Abstract
Background
This article describes a pilot study evaluating a novel liquid biopsy system for non‐small cell lung cancer (NSCLC) patients. The electric field‐induced release and measurement (EFIRM) method utilizes an electrochemical biosensor for detecting oncogenic mutations in biofluids.
Methods
Saliva and plasma of 17 patients were collected from three cancer centers prior to and after surgical resection. The EFIRM method was then applied to the collected samples to assay for exon 19 deletion and p.L858 mutations. EFIRM results were compared with cobas results of exon 19 deletion and p.L858 mutation detection in cancer tissues.
Results
The EFIRM method was found to detect exon 19 deletion with an area under the curve (AUC) of 1.0 in both saliva and plasma samples in lung cancer patients. For L858R mutation detection, the AUC of saliva was 1.0, while the AUC of plasma was 0.98. Strong correlations were also found between presurgery and post‐surgery samples for both saliva (0.86 for exon 19 and 0.98 for L858R) and plasma (0.73 for exon 19 and 0.94 for L858R).
Conclusion
Our study demonstrates the feasibility of utilizing EFIRM to rapidly, non‐invasively, and conveniently detect epidermal growth factor receptor mutations in the saliva of patients with NSCLC, with results corresponding perfectly with the results of cobas tissue genotyping.
Keywords: EGFR mutation, liquid biopsy, lung cancer, saliva diagnostics
Introduction
Lung cancer is the most common cancer and leading cause of cancer death in China and worldwide.1, 2, 3, 4, 5, 6, 7, 8 Non‐small cell lung cancer (NSCLC) constitutes 80% of all lung cancer cases, and is typically diagnosed at an advanced stage when survival rates are low.9, 10, 11, 12, 13, 14, 15 The discovery of a relationship between epidermal growth factor receptor (EGFR)‐activating mutations and EGFR‐tyrosine kinase inhibitors (TKI) has taken the treatment of patients with EGFR mutations into an era of precision medicine.16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29 The 2009 Iressa Pan‐Asia Study (IPASS) demonstrated that EGFR‐TKIs are superior to traditional chemotherapeutic agents for patients with EGFR mutant lung cancer, and the current clinical EGFR‐TKIs have become a standard first‐line treatment for patients with non‐squamous NSCLC carrying EGFR mutations.20 Genotyping for EGFR mutations in advanced non‐squamous NSCLC patients is highly recommended in current clinical practice.
Epidermal growth factor receptor testing for mutations is traditionally performed on tissues acquired by surgery or biopsy. However, the majority of these diagnoses are on late stage patients in poor physical condition. In these instances, the performance of biopsy or surgery is often impractical because of poor patient health and the risk of additional clinical complications. Moreover, a high proportion of NSCLC patients will develop EGFR‐TKI treatment resistance, and performing additional surgeries or biopsies to monitor for an additional change in EGFR mutation status would further increase the risk to patient health.21, 22, 23, 24, 25, 26 As a result of these limitations, explorations have been made into the feasibility of detecting actionable EGFR oncogenic mutations in biofluids, such as serum or plasma.3, 4, 5, 6, 7, 8
Recent emerging work from the University of California, Los Angeles (UCLA) has demonstrated the feasibility of performing saliva‐based EGFR mutation detection (SABER) based on a core technology called electric field‐induced release and measurement (EFIRM).21 This method is an electrochemical method based on immobilized nucleic acid probes for capturing mutated sequences and applying electric fields to facilitate the hybridization process. Because of the speed and simplicity of the method, EFIRM has the potential to be a suitable tool for oncogenic mutation monitoring in a clinical context. Recently, a randomized, blind study using SABER for NSCLC patients demonstrated high concordance with tumor tissue based genotyping.23 Here, we report the first clinical validation pilot study of EFIRM in Mainland China.
Methods
Patients and clinical specimens
Inpatients with suspicious lesions for lung cancer, treated at three large‐scale major hospitals in China, West China Hospital, Henan Cancer Hospital, and Jiangsu Cancer Hospital, were recruited as subjects for our study. Each study participant had 5 mL saliva and 10 mL blood collected twice on the day before surgery, the day after surgery, and approximately seven days after surgery. Blood plasma samples were collected by drawing blood into ethylenediaminetetraacetic acid tubes, centrifuging at 2500g for 10 minutes at −4°C, and then collecting the upper plasma layer. Samples were stored at −80 °C until analysis. As per previous studies in salivary biomarkers, saliva samples were kept on ice during collection from patients and then centrifuged at 2600g for 15 minutes at 4°C.22 The supernatant was removed from the pellet, treated with RNase inhibitor (Superase‐In, Ambion Inc., Austin, TX, USA), and stored at −80°C for future analyses. All subjects provided informed consent.
