Abstract
Transforming growth factors (TGFs) were discovered as activities that were secreted by cancer cells, and later by normal cells, and had the ability to phenotypically and reversibly transform immortalized fibroblasts. TGF-β distinguished itself from TGF-α because it did not bind to the same epidermal growth factor (EGF) receptor as TGF-α and, therefore, acted through different cell-surface receptors and signaling mediators. This review summarizes the discovery of TGF-β, the early developments in its molecular and biological characterization with its many biological activities in different cell and tissue contexts and its roles in disease, the realization that there is a family of secreted TGF-β-related proteins with many differentiation functions in development and activities in normal cell and tissue physiology, and the subsequent identification and characterization of the receptors and effectors that mediate TGF-β family signaling responses.
Pioneering work in the early 1980s led to the discovery of a remarkable range of biological activities for the TGF-β family of proteins and to the characterization of the receptors and signaling factors involved.
HISTORICAL CONTEXT
In the early 1970s, George Todaro and Robert Huebner, studying the transforming activity of C-type RNA tumor viruses, proposed the viral oncogene hypothesis, that is, that “oncogenes” were an important component of the virogene of RNA tumor viruses, whose expression was suppressed in “normal” cells, and that treatment of cells with carcinogens, mutagens, or radiation could activate expression of this viral information by inactivating a repressor (Todaro and Huebner 1972). A few years later, Michael Bishop and Harold Varmus, also studying the effects of RNA tumor viruses on malignant transformation, made the Nobel Prize–winning breakthrough that neoplastic transformation of a cell by the avian sarcoma virus was mediated by a single viral gene ([src] the “oncogene”), and most significantly that the product of the src gene, now known to be a cellular signaling mediator of many tyrosine kinase receptors, was a slightly modified analog of a normal cellular protein (Stehelin et al. 1976; Levinson et al. 1978). A few years later, they showed that the transforming retroviral oncogene, v-src, arose by transduction of the cellular c-src gene (Swanstrom et al. 1983).
Around the same time, based on results from a collaboration with the Nobel laureate Stanley Cohen, George Todaro suggested that murine and feline tumor viruses transform cells by inducing cells to secrete a molecule functionally related to epidermal growth factor (EGF), which acted on the same cells to affect their transformation (Todaro et al. 1976) through a process later termed autocrine secretion (Sporn and Todaro 1980). Along similar lines, the late Nobel laureate Robert Holley had proposed that one mechanism of acquisition of malignancy involved an increase in the availability of specific hormone (or growth factor) receptor sites on the cell membrane (Holley 1972) and that “transformed or malignant cells escape from normal growth controls by requiring less of such hormones or growth factors” (Holley 1975). The transforming growth factors (TGFs) described by De Larco and Todaro (1978) shared properties with the v-src gene, in that they were able to cause normal fibroblasts to form progressively growing colonies in soft agar (anchorage-independent growth), a property that was closely associated with the transformed phenotype in vivo (Todaro et al. 1979). In contrast with v-src and other oncogenes, however, transformation did not result from cell-intrinsic, genetic changes, but from secreted factors that did not affect the genotype. The term TGF was chosen for this activity because of the induction of a transformed phenotype, in both monolayer and soft agar cultures, of nontransformed cells, and because of the assumption, later proven to be incorrect (see below), that the factor was produced only by transformed (cancer) cells. This behavior was reversible on removal of the source of TGFs, leading to the suggestion that these factors acted as proximal effectors of transformation. Considering the highly visible discovery of oncogenes as the genetic basis of malignant transformation around that time, the notion of reversible transformation by secreted factors was met with skepticism.
DISCOVERY OF TGF-β
The TGF-β field began with three papers, one published in 1978 and two in 1981. First, De Larco and Todaro (1978) described the partial purification of polypeptide growth factors secreted by fibroblasts transformed by an RNA virus, Moloney sarcoma virus (MSV), which they called sarcoma growth factor (SGF). Although SGF bound to EGF receptors, it was distinguished from EGF (suggested to be the nontransforming product of “normal” cells) by its ability to transform normal fibroblasts in vitro in the soft agar colony-forming assay. That the secreted factor both bound to EGF receptors and could induce transformation was based on co-elution of these two activities using molecular sieve chromatography (De Larco and Todaro 1978). In 1981, however, it became apparent through the work in the Harold Moses laboratory, then at the Mayo Clinic (Moses et al. 1981), and the Michael Sporn and Anita Roberts laboratory at the National Cancer Institute (Roberts et al. 1981), that the EGF receptor competing activity could be separated from the soft agar colony-forming activity. Both groups pursued purification and characterization of factors with seemingly different transforming properties, dependent on the different cell lines used in their assays. These factors turned out to be the same TGF-β.
At the present time, TGF-β is seen as the prototype of a large family of secreted, dimeric growth factors and cytokines, encoded in humans and mice by 33 genes. TGF-β family proteins are now known to be critically involved in embryonic development, cell and tissue differentiation, tissue homeostasis in the adult, and in many disease states, as detailed in at least 70,000 publications at this point. This review will cover the early history of TGF-β in the context of general scientific knowledge at the time.
PURIFICATION AND CHARACTERIZATION OF TGF-β
Approach of the Sporn/Roberts Laboratory
In the late 1970s, the Sporn laboratory focused on the role of retinoids as chemopreventive agents. Based on the identification by the Todaro group of peptides thought to be proximal effectors of transformation, the two laboratories entered a collaboration to design a large-scale isolation of the transforming peptides with the ultimate goal of obtaining insights into the mechanisms of chemoprevention attributed to the retinoids. Because previous attempts at purification of SGF were based on the use of serum-free medium conditioned by MSV-transformed cells whose quantity was rather limited, the first breakthrough was the identification of a method that could be used to extract SGF directly from tumor cells. Using an acid–ethanol extraction procedure, previously described for the isolation of insulin from tissues and blood (Davoren 1962), and the formation of colonies in soft agar of “normal” NRK-2B clone 49F (NRK) fibroblasts as an assay, SGF-like activity was isolated from a variety of both virally or chemically transformed cells and tumors (Roberts et al. 1980). In attempts to purify SGF to homogeneity by successive chromatographic steps, it was noted, as mentioned, that the soft agar colony-forming response of NRK cells was lost, but could be restored by adding EGF to the assay (Roberts et al. 1981; Anzano et al. 1982). This led to the important finding that SGF, as well as colony-forming activities with similar biochemical properties isolated from a variety of non-neoplastic mouse, bovine, and human tissues, were actually comprised of two synergizing activities. One of these provided EGF receptor binding and was subsequently assigned to TGF-α, a member of the EGF family of growth factors, whereas the other one, subsequently named TGF-β, did not bind to EGF receptors, but was required to induce phenotypic transformation in combination with EGF. An interesting anecdote is that the assignment of “α” and “β” to these two TGFs was based on Todaro’s insistence that TGF-α, which he thought was uniquely expressed by tumor cells, represented the dominant transforming activity and that TGF-β, then known to be expressed in many normal tissues, served merely to modulate the process.
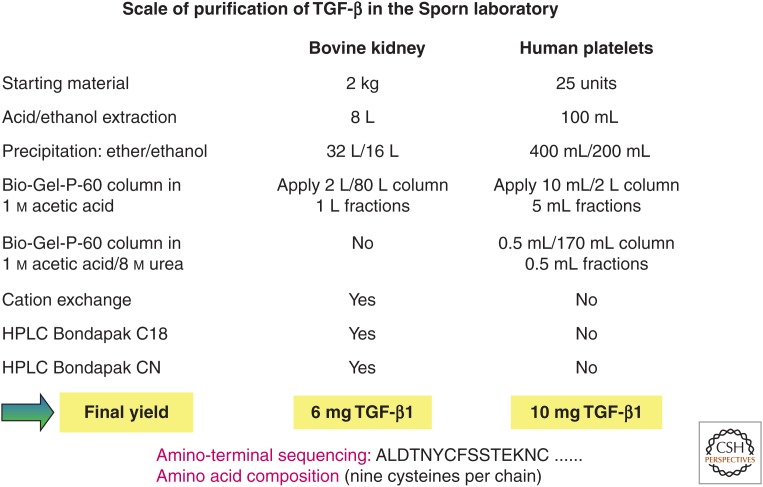
Advice from the late Nobel laureate Christian Anfinsen was to “think big” so that a purification scheme would ultimately yield sufficient quantities of material for amino acid sequencing and not simply show an “activity.” This was especially relevant because the soft agar colony-forming assay was 100-fold more sensitive than the detection of TGF-β protein on a silver-stained gel. Based on this advice, the Sporn/Roberts laboratory used normal tissues, including bovine kidneys, human placenta, and platelets, all available in larger quantities than transformed cells, for purification of TGF-β. The formation of colonies of NRK cells in soft agar in the presence of EGF (typically a 7-day assay) was used to monitor the purification, with image analysis to quantify the number of colonies above a designated size limit. Purification from bovine kidneys (Roberts et al. 1983a) or human placenta (Frolik et al. 1983) required very large-scale preparations and several chromatography steps (Fig. 1), whereas purification from human platelets (Assoian et al. 1983), which contain ∼100-fold greater activity, required only two chromatography steps to achieve homogeneity (Fig. 1). The colony-forming activity in the assay depended on the addition of EGF, confirming that these TGFs met the definition of a TGF-β used by this laboratory. The ED50 (half-maximal effective dose) of purified TGF-β in the NRK colony-forming assay was 2–3 pm in the presence of EGF. Significantly, the finding that TGF-β could be isolated from non-neoplastic tissues and especially from platelets (Childs et al. 1982; Assoian et al. 1983), strongly suggested normal physiological roles for this molecule, as subsequently shown in wound healing (Sporn et al. 1983) and development (Heine et al. 1987), and later in diseases such as fibrosis and cancer.
Figure 1.
Purification to homogeneity of transforming growth factor β (TGF-β) from bovine kidney and human platelets. Human platelets contain ∼100-fold greater amounts of TGF-β than most other normal tissues. The scale of purification from bovine kidneys (or placenta) was large, requiring volumes of solvents unlikely to be used in laboratories today, and requiring four chromatography steps. In contrast, purification of TGF-β to homogeneity from human platelets was achieved using just two chromatography steps on Bio-Gel P-60. These large-scale purification schemes enabled the first determination of the amino-terminal sequence and amino acid composition of TGF-β1. HPCL, High-performance liquid chromatography.
Approach of the Moses Laboratory
In the late 1970s, the Moses group was interested in the molecular changes associated with transformation of cells by chemical carcinogens. As was typical at the time, murine embryonic fibroblast cell lines served as model systems; methods for culturing nontransformed epithelial cells were not yet widely available. The Moses laboratory had shown that many of the phenotypic alterations observed in the chemically transformed AKR-MCA and C3H/MCA-58 cell lines could be accounted for by continuous growth factor stimulation (Moses et al. 1978, 1979). His group further found that the transformed cells had greatly diminished detectable EGF receptors relative to the nontransformed precursor cells, similar to the observations of De Larco and Todaro (1978) with MSV-transformed cells. As a result, the Moses laboratory pursued identification of SGF-like transforming activities by the chemically transformed cells. They used nontransformed AKR-2B fibroblasts, the progenitors of the chemically transformed AKR-MCA cells, as indicator cells, and this was different from the assays used by the Todaro and Sporn/Roberts laboratories, which used rat NRK fibroblasts. Soft agar–stimulating activity with a molecular sieve chromatography profile similar to that reported by De Larco and Todaro was detected in medium conditioned by the chemically transformed cells (Moses et al. 1981). Additional purification using ion exchange chromatography separated the EGF receptor–competing activity from soft agar growth–stimulating activity. However, in contrast to the findings of the Roberts group using NRK cells (Roberts et al. 1981), addition of EGF in the soft agar assay was not required to obtain maximum colony–stimulating activity of AKR-2B cells. This caused confusion leading to the assumption that the TGF identified by the Moses group might be different from that identified by the Sporn group, even to the extent that three types of TGFs were proposed to exist, that is, TGF-α, TGF-β, and TGF-γ (Roberts et al. 1983b). However, with exchange of reagents between the two laboratories, it was soon apparent that the requirement for EGF in the transformation assay reflected a difference among the reporter cell lines, with EGF needed for stimulation of colony formation by NRK but not for AKR-2B cells.
The Moses group proceeded to identify TGF-β activity in mouse embryos (Proper et al. 1982) and, more important, in serum (Childs et al. 1982). It was further shown that plasma lacked TGF-β activity and that the TGF activity in serum derived from platelets, which are an abundant source for purification of TGF-β (Childs et al. 1982). Platelets, as well as recombinant protein, are now the source of much of the commercially marketed TGF-β1.
Competitive Binding Assays Identify High-Affinity Cell-Surface Receptors
The first evidence for a high-affinity TGF-β receptor was provided by Joan Massagué in collaboration with Todaro (Massagué et al. 1982). Using preparations of SGF that were later shown to contain both TGF-β and TGF-α, they identified, by affinity cross-linking, a high-affinity 60-kD band, in addition to the 140- to 170-kD EGF receptor. Unlabeled EGF competed with 125I-labeled SGF binding to the latter band, but not the 60-kD band. The first competitive binding assays with Scatchard analyses for TGF-β receptors were reported in 1984 by the Sporn/Roberts and Moses laboratories working independently (Frolik et al. 1984; Tucker et al. 1984a). Both groups found very high-affinity binding sites, with dissociation constants of 25–40 pm and ∼10–30 receptors per cell. Subsequently, Massagué performed competitive binding assays with similar results. Using affinity cross-linking, his laboratory showed that there were three TGF-β binding species that are now known as the type I and type II receptors, and the type III (co)receptor, better known as betaglycan (Massagué and Like 1985). The development of competitive binding assays for TGF-β was critical in the demonstration that TGF-βs are inhibitors of cell proliferation, particularly in epithelial cells.
In addition to the type I and type II receptors, and the co-receptor betaglycan, other putative TGF-β receptors were reported, including a 70- to 74-kD band identified by 125I-TGF-β affinity cross-linking in GH3 pituitary cells, which was reported to interact with TGF-β, activin, and inhibin (Cheifetz et al. 1988). A 38-kD binding component was identified by similar means on choriocarcinoma cells (Mitchell et al. 1992). These and other putative TGF-β-binding “receptors” received little attention subsequent to the initial reports. On the other hand, the cell membrane TGF-β-binding component called TGF-β type V receptor was later identified as the low-density lipoprotein receptor-related protein (LRP-1) (Huang and Huang 2005). Additionally, the endothelial cell-surface protein endoglin was also shown to act as a TGF-β-binding co-receptor, of particular importance in endothelial cells (Cheifetz et al. 1992).
