Abstract
Background
Autophagy has been reported to increase in cancer cells after radiation. However, it remains unknown whether increased autophagy as a result of radiation affects DNA damage repair and sensitizes cancer cells. In this study, the radiosensitization effect of rapamycin, a mammalian target of rapamycin inhibitor that induces autophagy, on human lung adenocarcinoma A549 cells was investigated.
Methods
A549 cells were treated with different concentrations of rapamycin. Cell viability was evaluated by methyl‐thiazolyl‐tetrazolium assay. Survival fraction values of A549 cells after radiotherapy were detected by colony formation assay. Autophagosome was observed by a transmission electron microscope. Furthermore, Western blot was employed to examine alterations in autophagy protein LC3 and p62, DNA damage protein γ–H2AX, and DNA damage repair proteins Rad51, Ku70, and Ku80. Rad51, Ku70, and Ku80 messenger ribonucleic acid (mRNA) expression levels were examined by real‐time polymerase chain reaction.
Results
Rapamycin suppressed A549 cell proliferation in dose and time‐dependent manners. An inhibitory concentration (IC) 10 dose of rapamycin could induce autophagy in A549 cells. Rapamycin combined with radiation significantly decreased the colony forming ability of cells, compared with rapamycin or radiation alone. Rapamycin and radiation combined increased γ–H2AX expression levels and decreased Rad51 and Ku80 expression levels, compared with single regimens. However, rapamycin treatment did not induce any change in Rad51, Ku70, and Ku80 mRNA levels, regardless of radiation.
Conclusions
These findings indicate that increasing autophagy sensitizes lung cancer cells to radiation.
Keywords: DNA damage, lung cancer, radiotherapy, rapamycin
Introduction
Lung cancer has the highest incidence and mortality of all cancers. Radiotherapy is one of the main treatments for lung cancer and it has been well established that radiotherapy destroys cancer cells primarily by DNA single strand and DNA double strand breaks (DSBs).1 Unrepaired DSBs are detrimental to cancer cells, leading to cellular autophagy, apoptosis, and necrosis. However, radiation‐treated lung cancers have a high rate of relapse, because DSB repair limits the effectiveness of radiotherapy in cancer treatment. New strategies that sensitize cancer cells to radiotherapy need to be developed.2
DNA DSBs could be repaired through homologous recombination (HR) and non‐homologous end‐joining (NHEJ) pathways. HR‐mediated DNA repair is an error‐free mechanism that usually uses a sister chromatid as a template to accurately repair DNA, and this only occurs in cycling cells. In contrast, NHEJ‐mediated DNA repair is an error‐prone mechanism that does not need sequence homology to repair, and this occurs at any stage of the cell cycle.3 Many proteins are involved in the process of DNA damage repair, such as HR mediator Rad51 and NHEJ mediator DNA‐dependent protein kinase catalytic subunit (PKcs), Ku70, and Ku80.4 , 5 Changes in these protein levels in cells would affect repair efficiency. For example, the upregulation of Rad51 and Ku70/Ku80 is associated with poor prognosis in lung cancer, which may lead to the efficient repair of radiotherapy‐induced DNA damage.6, 7 This results in cellular resistance to radiotherapy and tumor cell clonogenic survival, which limit the outcome of radiotherapy. Consistently, the downregulation in Ku70 or Rad51 expression significantly sensitized lung cancer cells to radiotherapy, indicating that the inhibition of DNA damage repair in cancer cells is a promising approach for the treatment of lung cancer.7, 8
Autophagy is the process of self‐digestion, which degrades dysfunctional and useless cellular elements mediated by the actions of lysosomes.9 It is related to both cell survival and cell death, and is cellular context‐dependent. Most recently, a plethora of evidence has indicated that autophagy is tightly associated with cancers, and that this has become a new therapeutic target in cancer treatment.10, 11, 12 Rapamycin‐induced autophagy in MCF‐7 breast cancer cells delayed the attendance of Rad51 and breast cancer 1 (BRCA1) in DSBs upon radiotherapy, leading to the accumulation of DSBs and cell death.13 Similarly, the sensitivity of pancreatic carcinoma cell PC‐2 to radiotherapy was significantly enhanced post‐rapamycin treatments, and the upregulation of autophagy mediator Beclin‐1 sensitized lung cancer cells to irradiation.14 , 15 These findings suggest that autophagy may play an important role in cancer radiotherapy.
