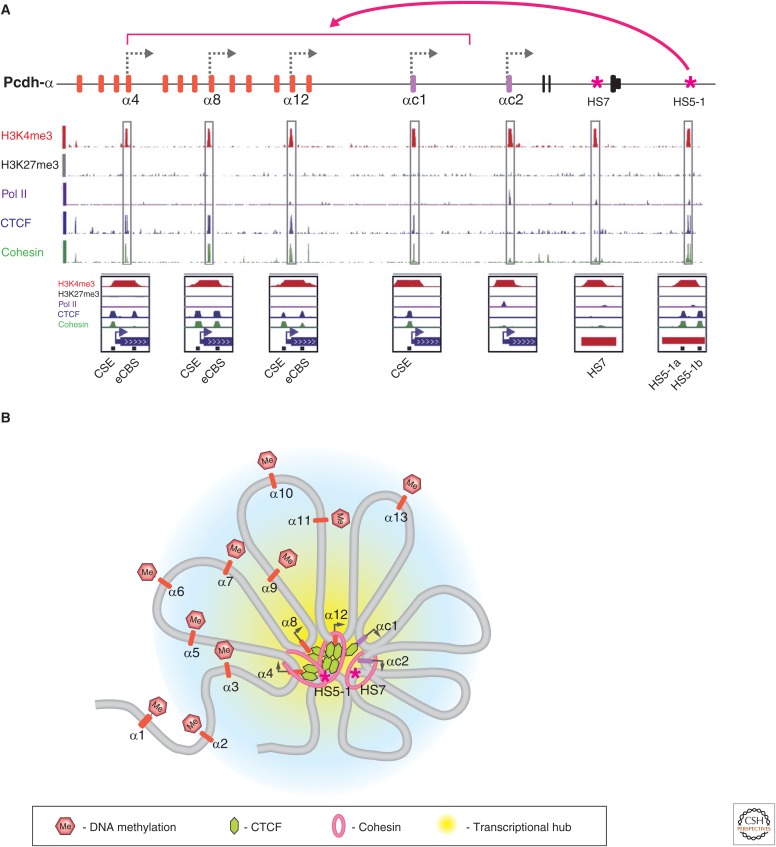
Summary of the epigenetic properties of the Pcdh-α cluster. (A) Summary of ChIP-seq (parallel sequencing technologies coupled to chromatin immunoprecipitation) analysis in a human diploid neuroblast cell line (SK-N-SH) that expresses only three variable Pcdh-α exons. There are two CTCF binding sites at each active variable exon (CSE and eCBS sequences) and two at the HS5-1 enhancer (HS5-1a and HS5-1b). (B) Summary of the epigenetic properties of active and silenced Pcdh-α exons on forming a 3D transcriptional hub, based on chromosome conformation capture (3C) data. Regions outside the central transcriptional hub are thought to be packaged in a repressive chromatin conformation (blue-shaded region), suggested by the DNA methylation of inactive Pcdh-α promoter regions. Active genes are thought to be concentrated in a transcriptional hub, aided by double-clamp binding of CTCF between the variable exons and the HS5-1 enhancer and cohesin.
The HS7 enhancer is also required for maximum levels of Pcdh-α expression in the brain, as its deletion results in a moderate down-regulation of Pcdh-α gene expression (Kehayova et al. 2011). This observation indicates that maximum levels of Pcdh-α gene expression require the combined activity of both the HS5-1 and HS7 enhancers. A simple model to explain the data is that all variable exon promoters have an equal chance of being activated, but a stochastic process selects one for activation (Fig. 4), the determining factor of which may be its interaction with HS7 and HS5-1.
Beyond the DNA sequence elements of the Pcdh gene clusters, recent studies have revealed that the random stochastic promoter choice correlates with a combination of epigenetic marks on DNA (i.e., DNA methylation) and histones (e.g., H3K4me3), and DNA-binding factors that affect higher-order chromatin structure. Because there is a mixed population of cells in the nervous system, and each cell expresses a distinct set of Pcdh isoforms, it is difficult to gain mechanistic insights into the regulation of Pcdh promoter choice in vivo. Progress, however, has been made using certain cell lines that stably express specific sets of Pcdhs to make correlations between DNA methylation, protein binding, chromatin modifications, and Pcdh isoform expression. For example, such studies have shown that the insulator binding protein, CTCF, and the cohesin complex subunit, Rad21, bind to two sites at active Pcdh promoters, the CSE upstream of the TSS and an exonic CTCF binding site (eCBS) downstream from TSS, and to two sites within the HS5-1 enhancer sequences (Golan-Mashiach et al. 2012; Monahan et al. 2012). This observation was corroborated by studies of a diploid human neuroblastoma cell line SK-N-SH, which stably expresses a small subset of Pcdhα isoforms (Fig. 5A) (Guo et al. 2012). The CTCF protein is known, among other functions, to promote long distance cis and trans genomic interactions (Phillips and Corces 2009), and Rad21 (a cohesin subunit) was recently shown to stabilize such interactions in a fashion similar to the stabilization that occurs for sister chromatid cohesion during mitosis (Kagey et al. 2010). Thus, the simultaneous presence of these two molecules on both the active variable promoters and a distant enhancer suggested that CTCF and cohesin may mediate enhancer/promoter interactions (see Kim et al. 2014 for further discussion). This does not explain, however, how only one, or sometimes two, of the 13 variable Pcdh-α promoters are selected for activation in individual cells. It is intriguing, though, that CTCF and the cohesin subunit Rad21 are only bound to promoters that are transcriptionally active, suggesting that initial choice may be accomplished by the selective binding of this complex to one of these competing promoters. In support of this idea, all the nontranscribed variable promoters are heavily methylated on CpG dinucleotides (Tasic et al. 2002; Kawaguchi et al. 2008). CpG methylation of CTCF-binding sites is known to abolish CTCF binding and, specifically, to CSE (Guo et al. 2012), in vitro, providing an epigenetic mechanism for promoter choice. Of course, in this case, the question is only moved a step back because promoter choice then becomes a question of how all but one variable promoter are methylated. However, the initial promoter choice is made, pharmacological inhibition of DNA methylation in cell lines that express only a subset of variable exons leads to reactivation of all the variable promoters that were normally silent in these cells (Kawaguchi et al. 2008). These results are equally consistent with an alternative mechanism, though, in which initially all the variable promoters are unmethylated and CTCF/Rad21 is bound, but only interaction with HS5-1 stabilizes this complex on the chosen promoter. The other variable promoters would eventually lose CTCF/Rad21 binding and they would become methylated, “locking” them into a permanent silent state without basal transcription activity. The silencing of the variable promoters not chosen could then be reinforced by heterochromatinization, as shown in olfactory sensory neurons (OSNs) by the enrichment of H3K9me3 and H4K20me3 marks on the silent variable exons (Magklara et al. 2011). Recently, a conditional knockout of the CTCF gene was used to show that the alternately expressed promoters require CTCF for normal levels of transcription in the cortex and hippocampus, whereas the biallelic promoters, ac2 and gc4, do not (Hirayama et al. 2012). This observation is consistent with the notion that CTCF/cohesin organizes the enhancer/promoter interactions in such a way that the alternately expressed promoters are chosen stochastically, in contrast to the biallelic exons.
The symmetric relationships between the two CTCF/cohesin binding sites in the promoter/exon and the HS5-1 enhancer suggests the interesting possibility that if the HS5-1 enhancer and promoter do interact through DNA looping, the CTCF/cohesin complex simultaneously binds to the enhancer and promoter to form a “double clamp” (i.e., two binding points between each DNA element and CTCF/cohesin complex), which may play a role in the epigenetic stabilization of promoter choice (Fig. 5B) (Guo et al. 2012; Monahan et al. 2012).
Long-range DNA-looping interactions in SK-N-SH cells between enhancers and promoters of the Pcdh-α cluster were detected using quantitative 3C assays (described in the text and Fig. 5 of Dekker and Misteli 2014). Interactions of varying strength were detected between both the HS7 and HS5-1 enhancers and the transcriptionally active promoters. The same experiments performed after small hairpin RNA knockdown of CTCF or the cohesin subunit Rad21 significantly decreased all of the enhancer/promoter DNA-looping interactions. These results, coupled to data indicating the requirement of both enhancers for maximal expression of Pcdh-α alternately expressed exons, suggest a complex mechanism of promoter choice; Guo et al. (2012) proposed that the DNA-looping interactions in the Pcdh-α gene cluster recruit CTCF-bound promoters to enhancers in an active “transcriptional hub” (Fig. 5B). This model is based on extensive and highly specific functional and physical interactions between promoters and enhancers, and the fact that the formaldehyde cross-linking used in the ChIP-seq and 3C studies would be expected to cross-link a large complex containing multiple enhancers and promoters. In this model, the DNA-looping interactions between HS5-1 and the promoters of α8 and α12 are formed by a double-clamping mechanism between the HS5-1a/HS5-1b sites of the enhancer and CSE/eCBS sites of alternate promoters (Fig. 5A). At the same time, HS5-1 must interact with αc1 because HS5-1 is required for its expression (Ribich et al. 2006; Kehayova et al. 2011). Also, HS7 must directly interact with αc2, as well as with the active alternate promoters and ubiquitous promoters, because this enhancer interacts with and is required for the maximum activation of these promoters (Kehayova et al. 2011). Finally, long-range DNA-looping interactions are formed between enhancers and active promoters. All of these observations are consistent with the model illustrated in Fig. 5B, in which the apparent simultaneous interactions between the two enhancers and multiple promoters leads to the formation of a large “transcriptional hub.”