DISCOVERY OF THE GROWTH-INHIBITORY EFFECTS OF TGF-β
The discovery of growth inhibition by TGF-β began with a publication in 1978 by Robert Holley (Holley et al. 1978). Following receipt of the Nobel Prize in Physiology or Medicine in 1968 for work on transfer RNA and the genetic code, Holley moved to the Salk Institute and redirected his efforts to growth control. He showed that BSC-1 cells, derived from African green monkey kidney tissue, secreted a peptide growth inhibitor, and proceeded to purify this growth inhibitor from medium conditioned by the BSC-1 cells. By early 1984, the Moses laboratory had derived data indicating that purified platelet-derived TGF-β could inhibit proliferation of AKR-2B cells in monolayer culture. After witnessing at a seminar that the BSC-1 growth inhibitor presented itself on a sodium dodecyl sulfate (SDS)-polyacrylamide gel as a 25-kD protein band under nonreducing conditions and a 12.5-kD band under reducing conditions, the Moses group hypothesized that the BSC-1 growth inhibitor may well be the same as TGF-β. A collaboration was established with Holley resulting in the exchange of reagents. It was shown that platelet-derived TGF-β and BSC-1 growth inhibitor had similar biological activities in stimulating colony formation of AKR-2B cells in soft agar, inhibiting proliferation of AKR-2B cells as well as BSC-1 and Mv1Lu cells in monolayer culture, and competing comparably for binding of TGF-β in the TGF-β receptor assay (Tucker et al. 1984b; Moses et al. 1985).
The Sporn/Roberts group at this same time had noted decreased proliferation of a number of cancer cell lines in soft agar with the addition of TGF-β, assuming that this decrease was the result of toxicity related to impurities in the TGF-β preparation. Following the presentation of some of the growth-inhibition data by Moses at a national meeting in the spring of 1984, the Sporn/Roberts group concluded that the decreased cell proliferation was the result of growth inhibition by TGF-β (Roberts et al. 1985). They also showed that the effects of TGF-β on anchorage-independent growth of cells could be modulated by other growth factors. As an example, Myc-1 cells formed large colonies in the presence of EGF, and formation of these colonies was strongly inhibited by addition of TGF-β. In contrast, identical concentrations of TGF-β enhanced colony formation of these same cells, when in the presence of platelet-derived growth factor (PDGF), which by itself had almost no colony-forming activity. Moreover, in monolayer culture, TGF-β inhibited the growth-promoting effects of PDGF on these same cells, emphasizing the highly contextual effects of TGF-β (Roberts et al. 1985).
It was then also important to evaluate whether TGF-β could inhibit the proliferation of normal cells in culture. Primary cultures of human foreskin keratinocytes were shown to be growth inhibited by TGF-β in a reversible manner, and to be arrested in the G1 phase of the cell cycle. In contrast, a squamous cell carcinoma cell line, derived from epithelial cells, was not inhibited by TGF-β (Shipley et al. 1986). Additionally, the Sporn/Roberts and Moses laboratories showed that several carcinoma cell lines were growth-inhibited by TGF-β, whereas others were not (Moses et al. 1985; Roberts et al. 1985). Numerous subsequent studies from several laboratories showed that TGF-β could inhibit a wide variety of epithelial cell types in culture, and that, in general, highly malignant carcinoma cells had lost the growth-inhibitory response. The notion that a growth factor could inhibit cell proliferation was at that time surprising and counterintuitive. Indeed, numerous growth factors had been identified in the 1970s and early 1980s, based on their abilities to stimulate cell proliferation, thus equating growth factors with growth stimulation. Additionally, transformation was inherently linked to increased cell proliferation. The notion of autocrine control of growth inhibition was novel.
The first demonstration that TGF-β could inhibit proliferation of epithelial cells in vivo was by Silberstein and Daniel (1987). They inserted slow-release pellets containing TGF-β into developing mouse mammary glands and observed marked inhibition of mammary ductal growth and morphogenesis. They further showed that this growth inhibition was reversible. Subsequently, systemically administered TGF-β1 was shown to inhibit the proliferative response of liver to partial hepatectomy (Russell et al. 1988). Additionally, transgenic mice overexpressing TGF-β1 in epithelial tissues also showed inhibition of proliferation in the mammary gland (Jhappan et al. 1993; Pierce et al. 1993) or skin (Cui et al. 1996; Fowlis et al. 1996). Numerous later studies involving transgenic expression of TGF-β, dominant-negative type II TGF-β receptors and conditional knockout of type II receptor expression supported the role of TGF-β as an endogenous negative regulator of proliferation of several cell types, including epithelial, endothelial, and hematopoietic cells.
By the late 1980s, many groups suspected that, even though TGF-β expression was shown to be up-regulated in cancers (Derynck et al. 1987), the TGF-β pathway may have a tumor-suppressive function because of its ability to inhibit cell proliferation. Thus, inactivation of the growth-inhibitory response to TGF-β would then initiate malignant transformation. However, solid evidence for tumor suppression by TGF-β signaling was not reported until 1995. Data supporting TGF-β’s tumor suppressor role were derived from transgenic mouse studies (Pierce et al. 1995) and through identification of inactivating mutations in the TGF-β type II receptor in human colon carcinoma cells with microsatellite instability (Markowitz et al. 1995). Subsequent studies of genetically modified mice and mutations in genes encoding TGF-β signaling mediators in human cancers came to support a major role for the TGF-β pathway in tumor suppression (reviewed in Akhurst and Derynck 2001). Additionally, however, a substantial body of evidence arose that TGF-β signaling facilitates tumor progression through effects on the carcinoma cells and on host cells and tumor stroma (reviewed in Derynck et al. 2001; Massagué 2008; Ikushima and Miyazono 2010; Pickup et al. 2013).
COMPLEMENTARY DNA (cDNA) CLONING AND CHARACTERIZATION OF PRE-PRO-TGF-β
cDNA Cloning of TGF-β1
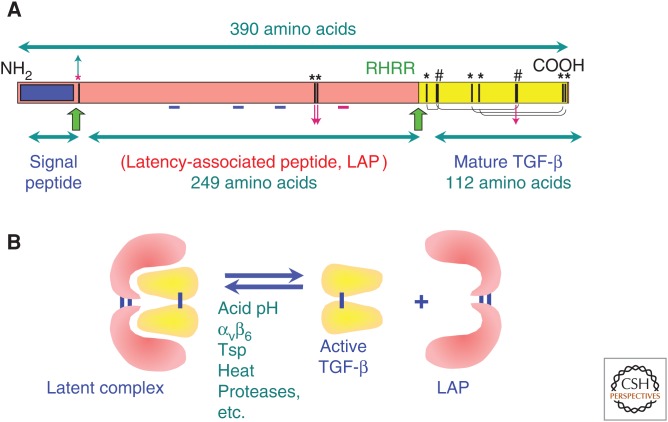
Searching for a new research area of interest, Rik Derynck, then at Genentech, became interested in the newly discovered TGF activity that was described to him by George Todaro over a pizza dinner at Frederick, Maryland, early in 1983. After all, the induction of reversible transformation by a secreted factor specifically made by tumor cells, even though controversial, was a remarkable observation, and, if indeed TGF exists, then one must be able to clone it. Although Derynck focused his efforts on the cDNA cloning of TGF-α (Derynck et al. 1984), Dave Goeddel, the head of the department, pointed him toward a recent paper from the Sporn/Roberts laboratory (Roberts et al. 1983a), biochemically showing that TGF-β was a dimeric protein, unlike TGF-α, with an amino-terminal sequence that was unrelated to TGF-α. Derynck first did not want to deal with it, being too busy with TGF-α cDNA cloning, but then contacted the Sporn group to affirm his interest in cloning TGF-β, which led to a formal collaboration. The Sporn/Roberts laboratory provided sufficient quantities of TGF-β isolated from human platelets (Assoian et al. 1983) for clostripain digestion, eventually resulting in the identification of a 55-residue-long contiguous amino-terminal amino acid sequence. Because the polymerase chain reaction had not yet been described (Mullis et al. 1986), a newly developed approach using long oligonucleotides as hybridization probes was used. As reliable amino acid sequence runs became available, two 44-residue-long oligonucleotides, designed based on a stretch of TGF-β sequence and the known bias of human codon usage, and a set of 16 14-mers complementary to all codons for amino acids 13–16 were used as hybridization probes against a human genomic DNA library. This led to the identification of an exon encoding amino acids 10–60 of mature TGF-β that then was used to screen a λgt10-based placenta cDNA library, and yielded a partial cDNA. Extensive screening of placenta and tumor cell cDNA libraries eventually led to a cDNA sequence of 2439 bp corresponding to the TGF-β precursor mRNA (Derynck et al. 1985). The deduced polypeptide sequence established that the purified biologically active TGF-β, now known as TGF-β1, was the carboxy-terminal 112 amino acid segment of a 390 amino acid precursor protein with an amino-terminal signal sequence and a long pro-sequence (Fig. 2). It was proposed, and subsequently verified, that the TGF-β monomer was cleaved from its precursor following a tetrabasic site, Arg-His-Arg-Arg. Later studies showed that furin, a member of the mammalian convertase family of endoproteases, is the likely protease required for processing TGF-β1 in vivo (Dubois et al. 1995). Subsequent cDNA cloning of TGF-β1 from mouse (Derynck et al. 1986), pig (Derynck and Rhee 1987), and monkey (Sharples et al. 1987) cells showed that the protein was highly conserved, that mature human and simian TGF-β1 differed by only one amino acid from murine TGF-β1, and that human and porcine TGF-β1 were identical. cDNA cloning of TGF-β and subsequent northern hybridizations also revealed that, unlike other growth factors that were typically expressed in specific cell types, TGF-β1 mRNA was synthesized by almost all normal and tumor cells examined, yet at higher levels by tumor cells (Derynck et al. 1985,1987), alluding to the broad spectrum of activities that were to be discovered. The cDNA cloning of TGF-α and TGF-β, first presented at a Cold Spring Harbor meeting in September 1984, led to the acceptance that TGFs did exist.
Figure 2.
Structure and function of pre-pro-transforming growth factor β (TGF-β). (A) Complementary DNA (cDNA) cloning of TGF-β showed that it derives from a large precursor with a signal peptide, the large pro-segment (latency-associated protein [LAP]), and the carboxy-terminal mature biologically active protein (in dimeric form). Large green arrows show processing sites, black bars and asterisks show the position of cysteine residues, and # shows two adjacent cysteine residues. The pattern of cross-linking of the four intrachain disulfides is shown in brackets. Red arrows (down) show the position of the interchain disulfide bridges, whereas a green arrow (up) shows the cysteine that links LAP to latent TGF-β-binding protein (LTBP). A blue underscore shows the position of glycosylation sites and a red underscore shows the position of the Arg-Gly-Asp (RGD)-integrin-binding sequence present in the LAPs of TGF-β1 and TGF-β3 but not the TGF-β2 LAP. (B) TGF-β is secreted from cells in a biologically inactive form (latent TGF-β), with noncovalent association of the dimeric LAP with the dimeric mature carboxy-terminal 112 amino acid TGF-β. Latent TGF-β can be activated by a variety of treatments in vitro resulting in dissociation of the LAP protein from mature TGF-β, thus unmasking its receptor-binding epitopes.
Concept of Growth Factor Latency
Even before TGF-β was cloned, it was seen as peculiar that TGF-β was secreted from cells in a biologically inactive form or complex that required activation by acidification (Lawrence et al. 1984; Pircher et al. 1984). This had initially not been noted because the isolation procedure started with the use of acid in the extraction, and subsequent purification steps were also performed under acidic conditions. Additional early studies showed that latent TGF-β could be activated not only by extremes of pH, but also by chaotropic agents, boiling, binding to α2-macroglobulin, or proteolysis using one of several proteases (Lawrence et al. 1985; O’Connor-McCourt and Wakefield 1987; Lyons et al. 1988; Sato et al. 1990). Moreover, because most cells can respond to TGF-β, physiological activation of this latent complex was increasingly seen as a key regulatory step in the control of its biological activity. The latency of TGF-β was subsequently revealed by Larry Gentry and Tony Purchio, and by Kohei Miyazono, then working with Calle Heldin, as well as Lalage Wakefield and Roberts, to result from noncovalent association of the mature TGF-β homodimer with the large dimerized pro-segment that has been renamed “latency-associated protein” (LAP), thus preventing binding of TGF-β to its receptors (Gentry et al. 1987; Miyazono et al. 1988; Wakefield et al. 1988). This “small latent complex” was also shown to bind to yet another and larger protein, latent TGF-β-binding protein (LTBP), a member of the fibrillin family of proteins, to direct extracellular localization of the latent complex (Miyazono et al. 1988, 1991; Wakefield et al. 1988; Kanzaki et al. 1990; Tsuji et al. 1990). The extensive sequence divergence among the three mammalian TGF-βs and the existence of four LTBPs suggest differences in extracellular targeting and activation mechanisms.
Expression of Recombinant TGF-β
Recombinant expression of TGF-β was reported 2 years after it was cloned (Gentry et al. 1987). Latent pre-pro-simian TGF-β was expressed in Chinese hamster ovary (CHO) cells, treated with methotrexate to amplify the coexpressed dihydrofolate reductase gene. One CHO clone showed TGF-β secretion exceeding by 50-fold the levels naturally secreted by cultured cells. The secreted recombinant TGF-β was latent, requiring acidification for biological activity. This observation led to the hypothesis that the precursor sequences might play a role in the activation process. Studies of recombinant TGF-β expression confirmed the processing sites of the signal peptide and the mature TGF-β that were predicted from the cDNA cloning of the TGF-β precursor (Gentry et al. 1988). These studies also showed that the precursor was cross-linked by disulfide bonding and likely to function as a dimer, and that glycosylation of TGF-β was restricted to the precursor domain (Fig. 2). Subsequent complementation assays showed that the TGF-β prosequence was also required for proper folding, disulfide formation, dimerization, and secretion of mature TGF-β (Gray and Mason 1990), and thus acts as a chaperone. Because TGF-β was shown not to be glycosylated, attempts were made to express the mature form of TGF-β in bacteria (Tuan et al. 1996). Bacterially expressed mature TGF-β was inactive, confirming the need of the precursor domains to assure proper disulfide bonding of the mature TGF-β, and required renaturation using the glutathione-redox method to endow some biological activity.
IDENTIFICATION OF TGF-β HOMOLOGS
Three TGF-βs
Following the identification of platelets as the most concentrated source of TGF-β, R&D Systems (Minneapolis, MN) chose porcine platelets to generate commercially available TGF-β, which made sense considering that porcine and human TGF-β had identical sequences (Derynck and Rhee 1987). They worked with the Moses laboratory, then at the Mayo Clinic, to learn purification techniques based on the method published by Assoian et al. (1983). Surprisingly, they identified three peaks of activity, subsequently found to be homodimeric TGF-β1, homodimeric TGF-β2 and a heterodimer, TGF-β1:TGF-β2 (Cheifetz et al. 1987). Although TGF-β2 had the identical size and biological activity of TGF-β1 in growth inhibition assays, it had a distinct amino-terminal amino acid sequence and was not recognized by antibodies raised against TGF-β1 sequences. It also showed differential binding efficiencies to the then putative three TGF-β receptor types, identified as the 65-kD type I receptor, the 85-kD type II receptor, and the 280-kD type III receptor or betaglycan. As later confirmed, once these TGF-β receptors had been cloned, TGF-β2 bound only poorly to the type II TGF-β receptor, but bound to betaglycan with similar affinity as TGF-β1 (Cheifetz et al. 1987; Lopez-Casillas et al. 1993). Independent studies in other laboratories identified TGF-β2 as a cartilage-inducing activity from bovine bone (Seyedin et al. 1987), a T-cell suppressor activity made by human glioblastoma cells (Wrann et al. 1987), and a growth inhibitor secreted by BSC-1 monkey kidney cells (Hanks et al. 1988). Subsequent cDNA cloning showed that TGF-β2 derives from a 412 amino acids precursor, and that its mature 112 amino acid sequence is 71% identical to TGF-β1 (de Martin et al. 1987; Marquardt et al. 1987).
