Abstract
All retroviruses package cellular RNAs into virions. Studies of murine leukemia virus (MLV) revealed that the major host cell RNAs encapsidated by this simple retrovirus were LTR retrotransposons and noncoding RNAs (ncRNAs). Several classes of ncRNAs appeared to be packaged by MLV shortly after synthesis, as precursors to tRNAs, small nuclear RNAs, and small nucleolar RNAs were all enriched in virions. To determine the extent to which the human immunodeficiency virus (HIV-1) packages similar RNAs, we used high-throughput sequencing to characterize the RNAs within infectious HIV-1 virions produced in CEM-SS T lymphoblastoid cells. We report that the most abundant cellular RNAs in HIV-1 virions are 7SL RNA and transcripts from numerous divergent and truncated members of the long interspersed element (LINE) and short interspersed element (SINE) families of retrotransposons. We also detected precursors to several tRNAs and small nuclear RNAs as well as transcripts derived from the ribosomal DNA (rDNA) intergenic spacers. We show that packaging of a pre-tRNA requires the nuclear export receptor Exportin 5, indicating that HIV-1 recruits at least some newly made ncRNAs in the cytoplasm. Together, our work identifies the set of RNAs packaged by HIV-1 and reveals that early steps in HIV-1 assembly intersect with host cell ncRNA biogenesis pathways.
Keywords: retrovirus, retrotransposons, pre-tRNAs, noncoding RNA precursors, nuclear export
INTRODUCTION
All retroviruses package specific host cell RNAs. These RNAs can account for up to 50% of the virion RNA by mass (Berkowitz et al. 1996). A cellular RNA packaged by all examined retroviruses is 7SL, a noncoding RNA (ncRNA) that functions in the cytoplasm as part of a signal recognition particle (SRP) (Bishop et al. 1970; Erikson et al. 1973; Onafuwa-Nuga et al. 2005, 2006; Eckwahl et al. 2016). All retroviruses also package the specific host tRNA needed to prime reverse transcription of their genomic RNA (gRNA) (Cen et al. 2002). For MLV, a combination of probing for candidate RNAs and deep sequencing revealed that the cellular RNAs in virions consist mostly of ncRNAs and endogenous retrovirus transcripts (Onafuwa-Nuga et al. 2005; Garcia et al. 2009; Eckwahl et al. 2015). In addition to 7SL, two classes of ncRNAs, called Y and vault RNAs, are encapsidated in one or more copies per virion (Onafuwa-Nuga et al. 2005; Garcia et al. 2009; Eckwahl et al. 2015). Many other ncRNAs, such as the spliceosomal U2 and U6 snRNAs, are packaged in sub-stoichiometric amounts (Onafuwa-Nuga et al. 2005; Garcia et al. 2009; Eckwahl et al. 2015). For HIV-1, tRNA profiling revealed that specific nonprimer tRNAs are enriched in virions (Pavon-Eternod et al. 2010), while deep sequencing of short (<40 nucleotides [nt]) RNAs revealed that some miRNAs are selectively packaged (Schopman et al. 2012). Several other ncRNAs, including 5S rRNA, U6 snRNA, and Y RNAs, have also been detected in HIV-1 virions (Houzet et al. 2007; Tian et al. 2007). However, a comprehensive analysis of the RNAs packaged by HIV-1 has not been reported.
Although the need for a specific packaged tRNA to prime reverse transcription was identified more than 40 yr ago (Dahlberg et al. 1974; Faras et al. 1974), the roles of other encapsidated RNAs are not well understood. One role could be to promote Gag oligomerization, an early step in virus assembly that occurs in cytosol, before arrival of Gag at the plasma membrane (Kutluay and Bieniasz 2010; Hendrix et al. 2015). Although oligomerization is enhanced by RNA (Campbell and Vogt 1995; Rein et al. 2011; O'Carroll et al. 2012; Sundquist and Krausslich 2012), virus-like particle (VLP) formation remains efficient when gRNA is absent or its packaging is prevented (Levin et al. 1974; Mann et al. 1983). Consistent with a role for cellular RNAs in nucleating virus assembly, in vivo crosslinking revealed that HIV-1 Gag associates primarily with host cell RNAs in cytosol (Kutluay et al. 2014). For some retroviruses, cellular RNA may contribute to scaffolding interactions between Gag molecules, since ribonuclease treatment disrupts MLV, but not HIV-1 cores (Campbell et al. 2001; Muriaux et al. 2001). Whether specific cellular RNAs normally fulfill these roles, or whether many packaged RNAs contribute, remains unknown.
Many ncRNAs appear to be recruited for packaging soon after synthesis, before binding their usual protein partners. For example, although 7SL is estimated to be present in 14 copies per HIV-1 virion, the SRP54 subunit is below the detection limit (Onafuwa-Nuga et al. 2006). Similarly, although 7SL, Y RNAs, and vault RNAs are all highly enriched in MLV virions, their protein partners (SRP19, Ro60, and TEP1, respectively) were not detected (Garcia et al. 2009; Eckwahl et al. 2015). Packaging of 7SL may occur before SRP19 binding, since overexpressing SRP19, the subunit that stabilizes a 7SL structure required for SRP54 association, significantly reduces 7SL packaging by HIV-1 (Wang et al. 2007; Bach et al. 2008; Apolonia et al. 2015). Moreover, although Y RNAs are unstable in cells lacking Ro60, Y RNA packaging by MLV remains efficient in these cells (Garcia et al. 2009), suggesting MLV recruits these RNAs prior to Ro60 binding. For MLV, high-throughput sequencing of virion RNA provided direct evidence that newly made ncRNAs were selectively encapsidated (Eckwahl et al. 2015). Specifically, 5′ and/or 3′ extended precursors to specific tRNAs, small nuclear RNAs (snRNAs) and small nucleolar RNAs (snoRNAs) were all strikingly enriched in virions, compared with the mature forms of these RNAs (Eckwahl et al. 2015).
Although many packaged RNAs, including 7SL, U6 snRNA, and pre-tRNAs, undergo ribonucleoprotein (RNP) assembly and/or RNA end maturation within nuclei (Hopper 2006), MLV recruits at least some of these RNAs in cytoplasm. Experiments in which siRNAs were used to deplete specific nuclear transport receptors revealed that packaging of pre-tRNAs and U6 snRNA by MLV required the export receptor Exportin-5 (XPO5) (Eckwahl et al. 2015). Moreover, depleting the cytoplasmic exoribonuclease DIS3L2 together with the nuclease complex known as the RNA exosome caused aberrant forms of several ncRNAs to accumulate in cells and virions, suggesting MLV recruits these RNAs from a pathway in which excess and/or aberrant newly made ncRNAs are exported to the cytoplasm for degradation (Eckwahl et al. 2015). For precursors to U2 spliceosomal snRNAs, which undergo nuclear export and re-import as part of their normal biogenesis (Matera and Wang 2014), virion packaging decreased when the snRNA-specific export adaptor PHAX was depleted and increased when snurportin, which adapts the immature snRNPs for nuclear re-import, was depleted (Eckwahl et al. 2015). Together, these studies indicated that MLV packages at least some newly made ncRNAs from the cytoplasm, in competition with their normal biogenesis, RNP assembly, and decay pathways.
To determine the extent to which the findings with MLV can be generalized to a complex retrovirus, we carried out high-throughput sequencing of RNA isolated from HIV-1 virions generated in an infected T cell line. Although 7SL RNA is the most abundant packaged host RNA, HIV-1 also packages large numbers of transcripts derived from truncated and divergent repeat elements and intronic sequences. Consistent with selective packaging of newly made RNAs, several precursors to specific tRNAs and spliceosomal snRNAs are enriched in virions, as are transcripts from ribosomal DNA (rDNA) intergenic spacer (IGS) regions. Since depleting the export receptor XPO5 decreases packaging of an intron-containing pre-tRNA, HIV-1 also packages at least some nascent ncRNAs from the cytoplasm. Our characterization of the HIV-1 “RNA packageome” confirms that retroviruses package newly made RNAs and sets the stage for elucidating the functional relevance of these RNAs to the HIV-1 lifecycle.
RESULTS
Characterizing the HIV-1 RNA packageome
To examine virion RNAs, we purified full-length infectious HIV-1 and used a cDNA synthesis strategy that captures a wide variety of RNAs in a single library (Eckwahl et al. 2015). To this end, HEK293T cells were transfected with the full-length molecular clone pNL4-3 (Adachi et al. 1986) and the resulting virus-containing media used to infect CEM-SS cells, a T lymphoblastoid cell line that has long been used to study HIV-1 replication (Nara et al. 1987). After chronic infection was established, virus was harvested from CEM-SS culture media and purified on iodixanol gradients. Virus-containing gradient fractions were identified by the presence of HIV-1 genomic RNA (gRNA) and 7SL RNA (Fig. 1A). To allow recovery of a broad range of RNAs, including mRNAs and ncRNAs with end modifications that prevent linker ligation, we fragmented the RNA before reverse transcription (Eckwahl et al. 2015). Although this cDNA strategy does not recover short (<50 nt) RNAs, an analysis of miRNA packaging by HIV-1 was reported (Schopman et al. 2012). As a control, we prepared cDNA from uninfected media that was harvested and fractionated in parallel (Fig. 1A).