However, information on autophagy and lung cancer are currently limited, and available information has shown contradicting results. For example, Cheng et al. demonstrated that p53 upregulation increased the sensitivity of lung adenocarcinoma cells to radiation, mediated by reduced autophagy.16 In contrast, Kim et al. reported that increased autophagy by mammalian target of rapamycin (mTOR) inhibitors sensitized lung cancer cells to radiation.17 This may indicate that autophagy has dual effects, in which cell survival is promoted and cell death is induced in cancer cells in a context‐dependent manner. Tumor cells would die if autophagy promotes the degradation of anti‐apoptotic proteins. Conversely, tumor cells would survive if autophagy promotes the degradation of pro‐apoptotic proteins.18 In the present study, we demonstrated that the induction of autophagy via rapamycin application sensitized lung cancer cell A549 to radiation, which was related to delayed DNA damage repair, along with downregulation in Rad51 and Ku80 expression.
Materials and methods
Cell culture and reagents
A549 cells (ATCC, Manassas, VA, USA) were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum (HyClone, Logan, UT, USA) containing penicillin (100 U/mL) and streptomycin (100 μg/mL), and incubated in a humidified incubator (95% air and 5% CO2) at 37°C. Rapamycin (Sigma‐Aldrich, St. Louis, MO, USA) was dissolved in dimethyl sulfoxide (DMSO).
Methyl‐thiazolyl‐tetrazolium assay
Cells were seeded in 96‐well plates. After culturing overnight, cells were treated with various concentrations of rapamycin (0, 0.1, 1, 10, 100, and 1000 nmol/L) for 24 hours, or exposed to 100 nmol/L of rapamycin at various time points (0, 12, 24, 36, and 48 hours). The medium was then exchanged with serum‐free medium containing 0.5 mg/mL of methyl‐thiazolyl‐tetrazolium (MTT), removed after four hours at 37°C, and formazan crystals were dissolved with DMSO. The amount of formazan was determined by measuring absorbance at 570 nm.
Western blot analysis
Total cell lysates were separated in 10% sodium dodecyl sulfate‐polyacrylamide gel electrophoresis. Proteins were transferred onto a nitrocellulose membrane, blocked with 5% nonfat milk for one hour at room temperature, and probed with a primary antibody overnight at 4°C. Primary antibodies used included anti‐LC3 (1:5000 dilution, Sigma), anti‐Rad51, anti‐Ku70/80, anti‐glyceraldehyde 3‐phosphate dehydrogenase (GAPDH; 1:1000, 1:500, and 1:1000 dilutions, respectively; Abcam, Cambridge, UK), and γ‐H2AX (1:1000 dilution, Cell Signaling Technologies, Danvers, MA, USA). The membranes were incubated with horseradish peroxidase‐conjugated secondary antibody at a dilution of 1:2000 for one hour at room temperature. Protein bands were visualized using ECL Western Blotting Detection Reagents (Millipore, Billerica, MA, USA) and exposed to an ECL Plus film (GE Healthcare, Piscataway, NJ, USA).
Real‐time polymerase chain reaction
Total RNA was extracted using Trizol (Ambio, Foster, CA, USA), according to the manufacturer's instructions. First‐strand cDNA was synthesized at a final volume of 20 μL using the Superscript III First‐Strand Synthesis System (Invitrogen, Carlsbad, CA, USA). Quantitative real‐time polymerase chain reaction (qRT‐PCR) analyses were performed using a SYBR Green mix in the Real‐Time PCR System (Applied Biosystems, Foster City, CA, USA). Twenty μL of PCR reaction was prepared as follows: 10 μL of 2 × SYBR Green PCR master mix, 0.4 μL of 10 μM of appropriate forward and reverse primers, 7.6 μL of RNase‐free water, and 2 μL of cDNA template. A negative control (no DNA template) was also performed for each master mix prepared. qRT‐PCR was performed for 10 minutes at 95°C, followed by 40 cycles of 15 seconds at 95°C and one minute at 60°C. A dissociation melting curve was generated using thermal conditions from 60°C to 95°C. An arbitrary unit was calculated by the comparative cycle threshold (CT) method, according to CT values of the internal control. Human GAPDH was used as a housekeeping internal reference for human messenger ribonucleic acid (mRNA). Primers used for RT‐PCR were Rad51, Ku70, Ku80, and GAPDH. The sequences for all primers are listed in Table 1.
Table 1.
