Abstract
Epigenetic modifications are involved in breast carcinogenesis. Identifying genes that are epigenetically silenced via methylation could select target patients for diagnostic as well as therapeutic potential. We assessed promoter methylation of breast cancer susceptibility gene 1 (BRCA1) and 17 Beta Hydroxysteroid Dehydrogenase Type 1 (17βHSD-1) in normal and cancer breast tissues of forty sporadic breast cancer (BC) cases using restriction enzyme based methylation-specific PCR (REMS-PCR). In cancerous tissues, BRCA1 and 17βHSD-1 were methylated in 42.5% and 97.5%, respectively, while normal tissues had 35% and 95% methylation, respectively. BRCA1 methylation in normal tissues was 12.2-fold more likely to associate with methylation in cancer tissues (p < 0.001). It correlated significantly with increased age at menopause, mitosis, the negative status of Her2, and the molecular subtype “luminal A” (p = 0.048, p = 0.042, p = 0.007, and p = 0.049, resp.). Methylation of BRCA1 and 17βHSD-1 related to luminal A subtype of breast cancer. Since a small proportion of normal breast epithelial cells had BRCA1 methylation, our preliminary findings suggest that methylation of BRCA1 may be involved in breast tumors initiation and progression; therefore, it could be used as a biomarker for the early detection of sporadic breast cancer. Methylation of 17βHSD-1 in normal and cancer tissue could save patients the long term use of adjuvant antiestrogen therapies.
1. Introduction
Breast cancer is a malignancy arising from the epithelial tissues that line the terminal ductal-lobular units of the breast [1, 2]. Breast cancer (BC) is the most common cancer in females worldwide [3]; BC is the leading cause of cancer death among females, with an estimated 1.7 million cases and 521,900 deaths in 2012; BC alone accounts for 25% of all cancer cases and 15% of all cancer deaths among females [4]. Based on data from the National Cancer Registry Program of Egypt (NCRP) in 2008–2011, BC is the most common malignancy among Egyptian females. It constituted 32.0% of all cancer cases [5]. According to GLOBOCAN 2012, the age-standardized incidence rate (ASR) of breast cancer was 42.3 per 100,000 Egyptian females, with a mortality rate of 17.4 per 100,000 females [4].
Many environmental factors combined with multiple genetic and epigenetic changes are involved in the onset and development of breast cancer [6, 7]. The carcinogenesis process is a multistep process during which genetic and epigenetic alterations accumulate in a cell, resulting in the progressive transformation of normal cells through steps of initiation, promotion, and progression into cancer cells [8]. Epigenetics are emerging as one of the most important events in carcinogenesis [9]. DNA methylation has an essential role in the regulation of gene expression in mammalian cells [10]. In normal cells, the majority of promoter cytosine phosphate guanosine (CpG) islands are protected from this epigenetic event; thus, they are unmethylated. Conversely, in cancer cells, several promoter CpG islands are hypermethylated and form a closed repressive chromatin configuration that affects the transcription initiation of the corresponding genes [11–13]. Moreover, promoter methylation is a common epigenetic mechanism to silence genes during breast cancer development [14].
Currently, genes that are epigenetically regulated via promoter methylation in breast cancer include cyclin-dependent kinase inhibitor (p16), breast cancer susceptibility gene 1 (BRCA1), estrogen receptor (ERα), progesterone receptor (PR), retinoic acid receptor-β2 (RARβ2), glutathione S-transferase p1 (GSTP1), E-cadherin, and tissue inhibitor of metalloproteinase 3 (TIMP3) [15]. The identification of these methylated promoters had significantly contributed to elucidating the altered molecular pathways in breast carcinoma and provided potential targets for molecular detection [16]. BRCA1 is a tumor suppressor gene that is involved in critical biological processes, including DNA damage repair, cell cycle control, and transcriptional regulation [17]. Silencing of BRCA1 gene via promoter hypermethylation is a common mechanism for its inactivation [18] ranging from 9 to 44% of sporadic breast cancers [19–21]. Breast cancer, with extensive hypermethylation in the BRCA1 promoter, correlates with a reduced BRCA1 expression [22]. In normal breast tissues, BRCA1 promoter methylation had been identified in 8.3–22% [23].
The growth of both normal and neoplastic mammary tissue is affected by a number of hormones especially estrogen, which exists in several forms, estrone (E1), estradiol (E2), estriol, estrone sulfate, and estradiol sulfate. E2 is the most biologically active form in the breast tissue. Increasing evidence indicates that intratumoral estrogens derived in situ are mitogenic; thus they promote BC progression, irrespective of the serum concentrations of ovarian estrogen [24, 25].
17 Beta Hydroxysteroid Dehydrogenases (17βHSDs) catalyze the interconversion of active and inactive forms of estrogens within tissues. 17 Beta Hydroxysteroid Dehydrogenase Type 1 (17βHSD-1) mainly converts E1 to the potent E2. The encoding gene (17βHSD-1) is located at 17q12–21, a region that often is rearranged in breast cancer [26]. In a study carried out by Gunnarsson et al., they found amplification of the encoding gene (17βHSD-1) in 14.5% of the breast tumors [27]. Meanwhile 17 Beta Hydroxysteroid Dehydrogenase Type 2 (17βHSD-2) catalyzes the conversion of E2 to E1, thereby reducing estradiol level and hence controlling its proliferative activity [28]. The encoding gene (17βHSD-2) is located at 16q24 and loss of heterozygosity (LOH) at this site is frequent and early event in breast cancer [29]. Interestingly, BRCA1 had been found to negatively affect estradiol activity by direct interaction with the estrogen receptor, thus controlling the proliferation caused by this steroid hormone [30].
Clinical interest in the treatment of tumors has gained increased impetus interest because the evidence on the use of novel therapeutic agents suggests that DNA promoter methylation is potentially reversible. This may thus allow for the development of future therapeutic interventions [31]. In the light of these evidences and because of the potential roles of estrogens in the early stages of human breast carcinogenesis, in the present study, we aimed to assess the promoter methylation status of both BRCA1 and 17βHSD-1 genes in the tumor and adjacent normal tissue from sporadic breast cancer patients to establish the role of epigenetic in regulating intratissue estrogen activity and thereby in the etiology of breast cancer.
2. Material and Methods
2.1. Tissue Specimen Collection
Surgically resected specimens were freshly obtained at the operation room from forty diagnosed primary breast carcinomas enrolled in the Surgical Oncology Unit in Suez Canal University Hospital during the period from 2013 to 2014. Their matching normal breast tissues, taken 3–5 cm away from the healthy safety margin of the site of the tumor in the same breast, were obtained to serve as controls. Patients receiving neoadjuvant chemotherapy and/or hormonal therapy were excluded. Harvested breast tissues were either divided for DNA isolation or kept in 10% neutral-buffered formalin for histopathological analysis. Labeled tissue sections were examined by a pathologist blinded to the identity of samples and only the researcher would know to whom it referred. Prior to surgical tumor removal, written informed approval consents were obtained from all participants included in the study according to the Ethics Committee of the Faculty of Medicine, Suez Canal University.