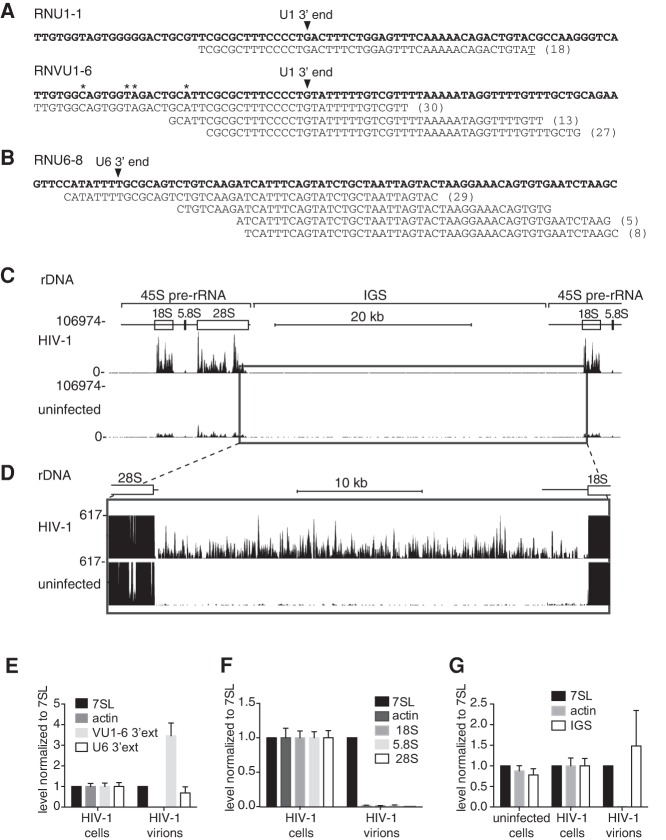
FIGURE 1.
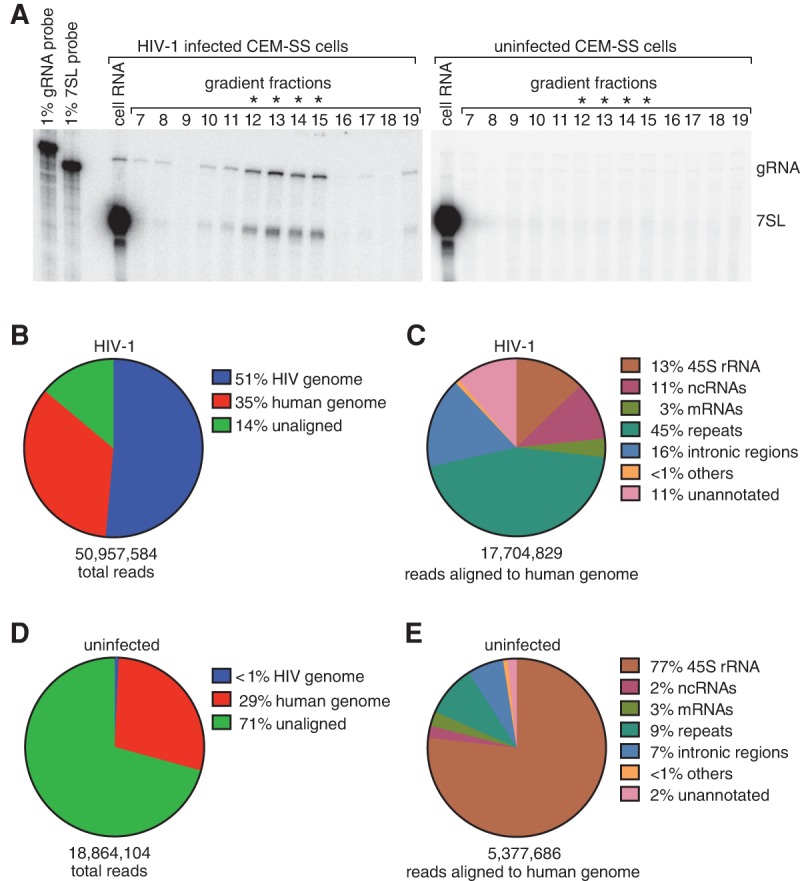
Characterizing the RNAs packaged by HIV-1. (A) After harvesting and concentrating by sedimentation, virions from HIV-infected (left panel) and control uninfected (right) CEM-SS cells were fractionated in iodixanol gradients. HIV-1-containing fractions were identified using RNase protection to detect HIV gRNA and 7SL RNA. Asterisks represent fractions used to prepare cDNA for RNA-seq. (B) Sequencing reads in the HIV-1 data set were aligned sequentially to the HIV-1 and human (hg19) genomes. The fraction of reads in each category is shown. (C) Overview of the alignment of reads from the HIV-1 data set to the human genome. RNAs in the “others” category include pseudogene transcripts and mitochondrial rRNA. (D) Alignment of reads from the uninfected control data set to the HIV and human genomes. (E) Overview of the alignment of reads from the control data set to the human genome.
Bioinformatic analyses of the virion data set revealed that ∼51% of reads mapped to the HIV-1 genome while ∼35% mapped to the human genome (Fig. 1B). Of the reads that mapped to the human genome, the largest categories were repeat element-derived RNAs (such as long interspersed elements [LINEs] and short interspersed elements [SINEs]), rRNAs, ncRNAs, and RNAs from intronic regions (Fig. 1C). Consistent with reports that mRNAs are minor virion components (Onafuwa-Nuga et al. 2006; Rulli et al. 2007), only 3% of reads derived from mRNA exons (Fig. 1C). Blasting of some of the virion-derived reads that failed to align to either the human or HIV-1 genome revealed that they mostly consisted of HIV-1 or human sequence containing more than the allowed number of nucleotide mismatches. In contrast, 71% of reads from the control library failed to align to either HIV-1 or human genomes and were largely bacterial in origin. Moreover, those reads from the control library that aligned to the human genome were largely derived from rRNA, a common contaminant (Fig. 1D,E).
7SL is the major host cell RNA in virions
To identify the most abundant RNAs in our data set, we used transcripts per million (TPM) to normalize for RNA length (Li et al. 2010; Patro et al. 2014). Using this metric, 7SL was the most abundant packaged RNA, with a TPM that was sixfold higher than that of HIV gRNA (Supplemental Table S1), consistent with the estimate that 7SL is packaged in sevenfold molar excess of gRNA (Onafuwa-Nuga et al. 2006). In addition to 7SL, we detected other host RNAs reported to be packaged, including 5S rRNA (Tian et al. 2007), U1 snRNA (Khan et al. 2007), U6 snRNA (Houzet et al. 2007), and Y RNAs (Khan et al. 2007; Tian et al. 2007). We also detected RNAs not reported previously, including Alu RNAs, LINEs, the vtRNA1-1 RNA subunit of cytoplasmic vault RNPs (Kickhoefer et al. 1998), and the primate-specific snaR-A RNA (Parrott et al. 2011). All these RNAs appeared to be packaged in far lower amounts than 7SL RNA (Supplemental Table S1). A biological replicate of the HIV-1 library yielded similar results (Supplemental Table S1). However, although the primer tRNA-Lys3/UUU is present in eight copies per virion (Huang et al. 1994), it was not abundant in our cDNA. This is consistent with reports that tRNAs, which are highly structured and contain modifications that can block reverse transcription, are poorly represented in most cDNA (Zheng et al. 2015). Short RNAs (such as tRNAs) may also be underrepresented in our protocol, since they are more likely to be fragmented to <50 nt.
To both confirm the RNA-seq data and determine the extent to which specific RNAs are selectively encapsidated, we performed Northern blotting. We compared the level of each RNA in virions with its level in an arbitrary amount of total cellular RNA. To allow comparisons between different RNAs, the enrichment in virions was normalized to 7SL RNA (Onafuwa-Nuga et al. 2005; Eckwahl et al. 2015). In contrast to MLV, where several host ncRNAs, such as Y RNAs and the mouse vault RNA, are similar to 7SL in virion enrichment (Onafuwa-Nuga et al. 2005; Eckwahl et al. 2015), all examined RNAs were far less enriched than either 7SL or the primer tRNALys. For example, although 5S rRNA and the U1 and U6 spliceosomal snRNAs are all abundant in cells, these RNAs were only detected in virions on long exposures of the blots and were at least 100-fold less enriched as 7SL (Fig. 2A,B and not shown). Similarly, although we could confirm the presence of vtRNA1-1 and snaR-A in virions by Northern blotting, these RNAs were at least 10-fold less enriched than 7SL (Fig. 2B). The four human Y RNAs were also poorly recruited, as hY1 and hY5 were not detected in virions and hY3 and hY5 were not enriched over their levels in uninfected cell media (Fig. 2C). Thus, although Y RNAs and vault RNA are both stoichiometric components of MLV virions (Garcia et al. 2009; Eckwahl et al. 2015) and U6 snRNA is present in 1 copy per RSV virion (Giles et al. 2004), these ncRNAs do not appear to be major components of our HIV-1 virions.
