Abstract
The nuclear THO and TREX-2 complexes are implicated in several steps of nuclear mRNP biogenesis, including transcription, 3′ end processing and export. In a recent genomic microscopy screen in Saccharomyces cerevisiae for mutants with constitutive stress granules, we identified that absence of THO and TREX-2 complex subunits leads to the accumulation of Pab1-GFP in cytoplasmic foci. We now show that these THO/TREX-2 mutant induced foci (“TT foci”) are not stress granules but instead are a mRNP granule containing poly(A)+ mRNA, some mRNP components also found in stress granules, as well several proteins involved in mRNA 3′ end processing and export not normally seen in stress granules. In addition, TT foci are resistant to cycloheximide-induced disassembly, suggesting the presence of mRNPs impaired for entry into translation. THO mutants also exhibit defects in normal stress granule assembly. Finally, our data also suggest that TT foci are targeted by autophagy. These observations argue that defects in nuclear THO and TREX-2 complexes can affect cytoplasmic mRNP function by producing aberrant mRNPs that are exported to cytosol, where they accumulate in TT foci and ultimately can be cleared by autophagy. This identifies a novel mechanism of quality control for aberrant mRNPs assembled in the nucleus.
Keywords: stress granules, THO complex, TREX-2 complex, mRNA, translation
INTRODUCTION
The biogenesis of mRNAs involves a series of nuclear processing and mRNA-protein (mRNP) remodeling events. Nascent transcripts must be 5′ capped, spliced, and terminated at the correct site, 3′ end processed and ultimately packaged into an mRNP capable of export to the cytoplasm. These events are tightly coordinated and subject to quality control pathways that recognize and degrade transcripts that have been improperly processed at any of these steps (Fasken and Corbett 2009; Moore and Proudfoot 2009; Porrua and Libri 2013; Schmid and Jensen 2013).
The nuclear THO complex connects and facilitates many of these processes, including transcription elongation and termination (Chávez et al. 2000; Huertas and Aguilera 2003; Rondón et al. 2003; Mason and Struhl 2005), 3′ end processing (Libri et al. 2002), and recruitment of export factors to nascent mRNA (Strässer et al. 2002). In S. cerevisiae, it is composed of four strongly interacting, but nonessential subunits (Mft1, Hpr1, Tho2, Thp2), two of which (Hpr1, Tho2) are conserved from yeast to humans. Tex1 has also been identified as a putative fifth subunit of this complex (Gewartowski et al. 2012).
THO mutants also exhibit defective 3′ end formation and polyadenylation. Studies of induced HSP104 expression indicate premature 3′ end truncation (Libri et al. 2002). Polyadenylation also appears impaired in THO mutants, which results from destabilization of Fip1, a cofactor necessary for full Pap1 poly(A) polymerase activity (Saguez et al. 2008). mRNAs such as HSP104 are then rapidly degraded in THO mutants by the nuclear exosome (Libri et al. 2002), with defective polyadenylation and/or 3′ end processing as reasons proposed to account for this effect (Saguez et al. 2008). However, a small population of THO-regulated mRNAs can escape such turnover, forming stalled mRNP complexes (termed “heavy chromatin”) at the 3′ end of genes that contain chromatin, polyadenylation factors, export factors, and nuclear pore components (Libri et al. 2002; Rougemaille et al. 2008; Qu et al. 2009). Indeed a low-abundance population of hyperadenylated transcripts is visible in THO mutants, which may result from sequestration of cleavage and polyadenylation factors on these stalled mRNPs (Qu et al. 2009). Thus, exosome-mediated decay and nuclear sequestration are two ways in which mRNAs misprocessed in THO mutants are prevented from exporting to the cytoplasm (Rougemaille et al. 2007).
THO mutants also exhibit mRNA export defects. This is suggested by the stalled export-factor containing mRNPs that accumulate at 3′ gene ends (see above), and by nuclear poly(A)+ accumulation in yeast (Strässer et al. 2002), Drosophila (Rehwinkel et al. 2004) and human cells (Chi et al. 2013). Studies of specific transcripts have also revealed nuclear accumulation in yeast and human cells (Libri et al. 2002; Rougemaille et al. 2008; Katahira et al. 2009; Guria et al. 2011). The THO complex aids binding of export factors Sub2/UAP56 and Yra1/Aly1 to mRNA (Strässer et al. 2002; Zenklusen et al. 2002) to form the transcription-export (TREX) complex. Yra1 in turn recruits the primary mRNA export protein Mex67/TAP, which can also be directly recruited by Hpr1, apparently bypassing Sub2 and Yra1 (Gwizdek et al. 2006). The THO complex also facilitates loading of the Serine/Argine rich (SR) proteins Gbp2 and Hrb1 (Hurt et al. 2004), which are implicated in quality control of spliced mRNAs, and in mRNA export via Mex67 recruitment (Hackmann et al. 2014). Thus, the THO complex may facilitate mRNA export in many ways.
The global effects of the THO complex on mRNA transcription, processing and export are not yet clear, but studies in several systems suggest specific transcripts may depend more on the THO complex than others. For example, knockdown of THO components in Drosophila resulted in <20% of the transcriptome showing significantly altered mRNA levels (Rehwinkel et al. 2004); in similar studies in mice, the value was only 3% (Guria et al. 2011). In mft1Δ yeast, around 400 genes were identified as putative THO complex mRNA targets, based on their fractionation in heavy chromatin complexes (Rougemaille et al. 2008). In these and other studies, mRNAs encoding heat shock proteins were particularly affected in terms of levels and export, though at least in yeast, not all heat shock protein mRNAs rely on THO for export (Rollenhagen et al. 2007). Other studies have suggested that highly expressed, long or G/C rich genes may be particularly susceptible to THO-mediated regulation of transcription elongation (Gómez-González et al. 2011). Finally, Hpr1 sumolyation may be key to the induction of acidic stress-responsive genes (Bretes et al. 2014). Taken together, it appears that THO-TREX complexes are not essential for processing and export of all mRNAs, but likely facilitate these processes, particularly on subsets of mRNAs under various growth and stress conditions.
The TREX-2 complex, which in yeast consists of Sac3, Thp1, Sus1, and Cdc31 is also conserved and also functions in linking transcription and mRNA export. Unlike the THO complex, the TREX-2 complex is mostly localized at nuclear pores (Fischer et al. 2002; Rodríguez-Navarro et al. 2004; Umlauf et al. 2013), where it can recruit transcriptionally active genes, partly via interactions with the SAGA histone acetylase complex (Rodríguez-Navarro et al. 2004) to facilitate their rapid and efficient export via a process termed “gene gating.” TREX-2 mutants often share similar phenotypes to THO mutants, including defects in transcription elongation, elevated recombination (González-Aguilera et al. 2008; Tous et al. 2011), and mRNA export defects (Strässer et al. 2002; Rodríguez-Navarro et al. 2004), although distinct genetic interactions with Sub2 are observed (González-Aguilera et al. 2008). How the THO and TREX-2 complexes interact to promote assembly of export-competent mRNPs remains unclear, and to date, no alterations in cytoplasmic mRNA metabolism have been described due to the presence of THO or TREX-2 mutations.
Previously, we identified THO and TREX-2 mutants in a screen for Pab1-GFP forming cytoplasmic foci, with the goal of identifying mutants that accumulated constitutive stress granules (Buchan et al. 2013). Stress granules are conserved, dynamic cytoplasmic assemblies of nontranslating mRNPs that rapidly form during conditions of stress when translation initiation becomes limiting and are otherwise absent during normal growth. They contain mRNA, translation initiation factors and various mRNA binding proteins and are thought to represent a pool of mRNPs stalled in the process of translation initiation (Anderson and Kedersha 2008; Buchan and Parker 2009). Stress granules interact and share mRNA/protein components with a second related mRNP granule, called P-bodies, which harbor more mRNA decay factors. Prior studies have demonstrated that mRNAs can transit back and forth between translation and these granule states (Brengues et al. 2005; Kedersha et al. 2005; Bhattacharyya et al. 2006), arguing for the existence of a cytoplasmic mRNP cycle. However, how newly exported mRNAs enter and affect this cycle is poorly understood.
Stress granules are of interest not only because of effects on mRNAs, but also because of roles in stress signaling pathways (Arimoto et al. 2008; Eisinger-Mathason et al. 2008; Takahara and Maeda 2012; Thedieck et al. 2013), and because of connections to degenerative diseases. Indeed, amyotrophic lateral sclerosis (ALS), frontotemporal lobar degeneration (FTLD), fragile X syndrome, and spinocerebellar ataxia-2 can result from mutations in stress granule proteins, which increase their ability to aggregate (Nonhoff et al. 2007; Didiot et al. 2009; Ito and Suzuki 2011; Kim et al. 2013). Additionally, a hallmark of ALS, FTLD, and some other degenerative diseases is the accumulation of “inclusion bodies” that compositionally resemble stress granules (Ginsberg et al. 1998; Ito and Suzuki 2011; Dewey et al. 2012). This has led to the hypothesis that aberrant persistence of stress granules might underpin pathogenesis in these diseases due to altered regulation of mRNAs, cell signaling, or defects in other processes affected by stress granule assembly (Ramaswami et al. 2013).
