Abstract
Background
The correlation between epidermal growth factor receptor‐tyrosine kinase inhibitors (EGFR‐TKIs) and EGFR sensitive mutation subtypes in advanced or metastatic non‐small cell lung cancer (NSCLC) remains uncertain. We performed this meta‐analysis to determine different clinical outcomes between patients with exon 19 deletion accepting EGFR‐TKI therapy compared with those with exon 21 L858R mutation.
Methods
PubMed and Web of Science were analyzed for eligible trials. Raw data were extracted to give pooled estimates of the effect of EGFR‐TKI therapy on objective response rate (ORR), one‐year progression‐free survival (PFS), and two‐year overall survival (OS).
Results
We identified 13 eligible trials involving 912 patients. Prospective meta‐analysis demonstrated that the ORR of the 19 deletion group was significantly higher than the 21 L858R mutation group (odds ratio [OR] 1.98, 95% confidence interval [CI] 1.18–3.33; P = 0.01), but no statistical significance between the one‐year PFS rate of the 19 deletion and 21 L858R groups (OR 1.44, 95% CI 0.96–2.18; P = 0.08) was found. However, retrospective meta‐analysis demonstrated that a significantly higher one‐year PFS rate was associated with the 19 deletion group (OR 1.73, 95% CI 1.17–2.56; P = 0.006). The two‐year survival rate of the 19 deletion group was significantly higher than the 21 L858R group (OR 5.27, 95 % CI 1.76–15.71; P = 0.003).
Conclusions
In advanced NSCLC patients, an exon 19 deleton may provide superior ORR, PFS, and OS after EGFR‐TKI treatment compared with an exon 21 L858R mutation.
Keywords: EGFR mutation, EGFR‐TKI, meta‐analysis, NSCLC
Introduction
Lung cancer is the leading cause of death from cancer.1 Non‐small‐cell lung cancer (NSCLC) accounts for more than 80% of all lung cancers. Unfortunately, few treatment options are available for patients with advanced or metastatic NSCLC.
In 2004, two pivotal studies proved that the somatic mutation in epidermal growth factor receptor (EGFR) can be detected in most patients with NSCLC, and was strongly relevant to high responsiveness to EGFR‐tyrosine kinase inhibitors (TKIs).2, 3 Small molecule EGFR‐TKIs were the first target drugs to be applied and widely used as a clinical treatment strategy for NSCLC. The two classical EGFR mutations are deletion of exon 19 (19del) and a single point mutation of L858R in exon 21 (21 L858R).4 Although patients with an EGFR mutation usually benefit more from EGFR‐TKI treatment than those with wild type, different subtypes of EGFR mutations respond to EGFR‐TKI treatment.5
Several studies have reported that advanced NSCLC patients with exon 19del accepting EGFR‐TKI therapy had a relatively longer progression‐free survival (PFS) and/or overall survival (OS) than those with an exon 21 L858R mutation.6, 7, 8, 9 However, this issue is controversial at present, as several studies have also shown no significant difference in EGFR‐TKI efficacy between the two subtypes of mutations.7, 10
Therefore, we performed this meta‐analysis to evaluate whether there is a correlation between the theraputic efficacy of EGFR‐TKIs and these two classical subtypes of EGFR mutations in advanced NSCLC. We also sought to accurately predict clinical prognosis in the patients who accepted EGFR‐TKI therapy.
Methods
Literature search
Two investigators independently searched eligible trials, and discrepancies were resolved by discussion between them. All relevant articles were identified with an English language search of PubMed and Web of Science using a combination of the keywords: “EGFR,” “EGF receptor,” “epidermal growth factor receptor,” “tyrosine kinase inhibitors” or “TKI,” and “non small cell lung cancer” or “NSCLC” from 2004–2015. The last search was performed on May 30, 2015, and studies were conducted using Prefered Reporting Items For Systematic Reviews and Meta‐analyses (PRISMA) standards.11
Study criteria
Studies were considered eligible if they met the following criteria: (i) prospective or retrospective clinical studies designed to investigate advanced or metastatic NSCLC (stage III–IV) with monotherapy of EGFR‐TKIs (gefitinib or erlotinib), first line or otherwise; (ii) patients harbored EGFR mutations (exon 19 del or 21 L858R); (iii) EGFR mutation was identified by tumor tissue samples rather than peripheral blood; (iv) all included patients were evaluated by treatment response, PFS, and OS (defined by Response Evaluation Criteria in Solid Tumors; complete and partial response were categorized to response, stable, or progressive); and (v) for the same study, only the most recent publication results were included.
Quality assessment and data extraction
Two investigators independently assessed the quality of studies using the Newcastle Ottawa Quality Assessment Scale, which was composed of eight items to assess patient selection, study comparability and outcome; the use of controversial literature was resolved by discussion with the third investigator in order to reach accordance.12
The following data were extracted from the included studies: (i) general information (lead author's name, publication time, and study type); (ii) features (treatment, treatment line, and sample size of EGFR mutation subtypes); and (iii) evaluation indexes (ORR, PFS, 1‐year PFS, OS and 2‐year OS; if the data couldn't be obtained directly, we calculated from the survival curve).