Shortly after the characterization of TGF-β2, cDNA cloning identified yet a third TGF-β, called TGF-β3, which was 72% identical to TGF-β1 and 76% identical to TGF-β2 (Derynck et al. 1988; ten Dijke et al. 1988). All nine cysteines in the 112–amino acid mature TGF-β forms were positionally conserved in the three TGF-βs. TGF-β3 was found to play a unique role in fusion of the palatal shelves (Proetzel et al. 1995) and, unlike TGF-β2, to bind the type II TGF-β receptor with similar affinity as TGF-β1. Although the three mature TGF-βs show 64%–82% sequence identity, their pro-domains are only 28%–45% identical, yet some sequence features are conserved. The three prodomains have conserved N-linked glycosylation sites, and, with the exception of the TGF-β2 pro-domain, an Arg-Gly-Asp (RGD) potential integrin binding site is apparent. In addition, each pro-region has three cysteines, two of which are involved in interchain disulfide bonds necessary for dimerization of the pro-region in the small latent TGF-β complex, whereas the third cross-links to its binding protein LTBP (Fig. 2). Mammalian cells encode only these three TGF-βs, encoded by three distinct genes (Table 1). Although the term TGF-β isoforms is often used, these three TGF-βs are not to be considered as isoforms derived from the same gene, but are encoded by different genes.
Table 1.
Comparative features of the TGF-β isoforms
| Number of amino acids | Chromosomal locus | ||||||
|---|---|---|---|---|---|---|---|
| Precursor | LAP | Mature | Tetrabasic site | Human | Mouse | mRNA (kb) | |
| TGF-β1 | 390 | 249 | 112 | RHRR | 19q13 | 7 | 2.5 |
| TGF-β2 | 414,442a | 281 | 112 | RKKR | 1q41 | 1 | 4.1,5.1,6.5 |
| TGF-β3 | 412 | 279 | 112 | RKKR | 14q23-4 | 12 | 3.0 |
aTGF-β2 has a prominent alternative spliced variation encoded by the 5.1-kb mRNA and is expressed only in certain tissues. A 29-amino acid insertion results in a latency-associated protein (LAP) with three additional cysteine residues, suggesting a more complex secondary structure (Webb et al. 1988).
The TGF-β “Superfamily”
Shortly after the cDNA cloning of TGF-β1, and at the time that TGF-β2 and -β3 were identified, unrelated cDNA cloning projects in other laboratories revealed the existence of other large precursors with sequence homology with TGF-β in their carboxy-terminal sequences and conservation of the last seven of TGF-β’s nine cysteines. Structural analyses later revealed that the positioning of these seven cysteines, which characterizes the class of TGF-β-related proteins, defines the “cystine knot” in the mature forms of these molecules (McDonald and Hendrickson 1993). The first two proteins so identified were the two polypeptides that form the disulfide-linked inhibin heterodimer, an inhibitor of follicle-stimulating hormone secretion (Mason et al. 1985). The alignment of the inhibin cDNA-encoded polypeptides, isolated by Tony Mason in the Seeburg laboratory at Genentech, with the TGF-β1 cDNA sequence followed a random evening encounter at Genentech, in which Mason discussed with Derynck the difficulties in cloning inhibin cDNAs that seemed to resemble similar frustrations with the TGF-β1 cDNA cloning. Half a year later, the cDNA-derived sequence for the Müllerian-inhibiting substance (MIS), which causes Müllerian duct regression in development, revealed again a large precursor sequence with a carboxy-terminal, TGF-β-related sequence (Cate et al. 1986), and the first role for a TGF-β-related protein in development. Again, half a year later, the DPP-C locus, which directs patterning in Drosophila was shown to encode a precursor with a carboxy-terminal TGF-β-related sequence, demonstrating the evolutionary ancestry of TGF-β proteins, and the first evidence that TGF-β-related proteins direct developmental patterning (Padgett et al. 1987). Later that year, Vg1 mRNA, which localizes to the vegetal hemisphere of Xenopus eggs, was also shown to encode a TGF-β-related protein as the carboxy-terminal fragment of a larger precursor, further expanding the TGF-β family, and further drawing attention to regulation of development, in this case mesoderm formation, by TGF-β-related proteins (Weeks and Melton 1987). Finally, the characterization, late in 1988, of the cartilage- and bone-inducing “bone morphogenetic protein (BMP),” isolated from bone, resulted in the characterization of cDNAs encoding the TGF-β-related BMP-2A (now known as BMP-2) and BMP-3, which induced cell differentiation, and formed the basis for the discovery of additional BMPs (Wozney et al. 1988).
The rapid succession of reports identifying TGF-β-related proteins, the revelation that they function in very diverse contexts and have multiple developmental roles in patterning, morphogenesis, and cell differentiation, and their evolutionary ancestry and conservation profoundly affected many fields, most notably developmental biology and cancer biology. These discoveries also generated enthusiasm for the notion that TGF-β-related proteins form a “superfamily” encoded by a “super gene family.” However, considering the similar organization of all encoded protein sequences as large precursors with a signal peptide, a large pro-segment and a carboxy-terminal mature TGF-β-related sequence, the TGF-β-related proteins constitute a family that, by analogy with other classes of proteins, does not meet the definition of a superfamily that groups proteins with divergent organization. We now know that the mouse and human genomes have 33 genes encoding precursor monomers of TGF-β-related proteins. With the existence of homodimeric and heterodimeric TGF-β-related proteins, the number and diversity of homo- and heterodimeric combinations with functional roles remains unclear. TGF-β family proteins play diverse roles in the control of cell proliferation, metabolism, and differentiation, and act as potent and often essential regulators of developmental processes. TGF-βs, BMPs, and activins (inhibin polypeptide homodimers) also emerged as regulators of disease pathogenesis and progression.
THE EXPANDING BIOLOGICAL ACTIVITIES OF TGF-β
TGF-β research in the latter half of the 1980s was characterized by discoveries of a plethora of biological effects on cells in culture. With the commercial availability of TGF-β, it had become apparent that most if not all cell types responded to TGF-β, while they also made and secreted TGF-β, thus allowing for autocrine stimulation. The many effects seen included regulation of cell proliferation, cell differentiation, extracellular matrix (ECM) production and remodeling, chemotaxis, lymphocyte function, and growth factor and hormone production. By the mid-1980s, it was widely accepted that TGF-β could either stimulate or inhibit cell proliferation depending on the cell type, and presence of other growth factors and culture conditions (discussed above). TGF-β also induced many other cell responses that appeared to also depend on the cell type and culture conditions. This led to an oversimplified and caricatural notion among some that TGF-β acted as a switch: It switched on what was off, and switched off what was on. In addition to being biologically confusing, this notion did not set an inviting stage to explore therapeutic modalities using TGF-β or TGF-β antagonists.
Given that TGF-β was expressed by many cell types found in all tissues examined and abundant in platelets, it was evident that the molecule must have physiological functions distinct from its transforming activity and roles in cancer. Early in vivo experiments in the Sporn/Roberts laboratory were based on the adage of Alexander Haddow (1972) that “the wound is a tumor that heals itself,” restated by Dvorak (1986) as “tumors are wounds that do not heal,” thus inviting the evaluation of a TGF in wounds. Release of TGF-β from Hunt–Shilling wire-mesh chambers implanted into the backs of rats profoundly stimulated a stromal response, including angiogenesis and ECM deposition (Sporn et al. 1983). Injection of TGF-β into the nape of the neck of newborn mice confirmed that TGF-β stimulated formation of granulation tissue, was chemotactic for inflammatory cells, and stimulated ECM production by fibroblasts (Roberts et al. 1986), thus demonstrating that it was active in multiple aspects of wound healing. These observations were confirmed and expanded in many follow-up studies using models of impaired healing (reviewed in Roberts 1995).
A molecular basis for these effects on wound healing and connective tissue deposition was provided by studies in cell culture (Fig. 3). TGF-β was shown to strongly induce the expression of ECM components, including fibronectin, certain collagens, chondroitin/dermatan sulfate, biglycan, decorin, osteopontin, osteonectin, tenascin, and thrombospondin (Massagué 1990). In addition, TGF-β stimulated the synthesis of inhibitors of matrix-degrading enzymes, including plasminogen activator inhibitor 1 (PAI-1) and a tissue inhibitor of matrix metalloproteinases (TIMPs) (Laiho et al. 1986; Overall et al. 1989). Conversely, TGF-β inhibited expression of matrix-degrading proteases such as collagenase, stromelysin, and plasminogen activator (Edwards et al. 1987; Kerr et al. 1988). Thus, the increase in inhibitors and decrease in matrix proteases caused increased ECM accumulation. TGF-β treatment was also shown to induce marked changes in integrin expression in several cell types resulting in altered cell binding to ECM components (Ignotz and Massagué 1987; Roberts et al. 1988; Heino et al. 1989). Further, TGF-β revealed itself as potently chemotactic for fibroblasts (Postlethwaite et al. 1987) and macrophages (Wahl et al. 1987), a function that likely contributes to the increased abundance of these cells at sites of increased TGF-β production such as wounds and cancers.
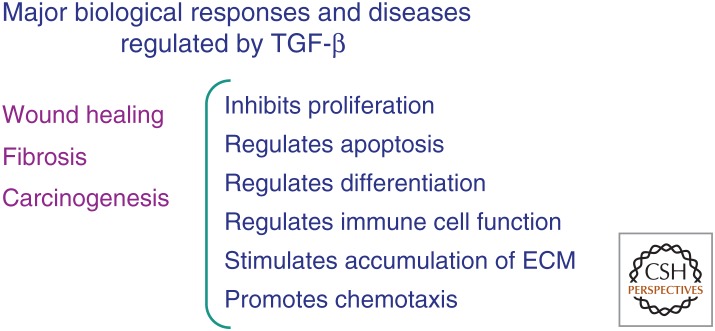
Figure 3.
Roles of transforming growth factor β (TGF-β) in pathophysiology. TGF-β plays prominent roles in wound healing, fibrosis, and carcinogenesis, as well as a host of other diseases. Listed on the right are various TGF-β-dependent cellular mechanisms that contribute to its effects. ECM, Extracellular matrix.
Around that time, collaborative studies between the laboratories of Sporn/Roberts and Toni Fauci at the National Institutes of Health revealed that TGF-β could inhibit the differentiation and functions of B and T lymphocytes as well as natural killer cells (Kehrl et al. 1986a,b; Rook et al. 1986). These unexpected findings with a factor that was thought to play a major role in cancers linked TGF-β to the immune system and provided the basis for the many subsequent studies that highlight the roles of TGF-β as a potent immunoregulator, controlling proliferation, differentiation, and function of most lymphocyte classes, as well as macrophages and dendritic cells, and a potent suppressor of immune surveillance (Letterio and Roberts 1998). These proposed roles of TGF-β were supported and in part elucidated by later studies using TGF-β1 knockout mouse (Shull et al. 1992; Kulkarni et al. 1993) that further characterized a role for TGF-β1 in autoimmunity (Letterio et al. 1994) and in immune tolerance (Letterio and Roberts 1998).
Additionally, steroidogenesis was reported to be inhibited by TGF-β in adrenocortical cells and Leydig cells, while it was stimulated in granulosa cells. TGF-β was also shown to stimulate expression of follicle-stimulating hormone by pituitary cells (Ying et al. 1986), whereas other studies revealed that TGF-β induces PDGF expression by fibroblasts (Leof et al. 1986). Consequently, TGF-β was now additionally seen as a mediator of hormonal control in different contexts, consistent with the discovery that the hormone inhibin is a TGF-β-related factor. Furthermore, it was now apparent that some activities of TGF-β occurred secondarily through the activation of expression of some hormones and growth factors.
Studies in the latter part of the 1980s also revealed that TGF-β has potent effects on differentiation of a wide variety of cell types. This stood in remarkable contrast with the “classical” growth factors, whose effects on cells were primarily seen to enhance proliferation, and were thought of primarily in the context of cancers. These effects on cell differentiation were especially remarkable considering TGF-β’s link to cell transformation. TGF-β was often seen to inhibit cell differentiation, for example, of preadipocytes, skeletal muscle myoblasts, muscle satellite cells, and osteoblasts, conceptually resembling inhibitory effects of TGF-β on immune cell functions. On the other hand, TGF-β stimulated differentiation of prechondrobasts, intestinal epithelial cells, keratinocytes, bronchial epithelial cells, and osteoblasts under some conditions. These findings led to the notion that TGF-β was a differentiation factor, in addition to a growth inhibitor, as summarized around that time by Massagué (1990). These initial observations, together with the realization that defects in developmental patterning and tissue differentiation were associated with dysregulation of TGF-β-related proteins, set the stage for a wide range of studies that defined the developmental control of tissue differentiation by TGF-β and TGF-β family proteins.
Expanding these observations on cell differentiation, the Derynck laboratory reported, in 1994, that TGF-β induces epithelial cells in culture to undergo a reversible transition into cells with mesenchymal appearance (Miettinen et al. 1994). Although this was consistent with observations by Ray Runyan that TGF-β is required for transition of endocardial cells into mesenchymal cells during heart valve formation (Potts et al. 1991), these results were nevertheless met with surprise, if not apprehension, primarily because of the overall notion that differentiation could not be redirected, and certainly not by one factor. Epithelial–mesenchymal transition (EMT) had been shown to occur in development (Hay 1995), but the underlying signaling mechanisms were not known. The induction of EMT by TGF-β in cell culture was the basis for many subsequent studies characterizing mechanisms and roles of TGF-β-induced, Smad-mediated gene reprogramming during EMT, and complementary roles of non-Smad pathways. As the roles of EMT in development, starting with mesoderm induction and gastrulation, became more apparent, many studies gradually revealed the developmental control of EMT by TGF-β-related proteins (reviewed in Lamouille et al. 2014).
The increasing realization in the late 1980s and early 1990s that TGF-β controls cell differentiation was complemented by in situ hybridization and immunohistochemical studies showing expression of all three TGF-βs during embryogenesis, each of these in specific patterns, both spatially and temporally (Heine et al. 1987; Lehnert and Akhurst 1988; Pelton et al. 1989, 1990a,b, 1991; Millan et al. 1991). These observations suggested specific roles for the individual TGF-βs in processes such as angiogenesis, cardiogenesis, palate formation, and osteogenesis, and provided the basis for subsequent studies on developmental defects resulting from targeted inactivation of the individual TGF-β genes in mice (Shull et al. 1992; Kulkarni et al. 1993; Proetzel et al. 1995; Sanford et al. 1997). Although Tgfb1−/− mice highlighted the roles of TGF-β1 in the control of the immune system and inflammation (Shull et al. 1992; Kulkarni et al. 1993), as well as angiogenesis, vasculogenesis, and hematopoiesis (Dickson et al. 1995), inactivation of TGF-β2 and TGF-β3 expression primarily resulted in defects in organs that develop through epithelial–mesenchymal tissue interactions (Proetzel et al. 1995; Sanford et al. 1997). These observations also revealed that, despite their very similar activities in cell culture, the three TGF-βs have highly divergent developmental roles.