2.2. Extraction of Genomic DNA
Genomic DNA was extracted from collected tissues using Qiagen Genomic DNA Purification Kit (cat # 51304; Qiagen, Hilden, Germany) according to the manufacturer's instructions. The quality of extracted genomic DNA was measured using the NanoDrop- (ND-) 1000 Spectrophotometer V3.1.0 (NanoDrop Technologies, Inc., Wilmington, DE, USA).
2.3. Restriction Enzyme Based Methylation-Specific Polymerase Chain Reaction (REMS-PCR)
Extracted DNA was digested using the fast restriction enzyme HpaII according to the manufacturer's instructions (Fast Digest®, Fermentas, CA, USA). The reaction mixture was incubated at 37°C in an oven for 1 hour followed by enzyme inactivation for 10 minutes at 90°C. Digested DNA aliquots were then PCR analyzed using BRCA1 and 17βHSD-1 specific primers [32] encompassing methylation-specific sites (Table 1) and the Taq PCR Master Mix Kit (cat # 201445; Qiagen, Hilden, Germany). A mock undigested sample containing 1 μL of nuclease-free water, instead of 1 μL of Fast Digest enzyme (HpaII), was used as a control. A sample of water instead of DNA was used as a negative control for each PCR. PCR was performed for 32 cycles using the following thermal cycling conditions: initial denaturation at 94°C for 5 minutes, denaturation at 94°C for 30 seconds, primer annealing at 55°C or 58°C for 30 seconds, extension at 72°C for 45 seconds, and a final extension at 72°C for 7 minutes. An attempt to perform BRCAI PCRs at higher annealing temperatures gave a total absence of bands. PCR products were detected using ethidium bromide-stained 2% agarose gels and bands were visualized under UV light. The presence of bands with sizes of 500 bp and 238 bp indicated methylation of BRCA1 and 17βHSD-1 promoters, respectively, while the absence of these bands indicated a lack of methylation (Figures 1 and 2).
Table 1.
Primers used for PCR amplification after HpaII digestion, along with the target band size and annealing temperatures.
Figure 1.
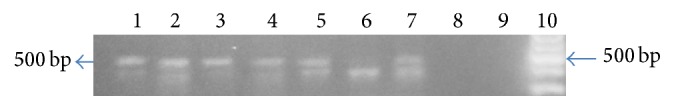
A representative gel picture of 2% agarose gel showing BRCA1 methylation status of two cases. Lanes 1, 2, 3, 4, 5, and 7: the presence of (500 bp) band indicates methylation of BRCA1 promoter in these specimens. Lanes 6 and 8: absence of (500 bp) band indicates the absence of methylation of BRCA1 promoter in these specimens. Lane 9: negative control. Lane 10: 100 bp DNA ladder. Lanes 1 and 5: undigested DNA of normal breast tissue specimens; lanes 2 and 6: digested DNA of normal breast tissue specimens; lanes 3 and 7: undigested DNA of cancerous breast tissue specimens; and lanes 4 and 8: digested DNA of cancerous breast tissue specimens.
Figure 2.
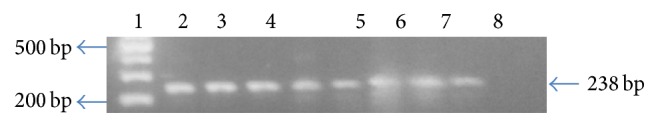
A representative gel picture of ethidium bromide-stained 2% agarose gel showing 17βHSD-1 methylation status of two cases. Lane 1: a 100 bp DNA ladder. Lanes 2, 3, 4, 5, 6, 7, 8, and 9: the presence of (238 bp) band indicates methylation of 17βHSD-1 promoter in these specimens. Lane 10: negative control. Lanes 2 and 6: undigested DNA of normal breast tissue specimens; lanes 3 and 7: digested DNA of normal breast tissue specimens; lanes 4 and 8: undigested DNA of cancerous breast tissue specimens; and lanes 5 and 9: digested DNA of cancerous breast tissue specimens.
2.4. Statistical Analysis
Coded collected data was analyzed using statistical package SPSS 16.0 for windows (SPSS, Chicago, IL, USA). Student's t-test was performed for statistical evaluation of quantitative variable between two independent groups in parametric data with p < 0.05 considered significant. Chi-square test, described in the form of frequency and percentages, was used to compare a qualitative variable between two independent groups.
3. Results
3.1. Association of BRCA1 and 17βHSD-1 Promoter Methylation with Clinicopathological Parameters
REMS-PCR showed PCR products of the expected sizes 500 bp and 238 bp for BRCA1 and 17βHSD-1 genes, respectively, in methylated samples, while the absence of these bands indicated a lack of methylation (Figures 1 and 2).
BRCA1 promoter was methylated in 42.5% of tumor tissue compared to 35% in control normal tissue (Figure 3). To our surprise, 17βHSD-1 promoter methylation was seen in 97.5% of tumor tissues as compared to 95% of neighboring normal tissue (Figure 4). We found a trend towards BRCA1 and 17βHSD-1 promoter hypermethylation in cancer tissue specimens of sporadic breast cancer patients compared with controls, although the difference was nonsignificant (p > 0.05).
Figure 3.
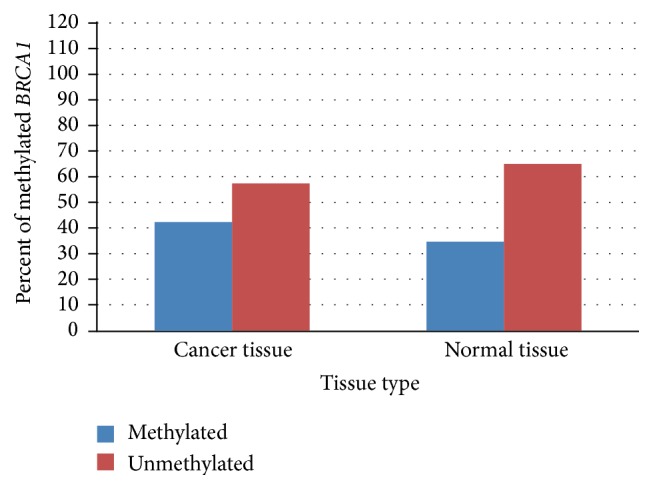
Difference between methylation status of BRCA1 promoter in cancer and normal tissue specimens.
Figure 4.
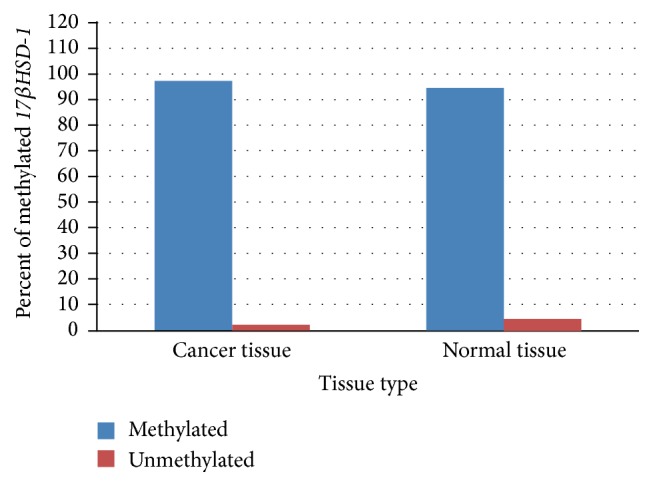
Difference between methylation status of 17βHSD-1 promoter in cancer and normal tissue specimens.