In summary, the regulation of Pcdh gene expression constitutes a complex regulatory system that involves unusual transcriptional and posttranscriptional events of paramount importance for the development and wiring of the nervous system. Although much has been learned since the discovery of this remarkable gene family, significant open questions, nonetheless, remain that likely involve epigenetic processes. For example, how does the splicing machinery recognize only the 5′ splice site proximal to active promoters, and ignore all of the downstream 5′-splice sites? This is likely to be a consequence of the organization of chromatin on the active and inactive variable region exons (Magklara et al. 2011; Monahan et al. 2012). Other questions include: At what point during neural differentiation does promoter choice occur, and how stable is the choice once it is made? As mentioned above, the specific pattern of Pcdh isoform expression in neuronal cell lines is stable over many generations, so, it is likely that an epigenetic mechanism is involved. However, given the elaborate enhancer/promoter interactions described above, the mechanism of epigenetic stability must be complex. Another question is: How can stochastic promoter choice and selective expression of individual Pcdh gene cluster occur at the same time? For example, Pcdh-α, but not -β or -γ, is expressed in the dorsal raphe and serotonergic system, but one presumes that stochastic choice still occurs in the Pcdh-α gene cluster (Katori et al. 2009). Finally, it will be of great interest to understand the organization of the proposed Pcdh transcription hub in individual neurons.
3. EPIGENETIC REGULATION OF OR CHOICE
Until recently, the H3K9me3 and H4K20me3 histone marks were considered hallmarks of constitutive heterochromatin described as occurring exclusively at pericentromeric and subtelomeric regions of the mammalian genome. These are primarily repetitive regions of the genome, with little abundance of genic regions. Epigenomic analysis of primary olfactory neurons, however, revealed an unexpected enrichment of H3K9me3 and H4K20me3 on ∼5% of mouse genes, namely, the OR genes (Magklara et al. 2011). Thus, this type of heterochromatin may actually be dynamic and directly involved in gene regulation, rather than solely to preserve genomic integrity and repress retrotransposons and other repetitive elements.
3.1. Anatomy and Genetics of the Olfactory System
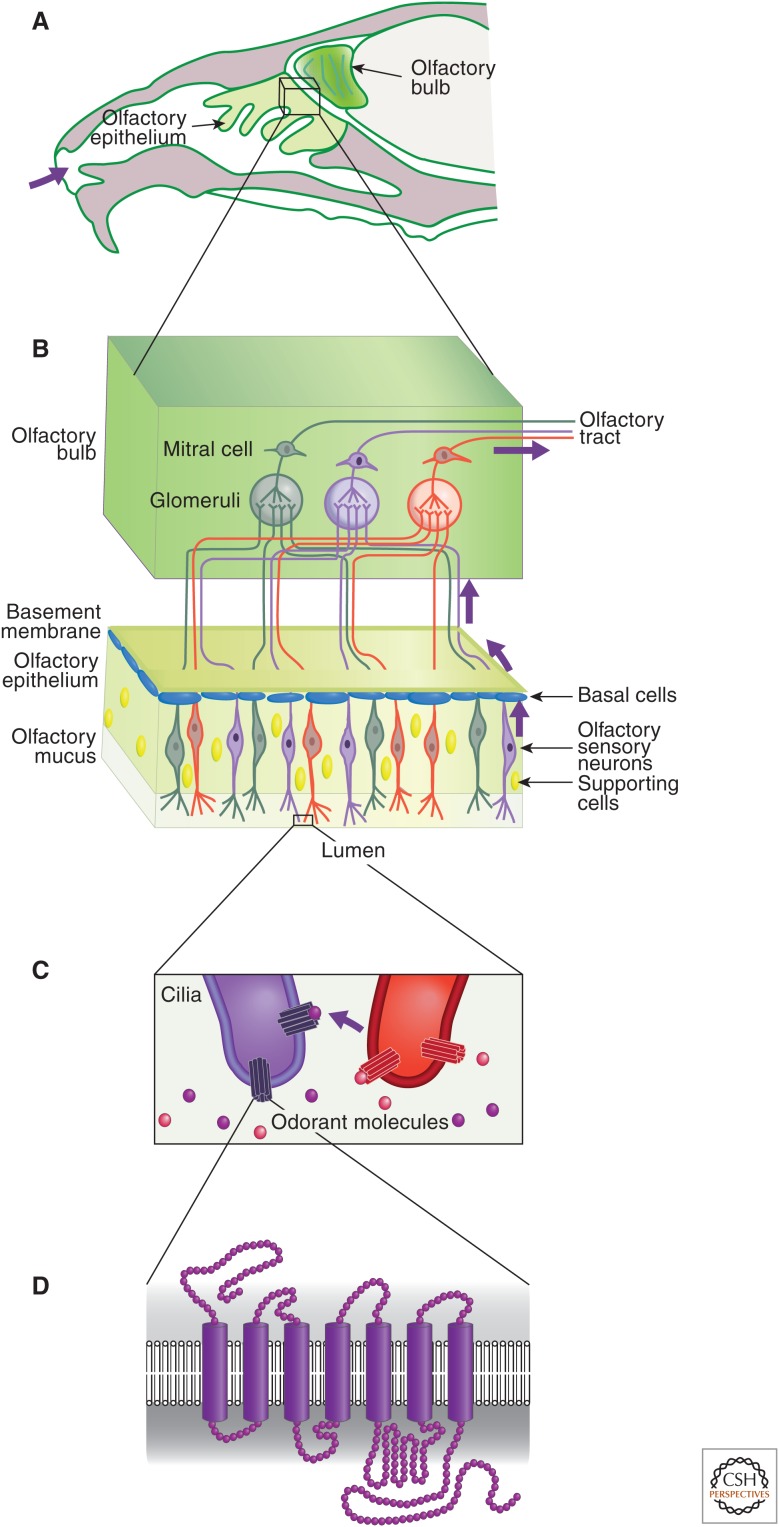
A description of the anatomy of the olfactory epithelium and bulb are helpful in understanding how this organ is functionally regulated at the genetic and epigenetic levels. In higher organisms, volatile odorants are detected by OSNs via G-coupled OR proteins, with seven membrane-spanning domains found on the surface (Fig. 6D) (Buck and Axel 1991). In mammals, these neurons reside in a specialized sensory organ called the main olfactory epithelium (MOE), which covers the lumen of the nasal cavity (Fig. 6A,B) (Axel 1995). The olfactory epithelium regenerates throughout life, and is composed of three main cell types organized in intermingling layers: basal cells, which are multipotent stem cells that give rise to all the other cell types of this sensory organ; OSNs, which are postmitotic neurons with odor-detection potential; and sustentacular (supporting) cells, which are nonneuronal cells that line the apical layer of the epithelium. The dendrites of the olfactory neurons terminate in cilia, which project beyond the apical layer of the MOE and contain ORs, and make contact with volatile odorants in the lumen of the nasal cavity (Fig. 6C).
Figure 6.
Introduction to olfactory receptors (ORs). The anatomy of the olfactory system is shown in increasing degrees of magnification from A to D, down to the depiction of ORs, which are G-protein-coupled receptors with seven transmembrane α-helices (D). Purple arrows throughout indicate the pathway of olfactory system transduction from the entrance of odorant molecules into the nasal cavity (A), their binding to ORs (C), and transduction of the signal through the olfactory system (B). (A) Head cross section in the mouse, indicating the olfactory tissues containing OSNs. The MOE is the olfactory sensory organ containing OSNs, basal pluripotent cells (blue), and sustentacular supporting cells (yellow nuclei). Each OSN expresses one type of OR (three types illustrated for simplicity as green, red, and purple) and the axons of OSNs expressing the same OR project to the same glomerulus in the olfactory bulb in which they synapse with interneuron/mitral cells to transmit the signal to the brain. Glomeruli map spatially into zones based on similarity of OR characteristics. (C) Enlargement of OSN cilia, depicting the single type of OR (red or purple), which binds a particular type of odorant molecule. (D) Structure of the seven transmembranes spanning the OR.