Saliva‐based epidermal growth factor receptor (EGFR) mutation detection in bodily fluids
Epidermal growth factor receptor mutations in serum and saliva were detected by EFIRM in blinded samples by trained laboratory personnel. Each plasma and saliva sample was measured in duplicate. Paired probes (capture and detector; TsingKe, Beijing, China) specific to the two TKI‐sensitive mutations were used: for the exon 19 deletion (19 del), a capture probe 5′‐TGT TGC TTC CTTGAT AGC GAC G‐3′ and a detector probe 5′‐GGA ATT TTA ACT TTC TCA CCT‐FITC‐3′; for the L858R point mutation, a capture probe: 5′‐CAG TTT GGC CCG CCC AAA ATC‐3′and detector probe: 5′‐TTG ACA TGC TGC GGT GTT TTC A‐FITC‐3′. The detector probes were labeled with fluorescein isothiocyanate. The EFIRM detection method involves four primary steps.21 First, copolymerization of capture probes with pyrrole onto the bare gold electrodes by applying a cyclic square wave electric. Second, hybridization of the samples with detector and capture probes. Third, the combination of anti‐fluorescein antibody conjugated to horseradish peroxidase (1:1000 dilution; Roche, Indianapolis, IN, USA). Fourth, chromogenesis by 3,3′, 5,5′‐tetramethylbenzidine substrate for horseradish peroxidase and measurement of the amperometric signal. The total detection time of the protocol was less than 10 minutes, and the procedure required 20 to 40 μL of the biological sample.
EGFR mutation detection in tumor tissues
Epidermal growth factor receptor mutations in tumor tissues were detected by cobas assay, which is an allele‐specific real‐time polymerase chain reaction (PCR) system that qualitatively measures the amplification of DNA to detect EGFR gene mutation from DNA derived from freshly frozen tissues. DNA from these tissue specimens was extracted according to the standard procedure delineated in the cobas DNA Sample Preparation Kit (Roche Molecular Systems Inc., Pleasanton, CA, USA).21 This processed involved first incubating the sample with protease and buffer ATL at 56°C until dissolution. Buffer AL was then added for an additional 10 minutes at 70°C. Next, DNA samples were collected by the DNeasy Mini spin column (Qiagen, Valencia, CA, USA). After DNA samples were collected, the amount of DNA was analyzed by spectrophotometer and adjusted to a concentration of 2 ng/μL. Finally, the prepared DNA was amplified and detected by cobas 4800 (Roche Molecular Systems Inc.) following the instructions for the cobas EGFR mutation test. The results were automatically reported in cobas 4800 software.
Statistical analysis
Receiver operating characteristic curves (ROC) and the area under the curve (AUC) with 95% confidence intervals (CI) were calculated to access the ability of EFIRM in detecting EGFR mutations. Correlation coefficients were calculated using Pearson parametric methods. All analyses were performed using SAS 9.2 (SAS Institute Inc., Cary, NC, USA).
Results
The clinical characteristics of patients
A total of 17 randomized patients with adenocarcinoma with EGFR mutation from three different cancer centers in China (10 from West China Hospital, 4 from Henan Cancer Hospital, and 3 from Jiangsu Cancer Hospital) were enrolled. The clinical characteristics of these patients, including gender, age, smoking status, cancer stage, and pathological type, are presented in Table 1.
Table 1.
