Abstract
The pervasive nature of bacterial recombination has become clear. Despite this, the population genetics of bacteria persist in being viewed as simple. Here, I argue against that characterization. After summarizing the history of the topic, I survey the evidence for remarkable and unexplained variation in recombination rate among and within bacterial species. I finally argue that despite recent assertions that recombination means bacterial genes are “public goods,” in bacteria the level of selection is the gene, and genes can be understood to have niches with dimensions including the other contents of the genome in which they find themselves.
Bacteria show remarkable and unexplained variation in recombination rates within and among species, making one question why they were ever considered “simple.”
The most outstanding feature of life’s history is a constant domination by bacteria.
—Stephen Jay Gould
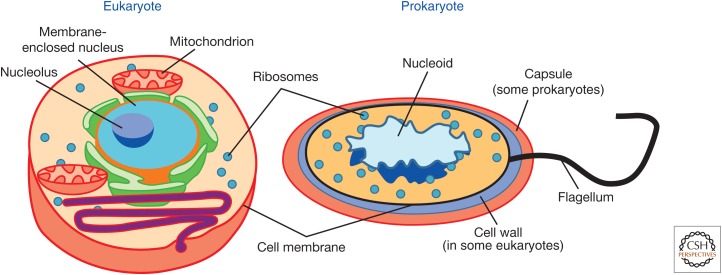
Bacteria are supposed to be simple. Toward the front of biology textbooks, we often find a diagram contrasting the eukaryotic and prokaryotic cell. Next to its larger relative, with its nucleus and organelles, the bacterial cell looks a little sad and featureless. A single, circular, haploid chromosome sits as a nucleoid, perhaps accompanied by a smaller plasmid (Fig. 1). The text makes it plain that, compared with eukaryotes, the genetics of bacteria are an easy affair. Genomes are smaller, with less selfish DNA, and evolution is clonal, or nearly so.
Figure 1.
A typical figure contrasting the complexity of the prokaryotic cell (left) with that of the eukaryote (right). This example is drawn from Wikimedia Commons, but similar images decorate many textbooks, with especially nice examples being found in Lodish (2003) among others. (From the Science Primer, a work of the National Center for Biotechnology Information, part of the National Institutes of Health. As a work of the U.S. Federal government, the image is in the public domain.)
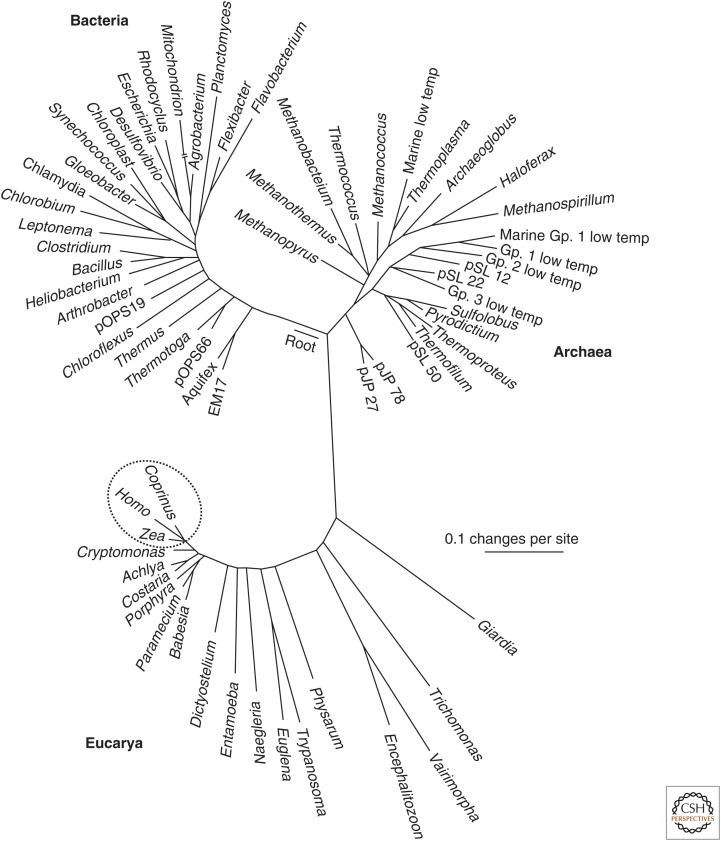
Except there is no such thing as a typical bacterium. We can get a hint of this from representations of the tree of life (Fig. 2). Let us set aside for the moment the controversies over that tree (Koonin and Wolf 2009; Ochman 2009), and note that almost all the diversity in this tree is microbial, with the things we can see with the naked eye being limited, in the example here, to just three taxa at one end: Homo, Zea mays, and Coprinus are separated by relatively short branches.
Figure 2.
The tree of life determined by analysis of 16S rRNA sequences. The three taxa circled are those representing plants, animals, and fungi. As can be seen, the great majority of diversity, at least as assessed by this metric, lies elsewhere. (From Pace 1997; modified, with permission, from The American Association for the Advancement of Science © 1997.)
Not only are the bacteria highly diverse when comparing species or genera, bacteria vary greatly in the genetic diversity maintained in populations. Some are monomorphic; Mycobacterium tuberculosis is the canonical example (Achtman 2008), whereas others such as Helicobacter pylori are extremely variable (Israel et al. 2001; Suerbaum and Achtman 2004). Contrary to the textbooks, bacteria can be diploid, albeit homozygous (Tobiason and Seifert 2010). In some cases, chromosomes and plasmids are linear (Hinnebusch and Tilly 1993) and among Spirochetes of the genus Borrelia, numerous. The genome can be made up of as many as 24 separate linear DNA molecules (Chaconas and Kobryn 2010). Plasmids need not be small—some are larger than the entire chromosome of many species (Rosenberg et al. 1981; Romanchuk et al. 2014). And, although bacteria do not engage in the reciprocal exchange of genetic material at meiosis, which is the case in sexually reproducing organisms, they have a multitude of mechanisms by which DNA from one lineage can find its way into another, in which it may have a major impact on fitness. As it has been memorably put “bacteria may not have sex often, but when they do, it can be really good …” (Johnsen et al. 2009). This bacterial “sex” or recombination is the subject of this essay.
As a result of its central role in the story of molecular biology, almost every microbiologist has probably at some point casually assumed that all bacteria are like Escherichia coli. It is sometimes a struggle to recall that it is only one species, and not necessarily typical. The pneumococcus (Streptococcus pneumoniae) is another important model organism that, unlike E. coli, is naturally competent, meaning it takes up DNA from its environment and is “transformed” by it (Griffith 1922; Claverys et al. 2009), and investigations into the mechanism by which it occurred led to the classic work showing that DNA is the genetic material (Avery et al. 1944). We can imagine a counterfactual history in which, instead of E. coli becoming the workhorse of molecular biology, the pneumococcus with its inbuilt ability to take up DNA had that honor. Microbiologists in this alternate universe might have considered transformation and competence to be the norm. Plasmids, which are scarce in the pneumococcus (Clewell 1981), might have been considered a peculiar aberration rather than a common feature of bacterial life. Eventually, as people examined more and more bacteria, we would have discovered the extraordinary diversity of the ways they generate the variation that is the raw material natural selection works on, and we would have wondered how anybody could consider them “simple.”
WHAT IS RECOMBINATION IN BACTERIA?
Despite the evident diversity of bacterial sexual processes, the ubiquity of bacteria in all ecological processes, their essential functions in our own microbiota and the fact they kill us in our millions, references (especially quantitative) to bacteria in the population genetics literature are remarkably scant. I have six excellent population genetics textbooks in easy reach of my desk (Crow and Kimura 1970; Gillespie 2004; Hartl and Clark 2007; Hamilton 2009; Charlesworth and Charlesworth 2010; Nielsen and Slatkin 2013), bacteria earn a specific reference in only three. Looking at the index of each of the three books that do mention bacteria or prokaryotes, we find a total of 12 pages. This is partially because the population genetics of bacteria, so far, as point mutations are concerned, are not fundamentally different from that of other organisms. They are just (mostly) haploid. The problem arises when we start considering multiple genes or genomes, at which point recombination becomes important.