We categorized our patients into two age groups below 50 years and above or equal to 50 years; there was a nonsignificant increase in BRCA1 promoter methylation of breast cancer tissue specimens in older women compared with younger patients (70.6% versus 29.4%, resp.) (Table 2). Similarly, 17βHSD-1 promoter methylation, in both study groups, was not associated with age (Table 3), indicating no dramatic effect of age on BRCA1 and 17βHSD-1 methylation status. Moreover, postmenopausal status was associated with a nonsignificant increased methylation of BRCA1 promoter in cancer tissue specimens compared with premenopausal females (70.6% versus 29.4%), respectively (Table 2). In contrast, the mean age at menopause was significantly higher among BRCA1-methylated than the BRCA1-unmethylated group in cancer tissue specimens, as shown in Table 2 (50.7 ± 3.1, 48.3 ± 3.4, resp.; p = 0.048). Finally, our results showed no statistically significant difference in BRCA1 promoter methylation in both study groups with tumor size, the number of positive nodes, or tumor stage (Table 2). However, BRCA1-methylated promoter in cancer tissues tended to be of a higher grade (82.4% methylated versus 69.6% unmethylated in grade 2 “tumors”).
Table 2.
Association of BRCA1 promoter methylation with clinicopathological parameters.
| Overall (N = 40) | % | BRCA1 methylation in normal tissues | BRCA1 methylation in cancer tissues | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Methylated (14) | Unmethylated (26) | p value | Methylated (17) | Unmethylated (23) | p value | |||||||
| Freq. | (%) | Freq. | (%) | Freq. | (%) | Freq. | (%) | |||||
| Age: | ||||||||||||
| <50 | 10 | 25% | 6 | (42.9) | 4 | (15.4) | 0.123 | 5 | (29.4) | 5 | (21.7) | 0.717 |
| ≥50 | 30 | 75% | 8 | (57.1) | 22 | (84.6) | 12 | (70.6) | 18 | (78.3) | ||
| Menopausal status: | ||||||||||||
| Premenopause | 7 | 17.5% | 5 | (35.7) | 2 | (7.7) | 5 | (29.4) | 2 | (8.7) | 0.113 | |
| Postmenopause | 33 | 82.5% | 9 | (64.3) | 24 | (92.3) | 12 | (70.6) | 21 | (91.3) | ||
| Age at menopause: | ||||||||||||
| ≤50 | 28 | 70.0% | 8 | (57.1) | 20 | (76.9) | 0.079 | 10 | (58.8) | 18 | (78.3) | 0.317 |
| >50 | 5 | 12.5% | 1 | (7.1) | 4 | (15.4) | 2 | (11.8) | 3 | (13.0) | ||
| Premenopausal | 7 | 17.5% | 5 | (35.7) | 2 | (7.7) | 5 | (29.4) | 2 | (8.7) | ||
| Mean ± SD | 49.2 ± 3.7 | 50.1 ± 2.8 | 48.8 ± 3.9 | 0.318 | 50.7 ± 3.1 | 48.3 ± 3.4 | 0.048∗ | |||||
| Tumor size: | ||||||||||||
| T1 | 4 | 10.0% | 2 | (14.3) | 2 | (7.7) | 0.478 | 1 | (5.9) | 3 | (13.0) | 0.800 |
| T2 | 28 | 70.0% | 11 | (78.6) | 17 | (65.4) | 13 | (76.5) | 15 | (65.2) | ||
| T3 | 4 | 10.0% | 1 | (7.1) | 3 | (11.5) | 2 | (11.8) | 2 | (8.7) | ||
| T4 | 4 | 10.0% | 0 | (0) | 4 | (15.4) | 1 | (5.9) | 3 | (13.0) | ||
| Lymph node status : | ||||||||||||
| N0 | 11 | 27.5% | 5 | (35.7) | 6 | (23.1) | 0.478 | 5 | (29.4) | 6 | (26.1) | 0.544 |
| N1 | 8 | 20.0% | 4 | (28.6) | 4 | (15.4) | 5 | (29.4) | 3 | (13.0) | ||
| N2 | 13 | 32.5% | 3 | (21.4) | 10 | (38.5) | 4 | (23.5) | 9 | (39.1) | ||
| N3 | 8 | 20.0% | 2 | (14.3) | 6 | (23.1) | 3 | (17.6) | 5 | (21.7) | ||
| Clinical stage: | ||||||||||||
| I | 2 | 5.0% | 2 | (14.3) | 0 | (0.0) | 0.124 | 1 | (5.9) | 1 | (4.3) | 0.940 |
| IIA | 10 | 25.0% | 4 | (28.6) | 6 | (23.1) | 4 | (23.5) | 6 | (26.1) | ||
| IIB | 13 | 32.5% | 6 | (42.9) | 7 | (26.9) | 7 | (41.2) | 6 | (26.1) | ||
| IIIA | 4 | 10.0% | 0 | (0.0) | 4 | (15.4) | 1 | (5.9) | 3 | (13.0) | ||
| IIIB | 3 | 7.5% | 0 | (0.0) | 3 | (11.5) | 1 | (5.9) | 2 | (8.7) | ||
| IIIC | 8 | 20.0% | 2 | (14.3) | 6 | (23.1) | 3 | (17.6) | 5 | (21.7) | ||
| Histological grade: | ||||||||||||
| G1 | 3 | 7.5% | 1 | (7.1) | 2 | (7.1) | 0.319 | 2 | (11.8) | 1 | (4.3) | 0.281 |
| G2 | 30 | 75.0% | 11 | (78.6) | 19 | (73.1) | 14 | (82.4) | 16 | (69.6) | ||
| G3 | 3 | 7.5% | 2 | (14.3) | 1 | (3.8) | 1 | (5.9) | 2 | (8.7) | ||
| No grade | 4 | 10.0% | 0 | (0.0) | 4 | (15.4) | 0 | (0.0) | 4 | (17.4) | ||
∗Chi-square exact test is statistically significant at 95% confidence level.
Table 3.