Survival curve data were preferably provided as survival data for a given period, for example, a one, two, three, or five‐year survival number/ratio. However, survival data needed to be directly extracted from the Kaplan‐Meier curve for two prospective and four retrospective studies. In this method, the x‐axis of the Kaplan–Meier curve was divided into given intervals equaling years of follow‐up, then a researcher read the survival ratio at the each time point using the GetData Graph Digitizer 2.2 (Informer Technologies Inc., Madrid, Spain).13 This method was considered weak but necessary, because some of the earlier studies did not provide survival ratio information at each time point. This weakness is shared by our analysis and some previously published meta‐analyses.
Statistical analysis
The meta‐analysis of the odds radio (OR) for objective responses and the one‐year PFS and two‐year OS rates were calculated using RevMan (Review Manager Version 5.3 for Windows, Cochrane Collaboration, Oxford, UK) and a pooled relative risk was calculated with 95% confidence intervals (CIs).
To undertake a random effects meta‐analysis, the standard errors of the study‐specific estimates are adjusted to incorporate a measure of the extent of variation or heterogeneity among the treatment effects observed in different studies. Statistical heterogeneity was evaluated using I 2 statistics and X 2‐based tests, and a P value of < 0.05 was considered statistically significant.
In the I 2 test, I 2 = 0% indicated no heterogeneity; I 2 (0%, 40%) indicated low heterogeneity; I 2 (40%, 60%) indicated moderate heterogeneity; I 2 (50%, 90%) indicated high heterogeneity; I 2 (75%, 100%) indicated maximum heterogeneity; and the X2 distribution test used a rejection region equal to 0.1.14
A funnel plot test was used to evaluate the existence of publication bias.
Results
As shown in Figure 1, we researched 706 studies from PubMed and 365 additional studies from Web of Science. After a screening process using PRISMA standards, we included seven prospective studies (508 patients with exon 19del and 354 with exon 21 L858R mutations) into a comprehensive meta‐analysis and included six retrospective studies for a supplementary meta‐analysis. The characteristics of the eligible studies are summarized in Tables 1 and 2.6,7,9,10, 15, 16, 17, 18, 19, 20, 21
Figure 1.
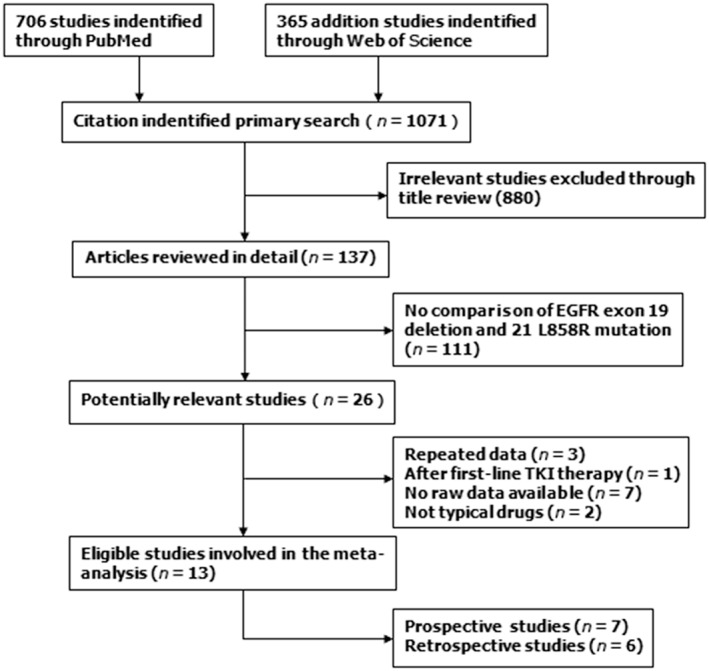
Flow diagram of patients included in the meta‐analysis. EGFR, epidermal growth factor receptor; TKI, tyrosine kinase inhibitor.
Table 1.