Here, we further analyzed the Pab1-GFP foci in THO/TREX-2 mutants to determine whether defects in THO or TREX-2 complexes led to the accumulation of constitutive stress granules. We demonstrate that THO/TREX-2 mutant induced foci (referred to as “TT foci”) are a new type of mRNP granule containing poly(A)+ mRNA, some stress granule mRNP components, and proteins involved in mRNA 3′ end processing and export that are not normally seen in stress granules. Unlike stress granules, TT foci are also resistant to cycloheximide-induced disassembly, suggesting TT foci mRNPs exchange poorly with polysomes. Finally, our data suggest that TT foci are targeted by autophagy. These results suggest that a downstream consequence of defects in THO/TREX-2 function is the export of defective mRNPs, which accumulate in TT foci, and that autophagic clearance of mRNP granules may be a broad phenomenon.
RESULTS
THO and TREX-2 null strains exhibit constitutive foci exhibiting multiple stress granule components
In a recent microscopy screen to examine stress granule and P-body assembly in various viable gene deletion strains under good growth conditions, all screened members of the related THO (HPR1, MFT1, THP2, THO2) and TREX-2 complexes (SAC3, THP1) exhibited Pab1-GFP foci in a modest but reproducible population of cells (Buchan et al. 2013). Unlike normal stress granules, these Pab1-GFP foci did not associate with P-bodies (Buchan et al. 2013) raising the possibility that they were different from stress granules. Thus, we performed several experiments to determine if these assemblies were in fact stress granules, an unrelated protein aggregate, or a variant of stress granules with some different properties.
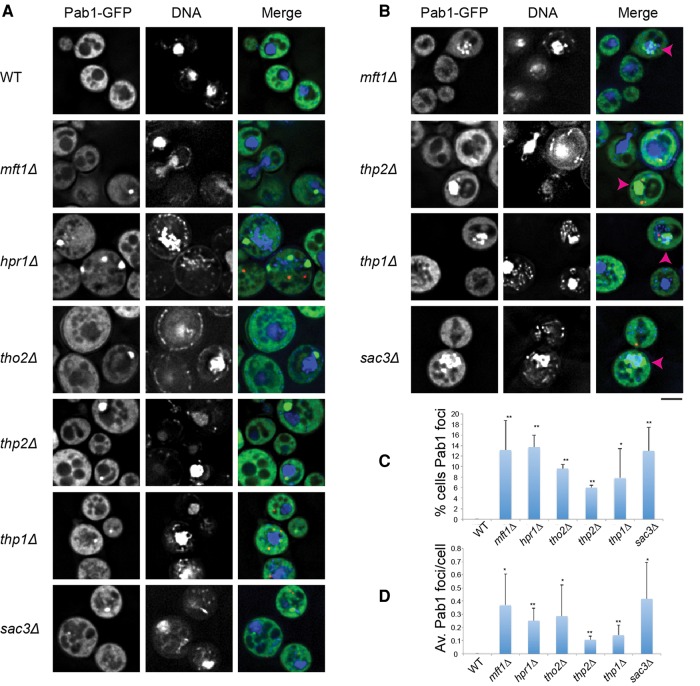
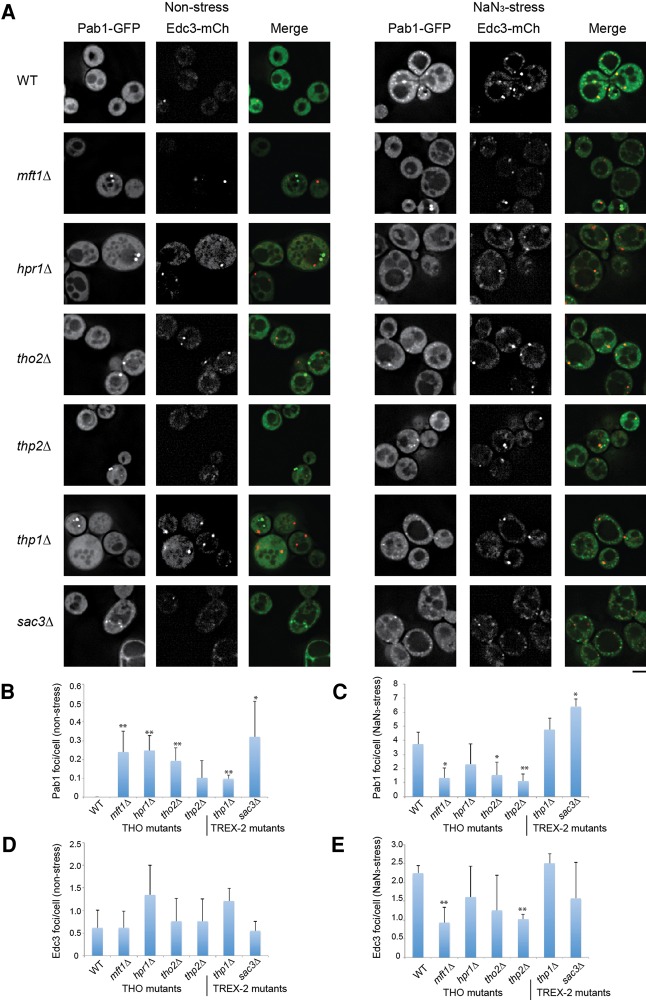
First, to better define the localization of all THO/TREX-2 foci, we examined Pab1 and Edc3 (a marker of P-bodies) localization in THO/TREX-2 null strains in the presence of Hoescht to stain DNA. As expected, most Pab1 foci were indeed cytoplasmic (Fig. 1A). We also observed occasional Pab1 aggregates that clearly occurred within the nucleus itself (Fig. 1B). Pab1 localization is thus altered in THO/TREX-2 null mutants, with cytoplasmic Pab1 foci the predominant novel feature.
FIGURE 1.
Deletion of THO and TREX-2 complex members leads to constitutive Pab1 foci under nonstress conditions. (A) Strains were transformed with pRP1657 expressing Pab1-GFP (stress granule marker) and Edc3-mCh (P-body marker; shown only in merges due to space constraints), grown to mid-log, incubated with hoescht DNA stain (10 µg/mL for 30 min), and examined. No difference in foci behavior was observed ± hoescht. (B) Examples of nuclear accumulation of Pab1, highlighted by pink arrowheads. Scale bar = 2.5 µM. (C) Average number of cells with Pab1-GFP (“TT”) foci. Data from three biological replicates (separate culture and transformant) with mean ± SD shown. Paired one-tailed Student's t-tests were conducted to assess significance of mutant data relative to WT; (*) P < 0.05, (**) P < 0.01. (D) Average number of Pab1-GFP (“TT”) foci/cell. Data from three biological replicates (separate cultures and transformant) with mean ± SD shown. Paired one-tailed Student's t-tests were conducted to assess significance of mutant data relative to WT; (*) P < 0.05, (**) P < 0.01
Although Pab1 is considered a core stress granule marker we determined if other core components of stress granules also assembled in TT foci. Using Ded1-mCh, Pub1-mCh, and Pbp1-mCh as additional markers of stress granules, we observed strong colocalization of Pab1-GFP foci with Pub1-mCh foci (90%–100%), Ded1-mCh foci (74.5%–100%), and slightly less colocalization with Pbp1-GFP (54%–100%, except for ∼29% in the hpr1Δ strain (Fig. 2; Supplemental Table S1). This suggests that TT foci do share some components with stress granules, but can vary in composition.
FIGURE 2.
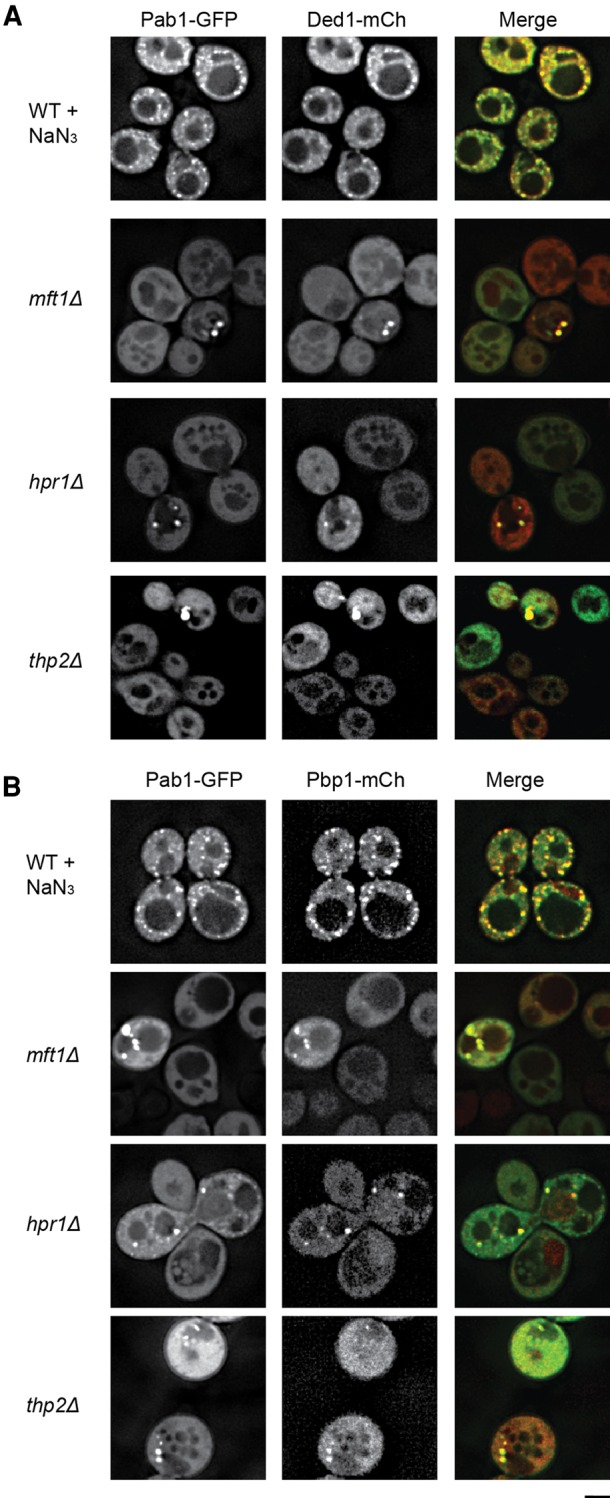
Constitutive Pab1 foci in THO and TREX-2 deletion strains exhibit compositional similarities to stress granules. (A) Strains were transformed with pRP2132 (Ded1-mCh) and pRP2478 (Pab1-GFP) and examined in mid-log growth; WT strains were subject to 30 min of 0.5% w/v NaN3 stress to induce stress granules as a positive control. Data representative of observations in two biological replicates (separate culture and transformant). (B) Strains were transformed with pRP1920 (Pbp1-mCh) and pRP2478 (Pab1-GFP) and examined as in A. Data representative of observations in two biological replicate experiments. Scale bar = 2.5 µM. Colocalization frequencies summarized in Supplemental Table S2.