FIGURE 2.
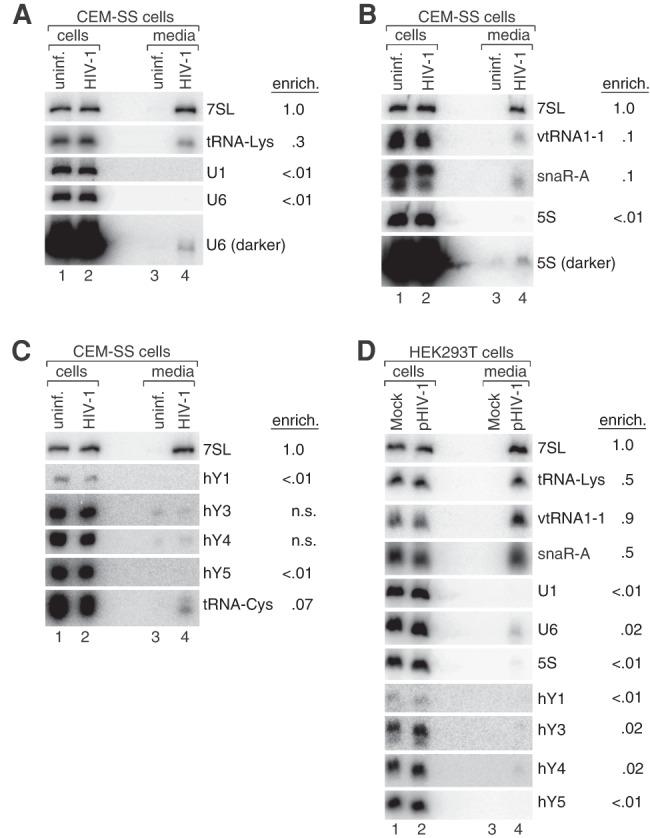
Most host cell ncRNAs are less enriched in virions than 7SL RNA. (A–D) Northern blotting was used to compare the levels of specific ncRNAs in virions (lane 4) with their levels in an arbitrary amount of cellular RNA (lane 2). As controls, we examined RNA from uninfected cells (lane 1) and from the culture media of these cells (lane 3). To facilitate comparisons between RNAs, the enrichment of each RNA was normalized to that of 7SL RNA. (A–C) Analyses of virus produced in infected and uninfected CEM-SS cells. (D) Analyses of virus produced by transfecting HEK293T cells with pNL4-3.
Because several of these ncRNAs were detected previously in HIV-1 virions produced by transfecting HEK293T cells with proviral clones (Houzet et al. 2007; Khan et al. 2007; Tian et al. 2007; Bach et al. 2008; Apolonia et al. 2015), we determined whether encapsidation of the RNAs increased under these conditions. After transfecting HEK293T cells with pNL4-3 and purifying the resulting virus, Northern blotting revealed that vtRNA1-1 and snaR-A were both more selectively packaged in these virions than in CEM-SS cell-derived virions (Fig. 2D). However, all other examined ncRNAs, including U1, U6, and 5S and the four Y RNAs, remained far less virion-enriched than 7SL. Thus, although the selectivity with which specific ncRNAs are encapsidated can vary when HIV-1 assembles in different cell types, 7SL RNA is the only examined ncRNA that is highly enriched in virions from both cell lines.
HIV-1 encapsidates transcripts from divergent and truncated repeat elements
Since reads derived from repeat element transcripts were abundant in our library (Fig. 1C), we determined which classes of elements were most represented. The largest number of reads derived from long interspersed elements (LINEs); however, other classes such as short interspersed elements (SINEs), long terminal repeat (LTR) retrotransposons, satellites, and DNA transposon transcripts were also present (Fig. 3A). Using TPM to normalize for RNA length (Supplemental Table S1), the major repeat RNAs were transcripts of Alu and LINE-1 (L1) elements. Human genomes contain more than 1 million Alu and 0.5 million L1 elements; however, most are not active in retrotransposition (Roy-Engel 2012). Although the two oldest Alu subfamilies (Alu J and AluS) can be active transcriptionally, only members of the youngest subfamily (AluY) are associated with insertions that cause human disease (Roy-Engel 2012). In the case of L1s, most genomic copies are truncated and/or contain inactivating mutations, and only 80–100 elements are capable of retrotransposition (Hancks and Kazazian 2012).
FIGURE 3.
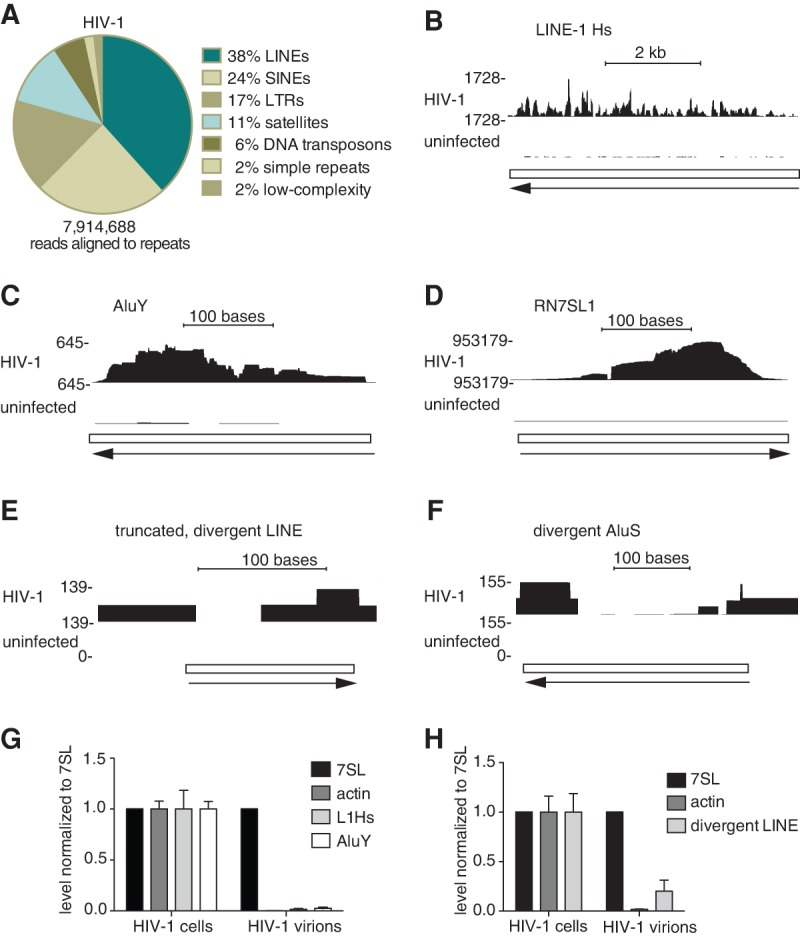
Packaging of transcripts from repetitive elements. (A) Composition of repeat element-derived reads in the HIV-1 data set. (B–D) Reads aligning to a full-length L1Hs (B; chr8:73787793–73793823), to a consensus AluY (C; chr14:31943010–31943320) and to RN7SL1, one of two genes encoding 7SL RNA (D; chr14:50,053,298–50,053,597) were visualized with the University of California at Santa Cruz (UCSC) genome browser. Arrows, transcription direction. Y-axis, read depth. (E,F) Reads aligning to a truncated and divergent LINE element (E; chr17:70,697,347–70,697,477) and a divergent AluSc element (F; chr2:141,573,687–141,573,984) were visualized using the UCSC genome browser. (G,H) RT-qPCR was used to determine the enrichment of consensus L1Hs and AluY transcripts (G) and the divergent LINE (H) in virions, compared to 7SL RNA. Virions were produced in CEM-SS cells. Actin, a nonselectively packaged mRNA (Onafuwa-Nuga et al. 2006), was a negative control.
Since our data set contained many reads derived from AluY elements and the youngest L1 elements, the human-specific L1s (L1Hs) (Supplemental Table S1), we determined if full-length, potentially active RNAs could be encapsidated selectively. First, to establish if reads covering the entire length of these elements were present, we mapped reads to the human genome using conditions where each read can align to all complementary sites (“multimapping”; see Materials and Methods). This revealed that our sequencing reads covered the full 6 kb of an active L1Hs element (Dombroski et al. 1993), while others mapped throughout a consensus AluY element (Fig. 3B,C). However, compared to the ∼1.8 million reads mapping to the two 7SL genes (Fig. 3D), relatively few reads mapped to these potentially active repeat elements, suggesting that full-length L1Hs and AluY RNAs were not abundant in virions. Instead, most LINE- and Alu-derived reads mapped to discrete sites on thousands of divergent and often truncated LINE and Alu elements (e.g., Fig. 3E,F).