Association of 17βHSD-1 promoter methylation with clinicopathological parameters.
| Overall (N = 40) | % | 17βHSD-1 methylation in normal tissues | 17βHSD-1 methylation in cancer tissues | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Methylated (38) | Unmethylated (2) | p value | Methylated (39) | Unmethylated (1) | p value | |||||||
| Freq. | (%) | Freq. | (%) | Freq. | (%) | Freq. | (%) | |||||
| Age: | ||||||||||||
| <50 | 10 | 25% | 5 | (29.4) | 5 | (21.7) | 0.717 | 9 | (23.1) | 1 | (100.0) | 0.250 |
| ≥50 | 30 | 75% | 12 | (70.6) | 18 | (78.3) | 30 | (76.9) | 0 | (0.0) | ||
| Menopausal status: | ||||||||||||
| Premenopause | 7 | 17.5% | 7 | (18.4) | 0 | (0.0) | 1.000 | 7 | (17.9) | 0 | (0.0) | 1.000 |
| Postmenopause | 33 | 82.5% | 31 | (81.6) | 2 | (100.0) | 32 | (82.1) | 1 | (100.0) | ||
| Age at menopause: | ||||||||||||
| ≤50 | 28 | 70.0% | 26 | (68.4) | 2 | (100.0) | 0.637 | 27 | (69.2) | 1 | (100.0) | 1.000 |
| >50 | 5 | 12.5% | 5 | (13.2) | 0 | (0.0) | 5 | (12.8) | 0 | (0.0) | ||
| Premenopausal | 7 | 17.5% | 7 | (18.4) | 0 | (0.0) | 7 | (17.9) | 0 | (0.0) | ||
| Mean ± SD | 49.2 ± 3.7 | 49.5 ± 3.4 | 44 ± 5.7 | 0.397 | 49.2 ± 3.7 | 45.0 | 0.242 | |||||
| Tumor size: | ||||||||||||
| T1 | 4 | 10.0% | 4 | (10.5) | 0 | (0.0) | 0.271 | 3 | (7.7) | 1 | (100.0) | 0.300 |
| T2 | 28 | 70.0% | 27 | (71.1) | 1 | (50.0) | 28 | (71.8) | 0 | (0.0) | ||
| T3 | 4 | 10.0% | 3 | (7.9) | 1 | (50.0) | 4 | (10.3) | 0 | (0.0) | ||
| T4 | 4 | 10.0% | 4 | (10.5) | 0 | (0.0) | 4 | (10.3) | 0 | (0.0) | ||
| Lymph node status: | ||||||||||||
| N0 | 11 | 27.5% | 11 | (28.9) | 0 | (0.0) | 0.368 | 11 | (28.2) | 0 | (0.0) | 0.400 |
| N1 | 8 | 20.0% | 7 | (18.4) | 1 | (50.0) | 7 | (17.9) | 1 | (100.0) | ||
| N2 | 13 | 32.5% | 13 | (34.2) | 0 | (0.0) | 13 | (33.3) | 0 | (0.0) | ||
| N3 | 8 | 20.0% | 7 | (18.4) | 1 | (50.0) | 8 | (20.5) | 0 | (0.0) | ||
| Clinical stage: | ||||||||||||
| I | 2 | 5.0% | 2 | (5.3) | 0 | (0.0) | 0.327 | 2 | (5.1) | 0 | (0.0) | 0.675 |
| IIA | 10 | 25.0% | 10 | (26.3) | 0 | (0.0) | 9 | (23.1) | 1 | (100.0) | ||
| IIB | 13 | 32.5% | 13 | (34.2) | 0 | (0.0) | 13 | (33.3) | 0 | (0.0) | ||
| IIIA | 4 | 10.0% | 3 | (7.9) | 1 | (50.0) | 4 | (10.3) | 0 | (0.0) | ||
| IIIB | 3 | 7.5% | 3 | (7.9) | 0 | (0.0) | 3 | (7.7) | 0 | (0.0) | ||
| IIIC | 8 | 20.0% | 7 | (18.4) | 1 | (50.0) | 8 | (20.5) | 0 | (0.0) | ||
| Histological grade: | ||||||||||||
| G1 | 3 | 7.5% | 3 | (7.9) | 0 | (0.0) | 0.873 | 3 | (7.7) | 0 | (0.0) | 1.000 |
| G2 | 30 | 75.0% | 28 | (73.7) | 2 | (100.0) | 29 | (74.4) | 1 | (100.0) | ||
| G3 | 3 | 7.5% | 3 | (7.9) | 0 | (0.0) | 3 | (7.7) | 0 | (0.0) | ||
| No grade | 4 | 10.0% | 4 | (10.5) | 0 | (0.0) | 4 | (10.3) | 0 | (0.0) | ||
Chi-square exact test is statistically significant at 95% confidence level.
When studying 17βHSD-1, promoter methylation did not associate with any clinicopathological characteristics in both study groups (Table 3). In addition, 17βHSD-1 promoter methylation did not correlate with lymph node status, clinical stage, or histological grade (Table 3).
3.2. Association of BRCA1 and 17βHSD-1 Promoter Methylation with Molecular Subtypes of Breast Cancer
Methylated BRCA1 in breast cancer specimens correlated with increased mitotic index, the negativity of Her2 receptors, and hence molecular subtype “luminal A” (p = 0.042, p = 0.007, and p = 0.049, resp.) (Table 4). Although methylation of this promoter tended to be higher in positive estrogen receptor specimens, this correlation was not significant (Table 4). 17βHSD-1 promoter methylation occurred with almost equal percentages when correlated with all molecular subtypes (Table 5). We observed a high trend towards 17βHSD-1 promoter methylation in the breast cancer tissue specimens in women who had positive estrogen, progesterone receptors, and negative Her2 (Table 5).
Table 4.
Association of BRCA1 promoter methylation with mitosis, immunohistochemistry, and molecular subtype of breast cancer.
| Overall (N = 40) | % | BRCA1 methylation in normal tissues | BRCA1 methylation in cancer tissues | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Methylated (14) | Unmethylated (26) | p value | Methylated (17) | Unmethylated (23) | p value | |||||||
| Freq. | (%) | Freq. | (%) | Freq. | (%) | Freq. | (%) | |||||
| Mitosis: | ||||||||||||
| 0 | 4 | 10.0% | 0 | (0) | 4 | (15.4) | 0.292 | 0 | (0.0) | 4 | (17.4) | 0.042∗ |
| 1 | 5 | 12.5% | 3 | (21.4) | 2 | (7.7) | 4 | (23.5) | 1 | (4.3) | ||
| 2 | 29 | 72.5% | 10 | (71.4) | 19 | (73.1) | 13 | (76.5) | 16 | (69.6) | ||
| 3 | 2 | 5.0% | 1 | (7.1) | 1 | (3.8) | 0 | (0.0) | 2 | (8.7) | ||
| Estrogen receptor: | ||||||||||||
| Positive | 27 | 67.5% | 10 | (71.4) | 17 | (65.4) | 0.912 | 13 | (76.5) | 14 | (60.9) | 0.568 |
| Negative | 7 | 17.5% | 2 | (14.3) | 5 | (19.2) | 2 | (11.8) | 5 | (21.7) | ||
| No available data | 6 | 15.0% | 2 | (14.3) | 4 | (15.4) | 2 | (11.8) | 4 | (17.4) | ||
| Progesterone receptor: | ||||||||||||
| Positive | 25 | 62.5% | 9 | (64.3) | 16 | (61.5) | 0.985 | 11 | (64.7) | 14 | (60.9) | 0.907 |
| Negative | 9 | 22.5% | 3 | (21.4) | 6 | (23.1) | 4 | (23.5) | 5 | (21.7) | ||
| No available data | 6 | 15.0% | 2 | (14.3) | 4 | (15.4) | 2 | (11.8) | 4 | (17.4) | ||
| Her2: | ||||||||||||
| Positive | 10 | 25.0% | 2 | (14.3) | 8 | (30.8) | 0.279 | 2 | (11.8) | 8 | (34.8) | 0.007∗ |
| Negative | 19 | 47.5% | 9 | (64.3) | 10 | (38.5) | 13 | (76.4) | 6 | (26.1) | ||
| No available data | 11 | 27.5% | 3 | (21.4) | 8 | (30.8) | 2 | (11.8) | 9 | (39.1) | ||
| Molecular subtype: | ||||||||||||
| Luminal A | 13 | 32.5% | 7 | (50.0) | 6 | (23.1) | 0.242 | 9 | (52.9) | 4 | (17.4) | 0.049∗ |
| Luminal B | 9 | 22.5% | 2 | (14.3) | 7 | (26.9) | 2 | (11.8) | 7 | (30.4) | ||
| Basal-like | 2 | 5.0% | 1 | (7.1) | 1 | (3.8) | 2 | (11.8) | 0 | (0.0) | ||
| Her2 | 1 | 2.5% | 1 | (7.1) | 0 | (0.0) | 0 | (0.0) | 1 | (4.3) | ||
| Others | 8 | 20.0% | 1 | (7.1) | 7 | (26.9) | 2 | (11.8) | 6 | (26.1) | ||
| No available data | 7 | 17.5% | 2 | (14.3) | 5 | (19.2) | 2 | (11.8) | 5 | (21.7) | ||
∗Chi-square exact test is statistically significant at 95% confidence level.