In the discussion that follows, I will use recombination as a catchall term for mechanisms that take genetic material from one genetic background and insert it into another. I broadly consider “genetic material” as anything contributing to a character state, including deletions as well as gene conversion and insertions. However, for context, we will briefly consider the classic division of recombination events into different types: homologous versus nonhomologous or illegitimate recombination, the latter often being termed horizontal gene transfer (HGT). It is helpful to distinguish between the processes by which DNA enters the cell, and what it does once inside. When thinking about DNA getting into a recipient cell we usually think of the three mechanisms of transformation, conjugation, and transduction. Each of these is then associated with the gain of particular classes of loci: the uptake of DNA from the environment by transformation is associated with homologous recombination and as a result tends to transfer DNA among close relatives, while unlike homologous recombination, the phage that mediate transduction can insert into the genome without regard for things like sequence identity. Conjugation is associated in the minds of many with plasmid transfer. These tidy divisions do not survive close inspection for very long.
WHEN IS HOMOLOGOUS RECOMBINATION NOT HOMOLOGOUS?
recA-mediated recombination is often described as “homologous.” The process involves the insertion of single-stranded DNA (ssDNA) into the genome through the formation of a heteroduplex, in which a high degree of sequence identity is of crucial importance. Although larger fragments are less efficiently transferred, roughly linearly declining with tract length, the efficiency of recombination drops much more rapidly with declining sequence identity; for taxa as varied as Streptococci, Bacillus, and E. coli, the efficiency of recombination consistently declines in log-linear fashion (Majewski and Cohan 1998, 1999). However, the importance of sequence identity is not constant across the inserted DNA; it is only within minimum efficiently processed segments (MEPS) at flanking regions of the inserted DNA. MEPS are tens of base pairs in length (specifically 26/27 for the recBC-dependent pathway in E. coli [Shen and Huang 1986]) with differences at the 5′ end being apparently less important than the 3′ (Sagi et al. 2006).
Much of the literature refers to recA as seeking “homology,” but this is obviously not necessarily the case. It seeks sequence identity, and homology is one way that this can be achieved. The confusion is understandable, but misleading. Given limits to DNA uptake (some of which, such as uptake sequences and pherotype, are discussed below), that mean the majority of DNA taken up is conspecific, the great majority of transfers are going to be of regions that are genuinely homologous. But the regions between the MEPS certainly need not be. For an example of this, consider the region in the pneumococcal genome that encodes the genes that make a complex polysaccharide capsule. The capsule is the major surface antigen of that pathogen, and the target of existing vaccines. These capsule loci are extremely variable, with >90 known that are distinguishable by serology. They lie in a specific region of the genome, flanked by the dexB and aliA loci (Bentley et al. 2006). These two genes, as well as the first few regulatory genes of the capsule locus proper, are highly conserved. Hence, they are a good source of MEPS for incoming DNA, and indeed recombination at this locus leads to changes in capsule and the resulting serotype (Coffey et al. 1998), with consequences including vaccine escape (Brueggemann et al. 2007; Croucher et al. 2011, 2013). Although the sequence identity that produces this fertile ground for recombination is undoubtedly the result of homology, this is not the case for all of the transferred loci that make up the capsule. The capsule genes contain multiple divergent and nonhomologous groups, including many glycosyltransferases, polysaccharide polymerases, and initial transferases, as well as the flippase that “flips” the complex final structure outside the cell once it has been built in the cytoplasm. The entire capsule locus varies in size from just >10 kb (serotype 3) to >30 kb (serotype 38) (Fig. 3). In the many cases known in which a pneumococcal lineage has changed its serotype through recombination, it makes little sense to think of the transferred segment as “homologous,” even if the function of the resulting structure is similar. Indeed, two serotypes make use of a different pathway entirely in capsule production and one of them (serotype 37) requires loci elsewhere in the chromosome (Dillard et al. 1995; Llull et al. 2001).
Figure 3.
Three of the >90 loci that encode serologically distinct capsules in the pneumococcus. The three capsules shown range from 13,844 to 30,298 bp in size. The three examples shown include a fraction of the genes contributing to pneumococcal serotype diversity. Loci are colored according to function and named according to homology groups, for details see the original publication (Bentley et al. 2006).
Sequence identity is required for recA-mediated recombination, and in the great majority of cases such identity will be the result of homology. But, although the majority of recombination events mediated by this mechanism will almost certainly transfer homologous loci between strains of the same species, this need not always be the case. We will return later to ask how much recombination might happen within clones, between isolates that are genetically very similar or identical, and that as a result is difficult or impossible to detect.
The ability of transducing phage to transfer genetic material among strains was recognized early on (Zinder and Lederberg 1952), and we now know of a bewildering array of mobile elements in many different classes, capable of mobilizing themselves and other genes (Darmon and Leach 2014). The various ways these elements insert into their recipient sequence means they are not limited by the strictures of sequence identity. But, provided the regions that flank the insertion site are sufficiently similar, there is no reason following insertion they may not then also be transferred by “homologous” recombination into lineages that lack them. The extent to which this happens in nature is not clear. One way to ask the question is to investigate whether there is evidence of phylogenetic incongruence among genes flanking the transferred element, such as has been reported in E. coli (Touchon et al. 2009). The amount that this has contributed to the observed distribution of elements in nature requires further and more systematic investigation.
In both transformation and transduction, there are multiple ways the donor DNA can enter the cell. Plasmids and other elements transferred by conjugation can encode the machinery for their own mobility. It is worth noting that plasmids are not the only things that can be transferred by this route. The large and growing numbers of integrative and conjugative elements (ICEs) also transfer by conjugation (Guglielmini et al. 2011). Further confusion in distinguishing homologous and nonhomologous recombination events arises from the observation of homology-facilitated illegitimate recombination (HFIR), in which homologous recombination occurs at one end of the inserted DNA, but not the other (Prudhomme et al. 2002; Harms et al. 2007).
Although the above sketches the complexity and some of the overlap between the canonical three modes of transfer, there is evidence for others that at present we can only guess at. A classic case is an Staphylococcus aureus clone generated by a recombination event 500 Mb in size, or nearly a quarter of the chromosome (Robinson and Enright 2004). Further study of population genomic data sets in the pneumococcus suggests that there is more than one mechanism contributing to the ongoing diversification, with short frequent inserts being distinct from large and infrequent ones (Mostowy et al. 2014). Finally, we have much to learn about the possible frequency and importance of small illegitimate recombination events (Overballe-Petersen et al. 2013).
HOW MUCH RECOMBINATION? VARIATION AMONG SPECIES
Although it is obvious that recombination can contribute to observed diversity, and the examples of horizontal transfer of resistance loci and other highly selected features makes it evident that it has actually performed so, it is a harder question to quantify how much of the variation we observe in nature has been produced by recombination. In particular, the extent of recombination at housekeeping loci was unknown until suitable data became available on allelic variation in natural populations.
A simple way to assess the contributions of recombination to a data set is to ask whether the character state at one locus is correlated with that at another: Are they in linkage disequilibrium (LD)? This can be quantified using the index of association (IA) (Brown et al. 1980). LD is important in eukaryotic genetics because it delivers information about the linkage between traits important in mapping the locations of different genes on the chromosome—loci further apart are more likely to lose any association in their character states because of an intervening crossing-over event. In eukaryotes, such events are coupled to reproduction and so happen at each generation. In contrast, bacteria undergo no such progress and for the most part recombination in which it does occur is more like gene conversion than meiosis. Hence, it was unsurprising that early studies in E. coli suggested that LD was extremely significant and consistent with “clonality” (Selander and Levin 1980).