Hpr1Δ Pab1-GFP foci contain several nuclear factors
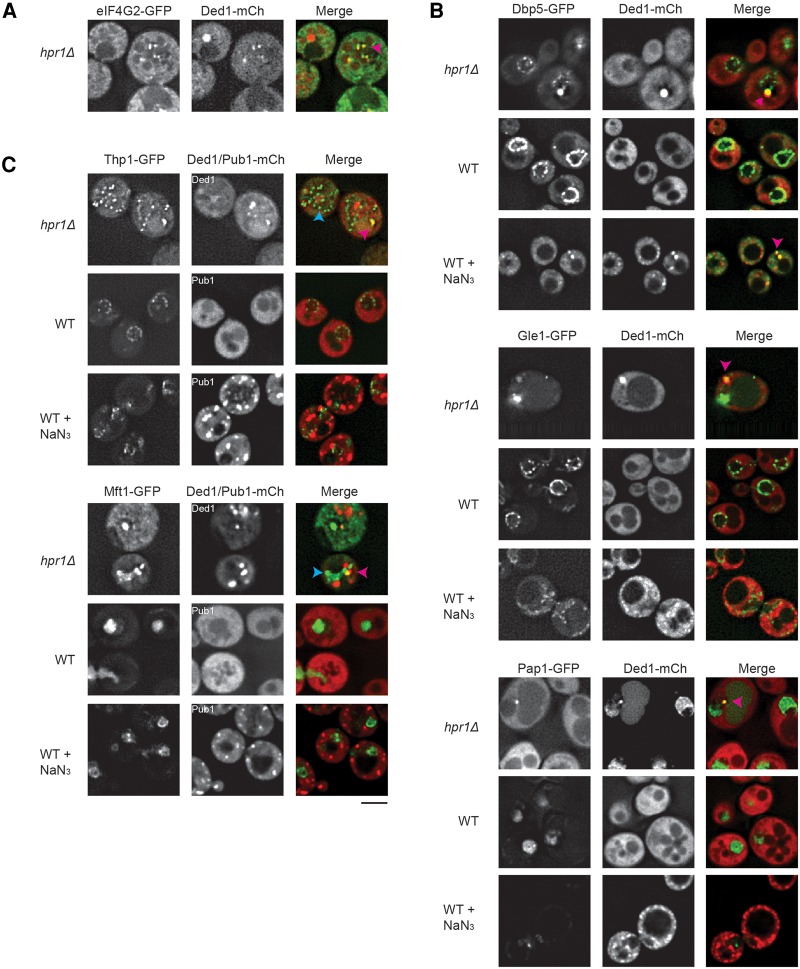
Since THO and TREX-2 complexes act at multiple stages of mRNP biogenesis and export, we hypothesized that the TT foci might consist of aberrant mRNPs produced in the THO or TREX-2 mutants that were exported to the cytosol but unable to enter translation. A prediction of this model is that proteins involved in 3′ end formation and export may be retained on the mRNA and accumulate in the TT foci. To test this possibility, the THO complex member HPR1 was knocked out of multiple yeast GFP strains, which feature a single ORF chromosomally tagged with GFP at the C-terminus. GFP strains were selected because of known roles in nuclear mRNP processing and export, including other THO and TREX-2 components. In addition, we used this approach to determine whether other components of stress granules were also in TT foci. For practical reasons, these strains were transformed with Ded1-mCh as an alternative marker of TT foci that form in the THO and TREX-2 mutants. Localization of each factor was also compared to that in WT cells under mid-log growth plus or minus stress (Fig. 3; Supplemental Table S2).
FIGURE 3.
Factors involved in export and 3′ end formation partially colocalize with hpr1Δ TT foci, or exhibit mislocalization in cells with such foci. (A) hpr1Δ eIF4G2-GFP strain transformed with pRP2132 (Ded1-mCh), examined under mid-log conditions. (B) hpr1Δ or WT GFP-tagged strains (Dbp5, Gle1, Pap1) were transformed with pRP2132 (Ded1-mCh) and examined under mid-log conditions ± NaN3 (30 min of 0.5% w/v). (C) As in B except Pub1-mCh (pRP2150) was utilized in place of Ded1-mCh in specific Mft1 or Thp1-GFP-tagged strains as indicated. In all hpr1Δ data sets above, pink arrowheads indicate colocalization of GFP-tagged proteins with Ded1-mCh; blue arrowheads indicate Ded1-distinct GFP foci. Data representative of observations in two biological replicates (separate culture and transformant). Scale bar = 2.5 µM. Colocalization frequencies summarized in Supplemental Table S2.
Several important observations were made in this hpr1Δ strain background. First, the initiation factor and stress granule component eIF4G2 formed cytoplasmic foci that showed strong overlap with Ded1 foci (Fig. 3A), showing that eIF4G2 is part of TT foci.
Second, Dbp5 (a DEAD-box helicase implicated in remodeling export mRNPs), Pap1 (poly(A) polymerase) and Gle1 (Dbp5 regulator) also formed cytoplasmic foci that showed partial overlap (68.5%, 58.3%, and 25.9%, respectively) with Ded1 foci (Fig. 3B; Supplemental Table S2). Interestingly, Dbp5 is observed in WT cell stress granules (Fig. 3) as well as in hpr1Δ Ded1-mCh foci, perhaps reflective of other documented roles in cytoplasmic function such as translation termination (Gross et al. 2007). Thus, Dbp5 is an additional components of TT foci shared with stress granules. In contrast, Pap1 and Gle1 are not observed in stress granules during NaN3 stress. Instead, Pap1 exhibits distinct relocalization to a single nuclear focus, whereas Gle1 becomes increasingly cytoplasmic in a diffuse manner. In summary, Pap1 and Gle1 are unique components of TT foci, highlighting their difference from stress granules.
A third interesting observation was that the THO subunit proteins Thp1 and Mft1 also formed cytoplasmic foci that showed weak overlap with Ded1 foci (19.3% and 9.6%, respectively), but did not localize to stress granules under stress conditions (Fig. 3C; Supplemental Table S2). The accumulation of Thp1 and Mft1 in at least some of TT foci seen in hpr1Δ strains provides additional evidence TT foci are distinct from stress granules.
In summary, there are clear differences in composition of stress granules versus hpr1Δ TT foci, including the unique localization of Pap1, Gle1 and to a lesser extent Thp1 and Mft1 in TT foci. These compositional differences, coupled with knowledge of THO-TREX-2 function, preliminarily suggest that THO/TREX-2 mutants may lead to accumulation in the cytoplasm of mRNPs defective in remodeling from nuclear and export processes.