The clinical characteristics of patients in the study
| Patient No. | Gender | Age | Smoking status | Histologic type | Stage† | EGFR status | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cobas | EFIRM | |||||||||
| Saliva | Plasma | |||||||||
| Before | After | Before | After | |||||||
| 1 | M | 76 | N | NSCLC | pT2bN0M0, IIA | No | No | No | No | No |
| 2 | M | 53 | Y | Adeno | pT2aN1M0, IIA | Ex19 del | Ex19 del | Ex19 del | Ex19 del | Ex19 del |
| 3 | M | 53 | Y | Squa | pT2bN0M1a, IV | No | No | No | No | No |
| 4 | M | 53 | Y | Adeno | pT2bN2M0, IIIA | Ex19 del | Ex19 del | Ex19 del | Ex19 del | Ex19 del |
| 5 | M | 65 | Y | Squa | pT3N0M0, IIB | No | No | No | No | No |
| 6 | F | 72 | N | NSCLC | pT3N0M0, IIB | G719X | No | No | No | No |
| 7 | M | 71 | Y | Adeno | PT2aN0M0, IB | L858R | L858R | L858R | L858R | L858R |
| 8 | M | 46 | Y | Adeno | pT3N3M0, IIIB | No | No | No | No | No |
| 9 | M | 57 | Y | Squa | pT4N0M0, IIIA | No | No | No | No | No |
| 10 | F | 61 | N | Adeno | pT2aN20M, IIIA | Ex19 del | NE | Ex19 del | NE | Ex19 del |
| 11 | M | 47 | Y | Adeno | pT2bN2M0, IIIA | No | No | No | No | No |
| 12 | M | 62 | Y | Squa | pT2aN0M0, IB | No | No | No | No | No |
| 13 | F | 51 | N | Adeno | pT2aN0M0, IIB | L858R | L858R | L858R | L858R | L858R |
| 14 | M | 71 | N | Squa | pT1aN1M0, IIA | No | No | No | No | No |
| 15 | M | 50 | N | Adeno | pT2N2M0, IIIA | No | No | No | L858R | L858R |
| 16 | M | 60 | Y | Squa | pT2N0M0, IB | No | No | No | No | No |
| 17 | M | 51 | N | Adeno | pT2N2M0, IIIA | L858R | L858R | L858R | L858R | L858R |
Stage was based on National Comprehensive Cancer Network guidelines 2014, V3.
Adeno, adenocarcinoma; EGFR, epidermal growth factor receptor; EFIRM, electric field‐induced release and measurement; F, female; M, male; NE, no samples; NSCLC, non‐small cell lung cancer; Squa, squamous lung cancer.
Electric field‐induced release and measurement detection of EGFR mutations in saliva and plasma
The strength of the amperometric currents detected by EFIRM represented the status of the EGFR mutation. The saliva samples of three patients taken prior to surgery showed significantly higher currents using exon 19 probes, which meant that they were detected to have EGFR exon 19 del mutations (Fig 1a; 85.3 ± 0.3 nA in the exon 19 del group vs. 51.2 ± 5.6 nA in the L858R mutant and 49.7 ± 7.4 nA in the wild‐type; P < 0.01). Similarly, three patients, whose saliva samples had higher current readouts on EFIRM using the L858R mutant probes, were detected to harbor EGFR L858R mutants (Fig 1b; 114.9 ± 26.5 nA in the L858R mutant group vs. 45.6 ± 0.2 nA in the exon 19 del and 53.6 ± 12.0 nA in the wild‐type; P < 0.01). This detection included the samples collected both before and after surgery to avoid the possibility of tumor cells or DNA dropping into the circulation system by outside forces, and the results showed high consistency (Figs 1a, 2a and 1b, 2b). ROC analysis indicated AUCs of 1 for the probe carrying the exon 19 del and L858R mutation (Figs 1c, 2c).
Figure 1.
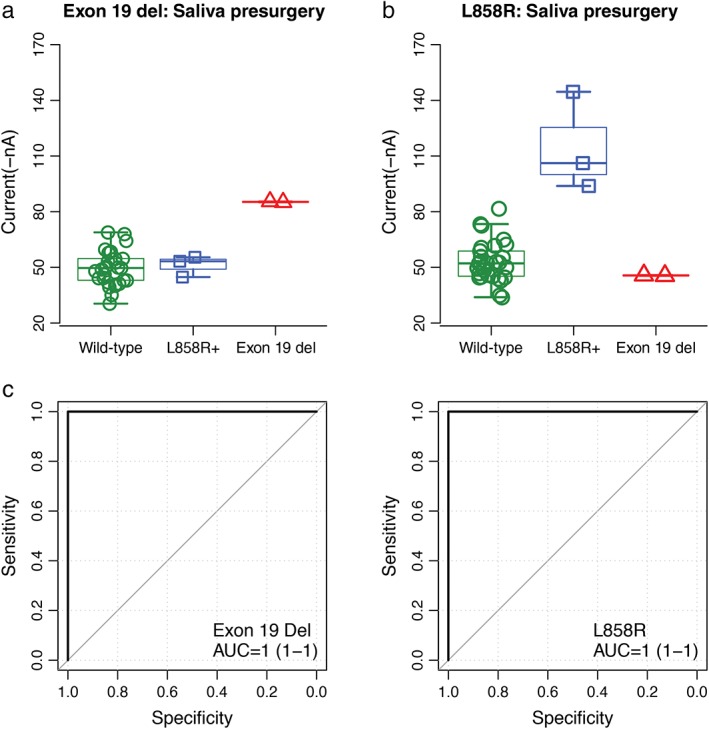
Blinded and randomized detection of epidermal growth factor receptor (EGFR) mutations by electric field‐induced release and measurement assay in saliva collected before surgery from patients with non‐small cell lung cancer: (a) the probe for EGFR exon 19 deletion in saliva; (b) the probe for L858R in saliva; and (c) receiver operating characteristic curves for detecting EGFR exon 19 deletion (area under the curve [AUC] = 1) and L858R mutations (AUC = 1), respectively.