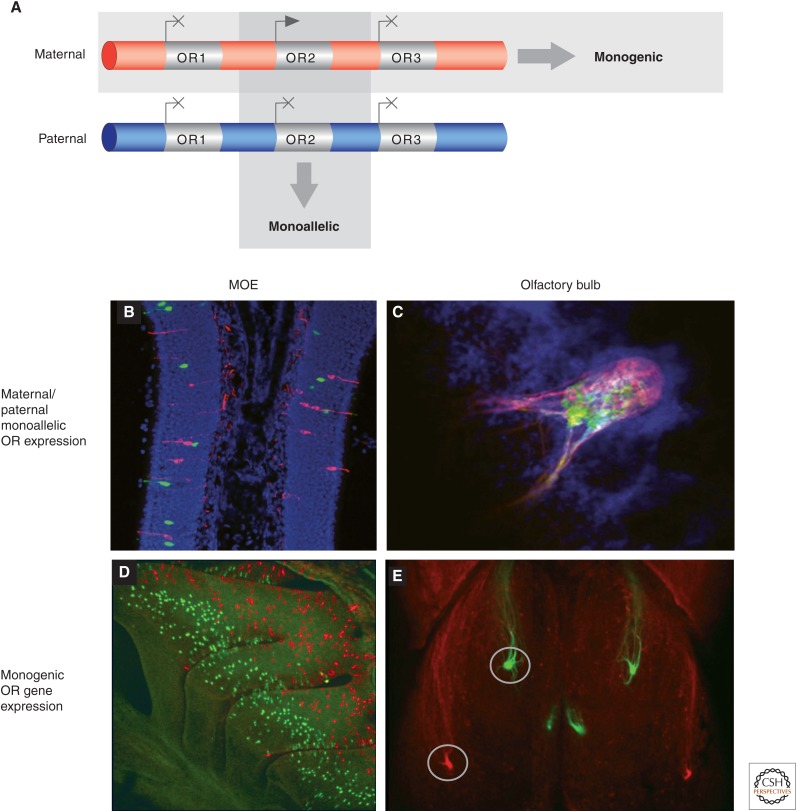
Most mammals have more than 1000 olfactory or odorant receptor genes, organized in genomic clusters of variable size (from two to about 200 OR genes), distributed across most chromosomes (Glusman et al. 2000; Zhang and Firestein 2002; Nei et al. 2008). The extraordinary number of genes dedicated to odor detection illustrates the importance of this chemosensory system in the survival and reproduction of most animal species. The question of how this myriad of OR genes are regulated in OSNs has intrigued researchers for a number of years. A striking characteristic of OR genes is that they are expressed in a mutually exclusive, monogenic, and monoallelic fashion; that is, each OSN expresses only one OR gene from a choice of more than 1000, from only one parental allele (Fig. 7A) (Chess et al. 1994; Shykind 2005). It is important to note that unlike parental imprinting, which is described in detail elsewhere in this collection (Barlow and Bartolomei 2014), maternal and paternal OR alleles are expressed in equal frequency overall in the olfactory epithelium, so, expression is termed “monoallelic” because both alleles are never coexpressed in the same neuron (Fig. 7B,C). This expression pattern is distinct from imprinting because the parent of origin does not determine which allele is used, and it differs from the Pcdh promoter choice because only one OR allele is expressed in an individual neuron, rather than one, two, or more isoforms that can be expressed independently from each parental copy from the Pcdh clustered genes. This expression pattern, known as the “one receptor per neuron” rule, is essential for the proper functioning of the olfactory system because the identity of the OR protein not only provides specificity in odorant detection, but also plays an instructive role in the guidance of its axon toward a specific glomerulus within the olfactory bulb (Fig. 7D,E) (Mombaerts et al. 1996; Wang et al. 1998; Barnea et al. 2004; Serizawa et al. 2006).
Figure 7.
Monogenic and monoallelic OR gene expression in the MOE and olfactory bulb. (A) A schematic representation of an OR gene cluster containing three genes, illustrating the monogenic and monoallelic nature of OR expression. (B,C) Immunofluorescence hybridization illustrates the monoallelic nature of OR gene expression. Maternal and paternal alleles of an OR gene are tagged with lacZ (red) and green fluorescent protein (GFP, green) by knocking-in two reporter genes. (B) A section of the MOE shows that there is a lack of coexpression of both parental alleles in a single OSN nucleus. (C) A section of the olfactory bulb shows that neurons expressing an OR from either the paternal or maternal allele coalesce in the same glomerulus. (D,E) OR expression is monogenic and expressed in a zonal fashion. Two different OR genes have been tagged with red fluorescent protein (red) and GFP (green), respectively. (D) Whole-mount imaging of the MOE shows the zonal distribution of expression between the two genes. (E) The axons of the neurons that express different OR genes coalesce in distinct and spatially separated glomeruli in the olfactory bulb. (Images kindly donated by Dr. Thomas Bozza.)
The glomerulus is a spherical structure located near the surface of the olfactory bulb, and it contains synapses between incoming OSN axons and the dendrites of mitral, tufted, and periglomerular cells residing in the olfactory bulb. If a neuron were to express more than one OR allele, it would respond inappropriately to different odorants, and would send signals to the wrong brain regions, resulting in sensory confusion and an olfactory system with impaired discriminatory power. Therefore, immense evolutionary pressure has resulted in perfecting this unique olfactory system and epigenetic mechanisms have evolved to safeguard the singularity of OR expression, as mentioned above (Fig. 4B). Simply stated, in the case of Pcdhs, the two chromosomes make independent stochastic choices, whereas in OR regulation, only one OR allele from only one cluster and one chromosome is expressed.
Some spatiotemporal specificity of OR gene regulation can be attributed to the promoter sequence signatures that ensure olfactory-restricted gene expression, through the binding of combinations of highly abundant transcription factors, such as Emx2, Lhx2, and Olf/Ebf (O/E) family members (Wang et al. 1997; Hirota and Mombaerts 2004; Rothman et al. 2005; McIntyre et al. 2008). Mapping of the TSS of OR genes revealed that they have extremely AT-rich sequences, with similar transcription factor-binding sites predicted ∼1 kb upstream of and ∼1 kb downstream from their TSS (Clowney et al. 2011; Plessy et al. 2011). Promoter signature variations ensure that each OR gene is subject to spatiotemporal restrictions visible as, at least, four broad “zones” of expression in the MOE, two of which are visible in Fig. 7D (Ressler et al. 1993; Vassar et al. 1993). This transcription factor–mediated control, however, is not sufficient to restrict OR activation to only one promoter per neuron. Thus, the fundamental question of how only one of the thousands of equivalent promoters is transcriptionally active in each OSN remains unanswered. In the mouse, there are approximately 1400 OR genes, of which about 1100 have detectable transcripts (Clowney et al. 2011) in the MOE. Identical paternal and maternal OR alleles are never coexpressed (i.e., its monoallelic property). A critical mechanism for this mutually exclusive expression pattern is the existence of a feedback signal that is generated by the expression of an OR protein to prevent the transcriptional activation of additional ORs (Serizawa et al. 2003; Lewcock and Reed 2004; Shykind et al. 2004; Nguyen et al. 2007). Lineage tracing experiments have shown that if a neuron makes a choice of a “nonfunctional” OR (because ∼20% of OR genes in the mouse are pseudogenes), the choice will be reset and the faulty OR allele replaced with a functional allele.
3.2. Epigenetic Regulation of Monoallelic OR Expression
Another mechanism that contributes to the specificity missing at the promoter level is the action of distant enhancer elements, which usually implicates a level of epigenetic regulation through chromatin marks and spatial organization (described further in Kim et al. 2014). Some distal OR enhancers have been described as being required for the proper expression of some OR genes (Serizawa et al. 2003; Khan et al. 2011). For example, the H enhancer element is necessary for the transcription of three ORs located 75 Kb downstream from its genomic location. This element can also drive the widespread expression of a transgenic OR when inserted proximal to its promoter (Serizawa et al. 2003). Moreover, chromosome conformation capture (3C and 4C), and two-color DNA fluorescence in situ hybridization (FISH) experiments, have shown that the H element interacts both in cis and trans (i.e., on the same and different chromosomes) with actively transcribed ORs (Lomvardas et al. 2006). However, the interaction of H with OR alleles in trans is not necessary for their expression (Fuss et al. 2007) and, based on results presented below, the trans interaction is most likely the consequence of the spatial segregation of active and silent OR alleles. A few ORs can, nonetheless, be expressed without distal regulatory elements as mini transgenes (minigenes) with <500 bp of DNA sequence upstream of the TSS (Vassalli et al. 2002; Rothman et al. 2005; Vassalli et al. 2011).
Transgenic ORs, in the form of minigenes or large yeast artificial chomosome (YAC)-based transgenes, are never coexpressed with endogenous ORs that share identical regulatory sequences. In addition, identical paternal and maternal OR alleles are never coexpressed. The fact that transgenic and endogenous ORs with the same regulatory sequences (Ebrahimi et al. 2000; Serizawa et al. 2000; Vassalli et al. 2002), or the identical paternal and maternal OR alleles, are never coexpressed in the same neuron (Chess et al. 1994; Feinstein and Mombaerts 2004) argues for a mechanism of gene silencing to achieve monoallelic expression. Based on experiments using transgenic OR minigenes, the coding sequence of ORs, specifically the second exons of these genes, appears to contain the sequence information necessary for the repression of transgenes (Nguyen et al. 2007). Thus, a simple solution for the maintenance of the monogenic and monoallelic OR expression is epigenetic silencing.