TT foci contain mRNA
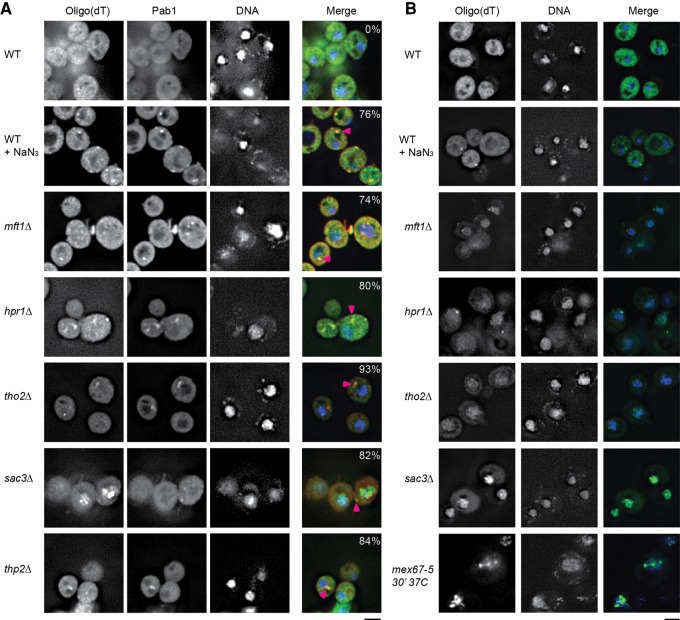
If TT foci represent pools of mRNPs they should include mRNAs. To determine this, we conducted FISH on THO and TREX-2 null strains and controls using an oligo(dT) probe to look for polyadenylated mRNA. In addition, we conducted immunofluorescence to probe for the localization of endogenous Pab1 foci in these strains. As controls we examined WT cells plus or minus NaN3 stress, which induces stress granules in yeast that were previously shown to harbor RNA with an RNA specific dye (RNAselect; data not shown). We also examined a MEX67 ts strain (mex67-5) as a FISH control, as this strain accumulates nuclear poly(A) signal at the nonpermissive temperature (Santos-Rosa et al. 1998).
The key observation was that in THO or TREX-2 null strains, Pab1 foci were observed which frequently colocalized with oligo(dT) (Fig. 4), suggesting that TT foci harbor mRNA, similar to stress granules, which also exhibited Pab1 and oligo(dT) foci overlap (Fig. 4A). As expected, WT cells fixed under normal growth conditions show mostly diffuse dT signal, albeit occasional small foci are visible; these could represent low levels of P-bodies or other structures where mRNA accumulate. Importantly, these WT cell foci showed no colocalization with Pab1 immunofluorescence, which remains diffuse under normal growth conditions (Fig. 4A). Strong nuclear accumulation of polyadenylated mRNA in the mex67-5 strain at the nonpermissive temperature was observed as expected (Fig. 4B), while THO and TREX-2 null strains also exhibited a similar, but less intense nuclear accumulation of polyadenylated mRNA, consistent with prior reports for a role of these complexes in export (Strässer et al. 2002). Notably, the extent of this nuclear signal was much reduced when FISH was combined with Pab1 immunofluorescence (cf. Fig. 4A and 4B), which may reflect loss of signal associated with additional wash steps in this combined procedure (also seen previously; Brengues and Parker 2007).
FIGURE 4.
THO and TREX-2 TT foci harbor mRNA. (A) FISH using an oligo(dT) probe, combined with Pab1 immunofluorescence was conducted as detailed in Materials and Methods. Cells were additionally mounted in DAPI-containing media to visualize DNA. WT cells were stressed with NaN3 as previously described. Pink arrowheads indicate colocalization of RNA and Pab1 signal. Percentage values indicate percentage of Pab1 foci overlapping with an oligo(dT) foci. Data representative of observations in two biological replicates (separate culture and transformant). Scale bar = 2.5 µM. (B) FISH was conducted as in A, but without Pab1 immunofluorescence. Notably, the nuclear accumulation of oligo(dT) signal in the nucleus was more apparent, suggesting loss of signal due to the immunofluorescence protocol. Mex67-5 was shifted to the nonpermissive temperature for 30 min prior to fixation. Scale bar = 2.5 µM
In summary, the presence of overlapping oligo(dT) and Pab1 foci in TT foci strengthens the interpretation of these foci as mRNP assemblies.
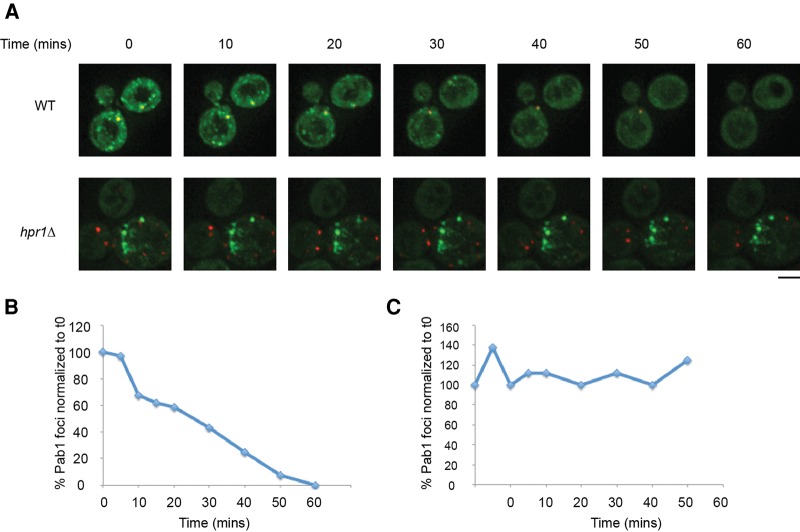
TT foci persist in the presence of cycloheximide
The hypothesis that TT foci arise from defective mRNPs unable to enter translation suggests they might be resistant to the addition of cycloheximide. This is based on previous observations that mRNAs within stress granules can exchange with polysomes and therefore, when cycloheximide is added to cells with stress granules, they disassemble as polysomes are stabilized (Kedersha et al. 2000; Sheth and Parker 2003; Brengues et al. 2005). As expected, we observed that in WT cells, preformed stress granules and P-bodies induced by NaN3 (Fig. 5; Supplemental movie 1) and glucose deprivation (not shown) can be disassembled by CHX addition in WT cells over a 1-h period, albeit P-bodies are not completely gone by this timepoint. In contrast, TT foci do not appreciably decline over this time period, albeit P-body levels did reduce, suggesting CHX was effective in these strains (Fig. 5; Supplemental movie 2–3). This resistance to CHX suggests that TT foci do not readily exchange their mRNPs with polysomes, and is consistent with the hypothesis that TT foci consist of mRNPs unable to enter translation.
FIGURE 5.
hpr1Δ TT foci are insensitive to cycloheximide, suggesting impaired exchange of resident mRNPs with polysomes. (A) WT and hpr1Δ null strains were transformed with pRP1657 and examined in midlog. WT strain was subject to 30 min of NaN3 stress (0.5% w/v) followed by 1 h of 100 µg/mL cycloheximide treatment. hpr1Δ strain was examined in midlog and exposed to an identical cycloheximide treatment. Data representative of observations in three biological replicates (separate culture and transformant). Scale bar = 2.5 µM. (B) Quantification of total Pab1 foci levels in WT cells shown in A, normalized to timepoint 0. (C) Quantification of total Pab1 foci levels in hpr1Δ cells shown in A, normalized to timepoint 0.
THO/TREX-2 mutants show alterations in stress granule assembly during stress
Under normal circumstances, stress granules are only observed during times of cellular stress that result in strong global repression of translation, a condition that also increases P-body levels. Since TT foci shared some components with stress granules we hypothesized they might reduce the concentration of stress granule components in the cytosol and thereby alter stress granule formation during a stress response.
We observed that, during NaN3 stress conditions in mid-log yeast, all THO mutants showed a modest impairment of stress granule assembly relative to a WT strain (Fig. 6A–C), although only mft1Δ, tho2Δ, and thp2Δ were statistically significant. In contrast, TREX-2 mutants (sac3Δ, thp1Δ) were not impaired in stress granule assembly; in fact sac3Δ exhibited a modest, but significant increase in stress granules compared to WT (Fig. 6A–C). P-bodies were more variable across all strains both with and without stress (Fig. 6D,E), albeit THO mutants were, like stress granules, generally impaired during stress relative to WT, whereas TREX-2 mutants showed no significant effect.
FIGURE 6.
THO, but not TREX-2 null strains, show moderate impairment of canonical stress granule induction. (A) WT and THO/TREX-2 null strains were transformed with pRP1657 and examined in midlog (left) or additionally subjected to NaN3 stress (0.5% w/v) for 30 min. (B) Quantitation of average number of stress granules/cell in control strains. (C) Quantitation of average number of stress granules/cell in NaN3 stressed strains. (D) Quantitation of average number of P-bodies/cell in control strains. (E) Quantitation of average number of P-bodies/cell in NaN3 stressed strains. For A–E, all data were collected from three biological replicates (separate culture and transformant) with mean ± SD shown. Paired one-tailed (B) or two-tailed (C–E) Student's t-tests were conducted to assess significance of mutant data relative to WT; (*) = P < 0.05; (**) = P < 0.01. Scale bar = 2.5 µM.