We used reverse transcription followed by quantitative PCR (RT-qPCR) to determine whether L1 and Alu transcripts were encapsidated selectively. Both L1 and AluY transcripts were only slightly more enriched in virions than actin mRNA, which is encapsidated nonselectively (Onafuwa-Nuga et al. 2006), and were far less enriched than 7SL (Fig. 3G). Attempts to detect transcripts from divergent and truncated elements were hampered by their low abundance in cells and by the fact that the nucleotide changes were mostly insufficient to unambiguously detect transcripts from specific loci. However, transcripts from at least one divergent LINE locus (Fig. 3E) were approximately 13-fold more enriched in virions than consensus L1Hs transcripts, although they were still fivefold less enriched than 7SL (compare Fig. 3H with 3G). Thus, while potentially active LINE and SINE transcripts are not selectively encapsidated, transcripts from at least some divergent and inactive elements may be enriched in virions.
HIV-1 packages pre-snRNAs, transcripts from rDNA intergenic spacers, and pre-tRNAs
Similar to MLV (Eckwahl et al. 2015), some reads in our HIV-1 data set were derived from ncRNA precursors that are usually rare in cells. For example, some reads originated from 3′ extended forms of the spliceosomal U1 snRNA. Human U1 snRNAs are encoded by a multicopy gene family located on the short arm of chromosome 1, while some pseudogenes and variant genes cluster on the long arm at 1q12-21 (Lindgren et al. 1985; O'Reilly et al. 2013). Our data set contained reads derived from both “wild-type” U1 genes (RNU1-1) and a variant locus (RNvU6-8) (Fig. 4A). Interestingly, both the wild-type and variant U1 RNAs appeared to be packaged as 3′ extended pre-snRNAs. Normally, following RNA polymerase II termination, nascent U1 RNAs are cleaved by the “integrator complex” to form precursors with ∼10 additional nt (Matera and Wang 2014). For both the wild-type and variant U1 transcripts, reads containing at least 30 nt beyond the mature 3′ end were present in virions, indicating they were derived from RNAs that had not undergone this cleavage event. We also detected reads derived from the U6 spliceosomal snRNA, which is transcribed by RNA polymerase III (Mroczek and Dziembowski 2013), that extend more than 60 nt past the termination site (arrow, Fig. 4B).
FIGURE 4.
HIV-1 selectively encapsidates pre-snRNAs and rDNA IGS transcripts. (A) Reads derived from transcription of the major U1 snRNA genes (RNU1-1; top group) and a variant locus (RNVU1-6; bottom group). The RNU1-1 and RNVU1-6 gene sequences are in bold type, with the mature U1 3′ end indicated by an arrow. Parentheses, number of identical reads. Asterisks represent nucleotides that differ from the wild-type U1 snRNA. Reads mapping to the RNU1-1 locus contain a nontemplated T at the 3′ end (underline). The extra T could derive from oligouridylation of pre-U1 snRNAs by terminal U transferases or could reflect the fact that only a fraction of the tandemly repeated U1 loci are part of the current human genome assembly. (B) Reads aligning to the U6 locus at chr14: 32,672,370–32,672,475. The RNU6-8 genomic sequence is on top (bold type). The arrow indicates the mature 3′ end. (C) Reads aligned to rDNA (chrUn_gl000220:105,424–157,100). The positions of two 45S rRNA precursors are indicated. The boxed region is shown in more detail in D. (D) Reads mapping to the rDNA IGS. To visualize the IGS transcripts, the read depth from the mature rRNAs was truncated. (E–G) RT-qPCR was used to determine the enrichment of 3′-extended U1 and U6 snRNAs (E), mature rRNAs (F), and IGS transcripts in virions produced in CEM-SS cells (G). All values were normalized to 7SL RNA. Actin, a randomly encapsidated mRNA.
We also obtained evidence that HIV-1 packages nascent transcripts from the rDNA locus. In humans, the ∼400 rRNA genes are organized in tandem repeats that encode 45S pre-rRNA, a ∼13 kb RNA polymerase I (Pol I) transcript that is matured to become 18S, 5.8S, and 28S rRNAs, followed by a ∼30-kb intergenic spacer (IGS) (Fig. 4C; Grummt et al. 1985). Although Pol I normally terminates several hundred bp downstream from the 28S rRNA 3′ end, read-through transcripts extending through the IGS become prominent during heat shock and several other forms of stress (Labhart and Reeder 1987; Parker and Bond 1989; Audas et al. 2012). Remarkably, in addition to many reads mapping to mature rRNAs, reads mapping throughout the IGS were detected in our virion data set but not in media from uninfected cells (Fig. 4D).
To determine whether the 3′ extended U snRNAs and rDNA IGS transcripts were selectively encapsidated, we performed RT-qPCR. Although mature U1 and U6 snRNAs were not enriched in virions (Fig. 2A), the 3′-extended form of the variant U1 snRNA was nearly 3.5-fold more enriched than 7SL RNA, while 3′ extended U6 snRNAs were nearly as enriched as 7SL RNA (Fig. 4E). Similarly, although mature 18S, 5.8S, and 28S rRNA are not enriched in virions (Fig. 4F), IGS transcripts are comparable to 7SL RNA in their virion enrichment (Fig. 4G). Notably, the levels of IGS transcripts were not significantly different between infected and uninfected cells, indicating these transcripts do not increase during HIV-1 infection (Fig. 4G).
HIV also selectively packages specific pre-tRNAs. As described for MLV (Eckwahl et al. 2015), some tRNA-derived reads in our data set contained 5′ or 3′ extensions and/or introns. Using Northern blotting, we demonstrated that the precursor form of tRNA-Ser-UGA (containing 5′ and 3′ extensions) was approximately one-third as enriched as 7SL RNA and far more enriched in virions than the mature tRNA (Fig. 5A). We also detected several intron-containing pre-tRNAs in virions produced by transfecting HEK293T cells with a HIV expression plasmid in which env was replaced by a puromycin cassette (pHIV-GPP; Fig. 5B–D). Although the pre-tRNAs are far less abundant in cellular RNA than the mature tRNAs, the primary transcripts (containing introns and 5′ and 3′ extensions) encoding tRNA-Ile-UAU, tRNA-Tyr-GUA, and Leu-CAA were all more selectively packaged than their respective mature versions (Fig. 5B–D). Consistent with encapsidation shortly after synthesis, the primary transcripts of tRNA-Tyr-GUA and tRNA-Leu-CAA were also packaged more selectively than processing intermediates that had undergone splicing but not end maturation (Fig. 5C,D). Additionally, although the cleaved tRNA-Leu-CAA 3′ half was below the level of detection in cellular RNA, it was present in virions (Fig. 5D).
FIGURE 5.
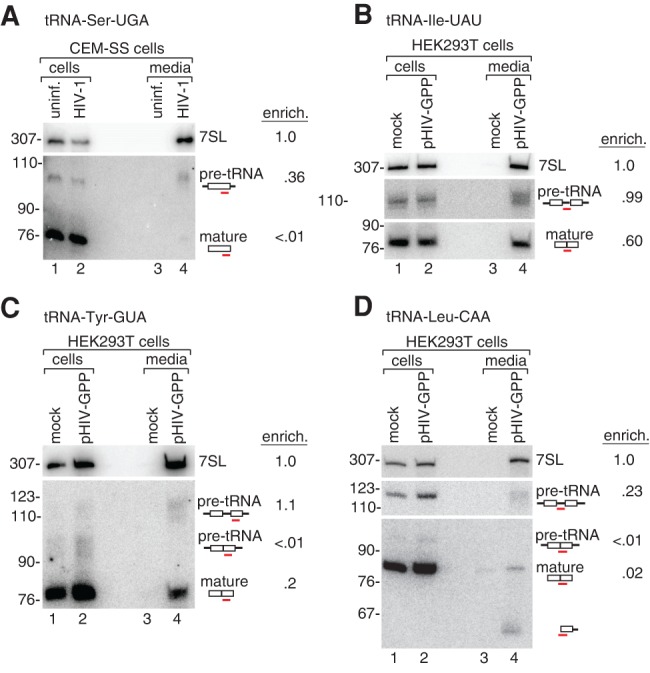
HIV-1 packages pre-tRNAs. (A) To determine whether specific pre-tRNAs were encapsidated selectively, Northern blotting was used to compare the levels of the precursor and mature forms of tRNA-Ser-UGA in RNA from HIV-infected CEM-SS cells (lane 2) and virions (lane 4). As controls, RNA from uninfected cells (lane 1) and culture media (lane 3) was examined. The probe detects sequences present in both the mature tRNA and its precursor. Virion enrichment was normalized to 7SL RNA. (B–D) Northern blotting was used to determine the levels of precursor and mature forms of tRNA-Ile-UAU (B), tRNA-Tyr-GUA (C), and tRNA-Leu-CAA (D) in cells (lane 2) and virions (lane 4) produced in HEK293T cells after transfection with a proviral plasmid lacking env. RNA from mock-transfected cells (lane 1) and media (lane 3) was also examined. Intron-containing forms of pre-tRNA-Ile-UAU (B) and pre-tRNA-Leu-CAA (D) were detected with oligonucleotides complementary to their intervening sequences (middle panels), while spliced forms were detected with oligonucleotides complementary to the spliced anticodon stem–loop (bottom panels). As the tRNA-Tyr-GUA probe is complementary to 3′ exon sequences, it detects both unspliced and spliced tRNAs. Consistent with a report that mature tRNA-Ile-UAU is highly enriched in HIV virions relative to its cellular concentration (Pavon-Eternod et al. 2010), we found that tRNA-Ile-UAU was more selectively incorporated into virions than the other mature tRNAs examined.