Sequences of primers used for real‐time polymerase chain reaction
| Genes | Forward primers | Reverse primers |
|---|---|---|
| GAPDH | 5′‐CAGAACATCATCCCTGCCTCTAC‐3′ | 5′‐TTGAAGTCAGAGGAGACCACCTG‐3′ |
| Rad51 | 5′‐CAACCCATTTCACGGTTAGAGC‐3′ | 5′‐GCTTTGGCTTCACTAATTCCCTT‐3′ |
| Ku70 | 5′‐ATGGCAACTCCAGAGCAGGTG‐3′ | 5′‐AGTGCTTGGTGAGGGCTTCCA‐3′ |
| Ku80 | 5′‐TGGTGCGGTCGGGGAATA‐3′ | 5′‐CAGAAAGGGGATTGTCAGTGC‐3′ |
GAPDH, glyceraldehyde 3‐phosphate dehydrogenase.
Clonogenic survival assay
The experiment was performed as previously described.14 In brief, A549 cells were seeded into wells of a six‐well plate with a concentration of 200 cells per well. After overnight incubation, cells were treated with 100 nmol/L of rapamycin for 24 hours and/or exposed to 4 Gy of radiation. The culture medium was changed every three days. All cells were allowed to grow for an additional 12 days to form colonies, and were stained with 0.1% crystal purple. Colonies with more than 50 cells were counted. Survival fraction (SF) was calculated according to the plating efficiency of the control group using GraphPad version 5.0 (GraphPad Software, La Jolla, CA, USA), and expressed as mean ± standard deviation (SD).
Transmission electron microscope
A549 cells were fixed in 2.5% glutaraldehyde and 4% paraformaldehyde at pH 7.4. Cells were embedded in Epon, stained with uranyl acetate, and processed as previously described.19 The sections (80 nm) were viewed under an electron microscope (JEM‐1400/1011, Jeol, Japan) at the core facility of Nanchang University.
Statistical analysis
Data were expressed as means ± SD and analyzed by analysis of variance (ANOVA). Differences were considered significant at P < 0.05. All analyses were performed using GraphPad Prism 5.0 software (San Diego, CA, USA).
Results
Rapamycin induced cytotoxicity in A549 cells
First, to examine the effects of rapamycin in A549 cells, the proliferation of cultured A549 cells was measured by MTT assay after 24 hours of exposure to rapamycin at various concentrations and exposure to 100 nmol/L of rapamycin at various time points. Rapamycin suppressed cell proliferation in dose and time‐dependent manners, as shown in Figure 1. For subsequent trials, inhibitory concentration (IC)10 values were measured to examine the suppression of cell proliferation in A549 cells. IC10 values were approximately 100 nmol/L for A549 cell exposure to rapamycin after 24 hours. We decided to adopt a dose of 100 nmol/L of rapamycin for subsequent experimental studies.
Figure 1.
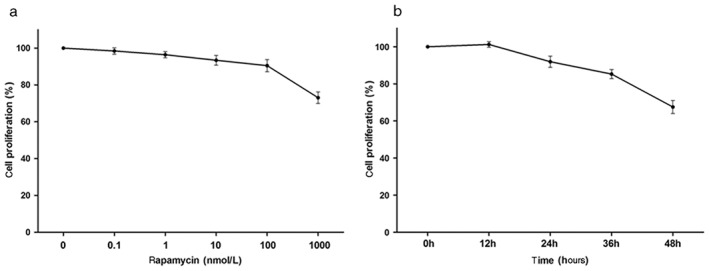
Rapamycin suppresses A549 cell proliferation. A549 cells were treated with varying concentrations of rapamycin for (a) 24 hours, and (b) at indicated time points with 100 nmol/L of rapamycin. Viability was detected by methyl‐thiazolyl‐tetrazolium assay. Data are representative of three independent experiments.
Reviewer 2 advised us to show the enhancement of radiation sensitivity by another drug, which would require the investigators to repeat the clonogenic assay with 1, 2, 4, 6, and 8 Gy of radiation. In our previous study, A549 cell survival was detected by colony formation assay after treatment with 0–12 Gy of radiation (Table 2). A549 cell proliferation was very low at a dose of > 4 Gy. Thus, the difference when clonogenic assay was repeated with 6 and 8 Gy of radiation combined with rapamycin was not significant.
Table 2.