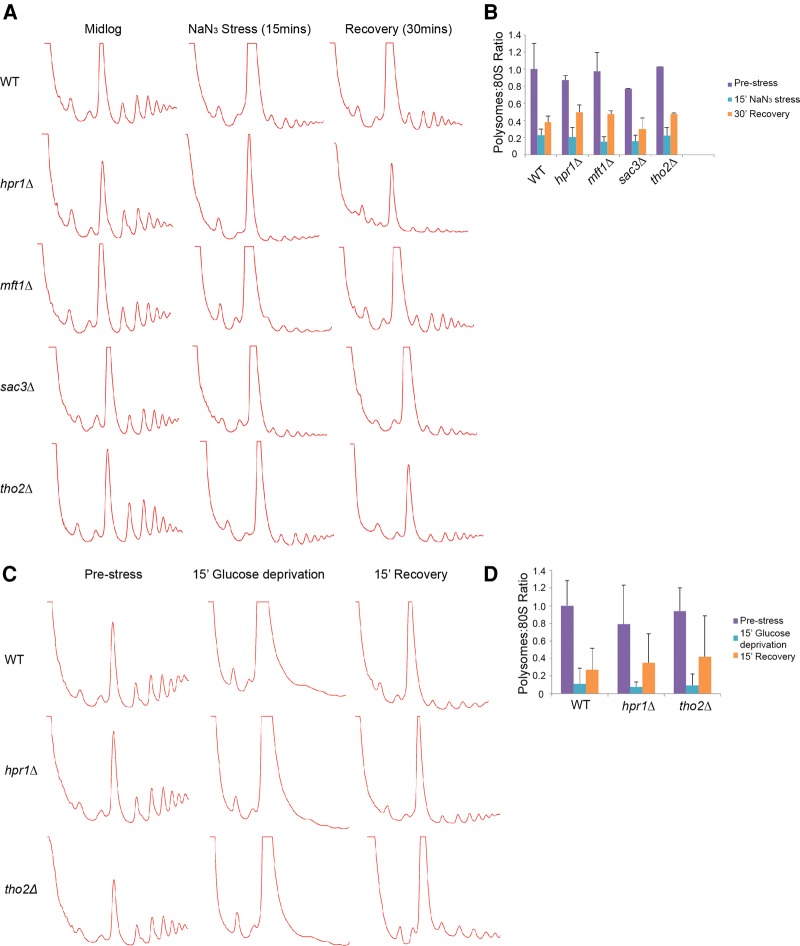
Failure to induce normal stress granule levels during stress in THO mutants could reflect a number of issues, one of which is inefficient repression of translation. To assess this, we conducted polysome analyses of WT and THO and TREX-2 mutant strains during mid-log growth, and after either a 15-min glucose repression or NaN3 stress. We also examined recovery of polysomes 15–30 min after these stresses had been removed, to assess if THO or TREX-2 mutations may affect translational re-entry. In no case did we observe significant deviations from wild-type responses (Fig. 7; Supplemental Fig. 1). This suggests that THO and TREX-2 mutations do not have a broad effect on stress-induced translational repression or translational re-entry following stress. It is thus possible that the defect seen in normal stress granule induction is due to interference by THO and TREX-2 mutations upon normal stress granule assembly mechanisms (see Discussion).
FIGURE 7.
THO and TREX-2 mutants do not show gross defects in translational repression or translational re-entry following stress. (A) WT BY4741 yeast, hpr1Δ, mft1Δ, tho2Δ (THO), and sac3Δ (TREX-2) strains were examined via polysome analysis in mid-log, during NaN3 stress (15 min 0.5% w/v), and following 30 min of recovery (media replacement). Data representative of observations in three biological replicates (separate culture and transformant). (B) Quantitation of the NaN3 polysome: 80S ratio before, during, and after stress. Mean ± SD shown. (C) WT BY4741 yeast, hpr1Δ and tho2Δ strains were examined as above, but following 15 min of glucose deprivation, and following 15 min of recovery. Data representative of observations in three biological replicates (separate culture and transformant). (D) Quantitation of the -Glucose polysome: 80S ratio before, during, and after stress. Mean ± SD shown. Additional glucose deprivation traces are present in Supplemental Figure 4.
TT foci can be cleared via autophagy
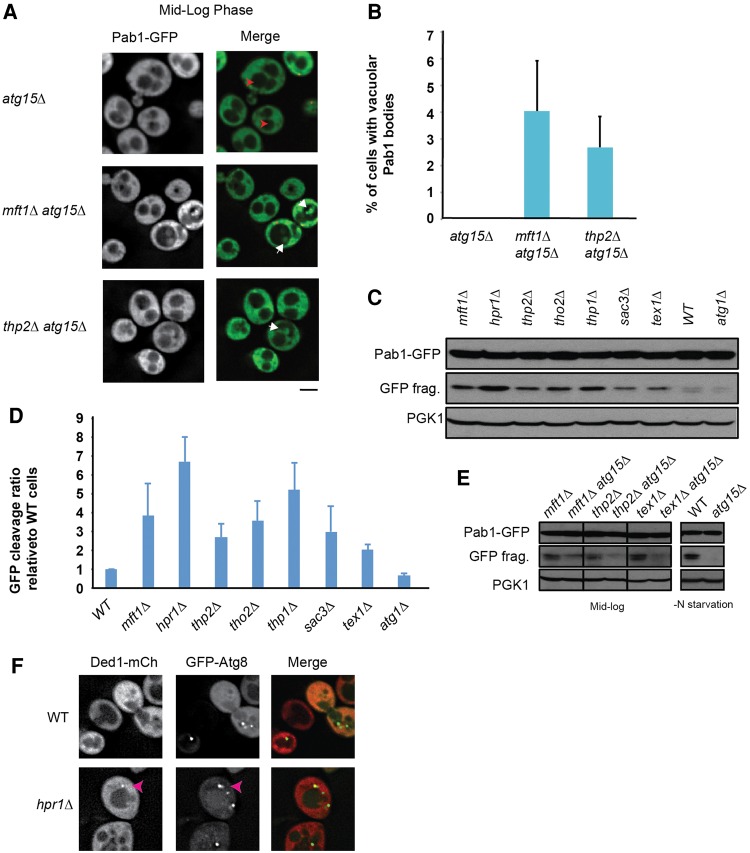
We recently described an autophagic pathway (“granulophagy”; Buchan et al. 2013) that can clear stress granules, most prevalently after the post-diauxic shift and/or following inactivation of the 5′–3′ mRNA decay. Since TT foci appeared resistant to CHX treatment, we hypothesized that they might also be cleared from the cytosol by autophagy. Consistent with this model, we occasionally observed Pab1-GFP localization within vacuoles in our THO and TREX-2 mutants during mid-log growth (e.g. mft1Δ Fig. 1A, hpr1Δ in Fig. 2B; Fig. 6, hpr1Δ, mft1Δ stress images, and thp2Δ nonstress image). To determine if TT foci are indeed cleared by autophagy we performed three experiments.
First, we deleted atg15Δ in a selection of THO mutant strains. Atg15 is a vacuolar lipase that aids in breakdown of autophagosomes that enter vacuoles (Epple et al. 2001), thus its absence facilitates visual detection of stress granule autophagic intermediates (Buchan et al. 2013). Therefore, an increase in detection of Pab1-GFP vacuolar signal in THO atg15Δ mutants versus an atg15Δ strain alone would support the hypothesis that TT foci are autophagic targets. We observed a small, but clear accumulation of vacuolar Pab1-GFP in mid-log growth in mft1Δ atg15Δ and thp2Δ atg15Δ yeast relative to atg15Δ yeast alone, where no such targeting was observed (consistent with absence of stress granules and granulophagy activity under this condition (Fig. 8A,B; Buchan et al. 2013).
FIGURE 8.
TT foci are cleared by autophagy. (A) Strains were transformed with pRP1657 and examined in mid-log or early stationary phase. White arrows indicate examples of Pab1 autophagic bodies within vacuoles; red arrows indicate empty vacuoles. Data representative of observations in three biological replicates (separate culture and transformant). Scale bar = 2.5 µM. (B) Quantitation of data in panel A; three biological replicates (separate cultures and transformants) with mean ± SD shown. (C) Pab1-GFP Western blot in mid-log cells; GFP fragment (GFP) indicative of autophagic turnover of Pab1-GFP. PGK1 serves as a loading control. Data representative of observations in three biological replicates (separate culture and transformant) (D) Quantitation of data in C; ratio of GFP fragment levels to full length were calculated for each strain and normalized to the WT ratio. Mean ± SD shown. (E) Loss of Atg15 impairs Pab1-GFP turnover both in THO-TREX-2 mutants and during nonselective autophagy (4h–N media). (F) GFP-Atg8 colocalizes with TT foci in mid-log hpr1Δ cells. Blue arrowhead indicates colocalization (see Supplemental Table S5 for quantitation).
A second method to assess autophagic targeting of components of TT foci, specifically in this case Pab1-GFP, is to utilize a Western blot GFP-fragment assay in which GFP-tagged proteins turned over by autophagy result in formation of a slow-to-degrade, characteristic GFP-free band (or bands) (Klionsky et al. 2012). We thus examined all THO and TREX-2 null strains in our study, as well as tex1Δ (putative THO component) for evidence of Pab1-GFP turnover via autophagic means in mid-log cells (Fig. 8C,D). As a positive control, we also examined Pab1-GFP turnover in WT cells subject to nitrogen starvation, which activates nonspecific autophagy (Fig. 8E). As a negative control, atg1Δ cells, which are strongly impaired in activating autophagy, were also examined (Fig. 8D). Supporting our microscopy data, we observed that all THO-TREX-2 mutants exhibited an increase in autophagic turnover of Pab1-GFP relative to WT strains (Fig. 8C,D). As expected, atg1Δ cells exhibited almost no GFP fragment accumulation, whereas WT cells exhibit robust GFP fragment accumulation during nitrogen deprivation (Fig. 8C–E). Notably, the presence of an atg15Δ deletion also inhibits fragment accumulation (Fig. 8E), presumably reflecting impaired autophagic turnover of Pab1-GFP due to its protection in vacuolar autophagic bodies. This further confirms the likelihood that THO and TREX-2 mutants make use of autophagy for clearance of TT foci components.