With TGF-β having been discovered as a factor that induces cell transformation, and the overall notion in the 1980s that growth factors act predominantly in cancers, thus contributing to cancer progression, many researchers aimed to elucidate roles of this newly discovered factor in cancer initiation and progression. As with other growth factors, much emphasis was on the control of cell proliferation by TGF-β, even though, and especially because, it rapidly became apparent that TGF-β has growth-inhibitory activities on epithelial, immune, and hematopoietic cells, which give rise to the bulk of cancers. As the other activities of TGF-β became apparent, researchers incorporated their results and views into a more complex picture of the roles of the apparently increased TGF-β signaling in cancers. Increased ECM protein expression in response to TGF-β was linked to increased ECM turnover in cancers. The chemotactic effects of TGF-β on fibroblasts and immune cells contributed to tumor stroma formation and the tumor microenvironment. Localized immunosuppression by TGF-β was seen to contribute to cancer growth and progression. TGF-β was also found to induce angiogenesis in cancers and to contribute to cancer-associated localized inflammation. Finally, EMT in response to increased TGF-β signaling was increasingly viewed as a key contributing event to carcinoma invasion and dissemination, and recently to the generation and properties of cancer stem cells (Derynck et al. 2001; Massagué 2008; Ikushima and Miyazono 2010; Katsuno et al. 2013).
Besides cancer, other pathological connections with dysregulated TGF-β production and responsiveness became gradually apparent. Most notably, the suppression of experimental glomerulonephritis by TGF-β antiserum (Border et al. 1990) provided the impetus for many subsequent studies highlighting the contributions of increased TGF-β signaling to different types of fibrosis. Again, increased TGF-β activity, TGF-β’s abilities to activate ECM protein expression, its potent chemotactic activities for fibroblasts and immune cells, and the induction of EMT by TGF-β became incorporated in the etiology and progression of fibrosis (Chapman 2011; Nieto 2011). In addition to fibrosis, several connective tissue and skeletal diseases were also seen to result from increased TGF-β signaling, which, in a number of cases, was associated with genetic mutations that lead to dysregulated TGF-β activation (Shore and Kaplan 2010; Doyle et al. 2012).
The sheer number and overwhelming variety of biological activities described for TGF-β during the 1980s was unprecedented in the growth-factor field, and attracted many investigators from different disciplines to the field. Although this generated rapid progress with new insights into the biology of TGF-β, one negative aspect of so many biological effects was that development of pharmacologic agents to target and modulate TGF-β actions by industry was discouraged for a number of years because of the potential plethora of side effects.
cDNA CLONING AND IDENTIFICATION OF TGF-β RECEPTORS AND SMADs
cDNA Cloning of the TGF-β Receptors
Once specific high-affinity receptors for TGF-β had been identified in 1984 (Massagué et al. 1982), the next goal was to characterize these with the ultimate aim to define the TGF-β signaling pathways that lead to the diversity of emerging biological activities. Using growth-factor-activated tyrosine kinase receptors as a reference point, it was rapidly concluded that the TGF-β receptors would act through a different class of receptors, because they induced growth inhibition, rather than growth stimulation, and many other responses hitherto not seen with receptor tyrosine kinases (RTKs). Furthermore, three types of receptors were consistently identified, with both the type I and type II receptors required for TGF-β responses, based on analyses of mutant cell lines (Laiho et al. 1991). This was again very different from RTKs. The task to purify or clone these receptors would, however, be challenging considering the very low levels of the TGF-β receptors at the cell surface (Frolik et al. 1984; Tucker et al. 1984a), when compared with average numbers for RTKs. Around 1987–1990, several laboratories started projects aimed at cDNA cloning of the receptors, using receptor purification or expression cDNA cloning approaches, in combination with radiolabeled ligand-binding assays.
The cDNA cloning of the first known receptor for a TGF-β family protein was achieved by Lawrence Mathews working in the laboratory of Wylie Vale (Mathews and Vale 1991). An expression cDNA cloning strategy in combination with 125I-activin binding to transfected cells allowed him to isolate cDNAs for the activin type II receptor, now known as ActRII, noting that it was likely a transmembrane serine/threonine kinase. The cytoplasmic domain of this receptor showed extensive sequence similarity to the transmembrane protein daf-1 in Caenorhabditis elegans, which had been cloned the year before, and was proposed as the first transmembrane serine/threonine kinase receptor with a ligand still to be identified (Georgi et al. 1990). The identification of ActRII was rapidly followed by the cDNA cloning of a related activin receptor, ActRIIB (Attisano et al. 1992; Mathews et al. 1992), and of alternatively spliced variants (Attisano et al. 1992). A similar expression cloning approach as for ActRII, but using 125I-TGF-β as ligand, resulted in the cDNA cloning of the TGF-β type II receptor, now known as TβRII, by the laboratories of Harvey Lodish and Bob Weinberg (Lin et al. 1992). Also, this receptor was shown to phosphorylate on serine and threonine. A few years later, the MIS binding type II receptor was cloned, based on its homology with previously isolated TGF-β family receptors (Baarends et al. 1994; di Clemente et al. 1994), and the type II BMP receptor BMPRII was cloned based on either its association with the type II TGF-β receptor in a yeast two-hybrid approach (Kawabata et al. 1995; Liu et al. 1995) or sequence homology using a polymerase chain reaction (PCR)-based strategy (Nohno et al. 1995; Rosenzweig et al. 1995). Five TGF-β family type II receptors are now known to be encoded by human cells.
cDNA cloning of the type I receptors was accomplished in large part using PCR-based strategies with oligonucleotide primers for conserved sequences in the ActRII, TβRII, and/or Daf1 receptors (Ebner et al. 1993a; He et al. 1993; ten Dijke et al. 1993). This strategy resulted in the identification of type I receptors with overall similarity to the type II receptors, easily recognizable by their distinct Gly-Ser (GS)-rich sequence immediately upstream of the kinase domains (Wrana et al. 1994a). As it was initially not clear what ligands they would bind, Peter ten Dijke coined the term ALK for “activin receptor-like kinase” for these receptors (ten Dijke et al. 1993), although various other names were chosen by other laboratories. Seven type I receptors, designated ALK1-7, are now known to be encoded by human cells (ten Dijke et al. 2000; ten Dijke and Hill, 2004), whereas ALK-8 was found in zebrafish (Bauer et al. 2001; Mintzer et al. 2001). With five type II and seven type I receptors, and a much larger number of ligands, the assignment of the type I receptors to individual ligands was, and continues to be, complex and tedious. It was proposed that the type II and type I receptors formed a heteromeric complex (Wrana et al. 1992), thus requiring the ligand binding and function of a type I receptor to be assessed in combination with the type II receptor. Furthermore, it became apparent that, at least in the case of TGF-β and activin, the type I receptors only bound ligand when combined with the appropriate ligand-binding type II receptor, which provides ligand specificity (Attisano et al. 1993; Ebner et al. 1993b). In contrast, the type II or type I BMP receptors, when expressed individually, generally showed only a weak affinity for BMPs, and only bind BMPs efficiently when coexpressed in heteromeric receptor combinations (Miyazono et al. 2001, 2010). Using a type II/type I receptor combinatorial strategy, TGF-β was shown to signal primarily through the type I receptor that is now known as TβRI or ALK-5 (Franzén et al. 1993, Bassing et al. 1994). The functional interdependence of the type II and type I receptors was subsequently shown to result from ligand-induced, type II receptor-mediated phosphorylation of the GS domain in the type I receptor that leads to activation of the type I receptor kinase (Wrana et al. 1994b). This dependence for signaling activation complemented the extracellular interdependence of the type II and type I receptors for productive ligand binding. We currently know many of the functional combinatorial type II/type I receptor interactions and the corresponding ligands that activate these complexes, but, considering the complexity of homomeric and heteromeric ligands, additional assignments will be forthcoming.
Finally, the type III TGF-β receptors, betaglycan and endoglin, differ from the other TGF-β family receptors in the sense that they do not directly initiate TGF-β signaling, but rather control ligand presentation to the type I and type II receptors (Bernabeu et al. 2009). Although betaglycan is in vivo primarily expressed in mesenchymal cells, endoglin expression is restricted to endothelial cells. The proteoglycan betaglycan was cloned by Fernando Lopez-Casillas, working with Joan Massagué, following protein purification and based on peptide sequences that allowed the generation of oligonucleotides as hybridization probes (Lopez-Casillas et al. 1991), and by Xiao-Fan Wang then working with Bob Weinberg using an expression cDNA cloning strategy (Wang et al. 1991). cDNA cloning of the glycoprotein endoglin was reported at a time that endoglin was only known to be a cell-surface protein specifically expressed in vascular endothelium (Gougos and Letarte 1990) and, thus, preceded the cDNA cloning of the first TGF-β family receptors and the realization that it participates in TGF-β signaling. Endoglin was subsequently found to bind TGF-β, allowing it to play a role in ligand presentation to type II/type I TGF-β receptor complexes (Cheifetz et al. 1992), and to link genetically with hereditary hemorrhagic telangiectasia type I (McAllister et al. 1994).
Discovery and Characterization of the Smad Pathway
At the time that the TGF-β receptors were cloned, little was known about any downstream mechanisms, and of course the next big step was to elucidate how TGF-β induces the many diverse responses. TGF-β had been shown to induce Ras and Erk mitogen-activated protein (MAP) kinase activation (Mulder and Morris 1992; Yan et al. 1994; Mucsi et al. 1996), but activation of this pathway was at odds with the substantial differences in responses when compared with RTKs that activate the Ras-Erk MAP kinase pathway. Additionally, TGF-β was shown to activate protein phosphatase 1 (Gruppuso et al. 1991) and to induce phosphorylation of the transcription factor cAMP response element-binding (CREB) protein (Kramer et al. 1991), but these findings again did not explain the major TGF-β responses. With the receptors in hand, several laboratories embarked on identifying associated proteins and kinase substrates for the TGF-β receptors. This led, within the next few years, to the identification of FKBP12 (Wang et al. 1996a; Chen et al. 1997) and the α subunit of farnesyltransferase (Ventura et al. 1996; Wang et al. 1996b) as associated proteins, and the translation initiation factor eIF2α/TRIP-1 (Chen et al. 1995) and Bα subunit of protein phosphase 2A (Griswold-Prenner et al. 1998) as kinase substrates, but these again were not seen as signaling effectors that mediate the predominant TGF-β responses. The identification by Matsumoto and colleagues of the “TGF-β-activated kinase 1” (TAK1) as a signaling mediator that is activated by TGF-β and BMP (Yamaguchi et al. 1995) generated considerable excitement. However, its identity as a MAPKK kinase that could activate MAP kinase pathways and the inability of TAK1 to activate TGF-β target gene responses raised the general question to what extent TAK1 contributed to TGF-β and BMP responses. Furthermore, TAK1 was subsequently also shown to be an effector of signaling in response to ligands that are not related to TGF-β and act through different receptor classes.
A major breakthrough toward the identification of TGF-β-activated signaling effectors came from genetic analyses of Drosophila and C. elegans. The Drosophila decapentaplegic (dpp) gene, which encodes the homolog of vertebrate BMP-2 and -4, was by then known to act through the type II receptor, Punt, and two type I receptors, Thickveins and Saxophone (Ruberte et al. 1995). A genetic screen for enhancers of weak dpp alleles in Bill Gelbart’s laboratory at Harvard yielded “mothers against dpp” (mad) and medea as genes that encode downstream components required for dpp signaling (Raftery et al. 1995), and revealed sequence similarity between Mad and three C. elegans genes (Sekelsky et al. 1995). Parallel studies in C. elegans had identified daf-4 as a BMP-2 and -4 homolog receptor (Estevez et al. 1993). Three genes, sma-2, -3, and -4, were shown to function in the same cells as daf-4, and to be required for daf-4 function (Savage et al. 1996). The identification of Mad in Drosophila and Sma proteins in C. elegans defined a new family of signaling mediators downstream from BMP-related proteins, and prompted an intense search by several groups for homologs in mammals. Within a year, five Mad homologs were identified in different laboratories resulting in a diverse set of names (Derynck and Zhang 1996). One of these, initially named human Mad4, was identical to DPC4 (“deleted in pancreatic carcinoma, locus 4”), whose gene had just been identified as a putative tumor suppressor gene with inactivating mutations in pancreas cancer (Hahn et al. 1996). The nomenclature was soon resolved by agreement of the involved investigators to use the term Smad as a combination of the Mad and Sma names (Derynck et al. 1996).
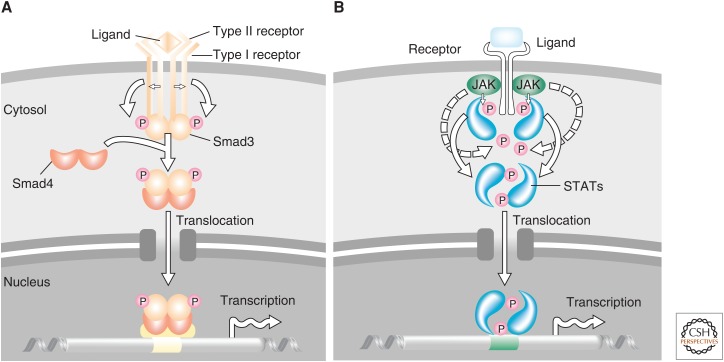
The assignment of Smads as effectors of TGF-β/activin or BMP ligands and key aspects of the overall signaling mechanism progressed at a frantically competitive pace in 1995–1996. Individual Smads, when overexpressed, were shown to mimic effects of corresponding ligands (Baker and Harland 1996; Graff et al. 1996; Liu et al. 1996; Zhang et al. 1996), and dominant-negative mutants were found to inhibit ligand-induced responses (Zhang et al. 1996). Ligand-induced receptor activation induced Smad phosphorylation (Eppert et al. 1996, Hoodless et al. 1996; Zhang et al. 1996), Smad3 was found to interact with the type I, but not the type II receptor (Zhang et al. 1996), and Smad4 cooperated with Smad3 (Zhang et al. 1996). Additionally, a carboxy-terminal fragment of Smad1 or Smad4 was shown to have transcription activation potential (Liu et al. 1996), and activin or BMP-2/-4-induced nuclear accumulation of Smad2 or Smad1, respectively (Baker and Harland 1996; Hoodless et al. 1996; Liu et al. 1996). These observations provided the basis for the model of Smad activation that conceptually paralleled the well-established model of signal transducers and activators of transcription (STAT) activation in response to cytokine receptors (Fig. 4). Thus, Smads were thought to interact with type I receptors and be activated through phosphorylation by the receptor complexes, form a complex with Smad4, translocate into the nucleus, and target regulatory gene sequences for direct transcriptional activation (Derynck and Zhang 1996). This model was rapidly refined (e.g., by the demonstration that the type I TGF-β receptor directly phosphorylates Smad2, and not Smad4) at two carboxy-terminal serines, thus leading to its activation (Hoodless et al. 1996; Macías-Silva et al. 1996), and that Smad4 cooperates with all receptor-activated Smads tested (Lagna et al. 1996; Zhang et al. 1997).
