Abstract
Synthetic gene switches are basic building blocks for the construction of complex gene circuits that transform mammalian cells into useful cell-based machines for next-generation biotechnological and biomedical applications. Ligand-responsive gene switches are cellular sensors that are able to process specific signals to generate gene product responses. Their involvement in complex gene circuits results in sophisticated circuit topologies that are reminiscent of electronics and that are capable of providing engineered cells with the ability to memorize events, oscillate protein production, and perform complex information-processing tasks. Microencapsulated mammalian cells that are engineered with closed-loop gene networks can be implanted into mice to sense disease-related input signals and to process this information to produce a custom, fine-tuned therapeutic response that rebalances animal metabolism. Progress in gene circuit design, in combination with recent breakthroughs in genome engineering, may result in tailored engineered mammalian cells with great potential for future cell-based therapies.
Engineered mammalian cells have many potential uses (e.g., disease diagnosis and treatment). Customized gene switches are key components of engineered cells; they enable the cells to sense and respond to specific signal inputs.
The engineering of mammalian cells holds great promise for cell-based technologies, such as stem cell reprogramming, biopharmaceutical manufacturing, and for cell-based therapies and diagnostics (Weber and Fussenegger 2012). The manipulation of complex, multicellular processes involving metabolic pathways, protein secretion, and production, and cell differentiation and development allows their precise control, improvement, and adaption and could promote the power of engineered mammalian cells in future cell-based applications. Central to synthetic biology-inspired engineering approaches are tailor-made gene controllers that enable cells to interact and respond to specific signal inputs and that can be rewired into programmable gene circuits to execute defined functions (Benenson 2012; Ausländer and Fussenegger 2013). When integrated into mammalian host cells, programmable gene circuits may enable the temporal control of stem cell reprogramming, advance cell-based therapies by arming autologous cells with closed-loop therapeutic gene circuits, and enhance the cellular production capacity in biopharmaceutical manufacturing.
Synthetic biology exploits the vast diversity of functional components found in the genetic material of all life forms. Because the cellular language of DNA, RNA, and protein is universal throughout all kingdoms of life, bioengineers can exchange these biological components between organisms to provide cells with a novel, customized function. For example, ligand-binding sensors are engineered to modulate diverse biological activities (Wieland and Fussenegger 2010; Ausländer and Fussenegger 2013), enzymes from different organisms are coupled to produce new biomolecules (Müller et al. 2012), and programmable DNA-binding proteins are harnessed for gene-editing purposes (Gaj et al. 2013). Moreover, we are beginning to create synthetic components that are not found in nature, expanding the set of appropriate biological components, and equipping engineered cells with special functionalities for useful applications. Synthetic nucleotides and amino acids are already being incorporated into cell-produced genomes (Malyshev et al. 2014) and proteins (Mandell et al. 2015; Rovner et al. 2015), respectively, and synthetic RNA sensors are being used to control gene expression in living mammalian cells (Culler et al. 2010; Saito et al. 2010; Ausländer et al. 2014c). However, the implementation of new biological components often requires precise connection and dosing of endogenous and synthetic components to assemble a functional system. The efficient engineering of mammalian cells remains a challenge because bioengineers often cannot predict the behavior of a cell on component integration. Additionally, technical barriers and an as-yet limited understanding of the entire living mammalian cell must be overcome.
Living cells continuously sense and respond to environmental and endogenous signals and drive cellular programs that adapt cellular behavior, for example, regulate cell death, fine-tune metabolic pathways, or differentiate into specified cell types. Fundamental to this adaptability is the dynamic regulation of gene-expression patterns. Mammalian cells use various complex mechanisms to influence the rates of transcription and translation of individual or multiple genes, enabling global or local alterations in cellular gene-expression patterns. Using an engineering approach, synthetic biology aims to control gene expression in a predictable manner by enabling bioengineers to program engineered cells to perform specific tasks. Central to this concept are programmable gene switches that are able to execute the gene regulatory function of heterologous or endogenous genes in a ligand-dependent manner (Benenson 2012; Ausländer and Fussenegger 2013). In principle, these gene switches can be exploited to execute information-processing tasks in living cells. Sensor units measure input signals and process the input state by rendering a defined output, that is, the gene-expression level. To run cellular programs that are more complex, multiple gene switches are combined to assemble gene circuits with specified network topologies. The design of such complex gene circuits is challenging because they must function precisely and specifically in the complicated cellular environment without influencing essential endogenous processes. For example, gene circuits are designed to improve, tune, or adapt the switching performance of target genes (Deans et al. 2007; Lapique and Benenson 2014; Prochazka et al. 2014) or to perform specific tasks such as biocomputing (Xie et al. 2011; Ausländer et al. 2012a) and oscillatory functions (Tigges et al. 2009), as well as recording environmental and endogenous events in synthetic memory devices (Burrill et al. 2012). Moreover, engineered cell–cell communication devices enable mammalian cells to interact and assemble synthetic multicellular systems (Bacchus et al. 2012). Basic gene switches and complex gene circuits have great potential for biomedical applications because they enable encapsulated, engineered cells to sense specific input signals in the body and to control the expression of specific outputs, that is, proteins, to initiate a therapeutic intervention (Ausländer et al. 2012b).
Here we describe a synthetic biology-inspired approach for engineering mammalian cells. We cover classic and emerging design strategies for synthetic gene switches and their different modes of action. In addition, we focus on different blueprints for connecting basic gene switches to build more complex gene circuits with preset network topologies. We also introduce advanced genome-engineering technologies that have the potential to revolutionize the manipulation and regulation of gene expression via the incorporation of synthetic DNA-encoded gene circuits. Finally, we review recent progress in cell-based applications in biomedicine and biotechnology and provide a perspective regarding future applications for mammalian cell–based technologies.
GENE SWITCHES
Gene expression in mammalian cells is divided into multiple steps and can be briefly described as follows: Transcription factors are recruited to regulatory sites to activate or repress the transcription of target genes, thereby controlling the production of nascent messenger RNA (mRNA) transcripts. In the nucleus, posttranscriptional modifications produce a mature mRNA molecule. A 5′ cap and 3′ poly(A) tail are attached to the nascent mRNA molecule, and introns are removed during the splicing process. Then mRNA is exported into the cytoplasm and subject to translation. In this section, we review synthetic gene switches that can intervene and control mammalian gene expression at different levels and that can serve as basic components for gene circuit design.
Transcriptional Gene Switches
In principle, a transcription factor consists of a DNA-binding domain (DBD) and a transcriptional modulator. When bound to the corresponding DNA target sequence, transcription factors recruit either positive or negative regulatory proteins, depending on the transcriptional modulator, to either induce or repress, respectively, the transcription of the target gene. The activity of transcription factors can be regulated by their availability at the DNA target site. Although transcription factors with a programmable DBD such as transcription activator-like effector (TALE) or zinc-finger (ZF) proteins cannot be controlled by a ligand, others possess ligand-responsive DBDs that modulate their affinity for a cognate DNA target sequence. Another mechanism to link signal sensing to a transcriptional response is based on the rewiring of endogenous signaling pathways that control the expression of target genes. For example, G protein–coupled receptor (GPCR)-dependent signaling pathways control the activity of transcription factors. Ligand binding to GPCRs induces signal transduction based on second messenger molecules and phosphorylation cascades that culminate in transcription factor activity. The number of divergent classes of transcription factors that have different characteristics with potential advantages or disadvantages for custom applications in mammalian cells is increasing.