Characteristics of included prospective studies for meta‐analysis
| Lead author (year) | Phase | Treatment line | Therapeutic regimen of TKI | Type of EGFR mutation | Sample size (TKI treatment) | ORR (%) | PFS (months) (95% CI) | OS (months) (95% CI) | Score |
|---|---|---|---|---|---|---|---|---|---|
| Sugio (2008) | Pro (II) | 1 | Gefitinib 250 mg/d, po | 19del | 7 | 71.4 | – | – | 6 |
| 21 L858R | 10 | 60 | – | – | |||||
| Kim (2011) | Pro (II) | Mix | Gefitinib 250 mg/d, po | 19del | 29 | 62.1 | 16.5 (12.5–19.3) | NA (24.9‐NA) | 7 |
| 21 L858R | 15 | 33.3 | 6.9 (3.6–13.3) | 16.2 (11.2–27.3) | |||||
| Iuchi (2013) | Pro (II) | 1 | Gefitinib 250 mg/d, po | 19del | 23 | 100 | 17.5 (13.9‐NA) | 30.3 | 7 |
| or Erlotinib 150 mg/d, po | 21 L858R | 15 | 80 | 10.2 (7.2‐NA) | 19.8 | ||||
| Yamada (2013) | Pro (II) | 2,3 | Erlotinib 150 mg/d, po | 19del | 19 | 47.4 | 8.0 (3.8–10.4) | – | 9 |
| 21 L858R | 7 | 71.4 | 11.6 (8.3–11.6) | – | |||||
| Goto (2013) | Pro (II) | Mix | Erlotinib 150 mg/d, po | 19del | 50 | 84 | 12.5 (10.3–16.6) | – | 7 |
| 21 L858R | 51 | 76 | 11.0 (6.9–15.2) | – | |||||
| Mitsudomi (2010) | Pro (III) | 1 | Gefitinib 250 mg/d, po | 19del | 50 | – | 9.0 (6.7–13.0) | – | 8 |
| 21 L858R | 36 | – | 9.6 (8.0–13.8) | – | |||||
| Maemondo (2010) | Pro, RCT | 1 | Gefitinib 250 mg/d, po | 19del | 58 | 82.8 | 11.5 | – | 9 |
| 21 L858R | 49 | 67.3 | 10.8 | – |
19del, exon 19 deletion; CI, confidence interval; EGFR, epidermal growth factor receptor; ORR, objective response rate; OS, overall survival; PFS, progression‐free survival; po, orally; Pro, prospective study; RCT, randomized controlled trial; TKI, tyrosine kinase inhibitor.
Table 2.
Characteristics of included retrospective studies for complementary meta analysis
| Lead author (year) | Treat line | Therapeutic regimen of TKI | Type of EGFR mutation | Sample size (TKI treatment) | ORR (%) | PFS (months) (95% CI) | OS (months) (95% CI) | Score |
|---|---|---|---|---|---|---|---|---|
| Won (2011) | Mix | Gefitinib 250 mg/d, po | 19del | 61 | 63.9 | 9.3 (7.5 to 11.1) | 17.7 (12.3 to 23.1) | 7 |
| or Erlotinib 150 mg/d, po | 21 L858R | 26 | 61.5 | 6.9 (4.1 to 9.7) | 20.5 (3.8–37.2) | |||
| Lee (2013) | 1 | Gefitinib 250 mg/d, po | 19del | 64 | 87.5 | 12.8 | 24.6 | 9 |
| or Erlotinib 150 mg/d, po | 21 L858R | 80 | 75.3 | 11.4 | 21 | |||
| Jackman (2006) | Mix | Gefitinib 250 mg/d, po | 19del | 22 | 73 | 24 | 38 | 6 |
| or Erlotinib 150 mg/d, po | 21 L858R | 10 | 50 | 10 | 17 | |||
| Yoshida (2007) | Mix | Gefitinib 250 mg/d, po | 19del | 8 | 87.5 | 7.8 (7.6‐NA) | – | 8 |
| 21 L858R | 13 | 92.3 | 6 (2.6 to 7.7) | – | ||||
| Igawa (2014) | Mix | Gefitinib 250 mg/d, po | 19del | 68 | 61.8 | 11.3 (8.0 to 14.6) | 32.2 (22.0 to 42.4) | 8 |
| 21 L858R | 56 | 58.9 | 9 (6.7 to 11.3) | 22.7 (9.4 to 36.0) | ||||
| Takano (2006) | Mix | Gefitinib 250 mg/d, po | 19del | 49 | 86 | 10.5 | 22 | 7 |
| 21 L858R | 36 | 67 | 7.4 | 12 |
19del, exon 19 deletion; CI, confidence interval; EGFR, epidermal growth factor receptor; NA, not available; ORR, objective response rate; OS, overall survival; PFS, progression‐free survival; po, orally; Pro, prospective study; RCT, randomized controlled trial; TKI, tyrosine kinase inhibitor.
Response rate
As Figure 2 shows, ORR data was available in six prospective trials, including five phase II and one randomized controlled trial (n = 333). Heterogeneity testing revealed a fixed effects model without significant heterogeneity (I 2 =10 %; P = 0.35). The ORR of the 19del group was significantly higher than the 21 L858R group (78.0% [145/186] vs. 68.0% [100/147], OR 1.98, 95% CI 1.18–3.33; P = 0.01; Fig 2).
Figure 2.
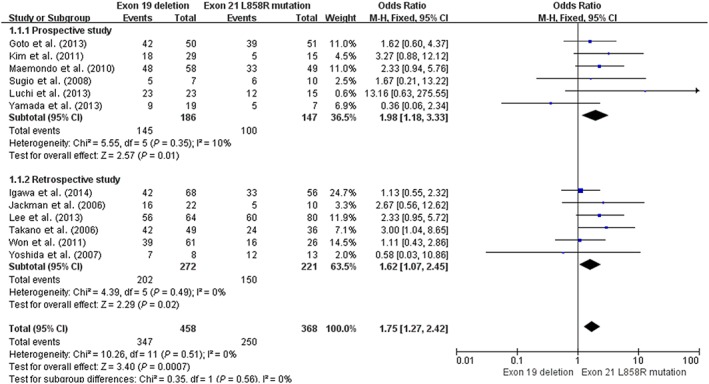
Forest plot of overall response rates of exon 19 deletion and 21 L858R mutation in prospective and retrospective studies. CI, confidence interval; M‐H, Mantel‐Haenszel.