This conclusion, however, misses the important fact that recombination in bacteria is fundamentally different to that found in organisms that undergo meiosis, with homologous recombination being equivalent to gene conversion rather than crossing over. Hence, we should not think of organisms as being divided into clonal and nonclonal, but instead recognize clonality as a matter of degree, based on the frequency with which recombination has contributed to the history of a population. This approach, taken in an extremely influential publication (Smith et al. 1993), was to ask how much recombination might be consistent with the superficial appearance of clonality. The answer turned out to be surprisingly high. Recombination could alter loci 10 to 20 times more often than mutation without removing significant LD. So, can we estimate how often a gene (or site) changes by recombination, relative to mutation? This is commonly written as r/m or the relative recombination rate.
Estimates of the r/m reveal a spectrum of clonality. At one end we have M. tuberculosis, which is considered to undergo recombination vanishingly infrequently, if at all, while at the other we have H. pylori, which has cleared the hurdle set by the IA and is panmictic (Smith et al. 2000). The study of recombination at the start of this century was greatly aided by the existence of large data sets of allelic variation, collected for the purposes of molecular epidemiology and focusing on housekeeping genes. This is important because, to assess baseline rates of recombination, we need to separate it from selection. There is a great deal of diversity and, not surprisingly, inferred recombination at or around many antigen genes, but this is skewed by the process of diversifying selection.
It is simple to estimate relative recombination rates from the allelic variation at multiple housekeeping loci. Briefly, if two strains are identical at all loci except one, but that one locus differs at multiple sites, then we can say with confidence that it is vanishingly unlikely that the slow random process of mutation would have generated this by chance, but left the other loci unscathed (note that if the locus in question was under strong diversifying selection, this would not be the case). Instead, it is more likely that such multiple changes were introduced by recombination. Cases in which the alleles differ at a single nucleotide may be caused by mutation, or possibly recombination with a very similar locus. The contribution of the latter can be estimated by examining whether the allele in question is found in other distantly related lineages (homoplasy). These approaches were used to estimate the rate of recombination relative to mutation (the r/m) for many species.
The results show the spectrum of clonality in more detail. S. aureus is revealed to be almost, but not quite, completely clonal. With an r/m of 0.7, it experiences recombination much less frequently that the important pathogens and components of the nasopharyngeal flora S. pneumoniae and Neisseria meningitidis, which were found to have r/m’s closer to 10 (the exact figures can differ slightly depending on the sample) (Feil et al. 1999, 2000; Feil and Spratt 2001; Feil and Enright 2004).
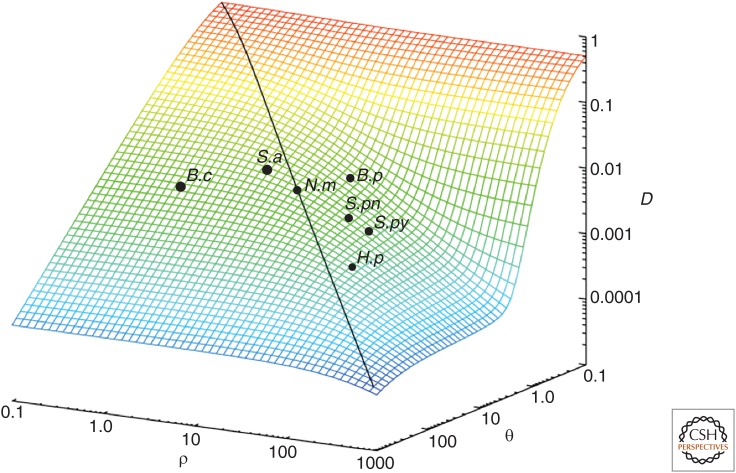
An alternative way of estimating recombination from a sample of allelic variation at multiple loci is to examine its impact on the distribution of allelic mismatches. In a sample from k loci, this is the proportion F of the sample that differs at i of k loci. It is intuitively easy to understand that, as recombination increases, the impact on this distribution will be to make it unlikely that two randomly picked isolates have no alleles in common at all; recombination should mean they share at least some variation with other members of the population, even if they are not closely related at other loci. This produces the following formula for the resulting equilibrium mismatch distribution of a sample (derived in Fraser et al. 2005):
where θ and ρ are, respectively, the population mutation and recombination rates. Fitting the observed distribution to the predictions of this formula produces estimates of the relative recombination rate that are independent of the empirical method described above and, comfortingly, the estimates from the two approaches are similar. Figure 4 is drawn from a publication that applies the method to a wider panel of species (Hanage et al. 2006), illustrating how much recombination rates can vary among species, and also the impact on the overall diversity observed in natural populations.
Figure 4.
The impact of homologous recombination on genotypic diversity. Density is measured here as D, the probability of randomly picking the same genotype twice. θ and ρ are, respectively, the population mutation and recombination rates. The positions of Streptococcus pneumoniae (S.pn), Neisseria meningitidis (N.m), Staphylococcus aureus (S.a), Helicobacter pylori (H.p), Streptococcus pyogenes (S.py), Burkholderia pseudomallei (B.p), and Bacillus cereus (B.c), based on parameters estimated from all data for those species in the corresponding multilocus sequence typing (MLST) databases, are shown. (From Hanage et al. 2006; modified, with permission, from the author.)
Here, diversity means the probability of two randomly sampled isolates being identical at all loci (Simpson’s D). This is important because people commonly assert that a lack of variation is a mark of clonality, but this is not the case. The confusion is linked to the fact that recombination is easier to observe in populations in which there is a large amount of variation to shuffle into different genomic backgrounds. Consider the example of Burkholderia pseudomallei, a soil saprophyte that shows little nucleotide diversity, but was nevertheless found to have the highest relative rate of recombination of the species considered in this study, an observation later confirmed and extended with genomic analyses (Pearson et al. 2009). This points to a recurrent issue with the study of (homologous) recombination. How much of it is undetectable? We can only, by definition, identify recombination where it has introduced variation (and sometimes not even then). A large amount of recombination may be within lineages, replacing like with like, and therefore undetectable.
Data sets comprising hundreds or thousands of bacterial genomes are now becoming common. Such population genomic samples have been especially valuable for studying recombination over the short term, within individual lineages. This is because in such samples it is relatively easy to detect recombination as tracts of anomalous sequence. In spirit, this approach has much in common with that of Feil et al. (1999, 2000), but including the entire genome. Once anomalous sequence has been removed, the remaining parts of the genome will be the clonal frame, in which variation has accumulated only by mutation, and this can be used to make a phylogeny. The programs CLONALFRAME (Didelot and Falush 2007; Marttinen et al. 2012), BRATNextGen (Marttinen et al. 2012), and GUBBINS (Croucher et al. 2015b) all follow this general approach. Although CLONALFRAME was the first to be developed, it was not designed to be used with whole genomes and, hence, can struggle with larger data sets. However, the other two follow the idea it pioneered, of identifying variation that appears to have originated outside a lineage and removing it. GUBBINS does this simply by identifying regions in an alignment, where there are more single nucleotide polymorphisms (SNPs) than would be expected given the assumption of an equal rate of mutation across the genome. The putative recombinant regions are removed, while the remainder are used to build a phylogeny, onto which recombination events can then be mapped. GUBBINS and CLONALFRAME make no effort to identify the origins of the recombined sequence. In contrast, BRATNextGen characterizes the variation across the sampled genomes in terms of the frequencies of different polymorphisms. This means that recombined regions can then be said to contain polymorphisms characteristic of another population in the sample (or an unsampled population). The tree constructed from the remaining clonal frame is similar to that obtained with the other two methods.
