Abstract
Increasing evidences have demonstrated that microRNAs (miRNAs) act an essential role in regulating tumor progression and metastasis. Previous miRNAs microarray data showed that hsa-miR-599 is lower expressed in hepatocellular carcinoma (HCC); however, the function and molecular mechanism of hsa-miR-599 on HCC has not been well illustrated. Here, we first analyzed the expression level of hsa-miR-599 in HCC tissues and cell lines by real-time reverse-transcription PCR (qRT-PCR). Interestingly, we found that hsa-miR-599 was significantly down-regulated in the examined HCC tissues and cell lines. Then cells proliferation, migration and invasion were assessed by MTT, wound-healing and trans-well assay respectively. The results showed that over-expression of hsa-miR-599 resulted in inhibited HCC cells proliferation, migration and invasion in vitro. In addition, dual-luciferase reporter assay, qRT-PCR and Western blot analyzes were used to confirm MYC (v-myc avian myelocytomatosis viral oncogene homolog) as a target gene of hsa-miR-599. MYC expression was up-regulated in HCC tissues and cell lines, and restoration of hsa-miR-599 could remarkably decreased the mRNA and protein levels of MYC. Moreover, over-expression of MYC partly reversed hsa-miR-599-mediated inhibition of HCC cells proliferation, migration and invasion in vitro. Taken together, our data demonstrate that hsa-miR-599 acts as a tumor suppressor and inhibits HCC cells proliferation, migration and invasion by partly targeting oncogenic MYC, which hints that hsa-miR-599 can be a diagnostic and therapeutic biomarker in HCC.
Keywords: Hsa-miR-599, hepatocellular carcinoma, suppressor, MYC
Introduction
Hepatocellular carcinoma (HCC) is the third most leading cause of cancer-associated death, and the sixth most common and aggressive malignancies all over the world [1,2]. Currently, the most effective treatment approaches for HCC are surgical resection or transplantation [3]. Although surgical and treatment measures have been improved over time, the 5-year survival rate of HCC patients is still only approximately 5% and exceed 600,000 people dying of HCC each year [4]. HCC is the result of a complicated, multi-step progresses associated with epigenetic and genetic changes [5]. Uncontrolled tumor metastasis, frequent intrahepatic propagation and extrahepatic progression are the mainly causes for HCC poor prognosis. Therefore, there is urgent to clarify the molecular mechanism of HCC and explore new therapeutic strategies.
MicroRNAs (miRNAs), a class of small non-coding RNAs of the length of 21 to 25 nucleotides that are responsible for posttranscriptional negatively regulation by binding to complementary sequences in the 3’ untranslated region (3’-UTR) of target RNAs [6,7]. MiRNAs has been reported to be participating in number of biological processes, such as differentiation, development, morphogenesis, and oncogenesis [8,9]. Growing evidences have revealed that miRNAs could play tumor suppressor or oncogene roles in various types of human cancers. For instance, microRNA-205 regulates ubiquitin specific peptidase 7 protein expression in hepatocellular carcinoma cells [10]; microRNA-365 inhibits growth, invasion and metastasis of malignant melanoma by targeting NRP1 expression [11]; miR-491 attenuates cancer stem cells-like properties of hepatocellular carcinoma by inhibition of GIT-1/NF-κB-mediated EMT [12]. These researches demonstrate that miRNAs could be used as biomarkers for diagnosis and prognosis prediction of tumors.
Previous studies have showed that a subset of miRNAs were deregulated in HCC by miRNA expression profiles [13,14]. Recently, several aberrantly expressed miRNAs have been demonstrated to regulate HCC cells proliferation and metastasis [15-17]. Hsa-miR-599 was a novel miRNA, which have not been reported in cancer. MiRNAs microarray data showed that hsa-miR-599 was down-regulated expressed in HCC [13,14].
In this study, we explored the potential roles and molecular mechanisms of hsa-miR-599 in HCC development. The results showed that hsa-miR-599 was lowly expressed in HCC tissues and cell lines, which function as a tumor-suppressor miRNA to restrain HCC cells proliferation, migration and invasion in vitro. MYC was a potential target of hsa-miR-599 based on bioinformatics analysis and luciferase reporter assay. Moreover, the negative regulation of MYC by hsa-miR-599 may partly account for hsa-miR-599-mediated inhibition of HCC cells proliferation, migration and invasion.
Materials and methods
HCC patients
Forty two paired HCC tissues and adjacent matched normal liver tissues were collected from Department of Gastroenterology, NanKai Hospital, including 24 cases of males and 18 cases of females, who underwent hepatectomy or partial hepatectomy between January 2009 and January 2013. Patients had not been accepted preoperative embolization or chemotherapy. The research was permitted by the Institutional Review Board of The NanKai Hospital. Written informed consent was also approved from all participant. All samples were frozen at -80°C immediately until further experiments.