A third and final method to assess if TT foci can undergo autophagy was to determine whether a subfraction of TT foci colocalized with markers of autophagosomes. To do this experiment we examined colocalization of TT foci in hpr1Δ, mft1Δ, and tho2Δ strains with the autophagosomal marker Atg8. Although colocalization of autophagy proteins with their substrates is often transient, in the absence of a block to autophagy, we still observed several cases of clear colocalization between TT foci (Ded1-mCh) and GFP-Atg8 (Fig. 8F). Specifically, in hpr1Δ, mft1Δ, and tho2Δ strains, 10.8%, 11.7%, and 5.8%, of Ded1-mCh foci, respectively, localized with Atg8 foci.
Taken together, these data suggest that TT foci can be targeted for autophagy, possibly via the same mechanism as stress granules (“granulophagy”). This may explain the absence of visible TT foci in most cells.
DISCUSSION
Several lines of evidence argue that defects in components of the THO or TREX-2 complexes lead to the accumulation of a novel form of cytoplasmic mRNP granule, referred to as TT foci. TT foci share similarity to stress granules since they contain poly(A)+ RNA as well as Pab1, Pub1, Ded1, Pbp1, and eIF4G2, all of which localize to yeast stress granules. However, TT foci are clearly different from normal stress granules. First, they contain additional proteins involved in 3′ end formation and export that are not normally seen in stress granules including Pap1 and Gle1 (and to a low extent other members of the THO and TREX-2 complexes; Mft1 and Thp1). Second, they persist in the presence of cycloheximide, suggesting that the mRNAs within these assemblies are not in dynamic exchange with a translating pool. Taken together, these observations argue that TT foci are distinct from stress granules.
One parsimonious explanation for the formation of TT foci is that they arise from aberrant mRNPs produced in the THO and TREX-2 mutants strains. This possibility is supported by evidence that THO and TREX-2 mutants have defects in mRNP biogenesis. For example, previous data suggest that specific mRNAs examined in yeast THO mutants suffer from premature 3′ end truncation and polyadenylation defects, some of which are degraded by the nuclear exosome. We hypothesize that a subpopulation of these aberrant mRNPs is exported to the cytosol in association with polyadenylation factors (Rougemaille et al. 2007). Export of aberrant mRNPs has been previously observed in other contexts; for example, mRNAs defective in splicing can escape nuclear quality control and are exported, and eventually degraded by the cytoplasmic mRNA decay machinery (Hilleren and Parker 2003). Similarly, capped and polyadenylated ncRNAs that arise as a result of cryptic RNAPII transcription from inter- and intragenic regions (cryptic unstable transcripts [CUTs] and stable uncharacterized transcripts [SUTS]) can in some cases (particularly for SUTs) successfully export to the cytoplasm where the cytoplasmic decay machinery is more important for their turnover (Thompson and Parker 2007; Marquardt et al. 2011). Interestingly, the Cole laboratory previously described aberrant cytoplasmic foci termed RNA export granules (REGs) that were spatially distinct from P-bodies (as TT foci are). These were observed in temperature sensitive DBP5 and MEX67 mutants (Scarcelli et al. 2008), and like TT foci, exhibited resistance to cycloheximide. The similarity of REGs to TT foci thus further hints at the idea that TT foci reflect a defect in exported mRNPs. Future work identifying genes that suppress or exacerbate assembly of TT foci, and conditions that may provoke their assembly in a WT context will be critical to understand the mechanism of TT formation. Defining the mRNP composition of these foci may also offer important insight into the cellular consequences of TT foci assembly.
A broader implication of this work, and that of others, is that cytoplasmic mRNA regulation can be perturbed in THO mutants. Specifically, we observe that stress granule assembly is perturbed in THO (but not TREX-2) mutants in response to NaN3 stress (Fig. 6), and in response to multiple stresses in an mft1Δ strain (Yang et al. 2014). Our polysome data (Fig. 7; Supplemental Fig. 1) suggests this is unlikely to be due to gross perturbations in translation repression or recovery following stress in THO/TREX-2 mutants, although stress granule assembly could still be impaired in principle by a failure to repress a subpopulation of mRNAs that our polysome assays would not detect. Alternatively, stress granule assembly factors could be sequestered within TT foci. However, since the percentage of cells with visible TT foci is usually low (6%–14%), sequestration in sub-microscopic TT foci, or enhanced clearance of sequestered stress granule assembly factors by autophagy (see below) could also be involved. Inactivation or altered expression of stress granule assembly factors could also explain the reduction in canonical stress granule assembly we observe in THO mutants. This could occur at multiple levels given the role of the THO complex in nuclear mRNA biogenesis and export, but could also involve altered cytoplasmic mRNA regulation as well. For instance, mft1Δ strains fail to up-regulate translation of an osmotic stress-responsive reporter mRNA during high salt stresses (Yang et al. 2014). Ultimately, global translation assays such as ribosome profiling will best address the question of do THO and TREX-2 proteins influence translation and translation repression of specific populations of mRNAs.
Four observations argue that TT foci formed in the THO/TREX-2 mutants can be, at least in part cleared from the cytosol by autophagy. First, in various THO/TREX-2 mutants we observe increased accumulation of Pab1-GFP in the vacuole. Second, in atg15Δ mutants, which trap autophagosomes in an intra-vacuolar state (Epple et al. 2003) we observe that defects in the THO or TREX-2 complex leads to an increase in Pab1-GFP vacuolar signal relative to WT strains (Fig. 8). Third, we also observe that autophagic degradation of Pab1-GFP, as assessed by a GFP fragment assay commonly used in the autophagy field (Klionsky et al. 2012), is increased in THO and TREX-2 mutant strains (Fig. 8). Finally, we observe colocalization of some TT foci with Atg8. Taken together, these observations strongly suggest that at least some TT foci are targeted for clearance by autophagy, perhaps by the same mechanisms that target stress granules for autophagy.
The clearance of aberrant mRNPs by autophagy may be a general mechanism the cell has evolved to deal with nonfunctional RNPs. Results herein suggest that aberrant mRNP foci resulting from THO/TREX-2 defects can be targeted by autophagy, in a manner reminiscent of stress granule clearance via granulophagy (Buchan et al. 2013). We suggest that autophagy may clear many RNP complexes from the cell that are unable to be utilized or degraded by other pathways. This general role of autophagy in RNA and RNP clearance could be significant in disease states both as autophagy generally decreases with age (Cuervo 2008; Rubinsztein et al. 2011), and due to mutations in autophagy factors that increase stress granules and may promote ALS and FTLD (Ramaswami et al. 2013). Thus an understanding of the roles of autophagy in both general RNA clearance and the clearance of aberrant RNPs will be an area of important future work.
MATERIALS AND METHODS
Yeast strains and growth conditions
The genotypes of strains used in this study are listed in Supplemental Table S3. Strains expressing proteins C-terminally tagged with GFP were obtained from the yeast GFP library (Huh et al. 2003). Hpr1Δ strains in various strain backgrounds were generated via integration of a hygromycin B cassette at the HPR1 locus via homologous recombination. Strains were grown on either YEPD or synthetic complete (SC) medium (0.17% yeast nitrogen base—Difco and 5 g/L Ammonium sulfate—Fisher) supplemented with appropriate amino acids and nucleotides; in both cases, 2% glucose (Glu) was used as a carbon source. Strains were grown at 30°C unless otherwise stated. Yeast strains were transformed via a standard lithium acetate technique. Nuclei were visualized using Hoescht 33342, incubated in cell media at a concentration of 0.5 mg/mL for 20–30 min. Glucose deprivation and NaN3 stresses were conducted as previously described (Buchan et al. 2011). Nitrogen starvation was conducted for 4 h in SC media lacking ammonium sulfate and amino acids.
Plasmids
Plasmids used in this study are listed in Supplemental Table S3. pRP2478 was generated by XbaI–SalI cleavage of Pab1-GFP, including promoter, ORF and terminator sequences, from pRP1363 (Brengues and Parker 2007), and ligation into similarly cut pRS415.
Microscopy and image analysis
Screening that initially identified THO/TREX-2 subunits as harboring constitutive stress granule-like foci has been previously described (Buchan et al. 2013). Analyses of protein localization, and assembly of stress granules or P-bodies during stress was conducted as previously described (Buchan et al. 2010), using a Deltavision RT or Deltavision Elite microscope, with a 100× objective, and Image J/Fiji (Schindelin et al. 2012) software. For all quantitated data sets with biological replicates (indicated in legend), a minimum of 50 cells per replicate were quantified. Protein localization and stress granule/P-bodies were scored in a blind manner (Buchan et al. 2010).