The results of such methods are subject to important caveats. All rely on identifying “anomalous” variation, although they differ in how they do so. We can easily see that they will struggle in species (or regions of the genome) that show little variation. We should recall that recombination is not the only means by which excess variation is generated. Antigen genes are typically subject to diversifying selection and hence contain an excess of polymorphisms that may or may not have been introduced by recombination. Although these methods can be used to generate an alignment “cleaned” of SNPs that may distort the true phylogeny, we should be cautious in assuming all such variation is the result of recombination. Methods that claim to identify the origins of recombined sequence will only do so if the origin is in the data set under analysis. And if there is more than one lineage that is a possible origin, there is no obvious way to distinguish among them.
VARIATION IN RECOMBINATION RATE
Using the same methods to examine other genomic data sets reveals a striking variation in the degree to which recombination has contributed to the history of a pneumococcal clone. A study of more than 600 genomes from pneumococci sampled from Massachusetts children found that the r/m, estimated as above, ranged from 34.06 to 0.06, with the majority being around 10. The highest was found in a lineage that had since vaccination become the most prevalent in carriage, and an important cause of invasive disease (Hanage et al. 2011). There was no obvious reason for the differences, although it is interesting to note that the lineage with the highest relative recombination rate was also resistant to multiple classes of antibiotics, an important cause of invasive disease, and had changed its major surface antigen by homologous recombination (allowing it to thrive in the presence of a vaccine that targets the antigen found among its ancestors). Such variation in recombination rate is not limited to pneumococcus and can happen even within a single closely related lineage. As noted previously, S. aureus is usually considered to recombine infrequently and be predominantly clonal, yet a study of genomes of the major MRSA lineage ST239 found great variation in the recombination observed in different sublineages associated with different geographic regions, and that this variation involved both core loci and accessory genes associated with mobile elements (Castillo-Ramirez et al. 2012).
To an extent, this should not be surprising. Transformation rates as measured in the laboratory are known to vary among strains. However, differences in the rates with which recombination impacts the genome are lineage specific and maintained such that more closely related strains are more likely to have a similar recombination rate. In most cases, the mechanisms behind the variation are not clear. There are multiple ways in which recombination rates might be influenced, and we can divide them by whether or not they arise from factors intrinsic to the organism, such as the process of DNA uptake and its insertion into the recipient’s genetic material, or factors related to ecology. The latter arise from the fact that in most cases the donor and recipient cells need to be in close proximity to one another, so organisms that inhabit different niches are unlikely to engage in the transfer of genetic material. In practice, this is hard to define with precision, because vectors such as transducing phage can in theory transfer material between organisms that inhabit different niches. An example of an apparent ecological barrier to recombination is that which exists between two distinct lineages of Campylobacter jejuni: ST45 and ST21. Although some C. jejuni show a clear association with specific host species, strains in these lineages are retrieved from chickens, cattle, and the environment, suggesting they are generalists. Despite this apparent niche overlap, there is scant evidence of any recombination between the two, although there is ample evidence for both of recombination with the other, host-restricted, lineages. ST21 and ST45 strains can be shown to recombine under laboratory conditions, and so the absence of recombination in nature has been taken to be evidence of an ecological barrier (Sheppard et al. 2014); the two lineages must not encounter each other often enough for recombination to occur, presumably because of some niche variation in addition to host tropism. The two lineages show marked differences in genes encoding vitamin B5 biosynthesis, and it is possible that these produce the effective ecological barrier between them (Sheppard et al. 2013). Inferring niche structure, through apparent barriers to recombination that cannot be explained by intrinsic factors, will be an interesting and unanticipated use for population genomic data.
Examples of “intrinsic” barriers include restriction modification systems (Oliveira et al. 2014), or responses to peptide hormones associated with quorum-sensing systems that govern the initiation of competence (Havarstein et al. 1997). However, it is easier to assert that these factors must limit the horizontal transfer of genes than to actually show that it has occurred. A recent exhaustive study of restriction modification systems in the pneumococcus found considerable variation among lineages, but no consistent association with recombination rates (Croucher et al. 2014). We also know that competence in pneumococcus is activated by binding of a peptide hormone to a two-component regulatory system encoded by the comCDE genes, and that these exist in multiple combinations of cognate hormones and receptors leading to different pherotypes. But whether these influence transfer in nature is controversial (Carrolo et al. 2009; Cornejo et al. 2010). Certainly, the relative rate of recombination does not substantially vary between the major pherotypes (Croucher et al. 2014).
Another intrinsic barrier, found in the Neisseria and Pasteurellaceae, is a bias of DNA uptake machinery to recognize and bind specific short motifs termed “uptake” sequences (Smith et al. 1999). Recombination occurs more efficiently with DNA containing these sequences, and they comprise ∼1% of the genome in those species that make use of them. As a result, these organisms preferentially take up conspecific DNA. Notably, this barrier is leaky, such that DNA from other species does occasionally make its way into the cell and becomes incorporated into the genome. Perhaps the best-known example of an intrinsic barrier, even if it is not often considered as such, is the steep decline in the efficiency of homologous recombination with sequence divergence of the flanking regions. This, however, will be no defense against transfer mediated by mobile elements.
The above has discussed ways in which rates of recombination between a specific donor and recipient may be impeded. But, it is possible for a formerly recombining taxon to lose this trait through lesions in genes governing the process. Two examples are noted in the PMEN-1 data set (Croucher et al. 2011). The frequency of such events is not known with any accuracy. Nor do we have a good understanding of the long-term future of a formerly recombining clone that loses this trait.
THE REASONS FOR VARIATION IN RECOMBINATION RATE
Mutation rates in bacteria seem to be relatively constant across taxa, with exceptions being organisms such as H. pylori that seem to lack typical machinery to repair errors (Drake 1999) (lesions in which also result in transient hypermutator lineages in many species [Sundin and Weigand 2007]). In contrast, the observed rates of intrinsic recombination vary greatly even within named species, for reasons that are not known. Given that the intrinsic rate of recombination is under the control of the cell, this should reflect natural selection. Mutation rates are controlled by selection, what about recombination?
The evolution of sexual reproduction has long troubled evolutionary biologists, who are confused as to why a process that lowers the probability of a successful gene getting into the next generation should be so pervasive. The processes of recombination that we are discussing here are often described as “bacterial sex” or “parasexual,” and although recombination is also an important part of sexual reproduction it is a mistake to draw the parallels too closely. The processes of horizontal transfer we have described here are not reciprocal but directed, and can both alter existing genes in a lineage and introduce wholly new ones to it (Narra and Ochman 2006). As such, they are fundamentally different from the replacement of homologous genes that is the outcome of meiosis. Homologous recombination in bacteria is more akin to gene conversion than the large crossover events we observe in sexually reproducing eukaryotes. And as noted above, homologous recombination is also capable of adding or removing genes. Although the outcome of recombination in bacteria and sexually reproducing eukaryotes is different, the homology of many of the proteins involved (such as recA) indicates some deep similarities.