3.3. The Chromatin Structure and Nuclear Organization of OR Clusters in OSN Development
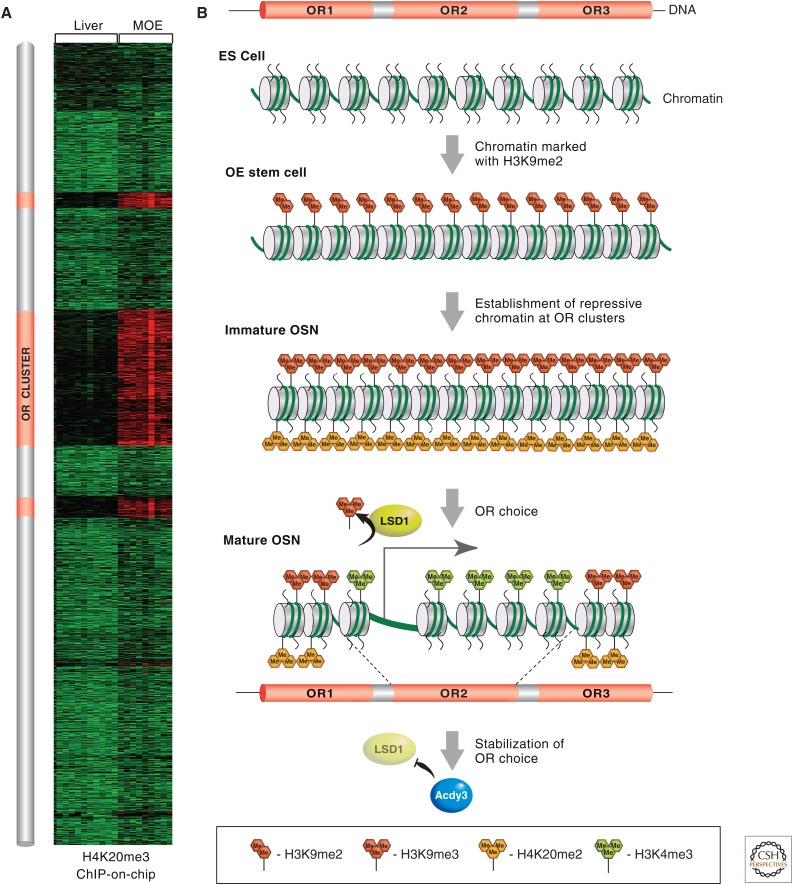
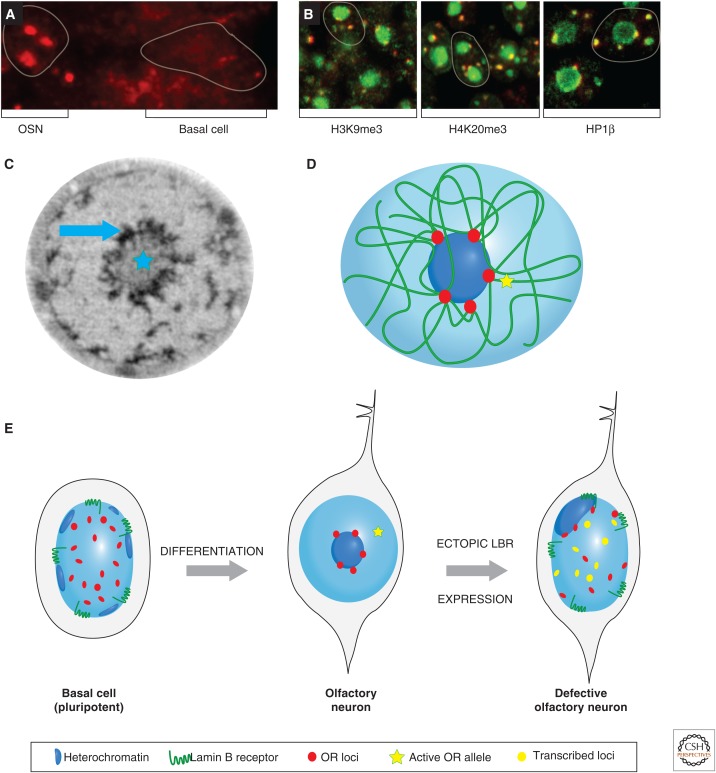
OR genes found in many cell lines have led to these regions being viewed as “epigenetic deserts,” based on numerous ChIP-on-chip or ChIP-seq experiments (Larson and Yuan 2010). This description is attributed to the lack of enrichment for any known chromatin modifications (Mikkelsen et al. 2007). However, ChIP-on-chip analysis with chromatin prepared from the MOE, the tissue in which OR genes are expressed, revealed the unexpected enrichment for H3K9me3 and H4K20me3 on ORs (Magklara et al. 2011). The presence of these heterochromatin marks is so specific to OR loci (and only a few other monoallelically expressed gene families, such as vomeronasal receptors) that genomic OR clusters can be identified solely based on the presence of H3K9me3 and H4K20me3 (Fig. 8A). Further analysis showed that H3K9 and H4K20 trimethylation of OR chromatin occurs in a differentiation-dependent and cell-type-specific manner. Specifically, in the pluripotent basal cells that support the constant regeneration of this tissue, because olfactory neurons have a finite life span, or in the case of injury, ORs are marked only with H3K9me2. On differentiation and commitment to the neuronal lineage, the trimethyl marks are deposited on ORs (Fig. 8B). Importantly, these marks are not restricted to promoters, as is the case for Polycomb-mediated repression, but spread over the entire gene into the intergenic regions between neighboring ORs, generating continuous genomic blocks of heterochromatin often >1 Mb long. The active allele in each mature OSN appears to be liberated from this epigenetic silencing with high enrichment for activating histone marks, such as H3K4me3, and depletion of H3K9me3 and H4K20me3 (Magklara et al. 2011). This epigenetic switch from repressive to activating histone modifications is restricted only to the allele that is transcriptionally active; ChIP-qPCR (quantitative polymerase chain reaction) experiments from fluorescence-activated cell-sorted neurons, which express a specific OR allele that is tagged by GFP, confirmed that neighboring ORs from the same cluster or the identical allele inherited from the other parent are epigenetically silenced as are the rest of the ORs.
Figure 8.
MOE-specific epigenetic regulation of OR gene choice during OSN development. (A) A heatmap depicting various enrichment levels for H4K20me3 on all the genes of mouse chromosome 2, from ChIP-on-chip analysis from MOE or liver. Red reflects high enrichment levels and green shows no enrichment. The three “red clusters” of high H4K20me3 levels coincide with the genomic coordinates of the OR clusters, schematically represented on this chromosome in orange to the left of the heatmap. (B) OR clusters start in early development (ES cells), devoid of most histone marks. At the onset of olfactory neurogenesis, OR clusters are marked with H3K9me2. Most OR genes then become marked by H3K9me3 and H4K20me3 during the transition from basal pluripotent cells to OSN. The OR allele that is chosen for expression (i.e., OR2) becomes liberated from its repressive chromatin marks and remodeled into an active chromatin configuration, aided by the action of the H3K9 demethylase, LSD1, and becomes positive for H3K4me3. Toward the end of the OSN differentiation pathway, the locking in of a single active OR gene is facilitated by the OR protein-induced expression of adenylyl cyclase III (Adcy3), which inhibits further LSD1 histone demethylation.
The marking of OR chromatin with H3K9me3 and H4K20me3 coincides with molecular and biochemical manifestations of heterochromatinization, such as recruitment of HP1β, reduced DNase I sensitivity, and altered sedimentation properties. Thus, the epigenetic silencing of OR loci could render OR promoters inaccessible to abundant transcription factors predicted to bind to them, potentially explaining why, in each OSN, the nonchosen OR genes and other parental alleles are completely silent at the transcriptional level. In agreement with this hypothesis, inserting a reporter transgene within the heterochromatic boundaries of an OR locus results in them being marked with H3K9me3 and H4K20me3, and leads to its expression in a zonal, sporadic, and monoallelic fashion that coincides with the neighboring OR gene (Pyrski et al. 2001; Magklara et al. 2011). In a rare showing of bona fide epigenetic regulation, this reporter transgene, which normally is expressed independently of Emx2, a transcription factor that activates OR transcription, becomes Emx2 dependent when inserted within OR heterochromatin (Magklara et al. 2011). Insertion of the same transgene into other parts of the genome results in its expression in most MOE neurons (Pyrski et al. 2001), suggesting that the epigenetic state of the neighboring OR influences the expression of the transgene, in a fashion similar to classic position effect variegation described in Elgin and Reuter (2013).
The timing of OR silencing, which occurs before OR gene activation, combined with the fact that the active OR allele is devoid of H3K9-methyl marks, immediately suggests that demethylation of H3K9 is a requirement for OR gene activation. Indeed, genetic experiments revealed that H3K9 demethylase Lsd1 is necessary for the initiation, but not the maintenance, of OR transcription (Fig. 8B) (Lyons et al. 2013). Moreover, timely down-regulation of Lsd1 in response to a signaling pathway triggered by OR expression (Dalton et al. 2013; Lyons et al. 2013) is necessary for the stabilization of OR choice, providing evidence for the paramount role of chromatin-mediated silencing and desilencing in OR gene regulation (Rodriguez 2013; Tan et al. 2013; Ferreira et al. 2014).
3.4. Spatial Organization of OR Genes
At the 3D level, nonexpressed OR genes converge in the nucleus into foci near the center of the nucleus. Recent experiments have shown that the epigenetic silencing of OR genes during neuronal differentiation in the MOE is coincident with widespread nuclear reorganization that culminates in intra- and interchromosomal association of hundreds of OR alleles within a few (approximately five) OR-selective heterochromatic foci (Fig. 9A) (Clowney et al. 2012). These aggregates are similar in size to the nucleolus and are frequently arranged on the periphery of the heterochromatic chromocenter of the olfactory nucleus, which primarily contains pericentromeric and centromeric repeats. Thus, OR foci represent nuclear territories highly enriched for heterochromatic markers, such as H3K9me3, H4K20me3 and HP1β (Fig. 9B), and are devoid of landmarks of euchromatin, such as Pol II, H3K4me3, and H3K27ac. The active OR allele in each OSN escapes these heterochromatic OR foci, but is found in transcriptionally competent nuclear territories with typically euchromatic epigenetic signatures (Clowney et al. 2012; Armelin-Correa et al. 2014) that also frequently contain the H enhancer (Lomvardas et al. 2006).