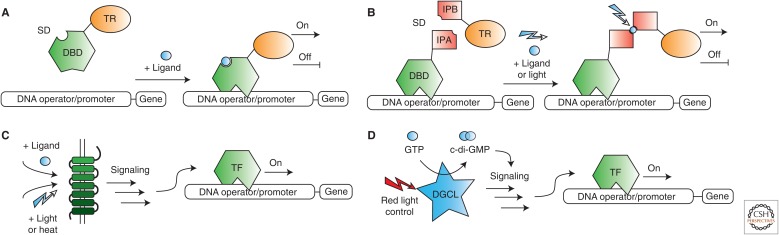
Prokaryotic regulator protein families are a widespread class of sensor proteins (e.g., >200,000 prokaryotic regulator protein-like sequences have been identified) that enable cells to adapt rapidly to environmental and metabolic changes by controlling the transcription of target genes depending on specific input signals (Ramos et al. 2005; Cuthbertson and Nodwell 2013). Most prokaryotic regulator proteins consist of an amino-terminal DBD that recognizes a unique DNA operator sequence and a carboxy-terminal sensory domain (SD) that specifically binds to a ligand molecule (Fig. 1A) (Cuthbertson and Nodwell 2013). Their diversity and sensory capacity render these proteins attractive for mammalian gene regulation and gene circuit design (Wieland and Fussenegger 2012). In a synthetic biology-inspired approach, prokaryotic regulator-based systems can be transferred easily into mammalian cells when DNA operator and protein sequence information and ligand identity are known. In brief, a transcriptional modulator (e.g., VP16, Krüppel-associated box [KRAB]) is fused to the transcriptional regulator (TR) protein, and DNA operator repeats are attached to suitable minimal or constitutive promoter sequences that drive target gene expression (Fig. 1A). Because the ligand-binding domain is part of the protein, the binding of the ligand to the regulator dimer leads to the dissociation of the operator–dimer complex, thus influencing gene-expression rates. Based on this design strategy, mammalian cells have been engineered with various synthetic systems to sense different ligand inputs (e.g., antibiotics, Gossen and Bujard 1992; Fussenegger et al. 2000; Urlinger et al. 2000; Weber et al. 2002, 2003; and small molecule ligands, Neddermann et al. 2003; Weber et al. 2006, 2009; Hartenbach et al. 2007; Gitzinger et al. 2009, 2012; Kemmer et al. 2010; Bacchus et al. 2013) and proven to be useful tools for diverse biological applications. Moreover, a synthetic system that enables dual-input gene-expression control was developed using a single engineered prokaryotic regulator protein fused to the transrepressor domain KRAB, termed KRAB–CbaR (Xie et al. 2014). The first input, benzoic acid, is able to induce target gene expression, whereas the second input, vanillic acid, leads to an increase in the affinity of KRAB–CbaR to the CbaR-specific DNA operator sequence, further enhancing target gene repression. Furthermore, chemical- or light-inducible dimerization systems (DSs) provide a second layer of gene control, for example, interaction partner A (IPA) is fused to the DBD of a prokaryotic regulator protein, and the interaction partner B (IPB) is connected to a TR, resulting in a split transcription factor (Fig. 1B). For example, the interaction of Arabidopsis thaliana–derived protein partners phytochrome-interacting factor 6 (PIF6) and phytochrome B (PhyB) can be triggered and reverted in a red- and far-red-controlled manner, respectively. This interaction has been combined with the prokaryotic repressor protein TetR to generate a synthetic light-dependent gene-regulation system (Müller et al. 2013a). The same approach was also used in the design of ultraviolet (UV)-responsive (Müller et al. 2013b) and fatty acid-inducible (Rössger et al. 2013b) gene-regulation systems. The following are characteristics of prokaryotic regulator-based systems: (1) orthogonality to mammalian cells, (2) high diversity of different prokaryotic regulators that bind to various ligands, (3) a plasticity simplifying gene switch design, (4) high specificity that enables the usage of multiple synthetic systems in single cells, and (5) robust switching performance in mammalian cells.
Figure 1.
Transcriptional gene switches. (A) Prokaryotic regulator proteins are fused to transcriptional regulator proteins and bind to DNA operator sequences to control the transcription of target genes in a ligand-responsive manner. (B) Combining prokaryotic regulator proteins with ligand- or light-induced dimerization systems (DSs) enables the signal-dependent recruitment of transcriptional regulator proteins. (C) Cell-surface-located G protein–coupled receptors (GPCRs) sense extracellular signals and trigger signal transduction via signaling pathways to control cellular processes such as gene transcription in target cells. (D) An engineered diguanylate cyclase (DGCL) synthesizes the second messenger c-di-GMP in a red-light-responsive manner, triggering a downstream signaling pathway and leading to the transcriptional activation of target genes. DBD, DNA-binding domain; SD, sensory domain; TR, transcriptional regulator; IPA, interaction partner A; IPB, interaction partner B; TF, transcription factor; GTP, guanosine triphosphate.
Another widespread family of ligand-responsive gene switches is based on a family of integral eukaryotic membrane proteins called GPCRs (<800 members in the human genome) (Heng et al. 2013). GPCRs detect, first, messengers (extracellular signal inputs) and activate a signaling cascade that includes the production of second messengers to transduce information into the cell and to modulate cellular processes such as gene transcription (Fig. 1C). Specified cell types in the human body use GPCRs to respond to changes in hormone levels, metabolite concentrations, and other disease-related ligand molecule alterations. Therefore, GPCRs are extraordinarily suited for sensing therapeutic-relevant signal molecules in the appropriate physiological or pathological range and are often used in mammalian cell–based biomedical applications. Many mammalian cell lines express the components required to assemble intracellular signaling pathways, making the rewiring of custom GPCR sensory proteins to synthetic promoters straightforward. A synthetic GPCR-based gene-regulation system consists of a constitutive GPCR-expression unit and a target gene-expression unit. The latter harbors a synthetic promoter sequence with cognate DNA response elements (CRE, SRE, NFAT-RE) that recruit signaling pathway-specific transcription factors. Thus far, synthetic GPCR-based gene control systems have been engineered that respond to metabolites, hormones, and drug ligands, including luteinizing hormone (Kemmer et al. 2011), histamine (Ausländer et al. 2014b), dopamine (Rössger et al. 2013a), and guanabenz (Ye et al. 2013), in addition to other signals, such as pH changes (Ausländer et al. 2014a), heat (Stanley et al. 2012), and blue light (Ye et al. 2011). The orthogonal usage of multiple GPCRs in individual cells is impeded if they use the same signaling pathway. However, GPCRs can also be engineered to rewire their signaling pathway to synthetic or alternative endogenous signaling pathways or to respond to synthetic ligands. For example, a set of GPCR-based designer receptors activated exclusively by designer drugs (DREADDs) have been engineered to activate GPCR signaling cascades selectively using the pharmacologically inert molecule clozapine-N-oxide (CNO) (Urban and Roth 2015). This tool has been used for modulating cell type-specific GPCRs in neuroscience (Wess et al. 2013) and for engineering a synthetic cell motility device in mammalian cells (Park et al. 2014). Another class of synthetic GPCR hybrids is based on so-called OptoXRs, which consist of engineered intracellular loops that are used to rewire GPCR signaling pathways, providing spatiotemporal light-responsive activation control (Airan et al. 2009). The following are characteristics of GPCR systems: (1) high compatibility with mammalian cells, (2) high diversity in sensing various disease-related ligands, (3) a simple gene switch design, (4) low connectivity of multiple GPCRs, and (5) robust switching performance in mammalian cells.
Stimulator of interferon genes (STING) is an endogenous, intracellular sensor protein that detects cytosolic DNA and intracellular pathogens by binding to cyclic dinucleotides (c-di-GMP, cGAMP). The activation of STING initiates a signaling cascade that leads to the nuclear translocation of interferon-regulatory factor 3 (IRF3), a transcription factor that specifically binds IRF3 DNA response elements to induce target gene expression. Although mammalian cells produce the cGAS protein, which synthesizes cGAMP on the detection of cytosolic DNA, c-di-GMP is only produced in specific prokaryotic cells. For example, the phototrophic bacterium Rhodobacter sphaeroides possesses a multidomain protein consisting of a diguanylate cyclase (DGCL) that synthesizes c-di-GMP, a phosphodiesterase (PDE) that breaks down c-di-GMP and a red-light sensor that controls the cyclase activity of DGCL. The integration of an engineered PDE-deficient variant of this protein into mammalian cells enables the red-light-responsive synthesis of c-di-GMP, thus rewiring the endogenous STING-dependent signaling pathway to activate synthetic target gene expression (Fig. 1D) (Folcher et al. 2014). This example shows the ability of synthetic biology-inspired concepts to enable the successful transfer and usage of prokaryotic second messenger molecules, such as c-di-GMP, in mammalian cells.