Together with reports that 7SL is packaged by HIV-1 without SRP54 (Onafuwa-Nuga et al. 2006), and that overexpression of SRP19 prevents 7SL encapsidation (Wang et al. 2007; Bach et al. 2008; Apolonia et al. 2015), our finding that HIV-1 selectively packages pre-snRNAs, nascent rDNA transcripts, and pre-tRNAs indicates that HIV-1, similar to MLV (Eckwahl et al. 2015), preferentially encapsidates newly made ncRNAs.
HIV packages some pre-tRNAs following nuclear export
Although HIV-1 assembly initiates in the cytoplasm and takes place largely at the plasma membrane (Kutluay and Bieniasz 2010), many packaged RNAs, including U6, nascent rDNA transcripts, and pre-tRNAs, normally reside in mammalian nuclei (Hopper 2006). Since MLV packages pre-tRNAs and U6 from a pathway in which newly made ncRNAs are exported to the cytoplasm for degradation (Eckwahl et al. 2015), we determined whether nuclear export was required for encapsidation of these same RNAs by HIV-1. Using siRNAs, we depleted HEK293T cells of Exportin-5 (XPO5), which functions as an export receptor for pre-miRNAs and other ncRNAs with a short 3′ overhang (Lee et al. 2011), and is required for packaging of pre-tRNAs and U6 snRNAs by MLV (Eckwahl et al. 2015). As a control, we depleted Exportin-T, the major export receptor for mature tRNA (Hopper 2006). After confirming that mRNA depletion was effective, we transfected the cells with the proviral plasmid lacking env, harvested the resulting virions and used Northern blotting and RT-qPCR to detect specific RNAs (Fig. 6A,B).
FIGURE 6.
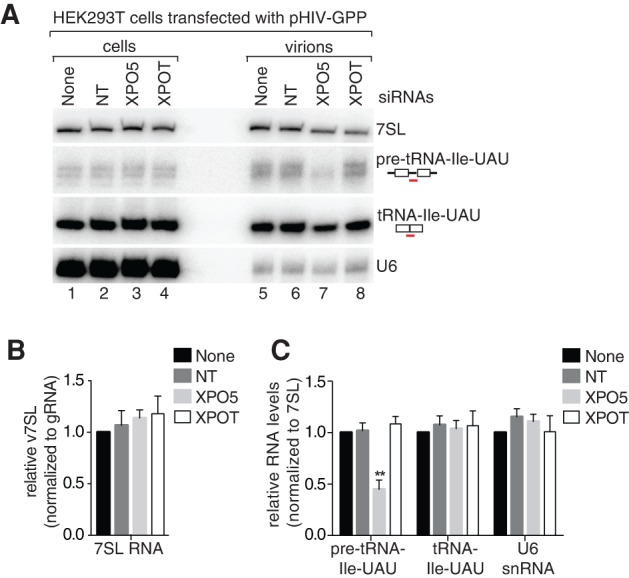
Packaging of some pre-tRNAs requires nuclear export. (A) HEK293T cells were transfected with siRNAs targeting XPOT, XPO5, or with control nontargeting (NT) RNAs. The following day, cells were transfected with pHIV-GPP and virions harvested 24–48 h afterward. RNA extracted from cells (lanes 1–4) and virus (lanes 5–8) was subjected to Northern blotting to detect the indicated RNAs. (B) The levels of 7SL RNA in virions were determined by RT-qPCR. Normalization was to HIVgRNA. Data are mean values from three biological replicates ± SEM. (C) Quantitation of A. Data are mean values from three biological replicates ± SEM. Loading was normalized to 7SL RNA. (*) P ≤ 0.05, (**) P ≤ 0.01, (***) P ≤ 0.001 (Student's t-test).
Similar to the results with MLV (Eckwahl et al. 2015), the ratio of 7SL RNA to gRNA in HIV-1 virions was unchanged upon depleting XPO5 or XPOT (Fig. 6A,B). Northern blotting revealed that packaging of the most easily detected pre-tRNA, pre-tRNA-Ile-UAU, decreased 2.2-fold when XPO5 was depleted (Fig. 6A,C). Attempts to determine if other pre-tRNAs decreased in virions were hampered by their low abundance. Surprisingly, while the amount of U6 snRNA packaged by MLV decreased twofold when XPO5 was depleted from infected mouse cells (Eckwahl et al. 2015), U6 snRNA packaging by HIV-1 was unaffected (Fig. 6A,C). Thus, either U6 snRNA utilizes different and/or redundant export factors in human cells, or HIV-1 acquires this RNA in nuclei. Nonetheless, our result that pre-tRNA-Ile-UAU packaging is reduced when XPO5 is depleted reveals that packaging of at least some pre-tRNAs occurs after nuclear export.
DISCUSSION
In this study, we used high-throughput sequencing to obtain a more complete description of the RNAs packaged by HIV-1. Using this approach, the major cellular RNAs in virions consist of 7SL and transcripts from repeat elements. Additionally, several RNA species that are rare in cells, such as pre-tRNAs and pre-snRNAs, were highly enriched in virions. Together with reports that 7SL RNA is recruited by both MLV and HIV-1 before assembling into mature SRP (Onafuwa-Nuga et al. 2006; Wang et al. 2007; Bach et al. 2008; Apolonia et al. 2015; Eckwahl et al. 2015) and that many nascent ncRNAs are enriched in MLV virions (Garcia et al. 2009; Eckwahl et al. 2015), our data suggest that the encapsidation of newly made RNAs may be a general property of retroviruses.
One theme that emerges from these studies is that forms of RNAs that are rare and difficult to detect in cells are strikingly enriched in virions. Some RNAs, such as U1 snRNAs that have not undergone 3′ cleavage, have only been detected previously when snRNP assembly was blocked with protein synthesis inhibitors (Yong et al. 2010) or the catalytic subunit of the integrator complex was depleted (Baillat et al. 2005). Other RNAs, such as transcripts from the rDNA IGS, have primarily been detected in response to environmental stressors such as heat-shock and acidosis (Labhart and Reeder 1987; Parker and Bond 1989; Audas et al. 2012). Our finding that these unusual RNAs do not increase in virus-infected cells (Fig. 4) underscores the selectivity of their virion packaging. Moreover, if 7SL RNA is indeed recruited into virions before assembling with most core SRP proteins (Onafuwa-Nuga et al. 2006; Wang et al. 2007; Bach et al. 2008; Apolonia et al. 2015; Eckwahl et al. 2015), retroviruses encapsidate a SRP biogenesis intermediate that has not otherwise been detected in cells.
Why might retroviruses package these very rare RNAs? There is much evidence that RNA binding promotes Gag oligomerization (Campbell and Vogt 1995; Rein et al. 2011; Hendrix et al. 2015); however, the identities of the responsible RNAs remain unknown. As noted previously (Eckwahl et al. 2015; Eckwahl et al. 2016), newly synthesized RNAs that have not bound their usual protein partners may be more likely than components of mature RNPs to have the 15–16 nt of protein-free RNA that is needed for Gag multimerization in vitro (Campbell and Rein 1999; Ma and Vogt 2002). In support of this idea, crosslinking experiments performed in living cells revealed that the major Gag-interacting RNAs in cytosol are host ncRNAs, particularly tRNAs and 7SL RNA (Kutluay et al. 2014). Although the tRNA crosslinks were largely dependent on Matrix, a Gag domain dispensable for forming oligomers, crosslinks to most other host RNAs were strongly reduced by mutations in nucleocapsid (NC), a domain important for oligomer formation (Kutluay et al. 2014; Hendrix et al. 2015).