Effect of different doses of radiation on A549 cell proliferation
| Group | IR | ||||||
|---|---|---|---|---|---|---|---|
| 0 Gy | 2 Gy | 4 Gy | 6 Gy | 8 Gy | 10 Gy | 12 Gy | |
| SF (%) | 100.00 | 68.15 ± 4.16* | 53.19 ± 3.86* | 29.16 ± 1.98* | 9.45 ± 1.21* | 2.98 ± 0.07* | 1.52 ± 0.23* |
P < 0.05. IR, irradiation; SF, survival fraction.
Rapamycin induced autophagy in A549 cells
Rapamycin is a common reagent used to induce autophagy. Therefore, we assessed whether rapamycin treatment induces autophagy in A549 cells. Electron microscopy was utilized to detect intracellular autophagosomes. Rare autophagosomes could be detected in vehicle treated cells, as shown in Figure 2. However, the exposure of A549 cells to 100 nmol/L of rapamycin (24 hours) or radiation (4 Gy) greatly increased the number of autophagosomes in the cytoplasm. The increment of autophagosomes further improved upon combining rapamycin with radiation in A549 cells. In addition, LC3 and p62 are the most common autophagic markers. Proteins were extracted after rapamycin and/or radiation treatment, and Western blot was performed to analyze LC3 and p62. Our results revealed that treatments with rapamycin/radiation significantly increased LC3II levels, leading to the increased ratio of LC3II/LC3I (Fig 3). Consistently, p62 expression decreased after rapamycin/radiation treatment, indicating that both rapamycin and radiation could induce cellular autophagy (Fig 3). Rapamycin and radiation combined further improved the number of autophagosomes.
Figure 2.
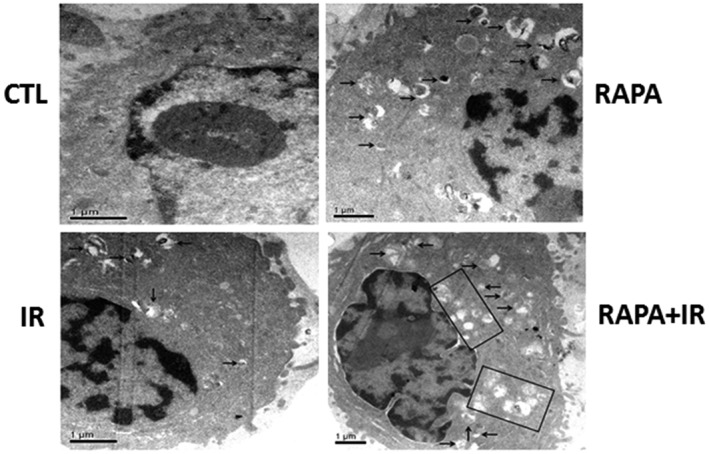
Rapamycin and radiation induce cellular autophagy. A549 cells were treated with 100 nmol/L of rapamycin (RAPA) for 24 hours and were subsequently exposed to 4 Gy of irradiation (IR). Cells were processed and observed under a transmission electron microscope (1000×). Arrows indicate intracellular autophagosomes. CTL, control.
Figure 3.
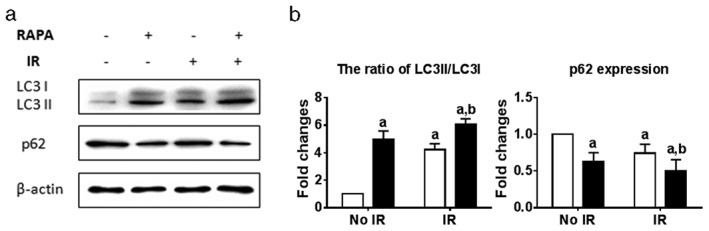
Rapamycin affected LC3II/I and p62 expression. Proteins were extracted after treatment with rapamycin and/or irradiation, and Western blot was performed to detect LC3 and p62. (a) Protein expression was quantitated using Adobe Photoshop CS4 software. (b) a: P < 0.05 versus No irradiation at 4 Gy (IR)+vehicle (Veh); b: P < 0.05 versus IR+Veh. RAPA, rapamycin at 100 nmol/L.