Because the aforementioned transcription factors bind to defined, invariable DNA sequences, correct insertion in proximity to promoter sequences is crucial, thus limiting their usage to regulate endogenous genes without laborious genetic engineering. Recent progress in genome engineering has produced advanced gene-editing systems capable of targeting and modifying any DNA sequence in the genome (Gaj et al. 2013). These systems are based on programmable, sequence-specific DNA-binding modules that can be equipped with nonspecific nucleases that cleave their target in proximity to the DNA-binding site. The three most popular gene-editing systems differ in their composition and characteristics. ZF proteins are commonly found in eukaryotes, and TALEs are derived from plant pathogens, for example, Xanthomonas (Gaj et al. 2013). To engineer these proteins and to determine their specific function, effector proteins such as nucleases or transcriptional regulators are fused to the customized DBD (Hsu et al. 2014). ZFs and TALEs consist of modular amino acid repeats that recognize either three (ZFs) or one (TALE) base pair(s) and that can be connected to one another to bind to DNA sequences of increased lengths. This modularity enables the engineering of specific ZFs or TALEs that bind to pre-defined sequences with high specificity within large genomes to execute defined functions. In contrast to ZFs and TALEs, the clustered regulatory interspaced short palindromic repeat (CRISPR)/CRISPR-associated (Cas)-based gene-editing system facilitates DNA recognition and binding using RNA (Doudna and Charpentier 2014). This system is derived from bacteria, in which it is involved in the cellular defense mechanism against viruses and plasmids. This system is based on the DNA endonuclease protein Cas9 and an engineered guide RNA (gRNA) that consists of two essential elements: a 20-nt-long RNA sequence that matches the target site and a structured RNA element that binds to the Cas9 effector protein and that facilitates DNA binding and cleavage. The recognition and binding of the gRNA is based on Watson–Crick base pairing, which allows for simple, predictable, and efficient reengineering of the gRNA to target any DNA sequence of interest. Moreover, the usage of multiple gRNAs enables multiplexed applications of the CRISPR/Cas system to manipulate distinct genomic sites simultaneously (Cong et al. 2013). The replacement of the nuclease domain with a transcriptional activator or repressor protein allows the three gene-editing systems to synthesize gene controllers with the potential ability to target any stretch of DNA sequence in the genome and thus to regulate the expression of any endogenous gene of interest (Doudna and Charpentier 2014). These systems, which have attracted great interest in synthetic biology, have been used to control heterologous gene expression and to design logic gates. Thus far, however, only a few options are available that enable the linkage of these synthetic gene controllers to sensor units to achieve ligand responsiveness. One option is to use chemical- or light-inducible DS, which consist of two proteins that interact in a ligand- or light-controllable manner (Fig. 1B). Coupling one of the DS proteins to a ZF or TALE and the other to a transcriptional regulatory protein results in ligand- or light-regulated recruitment of the transcriptional regulatory protein to the DNA-bound ZF or TALE and thus control of gene expression (Beerli et al. 2000; Polstein and Gersbach 2012; Konermann et al. 2013; Mercer et al. 2014). Based on the same concept, the CRISPR/Cas9 system has been engineered with light- or rapamycin-inducible dimerization domains enabling optogenetic or chemical control of RNA-guided gene regulation, respectively (Polstein and Gersbach 2015; Zetsche et al. 2015). The CRISPR/Cas9 system has been further adapted to allow for RNA-mediated recruiting of transcriptional regulators to specific genomic sites. In this adapted system, natural RNA-protein interactions are exploited by fusing different protein-binding RNA motifs to specific gRNAs that determine the genomic target locus (Zalatan et al. 2015). Because different transcriptional regulators can be fused to different RNA-binding proteins, simultaneous activation and repression of CRISPR-based gene regulation is possible. More recently, it has been shown that the CRISPR/Cas9 system is a highly flexible platform that accepts not only simple protein-binding aptamers but also much more complex RNA structures, such as long noncoding RNAs (lncRNAs) or fluorescent aptamers (Shechner et al. (2015). This powerful technology known as CRISPR-Display enables the expression of multiple gRNAs in single cells where each gRNA shows a different function. This enables, for example, the simultaneous activation and repression of multiple endogenous genes and at the same time the fluorescence-based visualization of genomic loci, thus providing new opportunities for basic research as well as synthetic biology.
Posttranscriptional Gene Switches
Transcriptional gene regulation relies on transcription factors that control the production of mRNAs and thus the amount of transcripts that are subject to translation. An additional layer of gene-expression control is provided by posttranscriptional regulatory modes that fine-tune and balance global gene expression. These control modes act on target mRNA transcripts and influence their stability, splicing, and translation. The versatility, modularity, and programmability of RNA render it attractive for the synthetic biology–inspired engineering of new functionalities (Saito and Inoue 2009; Grabow and Jaeger 2014). Synthetic RNA devices are engineered from basic RNA modules that provide diverse functionality. Natural and synthetic RNA aptamers bind to ligands with high specificity and give RNA the ability to sense small molecules, metabolites, and proteins (Famulok et al. 2007). Moreover, aptamers are often able to transmit the binding state via structural changes within the RNA molecule itself (Wunnicke et al. 2011). This ability can be exploited to manipulate the function of other RNA modules (e.g., short interfering RNAs [siRNAs] or ribozymes) in a ligand-responsive manner (Wieland and Fussenegger 2010). Endogenous gene-regulation pathways, such as RNA interference (RNAi) and mRNA splicing, can also be rewired and placed under the control of synthetic regulators.
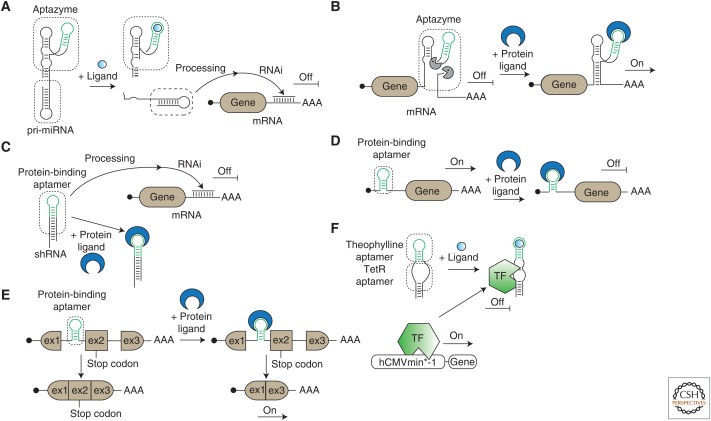
The mammalian RNAi pathway enables sequence-specific gene knockdown targeted by 21- to 23-nt RNA species called siRNAs (Mittal 2004). The programming of these siRNAs is simple and enables the targeting of virtually any mRNA sequence. Ligand-responsive ribozymes (aptazymes) have been fused to siRNA precursors to block siRNA maturation to construct ligand-responsive siRNA-based gene switches in mammalian cells (Fig. 2A) (Kumar et al. 2009; Nomura et al. 2012). The addition of the small-molecule ligand theophylline or guanine induces ribozyme self-cleavage and releases the respective siRNA precursor, which further matures into a functional siRNA molecule that can repress target gene expression. Ribozymes are potent building blocks for the engineering of RNA-based devices because of their efficient self-cleavage activity, which can be exploited to control the function of other RNA molecules. To regulate the self-cleavage activity of ribozymes by ligands, aptamers can be assembled into ribozymes, rendering them ligand-responsive molecules (Wieland et al. 2012). The integration of these engineered ribozymes into mRNA transcripts leads to ligand-dependent cleavage of the target mRNA and thus control over gene expression (Win and Smolke 2007; Ausländer et al. 2010; Nomura et al. 2012, 2013). Recently, a new design strategy for protein-responsive ribozymes and their implementation into gene circuits has been described (Fig. 2B) (Ausländer et al. 2014c). In another study, a novel design of aptamer–siRNA fusions enabled siRNA-processing control in mammalian cells (Fig. 2C) (Kashida et al. 2012). Protein-binding RNA aptamers (e.g., U1A protein-U1A RNA motif) have been engineered into siRNA precursors that can replace RNA stem loops that are essential for siRNA processing. Protein binding to the aptamer blocks efficient processing and thus impedes gene knockdown. Protein-responsive aptamers can also be integrated into the 5′ untranslated regions (UTRs) of mRNAs to control translational initiation in a protein-dependent manner (Fig. 2D) (Saito et al. 2010). This simple approach was first shown using the L7Ae protein-C/D box RNA motif, which was engineered into reporter mRNA transcripts. In subsequent studies, further advancements have been made to tune input–output functions and to use multiple protein-responsive aptamers for the robust control of endogenous pathways and gene circuits (Saito et al. 2011; Goldfless et al. 2012; Stapleton et al. 2012; Endo et al. 2013). In a similar approach, protein-binding aptamers (MS2, p65, p50, and β-catenin aptamers) have been introduced into the proximity of splicing recognition sites of an alternative exon element of mRNA precursors (Fig. 2E) (Culler et al. 2010). However, the alternative splicing pattern was influenced by the cognate protein ligands and varied in terms of exon inclusion or exclusion for different protein ligands. Because p65 and β-catenin proteins are downstream modulators from signaling pathways, alternative splicing regulation could be rewired using the cognate inducers tumor necrosis factor α (TNF-α) or LTD4. Aptamer–aptamer fusions have been developed to control the function of an engineered, bacterial repressor-based transcription factor (Fig. 2F) (Ausländer et al. 2011). The TetR-binding aptamer is able to bind to the DBD of the TetR-VP16 fusion protein, thereby releasing DNA operator-bound TetR and blocking gene expression. By rational design, the TetR-binding aptamer was fused to and placed under the control of a theophylline-binding aptamer, leading to an RNA-based transcriptional gene-regulation system.