The packaged RNAs could also have other roles. Encapsidation of the host cell restriction factors APOBEC3F and APOBEC3G into HIV-1 virions requires RNA, and it was shown recently that many cellular RNAs are capable of mediating packaging (Apolonia et al. 2015). Since, in vitro, APOBEC3G requires at least 10 nt of G-rich single-stranded RNA to form ternary complexes with NC (Bogerd and Cullen 2008), newly made ncRNAs could be the RNAs that have sufficient stretches of protein-free single-stranded RNA to confer packaging of APOBEC3 proteins in vivo. Packaging of newly synthesized RNAs could also potentially impact infectivity. For example, if, as we showed for MLV (Eckwahl et al. 2015), many host RNAs are recruited from a cytoplasmic decay pathway, these RNAs may have the accessible, protein-free 3′ ends that allow degradation by 3′ to 5′ exoribonucleases (Wolin et al. 2012). In this case, nascent RNAs could potentially function as decoys to prevent cytosolic exoribonucleases from degrading incoming gRNA. Finally, packaged RNAs that are RNA polymerase III transcripts, such as 7SL and pre-tRNAs, should contain 5′ triphosphates and thus could theoretically interact with cytosolic innate immune sensors such as RIG-I.
Remarkably, about half the cellular RNA packaged by HIV-1 derived from repeat sequences, particularly LINEs and SINEs. In contrast, although MLV packaged a similar fraction of repeat-derived RNAs (Eckwahl et al. 2015), most transcripts were members of the VL30 family of LTR retrotransposons, a class of endogenous retroviruses long known to be packaged by type C retroviruses (French and Norton 1997), and LINEs were not prominent. As there are considerable differences in the expression of repeat element transcripts between mice and humans, and also between different human tissues and cell types (Faulkner et al. 2009), we do not know if the observed variations in encapsidated RNAs result from differences in retrotransposon expression between the various producer cells or are inherent to the viruses studied. Because of difficulties in unambiguously detecting transcripts from divergent and truncated Alu and LINE elements, we also do not know the extent to which these transcripts are selectively recruited into virions, rather than nonselectively encapsidated. However, if transcripts from divergent retrotransposons resemble newly synthesized host ncRNAs in having stretches of accessible protein-free single-stranded RNA, they could function similarly to assist Gag multimerization.
Although some pre-tRNAs are packaged after nuclear export, we do not yet know where HIV-1 encounters other RNAs. SRP normally assembles with five of the six SRP subunits in nucleoli, after which this partly assembled complex is exported to the cytoplasm by CRM1 in yeast and by XPO5 in vertebrates (Politz et al. 2000; Grosshans et al. 2001; Takeiwa et al. 2015). As XPO5 depletion did not affect 7SL packaging (Fig. 6), one possibility is that HIV-1 encounters 7SL and other nascent RNAs in nuclei to form RNPs that are exported independently of XPO5. Possibly consistent with this idea, HIV-1 Gag has been detected in nuclei under some conditions, although whether HIV-1 Gag normally traverses the nucleus remains controversial (Stake et al. 2013). Importantly, 7SL RNA packaging into HIV-1 VLPs remains efficient when gRNA is absent (Onafuwa-Nuga et al. 2006), ruling out mechanisms in which the newly made RNA accesses the cytoplasm by base-pairing with gRNA. A final possibility is that 7SL, U6, and other newly made RNAs access the cytoplasm using an export receptor we have not yet tested and/or that they can utilize multiple receptors with overlapping specificities.
Lastly, our findings that 7SL is both the most abundantly packaged host RNA and is highly enriched relative to its cellular concentration raises the question as to whether the packaged 7SL confers some advantage during HIV-1 replication. Consistent with a functional role, 7SL encapsidation by HIV-1 involves specific RNA structural features and is also saturable, since truncated 7SL RNAs containing only the “Alu domain” compete with the endogenous full-length RNA for packaging (Keene and Telesnitsky 2012). Although 7SL packaging can be reduced by ∼90% without affecting virus production or infectivity, these studies involved virus produced in HEK293T cells and infectivity assays performed in HeLa cell lines (Wang et al. 2007; Apolonia et al. 2015). Thus, it remains possible that the packaged RNA contributes to HIV-1 replication in more physiologically relevant cell types or confers an advantage during some types of environmental stress. Moreover, our finding that several other host ncRNAs, such as vault RNA and snaR-A, are nearly as enriched as 7SL in virions produced in HEK293T cells raises the question as to whether these RNAs have specific roles when HIV-1 replicates in some cell types. Our identification of the RNAs packaged by HIV-1 lays the groundwork for experiments that address these questions.
MATERIALS AND METHODS
Cells and viruses
HEK293T cells were maintained in Dulbecco's modified Eagle's medium (Invitrogen) containing 10% fetal bovine serum, 100 U/mL penicillin–streptomycin and 2 mM L-glutamine. CEM-SS cells (Nara et al. 1987) were grown in Roswell Park Memorial Institute 1640 medium (Invitrogen) supplemented with 10% fetal bovine serum and 50 µg/mL gentamicin. After harvesting, media was passed through 0.2-μm filters and stored at −80°C until use.
To produce virus for infecting CEM-SS cells, HEK293T cells were cultured to 70% confluency in 10 cm2 dishes and transfected with 3 μg pNL4-3 DNA (Adachi et al. 1986) and polyethylenimine (PEI, PolySciences) as previously described (Keene et al. 2010). Virus-containing supernatant was harvested after 48 h. Virus was quantitated using a qPCR-based reverse transcriptase (RT) assay (Vermeire et al. 2012), using HIV-1-containing media with known concentrations of CA-p24 as standards. Afterward, virus equivalent to 1 ng CA-p24 was used to infect 2 × 106 CEM-SS cells. Establishment of chronically infected cultures was monitored using the qPCR-based RT assay. After virus concentrations plateaued, virus and cells were harvested. Filtered culture media were concentrated by centrifugation through a sucrose cushion and purified on a continuous gradient of 8%–22% iodixanol (Optiprep; Sigma-Aldrich) as previously described (Eckwahl et al. 2015).
To generate full-length infectious virus from HEK293T cells, these cells were transfected with either pNL4-3 or empty vector pCR3.1 (Invitrogen) using PEI and harvested 48 h later. Viral supernatants were concentrated and gradient-purified as above. To produce noninfectious virus in HEK293T cells under BSL-2 conditions, the plasmid pHIV-GPP (called HIV-1 NL4-3 helper construct in Lu et al. [2011]), an HIV-1 proviral clone in which env was replaced by a puromycin resistance cassette, was transfected using PEI as previously described (Keene et al. 2010). Virions were harvested between 24 and 48 h after transfection with pHIV-GPP, while cells were harvested after 48 h.
Library preparation and RNA-seq
After adding ERCC (External RNA Controls Consortium) RNA Spike-In Mix (Ambion, Life Technologies), RNA was fragmented and cDNA prepared as previously described (Eckwahl et al. 2015). Libraries were amplified with 20 PCR cycles and purified using the Nucleospin gel purification kit (Machery-Nagel). Samples were quantitated using the KAPA library quantification kit (Kapa Biosystems), spiked with 20% PhiX Sequencing Control (Invitrogen), and sequenced for 75 bases on an Illumina HiSeq 2500 at the Yale Center for Genome Analysis. After demultiplexing libraries based on the index sequences, adapters were removed (FASTX-toolkit) and reads aligned using the Burroughs-Wheeler Alignment (BWA) tool (Li and Durbin 2009). After filtering reads that mapped to PhiX and ERCC controls, the remaining reads were mapped to the HIV-1 genome (AF324493.2) and then to the human hg19 assembly. BEDTools (Quinlan and Hall 2010) was used to assign reads to genomic features, while Sailfish (Patro et al. 2014) was used to determine transcripts per million (TPM) (Li et al. 2010). To examine the extent to which full-length repeat transcripts could be present, reads were multimapped using Bowtie (Langmead et al. 2009), allowing ≤3 mismatches.
RNase protection, Northern analysis, and RT-qPCR
After extracting RNA in iodixanol gradient fractions using TRIzol (Invitrogen), RNase protection was carried out to identify HIV-containing fractions. To generate riboprobes to detect HIV-1 gRNA and 7SL RNA, plasmids pSKh72a and pSKh78 were linearized with BamH1 and transcribed with T7 polymerase (Promega) in the presence of [α-32P]rCTP. The resulting riboprobes are complementary to 201 nt of HIV-1 gag (CA/sp/NC) and nts C116–C217 in the S domain of 7SL RNA. RNase protection was carried out as previously described (Garcia et al. 2009).
For Northern analyses of RNAs present in infectious HIV-1 virions, RNA was prepared from iodixanol gradient fractions as described above. For experiments analyzing RNAs packaged by noninfectious HIV-1 produced in HEK293T cells, virus was concentrated by sedimenting the filtered media through a sucrose cushion as previously described (Eckwahl et al. 2015). Afterward, RNA was extracted from the viral pellets using TRIzol. Total RNA was prepared from ∼85% confluent cells using TRIzol. Afterward, RNAs were separated in 6% polyacrylamide-8.3 M urea gels and transferred to Hybond-N (Amersham) as previously described (Eckwahl et al. 2015). Blots were hybridized with [32P]-labeled oligonucleotides as previously described (Tarn et al. 1995). Probes are listed in Supplemental Table S2.