Table 5.
Association of 17βHSD-1 promoter methylation with mitosis, immunohistochemistry, and molecular subtype of breast cancer.
| Overall (N = 40) | % | 17βHSD-1 methylation in normal tissues | 17βHSD-1 methylation in cancer tissues | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Methylated (38) | Unmethylated (2) | p value | Methylated (39) | Unmethylated (1) | p value | |||||||
| Freq. | (%) | Freq. | (%) | Freq. | (%) | Freq. | (%) | |||||
| Mitosis: | ||||||||||||
| 0 | 4 | 10.0% | 4 | (10.5) | 0 | (0.0) | 0.850 | 4 | (10.3) | 0 | (0.0) | 1.000 |
| 1 | 5 | 12.5% | 5 | (13.2) | 0 | (0.0) | 5 | (12.8) | 0 | (0.0) | ||
| 2 | 29 | 72.5% | 27 | (71.1) | 2 | (100.0) | 28 | (71.8) | 1 | (100.0) | ||
| 3 | 2 | 5.0% | 2 | (5.3) | 0 | (0.0) | 2 | (5.1) | 0 | (0.0) | ||
| Estrogen receptor: | ||||||||||||
| Positive | 27 | 67.5% | 25 | (65.8) | 2 | (100.0) | 0.602 | 26 | (66.7) | 1 | (100.0) | 1.000 |
| Negative | 7 | 17.5% | 7 | (18.4) | 0 | (0.0) | 7 | (17.9) | 0 | (0.0) | ||
| No available data | 6 | 15.0% | 6 | (15.8) | 0 | (0.0) | 6 | (15.4) | 0 | (0.0) | ||
| Progesterone receptor: | ||||||||||||
| Positive | 25 | 62.5% | 23 | (60.5) | 2 | (100.0) | 0.532 | 25 | (64.1) | 0 | (0.0) | 0.375 |
| Negative | 9 | 22.5% | 9 | (23.7) | 0 | (0.0) | 8 | (20.5) | 1 | (100.0) | ||
| No available data | 6 | 15.0% | 6 | (15.8) | 0 | (0.0) | 6 | (15.4) | 0 | (0.0) | ||
| Her2: | ||||||||||||
| Positive | 10 | 25.0% | 10 | (26.3) | 0 | (0.0) | 0.312 | 10 | (25.6) | 0 | (0.0) | 1.000 |
| Negative | 19 | 47.5% | 17 | (44.7) | 2 | (100.0) | 18 | (46.2) | 1 | (100.0) | ||
| No available data | 11 | 27.5% | 11 | (29.0) | 0 | (0.0) | 11 | (28.2) | 0 | (0.0) | ||
| Molecular subtype: | ||||||||||||
| Luminal A | 13 | 32.5% | 11 | (28.9) | 2 | (100.0) | 0.120 | 13 | (32.5) | 0 | (0.0) | 0.174 |
| Luminal B | 9 | 22.5% | 9 | (23.7) | 0 | (0.0) | 9 | (22.5) | 0 | (0.0) | ||
| Basal-like | 2 | 5.0% | 2 | (5.3) | 0 | (0.0) | 2 | (5.1) | 0 | (0.0) | ||
| Her2 | 1 | 2.5% | 1 | (2.6) | 0 | (0.0) | 1 | (2.6) | 0 | (0.0) | ||
| Others | 8 | 20.0% | 8 | (21.1) | 0 | (0.0) | 7 | (17.9) | 1 | (100.0) | ||
| No available data | 7 | 17.5% | 7 | (18.4) | 0 | (0.0) | 7 | (17.9) | 0 | (0.0) | ||
Chi-square exact test is statistically significant at 95% confidence level.
3.3. The Concordant BRCA1 and 17βHSD-1 Promoter Methylation in Cancer and Normal Tissue Specimens
In addition, the concordant promoter methylation of the two studied genes was also investigated (Tables 6 and 7). BRCA1 promoter was methylated in both normal and cancer breast tissue specimens of 11 patients which was statistically significant (p < 0.001), while 20 cases showed combined unmethylation of BRCA1 promoter (Table 6).
Table 6.
Relation between methylation status of BRCA1 promoter in cancer tissue specimens and normal tissue specimens in 40 sporadic breast cancer patients.
| BRCA1 cancer tissues | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Methylated | Unmethylated | Total | p value | OR (95% CI) | |||||
| Freq. | (%) | Freq. | (%) | Freq. | % | ||||
|
BRCA1
normal tissues |
Methylated | 11 | (78.6) | 3 | (21.4) | 14 | 100.0 | 0.001∗ | 12.2 (2.5–58.7) |
| Unmethylated | 6 | (23.1) | 20 | (76.9) | 26 | 100.0 | |||
| Total | 17 | (42.5) | 23 | (57.5) | 40 | 100.0 | |||
∗Chi-square test is statistically significant at 95% confidence level.
Table 7.
Relation between methylation status of 17βHSD-1 promoter in cancer tissue specimens and normal tissue specimens in 40 sporadic breast cancer patients.
| 17βHSD-1 cancer tissues | ||||||||
|---|---|---|---|---|---|---|---|---|
| Methylated | Unmethylated | Total | p value | |||||
| Freq. | (%) | Freq. | (%) | Freq. | % | |||
|
17βHSD-1
normal tissues |
Methylated | 37 | (97.4) | 1 | (2.6) | 38 | 100.0 | 1.000 |
| Unmethylated | 2 | (100.0) | 0 | (0.0) | 2 | 100.0 | ||
| Total | 39 | (97.5) | 1 | (2.5) | 40 | 100.0 | ||
Chi-square test is statistically significant at 95% confidence level.
In the case of a 17βHSD-1 promoter, 37 tissue specimens had methylated normal and cancer tissues, while none of the studied tissues showed combined unmethylation of this gene promoter (Table 7).