Figure 2.
Posttranscriptional gene switches. (A) Aptazymes are fused to primary microRNA (pri-miRNA) molecules, enabling the ligand-responsive control of pri-miRNA processing and posttranscriptional target gene control. (B) Protein-responsive aptazymes are integrated into messenger RNAs (mRNAs) to regulate their stability, depending on the presence or absence of the protein ligand. (C) Protein binding to protein-binding aptamers that are integrated into small hairpin RNAs (shRNAs) inhibits shRNA processing and allows for protein-controlled target gene control. (D) Protein-binding aptamers integrated into the 5′ untranslated regions (UTRs) of mRNAs can be used to control translational initiation in a protein-dependent manner. (E) Integration of protein-binding aptamers into close proximity of splicing sites allows for protein-responsive alternative splicing regulation. (F) A TetR-binding aptamer is combined with a theophylline-responsive aptamer to enable the theophylline-dependent folding of the TetR-binding aptamer. When bound to its cognate aptamer, the TetR protein loses its DNA operator binding ability and influences gene expression at the transcriptional level. TF, Transcription factor; RNAi, RNA interference.
ENGINEERING GENE CIRCUITS
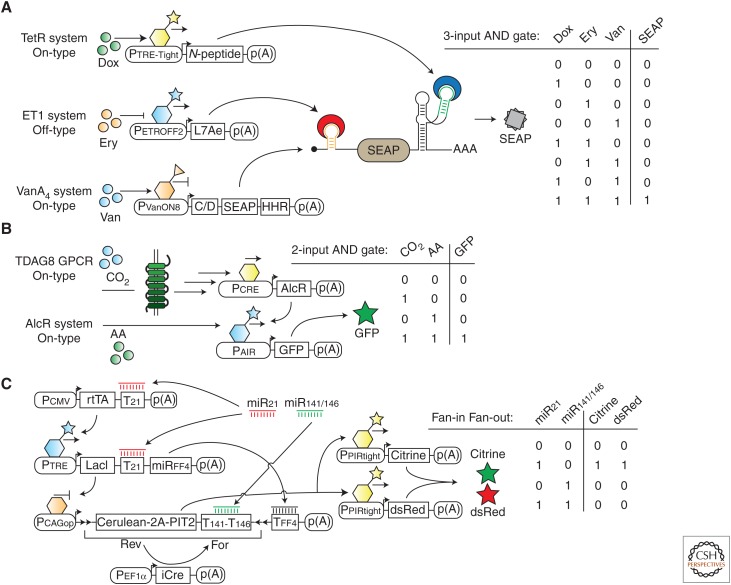
Synthetic biology is an engineering discipline that aims to find solutions for biological questions by creating and developing new biological machines and systems that are based on biomolecules and synthetic materials to perform customized tasks. Network abstractions of other engineering disciplines can be used to describe synthetic biology-inspired biological systems. For example, “circuit topology” specifies the interconnection of all components in a circuit and is commonly used in electronics. Circuit topology can also be applied in biology to show synthetic biological networks and to describe input–output functions. Gene circuits with different topologies, such as oscillators (Tigges et al. 2009), logic gates (Kramer et al. 2004; Rinaudo et al. 2007; Win and Smolke 2007, 2008; Ausländer et al. 2012a, 2014a), hysteresis (Kramer and Fussenegger 2005), band-pass (Greber and Fussenegger 2010), time delay (Weber et al. 2007; Lapique and Benenson 2014), bow-tie (Prochazka et al. 2014), and memory devices (Burrill et al. 2012) have already been constructed in living mammalian cells. Because the above-described synthetic gene switches are able to process input information and to produce a specific output, they are well-suited building blocks for the design of programmable gene circuits reminiscent of electronic circuits. In a seminal publication, Kramer et al. (2004) described the design of various gene circuits with two and three inputs that process a single output reporter protein and set the basis for programmable gene circuits. Bacterial repressor-based transcriptional systems were cascaded to imitate the input–output function of logic gates found in electronics. Different combinations of small molecule ligands controlled the DNA binding of synthetic repressor proteins, resulting in a specific output. Based on the concept of programmable gene circuits, many different biological types of engineering decision-making systems have been developed in mammalian cells. These systems differ in their component composition and complexity, input–output identity and performance characteristics. Gene circuits can also be based on the sole application of posttranscriptional gene switches such as engineered ligand-responsive ribozymes (Win and Smolke 2008). However, the combination of both transcriptional and posttranscriptional systems promises to advance gene circuit design because it enables gene control at both the DNA and RNA levels. The leakiness of a gene switch could be significantly improved by combining RNAi methods with common transcriptional control (Deans et al. 2007). The linkage of transcriptional and posttranscriptional control was also applied to design gene circuits that mimic half-adder circuits capable of performing basic arithmetic (Ausländer et al. 2012a) and showing oscillatory gene-expression behavior (Tigges et al. 2009) in mammalian cells. For the design of a three-input AND gate, an engineered protein-responsive hammerhead ribozyme that regulates mRNA stability was combined with a protein-responsive aptamer that blocks translation (Fig. 3A) (Ausländer et al. 2014c). Coupled to three different transcriptional systems, each of the three small molecule ligands controlled the production, the stability, or the translation of the reporter mRNA. This concept simplifies gene circuit design because it enables the combination of multiple gene switches to act on individual transcripts. The eligibility of a biological component for gene circuit design is determined by its performance, connectivity, programmability, and orthogonality. Specifically, synthetic gene switches must function in a complex cellular environment without influencing either endogenous or other synthetic components. In a recent publication, sets of different building blocks were used to resemble logic gates in mammalian cells (Fig. 3B) (Ausländer et al. 2014a). In particular, a GPCR-based CO2/H+-inducible gene-regulation system was combined with bacterial repressor- and protein dimerization–based systems to build gas-inducible gene circuits. Gene circuits have also been designed to detect siRNA states in mammalian cells (Rinaudo et al. 2007; Tigges et al. 2009; Leisner et al. 2010; Xie et al. 2011; Lapique and Benenson 2014; Prochazka et al. 2014). siRNA or miRNA target sites are integrated into synthetic components to process siRNA information logically, enabling either the programming of the resulting gene circuit by the exogenous application of siRNA molecules, the intracellular, ligand-inducible production of siRNA ligands or endogenous miRNA species. Gene circuit performance can be improved with the addition of regulatory connections and new circuit designs. Site-specific recombinases, which have the ability to excise, flip, or invert DNA sequences, have been established as powerful building blocks for genome engineering and transcriptional gene control in mammalian cells (Nern et al. 2011). Site-specific recombination sites are engineered to flank genes, promoters, or transcriptional blockers to influence gene expression and to introduce regulatory knots to decouple inputs and outputs (Lapique and Benenson 2014; Prochazka et al. 2014). When integrated into gene circuits in a bow-tie topology, recombinases can act as a “knot” to transmit the circuit’s information and thereby improve the performance of gene circuits significantly by reducing component leakage and input and output decoupling, as well as the timely production of individual components induced by gene flipping (Fig. 3C) (Prochazka et al. 2014). The production of an output protein within gene circuits is used to wire different gene switches within the circuit or to have a convenient readout protein for output quantification. Engineered mammalian cells harboring different gene circuits can communicate with each other when the sender cell population produces a signaling molecule that can be sensed by the receiver cell population. For example, sender cells can produce the enzyme tryptophan synthase that converts indole into l-tryptophan, which, in turn, can be detected by receiver cells that measure tryptophan levels using a transcriptional controller (Bacchus et al. 2012). The combination of this system with a second synthetic cell–cell communication system based on the communication molecule acetaldehyde resulted in a two-way multicellular communication network that could temporally program endothelial cells to form blood vessels. Although the above-described gene circuits are based on ligand-responsive gene switches, ligand-unresponsive components can also be connected to build gene networks that can be controlled by the presence or absence of their components. For example, TALE- or ZF-based transcription factors have been used to build basic logic gates in which component-encoding plasmids serve as input signals (Lohmueller et al. 2012; Lienert et al. 2013).