Rapamycin sensitized A549 cells to radiotherapy
In order to investigate whether rapamycin‐induced autophagy sensitizes A549 cells to radiation, cells were treated with 100 nmol/L of rapamycin for 24 hours and subsequently irradiated cells with 4 Gy of γ‐irradiation. Using the colony forming ability assay, both rapamycin and radiation demonstrated a modest decrease in cell survival fraction when the number of colonies were counted at day 12 after radiation (Fig 4). However, rapamycin administration significantly reduced survival fraction under radiation treatment (Fig 4). This suggests that rapamycin treatment sensitized A549 cells to radiation and increased cell death.
Figure 4.
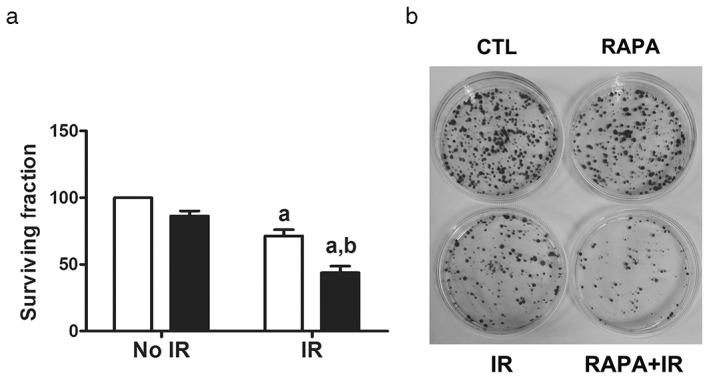
Rapamycin treatment sensitized A549 cells to radiation by clonogenic assay. A549 cells were incubated with vehicle or 100 nmol/L of rapamycin (RAPA) for 24 hours before exposure to 4 Gy of irradiation (IR). (a) Survival fraction of vehicle or rapamycin‐treated A549 cells after exposure to irradiation were analyzed by clonogenic assay and are presented as mean ± standard deviation of three independent experiments (P < 0.05 vs. No IR+vehicle [Veh]). (b) Representative photographs of clonogenic assay (P < 0.05 vs. IR+Veh; P < 0.05 vs. IR+Veh). CTL, control.
Rapamycin delayed DNA damage repair upon radiation
Radiotherapy effectiveness mainly depends on radiation‐induced DNA DSBs, leading to cellular apoptosis, autophagy, and cell death. γ–H2AX was phosphorylated immediately after radiation exposure and accumulated at DSB sites. Therefore, γ–H2AX expression was measured by Western blot at one and 24 hours after radiation (Fig 5). In control cells, a minimum level of γ–H2AX was detected. Conversely, γ–H2AX levels significantly increased at one hour after radiation, regardless of rapamycin treatment. The increment of radiation‐induced γ–H2AX gradually decreased 24 hours after radiation. However, the combination of rapamycin and radiation treatment significantly kept γ–H2AX levels elevated, compared with radiation alone at 24 hours after treatment (Fig 5). Results indicate that rapamycin treatment prolonged DNA damage after radiation, which led to failure in DNA damage repair and improved radiosensitization in cancer cells.
Figure 5.
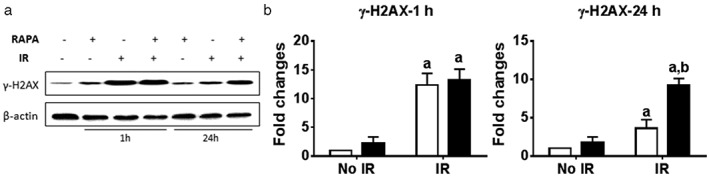
Rapamycin treatment delayed DNA damage repair after irradiation. A549 cells were incubated with vehicle or 100 nmol/L of rapamycin (RAPA) for 24 hours before exposure to 4 Gy of irradiation (IR). Proteins were extracted at one and 24 hours after IR. γ‐H2AX expression was measured by (a) Western blot (P < 0.05 vs. No IR+vehicle [Veh]) and (b) quantitated using Adobe Photoshop CS4 software (P < 0.05 vs. IR+Veh.)
Rapamycin decreased Rad51 and Ku80 expression in A549 cells
Our previous data revealed that low oxygen sensitized A549 cells to radiation, which was mediated by promoting autophagy formation. Current data indicate that rapamycin‐induced autophagy also sensitizes lung cancer cells to radiation. The question is how the induction of autophagy radiosensitizes cancer cells. According to our γ–H2AX data, we hypothesized that increased autophagy delayed radiation‐induced DNA damage repair. In order to test these possibilities, Rad51, Ku70, and Ku80 expression levels were measured because Rad51, Ku70, and Ku80 are important players in HR and NHEJ repair pathways. Radiation reduced the protein expression of Rad51, but not Ku70 and Ku80, as shown in Figure 6a,b. Importantly, rapamycin treatment significantly decreased the protein expression of Rad51 and Ku80, but not Ku70 after radiation exposure, suggesting that rapamycin‐induced autophagy inhibited the expression of Rad51 and Ku80, and negatively affected DNA damage repair in A549 cells.