Furthermore, a complementary meta‐analysis was performed on six retrospective studies (n = 493; Fig 2). The estimated proportion of the ORR in heterogeneity (I 2) was 0%, (P = 0.49). Patients with an exon 19del had a significantly higher ORR than patients with an exon 21 L858R mutation (74.3% [202/272] vs. 67.9% [150/221], OR 1.62, 95% CI 1.07–2.45; P = 0.02). The pooling of data from prospective and retrospective studies (n = 826) demonstrated a statistically significant ORR in patients with an exon 19del compared with those with an exon 21 L858R mutation (OR 1.75, 95% CI 1.27–2.42; P = 0.0007).
One‐year progression‐free survival rate
Six prospective trials (n = 401) included one‐year PFS data. The median PFS was 8.0–16.5 months in the 19del and 6.9–11.6 months in the 21 L858R group (Table 1). The estimated proportion of heterogeneity (I 2) between these six studies was 48% (P = 0.09) for one‐year PFS of the 19del group versus the 21 L858R group. As Figure 3 demonstrates, patients with 19del had higher one‐year PFS than patients with the 21 L858R mutation (44.1% [101/229] vs. 35.4% [61/172], OR 1.44, 95% CI 0.96–2.18; P = 0.08).
Figure 3.
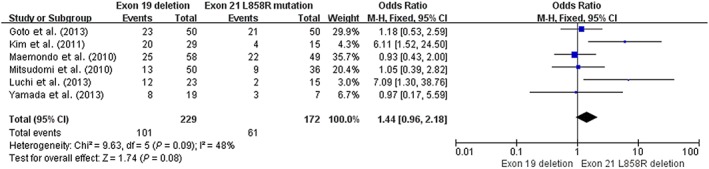
Forest plot of one‐year progression‐free survival rates of exon 19 deletion and 21 L858R mutation in prospective studies. CI, confidence interval; M‐H, Mantel‐Haenszel.
We conducted a supplementary meta‐analysis using five retrospective studies, as shown in Figure 4. Heterogeneity testing revealed that there was light heterogeneity (I 2 = 30%; P = 0.22), and patients with a 19del mutation had a significantly higher one‐year PFS rate than patients with an exon 21 L858R mutation (48.0% [118/264] vs. 32.2% [67/208], OR 1.73, 95% CI 1.17–2.56; P = 0.006).
Figure 4.
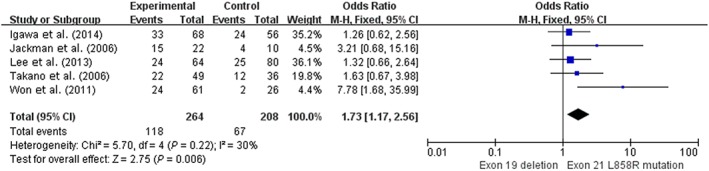
Forest plot of one‐year progression‐free survival rates of exon 19 deletion and 21 L858R mutation in retrospective studies. CI, confidence interval; M‐H, Mantel‐Haenszel.
Two‐year overall survival rate
The two‐year OS rate was available in two prospective trials (n = 82), both of which demonstrated that patients with a 19del mutation had a longer OS (24.9–30.3 months) than those with 21 L858R mutations (16.2–22.1 months; Table 1).16, 17 The I 2 statistic in the fixed effects model did not show significant heterogeneity (I 2 = 0 %; P = 0.52), and pooling analysis revealed that patients with an exon 19del mutation had a statistically significantly higher two‐year OS rate than patients with an exon 21 L858R mutation (55.8% [29/52] vs. 20% [6/30], OR 5.27, 95% CI 1.76–15.71; P = 0.003), as shown in Figure 5.
Figure 5.
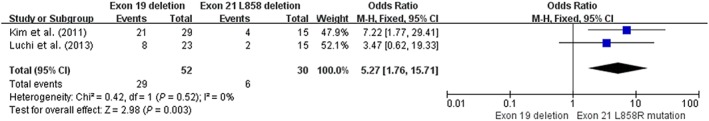
Forest plot of two‐year overall survival rates of exon 19 deletion and 21 L858R mutation in prospective studies. CI, confidence interval; M‐H, Mantel‐Haenszel.
Assessment of heterogeneity and publication bias
There was light statistical heterogeneity among the 13 trials and no publication bias for outcome measures, with asymmetrical appearance on funnel plot analysis of the relative ORR (Fig 6).
Figure 6.