Figure 4.
Initial working model of Smad signaling proposed in 1996. Following the identification of Smads as transforming growth factor β (TGF-β) family signaling effectors and several reports on how they might do so, this first model was proposed, based on these results and the perceived analogy with the established mode of signal transducers and activators of transcription (STAT) signaling. (From Derynck and Zhang 1996; reprinted, with permission, from the authors.)
How Smad complexes could activate transcription was again pursued at a high pace by several laboratories. An early report found Smad2 to interact with the Mix2 promoter through association with the DNA-binding transcription factor FAST1 (Chen et al. 1996), now known as FoxH1. However, Mad was subsequently shown to bind DNA directly, to a GC-rich sequence in the vestigial promoter (Kim et al. 1997), and a PCR-based selection procedure defined the palindromic sequence GTCTAGAC as the consensus “Smad-binding element” for Smad3 and Smad4 (Zawel et al. 1998). Therefore, it was concluded that receptor-activated Smads were DNA-binding transcription factors, similarly to STATs. On the other hand, the association and cooperation of Smad2 with FoxH1 at target promoters (Chen et al. 1996; Labbé et al. 1998), the association and cooperation of Smad3 with c-Jun/c-Fos at overlapping promoter sequences (Zhang et al. 1998), and the cooperation of Smad3 with the transcription factor TFE3 in the control of PAI-1 expression (Hua et al. 1998), which were revealed around the same time, all emphasized the need for cooperation of Smad complexes with high-affinity DNA-binding transcription factors. These and other observations led to the now generally accepted model that Smads activate target gene transcription, through physical association and functional cooperation with a variety of high-affinity DNA-binding transcription factors, thus enabling DNA binding to adjacent DNA sequences, and that Smad complexes use CBP or p300 as coactivators (Derynck et al. 1998). This model explained why, despite extensive efforts, no consensus TGF-β responsive DNA sequences could ever be found in TGF-β target genes, and illustrates the high versatility of Smad signaling dependent on the nature of the interacting transcription factor. This mode of cooperation also predicted direct cross-talk of Smads with other signaling pathways that target the Smad-associated transcription factors, thus providing a mechanistic basis for the context-dependence of the TGF-β responses. When this model was proposed in the context of an invited minireview for Cell, the editor, Ben Lewin, called Derynck to argue that this model was totally wrong, indicating that Smads were plain DNA-binding transcription factors with a DNA-binding consensus sequence (just like STATs), as just published in Molecular Cell (Zawel et al. 1998). After vigorous arguing, he very reluctantly agreed to have this model published. With an extensive body of ongoing research on many target genes in the next few years, Smads were shown to associate and cooperate with more than 100 DNA-binding transcription factors, depending on the nature of the target promoter, and with many co-repressors and coactivators that help define the amplitude of the TGF-β-induced transcription responses (Feng and Derynck 2005; Massagué et al. 2005).
Although we started understanding the mechanisms of TGF-β-induced, Smad-mediated activation of gene expression, these observations did not address how TGF-β family proteins induce repression of gene expression. With research on different target genes as model systems, it became subsequently apparent that also repression of gene expression was mediated by Smad complexes, thus fully attributing the control of gene expression by TGF-β family proteins to Smads. Smad3-mediated repression of myogenic transcription factor target genes was seen to result from direct physical interference by Smad3 with the formation of functional myogenic transcription factor complexes at E-box DNA sequences in target genes (Liu et al. 2001, 2004). In contrast, in TGF-β-induced repression of c-myc expression, Smad3/4 complexes were seen to associate with the DNA-binding E2F4/5 and DP1 transcription factors, and the co-repressor p107, thus enabling these complexes to bind at a composite Smad-E2F-binding site in the c-myc promoter, and to exert transcription repression through p107 (Chen et al. 2002). At the osteocalcin and Runx2 promoter, TGF-β-activated Smad3/4 complexes were seen to associate with Runx2 at a Runx2-binding DNA sequence and to directly recruit histone deacetylases (Alliston et al. 2001; Kang et al. 2005).
As these mechanistic insights were unfolding, two novel Smads were discovered that had a conserved transactivation domain that characterizes the Smads, but lacked the preceding amino-terminal Mad homology 1 (MH1) domain that is seen in all receptor-activated Smads and Smad4. These Smads, Smad6 and Smad7, with Dad as the Drosophila counterpart, were shown to associate with the type I receptors or other Smads, thus competitively preventing activation of the effector Smads, which is why they were named inhibitory Smads (Hayashi et al. 1997; Imamura et al. 1997; Nakao et al. 1997; Tsuneizumi et al. 1997; Hata et al. 1998). With Smad6 and Smad7 expression activated in response to several signaling pathways, it became apparent that the inhibitory Smads provide endogenous control of the level of Smad activation by TGF-β family proteins (ten Dijke and Hill 2004; Briones-Orta et al. 2011). Additional observations suggested that they may also directly repress transcription of target genes in the nucleus (Bai and Cao 2002; Grönroos et al. 2002).
With the overall mechanisms of Smad-mediated transcription control defined, at least in general terms, several laboratories have been focusing on the activation of non-Smad signaling pathways by TGF-β and TGF-β-related proteins. Indeed, TGF-β family proteins were shown to activate Erk, p38, and JNK MAP kinase pathways, as well as the PI3K-Akt-TOR pathway, and to act through TAK1, an upstream mediator of MAP kinase pathways (Derynck and Zhang 2003; Moustakas and Heldin 2005; Zhang 2009). The mechanisms through which TGF-β proteins induce activation of these pathways, or cross-talk with the Smad pathway, are being characterized, whereas other studies address how cross-talk between these pathways and the Smad pathway controls the cellular responses in cell differentiation and growth control and during development and in cancer.
SUMMARY
The pioneering efforts of the Todaro, Holley, Moses, Roberts/Sporn, Derynck, and Massagué laboratories (Fig. 5), resulting in the discovery and characterization of TGF-β1 in the early 1980s, have now led to the description of an almost unfathomable range of activities of TGF-β, and the identification of a large family of related growth and differentiation factors with important effects in development and adult physiology and disease pathogenesis. Many highly committed research groups have greatly contributed to our current mechanistic, structural, developmental, and physiological understanding of the biology of TGF-β and have made this into a highly exciting area of biology with substantial implications for our understanding of various diseases. The effects of TGF-β family proteins on many individual cell types and the very nature of Smad signaling, complemented by non-Smad pathways, emphasize and have made us appreciate the contextual nature of TGF-β effects at the cell, tissue, and organismal level. Certainly, 35 years after its discovery, the future of TGF-β research is bright and sure to yield exciting new insights and pharmacological approaches to manipulate this pathway for clinical benefit.
Figure 5.
Early contributors to the development of the transforming growth factor β (TGF-β) field. The picture taken in February 2006 shows (seated from left) Rik Derynck, Anita Roberts, Harold Moses, and (standing from left) Michael Sporn and Joan Massagué.
Footnotes
Editors: Rik Derynck and Kohei Miyazono
Additional Perspectives on The Biology of the TGF-β Family available at www.cshperspectives.org
REFERENCES
- Akhurst RJ, Derynck R. 2001. TGF-β signaling in cancer—A double-edged sword. Trends Cell Biol 11: S44–S51. [DOI] [PubMed] [Google Scholar]
- Alliston T, Choy L, Ducy P, Karsenty G, Derynck R. 2001. TGF-β-induced repression of CBFA1 by Smad3 decreases cbfa1 and osteocalcin expression and inhibits osteoblast differentiation. EMBO J 20: 2254–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzano MA, Roberts AB, Smith JM, Lamb LC, Sporn MB. 1982. Purification by reverse-phase high-performance liquid chromatography of an epidermal growth factor-dependent transforming growth factor. Anal Biochem 125: 217–224. [DOI] [PubMed] [Google Scholar]
- Assoian RK, Komoriya A, Meyers CA, Miller DM, Sporn MB. 1983. Transforming growth factor β in human platelets. Identification of a major storage site, purification, and characterization. J Biol Chem 258: 7155–7160. [PubMed] [Google Scholar]
- Attisano L, Wrana JL, Cheifetz S, Massagué J. 1992. Novel activin receptors: Distinct genes and alternative mRNA splicing generate a repertoire of serine/threonine kinase receptors. Cell 68: 97–108. [DOI] [PubMed] [Google Scholar]
- Attisano L, Cárcamo J, Ventura F, Weis FM, Massagué J, Wrana JL. 1993. Identification of human activin and TGF-β type I receptors that form heteromeric kinase complexes with type II receptors. Cell 75: 671–80. [DOI] [PubMed] [Google Scholar]
- Baarends WM, van Helmond MJ, Post M, van der Schoot PJ, Hoogerbrugge JW, de Winter JP, Uilenbroek JT, Karels B, Wilming LG, Meijers JH, et al. 1994. A novel member of the transmembrane serine/threonine kinase receptor family is specifically expressed in the gonads and in mesenchymal cells adjacent to the Müllerian duct. Development 120: 189–197. [DOI] [PubMed] [Google Scholar]
- Bai S, Cao X. 2002. A nuclear antagonistic mechanism of inhibitory Smads in transforming growth factor β signaling. J Biol Chem 277: 4176–4182. [DOI] [PubMed] [Google Scholar]
- Baker JC, Harland RM. 1996. A novel mesoderm inducer, Madr2, functions in the activin signal transduction pathway. Genes Dev 10: 1880–1889. [DOI] [PubMed] [Google Scholar]
- Bassing CH, Yingling JM, Howe DJ, Wang T, He WW, Gustafson ML, Shah P, Donahoe PK, Wang XF. 1994. A transforming growth factor β type I receptor that signals to activate gene expression. Science 263: 87–89. [DOI] [PubMed] [Google Scholar]
- Bauer H, Lele Z, Rauch GJ, Geisler R, Hammerschmidt M. 2001. The type I serine/threonine kinase receptor Alk8/Lost-a-fin is required for Bmp2b/7 signal transduction during dorsoventral patterning of the zebrafish embryo. Development 128: 849–858. [DOI] [PubMed] [Google Scholar]
- Bernabeu C, Lopez-Novoa JM, Quintanilla M. 2009. The emerging role of TGF-β superfamily coreceptors in cancer. Biochim Biophys Acta 1792: 954–973. [DOI] [PubMed] [Google Scholar]
- Border WA, Okuda S, Languino LR, Sporn MB, Ruoslahti E. 1990. Suppression of experimental glomerulonephritis by antiserum against transforming growth factor β1. Nature 346: 371–374. [DOI] [PubMed] [Google Scholar]
- Briones-Orta MA, Tecalco-Cruz AC, Sosa-Garrocho M, Caligaris C, Macías-Silva M. 2011. Inhibitory Smad7: Emerging roles in health and disease. Curr Mol Pharmacol 4: 141–153. [PubMed] [Google Scholar]
- Cate RL, Mattaliano RJ, Hession C, Tizard R, Farber NM, Cheung A, Ninfa EG, Frey AZ, Gash DJ, Chow EP, et al. 1986. Isolation of the bovine and human genes for Müllerian inhibiting substance and expression of the human gene in animal cells. Cell 45: 685–698. [DOI] [PubMed] [Google Scholar]
- Chapman HA. 2011. Epithelial–mesenchymal interactions in pulmonary fibrosis. Annu Rev Physiol 73: 413–435. [DOI] [PubMed] [Google Scholar]
- Cheifetz S, Weatherbee JA, Tsang ML, Anderson JK, Mole JE, Lucas R, Massagué J. 1987. The transforming growth factor β system, a complex pattern of cross-reactive ligands and receptors. Cell 48: 409–415. [DOI] [PubMed] [Google Scholar]
- Cheifetz S, Ling N, Guillemin R, Massagué J. 1988. A surface component on GH3 pituitary cells that recognizes transforming growth factor β, activin, and inhibin. J Biol Chem 263: 17225–17228. [PubMed] [Google Scholar]
- Cheifetz S, Bellón T, Calés C, Vera S, Bernabeu C, Massagué J, Letarte M. 1992. Endoglin is a component of the transforming growth factor β receptor system in human endothelial cells. J Biol Chem 267: 19027–19030. [PubMed] [Google Scholar]
- Chen RH, Miettinen PJ, Maruoka EM, Choy L, Derynck R. 1995. A WD-domain protein that is associated with and phosphorylated by the type II TGF-β receptor. Nature 377: 548–552. [DOI] [PubMed] [Google Scholar]
- Chen X, Rubock MJ, Whitman M. 1996. A transcriptional partner for MAD proteins in TGF-β signaling. Nature 383: 691–696 (Erratum in Nature384: 648). [DOI] [PubMed] [Google Scholar]
- Chen YG, Liu F, Massagué J. 1997. Mechanism of TGF-β receptor inhibition by FKBP12. EMBO J 16: 3866–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CR, Kang Y, Siegel PM, Massagué J. 2002. E2F4/5 and p107 as Smad cofactors linking the TGF-β receptor to c-myc repression. Cell 110: 19–32. [DOI] [PubMed] [Google Scholar]
- Childs CB, Proper JA, Tucker RF, Moses HL. 1982. Serum contains a platelet-derived transforming growth factor. Proc Natl Acad Sci 79: 5312–5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui W, Fowlis DJ, Bryson S, Duffie E, Ireland H, Balmain A, Akhurst RJ. 1996. TGF-β1 inhibits the formation of benign skin tumors, but enhances progression to invasive spindle carcinomas in transgenic mice. Cell 86: 531–542. [DOI] [PubMed] [Google Scholar]
- Davoren PR. 1962. The isolation of insulin from a single cat pancreas. Biochim Biophys Acta 63: 150–153. [DOI] [PubMed] [Google Scholar]