Figure 6.
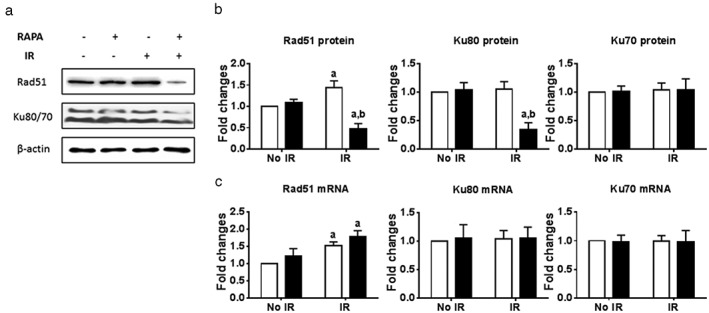
Rapamycin treatment decreased the protein expression of Rad51 and Ku80 after irradiation. A549 cells were incubated with vehicle or 100 nmol/L of rapamycin (RAPA) for 24 hours before exposure to 4 Gy of irradiation (IR). Proteins were extracted after four hours after irradiation. The expression of Rad51, Ku80, and Ku70 was measured by (a) Western blot and (b) quantitated using Adobe Photoshop CS4 software. RNA was extracted two hours after irradiation. The expression of Rad51, Ku80, and Ku70 was measured by real‐time polymerase chain reaction. a: P < 0.05 vs. No IR+vehicle (Veh); b: P < 0.05 vs. IR+Veh. CTL, control.
In order to further investigate how rapamycin treatment affects Rad51, Ku70, and Ku80 mRNA expression levels, RT‐PCR was used to detect mRNA expression. Although radiation at two and four hours increased Rad51 mRNA levels, rapamycin treatment did not induce any change in Rad51, Ku70, and Ku80 mRNA levels, regardless of radiation, suggesting that rapamycin and radiation treatment decreased the expression of Rad51 and Ku80 at a post‐translational level (Fig 6c).
Discussion
Radiation is frequently used to treat lung cancer. However, resistance to radiation in cancer cells highly limits clinical therapeutic efficiency. The major factor is that cancer cells can repair DNA DSBs induced by radiation.20 Although the mechanism of how cancer cells repair DNA damage via HR and NHEJ pathways has been extensively studied, strategies to overcome the radioresistance of cancer cells remain poorly developed. The reasons might be related to cellular apoptosis and autophagy.
Autophagy is a cellular self‐digestion process. Once intracellular proteins and organelles are damaged, degenerated, or useless, they are transported into lysosomes and digested by enzymes.21 This leads to an open question: whether increasing autophagy sensitizes cancer cells to radiotherapy and chemotherapy. Recently, investigators have paid more attention to the correlation between radiotherapy and autophagy. Studies have shown that the upregulation upstream factors of autophagy sensitize cancer cells to radiation. However, mechanisms that increase the sensitivity of cancer cells to autophagy and radiotherapy remain elusive. Furthermore, how autophagy affects radiation‐induced DNA damage and repair remains poorly understood.
It has been well established that autophagy can be induced by rapamycin treatment in various cells, including lung cancer cell A549. LC3 and p62 levels are often used to detect autophagy. When autophagy occurs, intracytoplasmic soluble LC3‐I interacts with phosphatidylethanolamine to form LC3‐II, leading to the reduction of LC3‐I. The activity of cellular autophagy can be reflected by the ratio of LC3‐II/LC3‐I, and p62/SQSTM1 decreases with protein degradation.22 In order to assess the effects of 100 nmol/L of rapamycin in lung cancer cell A549, we measured the expression levels of LC3 and p62 24 hours after rapamycin treatment. We first used electron microscopy, a surrogate assay, to detect intracellular autophagy upon rapamycin treatment. Our subsequent data demonstrated that rapamycin effectively increased LC3‐II and decreased LC3‐I, along with a decrease in p62 expression. These findings indicate that 100 nmol/L of rapamycin could upregulate autophagy activity in A549 cells.