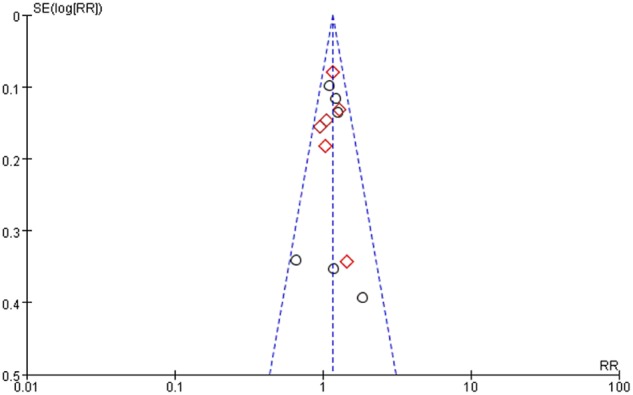
Funnel plot of the relative overall response rates.  , Prospective study;
, Prospective study;  , Retrospective study. RR, relative risk; SE, standard error.
, Retrospective study. RR, relative risk; SE, standard error.
Discussion
In 2014, two meta‐analyses of the correlation between EGFR‐TKIs and EGFR sensitive mutation subtypes in advanced or metastatic NSCLC were published. However, the two studies did not perfectly resolve the issue. Zhang et al. compared patients with an exon 19del who accepted EGFR‐TKI treatment with those with a 21 L858R mutation.24 Their results revealed significantly superior ORR, PFS, and OS in the 19del mutation group compared with the 21 L858R mutation group. However, their meta‐analysis lacked data, which led to incomplete results in the prospective subgroup meta‐analysis and indicated that PFS and OS rates in the 19del group had no significance. Wang et al.'s meta‐analysis was based on 22 studies published mainly during 2006–2011; as further studies have been published and renewed in the last three years, the quality of our data is superior. Their study focused on patients who received first‐line EGFR‐TKIs and demonstrated that patients with an exon 19del mutation have a longer PFS compared with those with an L858R mutation.25 They extracted hazard ratios (HR) for the PFS rate of TKIs/chemotherapy in 19del and 21 L858R subgroups and calculated the specific HR value to speculate the ratio of PFS, which means that a confounding bias may exist in their meta‐analysis. In our study, we performed a more comprehensive meta‐analysis, including evidence from both prospective and retrospective studies. In order to obtain an objective and accurately evaluated result and to assess the therapeutic effects of TKIs, we selected three indicators: an ORR indicated a short‐term effect, one‐year PFS a middle‐term effect, and two‐year OS a long‐term effect.
In our study, patients with an exon 19del had a significantly higher ORR after EGFR‐TKI therapy than patients with an exon 21 L858R mutation in both prospective (OR 1.98, 95% CI 1.18–3.33; P = 0.01) and retrospective subgroup analyses (OR 1.62, 95% CI 1.07–2.45; P = 0.02). Correspondingly, pooling the statistical results indicated a significant difference between the mutations (OR 1.75, 95% CI 1.27–2.42; P = 0.0007), suggesting that in advanced NSCLC, patients with an exon 19del usually experienced superior short‐term efficacy from EGFR‐TKIs than those with an exon 21 L858R mutation. Our meta‐analysis of the prospective subgroup indicated that the exon 19del group had a higher one‐year PFS than the 21 L858R mutation group, but the difference between them had no statistical significance (OR 1.44, 95% CI 0.96–2.18; P = 0.08). However, in retrospective subgroup meta‐analysis, the exon 19del group had a significantly higher one‐year PFS rate (OR 1.73, 95% CI 1.17–2.56; P = 0.006). Analyzing these six prospective studies individually, two studies demonstrated that the 21 L858R group had a longer PFS compared with the 19del group.18, 20 Thus, once the restrospective subgroup meta‐analysis was combined with the prospective meta‐analysis, it was clear that the results for the 19del group had been affected by the data from the two studies indicating that the 21 L858R group had a longer PFS. One of these, a phase II study of erlotinib, found PFS rates of the 19del (n = 19) and 21 L858R groups (n = 7) of 8.0 and 11.6 months, respectively (P = 0.1084), with no significant difference.18 This study had limited sufficient data, which might affect our result. The second study, WJTOG3405, was a phase III trial, in which the PFS rates of the 19del (n = 50) and 21 L858R groups (n = 36) were 9.0 versus 9.6 months, a difference with no statistical significance (P = 0.681).20 We consider that the WJTOG3405 result is not representative of phase III studies, as other phase III trials (OPTIMAL, IPASS, EURTAC, LUX‐Lung 3, and LUX‐Lung 6) have demonstrated that the HR of TKI/chemotherapy for PFS in the 19del group was lower than in 21 L858R group, suggesting that the PFS in patients with the 19del mutation was longer; however, we excluded these studies as raw data was not available.26 – 30 Considering these limitations, we reanalyzed the difference in one‐year PFS between the 19del and 21 L858R subgroups, leaving out the results of these two studies. Patients with 19del had higher one‐year PFS rates than patients with the 21 L858R mutation (50% [80/160] vs. 38.0% [49/129], OR 1.59, 95% CI 1.00–2.55; P = 0.05; Fig 7).
Figure 7.