There are multiple explanations for the prevalence of sex (an excellent summary is to be found in this introduction to a special symposium issue of American Naturalist on the topic [Otto 2009]), but two are of particular interest. The first is that recombination removes deleterious mutations and the second is the red queen hypothesis, in which recombination is a source of variation that accelerates adaptation, enabling survival in a rapidly changing environment. The contention that homologous recombination in bacteria is necessarily a source of variation is faulty. As shown in Figure 4, the contribution of recombination to diversity depends on how much diversity has arisen through mutation. Further, studies of how species clusters form in the presence of recombination show how it can prevent divergence (Fraser et al. 2007, 2009), because a divergent locus affected by recombination will most likely be replaced with variation that is typical of the species as a whole, a sort of “regression to the genome mean.” These simulations, however, consider the neutral situation. There is good reason to believe that in the case of adaptation, or recombination that might pick up loci under selection, the situation would be different (Levin and Cornejo 2009). Some empirical evidence for this is that, in pneumococcus, strains that harbor atypical variation at housekeeping loci that is consistent with a higher recombination rate across the genome are significantly more likely to be resistant to multiple classes of antibiotics (Hanage et al. 2009). Notably, the resistance mechanisms include those encoded by core loci such as penicillin binding proteins, but also those that are part of the accessory genome and carried on mobile elements (erythromycin and tetracycline resistance), suggesting a link between the uptake of mobile elements and homologous material elsewhere in the genome. Direct laboratory experiments have shown that recombination aids adaptation in H. pylori (Baltrus et al. 2008). This is consistent with theory; when homologous recombination, at rates typical of those observed in organisms like E. coli, H. influenzae, B. subtilis, or pneumococcus is introduced to models incorporating selection, it can accelerate adaptation (even if it incurs a fitness cost) (Levin and Cornejo 2009).
Hypotheses for the origins and maintenance of homologous recombination need not be mutually exclusive, and the two we have considered here are especially well matched. To recap, homologous recombination requires flanking regions of near complete sequence identity, but is unperturbed by the sequence in between. Hence, although it normally transfers homologous sequence, it can lead to acquisition of accessory genes, perhaps explaining the observations of resistance encoded by mobile elements, but associated with homologous recombination described above (Hanage et al. 2009). Although homologous recombination efficiently generates novel genetic combinations, it still requires the raw material of variation produced by other means. Finally, if mixing is random, recombination is most likely to replace a locus with one that is common and, because of this, it can limit diversification. Indeed, we do not know how much homologous recombination is invisible because the donor and recipients are identical.
Taken together, these features suggest a combination of a red queen hypothesis with the purging of deleterious variation, considered at the level of individual genes. Variable gene content is a feature of bacteria, and the genes in question may sometimes be deleterious and sometimes not. Recombination may be a response to this, such as in a case in which the transferred material is beneficial (such as macrolide resistance loci in the presence of a macrolide) and leads to relative success, it is rapidly spread to neighbors lacking it. In the situation in which a gene is acquired, which is deleterious, we might imagine a cost to resistance in the absence of drug then the gene could be deleted by homologous recombination with clone mates lacking it. Crucially, these neighbors will in most cases be identical at the loci encoding the recombination machinery. If homologous recombination is sufficiently frequent, it might act to both clear deleterious genetic material, and spread that which is beneficial, among strains that share appropriate competence genes, uptake sequences, and the like. The idea that homologous recombination might be an advantage in a situation in which the same gene can vary in selective value has conceptual similarities with phase variation or contingency loci (Moxon et al. 2006). Both are means of generating variation in the population, and both can generate a subpopulation that is maladapted. But for them to spread, the advantages must outweigh the costs (Wolf et al. 2005; Carja et al. 2014).
Although this and other hypotheses produce persuasive explanations for why homologous recombination is present, there is no satisfactory explanation for why it varies so much between species. If it aids adaptation in H. pylori, why would it not be similarly beneficial for other organisms to have the same very high rate of recombination? What is it about the biology of H. pylori that selected for this trait? The question is even more acute when concerning variation within species (or even individual lineages). What selective force leads isolates from different lineages of the same organism that inhabit (to our clumsy human eyes at least) the same niche and face the same challenges to consistently differ in their relative recombination rates by orders of magnitude?
RECOMBINATION: THE IMPLICATIONS
The presence of recombination is often taken to mean that representing the phylogenetic history of a lineage as a bifurcating tree is fundamentally wrong, with the alternative being a network representing the ancestral recombination graph. This objection can be overstated. If the contributions of recombination can be removed, the remaining clonal frame can be entirely legitimately used to infer a tree. In some cases, however, the clonal frame may be very small or nonexistent. The study of the pneumococcal PMEN-1 resistant clone, using population genomics to identify potentially recombining regions, found that >70% of the genome was inferred to have been replaced by recombination in at least one isolate (Croucher et al. 2011). This was estimated to have happened in ∼40 years. In a larger sample, or one with a common ancestor more distant in time, it is easy to imagine the clonal frame could shrink to nothing. This represents a horizon beyond which we are unable to make meaningful phylogenetic inferences about the strains within a species, and, in the cases of recombining organisms such as many of those discussed here, it is likely recent enough to pose a problem. For organisms like these, the precise relationships between deep branching major clades should be viewed with caution. This does not, however, necessarily impact the study of the relationships between species, as the great majority of recombination events are conspecific.
Having said that, given the great antiquity of life on earth, can we be certain that interspecific transfer has not happened at least once in the history of any locus in any bacteria? What does this mean for the tree of life, and to what extent has our view of biology been distorted by the acceptance of that tree? An important, even essential, tool to evolutionary biologists is homology, which is famously asserted to be indivisible. Two genes are either homologous or not. But this is nonsense as soon as we accept that sequence, rather than genes, is homologous. In fusion proteins, different parts of the genes have different ancestors and, hence, different homologs. Such events will be systematically undetected by methods that assess homology over the entirety of a locus, even though there is good evidence that once this requirement is relaxed previously undetected networks of gene remodeling are revealed (Haggerty et al. 2014).
The prevalence and importance of recombination at all time scales but the most recent has led some to decry “tree-thinking” and advocate for a view in which genes are “public goods” shared among organisms (McInerney et al. 2011). The grounds for this are that the universal nature of the genetic code means that any sequence is, in principle, interpretable and could be used by any organism—it is “nonexcludable.” Moreover, the presence of sequence in one organism does not deny it to others—a property described as “nonrival” but which has potentially confusing implications about competitive interactions. The terminology is borrowed from the theory of public goods in economics. This has some appeal, as it definitively uncouples the gene from its immediate genomic environment and encourages us to think of its potential value elsewhere. The proponents of the public goods hypothesis argue that many other theories, including the tree of life, selfish operons, and mobile elements, are regionalized instances embedded within the overarching framework they propose. However, it is not clear that satisfying the conditions of neither excluding nor rivaling in the terms above is sufficient to classify a gene or other sequence of DNA as a public good in anything other than a metaphorical sense. The framework also makes no mention of emergent properties that could violate the two conditions; a gene for a toxin, for instance, is excluded unless the strain harboring it also has the antidote. And, although the presence of a gene in one strain may not formally deny it to others, difference in overall fitness of those strains whether directly following from the gene or not would have the effect of doing so because one or the other would be outcompeted. Although public goods sound appealing, it is not easy to define a priori the public that benefits. While we can certainly point to widely distributed genes and infer that they have benefited the “public” that consists of the diverse genetic backgrounds in which they find themselves, this is a circular argument. Objections such as these can be argued as special, local cases of the overall theory, but a theory that can be made so flexible it explains everything has limitations as a guide for future research.
NICHES FOR GENES
The public goods hypothesis is a response to the evidence of widespread horizontal gene transfer, and when it comes to explaining this observation it does a very good job. A similar argument can be made, however, from an almost completely opposite perspective.
The public goods hypothesis is, at root, group selection. The idea that genes are good for some “public” implies that it is the public that benefits and, where evolution is concerned, that means selective benefit. The debates about the levels of selection have only brushed against microbiology, partially because the assumption of high levels of linkage makes it hard to consider a gene independently of its neighbors. In other fields of biology, selection at the level of the gene is widely accepted as a useful, perhaps the most useful, way to consider these questions (Okasha 2008). How might they be applied to bacteria?