Figure 3.
Engineered gene circuits. (A) The three ligand-responsive, transcriptional gene-control systems based on the transcriptional regulators TetR, ET1, and VanA4 are combined with two posttranscriptional gene switches that use C/D RNA motif-L7Ae protein and N-peptide-responsive ribozyme-N-peptide interactions to assemble a three-input AND gate. Mammalian cells engineered with this gene circuit can be programmed with the three ligands erythromycin (Ery), doxycycline (Dox), and vanilic acid (Van) to control output gene expression as shown in the truth table. (B) Design of a two-input AND gate that can be controlled by the two gaseous molecules carbon dioxide (CO2) and acetaldehyde (AA). Activation of the G protein–coupled receptor (GPCR) TDAG8 by low pH triggers a signaling pathway that leads to the expression of AlcR, which binds to its cognate promoter exclusively in the presence of AA, thus producing the output protein GFP. (C) Transcriptional activators (rtTA and PIT2) and repressors (LacI), in addition to the recombinase iCre, are connected to build a gene circuit using bow-tie architecture. Two miRNA inputs (miR21 and miR141/146) manipulate the gene circuit to express the two output proteins in the exclusive presence of miR21.
ENGINEERING MAMMALIAN CELLS FOR BIOMEDICAL APPLICATIONS
Reporter genes are used as output proteins for the design and characterization of new gene switches and circuits because they provide information regarding switching performance. The replacement of the reporter gene with output proteins that induce cell apoptosis or differentiation or that have a therapeutic effect allows for the use of gene circuit-engineered mammalian cells in biomedical applications. The design of a cell classifier circuit based on the input level detection of different miRNA species allows for the differentiation of different cell types (Xie et al. 2011). TetR- and LacI-based transcriptional regulation systems control the expression of mRNA transcripts that sense specific miRNA inputs and that are logically connected to produce the proapoptotic factor human Bcl-2-associated X protein (hBax). Cell death is only induced in cancer cells that express a specific miRNA pattern. Via a similar strategy, transcription factor patterns that are present only in cancerous cells can be detected by their cognate promoter target sequences (CXCL1, SSX1, and H2A1) to drive the expression of split transcription factor units (Gal4-Coh2 and DocS-VP16 fusion proteins) (Nissim and Bar-Ziv 2010). When both units are expressed, Coh2 and DocS heterodimerization results in upstream activation sequence (UAS)-binding of Gal4 and VP16-dependent activation of the adjacent promoter. The output protein thymidine kinase type 1 derived from the Herpes simplex virus converts the prodrug ganciclovir into a toxic molecule that induces cell death. A therapeutic application of the above-described circuits would require an efficient and complete delivery of all DNA components into a patient’s target cells.
In contrast, therapeutic cell implants consist of engineered mammalian cells that integrate into the metabolic processes of an organism such that these cells are able to produce a therapeutic effector (Ausländer et al. 2012b; Weber and Fussenegger 2012). Inert carriers such as alginate- or cellulose-based capsules permit the diffusion of nutrients and output molecules while protecting the engineered cells from host immune system attacks. Moreover, these capsules serve as an interface between bodily fluids and engineered cells and enable the detection of disease-related input signals and the incorporation of cell-produced therapeutic effectors into circulation. Encapsulated cells that contain ligand-responsive gene switches have shown the ability to regulate the production of therapeutic proteins in a ligand-dependent manner in mice. Although small molecules that are not found in an organism (e.g., antibiotics, Weber et al. 2002; and drugs, Ye et al. 2013) they must be invasively administered, gene-control systems based on radio waves (Stanley et al. 2012), light (Ye et al. 2011; Folcher et al. 2014; Kim et al. 2015), food additives (Gitzinger et al. 2012; Rössger et al. 2013b), and skin cream (Gitzinger et al. 2009), providing more convenient, noninvasive options to manipulate gene expression. Accordingly, recent work has shown the use of a brain–computer interface to control the illumination of red-light-emitting diodes (LEDs) to control gene expression in living cells (Folcher et al. 2014). These engineered mammalian cells contain a red-light-responsive gene-expression system that allows for the fine-tuning of target protein production.
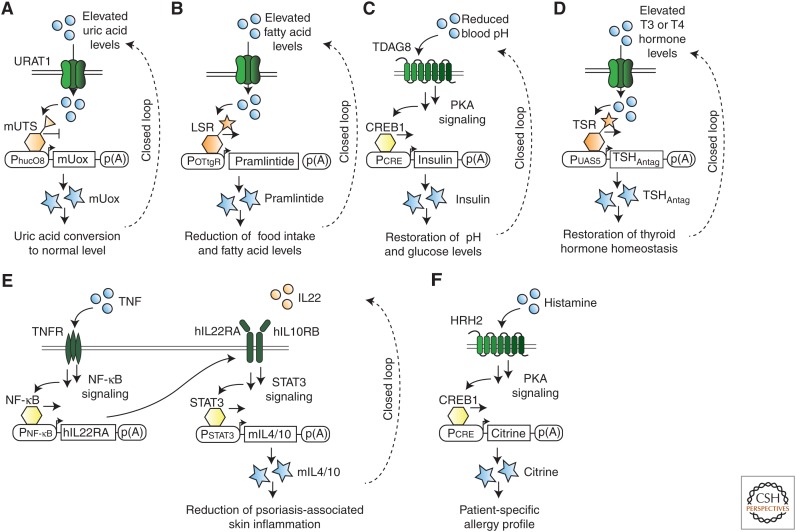
Human diseases often originate from an imbalance of metabolites, hormones, or other disease-related factors. Healthy individuals manage this balance via specified closed-loop circuits that are generally dysfunctional in sick patients. Prosthetic cell implants consist of engineered mammalian cells that can sense alterations in metabolite levels and, accordingly, that can produce a therapeutic response to counteract the disease (input) state, thus emulating closed-loop circuits (Ausländer et al. 2012b; Weber and Fussenegger 2012; Ausländer and Fussenegger 2013). Thus far, prosthetic cell implants have been developed to target (1) gouty arthritis, scoring elevated uric acid concentrations and producing a secreted version of the urate oxidase that converts uric acid to allantoin, thereby restoring uric acid homeostasis in the blood (Fig. 4A) (Kemmer et al. 2010); (2) obesity, measuring increased blood fatty acid levels and expressing the anorectic peptide hormone pramlintide, thereby reducing caloric food intake and body weight (Fig. 4B) (Rössger et al. 2013b); (3) diabetic ketoacidosis, sensing reduced blood pH levels and producing the hormone insulin, thereby restoring blood glucose and pH homeostasis (Fig. 4C) (Ausländer et al. 2014a); (4) Graves disease, sensing thyroid hormone levels and producing an engineered variant of the human thyroid-stimulating hormone (TSH), thereby restoring the thyrotrophic feedback control of the hypothalamus–pituitary–thyroid axis (Fig. 4D) (Saxena et al. 2016); and (5) psoriasis, sensing proinflammatory cytokines TNF as well as interleukin 22 (IL22) and producing anti-inflammatory cytokines IL4 and IL10 only in the presence of both input signals (AND-gate-expression logic), thereby preventing or attenuating psoriatic flares (Fig. 4E) (Schukur et al. 2015). In response to these disease states, specific therapeutic effectors are produced by the engineered cell implants to treat the respective medical condition in a closed-loop manner.