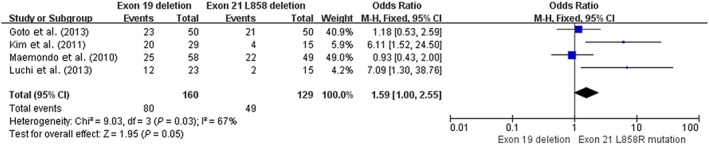
Forest plot of one‐year progression‐free survival rates of exon 19 deletion and 21 L858R mutation in prospective studies reanalyzed. CI, confidence interval; M‐H, Mantel‐Haenszel.
Although the evidence‐based lever of retrospective studies is relatively low, some retrospective studies have shown persuasive results. For example, a retrospective study published in the New England Journal of Medicine also revealed that patients with a 19del mutation experienced a longer PFS and OS than those with the 21 L858R, and the difference was statistically significant.31 Therefore, we arranged a retrospective subgroup meta‐analysis as a supplement to enlarge the sample size, which demonstrated a significantly higher one‐year PFS rate in the 19del group than the 21 L858R group.
In our meta‐analysis, the two‐year OS rate of the 19del group was significantly superior to the 21 L858R group (OR 5.27, 95% CI 1.76–15.71; P = 0.003), consistent with previous reports.8, 10, 31 In addition, the ORR, one‐year PFS, and two‐year OS rates suggest that patients harboring an exon 19del might display a higher sensitivity to EGFR‐TKI therapy than those harboring an exon 21 L858R point mutation; the difference in efficacy will become increasingly obvious over time and finally affect their survival.
However, advanced NSCLC patients with an exon 19del who did not accept EGFR‐TKI treatment failed to acquire such a superior prognosis to those with the 21 L858R mutation. The NEJ002 study showed that patients with an exon 19del had a higher ORR with gefitinib than patients with an L858R mutation (82.8% vs. 67.3%) and patients who were treated by carboplatin plus paclitaxel alone, who had a low ORR (30.5% vs. 30.3%).32 Shigematsu et al. reported that in NSCLC patients with these two subtypes of EGFR mutation who accepted surgical resection but not EGFR‐TKI, those with L858R had a relatively prolonged survival compared with patients with an exon 19del (P = 0.05).33
These results suggest that the different efficacies of EGFR‐TKI in patients with these two subtypes of sensitive mutation are likely related to a molecular biology mechanism: the EGFR exon 19del eliminates a leucine‐arginine‐glutamate‐alanine motif in the tyrosine kinase domain of EGFR, while an exon 21 L858R mutation performs a thymine‐to‐guanine transversion that results in arginine for leucine substitution at amino acid 858.34 Although no study has compared affinity when TKIs are combined with the exon 19del or 21 L858R mutation, Yun et al. reported that different mutations of EGFR resulted in different affinities for gefitinib and other TKIs.35 This suggests that the exon 19del of EGFR leads to a special structural change, which combines with TKIs faster than in the 21 L858R mutation. Zhu et al. obtained two stable cell lines expressing these two subtype mutations by transfection, and found that gefitinib inhibited the phosphorylation of EGFR, protein kinase B, and extracellular‐signal‐related‐kinase significantly greater in HEK293/19del cells than in HEK293/L858R cells, and the production of G1 arrest in exon 19del cells was higher than in L858R cells.36 Banno et al. used a methyl thiazolyl tetrazolium assay to examine sensitivities to AG1478 (a reversible EGFR‐TKI) and afatinib in NSCLC cell lines harboring EGFR exon 19del (PC‐9 and HCC827 cell lines) and 21 L858R mutations (11_18 cell line). The inhibitory concentration 50 of both AG1478 and afatinib of the PC‐9 and HCC827 cell lines were lower than that of the 11_18 cell line.37 These two studies suggest that cells harboring the exon 19del are more sensitive to EGFR‐TKIs than those with the 21 L858R mutation. Nevertheless, the mechanisms are still not well understood and require further examination.
Our meta‐analysis has several limitations. First, few studies have focused on the the prognostic value of different EGFR mutation types as to the efficacy of EGFR‐TKI, and some data came from subgroups, which might lead to incomplete results. Second, our meta‐analysis included seven retrospective studies. Although the evidence‐based lever of retrospective studies is low, we only employed it as a supplementary analysis, and the results supported those determined by prospective study analysis. Third, despite some heterogeneity between the studies, this was light (0–35%) and had no influence on the outcome of meta‐analysis.
Conclusion
For advanced NSCLC patients, exon 19del might provide superior ORR, PFS, and OS from EGFR‐TKI treatment compared with those with exon 21 L858R mutations. Therefore, it should be considered an essential factor in clinial EGFR‐TKI therapy.
Disclosure
No authors report any conflict of interest.
Acknowledgments
We appreciate the data provided by original authors. Thank you to all patients who took part in the trials and contributed to this research.