Oligo(dT) fluorescence in situ hybridization (FISH)
Five milliliters of yeast cultures (OD600 0.3–0.5) were incubated with 4% formaldehyde for 45 min, followed by two washes in KPi buffer (0.1 M KPi, pH 6.5), and one wash in wash buffer (1.2 M sorbitol in KPi buffer), followed by incubation at 30°C with gentle agitation in 1 mL spheroblasting buffer (100 µL 10 mg/mL zymolase solution (Zymoresearch), 100 µL vanadyl ribonucleoside complex (NEB), 2 µL β-mercaptoethanol (Sigma), and 3 µL RNase out (Invitrogen). Once adequately spheroblasted (∼90–120 min), cells were spun and washed twice at low speed in wash buffer, resuspended in 300 µL wash buffer, then spotted onto concanavilin A treated slides with “wells” demarked by hydrophobic coating (Tekdon), and allowed to adhere for 1 h. Wells were washed once for 5 min with wash buffer, KPi buffer, KPi buffer + 0.5% NP40 (to permeabilize cells), and KPi buffer, followed by addition of ice cold 70% ethanol and incubation in a humid chamber overnight at −20°C. The next day, wells were aspirated, rehydrated with two 5 min washes in 2× SSC, and one 10 min wash in 2× SSC/0.1%NP40/50% formamide. Prehybridization involved preparation of Eppendorfs for each well consisting of 12.5 µL of 2 mg/mL ssDNA and 6 µL of 4.8 mg/mL yeast tRNA being dried down, and resuspended in 15 µL of hybridization solution 1 (800 µL formamide, 10 µL 1 M NaP buffer, 90 µL dH2O). These were incubated at 100°C for 3 min, chilled on ice for 3 min, then added to each well, followed by addition of 15 µL of hybridization solution 2 to each well (25 µL formamide, 50 µL 20× SSC, 25 µL VRC, 10 µL 10 mg/mL BSA, 25 µL RNase out, and 115 µL H2O). Slides were incubated in a humid box at 37°C for at least 1 h, followed by hybridization, which repeats the prehybridization steps except that 10 µL of 250 nM oligo(dT)25-FITC LNA probe was additionally dried down in each Eppendorf prior to resuspension in hybridization solution 1. Incubation at 37°C in a humid box was done overnight. Wells were then twice washed in 2× SSC/0.1%NP40/50% formamide (15 min at 37°C), once in 2× SSC/0.1%NP40 (15 min at room temperature), twice in 1× SSC (15 min at room temperature), and twice in 1× PBS (5 min room temperature).
Yeast immunofluorescence
Following FISH, wells were washed in 30 µL PBS, blocked in PBS + 3% BSA (IgG free, Jackson Immunoresearch) for 1 h at room temperature. Wells were washed twice with PBS, then incubated with 1:50 dilution α-Pab1 rabbit sera (Cocalico Biologicals) in PBS + 3% BSA at room temperature for 3+ h. Wells were washed three times with PBS, then incubated with 1:200 donkey α-rabbit Fluor 647 (Santa Cruz), and left at room temperature for 1 h protected from light. Wells were washed five times with PBS (total 45 min), mounted with vectashield containing DAPI (Vector Labs), and stored at 4°C.
Polysome analysis
Polysome analyses were conducted as in Buchan et al. (2011) with the following minor difference. Cells were either harvested in midlog (OD600 0.3–0.6), after being subject to 15-min stresses (0.5% w/v NaN3, or glucose deprivation) or after a 15-min recovery period post stress (following replacement of stress media). Polysome traces were analyzed using Fiji (Schindelin et al. 2012), with a common baseline and vertical lines demarcating the 80S and polysome peak regions. Areas under these curves were calculated to obtain the polysome/80S ratio.
Western blotting/GFP-fragment autophagic turnover assay
Western blotting was conducted via standard methods. GFP was detected with Mouse α-GFP (Biolegend B34, previously Covance MMS-118P) at a 1:2000 dilution. Pgk1 was detected with Mouse α-Pgk1 (Abcam, ab113687) at a 1:10,000 dilution. Goat α-Mouse HRP (Santa Cruz) was used as a secondary antibody at a 1:5000 dilution. Blots were developed with the Supersignal West Dura kit (Thermo Scientific). Detection of GFP fragments as evidence of autophagic turnover of proteins has been described and validated as previously described in Klionsky et al. (2012).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We wish to thank members of the Buchan and Parker laboratories for useful discussions throughout the study, and the Thumm laboratory (University of Goettingen) for provision of the GFP-Atg8 construct. Research for this article was supported by the Howard Hughes Medical Institute (R.P.) and start-up funds from the University of Arizona (J.R.B.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.057224.116.
REFERENCES
- Anderson P, Kedersha N. 2008. Stress granules: the Tao of RNA triage. Trends Biochem Sci 33: 141–150. [DOI] [PubMed] [Google Scholar]
- Arimoto K, Fukuda H, Imajoh-Ohmi S, Saito H, Takekawa M. 2008. Formation of stress granules inhibits apoptosis by suppressing stress-responsive MAPK pathways. Nat Cell Biol 10: 1324–1332. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. 2006. Stress-induced reversal of microRNA repression and mRNA P-body localization in human cells. Cold Spring Harb Symp Quant Biol 71: 513–521. [DOI] [PubMed] [Google Scholar]
- Brengues M, Parker R. 2007. Accumulation of polyadenylated mRNA, Pab1p, eIF4E, and eIF4G with P-bodies in Saccharomyces cerevisiae. Mol Biol Cell 18: 2592–2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brengues M, Teixeira D, Parker R. 2005. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science 310: 486–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretes H, Rouviere JO, Leger T, Oeffinger M, Devaux F, Doye V, Palancade B. 2014. Sumoylation of the THO complex regulates the biogenesis of a subset of mRNPs. Nucleic Acids Res 42: 5043–5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Parker R. 2009. Eukaryotic stress granules: the ins and outs of translation. Mol Cell 36: 932–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Nissan T, Parker R. 2010. Analyzing P-bodies and stress granules in Saccharomyces cerevisiae. Methods Enzymol 470: 619–640. [DOI] [PubMed] [Google Scholar]
- Buchan JR, Yoon J-H, Parker R. 2011. Stress-specific composition, assembly and kinetics of stress granules in Saccharomyces cerevisiae. J Cell Sci 124: 228–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Kolaitis R-M, Taylor JP, Parker R. 2013. Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell 153: 1461–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chávez S, Beilharz T, Rondón AG, Erdjument-Bromage H, Tempst P, Svejstrup JQ, Lithgow T, Aguilera A. 2000. A protein complex containing Tho2, Hpr1, Mft1 and a novel protein, Thp2, connects transcription elongation with mitotic recombination in Saccharomyces cerevisiae. EMBO J 19: 5824–5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi B, Wang Q, Wu G, Tan M, Wang L, Shi M, Chang X, Cheng H. 2013. Aly and THO are required for assembly of the human TREX complex and association of TREX components with the spliced mRNA. Nucleic Acids Res 41: 1294–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo AM. 2008. Autophagy and aging: keeping that old broom working. Trends Genet 24: 604–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewey CM, Cenik B, Sephton CF, Johnson BA, Herz J, Yu G. 2012. TDP-43 aggregation in neurodegeneration: Are stress granules the key? Brain Res 1462: 16–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didiot M-C, Subramanian M, Flatter E, Mandel J-L, Moine H. 2009. Cells lacking the fragile X mental retardation protein (FMRP) have normal RISC activity but exhibit altered stress granule assembly. Mol Biol Cell 20: 428–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisinger-Mathason TSK, Andrade J, Groehler AL, Clark DE, Muratore-Schroeder TL, Pasic L, Smith JA, Shabanowitz J, Hunt DF, Macara IG, et al. 2008. Codependent functions of RSK2 and the apoptosis-promoting factor TIA-1 in stress granule assembly and cell survival. Mol Cell 31: 722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epple UD, Suriapranata I, Eskelinen EL, Thumm M. 2001. Aut5/Cvt17p, a putative lipase essential for disintegration of autophagic bodies inside the vacuole. J Bacteriol 183: 5942–5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epple UD, Eskelinen EL, Thumm M. 2003. Intravacuolar membrane lysis in Saccharomyces cerevisiae. Does vacuolar targeting of Cvt17/Aut5p affect its function? J Biol Chem 278: 7810–7821. [DOI] [PubMed] [Google Scholar]
- Fasken MB, Corbett AH. 2009. Mechanisms of nuclear mRNA quality control. RNA Biol 6: 237–241. [DOI] [PubMed] [Google Scholar]
- Fischer T, Strässer K, Rácz A, Rodriguez-Navarro S, Oppizzi M, Ihrig P, Lechner J, Hurt E. 2002. The mRNA export machinery requires the novel Sac3p-Thp1p complex to dock at the nucleoplasmic entrance of the nuclear pores. EMBO J 21: 5843–5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gewartowski K, Cuéllar J, Dziembowski A, Valpuesta JM. 2012. The yeast THO complex forms a 5-subunit assembly that directly interacts with active chromatin. Bioarchitecture 2: 134–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg SD, Galvin JE, Chiu TS, Lee VM, Masliah E, Trojanowski JQ. 1998. RNA sequestration to pathological lesions of neurodegenerative diseases. Acta Neuropathol 96: 487–494. [DOI] [PubMed] [Google Scholar]