Figure 9.
The spatial organization of OR gene clusters during OSN differentiation. (A) DNA FISH showing OR gene cluster distribution using a panOR probe (red) on MOE sections. The signal is diffuse at the basal pluripotent cells, but aggregated in neuronal nuclei (the nuclear borders are highlighted). (B) PanOR foci (red) are enriched at regions marked with H3K9me3, H4K20me3, and HP1β (green), suggesting that they are heterochromatic aggregates of the silent OR alleles. (C) Soft X-ray tomography (SXT) of an olfactory nucleus shows that compacted chromatin (highlighted by a blue star) is aggregated at the center of the nucleus. At the periphery of this heterochromatic core, SXT reveals even more compacted structures (pointed by blue arrow) that are OSN specific. (D) A schematic 3D nuclear landscape shows all the heterochromatic OR clusters converging, in cis and trans, to form silent foci incompatible with transcription. The active OR allele is physically separated from the inactive OR aggregates. This is based on epigenetic data obtained from ChIP-on-chip experiments depicted as heatmaps. (E) Prior to OSN differentiation, the precursor basal stem cell has a typical nuclear configuation with regard to heterochromatin with peripheral foci. The OR clusters are randomly distributed in nuclear space. Following differentiation, aggregation of all but one OR allele to heterochromatic foci surrounding the pericentromeric chromocenter of an OSN nucleus occurs. This serves the monogenic and monoallelic expression of these genes and depends on the down-regulation of lamin B receptor (LBR). Ectopic LBR expression in OSNs dissolves the OR foci, reverses the nuclear morphology, and causes the simultaneous expression of hundreds of ORs in a striking violation of the one receptor per neuron rule.
The fact that OR foci are incompatible with transcription or other nuclear processes is also suggested by measurements made with the novel imaging technique SXT. SXT is a high-resolution imaging method that relies on the same principle as medical X-ray imaging (Le Gros et al. 2009); as X rays penetrate a biological specimen (e.g., an olfactory nucleus or your broken hand), they are more efficiently absorbed in regions of concentrated organic material (i.e., compacted heterochromatin, in the case of the nucleus, or bone tissue, in the case of your hand). The fraction of X ray absorbed in different parts of the nucleus can be quantified and is higher in heterochromatic than euchromatic territories. SXT of olfactory nuclei reveals a unique nuclear architecture with high-absorbing heterochromatin in the center of the nucleus and even denser chromatin particles on the periphery of this heterochromatic core (Fig. 9C). Because these dense aggregates are OSN specific and have a nuclear arrangement and number similar to that of OR foci, we hypothesized that they correspond to the previously described aggregates revealed by the panOR FISH. Notably, these aggregates have X-ray absorption properties higher than pericentromeric heterochromatin (Clowney et al. 2012). Only sperm nuclei, in which protamines replace histones, have more compacted chromatin than these OSN-specific foci as determined by SXT.
Does the OSN-specific spatial aggregation of OR loci contribute to OR silencing and the monogenic expression of these genes? An unusual feature of OSN nuclei is that they have an “inside-out” nuclear morphology, whereby heterochromatin is aggregated in the center of the nucleus and euchromatin is peripheral, in sharp contrast to the textbook view of nuclear organization with heterochromatin restricted to the nuclear envelope. This inverse nuclear organization is actually found in other types of sensory neurons, such as photoreceptor neurons, in the retinal epithelium (Solovei et al. 2009). In olfactory neurons, the reason for the “collapse” of heterochomatin in the nuclear core is the lack of the nuclear envelope protein, LBR. In most cells, the amino terminus of LBR interacts with HP1 proteins and is shown to recruit heterochromatin to the nuclear periphery (e.g., basal cell in Fig. 9E) (Worman et al. 1988; Pyrpasopoulou et al. 1996; Hoffmann et al. 2002). A loss-of-function mutation in the LBR gene in non-OSN cells of the MOE (Shultz et al. 2003) causes pericentromeric heterochromatin to be localized to the nuclear core and ectopic aggregation of OR genes. Conversely, ectopic LBR expression in OSNs reverts the inside-out nuclear morphology by recruiting heterochromatin back to the nuclear envelop and allowing euchromatin to claim the interior parts of the nucleus as in most mammalian cell types. One consequence of this dramatic reorganization is that OR aggregation is perturbed and the OR foci are dissolved. From a biochemical perspective, this results in decompaction of OR heterochromatin as detected by increased DNase I sensitivity and reduced X-ray absorption using SXT imaging. These changes occur despite the fact that these genes remain marked by H3K9me3 and H4K20me3, suggesting that this epigenetic signature translates to an inaccessible chromatin structure only on proper secondary (intrachromosomal) and tertiary (interchromosomal) folding (Fig. 9D). Functionally (i.e., from a transcriptional perspective), LBR-induced disruption of OR foci results in coexpression of hundreds of OR alleles in each olfactory neuron and, thus, in the dramatic violation of the one receptor per neuron rule that describes the monogenic and monoallelic expression of OR genes (illustrated in the defective OSN cell, Fig. 9E) (Clowney et al. 2012).
In summary, OR choice is a complex process that relies on chromatin-mediated silencing and selective desilencing on one out of thousands of available alleles. Although the aforementioned data provide a conceptual framework for the regulatory principles of OR expression, major questions must be answered for a comprehensive understanding of this unique process. For example, there is no information on the source of “singularity” that activates only one OR allele at a time, or signaling pathway and the molecular targets of the feedback signal elicited by OR expression. Moreover, the identity of the histone methyltransferases and demethylases responsible for the silencing and activation of OR genes in not known and neither are the mechanisms by which they are recruited with such specificity on the OR loci. Finally, the mechanism responsible for the specific aggregation of silenced OR alleles and placement of the active OR allele outside of the repressive OR foci remains unknown. Of great interest will be to determine if the same methyltransferase activities responsible for the aggregation of pericentromeric heterochromatin (Pinheiro et al. 2012) are also responsible for the focal distribution of OR genes.
4. EPIGENETIC REGULATION OF OLFACTORY NEURON LIFE SPAN
Olfactory neurons, unlike most neurons, have a finite life span of ∼90 d and are constantly being replaced by neurogenesis that lasts through adulthood. OSNs, however, do not all have the same life span; their longevity is determined by the type of OR they express and abundance of its cognate odorant in the environment (Santoro and Dulac 2012). A key regulator of this process is, surprisingly, a previously uncharacterized histone variant named H2be. This histone variant differs by only five amino acids from the common H2b isoform and is expressed only in mature olfactory and vomeronasal (pheromone-detecting chemoreceptors) neurons. Although the molecular mechanism by which H2be affects longevity is not yet understood, it is striking that this histone variant cannot be acetylated or methylated on lysine 5, two posttranslational modifications associated with transcriptional elongation. As a result, olfactory neurons with high levels of H2be, which are particularly enriched at euchromatic parts of the genome, have low levels of methylated and acetylated lysine 5 of H2b. This is indicative of some kind of replication-independent mode of H2b substitution with H2be.
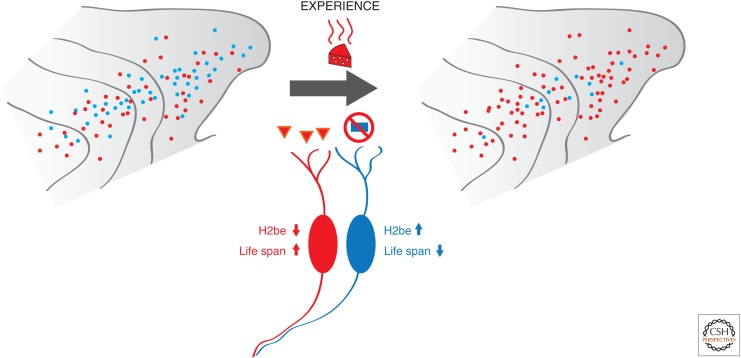
H2be has a sporadic expression pattern in the olfactory epithelium; some neurons have undetectable levels of this histone and others have moderate to high levels, based on immunofluorescence experiments. Strikingly, the identity of the receptor seems to determine the expression rates of H2be, given that the levels of H2be are the same among neurons that express the same OR gene. In fact, olfactory neurons with ORs that are frequently activated in a specific environment retain low levels of H2be in their nuclei (i.e., the red OSNs in Fig. 10). Loss- and gain-of-function experiments with knockout and knockin mice revealed that H2be promotes the apoptosis of understimulated olfactory neurons and reduces their life span. Thus, H2be acts as a sensor of neuronal stimulation and determines the longevity of the neuron; if a neuron is frequently activated, then H2be levels are low and the life span of that neuron is extended. If the neuron is dormant, then it will die faster. The benefit of this mechanism is obvious: Because ORs are stochastically chosen, many neurons might express receptors that are useless in a specific environment. If these neurons have a shorter life span, after several rounds of neurogenesis and OR choice, the olfactory epithelium will have a higher percentage of “useful” olfactory neurons and thus operate as a sensory organ that is better “tuned” to a specific odorant environment (Monahan and Lomvardas 2012; Santoro and Dulac 2012).