Figure 4.
Gene circuits for biomedical applications. (A) Closed-loop gene circuit designed to detect elevated uric acid levels. Uric acid is transported by URAT1 into the cell, where it binds to the engineered uric acid sensor mUTS. This interaction induces the transcription of the secreted enzyme mUox, leading to decreased uric acid levels and thereby closing the gene circuit loop. (B) Closed-loop gene circuit designed to detect elevated fatty acid levels. Fatty acids bind to the engineered fatty acid lipid-sensing receptor (LSR) and induce transcription of the secreted hormone pramlintide, leading to decreased fatty acid levels and thereby closing the gene circuit loop. (C) Closed-loop gene circuit designed to detect decreased blood pH levels. Protons bind to the G protein–coupled receptor (GPCR) TDAG8, triggering a signaling pathway that induces transcription of the secreted hormone insulin. This cascade results in restored blood pH levels, thereby closing the gene circuit loop. (D) Closed-loop gene circuit designed to detect elevated thyroid hormone levels in Graves disease. Thyroid hormones T3 and T4 are transported into the cell and bind the synthetic thyroid-sensing receptor (TSR) that induces transcription of the engineered thyroid-stimulating hormone (TSHAntag) receptor. TSHAntag competes with endogenous TSH receptor for thyroid hormone and autoantibody binding, thereby restoring the thyrotrophic feedback control of the hypothalamus–pituitary–thyroid axis. (E) Antipsoriatic cytokine converter, a closed-loop gene circuit converting the presence of proinflammatory cytokines tumor necrosis factor (TNF) and interleukin 22 (IL22) levels to production of the antiinflammatory cytokines IL4 and IL10 with AND-gate-expression logic. TNF activates the endogenous TNF receptor on the cell surface, and induces the NF-κB signaling pathway. This leads to the expression of the hIL22RA, which heterodimerizes with endogenous hIL10RB to form a functional IL22 receptor. In the presence of IL22, IL22 receptor activates the STAT3 signaling pathway that drives the expression of the secreted, antiinflammatory cytokines mIL4 and mIL10. These cytokines attenuate psoriasis-associated skin inflammations. (F) Gene circuit designed to sense blood-derived histamine. Histamine binds to the GPCR HRH2 and triggers a signaling pathway that induces the transcription of a reporter protein that enables the readout of allergic reactions using engineered mammalian cells. TNFR, Tumor necrosis factor receptor; PKA, protein kinase A.
Engineered cells are useful for both therapeutic and diagnostic applications because they are able to sense disease-relevant molecules in the physiological range. A prime example is the design of histamine-responsive mammalian cells that can be used to profile the histamine levels of allergen-treated immune effector cells in human whole blood (Fig. 4F) (Ausländer et al. 2014b). These mammalian cells have been engineered with the histamine-sensing GPCR HRH2 and with a cognate synthetic response cassette that consists of an HRH2 signaling–induced promoter and a reporter gene for simple readout. By treating human blood with a variety of allergens and by subsequently detecting immune cell–derived histamine using these engineered cells, researchers were able to generate personalized allergy profiles of human patients.
CONCLUDING REMARKS
The design of novel, synthetic gene controllers is of central importance in synthetic biology because they are the basic building blocks for the construction of complex gene circuits. Artificial gene circuits transcend the usage of mere transcriptional regulators and combine RNA- and protein-based switches to increase their complexity and functionality. Using the design of cell–cell communication systems, circuits that are more complex are envisaged to distribute complexity into simpler tasks that, when combined, form designer cell implants capable of autonomously sensing and treating multiple diseases. Engineered mammalian cells have also proven to be useful for diagnostic purposes because of their ability to detect disease-relevant molecules in the physiological range. Recent advancements in genome-editing technologies may soon enable the stable integration of multicomponent gene circuits, resulting in next-generation engineered mammalian cells with the potential to accelerate the transfer of cell-based treatment strategies into clinical settings.
ACKNOWLEDGMENTS
We thank David Ausländer for generous advice and comments on the manuscript.
Footnotes
Editors: Daniel G. Gibson, Clyde A. Hutchison III, Hamilton O. Smith, and J. Craig Venter
Additional Perspectives on Synthetic Biology available at www.cshperspectives.org
REFERENCES
- Airan RD, Thompson KR, Fenno LE, Bernstein H, Deisseroth K. 2009. Temporally precise in vivo control of intracellular signalling. Nature 458: 1025–1029. [DOI] [PubMed] [Google Scholar]
- Ausländer S, Fussenegger M. 2013. From gene switches to mammalian designer cells: Present and future prospects. Trends Biotechnol 31: 155–168. [DOI] [PubMed] [Google Scholar]
- Ausländer S, Ketzer P, Hartig JS. 2010. A ligand-dependent hammerhead ribozyme switch for controlling mammalian gene expression. Mol Biosyst 6: 807–814. [DOI] [PubMed] [Google Scholar]
- Ausländer D, Wieland M, Ausländer S, Tigges M, Fussenegger M. 2011. Rational design of a small molecule-responsive intramer controlling transgene expression in mammalian cells. Nucleic Acids Res 39: e155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausländer S, Ausländer D, Müller M, Wieland M, Fussenegger M. 2012a. Programmable single-cell mammalian biocomputers. Nature 487: 123–127. [DOI] [PubMed] [Google Scholar]
- Ausländer S, Wieland M, Fussenegger M. 2012b. Smart medication through combination of synthetic biology and cell microencapsulation. Metab Eng 14: 252–260. [DOI] [PubMed] [Google Scholar]
- Ausländer D, Ausländer S, Charpin-El Hamri G, Sedlmayer F, Müller M, Frey O, Hierlemann A, Stelling J, Fussenegger M. 2014a. A synthetic multifunctional mammalian pH sensor and CO2 transgene-control device. Mol Cell 55: 397–408. [DOI] [PubMed] [Google Scholar]
- Ausländer D, Eggerschwiler B, Kemmer C, Geering B, Ausländer S, Fussenegger M. 2014b. A designer cell-based histamine-specific human allergy profiler. Nat Commun 5: 4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausländer S, Stucheli P, Rehm C, Ausländer D, Hartig JS, Fussenegger M. 2014c. A general design strategy for protein-responsive riboswitches in mammalian cells. Nat Method 11: 1154–1160. [DOI] [PubMed] [Google Scholar]
- Bacchus W, Lang M, El-Baba MD, Weber W, Stelling J, Fussenegger M. 2012. Synthetic two-way communication between mammalian cells. Nat Biotechnol 30: 991–996. [DOI] [PubMed] [Google Scholar]
- Bacchus W, Weber W, Fussenegger M. 2013. Increasing the dynamic control space of mammalian transcription devices by combinatorial assembly of homologous regulatory elements from different bacterial species. Metab Eng 15: 144–150. [DOI] [PubMed] [Google Scholar]
- Beerli RR, Schopfer U, Dreier B, Barbas CF III. 2000. Chemically regulated zinc finger transcription factors. J Biol Chem 275: 32617–32627. [DOI] [PubMed] [Google Scholar]
- Benenson Y. 2012. Biomolecular computing systems: Principles, progress and potential. Nat Rev Genet 13: 455–468. [DOI] [PubMed] [Google Scholar]
- Burrill DR, Inniss MC, Boyle PM, Silver PA. 2012. Synthetic memory circuits for tracking human cell fate. Genes Dev 26: 1486–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culler SJ, Hoff KG, Smolke CD. 2010. Reprogramming cellular behavior with RNA controllers responsive to endogenous proteins. Science 330: 1251–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuthbertson L, Nodwell JR. 2013. The TetR family of regulators. Microbiol Mol Biol Rev 77: 440–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deans TL, Cantor CR, Collins JJ. 2007. A tunable genetic switch based on RNAi and repressor proteins for regulating gene expression in mammalian cells. Cell 130: 363–372. [DOI] [PubMed] [Google Scholar]