It has been shown that radiotherapy could induce autophagy in cancer cells, which may positively affect the effectiveness of radiotherapy. Using inhibitor NVP‐BEZ235 of the phosphoinositide 3‐kinase‐mTOR signaling pathway, Kuger et al. revealed that NVP‐BEZ235 treatment sensitized tumor cells to radiation through the induction of autophagy.23 This may lead to G2/M phase arrest, delay in DNA damage repair, and an increase in apoptosis. Furthermore, Fujiwara et al. elucidated that protein kinase B inhibitors induced cellular autophagy, but not apoptosis, to play a role in anticancer and radiosensitizing effects.24 In order to further investigate the effects of autophagy in lung cancer under radiation, we used A549 cells to test their ability to form colonies in vitro after rapamycin and/or radiation treatment. As expected, the survival fraction after radiation and rapamycin treatment dramatically decreased compared with radiation or rapamycin alone, indicating that rapamycin has a radiosensitizing effect on A549 cells.
The capacity to repair DNA DSBs is densely correlated to radiosensitization in cancer cells. However, it remains elusive whether autophagy radiosensitizes tumor cells by inhibiting DNA damage repair. Evidence in yeast has shown that autophagy could block the homology recombination pathway through the degradation of DSB‐related enzymes, such as choline transporter‐like and exonuclease 1.25 Our data indicates that increased autophagy activity by rapamycin treatment prolonged phosphorylated γ‐H2AX formation and delayed the repair of radiation‐induced DSBs, which might synthesize radiation effects in lung cancer cells. Consistently, other studies have revealed that decreased levels of DNA damage repair‐related proteins sensitized cancer cells to radiotherapy. For example, Zhou et al. downregulated E3 ubiquitin ligase RNF8 and decreased the expression of Rad51, resulting in the sensitization of A549 cells to radiation.26 Belenkov et al. decreased the expression of Ku80 to radiosensitize M059K glioma cells by inhibiting DNA damage repair.27 Using breast cancer cell line MCF‐7 as an in vitro model, Chen et al. demonstrated that rapamycin could increase radiosensitization by decreasing the accumulation of Rad51 to radiation‐induced DSBs.13 Intriguingly, our data suggests that rapamycin treatment triggered cellular autophagy and decreased the protein expression of Rad51 and Ku80 to inhibit DSB repair with increasing radiosensitivity in A549 cells. However, rapamycin treatment did not affect Rad51, Ku70, and Ku80 mRNA expression levels.
These findings indicate that rapamycin regulated the expression of Rad51 and Ku80 at post‐translational levels. A more detailed investigation on how rapamycin treatment regulates the expression levels of these proteins is required to instruct radiotherapeutic studies.
Disclosure
No authors report any conflict of interest.
Acknowledgment
This work was supported by the National Natural Science Foundation of China (Grant No. 81560379, 81460292 and 81260288).
References
- 1. Spalding AC, Lawrence TS. New and emerging radiosensitizers and radioprotectors. Cancer Invest 2006; 24: 444–456. [DOI] [PubMed] [Google Scholar]
- 2. White RR, Sung P, Vestal CG, Benedetto G, Cornelio N, Richardson C. Double‐strand break repair by interchromosomal recombination: An in vivo repair mechanism utilized by multiple somatic tissues in mammals. PLoS ONE 2013; 8 (12): e84379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sorensen CS, Hansen LT, Dziegielewski J et al The cell‐cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair. Nat Cell Biol 2005; 7: 195–201. [DOI] [PubMed] [Google Scholar]
- 4. Klein HL. The consequences of Rad51 overexpression for normal and tumor cells. DNA Repair (Amst) 2008; 7: 686–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Burma S, Chen BP, Chen DJ. Role of non‐homologous end joining (NHEJ) in maintaining genomic integrity. DNA Repair (Amst) 2006; 5: 1042–1048. [DOI] [PubMed] [Google Scholar]
- 6. Qiao GB, Wu YL, Yang XN et al High‐level expression of Rad51 is an independent prognostic marker of survival in non‐small‐cell lung cancer patients. Br J Cancer 2005; 93: 137–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Omori S, Takiguchi Y, Suda A et al Suppression of a DNA double‐strand break repair gene, Ku70, increases radio‐ and chemosensitivity in a human lung carcinoma cell line. DNA Repair (Amst) 2002; 1: 299–310. [DOI] [PubMed] [Google Scholar]