3.4. Combined BRCA1 and 17βHSD-1 Promoter Methylation in Both Study Groups
Among the studied samples, 45% of cancer tissues and 35% of normal tissues had combined BRCA1 and 17βHSD-1 promoter methylation (Table 8). However, there was no significant relation between combined BRCA1 and 17βHSD-1 promoter methylation and type of tissue. The odds ratio = 1.5; 95% confidence interval = 0.6–3.7 and p = 0.361.
Table 8.
Relation between combined BRCA1 and 17βHSD-1 promoter methylation and the type of breast tissues of 40 sporadic breast cancer patients.
| Combined BRCA1 and 17βHSD-1 promoter methylation | ||||||||
|---|---|---|---|---|---|---|---|---|
| Overall (N = 40) | Methylated | Unmethylated | p value | OR (95% CI) | ||||
| Freq. | % | Freq. | (%) | Freq. | (%) | |||
| Type of tissue | ||||||||
| Cancer tissues | 40 | 100.0% | 18 | (45.0) | 22 | (55.0) | 0.361 | 1.5 (0.6–3.7) |
| Normal tissues | 40 | 100.0% | 14 | (35.0) | 26 | (65.0) | ||
| Total | 80 | 100.0% | 32 | (40.0) | 48 | (60.0) | ||
Chi-square test is statistically significant at 95% confidence level.
4. Discussion
BRCA1 promoter hypermethylation is implicated as one of the mechanisms of loss of gene expression [33]. It was identified in 9–44% of sporadic breast cancers [19–21]. The mitogenic effect of estradiol on breast epithelium is counteracted by the upregulation of BRCA1 expression, which in turn exerts a negative feedback effect on estradiol action by direct interaction with estrogen receptor [30]. Hence, transcriptional inactivation of BRCA1 due to methylation may fail to decrease estradiol activity resulting in increased breast tissue proliferation. Despite the ample studies on the role of BRCA1 in BC and its epigenetic modification, nevertheless, its association with the estrogen level regulating gene 17βHSD-1 has not been conducted. In this study, we investigated the methylation status of BRCA1 and 17βHSD-1 in sporadic breast cancer Egyptian patients and correlated the findings to those in normal breast tissues.
BRCA1 methylation percentage in our study is near to the upper end of previously reported frequencies for this alteration in sporadic breast cancer [19–21]. Hsu et al. data had hypermethylation of the BRCA1 in 56% (78 of 139) of Taiwanese women with early-stage sporadic breast carcinomas, which is significantly higher than previously reported frequencies for this alteration in sporadic breast tumors [34]. The incidence of BRCA1 methylation has previously been reported to be higher in breast tumors of infiltrating ductal type suggesting that it might play a role in breast carcinogenesis [19]. In other studies, BRCA1 existed in even lower percentages, 9–32%, in sporadic breast cancer [33]. This variation may be due to several factors: first factor is the methylation assay or the analysis method used [35], bisulfite method [15, 19, 36, 37], and genomic sequencing [22, 38, 39]; secondly, MSP detects differential methylation status by amplification of bisulfite-treated DNA with primers specific for methylated versus unmethylated DNA [40]. CpG sites residing within the primer sets were used as a proxy for the methylation status of the region of interest. Although most published studies mentioned above [15, 19, 36, 37] used MSP, the primer sequences and target regions varied from study to study. Finally, contaminating unmethylated normal tissue may occur during tissue dissection that might attenuate the methylation levels of the tumor tissue.
A small population of apparently normal breast epithelial cells could harbor BRCA1 promoter methylation in patients' samples with BRCA1-methylated tumors [41, 42], but not in those with BRCA1-unmethylated tumors. BRCA1 promoter was methylated in both normal and cancer breast tissue specimens of 11 patients which was statistically significant (p < 0.001), while 20 cases showed combined unmethylation of BRCA1 promoter. In the case of 17βHSD-1 promoter, 37 tissue specimens had methylated normal and cancer tissues, while none of the studied tissues showed combined unmethylation of this gene promoter.
BRCA1 methylation was detected in 76.5% of cancer tissues with positive ER and 64.7% with positive PR; on the other hand, methylation was significantly (p = 0.007) higher among tissues with negative Her2 (68.4%) than those with positive Her2, while 17βHSD-1 promoter methylation was found in cancer tissue specimens in women who had positive ER (66.7%), positive PR (64.1%), and negative Her2 (46.2%).
Although we could not have complete data for the ER, PR, and Her2 receptor staining status for the entire studied sample size, our data showed that “luminal A” molecular subtype, defined as being positive for both ER and PR but negative for Her2 receptor, had the highest level of promoter methylation. In particular, 17βHSD-1 showed the highest association with “luminal A” subtype. Similarly, methylation of BRCA1 promoter correlated significantly with “luminal A” subtype and was evenly distributed among other molecular subtypes. Our finding that 17βHSD-1 is associated with the luminal subtypes A and B is surprising since it has been shown that this enzyme activity correlates with the positivity of both ER and PR [28]. Methylation of 17βHSD-1 in both normal and cancer tissue specimens directs the attentions towards saving those patients from the long term use of adjuvant antiestrogen therapies. Although our results indicated that BRCA1 promoter methylation did not correlate significantly with triple-negative breast cancer (p = 0.174), Bal et al. showed that BRCA1 promoter methylation correlated with decreased expression of ER and basal-like phenotype [33]. More than half of the patients with the BRCA1 mutation had triple-negative breast cancer, and they also shared common clinical and pathological features [43]. However, a significant portion of triple-negative breast cancer patients does not carry BRCA1 mutations. In studies of US and British women, triple-negative/basal-like tumors appeared to be more common among black women (especially before menopause) compared to white women [44, 45].
Theoretically, unmethylated/hypomethylated 17βHSD-1 should increase the levels of active estradiol and the risk of BC. Contrary to this, we observed that methylation of 17βHSD-1 was seen in 97.5% of tumor tissue compared to 95% of neighboring normal tissue. Some genes are known to alter their methylation status with age and these are usually tissue-specific [33]. Our results showed that the mean age of sporadic breast cancer patients with 17βHSD-1 promoters methylation group (56.0 ± 9.3 years) was slightly higher than unmethylated group (49.0 years), although the relation between age and 17βHSD-1 methylation was statistically insignificant. The present study, therefore, indicates that the methylation status of 17βHSD-1 may be age-specific; alteration of this methylation pattern of the 17βHSD-1 gene in breast tissue may play an important role in BC progression.
Our preliminary results presented here only demonstrate the association of BRCA1 promoter methylation between tumors and normal breast tissues. Direct evidence shows that progression from BRCA1-methylated normal breast epithelial cells to BRCA1-methylated breast cancer needs to be investigated in future studies. The high methylation of 17βHSD-1 promoter in both normal and cancer breast tissues rules out the biological significance of this epigenetic modification in distinguishing normal and cancer tissues. Given the fact that breast tumors express high heterogeneity, one limitation in explaining this finding is the small sample size of our studied population. Suzuki et al. showed that 17βHSD-1 negative breast tissues are less differentiated; hence, they escape normal regulation of proliferation; thus, it is possible that the reduced expression of this enzyme via hypermethylation in normal tissue reflects an increased carcinogenic potential in normal breast tissues harboring a nearby cancer tumor [28]. In addition, based on the fact that the activity of both 17βHSD-2 and 17βHSD-1 controls the in situ level of breast estrogen, it is possible that 17βHSD-2 activity is dominating the control of estrogen level in the 17βHSD-1 hypermethylated tumors and it would be beneficial to evaluate the methylation status of this gene especially in breast cancer hypermethylated 17βHSD-1 tissues [46]. Furthermore, we found that methylation of 17βHSD-1 in both normal and cancer tissue specimens may direct the attentions towards saving those patients from the long term use of adjuvant antiestrogen therapies, for example, tamoxifen, especially in ER+. Increased understanding of the genetic/epigenetic abnormality in the pathogenesis of breast cancer is crucial and may provide a basis for detection and treatment. This study highlights the frequent promoter methylation of BRCA1 and its prognostic significance, irrespective of BRCA1 gene mutation in Egyptian patients with early-stage breast cancer.