Figure 10.
The H2be histone variant in OSNs. The figure summarizes results of experimental manipulations showing that olfactory experience regulates the expression levels of H2be and dictates the longevity of olfactory neurons. Neurons frequently activated (in red) do not accumulate H2be in the chromatin of OR clusters and live longer, eventually dominating the MOE.
The unexpected discovery that a histone variant with extreme tissue specificity determines the life span of olfactory neurons in an activity-dependent manner provides an elegant showing of the diversity of epigenetic processes that contribute to the development and function of the nervous system. In addition to the regulation of H2be expression, neuronal activity may directly affect the stability of OR choice because Lsd1 down-regulation requires OR-induced expression of Adcy3 (Lyons et al. 2013), the major contributor of cAMP, in response to OR activation by odorants. Thus, the olfactory system, with its extreme regulatory requirements, provides an excellent model system for the study of the interplay between neuronal activity and epigenetic processes that control nuclear plasticity and regulate neuronal longevity.
5. SUMMARY AND PERSPECTIVE
In this article, we provided four specific examples of epigenetic processes involved in the development and wiring of the nervous system. There are, of course, other epigenetic processes involved in the nervous system in both vertebrates and invertebrates. For this, we point you to several excellent reviews and papers therein (Dulac 2010; Zocchi and Sassone-Corsi 2010; Qureshi and Mehler 2012; Russo and Nestler 2013). In three of the cases covered in this article, the cell under study has to “make” decisions that will determine its developmental fate and function: choice between neurogenesis or gliogenesis, choice between a combination of Pcdh isoforms, and choice of a single OR allele. In all three cases, a choice is determined by the balance between epigenetic silencing and activation, but the mechanistic details differ depending on specific regulatory needs. In the case of neural precursor cells, in which this choice is determined by extrinsic signals and thus is highly regulated during development, there is a balance between Polycomb and Trithorax complexes modulated by Dnmt3a activity. In the case of Pcdh and olfactory gene choice, however, which are seemingly stochastic and the goal is to achieve maximum diversity of expression programs rather than specific transcriptional outcomes, different, unknown, epigenetic regulators are involved, which assure that only one of multiple promoters will become accessible to the transcription machinery. On the other hand, in the case of the histone variant H2be, the role of this protein is not to determine a developmental decision or make a transcriptional choice, but to affect the longevity of the neuron through mechanisms that are not yet understood. Most certainly, novel epigenetic mechanisms will emerge from the study of other neuronal processes, such as learning and memory, neuronal plasticity, critical periods, neurodegeneration, and replacement of neurons during adult neurogenesis. In all of these cases, a common question that needs to be answered is how external input received by different neuronal populations translates into specific epigenetic changes resulting in transcriptional and physiological output.
Footnotes
Editors: C. David Allis, Marie-Laure Caparros, Thomas Jenuwein, Danny Reinberg, and Monika Lachner
Additional Perspectives on Epigenetics available at www.cshperspectives.org
REFERENCES
*Reference is also in this subject collection.
- Alvarez-Buylla A, Garcia-Verdugo JM. 2002. Neurogenesis in adult subventricular zone. J Neurosci 22: 629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Buylla A, Lim DA. 2004. For the long run: Maintaining germinal niches in the adult brain. Neuron 41: 683–686. [DOI] [PubMed] [Google Scholar]
- Armelin-Correa LM, Gutiyama LM, Brandt DYC, Malnic B. 2014. Nuclear compartmentalization of odorant receptor genes. Proc Natl Acad Sci 111: 2782–2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axel R. 1995. The molecular logic of smell. Sci Am 273: 154–159. [DOI] [PubMed] [Google Scholar]
- *.Barlow DP, Bartolomei MS. 2014. Genomic imprinting in mammals. Cold Spring Harb Perspect Biol doi 6: a018382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnea G, O’Donnell S, Mancia F, Sun X, Nemes A, Mendelsohn M, Axel R. 2004. Odorant receptors on axon termini in the brain. Science 304: 1468. [DOI] [PubMed] [Google Scholar]
- *.Becker PB, Workman JL. 2013. Nucleosome remodeling and epigenetics. Cold Spring Harb Perspect Biol 5: a017905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326. [DOI] [PubMed] [Google Scholar]
- Bonn S, Seeburg PH, Schwarz MK. 2007. Combinatorial expression of α- and γ-protocadherins alters their presenilin-dependent processing. Mol Cell Biol 27: 4121–4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Brockdorff N, Turner BM. 2014. Dosage compensation in mammals. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan SM, Schalm SS, Maniatis T. 2010. Proteolytic processing of protocadherin proteins requires endocytosis. Proc Natl Acad Sci 107: 17774–17779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck L, Axel R. 1991. A novel multigene family may encode odorant receptors: A molecular basis for odor recognition. Cell 65: 175–187. [DOI] [PubMed] [Google Scholar]
- Burgold T, Spreafico F, De Santa F, Totaro MG, Prosperini E, Natoli G, Testa G. 2008. The histone H3 lysine 27-specific demethylase Jmjd3 is required for neural commitment. PLoS ONE 3: e3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Busslinger M, Tarakhovsky A. 2014. Epigenetic control of immunity. Cold Spring Harb Perspect Biol 6: a019307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron S, Rao Y. 2010. Molecular mechanisms of tiling and self-avoidance in neural development. Mol Brain 3: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WV, Maniatis T. 2013. Clustered protocadherins. Development 140: 3297–3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chess A. 2013. Random and non-random monoallelic expression. Neuropsychopharmacology 38: 55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chess A, Simon I, Cedar H, Axel R. 1994. Allelic inactivation regulates olfactory receptor gene expression. Cell 78: 823–834. [DOI] [PubMed] [Google Scholar]
- Clowney EJ, Magklara A, Colquitt BM, Pathak N, Lane RP, Lomvardas S. 2011. High-throughput mapping of the promoters of the mouse olfactory receptor genes reveals a new type of mammalian promoter and provides insight into olfactory receptor gene regulation. Genome Res 21: 1249–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clowney EJ, LeGros MA, Mosley CP, Clowney FG, Markenskoff-Papadimitriou EC, Myllys M, Barnea G, Larabell CA, Lomvardas S. 2012. Nuclear aggregation of olfactory receptor genes governs their monogenic expression. Cell 151: 724–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton RP, Lyons DB, Lomvardas S. 2013. Co-opting the unfolded protein response to elicit olfactory receptor feedback. Cell 155: 321–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Dekker J, Misteli T. 2014. Long-range chromatin interactions. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doetsch F, Caille I, Lim DA, Garcia-Verdugo JM, Alvarez-Buylla A. 1999. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell 97: 703–716. [DOI] [PubMed] [Google Scholar]
- Dulac C. 2010. Brain function and chromatin plasticity. Nature 465: 728–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahimi FA, Edmondson J, Rothstein R, Chess A. 2000. YAC transgene-mediated olfactory receptor gene choice. Dev Dyn 217: 225–231. [DOI] [PubMed] [Google Scholar]
- *.Elgin SCR, Reuter G. 2013. Position-effect variegation, heterochromatin formation, and gene silencing in Drosophila. Cold Spring Harb Perspect Biol 5: a017780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esumi S, Kakazu N, Taguchi Y, Hirayama T, Sasaki A, Hirabayashi T, Koide T, Kitsukawa T, Hamada S, Yagi T. 2005. Monoallelic yet combinatorial expression of variable exons of the protocadherin-α gene cluster in single neurons. Nat Genet 37: 171–176. [DOI] [PubMed] [Google Scholar]
- Fass DM, Schroeder FA, Perlis RH, Haggarty SJ. 2014. Epigenetic mechanisms in mood disorders: Targeting neuroplasticity. Neuroscience 264C: 112–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinstein P, Mombaerts P. 2004. A contextual model for axonal sorting into glomeruli in the mouse olfactory system. Cell 117: 817–831. [DOI] [PubMed] [Google Scholar]
- Ferreira T, Wilson SR, Choi YG, Risso D, Dudoit S, Speed TP, Ngai J. 2014. Silencing of odorant receptor genes by G protein βγ signaling ensures the expression of one odorant receptor per olfactory sensory neuron. Neuron 81: 847–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuss SH, Omura M, Mombaerts P. 2007. Local and cis effects of the H element on expression of odorant receptor genes in mouse. Cell 130: 373–384. [DOI] [PubMed] [Google Scholar]