For experiments examining ncRNA levels by RT-qPCR, RNA was treated with DNase I (Roche), primed with random hexamers (Promega) and reverse-transcribed using SuperScript III with an annealing temperature of 65°C. To measure mRNA levels after siRNA-depletions, DNase-treated RNA was reverse-transcribed using the iScript cDNA synthesis kit (Bio-Rad). qPCR was carried out using a SYBR Green real-time PCR master mix (Bio-Rad). Primers for RT-qPCR are listed in Supplemental Table S2. The primers used to detect consensus L1Hs were described (Wissing et al. 2012). The primers used to detect AluY sequences were modified from previously described primers (Macia et al. 2011).
siRNA transfections
Smartpool siRNAs for XPOT, XPO5, and nontargeting control siRNA #3 were from Dharmacon. siRNA transfections were carried out with Lipofectamine RNAiMAX (Invitrogen) and 20 nM siRNA as previously described (Eckwahl et al. 2015). After 24 h, cells were transfected with pHIV-GPP using PEI as previously described (Keene et al. 2010). Virions were harvested between 24 and 48 h after transfection with pHIV-GPP, while cells were harvested after 48 h.
DATA DEPOSITION
The sequencing data have been deposited in the NCBI Sequence Read Archive (SRA) under accession number SRP074288.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Silas Johnson for preparing HIV-1 RNA in the early stages of this work and Walther Mothes, Soyeong Sim, and Xinguo Chen for much helpful advice. M.J.E. was a National Science Foundation Graduate Research Fellow. This work was supported by National Institutes of Health Training Grant T32 GM007223 (M.J.E.), National Institutes of Health grant P50 GM103297 to A.T. and P.D.B, Collaborative Development Grant from the Center for RNA Studies (P50 GM103297), National Institutes of Health grant R21 AI116381, and a Pilot Grant from Yale Cancer Center to S.L.W.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.057299.116.
REFERENCES
- Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, Martin MA. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol 59: 284–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apolonia L, Schulz R, Curk T, Rocha P, Swanson CM, Schaller T, Ule J, Malim MH. 2015. Promiscuous RNA binding ensures effective encapsidation of APOBEC3 proteins by HIV-1. PLoS Pathog 11: e1004609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audas TE, Jacob MD, Lee S. 2012. Immobilization of proteins in the nucleolus by ribosomal intergenic spacer noncoding RNA. Mol Cell 45: 147–157. [DOI] [PubMed] [Google Scholar]
- Bach D, Peddi S, Mangeat B, Lakkaraju A, Strub K, Trono D. 2008. Characterization of APOBEC3G binding to 7SL RNA. Retrovirology 5: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillat D, Hakimi MA, Naar AM, Shilatifard A, Cooch N, Shiekhattar R. 2005. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell 123: 265–276. [DOI] [PubMed] [Google Scholar]
- Berkowitz R, Fisher J, Goff SP. 1996. RNA packaging. Curr Top Microbiol Immunol 214: 177–218. [DOI] [PubMed] [Google Scholar]
- Bishop JM, Levinson WE, Sullivan D, Fanshier L, Quintrell N, Jackson J. 1970. The low molecular weight RNAs of Rous sarcoma virus. II. The 7 S RNA. Virology 42: 927–937. [DOI] [PubMed] [Google Scholar]
- Bogerd HP, Cullen BR. 2008. Single-stranded RNA facilitates nucleocapsid: APOBEC3G complex formation. RNA 14: 1228–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell S, Rein A. 1999. In vitro assembly properties of human immunodeficiency virus type 1 Gag protein lacking the p6 domain. J Virol 73: 2270–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell S, Vogt VM. 1995. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J Virol 69: 6487–6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell S, Fisher RJ, Towler EM, Fox S, Issaq HJ, Wolfe T, Phillips LR, Rein A. 2001. Modulation of HIV-like particle assembly in vitro by inositol phosphates. Proc Natl Acad Sci 98: 10875–10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cen S, Javanbakht H, Kim S, Shiba K, Craven R, Rein A, Ewalt K, Schimmel P, Musier-Forsyth K, Kleiman L. 2002. Retrovirus-specific packaging of aminoacyl-tRNA synthetases with cognate primer tRNAs. J Virol 76: 13111–13115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlberg JE, Sawyer RC, Taylor JM, Faras AJ, Levinson WE, Goodman HM, Bishop JM. 1974. Transcription of DNA from the 70S RNA of Rous sarcoma virus. I. Identification of a specific 4S RNA which serves as primer. J Virol 13: 1126–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombroski BA, Scott AF, Kazazian HH Jr. 1993. Two additional potential retrotransposons isolated from a human L1 subfamily that contains an active retrotransposable element. Proc Natl Acad Sci 90: 6513–6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckwahl MJ, Sim S, Smith D, Telesnitsky A, Wolin SL. 2015. A retrovirus packages nascent host noncoding RNAs from a novel surveillance pathway. Genes Dev 29: 646–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckwahl MJ, Telesnitsky A, Wolin SL. 2016. Host RNA packaging by retroviruses: a newly synthesized story. MBio 7: e02025–e02015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erikson E, Erikson RL, Henry B, Pace NR. 1973. Comparison of oligonucleotides produced by RNase T1 digestion of 7 S RNA from avian and murine oncornaviruses and from uninfected cells. Virology 53: 40–46. [DOI] [PubMed] [Google Scholar]
- Faras AJ, Dahlberg JE, Sawyer RC, Harada F, Taylor JM, Levinson WE, Bishop JM, Goodman HM. 1974. Transcription of DNA from the 70S RNA of Rous sarcoma virus. II. Structure of a 4S RNA primer. J Virol 13: 1134–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner GJ, Kimura Y, Daub CO, Wani S, Plessy C, Irvine KM, Schroder K, Cloonan N, Steptoe AL, Lassmann T, et al. 2009. The regulated retrotransposon transcriptome of mammalian cells. Nat Genet 41: 563–571. [DOI] [PubMed] [Google Scholar]
- French NS, Norton JD. 1997. Structure and functional properties of mouse VL30 retrotransposons. Biochim Biophys Acta 1352: 33–47. [DOI] [PubMed] [Google Scholar]
- Garcia EL, Onafuwa-Nuga A, Sim S, King SR, Wolin SL, Telesnitsky A. 2009. Packaging of host mY RNAs by murine leukemia virus may occur early in Y RNA biogenesis. J Virol 83: 12526–12534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles K, Caputi M, Beemon KL. 2004. Packaging and reverse transcription of snRNAs by retroviruses may generate pseudogenes. RNA 10: 299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosshans H, Deinert K, Hurt E, Simos G. 2001. Biogenesis of the signal recognition particle (SRP) involves import of SRP proteins into the nucleolus, assembly with the SRP-RNA, and Xpo1p-mediated export. J Cell Biol 153: 745–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt I, Maier U, Ohrlein A, Hassouna N, Bachellerie JP. 1985. Transcription of mouse rDNA terminates downstream of the 3′ end of 28S RNA and involves interaction of factors with repeated sequences in the 3′ spacer. Cell 43: 801–810. [DOI] [PubMed] [Google Scholar]
- Hancks DC, Kazazian HH Jr. 2012. Active human retrotransposons: variation and disease. Curr Opin Genet Dev 22: 191–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix J, Baumgartel V, Schrimpf W, Ivanchenko S, Digman MA, Gratton E, Krausslich HG, Muller B, Lamb DC. 2015. Live-cell observation of cytosolic HIV-1 assembly onset reveals RNA-interacting Gag oligomers. J Cell Biol 210: 629–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper AK. 2006. Cellular dynamics of small RNAs. Crit Rev Biochem Mol Biol 41: 3–19. [DOI] [PubMed] [Google Scholar]
- Houzet L, Paillart JC, Smagulova F, Maurel S, Morichaud Z, Marquet R, Mougel M. 2007. HIV controls the selective packaging of genomic, spliced viral and cellular RNAs into virions through different mechanisms. Nucleic Acids Res 35: 2695–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Mak J, Cao Q, Li Z, Wainberg MA, Kleiman L. 1994. Incorporation of excess wild-type and mutant tRNALys3 into human immunodeficiency virus type 1. J Virol 68: 7676–7683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene SE, Telesnitsky A. 2012. cis-Acting determinants of 7SL RNA packaging by HIV-1. J Virol 86: 7934–7942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene SE, King SR, Telesnitsky A. 2010. 7SL RNA is retained in HIV-1 minimal virus-like particles as an S-domain fragment. J Virol 84: 9070–9077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MA, Goila-Gaur R, Opi S, Miyagi E, Takeuchi H, Kao S, Strebel K. 2007. Analysis of the contribution of cellular and viral RNA to the packaging of APOBEC3G into HIV-1 virions. Retrovirology 4: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kickhoefer VA, Rajavel KS, Scheffer GL, Dalton WS, Scheper RJ, Rome LH. 1998. Vaults are up-regulated in multidrug-resistant cancer cell lines. J Biol Chem 273: 8971–8974. [DOI] [PubMed] [Google Scholar]