- Gómez-González B, García-Rubio M, Bermejo R, Gaillard H, Shirahige K, Marín A, Foiani M, Aguilera A. 2011. Genome-wide function of THO/TREX in active genes prevents R-loop-dependent replication obstacles. EMBO J 30: 3106–3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Aguilera C, Tous C, Gómez-González B, Huertas P, Luna R, Aguilera A. 2008. The THP1-SAC3-SUS1-CDC31 complex works in transcription elongation-mRNA export preventing RNA-mediated genome instability. Mol Biol Cell 19: 4310–4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross T, Siepmann A, Sturm D, Windgassen M, Scarcelli JJ, Seedorf M, Cole CN, Krebber H. 2007. The DEAD-box RNA helicase Dbp5 functions in translation termination. Science 315: 646–649. [DOI] [PubMed] [Google Scholar]
- Guria A, Tran DDH, Ramachandran S, Koch A, El Bounkari O, Dutta P, Hauser H, Tamura T. 2011. Identification of mRNAs that are spliced but not exported to the cytoplasm in the absence of THOC5 in mouse embryo fibroblasts. RNA 17: 1048–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwizdek C, Iglesias N, Rodriguez MS, Ossareh-Nazari B, Hobeika M, Divita G, Stutz F, Dargemont C. 2006. Ubiquitin-associated domain of Mex67 synchronizes recruitment of the mRNA export machinery with transcription. Proc Natl Acad Sci 103: 16376–16381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackmann A, Wu H, Schneider U-M, Meyer K, Jung K, Krebber H. 2014. Quality control of spliced mRNAs requires the shuttling SR proteins Gbp2 and Hrb1. Nat Commun 5: 3123. [DOI] [PubMed] [Google Scholar]
- Hilleren PJ, Parker R. 2003. Cytoplasmic degradation of splice-defective pre-mRNAs and intermediates. Mol Cell 12: 1453–1465. [DOI] [PubMed] [Google Scholar]
- Huertas P, Aguilera A. 2003. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol Cell 12: 711–721. [DOI] [PubMed] [Google Scholar]
- Huh W-K, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. 2003. Global analysis of protein localization in budding yeast. Nature 425: 686–691. [DOI] [PubMed] [Google Scholar]
- Hurt E, Luo M-J, Röther S, Reed R, Strässer K. 2004. Cotranscriptional recruitment of the serine-arginine-rich (SR)-like proteins Gbp2 and Hrb1 to nascent mRNA via the TREX complex. Proc Natl Acad Sci 101: 1858–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito D, Suzuki N. 2011. Conjoint pathologic cascades mediated by ALS/FTLD-U linked RNA-binding proteins TDP-43 and FUS. Neurology 77: 1636–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katahira J, Inoue H, Hurt E, Yoneda Y. 2009. Adaptor Aly and co-adaptor Thoc5 function in the Tap-p15-mediated nuclear export of HSP70 mRNA. EMBO J 28: 556–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Cho MR, Li W, Yacono PW, Chen S, Gilks N, Golan DE, Anderson P. 2000. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J Cell Biol 151: 1257–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P. 2005. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol 169: 871–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Kim NC, Wang Y-D, Scarborough EA, Moore J, Diaz Z, MacLea KS, Freibaum B, Li S, Molliex A, et al. 2013. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature 495: 467–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, et al. 2012. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 8: 445–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libri D, Dower K, Boulay J, Thomsen R, Rosbash M, Jensen TH. 2002. Interactions between mRNA export commitment: 3′-end quality control, and nuclear degradation. Mol Cell Biol 22: 8254–8266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquardt S, Hazelbaker DZ, Buratowski S. 2011. Distinct RNA degradation pathways and 3′ extensions of yeast non-coding RNA species. Transcription 2: 145–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason PB, Struhl K. 2005. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol Cell 17: 831–840. [DOI] [PubMed] [Google Scholar]
- Moore MJ, Proudfoot NJ. 2009. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell 136: 688–700. [DOI] [PubMed] [Google Scholar]
- Nonhoff U, Ralser M, Welzel F, Piccini I, Balzereit D, Yaspo M-L, Lehrach H, Krobitsch S. 2007. Ataxin-2 interacts with the DEAD/H-box RNA helicase DDX6 and interferes with P-bodies and stress granules. Mol Biol Cell 18: 1385–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrua O, Libri D. 2013. RNA quality control in the nucleus: the Angels’ share of RNA. Biochim Biophys Acta 1829: 604–611. [DOI] [PubMed] [Google Scholar]
- Qu X, Lykke-Andersen S, Nasser T, Saguez C, Bertrand E, Jensen TH, Moore C. 2009. Assembly of an export-competent mRNP is needed for efficient release of the 3′-end processing complex after polyadenylation. Mol Cell Biol 29: 5327–5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswami M, Taylor JP, Parker R. 2013. Altered ribostasis: RNA-protein granules in degenerative disorders. Cell 154: 727–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel J, Herold A, Gari K, Köcher T, Rode M, Ciccarelli FL, Wilm M, Izaurralde E. 2004. Genome-wide analysis of mRNAs regulated by the THO complex in Drosophila melanogaster. Nat Struct Mol Biol 11: 558–566. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Navarro S, Fischer T, Luo M-J, Antúnez O, Brettschneider S, Lechner J, Pérez-Ortín JE, Reed R, Hurt E. 2004. Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell 116: 75–86. [DOI] [PubMed] [Google Scholar]
- Rollenhagen C, Hodge CA, Cole CN. 2007. Following temperature stress, export of heat shock mRNA occurs efficiently in cells with mutations in genes normally important for mRNA export. Eukaryot Cell 6: 505–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondón AG, Jimeno S, García-Rubio M, Aguilera A. 2003. Molecular evidence that the eukaryotic THO/TREX complex is required for efficient transcription elongation. J Biol Chem 278: 39037–39043. [DOI] [PubMed] [Google Scholar]
- Rougemaille M, Gudipati RK, Olesen JR, Thomsen R, Seraphin B, Libri D, Jensen TH. 2007. Dissecting mechanisms of nuclear mRNA surveillance in THO/sub2 complex mutants. EMBO J 26: 2317–2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougemaille M, Dieppois G, Kisseleva-Romanova E, Gudipati RK, Lemoine S, Blugeon C, Boulay J, Jensen TH, Stutz F, Devaux F, et al. 2008. THO/Sub2p functions to coordinate 3′-end processing with gene-nuclear pore association. Cell 135: 308–321. [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC, Mariño G, Kroemer G. 2011. Autophagy and aging. Cell 146: 682–695. [DOI] [PubMed] [Google Scholar]
- Saguez C, Schmid M, Olesen JR, Ghazy MAE-H, Qu X, Poulsen MB, Nasser T, Moore C, Jensen TH. 2008. Nuclear mRNA surveillance in THO/sub2 mutants is triggered by inefficient polyadenylation. Mol Cell 31: 91–103. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H, Moreno H, Simos G, Segref A, Fahrenkrog B, Panté N, Hurt E. 1998. Nuclear mRNA export requires complex formation between Mex67p and Mtr2p at the nuclear pores. Mol Cell Biol 18: 6826–6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarcelli JJ, Viggiano S, Hodge CA, Heath CV, Amberg DC, Cole CN. 2008. Synthetic genetic array analysis in Saccharomyces cerevisiae provides evidence for an interaction between RAT8/DBP5 and genes encoding P-body components. Genetics 179: 1945–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Jensen TH. 2013. Transcription-associated quality control of mRNP. Biochim Biophys Acta 1829: 158–168. [DOI] [PubMed] [Google Scholar]
- Sheth U, Parker R. 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300: 805–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strässer K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez-Navarro S, Rondón AG, Aguilera A, Struhl K, Reed R, et al. 2002. TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417: 304–308. [DOI] [PubMed] [Google Scholar]
- Takahara T, Maeda T. 2012. Transient sequestration of TORC1 into stress granules during heat stress. Mol Cell 47: 242–252. [DOI] [PubMed] [Google Scholar]
- Thedieck K, Holzwarth B, Prentzell MT, Boehlke C, Kläsener K, Ruf S, Sonntag AG, Maerz L, Grellscheid S-N, Kremmer E, et al. 2013. Inhibition of mTORC1 by astrin and stress granules prevents apoptosis in cancer cells. Cell 154: 859–874. [DOI] [PubMed] [Google Scholar]
- Thompson DM, Parker R. 2007. Cytoplasmic decay of intergenic transcripts in Saccharomyces cerevisiae. Mol Cell Biol 27: 92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tous C, Rondón AG, García-Rubio M, González-Aguilera C, Luna R, Aguilera A. 2011. A novel assay identifies transcript elongation roles for the Nup84 complex and RNA processing factors. EMBO J 30: 1953–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umlauf D, Bonnet J, Waharte F, Fournier M, Stierle M, Fischer B, Brino L, Devys D, Tora L. 2013. The human TREX-2 complex is stably associated with the nuclear pore basket. J Cell Sci 126: 2656–2667. [DOI] [PubMed] [Google Scholar]
- Yang X, Shen Y, Garre E, Hao X, Krumlinde D, Cvijović M, Arens C, Nyström T, Liu B, Sunnerhagen P. 2014. Stress granule-defective mutants deregulate stress responsive transcripts. PLoS Genet 10: e1004763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenklusen D, Vinciguerra P, Wyss J-C, Stutz F. 2002. Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p. Mol Cell Biol 22: 8241–8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.