Figure 2.
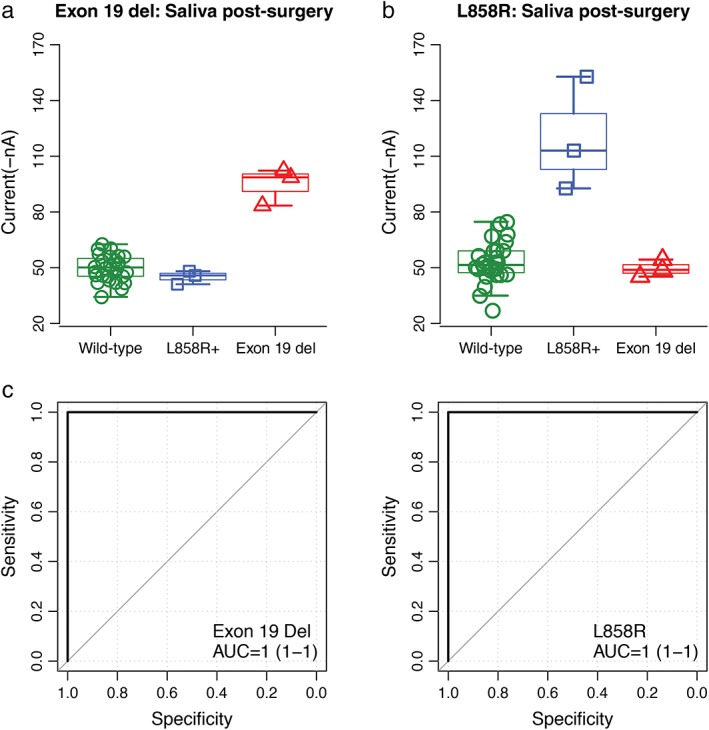
Blinded and randomized detection of epidermal growth factor receptor (EGFR) mutations by electric field‐induced release and measurement assay in saliva collected after surgery from patients with non‐small cell lung cancer: (a) the probe for EGFR exon 19 deletion in saliva; (b) the probe for L858R in saliva; and (c) receiver operating characteristic curves for detecting EGFR exon 19 deletion (area under the curve [AUC]= 1) and L858R mutations (AUC = 1), respectively.
Plasma samples were collected before and after surgery by EFIRM, except in one patient who only had samples taken after surgery. The results showed three exon 19 deletions (Fig 3a, 85.6 ± 9.6 nA in the exon 19 del group vs. 48.9 ± 8.0 nA in the L858R mutant and 51.1 ± 9.5 nA in the wild‐type; P = 0.001) and four L858R mutations (Fig 4a, 111.4 ± 27.8 nA in the L858R mutant group vs. 46.2 ± 0.7 nA in the exon 19 del and 51.9 ± 11.8 nA in the wild‐type; P < 0.001) before surgery. Similar results were obtained from plasma collected after surgery (Fig 3b, 92.6 ± 18.8 nA in the exon 19 del group vs. 52.9 ± 6.1 nA in the L858R mutant and 51.8 ± 5.4 nA in the wild‐type; P < 0.001; Fig 4b, 108.9 ± 22.2 in the L858R mutant group vs. 45.9 ± 1.7 nA in the exon 19 del and 52.4 ± 13.6 nA in the wild‐type; P < 0.001). The AUCs are shown in Fig 4c, respectively.
Figure 3.