- Doudna JA, Charpentier E. 2014. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346: 1258096. [DOI] [PubMed] [Google Scholar]
- Endo K, Hayashi K, Inoue T, Saito H. 2013. A versatile cis-acting inverter module for synthetic translational switches. Nat Commun 4: 2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Famulok M, Hartig JS, Mayer G. 2007. Functional aptamers and aptazymes in biotechnology, diagnostics, and therapy. Chem Rev 107: 3715–3743. [DOI] [PubMed] [Google Scholar]
- Folcher M, Oesterle S, Zwicky K, Thekkottil T, Heymoz J, Hohmann M, Christen M, Daoud El-Baba M, Buchmann P, Fussenegger M. 2014. Mind-controlled transgene expression by a wireless-powered optogenetic designer cell implant. Nat Commun 5: 5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fussenegger M, Morris RP, Fux C, Rimann M, von Stockar B, Thompson CJ, Bailey JE. 2000. Streptogramin-based gene regulation systems for mammalian cells. Nat Biotechnol 18: 1203–1208. [DOI] [PubMed] [Google Scholar]
- Gaj T, Gersbach CA, Barbas CF III. 2013. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol 31: 397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitzinger M, Kemmer C, El-Baba MD, Weber W, Fussenegger M. 2009. Controlling transgene expression in subcutaneous implants using a skin lotion containing the apple metabolite phloretin. Proc Natl Acad Sci 106: 10638–10643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitzinger M, Kemmer C, Fluri DA, El-Baba MD, Weber W, Fussenegger M. 2012. The food additive vanillic acid controls transgene expression in mammalian cells and mice. Nucleic Acids Res 40: e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfless SJ, Belmont BJ, de Paz AM, Liu JF, Niles JC. 2012. Direct and specific chemical control of eukaryotic translation with a synthetic RNA-protein interaction. Nucleic Acids Res 40: e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen M, Bujard H. 1992. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci 89: 5547–5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabow WW, Jaeger L. 2014. RNA self-assembly and RNA nanotechnology. Acc Chem Res 47: 1871–1880. [DOI] [PubMed] [Google Scholar]
- Greber D, Fussenegger M. 2010. An engineered mammalian band-pass network. Nucleic Acids Res 38: e174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartenbach S, Daoud-El Baba M, Weber W, Fussenegger M. 2007. An engineered l-arginine sensor of Chlamydia pneumoniae enables arginine-adjustable transcription control in mammalian cells and mice. Nucleic Acids Res 35: e136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng BC, Aubel D, Fussenegger M. 2013. An overview of the diverse roles of G-protein coupled receptors (GPCRs) in the pathophysiology of various human diseases. Biotechnol Adv 31: 1676–1694. [DOI] [PubMed] [Google Scholar]
- Hsu PD, Lander ES, Zhang F. 2014. Development and applications of CRISPR-Cas9 for genome engineering. Cell 157: 1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashida S, Inoue T, Saito H. 2012. Three-dimensionally designed protein-responsive RNA devices for cell signaling regulation. Nucleic Acids Res 40: 9369–9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemmer C, Gitzinger M, Daoud-El Baba M, Djonov V, Stelling J, Fussenegger M. 2010. Self-sufficient control of urate homeostasis in mice by a synthetic circuit. Nat Biotechnol 28: 355–360. [DOI] [PubMed] [Google Scholar]
- Kemmer C, Fluri DA, Witschi U, Passeraub A, Gutzwiller A, Fussenegger M. 2011. A designer network coordinating bovine artificial insemination by ovulation-triggered release of implanted sperms. J Control Release 150: 23–29. [DOI] [PubMed] [Google Scholar]
- Kim T, Folcher M, Doaud-El Baba M, Fussenegger M. 2015. A synthetic erectile optogenetic stimulator enabling blue-light-inducible penile erection. Angew Chem Int Ed Engl 54: 5933–5938. [DOI] [PubMed] [Google Scholar]
- Konermann S, Brigham MD, Trevino AE, Hsu PD, Heidenreich M, Cong L, Platt RJ, Scott DA, Church GM, Zhang F. 2013. Optical control of mammalian endogenous transcription and epigenetic states. Nature 500: 472–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer BP, Fussenegger M. 2005. Hysteresis in a synthetic mammalian gene network. Proc Natl Acad Sci 102: 9517–9522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer BP, Fischer C, Fussenegger M. 2004. BioLogic gates enable logical transcription control in mammalian cells. Biotechnol Bioeng 87: 478–484. [DOI] [PubMed] [Google Scholar]
- Kumar D, An CI, Yokobayashi Y. 2009. Conditional RNA interference mediated by allosteric ribozyme. J Am Chem Soc 131: 13906–13907. [DOI] [PubMed] [Google Scholar]
- Lapique N, Benenson Y. 2014. Digital switching in a biosensor circuit via programmable timing of gene availability. Nat Chem Biol 10: 1020–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leisner M, Bleris L, Lohmueller J, Xie Z, Benenson Y. 2010. Rationally designed logic integration of regulatory signals in mammalian cells. Nat Nanotechnol 5: 666–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lienert F, Torella JP, Chen JH, Norsworthy M, Richardson RR, Silver PA. 2013. Two- and three-input TALE-based AND logic computation in embryonic stem cells. Nucleic Acids Res 41: 9967–9975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmueller JJ, Armel TZ, Silver PA. 2012. A tunable zinc finger-based framework for Boolean logic computation in mammalian cells. Nucleic Acids Res 40: 5180–5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malyshev DA, Dhami K, Lavergne T, Chen T, Dai N, Foster JM, Correa IR Jr, Romesberg FE. 2014. A semi-synthetic organism with an expanded genetic alphabet. Nature 509: 385–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandell DJ, Lajoie MJ, Mee MT, Takeuchi R, Kuznetsov G, Norville JE, Gregg CJ, Stoddard BL, Church GM. 2015. Biocontainment of genetically modified organisms by synthetic protein design. Nature 518: 55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer AC, Gaj T, Sirk SJ, Lamb BM, Barbas CF 3rd. 2014. Regulation of endogenous human gene expression by ligand-inducible TALE transcription factors. ACS Synth Biol 3: 723–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal V. 2004. Improving the efficiency of RNA interference in mammals. Nat Rev Genet 5: 355–365. [DOI] [PubMed] [Google Scholar]
- Müller M, Ausländer S, Ausländer D, Kemmer C, Fussenegger M. 2012. A novel reporter system for bacterial and mammalian cells based on the non-ribosomal peptide indigoidine. Met Eng 14: 325–335. [DOI] [PubMed] [Google Scholar]
- Müller K, Engesser R, Metzger S, Schulz S, Kampf MM, Busacker M, Steinberg T, Tomakidi P, Ehrbar M, Nagy F, et al. 2013a. A red/far-red light-responsive bi-stable toggle switch to control gene expression in mammalian cells. Nucleic Acids Res 41: e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller K, Engesser R, Schulz S, Steinberg T, Tomakidi P, Weber CC, Ulm R, Timmer J, Zurbriggen MD, Weber W. 2013b. Multi-chromatic control of mammalian gene expression and signaling. Nucleic Acids Res 41: e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neddermann P, Gargioli C, Muraglia E, Sambucini S, Bonelli F, De Francesco R, Cortese R. 2003. A novel, inducible, eukaryotic gene expression system based on the quorum-sensing transcription factor TraR. EMBO Rep 4: 159–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nern A, Pfeiffer BD, Svoboda K, Rubin GM. 2011. Multiple new site-specific recombinases for use in manipulating animal genomes. Proc Natl Acad Sci 108: 14198–14203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissim L, Bar-Ziv RH. 2010. A tunable dual-promoter integrator for targeting of cancer cells. Mol Syst Biol 6: 444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura Y, Kumar D, Yokobayashi Y. 2012. Synthetic mammalian riboswitches based on guanine aptazyme. Chem Commun (Camb) 48: 7215–7217. [DOI] [PubMed] [Google Scholar]