A major difference between bacteria and eukaryotes is their variation in gene content. Any theory should be able to deal with this and it certainly works well with the public goods framework. However, it can also be captured by a variant of gene level selection. In ecology, Hutchinson’s niche refers to the idea that for any organism there is a particular notional hypervolume in “resource space,” meaning the circumstances in which an organism finds itself, including things like temperature or salinity as well as more obvious resources like food but excluding competing species in which it can survive and reproduce (Hutchinson 1959). It is easy to see how this could be extended to apply to a gene. Consider the example of macrolide resistance. This phenotype is often encoded by efflux pumps. The genes encoding these pumps have a higher fitness in the presence of antibiotics, and this might be considered the gene’s niche. We can also consider the genome as part of the gene’s environment—because it requires other genes to exist just as other genes will need it, in the presence of macrolides at least. Previous work has suggested that a genome could be a “home” for new genes (Daubin and Ochman 2004), and this extends that concept to suggest that all genes can be viewed in this way, but some genes are more established than others. The genomic niches of such genes could be described as “broad,” meaning that they can be expressed in many different genomes with minimal impact on fitness (core loci conserved in multiple species may be an example) or “narrow” meaning that there are only a few environments in which they will persist (such as resistance genes that can exact a cost in the absence of drug). Niche breadth will also be affected by whether a gene can be readily incorporated into a cell’s existing metabolic framework and other possible epistatic effects (Croucher et al. 2015a).
In this view, bacteria are bags of genes brought together for transient mutual benefit. How transient exactly that might be will vary with some loci only being present for a short while, before being lost, while others will form strong connections and produce a stable ecosystem of interactions that we recognize as a core genome. Like the public goods hypothesis, this hypothesis explains the observed distributions of genes, but with reference to selection not economics. The resulting metaphor is for the genome as an ecosystem, which can include all manner of interactions from direct competition to commensalism and mutualism. However, although ecosystems can appear stable and include symbioses, they are produced by selection, red in tooth and claw.
RECOMBINATION REDUX
Although we have here discussed recombination or horizontal gene transfer in Bacteria, in fact, these processes occur not only between species but kingdoms of life. We have long accepted the endosymbiont origins of mitochondria and chloroplasts from free-living bacteria, and the subsequent transfer of genetic material from the ancestral eubacteria to the eukaryotic nucleus. So, we should not be surprised by the observation that similar transfers have been found from intracellular Wolbachia parasites to their arthropod hosts (Dunning Hotopp et al. 2007). The existence of “bacterial” DNA in the arthropod genome went unnoticed at first because of the assumption that this was not possible, leading to the systematic removal of “bacterial” DNA from the initial analysis as the result of contamination. Transfer of genetic material in the other direction is also possible: Rhizobium radiobacter (the bacterium formerly known as Agrobacterium tumefaciens) is so efficient at the process that it is used to genetically engineer its plant hosts (Zambryski et al. 1983). There are also reported cases of apparently human DNA (a LINE element) appearing in the genome of the notoriously recombinogenic pathogen Neisseria gonorrhoeae (Anderson and Seifert 2011).
Together with other putative cases of transfer from hosts to infecting species (Pombert et al. 2015), and a suggestion that more than 100 human genes may have originated in other species (Crisp et al. 2015), it seems that horizontal transfer is not just for bacteria. However, a cautionary note is important, as we can produce a signal that looks like horizontal transfer by multiple means, including, but not limited to, restricted sampling of phylogenetic diversity combined with the loss of the gene from some taxa. Eukaryote biologists have also long known that interspecific recombination is more frequent than might be expected from a strict definition of the biological species concept. In one particularly nice example, sympatric species of Heliconius butterflies were found to show similar wing patterns—but the wing pattern within each species varied over the geographic range. So, butterflies would more closely resemble local members of the other species than samples of their own species from more remote locations. Genomic analysis showed this to be down to the transfer between the species of the genes controlling wing pattern (The Heliconius Genome Consortium 2012).
The bdelloid rotifers were memorably described by John Maynard Smith as an “evolutionary scandal,” because of their evident antiquity as a lineage, combined with the lack of evidence for sexual reproduction over that history. Sexual reproduction, at least among eukaryotes, has been considered to be something like essential. Yet, here is a taxa that seems to have dispensed with it entirely and suffered no ill effects. However, bdelloid rotifers do engage in the transfer of genetic material, with horizontal gene transfer being an especially interesting feature, coupled with extensive gene conversion and genomic rearrangements, and an estimated 8% of genes being of “nonmetazoan origin” (Flot et al. 2013). This should put into perspective our difficulty in concocting a coherent picture of bacterial evolution; one that accounts for recombination and horizontal gene transfer in a quantitative fashion. These are not problems unique to the bacteria. In fact, it is a problem of life itself.
ACKNOWLEDGMENTS
I thank Esther Robinson for suggesting the elegant notion of “regression to the genome mean,” and Dave Baltrus for his insightful and valuable discussions.
Footnotes
Editor: Howard Ochman
Additional Perspectives on Microbial Evolution available at www.cshperspectives.org
REFERENCES
- Achtman M. 2008. Evolution, population structure, and phylogeography of genetically monomorphic bacterial pathogens. Annu Rev Microbiol 62: 53–70. [DOI] [PubMed] [Google Scholar]
- Anderson MT, Seifert HS. 2011. Neisseria gonorrhoeae and humans perform an evolutionary LINE dance. Mob Genet Elements 1: 85–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery OT, Macleod CM, McCarty M. 1944. Studies on the chemical nature of the substance inducing transformation of pneumococcal types: Induction of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type III. J Exp Med 79: 137–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltrus DA, Guillemin K, Phillips PC. 2008. Natural transformation increases the rate of adaptation in the human pathogen Helicobacter pylori. Evolution 62: 39–49. [DOI] [PubMed] [Google Scholar]
- Bentley SD, Aanensen DM, Mavroidi A, Saunders D, Rabbinowitsch E, Collins M, Donohoe K, Harris D, Murphy L, Quail MA, et al. 2006. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet 2: e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AH, Feldman MW, Nevo E. 1980. Multilocus structure of natural populations of Hordeum spontaneum. Genetics 96: 523–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brueggemann AB, Pai R, Crook DW, Beall B. 2007. Vaccine escape recombinants emerge after pneumococcal vaccination in the United States. PLoS Pathog 3: e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carja O, Liberman U, Feldman MW. 2014. Evolution in changing environments: Modifiers of mutation, recombination, and migration. Proc Natl Acad Sci 111: 17935–17940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrolo M, Pinto FR, Melo-Cristino J, Ramirez M. 2009. Pherotypes are driving genetic differentiation within Streptococcus pneumoniae. BMC Microbiol 9: 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo-Ramirez S, Corander J, Marttinen P, Aldeljawi M, Hanage WP, Westh H, Boye K, Gulay Z, Bentley SD, Parkhill J, et al. 2012. Phylogeographic variation in recombination rates within a global clone of methicillin-resistant Staphylococcus aureus. Genome Biol 13: R126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaconas G, Kobryn K. 2010. Structure, function, and evolution of linear replicons in Borrelia. Annu Rev Microbiol 64: 185–202. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D. 2010. Elements of evolutionary genetics. Roberts and Company, Greenwood Village, CO. [Google Scholar]