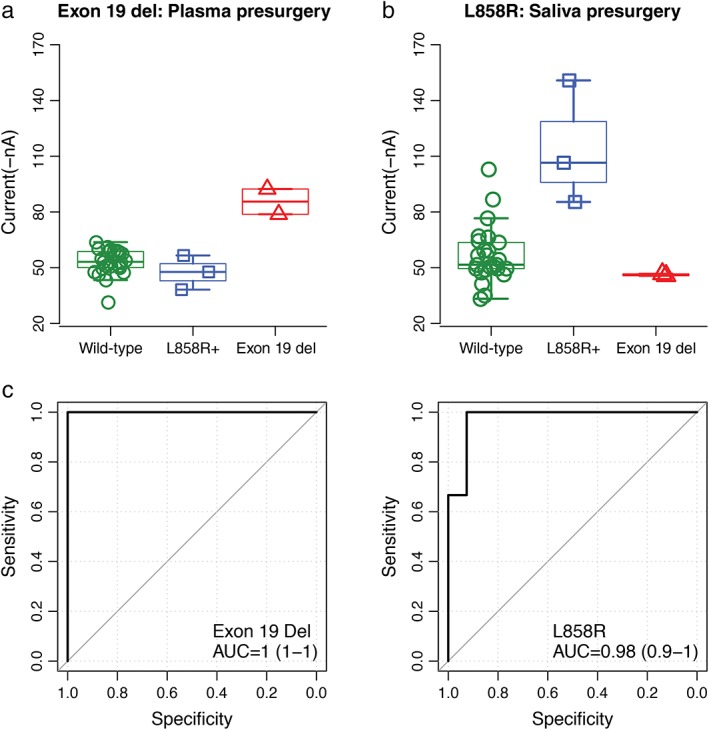
Blinded and randomized detection of epidermal growth factor receptor (EGFR) mutations by electric field‐induced release and measurement assay in plasma collected before surgery from patients with non‐small cell lung cancer: (a) the probe for EGFR exon 19 deletion; (b) the probe for L858R; and (c) receiver operating characteristic curves for detecting EGFR exon 19 deletion (area under the curve [AUC]= 1) and L858R mutations (AUC = 0.98), respectively.
Figure 4.
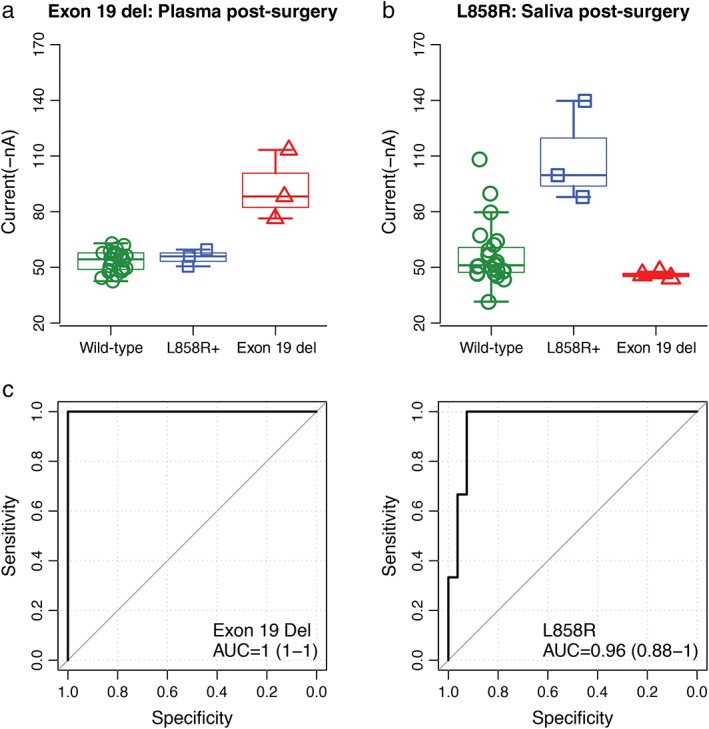
Blinded and randomized detection of epidermal growth factor receptor (EGFR) mutations by electric field‐induced release and measurement assay in plasma collected after surgery from patients with non‐small cell lung cancer: (a) the probe for EGFR exon 19 deletion; (b) the probe for L858R; and (c) receiver operating characteristic curves for detecting EGFR exon 19 deletion (area under the curve [AUC = 1) and L858R mutations (AUC = 0.96), respectively.