- 8. Sak A, Stueben G, Groneberg M, Böcker W, Stuschke M. Targeting of Rad51‐dependent homologous recombination: Implications for the radiation sensitivity of human lung cancer cell lines. Br J Cancer 2005; 92: 1089–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell 2008; 132: 27–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Scott RC, Juhász G, Neufeld TP. Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. CurrBiol 2007; 17: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lu J, Shen Y, Qian HY et al Effects of mild hypothermia on the ROS and expression of caspase‐3 mRNA and LC3 of hippocampus nerve cells in rats after cardiopulmonary resuscitation. World J Emerg Med 2014; 5: 298–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen T, Stephens PA, Middleton FK, Curtin NJ. Targeting the S and G2 checkpoint to treat cancer. Drug Discov Today 2012; 17: 194–202. [DOI] [PubMed] [Google Scholar]
- 13. Chen H, Ma Z, Vanderwaal RP et al The mTOR inhibitor rapamycin suppresses DNA double‐strand break repair. Radiat Res 2011; 175: 214–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dai ZJ, Gao J, Kang HF et al Targeted inhibition of mammalian target of rapamycin (mTOR) enhances radiosensitivity in pancreatic carcinoma cells. Drug Des Devel Ther 2013; 7: 149–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chang SH, Minai‐Tehrani A, Shin JY et al Beclin1‐induced autophagy abrogates radioresistance of lung cancer cells by suppressing osteopontin. J Radiat Res 2012; 53: 422–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cheng G, Kong D, Hou X et al The tumor suppressor, p53, contributes to radiosensitivity of lung cancer cells by regulating autophagy and apoptosis. Cancer Biother Radiopharm 2013; 28: 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim EJ, Jeong JH, Bae S, Kang S, Kim CH, Lim YB. mTOR inhibitors radiosensitize PTEN‐deficient non‐small‐cell lung cancer cells harboring an EGFR activating mutation by inducing autophagy. J Cell Biochem 2013; 114: 1248–1256. [DOI] [PubMed] [Google Scholar]
- 18. Vessoni AT, Filippi‐Chiela EC, Menck CF, Lenz G. Autophagy and genomic integrity. Cell Death Differ 2013; 20: 1444–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hao J, Pei Y, Ji G, Li W, Feng S, Qiu S. Autophagy is induced by 3beta‐O‐succinyl‐lupeol (LD9‐4) in A549 cells via up‐regulation of Beclin 1 and down‐regulation mTOR pathway. Eur J Pharmacol 2011; 670: 29–38. [DOI] [PubMed] [Google Scholar]
- 20. Kuefner MA, Brand M, Engert C, Schwab SA, Uder M. Radiation induced DNA double‐strand breaks in radiology. Rofo 2015; 187: 872–878. [DOI] [PubMed] [Google Scholar]
- 21. White E. The role for autophagy in cancer. J Clin Invest 2015; 125: 42–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bjorkoy G, Lamark T, Brech A et al p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin‐induced cell death. J Cell Biol 2005; 171: 603–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kuger S, Graus D, Brendtke R et al Radiosensitization of glioblastoma cell lines by the dual PI3K and mTOR inhibitor NVP‐BEZ235 depends on drug‐irradiation schedule. Transl Oncol 2013; 6: 169–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fujiwara K, Iwado E, Mills GB, Sawaya R, Kondo S, Kondo Y. Akt inhibitor shows anticancer and radiosensitizing effects in malignant glioma cells by inducing autophagy. Int J Oncol 2007; 31: 753–760. [PubMed] [Google Scholar]
- 25. Robert T, Vanoli F, Chiolo I et al HDACs link the DNA damage response, processing of double‐strand breaks and autophagy. Nature 2011; 471: 74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhou H, Mu X, Chen J et al RNAi silencing targeting RNF8 enhances radiosensitivity of a non‐small cell lung cancer cell line A549. Int J Radiat Biol 2013; 89: 708–715. [DOI] [PubMed] [Google Scholar]
- 27. Belenkov AI, Paiement JP, Panasci LC, Monia BP, Chow TY. An antisense oligonucleotide targeted to human Ku86 messenger RNA sensitizes M059K malignant glioma cells to ionizing radiation, bleomycin, and etoposide but not DNA cross‐linking agents. Cancer Res 2002; 62: 5888–5896. [PubMed] [Google Scholar]