- Glusman G, Bahar A, Sharon D, Pilpel Y, White J, Lancet D. 2000. The olfactory receptor gene superfamily: Data mining, classification, and nomenclature. Mamm Genome 11: 1016–1023. [DOI] [PubMed] [Google Scholar]
- Golan-Mashiach M, Grunspan M, Emmanuel R, Gibbs-Bar L, Dikstein R, Shapiro E. 2012. Identification of CTCF as a master regulator of the clustered protocadherin genes. Nucleic Acids Res 40: 3378–3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Grossniklaus U, Paro R. 2014. Transcriptional silencing by Polycomb-group proteins. Cold Spring Harb Perspect Biol 6: a019331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grueber WB, Sagasti A. 2010. Self-avoidance and tiling: Mechanisms of dendrite and axon spacing. Cold Spring Harb Perspect Biol 2: a001750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Monahan K, Wu H, Gertz J, Varley KE, Li W, Myers RM, Maniatis T, Wu Q. 2012. CTCF/cohesin-mediated DNA looping is required for protocadherin α promoter choice. Proc Natl Acad Sci 109: 21081–21086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas IG, Frank M, Véron N, Kemler R. 2005. Presenilin-dependent processing and nuclear function of γ-protocadherins. J Biol Chem 280: 9313–9319. [DOI] [PubMed] [Google Scholar]
- Hambsch B, Grinevich V, Seeburg PH, Schwarz MK. 2005. γ-Protocadherins, presenilin-mediated release of C-terminal fragment promotes locus expression. J Biol Chem 280: 15888–15897. [DOI] [PubMed] [Google Scholar]
- Han MH, Lin C, Meng S, Wang X. 2010. Proteomics analysis reveals overlapping functions of clustered protocadherins. Mol Cell Proteomics 9: 71–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirabayashi Y, Gotoh Y. 2010. Epigenetic control of neural precursor cell fate during development. Nat Rev Neurosci 11: 377–388. [DOI] [PubMed] [Google Scholar]
- Hirabayashi Y, Suzki N, Tsuboi M, Endo TA, Toyoda T, Shinga J, Koseki H, Vidal M, Gotoh Y. 2009. Polycomb limits the neurogenic competence of neural precursor cells to promote astrogenic fate transition. Neuron 63: 600–613. [DOI] [PubMed] [Google Scholar]
- Hirayama T, Tarusawa E, Yoshimura Y, Galjart N, Yagi T. 2012. CTCF is required for neural development and stochastic expression of clustered Pcdh genes in neurons. Cell Rep 2: 345–357. [DOI] [PubMed] [Google Scholar]
- Hirota J, Mombaerts P. 2004. The LIM-homeodomain protein Lhx2 is required for complete development of mouse olfactory sensory neurons. Proc Natl Acad Sci 101: 8751–8755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann K, Dreger CK, Olins AL, Olins DE, Shultz LD, Lucke B, Karl H, Kaps R, Muller D, Vaya A, et al. 2002. Mutations in the gene encoding the lamin B receptor produce an altered nuclear morphology in granulocytes (Pelger-Huet anomaly). Nat Genet 31: 410–414. [DOI] [PubMed] [Google Scholar]
- Jobe EM, McQuate AL, Zhao X. 2012. Crosstalk among epigenetic pathways regulates neurogenesis. Front Neurosci 6: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. 2010. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467: 430–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko R, Kawaguchi M, Toyama T, Taguchi Y, Yagi T. 2009. Expression levels of Protocadherin-α transcripts are decreased by nonsense-mediated mRNA decay with frameshift mutations and by high DNA methylation in their promoter regions. Gene 430: 86–94. [DOI] [PubMed] [Google Scholar]
- Katori S, Hamada S, Noguchi Y, Fukuda E, Yamamoto T, Yamamoto H, Hasegawa S, Yagi T. 2009. Protocadherin-α family is required for serotonergic projections to appropriately innervate target brain areas. J Neurosci 29: 9137–9147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi M, Toyama T, Kaneko R, Hirayama T, Kawamura Y, Yagi T. 2008. Relationship between DNA methylation states and transcription of individual isoforms encoded by the protocadherin-α gene cluster. J Biol Chem 283: 12064–12075. [DOI] [PubMed] [Google Scholar]
- Kehayova P, Monahan K, Chen W, Maniatis T. 2011. Regulatory elements required for the activation and repression of the protocadherin-α gene cluster. Proc Natl Acad Sci 108: 17195–17200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M, Vaes E, Mombaerts P. 2011. Regulation of the probability of mouse odorant receptor gene choice. Cell 147: 907–921. [DOI] [PubMed] [Google Scholar]
- *.Kim T-K, Hemberg M, Gray JM. 2014. Enhancer RNAs: A class of long noncoding RNAs synthesized at enhancers. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Kingston RE, Tamkun JW. 2014. Transcriptional regulation by trithorax-group proteins. Cold Spring Harb Perspect Biol 6: a019349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. 2009. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324: 929–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson JL, Yuan GC. 2010. Epigenetic domains found in mouse embryonic stem cells via a hidden Markov model. BMC Bioinformatics 11: 557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre JL, Kostadinov D, Chen WV, Maniatis T, Sanes JR. 2012. Protocadherins mediate dendritic self-avoidance in the mammalian nervous system. Nature 488: 517–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Gros MA, McDermott G, Uchida M, Knoechel CG, Larabell CA. 2009. High-aperture cryogenic light microscopy. J Microsc 235: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewcock JW, Reed RR. 2004. A feedback mechanism regulates monoallelic odorant receptor expression. Proc Natl Acad Sci 101: 1069–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Li E, Zhang Y. 2014. DNA methylation in mammals. Cold Spring Harb Perspect Biol 6: a019133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim DA, Huang YC, Swigut T, Mirick AL, Garcia-Verdugo JM, Wysocka J, Ernst P, Alvarez-Buylla A. 2009. Chromatin remodelling factor Mll1 is essential for neurogenesis from postnatal neural stem cells. Nature 458: 529–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R. 2006. Interchromosomal interactions and olfactory receptor choice. Cell 126: 403–413. [DOI] [PubMed] [Google Scholar]
- Lunyak VV, Rosenfeld MG. 2005. No rest for REST: REST/NRSF regulation of neurogenesis. Cell 121: 499–501. [DOI] [PubMed] [Google Scholar]
- Lyons DB, Allen WE, Goh T, Tsai L, Barnea G, Lomvardas S. 2013. An epigenetic trap stabilizes singular olfactory receptor expression. Cell 154: 325–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DK, Marchetto MC, Guo JU, Ming GL, Gage FH, Song H. 2010. Epigenetic choreographers of neurogenesis in the adult mammalian brain. Nat Neurosci 13: 1338–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magklara A, Yen A, Colquitt BM, Clowney EJ, Allen W, Markenscoff-Papadimitriou E, Evans ZA, Kheradpour P, Mountoufaris G, Carey C, et al. 2011. An epigenetic signature for monoallelic olfactory receptor expression. Cell 145: 555–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre JC, Bose SC, Stromberg AJ, McClintock TS. 2008. Emx2 stimulates odorant receptor gene expression. Chem Senses 33: 825–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellen M, Ayata P, Dewell S, Kriaucionis S, Heintz N. 2012. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell 151: 1417–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mombaerts P, Wang F, Dulac C, Chao SK, Nemes A, Mendelsohn M, Edmondson J, Axel R. 1996. Visualizing an olfactory sensory map. Cell 87: 675–686. [DOI] [PubMed] [Google Scholar]
- Monahan K, Lomvardas S. 2012. How keeping active pays off in the olfactory system. eLife 1: e00326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monahan K, Rudnick ND, Kehayova PD, Pauli F, Newberry KM, Myers RM, Maniatis T. 2012. Role of CCCTC binding factor (CTCF) and cohesin in the generation of single-cell diversity of protocadherin-α gene expression. Proc Natl Acad Sci 109: 9125–9130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munzel M, Globisch D, Bruckl T, Wagner M, Welzmiller V, Michalakis S, Muller M, Biel M, Carell T. 2010. Quantification of the sixth DNA base hydroxymethylcytosine in the brain. Angew Chem Int Ed Engl 49: 5375–5377. [DOI] [PubMed] [Google Scholar]
- Nei M, Niimura Y, Nozawa M. 2008. The evolution of animal chemosensory receptor gene repertoires: Roles of chance and necessity. Nat Rev 9: 951–963. [DOI] [PubMed] [Google Scholar]
- Nestler EJ. 2014. Epigenetic mechanisms of drug addiction. Neuropharmacology 76 Pt B: 259–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen MQ, Zhou Z, Marks CA, Ryba NJ, Belluscio L. 2007. Prominent roles for odorant receptor coding sequences in allelic exclusion. Cell 131: 1009–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olynik BM, Rastegar M. 2012. The genetic and epigenetic journey of embryonic stem cells into mature neural cells. Front Genet 3: 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JE, Corces VG. 2009. CTCF: Master weaver of the genome. Cell 137: 1194–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro I, Margueron R, Shukeir N, Eisold M, Fritzsch C, Richter FM, Mittler G, Genoud C, Goyama S, Kurokawa M, et al. 2012. Prdm3 and Prdm16 are H3K9me1 methyltransferases required for mammalian heterochromatin integrity. Cell 150: 948–960. [DOI] [PubMed] [Google Scholar]