- De Larco JE, Todaro GJ. 1978. Growth factors from murine sarcoma virus-transformed cells. Proc Natl Acad Sci 75: 4001–4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Martin R, Haendler B, Hofer-Warbinek R, Gaugitsch H, Wrann M, Schlüsener H, Seifert JM, Bodmer S, Fontana A, Hofer E. 1987. Complementary DNA for human glioblastoma-derived T cell suppressor factor, a novel member of the transforming growth factor β gene family. EMBO J 6: 3673–3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derynck R, Rhee L. 1987. Sequence of the porcine transforming growth factor β precursor. Nucleic Acids Res 15: 3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derynck R, Zhang Y. 1996. Intracellular signalling: The Mad way to do it. Curr Biol 6: 1226–1229. [DOI] [PubMed] [Google Scholar]
- Derynck R, Zhang YE. 2003. Smad-dependent and Smad-independent pathways in TGF-β family signalling. Nature 425: 577–584. [DOI] [PubMed] [Google Scholar]
- Derynck R, Roberts AB, Winkler ME, Chen EY, Goeddel DV. 1984. Human transforming growth factor α: Precursor structure and expression in E. coli. Cell 38: 287–297. [DOI] [PubMed] [Google Scholar]
- Derynck R, Jarrett JA, Chen EY, Eaton DH, Bell JR, Assoian RK, Roberts AB, Sporn MB, Goeddel DV. 1985. Human transforming growth factor β complementary DNA sequence and expression in normal and transformed cells. Nature 316: 701–705. [DOI] [PubMed] [Google Scholar]
- Derynck R, Jarrett JA, Chen EY, Goeddel DV. 1986. The murine transforming growth factor β precursor. J Biol Chem 261: 4377–4379. [PubMed] [Google Scholar]
- Derynck R, Goeddel DV, Ullrich A, Gutterman JU, Williams RD, Bringman TS, Berger WH. 1987. Synthesis of messenger RNAs for transforming growth factors α and β and the epidermal growth factor receptor by human tumors. Cancer Res 47: 707–712. [PubMed] [Google Scholar]
- Derynck R, Lindquist PB, Lee A, Wen D, Tamm J, Graycar JL, Rhee L, Mason AJ, Miller DA, Coffey RJ, et al. 1988. A new type of transforming growth factor β, TGF-β3. EMBO J 7: 3737–3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derynck R, Gelbart WM, Harland RM, Heldin CH, Kern SE, Massagué J, Melton DA, Mlodzik M, Padgett RW, Roberts AB, et al. 1996. Nomenclature: Vertebrate mediators of TGF-β family signals. Cell 87: 173. [DOI] [PubMed] [Google Scholar]
- Derynck R, Zhang Y, Feng XH. 1998. Smads: Transcriptional activators of TGF-β responses. Cell 95: 737–740. [DOI] [PubMed] [Google Scholar]
- Derynck R, Akhurst RJ, Balmain A. 2001. TGF-β signaling in tumor suppression and cancer progression. Nat Genet 29: 117–129. [DOI] [PubMed] [Google Scholar]
- Dickson MC, Martin JS, Cousins FM, Kulkarni AB, Karlsson S, Akhurst RJ. 1995. Defective haematopoiesis and vasculogenesis in transforming growth factor β1 knock out mice. Development 121: 1845–1854. [DOI] [PubMed] [Google Scholar]
- di Clemente N, Wilson C, Faure E, Boussin L, Carmillo P, Tizard R, Picard JY, Vigier B, Josso N, Cate R. 1994. Cloning, expression, and alternative splicing of the receptor for anti-Müllerian hormone. Mol Endocrinol 8: 1006–1020. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Gerber EE, Dietz HC. 2012. Matrix-dependent perturbation of TGF-β signaling and disease. FEBS Lett 586: 2003–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois CM, Laprise MH, Blanchette F, Gentry LE, Leduc R. 1995. Processing of transforming growth factor β1 precursor by human furin convertase. J Biol Chem 270: 10618–10624. [DOI] [PubMed] [Google Scholar]
- Dvorak HF. 1986. Tumors: Wounds that do not heal. Similarities between tumor stroma generation and wound healing. N Engl J Med 315: 1650–1659. [DOI] [PubMed] [Google Scholar]
- Ebner R, Chen RH, Shum L, Lawler S, Zioncheck TF, Lee A, Lopez AR, Derynck R. 1993a. Cloning of a type I TGF-β receptor and its effect on TGF-β binding to the type II receptor. Science 260: 1344–1348. [DOI] [PubMed] [Google Scholar]
- Ebner R, Chen RH, Lawler S, Zioncheck T, Derynck R. 1993b. Determination of type I receptor specificity by the type II receptors for TGF-β or activin. Science 262: 900–902. [DOI] [PubMed] [Google Scholar]
- Edwards DR, Murphy G, Reynolds JJ, Whitham SE, Docherty AJ, Angel P, Heath JK. 1987. Transforming growth factor β modulates the expression of collagenase and metalloproteinase inhibitor. EMBO J 6: 1899–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppert K, Scherer SW, Ozcelik H, Pirone R, Hoodless P, Kim H, Tsui LC, Bapat B, Gallinger S, Andrulis IL, et al. 1996. MADR2 maps to 18q21 and encodes a TGF-β-regulated MAD-related protein that is functionally mutated in colorectal carcinoma. Cell 86: 543–552. [DOI] [PubMed] [Google Scholar]
- Estevez M, Attisano L, Wrana JL, Albert PS, Massagué J, Riddle DL. 1993. The daf-4 gene encodes a bone morphogenetic protein receptor controlling C. elegans dauer larva development. Nature 365: 644–649. [DOI] [PubMed] [Google Scholar]
- Feng XH, Derynck R. 2005. Specificity and versatility in TGF-β signaling through Smads. Annu Rev Cell Dev Biol 21: 659–693. [DOI] [PubMed] [Google Scholar]
- Fowlis DJ, Cui W, Johnson SA, Balmain A, Akhurst RJ. 1996. Altered epidermal cell growth control in vivo by inducible expression of transforming growth factor β1 in the skin of transgenic mice. Cell Growth Differ 7: 679–687. [PubMed] [Google Scholar]
- Franzén P, ten Dijke P, Ichijo H, Yamashita H, Schulz P, Heldin CH, Miyazono K. 1993. Cloning of a TGF-β type I receptor that forms a heteromeric complex with the TGF-β type II receptor. Cell 75: 681–692. [DOI] [PubMed] [Google Scholar]
- Frolik CA, Dart LL, Meyers CA, Smith DM, Sporn MB. 1983. Purification and initial characterization of a type β transforming growth factor from human placenta. Proc Natl Acad Sci 80: 3676–3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolik CA, Wakefield LM, Smith DM, Sporn MB. 1984. Characterization of a membrane receptor for transforming growth factor β in normal rat kidney fibroblasts. J Biol Chem 259: 10995–11000. [PubMed] [Google Scholar]
- Gentry LE, Webb NR, Lim GJ, Brunner AM, Ranchalis JE, Twardzik DR, Lioubin MN, Marquardt H, Purchio AF. 1987. Type 1 transforming growth factor β: Amplified expression and secretion of mature and precursor polypeptides in Chinese hamster ovary cells. Mol Cell Biol 7: 3418–3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentry LE, Lioubin MN, Purchio AF, Marquardt H. 1988. Molecular events in the processing of recombinant type 1 pre-pro-transforming growth factor β to the mature polypeptide. Mol Cell Biol 8: 4162–4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgi LL, Albert PS, Riddle DL. 1990. daf-1, a C. elegans gene controlling dauer larva development, encodes a novel receptor protein kinase. Cell 61: 635–645. [DOI] [PubMed] [Google Scholar]
- Gougos A, Letarte M. 1990. Primary structure of endoglin, an RGD-containing glycoprotein of human endothelial cells. J Biol Chem 265: 8361–8364. [PubMed] [Google Scholar]
- Graff JM, Bansal A, Melton DA. 1996. Xenopus Mad proteins transduce distinct subsets of signals for the TGF-β superfamily. Cell 85: 479–487. [DOI] [PubMed] [Google Scholar]
- Gray AM, Mason AJ. 1990. Requirement for activin A and transforming growth factor β1 pro-regions in homodimer assembly. Science 247: 1328–1330. [DOI] [PubMed] [Google Scholar]
- Griswold-Prenner I, Kamibayashi C, Maruoka EM, Mumby MC, Derynck R. 1998. Physical and functional interactions between type I transforming growth factor β receptors and Bα, a WD-40 repeat subunit of phosphatase 2A. Mol Cell Biol 18: 6595–6604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grönroos E, Hellman U, Heldin CH, Ericsson J. 2002. Control of Smad7 stability by competition between acetylation and ubiquitination. Mol Cell 10: 483–493. [DOI] [PubMed] [Google Scholar]
- Gruppuso PA, Mikumo R, Brautigan DL, Braun L. 1991. Growth arrest induced by transforming growth factor β1 is accompanied by protein phosphatase activation in human keratinocytes. J Biol Chem 266: 3444–3448. [PubMed] [Google Scholar]
- Haddow A. 1972. Molecular repair, wound healing, and carcinogenesis: Tumor production a possible overhealing? Adv Cancer Res 16: 181–234. [DOI] [PubMed] [Google Scholar]
- Hahn SA, Schutte M, Hoque AT, Moskaluk CA, da Costa LT, Rozenblum E, Weinstein CL, Fischer A, Yeo CJ, Hruban RH, et al. 1996. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science 271: 350–353. [DOI] [PubMed] [Google Scholar]
- Hanks SK, Armour R, Baldwin JH, Maldonado F, Spiess J, Holley RW. 1988. Amino acid sequence of the BSC-1 cell growth inhibitor (polyergin) deduced from the nucleotide sequence of the cDNA. Proc Natl Acad Sci 85: 79–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata A, Lagna G, Massagué J, Hemmati-Brivanlou A. 1998. Smad6 inhibits BMP/Smad1 signaling by specifically competing with the Smad4 tumor suppressor. Genes Dev 12: 186–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay ED. 1995. An overview of epithelio-mesenchymal transformation. Acta Anat (Basel) 154: 8–20. [DOI] [PubMed] [Google Scholar]
- Hayashi H, Abdollah S, Qiu Y, Cai J, Xu YY, Grinnell BW, Richardson MA, Topper JN, Gimbrone MA Jr, Wrana JL, et al. 1997. The MAD-related protein Smad7 associates with the TGF-β receptor and functions as an antagonist of TGF-β signaling. Cell 89: 1165–1173. [DOI] [PubMed] [Google Scholar]
- He WW, Gustafson ML, Hirobe S, Donahoe PK. 1993. Developmental expression of four novel serine/threonine kinase receptors homologous to the activin/transforming growth factor β type II receptor family. Dev Dyn 196: 133–142. [DOI] [PubMed] [Google Scholar]
- Heine U, Munoz EF, Flanders KC, Ellingsworth LR, Lam HY, Thompson NL, Roberts AB, Sporn MB. 1987. Role of transforming growth factor β in the development of the mouse embryo. J Cell Biol 105: 2861–2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heino J, Ignotz RA, Hemler ME, Crouse C, Massagué J. 1989. Regulation of cell adhesion receptors by transforming growth factor β. Concomitant regulation of integrins that share a common β1 subunit. J Biol Chem 264: 380–388. [PubMed] [Google Scholar]
- Holley RW. 1972. A unifying hypothesis concerning the nature of malignant growth. Proc Natl Acad Sci 69: 2840–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holley RW. 1975. Control of growth of mammalian cells in culture. Nature 258: 487–490. [DOI] [PubMed] [Google Scholar]
- Holley RW, Armour R, Baldwin JH. 1978. Density-dependent regulation of growth of BSC-1 cells in cell culture: Growth inhibitors formed by the cells. Proc Natl Acad Sci 75: 1864–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoodless PA, Haerry T, Abdollah S, Stapleton M, O’Connor MB, Attisano L, Wrana JL. 1996. MADR1, a MAD-related protein that functions in BMP2 signaling pathways. Cell 85: 489–500. [DOI] [PubMed] [Google Scholar]
- Hua X, Liu X, Ansari DO, Lodish HF. 1998. Synergistic cooperation of TFE3 and smad proteins in TGF-β-induced transcription of the plasminogen activator inhibitor-1 gene. Genes Dev 12: 3084–3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SS, Huang JS. 2005. TGF-β control of cell proliferation. J Cell Biochem 96: 447–462. [DOI] [PubMed] [Google Scholar]
- Ignotz RA, Massagué J. 1987. Cell adhesion protein receptors as targets for transforming growth factor β action. Cell 51: 189–197. [DOI] [PubMed] [Google Scholar]
- Ikushima H, Miyazono K. 2010. TGF-β signalling: A complex web in cancer progression. Nat Rev Cancer 10: 415–424. [DOI] [PubMed] [Google Scholar]
- Imamura T, Takase M, Nishihara A, Oeda E, Hanai J, Kawabata M, Miyazono K. 1997. Smad6 inhibits signalling by the TGF-β superfamily. Nature 389: 622–626. [DOI] [PubMed] [Google Scholar]
- Jhappan C, Geiser AG, Kordon EC, Bagheri D, Hennighausen L, Roberts AB, Smith GH, Merlino G. 1993. Targeting expression of a transforming growth factor β1 transgene to the pregnant mammary gland inhibits alveolar development and lactation. EMBO J 12: 1835–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JS, Alliston T, Delston R, Derynck R. 2005. Repression of Runx2 function by TGF-β through recruitment of class II histone deacetylases by Smad3. EMBO J 24: 2543–2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanzaki T, Olofsson A, Morén A, Wernstedt C, Hellman U, Miyazono K, Claesson-Welsh L, Heldin CH. 1990. TGF-β1 binding protein: A component of the large latent complex of TGF-β1 with multiple repeat sequences. Cell 61: 1051–1061. [DOI] [PubMed] [Google Scholar]
- Katsuno Y, Lamouille S, Derynck R. 2013. TGF-β signaling and epithelial-mesenchymal transition in cancer progression. Curr Opin Oncol 25: 76–84. [DOI] [PubMed] [Google Scholar]
- Kawabata M, Chytil A, Moses HL. 1995. Cloning of a novel type II serine/threonine kinase receptor through interaction with the type I transforming growth factor β receptor. J Biol Chem 270: 5625–5630. [DOI] [PubMed] [Google Scholar]
- Kehrl JH, Roberts AB, Wakefield LM, Jakowlew S, Sporn MB, Fauci AS. 1986a. Transforming growth factor β is an important immunomodulatory protein for human B lymphocytes. J Immunol 137: 3855–3860. [PubMed] [Google Scholar]
- Kehrl JH, Wakefield LM, Roberts AB, Jakowlew S, Alvarez-Mon M, Derynck R, Sporn MB, Fauci AS. 1986b. Production of transforming growth factor β by human T lymphocytes and its potential role in the regulation of T cell growth. J Exp Med 163: 1037–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr LD, Olashaw NE, Matrisian LM. 1988. Transforming growth factor β1 and cAMP inhibit transcription of epidermal growth factor- and oncogene-induced transin RNA. J Biol Chem 263: 16999–17005. [PubMed] [Google Scholar]
- Kim J, Johnson K, Chen HJ, Carroll S, Laughon A. 1997. Drosophila Mad binds to DNA and directly mediates activation of vestigial by Decapentaplegic. Nature 388: 304–308. [DOI] [PubMed] [Google Scholar]
- Kramer IM, Koornneef I, de Laat SW, van den Eijnden-van Raaij AJ. 1991. TGF-β1 induces phosphorylation of the cyclic AMP responsive element binding protein in ML-CCl64 cells. EMBO J 10: 1083–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni AB, Huh CG, Becker D, Geiser A, Lyght M, Flanders KC, Roberts AB, Sporn MB, Ward JM, Karlsson S. 1993. Transforming growth factor β1 null mutation in mice causes excessive inflammatory response and early death. Proc Natl Acad Sci 90: 770–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbé E, Silvestri C, Hoodless PA, Wrana JL, Attisano L. 1998. Smad2 and Smad3 positively and negatively regulate TGF-β-dependent transcription through the forkhead DNA-binding protein FAST2. Mol Cell 2:109–120. [DOI] [PubMed] [Google Scholar]