- Claverys JP, Martin B, Polard P. 2009. The genetic transformation machinery: Composition, localization, and mechanism. FEMS Microbiol Rev 33: 643–656. [DOI] [PubMed] [Google Scholar]
- Clewell DB. 1981. Plasmids, drug resistance, and gene transfer in the genus Streptococcus. Microbiol Rev 45: 409–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffey TJ, Enright MC, Daniels M, Morona JK, Morona R, Hryniewicz W, Paton JC, Spratt BG. 1998. Recombinational exchanges at the capsular polysaccharide biosynthetic locus lead to frequent serotype changes among natural isolates of Streptococcus pneumoniae. Mol Microbiol 27: 73–83. [DOI] [PubMed] [Google Scholar]
- Cornejo OE, McGee L, Rozen DE. 2010. Polymorphic competence peptides do not restrict recombination in Streptococcus pneumoniae. Mol Biol Evol 27: 694–702. [DOI] [PubMed] [Google Scholar]
- Crisp A, Boschetti C, Perry M, Tunnacliffe A, Micklem G. 2015. Expression of multiple horizontally acquired genes is a hallmark of both vertebrate and invertebrate genomes. Genome Biol 16: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croucher NJ, Harris SR, Fraser C, Quail MA, Burton J, van der Linden M, McGee L, von Gottberg A, Song JH, Ko KS, et al. 2011. Rapid pneumococcal evolution in response to clinical interventions. Science 331: 430–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croucher NJ, Finkelstein JA, Pelton SI, Mitchell PK, Lee GM, Parkhill J, Bentley SD, Hanage WP, Lipsitch M. 2013. Population genomics of post-vaccine changes in pneumococcal epidemiology. Nat Genet 45: 656–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croucher NJ, Coupland PG, Stevenson AE, Callendrello A, Bentley SD, Hanage WP. 2014. Diversification of bacterial genome content through distinct mechanisms over different timescales. Nat Commun 5: 5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croucher NJ, Kagedan L, Thompson CM, Parkhill J, Bentley SD, Finkelstein JA, Lipsitch M, Hanage WP. 2015a. Selective and genetic constraints on pneumococcal serotype switching. PLoS Genet 11: e1005095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, Parkhill J, Harris SR. 2015b. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res 43: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow JF, Kimura M. 1970. An introduction to population genetics theory. Harper & Row, New York. [Google Scholar]
- Darmon E, Leach DR. 2014. Bacterial genome instability. Microbiol Mol Biol Rev 78: 1–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubin V, Ochman H. 2004. Bacterial genomes as new gene homes: The genealogy of ORFans in E. coli. Genome Res 14: 1036–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didelot X, Falush D. 2007. Inference of bacterial microevolution using multilocus sequence data. Genetics 175: 1251–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillard JP, Vandersea MW, Yother J. 1995. Characterization of the cassette containing genes for type 3 capsular polysaccharide biosynthesis in Streptococcus pneumoniae. J Exp Med 181: 973–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake JW. 1999. The distribution of rates of spontaneous mutation over viruses, prokaryotes, and eukaryotes. Ann NY Acad Sci 870: 100–107. [DOI] [PubMed] [Google Scholar]
- Dunning Hotopp JC, Clark ME, Oliveira DC, Foster JM, Fischer P, Munoz Torres MC, Giebel JD, Kumar N, Ishmael N, Wang S, et al. 2007. Widespread lateral gene transfer from intracellular bacteria to multicellular eukaryotes. Science 317: 1753–1756. [DOI] [PubMed] [Google Scholar]
- Feil EJ, Enright MC. 2004. Analyses of clonality and the evolution of bacterial pathogens. Curr Opin Microbiol 7: 308–313. [DOI] [PubMed] [Google Scholar]
- Feil EJ, Spratt BG. 2001. Recombination and the population structures of bacterial pathogens. Annu Rev Microbiol 55: 561–590. [DOI] [PubMed] [Google Scholar]
- Feil EJ, Maiden MCJ, Achtman M, Spratt BG. 1999. The relative contributions of recombination and mutation to the divergence of clones of Neisseria meningitidis. Mol Biol Evol 16: 1496–1502. [DOI] [PubMed] [Google Scholar]
- Feil EJ, Smith JM, Enright MC, Spratt BG. 2000. Estimating recombinational parameters in Streptococcus pneumoniae from multilocus sequence typing data. Genetics 154: 1439–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flot JF, Hespeels B, Li X, Noel B, Arkhipova I, Danchin EG, Hejnol A, Henrissat B, Koszul R, Aury JM, et al. 2013. Genomic evidence for ameiotic evolution in the bdelloid rotifer Adineta vaga. Nature 500: 453–457. [DOI] [PubMed] [Google Scholar]
- Fraser C, Hanage WP, Spratt BG. 2005. Neutral microepidemic evolution of bacterial pathogens. Proc Natl Acad Sci 102: 1968–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser C, Hanage WP, Spratt BG. 2007. Recombination and the nature of bacterial speciation. Science 315: 476–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser C, Alm EJ, Polz MF, Spratt BG, Hanage WP. 2009. The bacterial species challenge: Making sense of genetic and ecological diversity. Science 323: 741–746. [DOI] [PubMed] [Google Scholar]
- Gillespie JH. 2004. Population genetics: A concise guide. Johns Hopkins University Press, Baltimore, MD. [Google Scholar]
- Griffith F. 1922. Types of pneumococci obtained from cases of lobar pneumonia. In Reports on public health and medical subjects, No 13. Bacteriological studies, pp. 1–13. Ministry of Health, London. [Google Scholar]
- Guglielmini J, Quintais L, Garcillan-Barcia MP, de la Cruz F, Rocha EP. 2011. The repertoire of ICE in prokaryotes underscores the unity, diversity, and ubiquity of conjugation. PLoS Genet 7: e1002222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haggerty LS, Jachiet PA, Hanage WP, Fitzpatrick DA, Lopez P, O’Connell MJ, Pisani D, Wilkinson M, Bapteste E, McInerney JO. 2014. A pluralistic account of homology: Adapting the models to the data. Mol Biol Evol 31: 501–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton MB. 2009. Population genetics. Wiley-Blackwell, Chichester, UK. [Google Scholar]
- Hanage WP, Fraser C, Spratt BG. 2006. The impact of homologous recombination on the generation of diversity in bacteria. J Theor Biol 239: 210–219. [DOI] [PubMed] [Google Scholar]
- Hanage WP, Fraser C, Tang J, Connor TR, Corander J. 2009. Hyper-recombination, diversity, and antibiotic resistance in pneumococcus. Science 324: 1454–1457. [DOI] [PubMed] [Google Scholar]
- Hanage WP, Bishop CJ, Lee GM, Lipsitch M, Stevenson A, Rifas-Shiman SL, Pelton SI, Huang SS, Finkelstein JA. 2011. Clonal replacement among 19A Streptococcus pneumoniae in Massachusetts, prior to 13 valent conjugate vaccination. Vaccine 29: 8877–8881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms K, Schon V, Kickstein E, Wackernagel W. 2007. The RecJ DNase strongly suppresses genomic integration of short but not long foreign DNA fragments by homology-facilitated illegitimate recombination during transformation of Acinetobacter baylyi. Mol Microbiol 64: 691–702. [DOI] [PubMed] [Google Scholar]
- Hartl DL, Clark AG. 2007. Principles of population genetics. Sinauer Associates, Sunderland, MA. [Google Scholar]
- Havarstein LS, Hakenbeck R, Gaustad P. 1997. Natural competence in the genus Streptococcus: Evidence that streptococci can change pherotype by interspecies recombinational exchanges. J Bacteriol 179: 6589–6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch J, Tilly K. 1993. Linear plasmids and chromosomes in bacteria. Mol Microbiol 10: 917–922. [DOI] [PubMed] [Google Scholar]