References
- 1. Siegel R, Ma J, Zou Z, Ahmedin JP et al Cancer statistics, 2014. CA Cancer J Clin 2014; 64: 9–29. [DOI] [PubMed] [Google Scholar]
- 2. Paez JG, Jänne PA, Lee JC et al EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science 2004; 304: 1497–1500. [DOI] [PubMed] [Google Scholar]
- 3. Lynch TJ, Bell DW, Sordella R et al Activating mutations in the epidermal growth factor receptor underlying responsiveness of non‐small‐cell lung cancer to gefitinib. N Engl J Med 2004; 350: 2129–2139. [DOI] [PubMed] [Google Scholar]
- 4. Kosaka T, Yatabe Y, Endoh H, Kuwano H, Takahashi T , Mitsudomi T. Mutations of the epidermal growth factor receptor gene in lung cancer: Biological and clinical implications. Cancer Res 2004; 64: 8919–8923. [DOI] [PubMed] [Google Scholar]
- 5. Gazdar AF. Activating and resistance mutations of EGFR in non‐small‐cell lung cancer: Role in clinical response to EGFR tyrosine kinase inhibitors. Oncogene 2009; 28 (Suppl 1): S24–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jackman DM, Yeap BY, Sequist LV et al Exon 19 deletion mutations of epidermal growth factor receptor are associated with prolonged survival in non‐small cell lung cancer patients treated with gefitinib or erlotinib. Clin Cancer Res 2006; 12: 3908–3914. [DOI] [PubMed] [Google Scholar]
- 7. Takano T, Ohe Y, Tsuta K. Epidermal growth factor receptor mutation detection using high‐resolution melting analysis predicts outcomes in patients with advanced non small cell lung cancer treated with gefitinib. Clin Cancer Res 2007; 13: 5385–5390. [DOI] [PubMed] [Google Scholar]
- 8. Riely GJ, Pao W, Pham D et al Clinical course of patients with non‐small cell lung cancer and epidermal growth factor receptor exon 19 and exon 21 mutations treated with gefitinib or erlotinib. Clin Cancer Res 2006; 12: 839–844. [DOI] [PubMed] [Google Scholar]
- 9. Won YW, Han JY, Lee GK et al Comparison of clinical outcome of patients with non‐small‐cell lung cancer harbouring epidermal growth factor receptor exon 19 or exon 21 mutations. J Clin Pathol 2011; 64: 947–952. [DOI] [PubMed] [Google Scholar]
- 10. Lee VH, Tin VP, Choy TS et al Association of exon 19 and 21 EGFR mutation patterns with treatment outcome after first‐line tyrosine kinase inhibitor in metastatic non‐small‐cell lung cancer. J Thorac Oncol 2013; 8: 1148–1155. [DOI] [PubMed] [Google Scholar]
- 11. Moher D, Liberati A, Tetzlaff J , Altman DG, PRISMA Group. Preferred reporting items for systematic reviews and meta‐analyses: The PRISMA statement. J Clin Epidemiol 2009; 62: 1006–1012. [DOI] [PubMed] [Google Scholar]
- 12. Stang A. Critical evaluation of the Newcastle‐Ottawa scale for the assessment of the quality of nonrandomized studies in meta‐analyses. Eur J Epidemiol 2010; 25: 603–605. [DOI] [PubMed] [Google Scholar]
- 13. Parmar MK, Torri V, Stewart L. Extracting summary statistics to perform meta‐analyses of the published literature for survival endpoints. Stat Med 1998; 17: 2815–2834. [DOI] [PubMed] [Google Scholar]
- 14. Higgins JPT, Green S. Cochrane Handbook for Systematic Reviews of Interventions, version 5.1.0 [updated March 2011] The Cochrane Collaboration [Cited 27 Feb 2016.] Available from URL: http://www.cochrane‐handbook.org
- 15. Sugio K, Uramoto H, Onitsuka T et al Prospective phase II study of gefitinib in non‐small cell lung cancer with epidermal growth factor receptor gene mutations. Lung Cancer 2009; 64: 314–318. [DOI] [PubMed] [Google Scholar]
- 16. Kim DW, Lee SH, Lee JS et al A multicenter phase II study to evaluate the efficacy and safety of gefitinib as first‐line treatment for Korean patients with advanced pulmonary adenocarcinonna harboring EGFR mutations. Lung Cancer 2011; 71: 65–69. [DOI] [PubMed] [Google Scholar]
- 17. Iuchi T, Shingyoji M, Sakaida T et al Phase II trial of gefitinib alone without radiation therapy for Japanese patients with brain metastases from EGFR‐mutant lung adenocarcinoma. Lung Cancer 2013; 82: 282–287. [DOI] [PubMed] [Google Scholar]
- 18. Yamada K, Takayama K, Kawakami S et al Phase II trial of erlotinib for Japanese patients with previously treated non‐small‐cell lung cancer harboring EGFR mutations: Results of Lung Oncology Group in Kyushu (LOGiK0803). Jpn J Clin Oncol 2013; 43: 629–635. [DOI] [PubMed] [Google Scholar]
- 19. Goto K, Nishio M, Yamamoto N et al A prospective, phase II, open‐label study (JO22903) of first‐line erlotinib in Japanese patients with epidermal growth factor receptor (EGFR) mutation‐positive advanced non‐small‐cell lung cancer (NSCLC). Lung Cancer 2013; 82: 109–114. [DOI] [PubMed] [Google Scholar]