- Lagna G, Hata A, Hemmati-Brivanlou A, Massagué J. 1996. Partnership between DPC4 and SMAD proteins in TGF-β signalling pathways. Nature 383: 832–836. [DOI] [PubMed] [Google Scholar]
- Laiho M, Saksela O, Andreasen PA, Keski-Oja J. 1986. Enhanced production and extracellular deposition of the endothelial-type plasminogen activator inhibitor in cultured human lung fibroblasts by transforming growth factor β. J Cell Biol 103: 2403–2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laiho M, Weis FMB, Boyd FT, Ignotz RA, Massagué J. 1991. Responsiveness to transforming growth factor β restored by complementation between cells defective in TGF-β receptors I and II. J Biol Chem 286: 9106–9112. [PubMed] [Google Scholar]
- Lamouille S, Xu J, Derynck R. 2014. Molecular mechanisms of epithelial–mesenchymal transition. Nat Rev Mol Cell Biol 15: 178–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence DA, Pircher R, Kryceve-Martinerie C, Jullien P. 1984. Normal embryo fibroblasts release transforming growth factors in a latent form. J Cell Physiol 121: 184–188. [DOI] [PubMed] [Google Scholar]
- Lawrence DA, Pircher R, Jullien P. 1985. Conversion of a high molecular weight latent β-TGF from chicken embryo fibroblasts into a low molecular weight active β-TGF under acidic conditions. Biochem Biophys Res Commun 133: 1026–1034. [DOI] [PubMed] [Google Scholar]
- Lehnert SA, Akhurst RJ. 1988. Embryonic expression pattern of TGF-β type-1 RNA suggests both paracrine and autocrine mechanisms of action. Development 104: 263–273. [DOI] [PubMed] [Google Scholar]
- Leof EB, Proper JA, Goustin AS, Shipley GD, DiCorleto PE, Moses HL. 1986. Induction of c-sis mRNA and activity similar to platelet-derived growth factor by transforming growth factor β: A proposed model for indirect mitogenesis involving autocrine activity. Proc Natl Acad Sci 83: 2453–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letterio JJ, Roberts AB. 1998. Regulation of immune responses by TGF-β. Annu Rev Immunol 16: 137–161. [DOI] [PubMed] [Google Scholar]
- Letterio JJ, Geiser AG, Kulkarni AB, Roche NS, Sporn MB, Roberts AB. 1994. Maternal rescue of transforming growth factor β1 null mice. Science 264: 1936–1938. [DOI] [PubMed] [Google Scholar]
- Levinson AD, Oppermann H, Levintow L, Varmus HE, Bishop JM. 1978. Evidence that the transforming gene of avian sarcoma virus encodes a protein kinase associated with a phosphoprotein. Cell 15: 561–572. [DOI] [PubMed] [Google Scholar]
- Lin HY, Wang XF, Ng-Eaton E, Weinberg RA, Lodish HF. 1992. Expression cloning of the TGF-β type II receptor, a functional transmembrane serine/threonine kinase. Cell 68: 775–785. [DOI] [PubMed] [Google Scholar]
- Liu F, Ventura F, Doody J, Massagué J. 1995. Human type II receptor for bone morphogenic proteins (BMPs): Extension of the two-kinase receptor model to the BMPs. Mol Cell Biol 15: 3479–3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Hata A, Baker JC, Doody J, Cárcamo J, Harland RM, Massagué J. 1996. A human Mad protein acting as a BMP-regulated transcriptional activator. Nature 381: 620–623. [DOI] [PubMed] [Google Scholar]
- Liu D, Black BL, Derynck R. 2001. TGF-β inhibits muscle differentiation through functional repression of myogenic transcription factors by Smad3. Genes Dev 15: 2950–2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Kang JS, Derynck R. 2004. TGF-β-activated Smad3 represses MEF2-dependent transcription in myogenic differentiation. EMBO J 23: 1557–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Casillas F, Cheifetz S, Doody J, Andrés JL, Lane WS, Massagué J. 1991. Structure and expression of the membrane proteoglycan betaglycan, a component of the TGF-β receptor system. Cell 67: 785–795. [DOI] [PubMed] [Google Scholar]
- Lopez-Casillas F, Wrana JL, Massagué J. 1993. Betaglycan presents ligand to the TGF-β signaling receptor. Cell 73: 1435–1444. [DOI] [PubMed] [Google Scholar]
- Lyons RM, Keski-Oja J, Moses HL. 1988. Proteolytic activation of latent transforming growth factor β from fibroblast-conditioned medium. J Cell Biol 106: 1659–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macías-Silva M, Abdollah S, Hoodless PA, Pirone R, Attisano L, Wrana JL. 1996. MADR2 is a substrate of the TGF-β receptor and its phosphorylation is required for nuclear accumulation and signaling. Cell 87: 1215–1224. [DOI] [PubMed] [Google Scholar]
- Markowitz S, Wang J, Myeroff L, Parsons R, Sun L, Lutterbaugh J, Fan RS, Zborowska E, Kinzler KW, Vogelstein B. 1995. Inactivation of the type II TGF-β receptor in colon cancer cells with microsatellite instability. Science 268: 1336–1338. [DOI] [PubMed] [Google Scholar]
- Marquardt H, Lioubin MN, Ikeda T. 1987. Complete amino acid sequence of human transforming growth factor type β2. J Biol Chem 262: 12127–12131. [PubMed] [Google Scholar]
- Mason AJ, Hayflick JS, Ling N, Esch F, Ueno N, Ying SY, Guillemin R, Niall H, Seeburg PH. 1985. Complementary DNA sequences of ovarian follicular fluid inhibin show precursor structure and homology with transforming growth factor β. Nature 318: 659–663. [DOI] [PubMed] [Google Scholar]
- Massagué J. 1990. The transforming growth factor β family. Annu Rev Cell Biol 6: 597–641. [DOI] [PubMed] [Google Scholar]
- Massagué J. 2008. TGF-β in cancer. Cell 134: 215–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué J, Like B. 1985. Cellular receptors for type β transforming growth factor. Ligand binding and affinity labeling in human and rodent cell lines. J Biol Chem 260: 2636–2645. [PubMed] [Google Scholar]
- Massagué J, Czech MP, Iwata K, De Larco JE, Todaro GJ. 1982. Affinity labeling of a transforming growth factor receptor that does not interact with epidermal growth factor. Proc Natl Acad Sci 79: 6822–6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué J, Seoane J, Wotton D. 2005. Smad transcription factors. Genes Dev 19: 2783–2810. [DOI] [PubMed] [Google Scholar]
- Mathews LS, Vale WW. 1991. Expression cloning of an activin receptor, a predicted transmembrane serine kinase. Cell 65: 973–982. [DOI] [PubMed] [Google Scholar]
- Mathews LS, Vale WW, Kintner CR. 1992. Cloning of a second type of activin receptor and functional characterization in Xenopus embryos. Science 255: 1702–1705. [DOI] [PubMed] [Google Scholar]
- McAllister KA, Grogg KM, Johnson DW, Gallione CJ, Baldwin MA, Jackson CE, Helmbold EA, Markel DS, McKinnon WC, Murrell J, et al. 1994. Endoglin, a TGF-β binding protein of endothelial cells, is the gene for hereditary haemorrhagic telangiectasia type 1. Nat Genet 8: 345–351. [DOI] [PubMed] [Google Scholar]
- McDonald NQ, Hendrickson WA. 1993. A structural superfamily of growth factors containing a cystine knot motif. Cell 73: 421–424. [DOI] [PubMed] [Google Scholar]
- Miettinen PJ, Ebner R, Lopez AR, Derynck R. 1994. TGF-β induced transdifferentiation of mammary epithelial cells to mesenchymal cells: Involvement of type I receptors. J Cell Biol 127: 2021–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millan FA, Denhez F, Kondaiah P, Akhurst RJ. 1991. Embryonic gene expression patterns of TGF-β1, β2 and β3 suggest different developmental functions in vivo. Development 111: 131–143. [DOI] [PubMed] [Google Scholar]
- Mintzer KA, Lee MA, Runke G, Trout J, Whitman M, Mullins MC. 2001. Lost-a-fin encodes a type I BMP receptor, Alk8, acting maternally and zygotically in dorsoventral pattern formation. Development 128: 859–869. [DOI] [PubMed] [Google Scholar]
- Mitchell EJ, Lee K, O’Connor-McCourt MD. 1992. Characterization of transforming growth factor β (TGF-β) receptors on BeWo choriocarcinoma cells including the identification of a novel 38-kDa TGF-β binding glycoprotein. Mol Biol Cell 3: 1295–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazono K, Hellman U, Wernstedt C, Heldin CH. 1988. Latent high molecular weight complex of transforming growth factor β1. Purification from human platelets and structural characterization. J Biol Chem 263: 6407–6415. [PubMed] [Google Scholar]
- Miyazono K, Olofsson A, Colosetti P, Heldin CH. 1991. A role of the latent TGF-β1-binding protein in the assembly and secretion of TGF-β1. EMBO J 10: 1091–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazono K, Kusanagi K, Inoue H. 2001. Divergence and convergence of TGF-β/BMP signaling. J Cell Physiol 187: 265–276. [DOI] [PubMed] [Google Scholar]
- Miyazono K, Kamiya Y, Morikawa M. 2010. Bone morphogenetic protein receptors and signal transduction. J Biochem 147: 35–51. [DOI] [PubMed] [Google Scholar]
- Moses HL, Proper JA, Volkenant ME, Wells DJ, Getz MJ. 1978. Mechanism of growth arrest of chemically transformed cells in culture. Cancer Res 38: 2807–2812. [PubMed] [Google Scholar]
- Moses HL, Wells DJ, Swartzendruber DE, Getz MJ. 1979. Comparison of RNA metabolism in G1-arrested and stimulated nontransformed and chemically transformed mouse embryo cells in culture. Cancer Res 39: 4516–4524. [PubMed] [Google Scholar]
- Moses HL, Branum EL, Proper JA, Robinson RA. 1981. Transforming growth factor production by chemically transformed cells. Cancer Res 41: 2842–2848. [PubMed] [Google Scholar]
- Moses HL, Tucker RF, Leof EB, Coffey RJ Jr, Halper J, Shipley GD. 1985. Type β transforming growth factor is a growth stimulator and a growth inhibitor. Cancer Cells 3: 65–71. [Google Scholar]
- Moustakas A, Heldin CH. 2005. Non-Smad TGF-β signals. J Cell Sci 118: 3573–3584. [DOI] [PubMed] [Google Scholar]
- Mucsi I, Skorecki KL, Goldberg HJ. 1996. Extracellular signal-regulated kinase and the small GTP-binding protein, Rac, contribute to the effects of transforming growth factor β1 on gene expression. J Biol Chem 271: 16567–16572. [DOI] [PubMed] [Google Scholar]
- Mulder KM, Morris SL. 1992. Activation of p21ras by transforming growth factor β in epithelial cells. J Biol Chem 267: 5029–5031. [PubMed] [Google Scholar]
- Mullis K, Faloona F, Scharf S, Saiki R, Horn G, Erlich H. 1986. Specific enzymatic amplification of DNA in vitro: The polymerase chain reaction. Cold Spring Harb Symp Quant Biol 51: 263–273. [DOI] [PubMed] [Google Scholar]
- Nakao A, Afrakhte M, Morén A, Nakayama T, Christian JL, Heuchel R, Itoh S, Kawabata M, Heldin NE, Heldin CH, et al. 1997. Identification of Smad7, a TGF-β-inducible antagonist of TGF-β signalling. Nature 389: 631–635. [DOI] [PubMed] [Google Scholar]
- Nieto MA. 2011. The ins and outs of the epithelial to mesenchymal transition in health and disease. Annu Rev Cell Dev Biol 27: 347–376. [DOI] [PubMed] [Google Scholar]
- Nohno T, Ishikawa T, Saito T, Hosokawa K, Noji S, Wolsing DH, Rosenbaum JS. 1995. Identification of a human type II receptor for bone morphogenetic protein-4 that forms differential heteromeric complexes with bone morphogenetic protein type I receptors. J Biol Chem 270: 22522–22526. [DOI] [PubMed] [Google Scholar]
- O’Connor-McCourt MD, Wakefield LM. 1987. Latent transforming growth factor β in serum. A specific complex with α2-macroglobulin. J Biol Chem 262: 14090–14099. [PubMed] [Google Scholar]
- Overall CM, Wrana JL, Sodek J. 1989. Independent regulation of collagenase, 72-kDa progelatinase, and metalloendoproteinase inhibitor expression in human fibroblasts by transforming growth factor β. J Biol Chem 264: 1860–1869. [PubMed] [Google Scholar]
- Padgett RW, St Johnston RD, Gelbart WM. 1987. A transcript from a Drosophila pattern gene predicts a protein homologous to the transforming growth factor β family. Nature 325: 81–84. [DOI] [PubMed] [Google Scholar]
- Pelton RW, Nomura S, Moses HL, Hogan BL. 1989. Expression of transforming growth factor β2 RNA during murine embryogenesis. Development 106: 759–767. [DOI] [PubMed] [Google Scholar]
- Pelton RW, Dickinson ME, Moses HL, Hogan BL. 1990a. In situ hybridization analysis of TGF-β3 RNA expression during mouse development: Comparative studies with TGF-β1 and β2. Development 110: 609–620. [DOI] [PubMed] [Google Scholar]
- Pelton RW, Hogan BL, Miller DA, Moses HL. 1990b. Differential expression of genes encoding TGFs β1, β2, and β3 during murine palate formation. Dev Biol 141: 456–460. [DOI] [PubMed] [Google Scholar]
- Pelton RW, Saxena B, Jones M, Moses HL, Gold LI. 1991. Immunohistochemical localization of TGF-β1, TGF-β2, and TGF-β3 in the mouse embryo: Expression patterns suggest multiple roles during embryonic development. J Cell Biol 115: 1091–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickup M, Novitskiy S, Moses HL. 2013. The roles of TGFβ in the tumour microenvironment. Nat Rev Cancer 13: 788–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce DF Jr, Johnson MD, Matsui Y, Robinson SD, Gold LI, Purchio AF, Daniel CW, Hogan BL, Moses HL. 1993. Inhibition of mammary duct development but not alveolar outgrowth during pregnancy in transgenic mice expressing active TGF-β1. Genes Dev 7: 2308–2317. [DOI] [PubMed] [Google Scholar]
- Pierce DF Jr, Gorska AE, Chytil A, Meise KS, Page DL, Coffey RJ Jr, Moses HL. 1995. Mammary tumor suppression by transforming growth factor β1 transgene expression. Proc Natl Acad Sci 92: 4254–4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pircher R, Lawrence DA, Jullien P. 1984. Latent β-transforming growth factor in nontransformed and Kirsten sarcoma virus-transformed normal rat kidney cells, clone 49F. Cancer Res 44: 5538–5543. [PubMed] [Google Scholar]
- Postlethwaite AE, Keski-Oja J, Moses HL, Kang AH. 1987. Stimulation of the chemotactic migration of human fibroblasts by transforming growth factor β. J Exp Med 165: 251–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts JD, Dagle JM, Walder JA, Weeks DL, Runyan RB. 1991. Epithelial-mesenchymal transformation of embryonic cardiac endothelial cells is inhibited by a modified antisense oligodeoxynucleotide to transforming growth factor β3. Proc Natl Acad Sci 88: 1516–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proetzel G, Pawlowski SA, Wiles MV, Yin M, Boivin GP, Howles PN, Ding J, Ferguson MW, Doetschman T. 1995. Transforming growth factor β3 is required for secondary palate fusion. Nat Genet 11: 409–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proper JA, Bjornson CL, Moses HL. 1982. Mouse embryos contain polypeptide growth factor(s) capable of inducing a reversible neoplastic phenotype in nontransformed cells in culture. J Cell Physiol 110: 169–174. [DOI] [PubMed] [Google Scholar]