- Hutchinson GE. 1959. Homage to Santa Rosalia; or, why are there so many kinds of animals? Am Nat 93: 145–159. [Google Scholar]
- Israel DA, Salama N, Krishna U, Rieger UM, Atherton JC, Falkow S, Peek RM Jr. 2001. Helicobacter pylori genetic diversity within the gastric niche of a single human host. Proc Natl Acad Sci 98: 14625–14630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsen PJ, Dubnau D, Levin BR. 2009. Episodic selection and the maintenance of competence and natural transformation in Bacillus subtilis. Genetics 181: 1521–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Wolf YI. 2009. The fundamental units, processes and patterns of evolution, and the tree of life conundrum. Biol Direct 4: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin BR, Cornejo OE. 2009. The population and evolutionary dynamics of homologous gene recombination in bacterial populations. PLoS Genet 5: e1000601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llull D, Garcia E, Lopez R. 2001. Tts, a processive β-glucosyltransferase of Streptococcus pneumoniae, directs the synthesis of the branched type 37 capsular polysaccharide in Pneumococcus and other Gram-positive species. J Biol Chem 276: 21053–21061. [DOI] [PubMed] [Google Scholar]
- Lodish HF. 2003. Molecular cell biology. W.H. Freeman, New York. [Google Scholar]
- Majewski J, Cohan FM. 1998. The effect of mismatch repair and heteroduplex formation on sexual isolation in Bacillus. Genetics 148: 13–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewski J, Cohan FM. 1999. DNA sequence similarity requirements for interspecific recombination in Bacillus. Genetics 153: 1525–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marttinen P, Hanage WP, Croucher NJ, Connor TR, Harris SR, Bentley SD, Corander J. 2012. Detection of recombination events in bacterial genomes from large population samples. Nucleic Acids Res 40: e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerney JO, Pisani D, Bapteste E, O’Connell MJ. 2011. The public goods hypothesis for the evolution of life on Earth. Biol Direct 6: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostowy R, Croucher NJ, Hanage WP, Harris SR, Bentley S, Fraser C. 2014. Heterogeneity in the frequency and characteristics of homologous recombination in pneumococcal evolution. PLoS Genet 10: e1004300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moxon R, Bayliss C, Hood D. 2006. Bacterial contingency loci: The role of simple sequence DNA repeats in bacterial adaptation. Ann Rev Genet 40: 307–333. [DOI] [PubMed] [Google Scholar]
- Narra HP, Ochman H. 2006. Of what use is sex to bacteria? Curr Biol 16: R705–R710. [DOI] [PubMed] [Google Scholar]
- Nielsen R, Slatkin M. 2013. An introduction to population genetics: Theory and applications. Sinauer Associates, Sunderland, MA. [Google Scholar]
- Ochman H. 2009. Radical views of the tree of life. Environ Microbiol 11: 731–732. [DOI] [PubMed] [Google Scholar]
- Okasha S. 2008. Evolution and the levels of selection. Clarendon Press, New York. [Google Scholar]
- Oliveira PH, Touchon M, Rocha EP. 2014. The interplay of restriction-modification systems with mobile genetic elements and their prokaryotic hosts. Nucleic Acids Res 42: 10618–10631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto SP. 2009. The evolutionary enigma of sex. Am Nat 174: S1–S14. [DOI] [PubMed] [Google Scholar]
- Overballe-Petersen S, Harms K, Orlando LA, Mayar JV, Rasmussen S, Dahl TW, Rosing MT, Poole AM, Sicheritz-Ponten T, Brunak S, et al. 2013. Bacterial natural transformation by highly fragmented and damaged DNA. Proc Natl Acad Sci 110: 19860–19865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace NR. 1997. A molecular view of microbial diversity and the biosphere. Science 276: 734–740. [DOI] [PubMed] [Google Scholar]
- Pearson T, Giffard P, Beckstrom-Sternberg S, Auerbach R, Hornstra H, Tuanyok A, Price EP, Glass MB, Leadem B, Beckstrom-Sternberg JS, et al. 2009. Phylogeographic reconstruction of a bacterial species with high levels of lateral gene transfer. BMC Biol 7: 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pombert JF, Haag KL, Beidas S, Ebert D, Keeling PJ. 2015. The Ordospora colligata genome: Evolution of extreme reduction in microsporidia and host-to-parasite horizontal gene transfer. MBio 6: e02400-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudhomme M, Libante V, Claverys JP. 2002. Homologous recombination at the border: insertion-deletions and the trapping of foreign DNA in Streptococcus pneumoniae. Proc Natl Acad Sci 99: 2100–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DA, Enright MC. 2004. Evolution of Staphylococcus aureus by large chromosomal replacements. J Bacteriol 186: 1060–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanchuk A, Jones CD, Karkare K, Moore A, Smith BA, Jones C, Dougherty K, Baltrus DA. 2014. Bigger is not always better: Transmission and fitness burden of ∼1 MB Pseudomonas syringae megaplasmid pMPPla107. Plasmid 73: 16–25. [DOI] [PubMed] [Google Scholar]
- Rosenberg C, Boistard P, Denarie J, Casse-Delbart F. 1981. Genes controlling early and late functions in symbiosis are located on a megaplasmid in Rhizobium meliloti. Mol Gen Genet 184: 326–333. [DOI] [PubMed] [Google Scholar]
- Sagi D, Tlusty T, Stavans J. 2006. High fidelity of RecA-catalyzed recombination: A watchdog of genetic diversity. Nucleic Acids Res 34: 5021–5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selander RK, Levin BR. 1980. Genetic diversity and structure in Escherichia coli populations. Science 210: 545–547. [DOI] [PubMed] [Google Scholar]
- Shen P, Huang HV. 1986. Homologous recombination in Escherichia coli: Dependence on substrate length and homology. Genetics 112: 441–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard SK, Didelot X, Meric G, Torralbo A, Jolley KA, Kelly DJ, Bentley SD, Maiden MC, Parkhill J, Falush D. 2013. Genome-wide association study identifies vitamin B5 biosynthesis as a host specificity factor in Campylobacter. Proc Natl Acad Sci 110: 11923–11927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard SK, Cheng L, Meric G, de Haan CP, Llarena AK, Marttinen P, Vidal A, Ridley A, Clifton-Hadley F, Connor TR, et al. 2014. Cryptic ecology among host generalist Campylobacter jejuni in domestic animals. Mol Ecol 23: 2442–2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JM, Smith NH, O’Rourke M, Spratt BG. 1993. How clonal are bacteria? Proc Natl Acad Sci 90: 4384–4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith HO, Gwinn ML, Salzberg SL. 1999. DNA uptake signal sequences in naturally transformable bacteria. Res Microbiol 150: 603–616. [DOI] [PubMed] [Google Scholar]
- Smith J, Feil EJ, Smith NH. 2000. Population structure and evolutionary dynamics of pathogenic bacteria. Bioessays 22: 1115–1122. [DOI] [PubMed] [Google Scholar]
- Suerbaum S, Achtman M. 2004. Helicobacter pylori: Recombination, population structure and human migrations. Int J Med Microbiol 294: 133–139. [DOI] [PubMed] [Google Scholar]
- Sundin GW, Weigand MR. 2007. The microbiology of mutability. FEMS Microbiol Lett 277: 11–20. [DOI] [PubMed] [Google Scholar]
- The Heliconius Genome Consortium. 2012. Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature 487: 94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobiason DM, Seifert HS. 2010. Genomic content of Neisseria species. J Bacteriol 192: 2160–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touchon M, Hoede C, Tenaillon O, Barbe V, Baeriswyl S, Bidet P, Bingen E, Bonacorsi S, Bouchier C, Bouvet O, et al. 2009. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet 5: e1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf DM, Vazirani VV, Arkin AP. 2005. Diversity in times of adversity: Probabilistic strategies in microbial survival games. J Theor Biol 234: 227–253. [DOI] [PubMed] [Google Scholar]
- Zambryski P, Joos H, Genetello C, Leemans J, Montagu MV, Schell J. 1983. Ti plasmid vector for the introduction of DNA into plant cells without alteration of their normal regeneration capacity. EMBO J 2: 2143–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinder ND, Lederberg J. 1952. Genetic exchange in Salmonella. J Bacteriol 64: 679–699. [DOI] [PMC free article] [PubMed] [Google Scholar]