In conclusion, we showed that a significant proportion of patients with BRCA1-methylated tumors harbored BRCA1 promoter methylation in normal breast tissues and that 17βHSD-1 methylation was observed in the normal tissues of the 17βHSD-1 promoter methylation status of the tumors. This suggests a possibility that a small proportion of the epithelial cells with BRCA1 promoter methylation can be precursor cells from which BRCA1-methylated breast tumors originate. Although our preliminary results presented here need to be validated by future studies, they may provide further insight into the different roles of promoter methylation of these genes in breast carcinogenesis.
Acknowledgments
The authors are indebted to the participants for their utmost cooperation.
Competing Interests
The authors declare that they have no competing interests.
References
- 1.Sainsbury J. R. C., Anderson T. J., Morgan D. A. L. ABC of breast diseases: breast cancer. British Medical Journal. 2000;321(7263):745–750. doi: 10.1136/bmj.321.7263.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gudjonsson T., Adriance M. C., Sternlicht M. D., Petersen O. W., Bissell M. J. Myoepithelial cells: their origin and function in breast morphogenesis and neoplasia. Journal of Mammary Gland Biology and Neoplasia. 2005;10(3):261–272. doi: 10.1007/s10911-005-9586-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeSantis C. E., Lin C. C., Mariotto A. B., et al. Cancer treatment and survivorship statistics, 2014. CA: A Cancer Journal for Clinicians. 2014;64(4):252–271. doi: 10.3322/caac.21235. [DOI] [PubMed] [Google Scholar]
- 4.Torre L. A., Bray F., Siegel R. L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics, 2012. CA: Cancer Journal for Clinicians. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 5.Ibrahim A. S., Khaled H. M., Mikhail N. N., Baraka H., Kamel H. Cancer incidence in Egypt: results of the national population-based cancer registry program. Journal of Cancer Epidemiology. 2014;2014:18. doi: 10.1155/2014/437971.437971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sadikovic B., Al-Romaih K., Squire J. A., Zielenska M. Cause and consequences of genetic and epigenetic alterations in human cancer. Current Genomics. 2008;9(6):394–408. doi: 10.2174/138920208785699580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brena R. M., Costello J. F. Genome—epigenome interactions in cancer. Human Molecular Genetics. 2007;16(1):R96–R105. doi: 10.1093/hmg/ddm073. [DOI] [PubMed] [Google Scholar]
- 8.Esteller M. CpG island hypermethylation and tumor suppressor genes: a booming present, a brighter future. Oncogene. 2002;21(35):5427–5440. doi: 10.1038/sj.onc.1205600. [DOI] [PubMed] [Google Scholar]
- 9.Baylin S. B., Herman J. G. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends in Genetics. 2000;16(4):168–174. doi: 10.1016/s0168-9525(99)01971-x. [DOI] [PubMed] [Google Scholar]
- 10.Eden S., Cedar H. Role of DNA methylation in the regulation of transcription. Current Opinion in Genetics and Development. 1994;4(2):255–259. doi: 10.1016/s0959-437x(05)80052-8. [DOI] [PubMed] [Google Scholar]
- 11.Baylin S. B., Ohm J. E. Epigenetic gene silencing in cancer—a mechanism for early oncogenic pathway addiction? Nature Reviews Cancer. 2006;6(2):107–116. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- 12.Widschwendter M., Apostolidou S., Raum E., et al. Epigenotyping in peripheral blood cell DNA and breast cancer risk: a proof of principle study. PLoS ONE. 2008;3(7) doi: 10.1371/journal.pone.0002656.e2656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esteller M., Corn P. G., Baylin S. B., Herman J. G. A gene hypermethylation profile of human cancer. Cancer Research. 2001;61(8):3225–3229. [PubMed] [Google Scholar]
- 14.Suijkerbuijk K. P., Fackler M. J., Sukumar S., et al. Methylation is less abundant in BRCA1-associated compared with sporadic breast cancer. Annals of Oncology. 2008;19(11):1870–1874. doi: 10.1093/annonc/mdn40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Catteau A., Morris J. R. BRCA1 methylation: a significant role in tumour development? Seminars in Cancer Biology. 2002;12(5):359–371. doi: 10.1016/s1044-579x(02)00056-1. [DOI] [PubMed] [Google Scholar]
- 16.Karray-Chouayekh S., Trifa F., Khabir A., et al. Clinical significance of epigenetic inactivation of hMLH1 and BRCA1 in tunisian patients with invasive breast carcinoma. Journal of Biomedicine and Biotechnology. 2009;2009:7. doi: 10.1155/2009/369129.369129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andrieu N., Goldgar D. E., Easton D. F., et al. Pregnancies, breast-feeding, and breast cancer risk in the International BRCA1/2 Carrier Cohort Study (IBCCS) Journal of the National Cancer Institute. 2006;98(8):535–544. doi: 10.1093/jnci/djj132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rice J. C., Massey-Brown K. S., Futscher B. W. Aberrant methylation of the BRCA1 CpG island promoter is associated with decreased BRCA1 mRNA in sporadic breast cancer cells. Oncogene. 1998;17(14):1807–1812. doi: 10.1038/sj.onc.1202086. [DOI] [PubMed] [Google Scholar]
- 19.Birgisdottir V., Stefansson O. A., Bodvarsdottir S. K., Hilmarsdottir H., Jonasson J. G., Eyfjord J. E. Epigenetic silencing and deletion of the BRCA1 gene in sporadic breast cancer. Breast Cancer Research. 2006;8(4, article R38) doi: 10.1186/bcr1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Al-Moghrabi N., Al-Qasem A. J. S., Aboussekhra A. Methylation-related mutations in the BRCA1 promoter in peripheral blood cells from cancer-free women. International Journal of Oncology. 2011;39(1):129–135. doi: 10.3892/ijo.2011.1021. [DOI] [PubMed] [Google Scholar]
- 21.Butcher D. T., Rodenhiser D. I. Epigenetic inactivation of BRCA1 is associated with aberrant expression of CTCF and DNA methyltransferase (DNMT3B) in some sporadic breast tumours. European Journal of Cancer. 2007;43(1):210–219. doi: 10.1016/j.ejca.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 22.Ali A. B., Iau P. T. C., Sng J.-H. Cancer-specific methylation in the BRCA1 promoter in sporadic breast tumours. Medical Oncology. 2011;28(1):64–66. doi: 10.1007/s12032-010-9438-y. [DOI] [PubMed] [Google Scholar]