- Raftery LA, Twombly V, Wharton K, Gelbart WM. 1995. Genetic screens to identify elements of the decapentaplegic signaling pathway in Drosophila. Genetics 139: 241–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AB. 1995. Transforming growth factor β: Activity and efficacy in animal models of wound healing. Wound Repair Regen 3: 408–418. [DOI] [PubMed] [Google Scholar]
- Roberts AB, Lamb LC, Newton DL, Sporn MB, De Larco JE, Todaro GJ. 1980. Transforming growth factors: Isolation of polypeptides from virally and chemically transformed cells by acid/ethanol extraction. Proc Natl Acad Sci 77: 3494–3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AB, Anzano MA, Lamb LC, Smith JM, Sporn MB. 1981. New class of transforming growth factors potentiated by epidermal growth factor: Isolation from non-neoplastic tissues. Proc Natl Acad Sci 78: 5339–5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AB, Anzano MA, Meyers CA, Wideman J, Blacher R, Pan YC, Stein S, Lehrman SR, Smith JM, Lamb LC, et al. 1983a. Purification and properties of a type β transforming growth factor from bovine kidney. Biochemistry 22: 5692–5698. [DOI] [PubMed] [Google Scholar]
- Roberts AB, Frolik CA, Anzano MA, Sporn MB. 1983b. Transforming growth factors from neoplastic and nonneoplastic tissues. Fed Proc 42: 2621–2626. [PubMed] [Google Scholar]
- Roberts AB, Anzano MA, Wakefield LM, Roche NS, Stern DF, Sporn MB. 1985. Type β transforming growth factor: A bifunctional regulator of cellular growth. Proc Natl Acad Sci 82: 119–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AB, Sporn MB, Assoian RK, Smith JM, Roche NS, Wakefield LM, Heine UI, Liotta LA, Falanga V, Kehrl JH, et al. 1986. Transforming growth factor type β: Rapid induction of fibrosis and angiogenesis in vivo and stimulation of collagen formation in vitro. Proc Natl Acad Sci 83: 4167–4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts CJ, Birkenmeier TM, McQuillan JJ, Akiyama SK, Yamada SS, Chen WT, Yamada KM, McDonald JA. 1988. Transforming growth factor β stimulates the expression of fibronectin and of both subunits of the human fibronectin receptor by cultured human lung fibroblasts. J Biol Chem 263: 4586–4592. [PubMed] [Google Scholar]
- Rook AH, Kehrl JH, Wakefield LM, Roberts AB, Sporn MB, Burlington DB, Lane HC, Fauci AS. 1986. Effects of transforming growth factor β on the functions of natural killer cells: Depressed cytolytic activity and blunting of interferon responsiveness. J Immunol 136: 3916–3920. [PubMed] [Google Scholar]
- Rosenzweig BL, Imamura T, Okadome T, Cox GN, Yamashita H, ten Dijke P, Heldin CH, Miyazono K. 1995. Cloning and characterization of a human type II receptor for bone morphogenetic proteins. Proc Natl Acad Sci 92: 7632–7636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruberte E, Marty T, Nellen D, Affolter M, Basler K. 1995. An absolute requirement for both the type II and type I receptors, punt and thick veins, for dpp signaling in vivo. Cell 80: 889–897. [DOI] [PubMed] [Google Scholar]
- Russell WE, Coffey RJ Jr, Ouellette AJ, Moses HL. 1988. Type β transforming growth factor reversibly inhibits the early proliferative response to partial hepatectomy in the rat. Proc Natl Acad Sci 85: 5126–5130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford LP, Ormsby I, Gittenberger-de Groot AC, Sariola H, Friedman R, Boivin GP, Cardell EL, Doetschman T. 1997. TGF-β2 knockout mice have multiple developmental defects that are non-overlapping with other TGF-β knockout phenotypes. Development 124: 2659–2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Tsuboi R, Lyons R, Moses H, Rifkin DB. 1990. Characterization of the activation of latent TGF-β by co-cultures of endothelial cells and pericytes or smooth muscle cells: A self-regulating system. J Cell Biol 111: 757–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage C, Das P, Finelli AL, Townsend SR, Sun CY, Baird SE, Padgett RW. 1996. Caenorhabditis elegans genes sma-2, sma-3, and sma-4 define a conserved family of transforming growth factor β pathway components. Proc Natl Acad Sci 93: 790–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekelsky JJ, Newfeld SJ, Raftery LA, Chartoff EH, Gelbart WM. 1995. Genetic characterization and cloning of mothers against dpp, a gene required for decapentaplegic function in Drosophila melanogaster. Genetics 139: 1347–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyedin SM, Segarini PR, Rosen DM, Thompson AY, Bentz H, Graycar J. 1987. Cartilage-inducing factor-B is a unique protein structurally and functionally related to transforming growth factor β. J Biol Chem 262: 1946–1949. [PubMed] [Google Scholar]
- Sharples K, Plowman GD, Rose TM, Twardzik DR, Purchio AF. 1987. Cloning and sequence analysis of simian transforming growth factor β cDNA. DNA 6: 239–244. [DOI] [PubMed] [Google Scholar]
- Shipley GD, Pittelkow MR, Wille JJ Jr, Scott RE, Moses HL. 1986. Reversible inhibition of normal human prokeratinocyte proliferation by type β transforming growth factor-growth inhibitor in serum-free medium. Cancer Res 46: 2068–2071. [PubMed] [Google Scholar]
- Shore EM, Kaplan FS. 2010. Inherited human diseases of heterotopic bone formation. Nat Rev Rheumatol 6: 518–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shull MM, Ormsby I, Kier AB, Pawlowski S, Diebold RJ, Yin M, Allen R, Sidman C, Proetzel G, Calvin D, et al. 1992. Targeted disruption of the mouse transforming growth factor β1 gene results in multifocal inflammatory disease. Nature 359: 693–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberstein GB, Daniel CW. 1987. Reversible inhibition of mammary gland growth by transforming growth factor β. Science 237: 291–293. [DOI] [PubMed] [Google Scholar]
- Sporn MB, Todaro GJ. 1980. Autocrine secretion and malignant transformation of cells. N Engl J Med 303: 878–880. [DOI] [PubMed] [Google Scholar]
- Sporn MB, Roberts AB, Shull JH, Smith JM, Ward JM, Sodek J. 1983. Polypeptide transforming growth factors isolated from bovine sources and used for wound healing in vivo. Science 219: 1329–1331. [DOI] [PubMed] [Google Scholar]
- Stehelin D, Varmus HE, Bishop JM, Vogt PK. 1976. DNA related to the transforming gene(s) of avian sarcoma viruses is present in normal avian DNA. Nature 260: 170–173. [DOI] [PubMed] [Google Scholar]
- Swanstrom R, Parker RC, Varmus HE, Bishop JM. 1983. Transduction of a cellular oncogene: The genesis of Rous sarcoma virus. Proc Natl Acad Sci 80: 2519–2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Dijke P, Hill CS. 2004. New insights into TGF-β-Smad signalling. Trends Biochem Sci 29: 265–273. [DOI] [PubMed] [Google Scholar]
- ten Dijke P, Hansen P, Iwata KK, Pieler C, Foulkes JG. 1988. Identification of another member of the transforming growth factor type β gene family. Proc Natl Acad Sci 85: 4715–4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Dijke P, Ichijo H, Franzén P, Schulz P, Saras J, Toyoshima H, Heldin CH, Miyazono K. 1993. Activin receptor-like kinases: A novel subclass of cell-surface receptors with predicted serine/threonine kinase activity. Oncogene 8: 2879–2887. [PubMed] [Google Scholar]
- ten Dijke P, Miyazono K, Heldin CH. 2000. Signaling inputs converge on nuclear effectors in TGF-β signaling. Trends Biochem Sci 25: 64–70. [DOI] [PubMed] [Google Scholar]
- Todaro GJ, Huebner RJ. 1972. N.A.S. symposium: New evidence as the basis for increased efforts in cancer research. Proc Natl Acad Sci 69: 1009–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todaro GJ, De Larco JE, Cohen S. 1976. Transformation by murine and feline sarcoma viruses specifically blocks binding of epidermal growth factor to cells. Nature 264: 26–31. [DOI] [PubMed] [Google Scholar]
- Todaro GJ, De Larco JE, Marquardt H, Bryant ML, Sherwin SA, Sliski AH. 1979. Polypeptide growth factors produced by tumor cells and virus-transformed cells: A possible growth advantage for the producer cells. In Hormones and cell culture. Cell proliferation conference, Vol. 6, pp. 113–127. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Tsuji T, Okada F, Yamaguchi K, Nakamura T. 1990. Molecular cloning of the large subunit of transforming growth factor type β masking protein and expression of the mRNA in various rat tissues. Proc Natl Acad Sci 87: 8835–8839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuneizumi K, Nakayama T, Kamoshida Y, Kornberg TB, Christian JL, Tabata T. 1997. Daughters against dpp modulates dpp organizing activity in Drosophila wing development. Nature 389: 627–631. [DOI] [PubMed] [Google Scholar]
- Tuan TL, Cheung DT, Wu LT, Yee A, Gabriel S, Han B, Morton L, Nimni ME, Hall FL. 1996. Engineering, expression and renaturation of targeted TGF-β fusion proteins. Connect Tissue Res 34: 1–9. [DOI] [PubMed] [Google Scholar]
- Tucker RF, Branum EL, Shipley GD, Ryan RJ, Moses HL. 1984a. Specific binding to cultured cells of 125I-labeled type β transforming growth factor from human platelets. Proc Natl Acad Sci 81: 6757–6761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker RF, Shipley GD, Moses HL, Holley RW. 1984b. Growth inhibitor from BSC-1 cells closely related to platelet type β transforming growth factor. Science 226: 705–707. [DOI] [PubMed] [Google Scholar]
- Ventura F, Liu F, Doody J, Massagué J. 1996. Interaction of transforming growth factor-β receptor I with farnesyl-protein transferase-α in yeast and mammalian cells. J Biol Chem 271: 13931–13934. [DOI] [PubMed] [Google Scholar]
- Wahl SM, Hunt DA, Wakefield LM, McCartney-Francis N, Wahl LM, Roberts AB, Sporn MB. 1987. Transforming growth factor type β induces monocyte chemotaxis and growth factor production. Proc Natl Acad Sci 84: 5788–5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakefield LM, Smith DM, Flanders KC, Sporn MB. 1988. Latent transforming growth factor β from human platelets. A high molecular weight complex containing precursor sequences. J Biol Chem 263: 7646–7654. [PubMed] [Google Scholar]
- Wang XF, Lin HY, Ng-Eaton E, Downward J, Lodish HF, Weinberg RA. 1991. Expression cloning and characterization of the TGF-β type III receptor. Cell 67: 797–805. [DOI] [PubMed] [Google Scholar]
- Wang T, Li BY, Danielson PD, Shah PC, Rockwell S, Lechleider RJ, Martin J, Manganaro T, Donahoe PK. 1996a. The immunophilin FKBP12 functions as a common inhibitor of the TGF-β family type I receptors. Cell 86: 435–444. [DOI] [PubMed] [Google Scholar]
- Wang T, Danielson PD, Li BY, Shah PC, Kim SD, Donahoe PK. 1996b. The p21RAS farnesyltransferase α subunit in TGF-β and activin signaling. Science 271: 1120–1122. [DOI] [PubMed] [Google Scholar]
- Webb NR, Madisen L, Rose TM, Purchio AF. 1988. Structural and sequence analysis of TGF-β2 cDNA clones predicts two different precursor proteins produced by alternative mRNA splicing. DNA 7: 493–497. [DOI] [PubMed] [Google Scholar]
- Weeks DL, Melton DA. 1987. A maternal mRNA localized to the vegetal hemisphere in Xenopus eggs codes for a growth factor related to TGF-β. Cell 51: 861–867. [DOI] [PubMed] [Google Scholar]
- Wozney JM, Rosen V, Celeste AJ, Mitsock LM, Whitters MJ, Kriz RW, Hewick RM, Wang EA. 1988. Novel regulators of bone formation: Molecular clones and activities. Science 242: 1528–1534. [DOI] [PubMed] [Google Scholar]
- Wrana JL, Attisano L, Cárcamo J, Zentella A, Doody J, Laiho M, Wang XF, Massagué J. 1992. TGF-β signals through a heteromeric protein kinase receptor complex. Cell 71: 1003–1014. [DOI] [PubMed] [Google Scholar]
- Wrana JL, Tran H, Attisano L, Arora K, Childs SR, Massagué J, O’Connor MB. 1994a. Two distinct transmembrane serine/threonine kinases from Drosophila melanogaster form an activin receptor complex. Mol Cell Biol 14: 944–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrana JL, Attisano L, Wieser R, Ventura F, Massagué J. 1994b. Mechanism of activation of the TGF-β receptor. Nature 370: 341–347. [DOI] [PubMed] [Google Scholar]
- Wrann M, Bodmer S, de Martin R, Siepl C, Hofer-Warbinek R, Frei K, Hofer E, Fontana A. 1987. T cell suppressor factor from human glioblastoma cells is a 12.5-kd protein closely related to transforming growth factor β. EMBO J 6: 1633–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi K, Shirakabe K, Shibuya H, Irie K, Oishi I, Ueno N, Taniguchi T, Nishida E, Matsumoto K. 1995. Identification of a member of the MAPKKK family as a potential mediator of TGF-β signal transduction. Science 270: 2008–2011. [DOI] [PubMed] [Google Scholar]
- Yan Z, Winawer S, Friedman E. 1994. Two different signal transduction pathways can be activated by transforming growth factor β1 in epithelial cells. J Biol Chem 269: 13231–13237. [PubMed] [Google Scholar]
- Ying SY, Becker A, Ling N, Ueno N, Guillemin R. 1986. Inhibin and β type transforming growth factor (TGF-β) have opposite modulating effects on the follicle stimulating hormone (FSH)-induced aromatase activity of cultured rat granulosa cells. Biochem Biophys Res Commun 136: 969–975. [DOI] [PubMed] [Google Scholar]
- Zawel L, Dai JL, Buckhaults P, Zhou S, Kinzler KW, Vogelstein B, Kern SE. 1998. Human Smad3 and Smad4 are sequence-specific transcription activators. Mol Cell 1: 611–617. [DOI] [PubMed] [Google Scholar]
- Zhang YE. 2009. Non-Smad pathways in TGF-β signaling. Cell Res 19: 128–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Feng XH, Wu R, Derynck R. 1996. Receptor-associated Mad homologues synergize as effectors of the TGF-β response. Nature 383: 168–172. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Musci T, Derynck R. 1997. The tumor suppressor Smad4/DPC 4 as a central mediator of Smad function. Curr Biol 7: 270–276. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Feng XH, Derynck R. 1998. Smad3 and Smad4 cooperate with c-Jun/c-Fos to mediate TGF-β-induced transcription. Nature 394: 909–913 (Erratum in Nature396: 491). [DOI] [PubMed] [Google Scholar]