- Kutluay SB, Bieniasz PD. 2010. Analysis of the initiating events in HIV-1 particle assembly and genome packaging. PLoS Pathog 6: e1001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutluay SB, Zang T, Blanco-Melo D, Powell C, Jannain D, Errando M, Bieniasz PD. 2014. Global changes in the RNA binding specificity of HIV-1 gag regulate virion genesis. Cell 159: 1096–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labhart P, Reeder RH. 1987. Heat shock stabilizes highly unstable transcripts of the Xenopus ribosomal gene spacer. Proc Natl Acad Sci 84: 56–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Jiko C, Yamashita E, Tsukihara T. 2011. Selective nuclear export mechanism of small RNAs. Curr Opin Struct Biol 21: 101–108. [DOI] [PubMed] [Google Scholar]
- Levin JG, Grimley PM, Ramseur JM, Berezesky IK. 1974. Deficiency of 60 to 70S RNA in murine leukemia virus particles assembled in cells treated with actinomycin D. J Virol 14: 152–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Ruotti V, Stewart RM, Thomson JA, Dewey CN. 2010. RNA-Seq gene expression estimation with read mapping uncertainty. Bioinformatics 26: 493–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgren V, Bernstein LB, Weiner AM, Francke U. 1985. Human U1 small nuclear RNA pseudogenes do not map to the site of the U1 genes in 1p36 but are clustered in 1q12-q22. Mol Cell Biol 5: 2172–2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K, Heng X, Garyu L, Monti S, Garcia EL, Kharytonchyk S, Dorjsuren B, Kulandaivel G, Jones S, Hiremath A, et al. 2011. NMR detection of structures in the HIV-1 5′-leader RNA that regulate genome packaging. Science 334: 242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma YM, Vogt VM. 2002. Rous sarcoma virus Gag protein-oligonucleotide interaction suggests a critical role for protein dimer formation in assembly. J Virol 76: 5452–5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macia A, Munoz-Lopez M, Cortes JL, Hastings RK, Morell S, Lucena-Aguilar G, Marchal JA, Badge RM, Garcia-Perez JL. 2011. Epigenetic control of retrotransposon expression in human embryonic stem cells. Mol Cell Biol 31: 300–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann R, Mulligan RC, Baltimore D. 1983. Construction of a retrovirus packaging mutant and its use to produce helper-free defective retrovirus. Cell 33: 153–159. [DOI] [PubMed] [Google Scholar]
- Matera AG, Wang Z. 2014. A day in the life of the spliceosome. Nat Rev Mol Cell Biol 15: 108–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mroczek S, Dziembowski A. 2013. U6 RNA biogenesis and disease association. Wiley Interdiscip Rev RNA 4: 581–592. [DOI] [PubMed] [Google Scholar]
- Muriaux D, Mirro J, Harvin D, Rein A. 2001. RNA is a structural element in retrovirus particles. Proc Natl Acad Sci 98: 5246–5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nara PL, Hatch WC, Dunlop NM, Robey WG, Arthur LO, Gonda MA, Fischinger PJ. 1987. Simple, rapid, quantitative, syncytium-forming microassay for the detection of human immunodeficiency virus neutralizing antibody. AIDS Res Hum Retroviruses 3: 283–302. [DOI] [PubMed] [Google Scholar]
- O'Carroll IP, Crist RM, Mirro J, Harvin D, Soheilian F, Kamata A, Nagashima K, Rein A. 2012. Functional redundancy in HIV-1 viral particle assembly. J Virol 86: 12991–12996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onafuwa-Nuga AA, King SR, Telesnitsky A. 2005. Nonrandom packaging of host RNAs in Moloney murine leukemia virus. J Virol 79: 13528–13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onafuwa-Nuga AA, Telesnitsky A, King SR. 2006. 7SL RNA, but not the 54-kd signal recognition particle protein, is an abundant component of both infectious HIV-1 and minimal virus-like particles. RNA 12: 542–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Reilly D, Dienstbier M, Cowley SA, Vazquez P, Drozdz M, Taylor S, James WS, Murphy S. 2013. Differentially expressed, variant U1 snRNAs regulate gene expression in human cells. Genome Res 23: 281–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker KA, Bond U. 1989. Analysis of pre-rRNAs in heat-shocked HeLa cells allows identification of the upstream termination site of human polymerase I transcription. Mol Cell Biol 9: 2500–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrott AM, Tsai M, Batchu P, Ryan K, Ozer HL, Tian B, Mathews MB. 2011. The evolution and expression of the snaR family of small non-coding RNAs. Nucleic Acids Res 39: 1485–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R, Mount SM, Kingsford C. 2014. Sailfish enables alignment-free isoform quantification from RNA-seq reads using lightweight algorithms. Nat Biotechnol 32: 462–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavon-Eternod M, Wei M, Pan T, Kleiman L. 2010. Profiling non-lysyl tRNAs in HIV-1. RNA 16: 267–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politz JC, Yarovoi S, Kilroy SM, Gowda K, Zwieb C, Pederson T. 2000. Signal recognition particle components in the nucleolus. Proc Natl Acad Sci 97: 55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rein A, Datta SA, Jones CP, Musier-Forsyth K. 2011. Diverse interactions of retroviral Gag proteins with RNAs. Trends Biochem Sci 36: 373–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy-Engel AM. 2012. LINEs, SINEs and other retroelements: do birds of a feather flock together? Front Biosci (Landmark Ed) 17: 1345–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rulli SJ Jr, Hibbert CS, Mirro J, Pederson T, Biswal S, Rein A. 2007. Selective and nonselective packaging of cellular RNAs in retrovirus particles. J Virol 81: 6623–6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopman NC, van Montfort T, Willemsen M, Knoepfel SA, Pollakis G, van Kampen A, Sanders RW, Haasnoot J, Berkhout B. 2012. Selective packaging of cellular miRNAs in HIV-1 particles. Virus Res 169: 438–447. [DOI] [PubMed] [Google Scholar]
- Stake MS, Bann DV, Kaddis RJ, Parent LJ. 2013. Nuclear trafficking of retroviral RNAs and Gag proteins during late steps of replication. Viruses 5: 2767–2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundquist WI, Krausslich HG. 2012. HIV-1 assembly, budding, and maturation. Cold Spring Harb Perspect Med 2: a006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeiwa T, Taniguchi I, Ohno M. 2015. Exportin-5 mediates nuclear export of SRP RNA in vertebrates. Genes Cells 20: 281–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarn WY, Yario TA, Steitz JA. 1995. U12 snRNA in vertebrates: evolutionary conservation of 5′ sequences implicated in splicing of pre-mRNAs containing a minor class of introns. RNA 1: 644–656. [PMC free article] [PubMed] [Google Scholar]
- Tian C, Wang T, Zhang W, Yu XF. 2007. Virion packaging determinants and reverse transcription of SRP RNA in HIV-1 particles. Nucleic Acids Res 35: 7288–7302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeire J, Naessens E, Vanderstraeten H, Landi A, Iannucci V, Van Nuffel A, Taghon T, Pizzato M, Verhasselt B. 2012. Quantification of reverse transcriptase activity by real-time PCR as a fast and accurate method for titration of HIV, lenti- and retroviral vectors. PLoS One 7: e50859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Tian C, Zhang W, Luo K, Sarkis PT, Yu L, Liu B, Yu Y, Yu XF. 2007. 7SL RNA mediates virion packaging of the antiviral cytidine deaminase APOBEC3G. J Virol 81: 13112–13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissing S, Munoz-Lopez M, Macia A, Yang Z, Montano M, Collins W, Garcia-Perez JL, Moran JV, Greene WC. 2012. Reprogramming somatic cells into iPS cells activates LINE-1 retroelement mobility. Hum Mol Genet 21: 208–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolin SL, Sim S, Chen X. 2012. Nuclear noncoding RNA surveillance: is the end in sight? Trends Genet 28: 306–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong J, Kasim M, Bachorik JL, Wan L, Dreyfuss G. 2010. Gemin5 delivers snRNA precursors to the SMN complex for snRNP biogenesis. Mol Cell 38: 551–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G, Qin Y, Clark WC, Dai Q, Yi C, He C, Lambowitz AM, Pan T. 2015. Efficient and quantitative high-throughput tRNA sequencing. Nat Methods 12: 835–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.