Cobas detection of EGFR mutations in tumor tissues
As a result of the approval of cobas for clinical use by the United States Food and Drug Administration and the China Food and Drug Administration, cobas assay is used as the gold standard method of EGFR mutation detection. We detected 17 tissue samples and found six samples harboring EGFR mutation (35.3%; 3 exon 19 del and 3 L858R mutations). We compared the results from EFIRM assay using saliva and plasma with the cobas assay using tumor tissues to evaluate the sensitivity and specificity of EFIRM assay in detecting EGFR mutation. The saliva and plasma samples of three patients that were identified to harbor EGFR exon 19 del with EFIRM were confirmed by cobas assay. Furthermore, these three patients with EGFR L858R mutations also perfectly matched the mutation results for saliva, plasma, and tissue samples. The other 11 patients were EGFR wild‐type in both saliva and tissue samples, which meant that there was a single false positive L858R mutation in plasma sample detection by EFIRM. The specificity and sensitivity was 90.9% and 100%, respectively, by plasma. In addition, all of the patients with EGFR mutation were histologically diagnosed with adenocarcinoma (3 stage III, 2 stage II, and 1 stage I), which indicated that EFIRM assay could maintain a high sensitivity in different cancer stages.
The correlation of EGFR mutation detected before and after surgery
We compared the results from the saliva and plasma collected before and after surgery to evaluate whether surgery affected EGFR mutation status. The strength of the amperometric currents in the saliva samples collected before surgery correlated with those after surgery using the L858R and exon 19 probes (Table 2, R = 0.86 in exon 19 del; R = 0.98 in L858R). Similar results were obtained in plasma samples (Table 2, R = 0.73 in exon 19 del; R = 0.94 in L858R). Additionally, regardless of whether samples were collected before or after surgery, the amperometric currents between saliva and plasma had a strong correlation (Table 2, before surgery: R = 0.87 in exon 19 del and R = 0.67 in L858R; after surgery: R = 0.79 in exon 19 del and R = 0.70 in L858R).
Table 2.
The correlation of EGFR mutation detected before and after surgery or biopsy
| Probe type | Pearson | |
|---|---|---|
| Saliva: pre/post surgery | ||
| Exon 19 del | 0.86 | |
| L858 | 0.98 | |
| Plasma: pre/post surgery | ||
| Exon 19 del | 0.73 | |
| L858 | 0.94 | |
| Pre surgery: saliva/plasma | ||
| Exon 19 del | 0.87 | |
| L858 | 0.67 | |
| Post surgery: saliva/plasma | ||
| Exon 19 del | 0.79 | |
| L858 | 0.70 |
EGFR, epidermal growth factor receptor.
Discussion
Liquid biopsy to detect actionable mutations in NSCLC in clinical practice is primarily based on digital droplet PCR (ddPCR) and/or next generation sequencing technologies with performance at ~70–80% concordance with tissue‐based genotyping. Technologies that will permit 100% concordance detection (sensitivity) will provide the ultimate complementation to the tumor‐specific fingerprint (specificity) for unambiguous detection of the tumor in a non‐invasive setting.3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14 To our knowledge, this is the first pilot study aimed at evaluating a novel saliva‐based EGFR mutation detection system for lung cancer in Mainland China. Our findings clearly show that saliva and plasma assays conducted using the EFIRM method have high concordance with biopsy tissue‐based testing for lung cancer EGFR mutation status. EFIRM liquid biopsy is accurate, non‐invasive, and rapid. It is can be used in continuous monitoring EGFR mutation status during EGFR‐TKI treatment.
We demonstrated that the amperometric currents of the EFIRM signals from saliva were highly correlated with those from plasma using the probe designed for p.E746‐A750del and p.L858R, regardless of whether the sample was collected presurgery or post‐surgery (0.70s range). These results suggest that the amount of EGFR mutant DNA in saliva and plasma is correlated. This may have a mechanistic implication, suggesting a link between the peripheral circulatory system and the salivary glands. Previous studies of this link have also found similarly strong correlations.22 However, the precise mechanism by which lung cancer cells release the EGFR mutant DNA and disperse it via the blood to distal sites remains to be established.
Another interesting finding in our study was the strong correlations found between presurgery and post‐surgery saliva assays (0.86 and 0.98) and plasma (0.73 and 0.94). This data implies that surgery may not influence EGFR mutant DNA status in saliva or plasma. Saliva collected after biopsy may result in tumor cells or cell‐free DNA that make their way into the saliva after tissue disruption, demonstrating that the detection of EGFR mutation in saliva was not a result of biopsy materials leaking into saliva.