- 20. Mitsudomi T, Morita S, Yatabe Y et al Gefitinib versus cisplatin plus docetaxel in patients with non‐small‐cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): An open label, randomised phase 3 trial. Lancet Oncol 2010; 11: 121–128. [DOI] [PubMed] [Google Scholar]
- 21. Maemondo M, Inoue A, Kobayashi K et al Gefitinib or chemotherapy for non‐small‐cell lung cancer with mutated EGFR. N Engl J Med 2010; 362: 2380–2388. [DOI] [PubMed] [Google Scholar]
- 22. Yoshida K, Yatabe Y, Park JY et al Prospective validation for prediction of gefitinib sensitivity by epidermal growth factor receptor gene mutation in patients with non‐small cell lung cancer. J Thorac Oncol 2007; 2: 22–28. [PubMed] [Google Scholar]
- 23. Igawa S, Kasajima M, Ishihara M et al Comparison of the efficacy of gefitinib in patients with non‐small cell lung cancer according to the type of epidermal growth factor receptor mutation. Oncology 2014; 87: 215–223. [DOI] [PubMed] [Google Scholar]
- 24. Zhang Y, Sheng J, Kang S et al Patients with exon 19 deletion were associated with longer progression‐free survival compared to those with L858R mutation after first‐line EGFR‐TKIs for advanced non‐small cell lung cancer: A meta‐analysis. PLoS ONE 2014; 9 (9): e107161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang H, Huang J, Yu X et al Different efficacy of EGFR tyrosine kinase inhibitors and prognosis in patients with subtypes of EGFR‐mutated advanced non‐small cell lung cancer: A meta‐analysis. (Published erratum appears in J Cancer Res Clin Oncol 2014; 140: 1911) J Cancer Res Clin Oncol 2014; 140: 1901–1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhou C, Wu YL, Chen G et al Erlotinib versus chemotherapy as first‐line treatment for patients with advanced EGFR mutation‐positive non‐small‐cell lung cancer (OPTIMAL, CTONG‐0802): A multicentre, open‐label, randomised, phase 3 study. Lancet Oncol 2011; 12: 735–742. [DOI] [PubMed] [Google Scholar]
- 27. Fukuoka M, Wu YL, Thongprasert S et al Biomarker analyses and final overall survival results from a phase III, randomized, open‐label, first‐line study of gefitinib versus carboplatin/paclitaxel in clinically selected patients with advanced non‐small‐cell lung cancer in Asia (IPASS). J Clin Oncol 2011; 29: 2866–2874. [DOI] [PubMed] [Google Scholar]
- 28. Rosell R, Carcereny E, Gervais R et al Erlotinib versus standard chemotherapy as first‐line treatment for European patients with advanced EGFR mutation‐positive non‐small‐cell lung cancer (EURTAC): A multicentre, open‐label, randomised phase 3 trial. Lancet Oncol 2012; 13: 239–246. [DOI] [PubMed] [Google Scholar]
- 29. Köhler J, Schuler M. LUX‐Lung 3: Redundancy, toxicity or a major step forward? Afatinib as front‐line therapy for patients with metastatic EGFR‐mutated lung cancer. Future Oncol 2014; 10: 533–540. [DOI] [PubMed] [Google Scholar]
- 30. Wu YL, Zhou C, Hu CP et al Afatinib versus cisplatin plus gemcitabine for first‐line treatment of Asian patients with advanced non‐small‐cell lung cancer harbouring EGFR mutations (LUX‐Lung 6): An open‐label, randomised phase 3 trial. Lancet Oncol 2014; 15: 213–222. [DOI] [PubMed] [Google Scholar]
- 31. Rosell R, Moran T, Queralt C et al Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med 2009; 361: 958–967. [DOI] [PubMed] [Google Scholar]
- 32. Fukuhara T, Maemondo M, Inoue A et al Factors associated with a poor response to gefitinib in the NEJ002 study: Smoking and the L858R mutation. Lung Cancer 2015; 88: 181–186. [DOI] [PubMed] [Google Scholar]
- 33. Shigematsu H, Lin L, Takahashi T et al Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst 2005; 97: 339–346. [DOI] [PubMed] [Google Scholar]
- 34. Pan Q, Pao W, Ladanyi M. Rapid polymerase chain reaction‐based detection of epidermal growth factor receptor gene mutations in lung adenocarcinomas. J Mol Diagn 2005; 7: 396–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yun CH, Boggon TJ, Li Y et al Structures of lung cancer‐derived EGFR mutants and inhibitor complexes: Mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell 2007; 11: 217–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhu JQ, Zhong WZ, Zhang GC et al Better survival with EGFR exon 19 than exon 21 mutations in gefitinib‐treated non‐small cell lung cancer patients is due to differential inhibition of downstream signals. Cancer Lett 2008; 265: 307–317. [DOI] [PubMed] [Google Scholar]
- 37. Banno E, Togashi Y, Kobayashi Y, Hayashi H, Mitsudomi T, Nishio K et al Afatinib is especially effective against non‐small cell lung cancer carrying an EGFR exon 19 deletion. Anticancer Res 2015; 35: 2005–2008. [PubMed] [Google Scholar]