- 23.Dumitrescu R. G., Marian C., Krishnan S. S., et al. Familial and racial determinants of tumor suppressor genes promoter hypermethylation in breast tissues from healthy women. Journal of Cellular and Molecular Medicine. 2010;14(6):1468–1475. doi: 10.1111/j.1582-4934.2009.00924.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sasano H., Suzuki T., Takeyama J., et al. 17-Beta-hydroxysteroid dehydrogenase in human breast and endometrial carcinoma. A new development in intracrinology. Oncology. 2000;59(1):5–12. doi: 10.1159/000055281. [DOI] [PubMed] [Google Scholar]
- 25.Sasano H., Suzuki T., Nakata T., Moriya T. New development in intracrinology of breast carcinoma. Breast Cancer. 2006;13(2):129–136. doi: 10.2325/jbcs.13.129. [DOI] [PubMed] [Google Scholar]
- 26.Kauraniemi P., Bärlund M., Monni O., Kallioniemi A. New amplified and highly expressed genes discovered in the ERBB2 amplicon in breast cancer by cDNA microarrays. Cancer Research. 2001;61(22):8235–8240. [PubMed] [Google Scholar]
- 27.Gunnarsson C., Ahnström M., Kirschner K., et al. Amplification of HSD17B1 and ERBB2 in primary breast cancer. Oncogene. 2003;22(1):34–40. doi: 10.1038/sj.onc.1206078. [DOI] [PubMed] [Google Scholar]
- 28.Suzuki T., Moriya T., Ariga N., Kaneko C., Kanazawa M., Sasano H. 17β-hydroxysteroid dehydrogenase type 1 and type 2 in human breast carcinoma: a correlation to clinicopathological parameters. British Journal of Cancer. 2000;82(3):518–523. doi: 10.1054/bjoc.1999.0956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cleton-Jansen A.-M., Callen D. F., Seshadri R., et al. Loss of heterozygosity mapping at chromosome arm 16q in 712 breast tumors reveals factors that influence delineation of candidate regions. Cancer Research. 2001;61(3):1171–1177. [PubMed] [Google Scholar]
- 30.Magdinier F., Billard L.-M., Wittmann G., et al. Regional methylation of the 5′ end CpG island of BRCA1 is associated with reduced gene expression in human somatic cells. The FASEB Journal. 2000;14(11):1585–1594. doi: 10.1096/fj.14.11.1585. [DOI] [PubMed] [Google Scholar]
- 31.Horan M. P. eLS. New York, NY, USA: John Wiley & Sons; 2011. Methylation-mediated transcriptional silencing in tumorigenesis. [Google Scholar]
- 32.Bhavani V., Srinivasulu M., Ahuja Y. R., Hasan Q. Role of BRCA1, HSD17B1 and HSD17B2 methylation in breast cancer tissue. Cancer Biomarkers. 2009;5(4-5):207–213. doi: 10.3233/CBM-2009-0105. [DOI] [PubMed] [Google Scholar]
- 33.Bal A., Verma S., Joshi K., et al. BRCA1-methylated sporadic breast cancers are BRCA-like in showing a basal phenotype and absence of ER expression. Virchows Archiv. 2012;461(3):305–312. doi: 10.1007/s00428-012-1286-z. [DOI] [PubMed] [Google Scholar]
- 34.Hsu N. C., Huang Y. F., Yokoyama K. K., Chu P. Y., Chen F. M., Hou M. F. Methylation of BRCA1 promoter region is associated with unfavorable prognosis in women with early-stage breast cancer. PLoS ONE. 2013;8(2):p. 6. doi: 10.1371/journal.pone.0056256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bianco T., Chenevix-Trench G., Walsh D. C. A., Cooper J. E., Dobrovic A. Tumour-specific distribution of BRCA1 promoter region methylation supports a pathogenetic role in breast and ovarian cancer. Carcinogenesis. 2000;21(2):147–151. doi: 10.1093/carcin/21.2.147. [DOI] [PubMed] [Google Scholar]
- 36.Esteller M., Silva J. M., Dominguez G., et al. Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. Journal of the National Cancer Institute. 2000;92(7):564–569. doi: 10.1093/jnci/92.7.564. [DOI] [PubMed] [Google Scholar]
- 37.Mirza S., Sharma G., Prasad C. P., et al. Promoter hypermethylation of TMS1, BRCA1, ERα and PRB in serum and tumor DNA of invasive ductal breast carcinoma patients. Life Sciences. 2007;81(4):280–287. doi: 10.1016/j.lfs.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 38.Mancini D. N., Rodenhiser D. I., Ainsworth P. J., et al. CpG methylation within the 5' regulatory region of the BRCA1 gene is tumor specific and includes a putative CREB binding site. Oncogene. 1998;16(9):1161–1169. doi: 10.1038/sj.onc.1201630. [DOI] [PubMed] [Google Scholar]
- 39.Cai F., Ge I., Wang M., Biskup E., Lin X., Zhong X. Pyrosequencing analysis of BRCA1 methylation level in breast cancer cells. Tumor Biology. 2014;35(4):3839–3844. doi: 10.1007/s13277-013-1508-2. [DOI] [PubMed] [Google Scholar]
- 40.Herman J. G., Graff J. R., Myöhänen S., Nelkin B. D., Baylin S. B. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(18):9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Snell C., Krypuy M., Wong E. M., Loughrey M. B., Dobrovic A. BRCA1 promoter methylation in peripheral blood DNA of mutation negative familial breast cancer patients with a BRCA1 tumour phenotype. Breast Cancer Research. 2008;10(1, article R12) doi: 10.1186/bcr1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Otani Y., Miyake T., Kagara N., et al. BRCA1 promoter methylation of normal breast epithelial cells as a possible precursor for BRCA1-methylated breast cancer. Cancer Science. 2014;105(10):1369–1376. doi: 10.1111/cas.12506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rennert G., Bisland-Naggan S., Barnett-Griness O., et al. Clinical outcomes of breast cancer in carriers of BRCA1 and BRCA2 mutations. New England Journal of Medicine. 2007;357(2):115–123. doi: 10.1056/NEJMoa070608. [DOI] [PubMed] [Google Scholar]
- 44.Sineshaw H. M., Gaudet M., Ward E. M., et al. Association of race/ethnicity, socioeconomic status, and breast cancer subtypes in the National Cancer Data Base (2010-2011) Breast Cancer Research and Treatment. 2014;145(3):753–763. doi: 10.1007/s10549-014-2976-9. [DOI] [PubMed] [Google Scholar]
- 45.Howlader N., Altekruse S. F., Li C. I., et al. US incidence of breast cancer subtypes defined by joint hormone receptor and HER2 status. Journal of the National Cancer Institute. 2014;106(5) doi: 10.1093/jnci/dju055.dju055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gunnarsson C., Hellqvist E., Stål O. 17β-hydroxysteroid dehydrogenases involved in local oestrogen synthesis have prognostic significance in breast cancer. British Journal of Cancer. 2005;92(3):547–552. doi: 10.1038/sj.bjc.6602375. [DOI] [PMC free article] [PubMed] [Google Scholar]