Although this cohort of tests was only conducted on a small scale, there were still overlapping current values of EGFR wild type and mutation type at exon 21 L858R in plasma, which may have led to false‐positive results. Previous studies have demonstrated a similar phenomenon at L858R with an unclear threshold between wild type and mutation type.22 A large prospective study will be conducted to determine the adequate and optimal estimate of a threshold to decide the balance between false positive and false negative rates.
Electric field‐induced release and measurement can detect EGFR mutations in liquid biopsy from NSCLC patients, but at present this work has mainly focused on EGFR L858R and exon 19 del mutations. Because the second T790M EGFR mutation accounts for half of the TKI‐resistant cases, monitoring the T790M mutation would be useful to estimate EGFR‐TKI resistance. Therefore, development of a new probe designed for EFIRM based on T790M mutation is necessary, and this approach, as with other liquid biopsy‐based EGFR mutation assays (such as circulating tumor cells or cell‐free circulating tumor DNA), might be a valid noninvasive alternative to biopsy, as well as a better tailoring of individualized precision therapy by monitoring the response to treatment in NSCLC patients.
In conclusion, our results provide evidence that EFIRM detection of EGFR mutation in the saliva of patients with lung cancer is accurate, non‐invasive, and rapid.
Disclosure
David Wong is a co‐founder of RNAmeTRIX Inc., a molecular diagnostic company. He holds equity in RNAmeTRIX, and serves as a company Director and Scientific Advisor. The University of California also holds equity in RNAmeTRIX. Intellectual property invented by David Wong and patented by the University of California has been licensed to RNAmeTRIX.
Acknowledgments
This work was partly supported by grants from the National “863” Major Project of China (No. 2012AA02A201 and No. 2012AA02A502), the National Natural Science Foundation of China (No. 81572288) and the Ministry of Education Fund Priority to the Development of Instructions of Higher Leading Doctoral Degree Field (No. 20131202130001). We thank the patients for their participation in the study.
References
- 1. Chen W, Zheng R, Zhang S, Zhao P, Zeng H, Zou X. Report of cancer incidence and mortality in China, 2010. Ann Transl Med 2014; 2 (7): 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet‐Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin 2015; 65: 87–108. [DOI] [PubMed] [Google Scholar]
- 3. Sueoka‐Aragane N, Katakami N, Satouchi M et al Monitoring EGFR T790M with plasma DNA from lung cancer patients in a prospective observational study. Cancer Sci 2015; 107: 162–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Seki Y, Fujiwara Y, Kohno T et al Picoliter‐droplet digital polymerase chain reaction‐based analysis of cell‐free plasma DNA to assess EGFR mutations in lung adenocarcinoma that confer resistance to tyrosine‐kinase inhibitors. Oncologist 2016; 21: 156–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cohen V, Agulnik JS, Jarry J et al Evaluation of denaturing high‐performance liquid chromatography as a rapid detection method for identification of epidermal growth factor receptor mutations in nonsmall‐cell lung cancer. Cancer 2006; 107: 2858–65. [DOI] [PubMed] [Google Scholar]
- 6. Kawahara A, Fukumitsu C, Taira T et al Epidermal growth factor receptor mutation status in cell‐free DNA supernatant of bronchial washings and brushings. Cancer Cytopathol 2015; 123: 620–8. [DOI] [PubMed] [Google Scholar]
- 7. Uchida J, Kato K, Kukita Y et al Diagnostic accuracy of noninvasive genotyping of EGFR in lung cancer patients by deep sequencing of plasma cell‐free DNA. Clin Chem 2015; 61: 1191–6. [DOI] [PubMed] [Google Scholar]
- 8. Qian X, Wang S, Shen Y et al [Methodology comparison and influence factors analysis of epidermal growth factor receptor mutation detection]. Zhonghua Yi Xue Za Zhi 2015; 95: 106–11. (In Chinese.). [PubMed] [Google Scholar]
- 9. Xu H, Sun W, Zhang G, Cheng Y. Detection of epidermal growth factor receptor mutation in non‐small‐cell lung carcinoma using cytological and histological specimens. J BUON 2015; 20 (1): 142–5. [PubMed] [Google Scholar]