- Nomura Y, Zhou L, Miu A, Yokobayashi Y. 2013. Controlling mammalian gene expression by allosteric hepatitis delta virus ribozymes. ACS Synth Biol 2: 684–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JS, Rhau B, Hermann A, McNally KA, Zhou C, Gong D, Weiner OD, Conklin BR, Onuffer J, Lim WA. 2014. Synthetic control of mammalian-cell motility by engineering chemotaxis to an orthogonal bioinert chemical signal. Proc Natl Acad Sci 111: 5896–5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polstein LR, Gersbach CA. 2012. Light-inducible spatiotemporal control of gene activation by customizable zinc finger transcription factors. J Am Chem Soc 134: 16480–16483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polstein LR, Gersbach CA. 2015. A light-inducible CRISPR-Cas9 system for control of endogenous gene activation. Nat Chem Biol 11: 198–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prochazka L, Angelici B, Haefliger B, Benenson Y. 2014. Highly modular bow-tie gene circuits with programmable dynamic behaviour. Nat Commun 5: 4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos JL, Martinez-Bueno M, Molina-Henares AJ, Teran W, Watanabe K, Zhang X, Gallegos MT, Brennan R, Tobes R. 2005. The TetR family of transcriptional repressors. Microbiol Mol Biol Rev 69: 326–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaudo K, Bleris L, Maddamsetti R, Subramanian S, Weiss R, Benenson Y. 2007. A universal RNAi-based logic evaluator that operates in mammalian cells. Nat Biotechnol 25: 795–801. [DOI] [PubMed] [Google Scholar]
- Rössger K, Charpin-El Hamri G, Fussenegger M. 2013a. Reward-based hypertension control by a synthetic brain-dopamine interface. Proc Natl Acad Sci 110: 18150–18155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rössger K, Charpin-El-Hamri G, Fussenegger M. 2013b. A closed-loop synthetic gene circuit for the treatment of diet-induced obesity in mice. Nat Commun 4: 2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovner AJ, Haimovich AD, Katz SR, Li Z, Grome MW, Gassaway BM, Amiram M, Patel JR, Gallagher RR, Rinehart J, et al. 2015. Recoded organisms engineered to depend on synthetic amino acids. Nature 518: 89–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito H, Inoue T. 2009. Synthetic biology with RNA motifs. Int J Biochem Cell Biol 41: 398–404. [DOI] [PubMed] [Google Scholar]
- Saito H, Kobayashi T, Hara T, Fujita Y, Hayashi K, Furushima R, Inoue T. 2010. Synthetic translational regulation by an L7Ae–kink-turn RNP switch. Nat Chem Biol 6: 71–78. [DOI] [PubMed] [Google Scholar]
- Saito H, Fujita Y, Kashida S, Hayashi K, Inoue T. 2011. Synthetic human cell fate regulation by protein-driven RNA switches. Nat Commun 2: 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena P, Charpin-El Hamri G, Folcher M, Zulewski H, Fussenegger M. 2016. Synthetic gene network restoring endogenous pituitary-thyroid feedback control in experimental Graves’ disease. Proc Natl Acad Sci 113: 1244–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schukur L, Geering B, Charpin-El Hamri G, Fussenegger M. 2015. Implantable synthetic cytokine converter cells with AND-gate logic treat experimental psoriasis. Sci Transl Med 7: 318ra201. [DOI] [PubMed] [Google Scholar]
- Shechner DM, Hacisuleyman E, Younger ST, Rinn JL. 2015. Multiplexable, locus-specific targeting of long RNAs with CRISPR-Display. Nat Methods 12: 664–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley SA, Gagner JE, Damanpour S, Yoshida M, Dordick JS, Friedman JM. 2012. Radio-wave heating of iron oxide nanoparticles can regulate plasma glucose in mice. Science 336: 604–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton JA, Endo K, Fujita Y, Hayashi K, Takinoue M, Saito H, Inoue T. 2012. Feedback control of protein expression in mammalian cells by tunable synthetic translational inhibition. ACS Synth Biol 1: 83–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tigges M, Marquez-Lago TT, Stelling J, Fussenegger M. 2009. A tunable synthetic mammalian oscillator. Nature 457: 309–312. [DOI] [PubMed] [Google Scholar]
- Urban DJ, Roth BL. 2015. DREADDs (designer receptors exclusively activated by designer drugs): Chemogenetic tools with therapeutic utility. Annu Rev Pharmacol Toxicol 55: 399–417. [DOI] [PubMed] [Google Scholar]
- Urlinger S, Baron U, Thellmann M, Hasan MT, Bujard H, Hillen W. 2000. Exploring the sequence space for tetracycline-dependent transcriptional activators: Novel mutations yield expanded range and sensitivity. Proc Natl Acad Sci 97: 7963–7968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber W, Fussenegger M. 2012. Emerging biomedical applications of synthetic biology. Nat Rev Genet 13: 21–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber W, Fux C, Daoud-el Baba M, Keller B, Weber CC, Kramer BP, Heinzen C, Aubel D, Bailey JE, Fussenegger M. 2002. Macrolide-based transgene control in mammalian cells and mice. Nat Biotechnol 20: 901–907. [DOI] [PubMed] [Google Scholar]
- Weber W, Schoenmakers R, Spielmann M, El-Baba MD, Folcher M, Keller B, Weber CC, Link N, van de Wetering P, Heinzen C, et al. 2003. Streptomyces-derived quorum-sensing systems engineered for adjustable transgene expression in mammalian cells and mice. Nucleic Acids Res 31: e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber W, Link N, Fussenegger M. 2006. A genetic redox sensor for mammalian cells. Metab Eng 8: 273–280. [DOI] [PubMed] [Google Scholar]
- Weber W, Kramer BP, Fussenegger M. 2007. A genetic time-delay circuitry in mammalian cells. Biotechnol Bioeng 98: 894–902. [DOI] [PubMed] [Google Scholar]
- Weber W, Lienhart C, Baba MD, Fussenegger M. 2009. A biotin-triggered genetic switch in mammalian cells and mice. Metab Eng 11: 117–124. [DOI] [PubMed] [Google Scholar]
- Wess J, Nakajima K, Jain S. 2013. Novel designer receptors to probe GPCR signaling and physiology. Trends Pharmacol Sci 34: 385–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland M, Fussenegger M. 2010. Ligand-dependent regulatory RNA parts for synthetic biology in eukaryotes. Curr Opin Biotechnol 21: 760–765. [DOI] [PubMed] [Google Scholar]
- Wieland M, Fussenegger M. 2012. Engineering molecular circuits using synthetic biology in mammalian cells. Annu Rev Chem Biomol Eng 3: 209–234. [DOI] [PubMed] [Google Scholar]
- Wieland M, Ausländer D, Fussenegger M. 2012. Engineering of ribozyme-based riboswitches for mammalian cells. Methods 56: 351–357. [DOI] [PubMed] [Google Scholar]
- Win MN, Smolke CD. 2007. A modular and extensible RNA-based gene-regulatory platform for engineering cellular function. Proc Natl Acad Sci 104: 14283–14288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Win MN, Smolke CD. 2008. Higher-order cellular information processing with synthetic RNA devices. Science 322: 456–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunnicke D, Strohbach D, Weigand JE, Appel B, Feresin E, Suess B, Müller S, Steinhoff HJ. 2011. Ligand-induced conformational capture of a synthetic tetracycline riboswitch revealed by pulse EPR. RNA 17: 182–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Wroblewska L, Prochazka L, Weiss R, Benenson Y. 2011. Multi-input RNAi-based logic circuit for identification of specific cancer cells. Science 333: 1307–1311. [DOI] [PubMed] [Google Scholar]
- Xie M, Ye H, Hamri GC, Fussenegger M. 2014. Antagonistic control of a dual-input mammalian gene switch by food additives. Nucleic Acids Res 42: e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H, Daoud-El Baba M, Peng RW, Fussenegger M. 2011. A synthetic optogenetic transcription device enhances blood-glucose homeostasis in mice. Science 332: 1565–1568. [DOI] [PubMed] [Google Scholar]
- Ye H, Charpin-El Hamri G, Zwicky K, Christen M, Folcher M, Fussenegger M. 2013. Pharmaceutically controlled designer circuit for the treatment of the metabolic syndrome. Proc Natl Acad Sci 110: 141–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalatan JG, Lee ME, Almeida R, Gilbert LA, Whitehead EH, La Russa M, Tsai JC, Weissman JS, Dueber JE, Qi LS, et al. 2015. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell 160: 339–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, Volz SE, Zhang F. 2015. A split-Cas9 architecture for inducible genome editing and transcription modulation. Nat Biotechnol 33: 139–142. [DOI] [PMC free article] [PubMed] [Google Scholar]