- Plessy C, Pascarella G, Bertin N, Akalin A, Carrieri C, Vassalli A, Lazarevic D, Severin J, Vlachouli C, Simone R, et al. 2011. Promoter architecture of mouse olfactory receptor genes. Genome Res 22: 486–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyrpasopoulou A, Meier J, Maison C, Simos G, Georgatos SD. 1996. The lamin B receptor (LBR) provides essential chromatin docking sites at the nuclear envelope. EMBO J 15: 7108–7119. [PMC free article] [PubMed] [Google Scholar]
- Pyrski M, Xu Z, Walters E, Gilbert DJ, Jenkins NA, Copeland NG, Margolis FL. 2001. The OMP-lacZ transgene mimics the unusual expression pattern of OR-Z6, a new odorant receptor gene on mouse chromosome 6: Implication for locus-dependent gene expression. J Neurosci 21: 4637–4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi IA, Mehler MF. 2012. Emerging roles of non-coding RNAs in brain evolution, development, plasticity and disease. Nat Rev Neurosci 13: 528–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss K, Maretzky T, Haas IG, Schulte M, Ludwig A, Frank M, Saftig P. 2006. Regulated ADAM10-dependent ectodomain shedding of γ-protocadherin C3 modulates cell–cell adhesion. J Biol Chem 281: 21735–21744. [DOI] [PubMed] [Google Scholar]
- Ressler KJ, Sullivan SL, Buck LB. 1993. A zonal organization of odorant receptor gene expression in the olfactory epithelium. Cell 73: 597–609. [DOI] [PubMed] [Google Scholar]
- Ribich S, Tasic B, Maniatis T. 2006. Identification of long-range regulatory elements in the protocadherin-α gene cluster. Proc Natl Acad Sci 103: 19719–19724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccio A. 2010. Dynamic epigenetic regulation in neurons: Enzymes, stimuli and signaling pathways. Nat Neurosci 13: 1330–1337. [DOI] [PubMed] [Google Scholar]
- Rodriguez I. 2013. Singular expression of olfactory receptor genes. Cell 155: 274–277. [DOI] [PubMed] [Google Scholar]
- Rothman A, Feinstein P, Hirota J, Mombaerts P. 2005. The promoter of the mouse odorant receptor gene M71. Mol Cell Neurosci 28: 535–546. [DOI] [PubMed] [Google Scholar]
- Rudenko A, Tsai LH. 2014. Epigenetic regulation in memory and cognitive disorders. Neuroscience 264C: 51–63. [DOI] [PubMed] [Google Scholar]
- Russo SJ, Nestler EJ. 2013. The brain reward circuitry in mood disorders. Nat Rev Neurosci 14: 609–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro SW, Dulac C. 2012. The activity-dependent histone variant H2BE modulates the life span of olfactory neurons. eLife 1: e00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalm SS, Ballif BA, Buchanan SM, Phillips GR, Maniatis T. 2010. Phosphorylation of protocadherin proteins by the receptor tyrosine kinase Ret. Proc Natl Acad Sci 107: 13894–13899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmucker D, Chen B. 2009. Dscam and DSCAM: Complex genes in simple animals, complex animals yet simple genes. Genes Dev 23: 147–156. [DOI] [PubMed] [Google Scholar]
- Schmucker D, Clemens JC, Shu H, Worby CA, Xiao J, Muda M, Dixon JE, Zipursky SL. 2000. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell 101: 671–684. [DOI] [PubMed] [Google Scholar]
- Schreiner D, Weiner JA. 2010. Combinatorial homophilic interaction between γ-protocadherin multimers greatly expands the molecular diversity of cell adhesion. Proc Natl Acad Sci 107: 14893–14898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serizawa S, Ishii T, Nakatani H, Tsuboi A, Nagawa F, Asano M, Sudo K, Sakagami J, Sakano H, Ijiri T, et al. 2000. Mutually exclusive expression of odorant receptor transgenes. Nat Neurosci 3: 687–693. [DOI] [PubMed] [Google Scholar]
- Serizawa S, Miyamichi K, Nakatani H, Suzuki M, Saito M, Yoshihara Y, Sakano H. 2003. Negative feedback regulation ensures the one receptor-one olfactory neuron rule in mouse. Science 302: 2088–2094. [DOI] [PubMed] [Google Scholar]
- Serizawa S, Miyamichi K, Takeuchi H, Yamagishi Y, Suzuki M, Sakano H. 2006. A neuronal identity code for the odorant receptor-specific and activity-dependent axon sorting. Cell 127: 1057–1069. [DOI] [PubMed] [Google Scholar]
- *.Seto E, Yoshida M. 2014. Erasers of histone acetylation: The histone deacetylase enzymes. Cold Spring Harb Perspect Biol 6: a018713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shultz LD, Lyons BL, Burzenski LM, Gott B, Samuels R, Schweitzer PA, Dreger C, Herrmann H, Kalscheuer V, Olins AL, et al. 2003. Mutations at the mouse ichthyosis locus are within the lamin B receptor gene: A single gene model for human Pelger-Huet anomaly. Hum Mol Genet 12: 61–69. [DOI] [PubMed] [Google Scholar]
- Shykind BM. 2005. Regulation of odorant receptors: One allele at a time. Hum Mol Genet 14: R33–R39. [DOI] [PubMed] [Google Scholar]
- Shykind BM, Rohani SC, O’Donnell S, Nemes A, Mendelsohn M, Sun Y, Axel R, Barnea G. 2004. Gene switching and the stability of odorant receptor gene choice. Cell 117: 801–815. [DOI] [PubMed] [Google Scholar]
- Solovei I, Kreysing M, Lanctot C, Kosem S, Peichl L, Cremer T, Guck J, Joffe B. 2009. Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell 137: 356–368. [DOI] [PubMed] [Google Scholar]
- Suo L, Lu H, Ying G, Capecchi MR, Wu Q. 2012. Protocadherin clusters and cell adhesion kinase regulate dendrite complexity through Rho GTPase. J Mol Cell Biol 4: 362–376. [DOI] [PubMed] [Google Scholar]
- Szulwach KE, Li X, Li Y, Song CX, Wu H, Dai Q, Irier H, Upadhyay AK, Gearing M, Levey AI, et al. 2011. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat Neurosci 14: 1607–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L, Zong C, Xie XS. 2013. Rare event of histone demethylation can initiate singular gene expression of olfactory receptors. Proc Natl Acad Sci 110: 21148–21152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasic B, Nabholz CE, Baldwin KK, Kim Y, Rueckert EH, Ribich SA, Cramer P, Wu Q, Axel R, Maniatis T. 2002. Promoter choice determines splice site selection in protocadherin α and γ pre-mRNA splicing. Mol Cell 10: 21–33. [DOI] [PubMed] [Google Scholar]
- Thu CA, Chen WV, Rubinstein R, Chevee M, Wolcott HN, Felsovalyl KO, Tapia JC, Shapiro L, Honig B, Maniatis T. 2014. Single-cell identity generated by combinatorial homophilic interactions between α, β, γ protocadherins. Cell 158: 1045–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyssowski K, Kishi Y, Gotoh Y. 2014. Chromatin regulators of neural development. Neuroscience 264C: 4–16. [DOI] [PubMed] [Google Scholar]
- Vassalli A, Rothman A, Feinstein P, Zapotocky M, Mombaerts P. 2002. Minigenes impart odorant receptor-specific axon guidance in the olfactory bulb. Neuron 35: 681–696. [DOI] [PubMed] [Google Scholar]
- Vassalli A, Feinstein P, Mombaerts P. 2011. Homeodomain binding motifs modulate the probability of odorant receptor gene choice in transgenic mice. Mol Cell Neurosci 46: 381–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassar R, Ngai J, Axel R. 1993. Spatial segregation of odorant receptor expression in the mammalian olfactory epithelium. Cell 74: 309–318. [DOI] [PubMed] [Google Scholar]
- Wang SS, Tsai RY, Reed RR. 1997. The characterization of the Olf-1/EBF-like HLH transcription factor family: Implications in olfactory gene regulation and neuronal development. J Neurosci 17: 4149–4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Nemes A, Mendelsohn M, Axel R. 1998. Odorant receptors govern the formation of a precise topographic map. Cell 93: 47–60. [DOI] [PubMed] [Google Scholar]
- Worman HJ, Yuan J, Blobel G, Georgatos SD. 1988. A lamin B receptor in the nuclear envelope. Proc Natl Acad Sci 85: 8531–8534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Maniatis T. 1999. A striking organization of a large family of human neural cadherin-like cell adhesion genes. Cell 97: 779–790. [DOI] [PubMed] [Google Scholar]
- Wu Q, Maniatis T. 2000. Large exons encoding multiple ectodomains are a characteristic feature of protocadherin genes. Proc Natl Acad Sci 97: 3124–3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Zhang T, Cheng JF, Kim Y, Grimwood J, Schmutz J, Dickson M, Noonan JP, Zhang MQ, Myers RM, et al. 2001. Comparative DNA sequence analysis of mouse and human protocadherin gene clusters. Genome Res 11: 389–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Coskun V, Tao J, Xie W, Ge W, Yoshikawa K, Li E, Zhang Y, Sun YE. 2010. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science 329: 444–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi T. 2012. Molecular codes for neuronal individuality and cell assembly in the brain. Front Mol Neurosci 5: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi T. 2013. Genetic basis of neuronal individuality in the mammalian brain. J Neurogenet 27: 97–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Firestein S. 2002. The olfactory receptor gene superfamily of the mouse. Nat Neurosci 5: 124–133. [DOI] [PubMed] [Google Scholar]
- Zipursky SL, Grueber WB. 2013. The molecular basis of self-avoidance. Annu Rev Neurosci 36: 547–568. [DOI] [PubMed] [Google Scholar]
- Zipursky SL, Sanes JR. 2010. Chemoaffinity revisited: Dscams, protocadherins, and neural circuit assembly. Cell 143: 343–353. [DOI] [PubMed] [Google Scholar]
- Zocchi L, Sassone-Corsi P. 2010. Joining the dots: From chromatin remodeling to neuronal plasticity. Curr Opin Neurobiol 20: 432–440. [DOI] [PMC free article] [PubMed] [Google Scholar]