- 10. Zhu G, Ye X, Dong Z et al Highly sensitive droplet digital PCR method for detection of EGFR‐activating mutations in plasma cell‐free DNA from patients with advanced non‐small cell lung cancer. J Mol Diagn 2015; 17: 265–72. [DOI] [PubMed] [Google Scholar]
- 11. Lewandowska MA, Czubak K, Klonowska K, Jozwicki W, Kowalewski J, Kozlowski P. The use of a two‐tiered testing strategy for the simultaneous detection of small EGFR mutations and EGFR amplification in lung cancer. PLoS one 2015; 10 (2): e0117983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lin J, Gu Y, Du R, Deng M, Lu Y, Ding Y. Detection of EGFR mutation in supernatant, cell pellets of pleural effusion and tumor tissues from non‐small cell lung cancer patients by high resolution melting analysis and sequencing. Int J Clin Exp Pathol 2014; 7: 8813–22. [PMC free article] [PubMed] [Google Scholar]
- 13. Takata M, Chikumi H, Matsunami K et al A new rapid method for detecting epidermal growth factor receptor mutations in non‐small cell lung cancer. Oncol Rep 2015; 33: 1040–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Romanus D, Cardarella S, Cutler D, Landrum MB, Lindeman NI, Gazelle GS. Cost‐effectiveness of multiplexed predictive biomarker screening in non‐small‐cell lung cancer. J Thorac Oncol 2015; 10: 586–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lozano MD, Labiano T, Echeveste J et al Assessment of EGFR and KRAS mutation status from FNAs and core‐needle biopsies of non‐small cell lung cancer. Cancer Cytopathol 2015; 123: 230–6. [DOI] [PubMed] [Google Scholar]
- 16. Tseng JS, Yang TY, Tsai CR et al Dynamic plasma EGFR mutation status as a predictor of EGFR‐TKI efficacy in patients with EGFR‐mutant lung adenocarcinoma. J Thorac Oncol 2015; 10: 603–10. [DOI] [PubMed] [Google Scholar]
- 17. Scarpino S, Pulcini F, Di Napoli A, Giubettini M, Ruco L. EGFR mutation testing in pulmonary adenocarcinoma: Evaluation of tumor cell number and tumor percent in paraffin sections of 120 small biopsies. Lung Cancer 2015; 87: 8–13. [DOI] [PubMed] [Google Scholar]
- 18. Sueoka‐Aragane N, Sato A, Kobayashi N et al Correlation between plasma DNA and tumor status in an animal model. PLoS One 2014; 9 (12): e111881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Deeb KK, Hohman CM, Risch NF, Metzger DJ, Starostik P. Routine clinical mutation profiling of non‐small cell lung cancer using next‐generation sequencing. Arch Pathol Lab Med 2015; 139: 913–21. [DOI] [PubMed] [Google Scholar]
- 20. Mok TS, Wu YL, Thongprasert S et al Gefitinib or carboplatin‐paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009; 361: 947–57. [DOI] [PubMed] [Google Scholar]
- 21. Niu FY, Wu YL. Novel agents and strategies for overcoming EGFR TKIs resistance. Exp Hematol Oncol 2014; 3: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wei F, Lin CC, Joon A et al Noninvasive saliva‐based EGFR gene mutation detection in patients with lung cancer. Am J Respir Crit Care Med 2014; 190: 1117–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Skog J, Würdinger T, van Rijn S et al Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nature Cell Biol 2008; 10: 1470–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li Y, St John MA, Zhou X et al Salivary transcriptome diagnostics for oral cancer detection. Clin Cancer Res 2004; 10: 8442–50. [DOI] [PubMed] [Google Scholar]
- 25. Kimura H, Ohira T, Uchida O et al Analytical performance of the cobas EGFR mutation assay for Japanese non‐small‐cell lung cancer. Lung Cancer 2014; 83: 329–33. [DOI] [PubMed] [Google Scholar]
- 26. Sandri MT, Zorzino L, Cassatella MC et al Changes in circulating tumor cell detection in patients with localized breast cancer before and after surgery. Ann Surg Oncol 2010; 17: 1539–45. [DOI] [PubMed] [Google Scholar]
- 27. Biggers B, Knox S, Grant M et al Circulating tumor cells in patients undergoing surgery for primary breast cancer: Preliminary results of a pilot study. Ann Surg Oncol 2009; 16: 969–71. [DOI] [PubMed] [Google Scholar]
- 28. Ma C, Wei S, Song Y. T790M and acquired resistance of EGFR TKI: A literature review of clinical reports. J Thorac Dis 2011; 3: 10–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pao W, Miller VA, Politi KA et al Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med 2005; 2 (3): e73. [DOI] [PMC free article] [PubMed] [Google Scholar]