Cell lines and cell culture
Human HCC cell lines HepG2, Bel7402, SMMC7721 and Huh7 as well as normal liver epithelial cell L02 were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). Cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS) at 37°C and supplemented with 5% CO2 in a humidified chamber.
Real-time reverse-transcription PCR (qRT-PCR)
For miRNA analysis, total RNA was extracted from fresh HCC samples and cell lines by using TRIzol reagent (Invitrogen, Carlsbad, CA, SUA) according to the manufacturer’s instructions. cDNA was synthesized with 1 µg of total RNA by using the One Step Prime script miRNA cDNA Synthesis Kit (Qiagen, Valencia, CA, USA). qRT-PCR assay was performed using SYBR Green Premix Ex Taq II (Takara, Dalian, Japan) on Light Cycler 480 II (Roche, Rotkreuz, Switzerland). The U6 small nuclear RNA were used as endogenous control. The primers used for the amplification were as follows: hsa-miR-599: 5’-TAAGCTGACATGGGACAGGGAT-3’ (forward), miScript SYBR Green PCR kit Universal Primer (reverse); U6: 5’-CTCGCTTCGGCAGCACA-3’ (forward), 5’-ACGCTTCACGAATTTGCGT-3’ (reverse).
For MYC analysis, cDNA was synthesized using the PrimeScript RT Reagent Kit (Takara, Dalian, Japan). qRT-PCR assay was also applied with SYBR Green Premix Ex Taq II (Takara, Dalian, Japan) on Light Cycler 480 II. ACTB was used as an internal control. The primer sequences of MYC and ACTB were as follows: MYC (sense, 5’-GGAGGAACAAGAAGATGAGGAAGAA-3’ and anti-sense: 5’-AGGACCAGTGGGCTGTGAGGAG-3’); ACTB (sense: 5’-GACTTAGTTGCGTTACACCCTTTCT-3’ and anti-sense: 5’-ACTGCTGTCACCTTCACCGTTC-3’). All reactions were repeated in triplicate and the fold changes of genes were calculated by the 2-ΔΔCt method [18].
Cell transfection
Hsa-miR-599 mimics and negative control mimics (NC), were bought in Shanghai GenePharma (Shanghai, China). The sequences of hsa-miR-599 mimics: 5’-CUGUCCACAGUGUGUUUGAUAAG-3’, negative control mimics: 5’-CAGUACUUUUGUGUAGUACAA-3’. MYC over-expression plasmid (pCMV-HA-MYC) and corresponding empty vector (pCMV-HA) were obtained from life technology (Invitrogen, Carlsbad, CA, SUA). Transfection was conducted with Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocols. Total RNA and protein were extracted as previous described after 48 h transfection. Then, transfection efficiency results were analyzed in every experiment by qRT-PCR and Western blot after 48 h transfection.
Cell proliferation assay
The proliferation ability of HepG2 and Bel7402 cells was detected using the MTT (3-(4,5-dimethyl-thiazol-2-y1) 2,5-diphenyl tetrazolium bromide) assay. HepG2 and Bel7402 cells were seeded into 96-well plates at 5×104 cells per well at 24 h prior to transfection. Then, cells were washed with PBS and incubated in 30 μl of 5mg/ml MTT reagent (Invitrogen, Carlsbad, CA, USA) for 3~4 h after 0, 24, 48, 72 and 96 h transfection. The number of cells per well were measured by the absorbance (480 nm) using the microplate reader Thermo Plate (Bio-Rad, Hercules, CA, USA) at the indicated time points.
Cell migration and invasion assay
Cell migration was measured using migration scratch assay. About 8×105 cells were seeded into 6-well plates and transfected with hsa-miR-599 mimics or NC and pCMV-HA-MYC or pCMV-HA after 24 h. After 6 h of transfection, scratches was made by using a sterile 20 µl pipette tip. The migration distances of cells were measured at 0 and 24 h after the scratches were made. Three visual fields (×200) were randomly selected from each scratch, and the migration distances were counted via a light microscope (Olympus, Tokyo, Japan).
Cell invasion were assayed using transwell assay. For the invasion assay, transwell chamber (Becton-Dickinson Biosciences, MA, USA) was coated with 30 μl Matrigel and placed into 24-well plates and incubated for 30 min at 37°C. After 48 h of transfection, HepG2 and Bel7402 cells were seeded into chambers at the density of 2×105 cells per well and cultured with DMEM supplemented with 2% serum. 48 h later, the invaded cells were fixed with 100% methanol for 30 min and stained with crystal violet for 20 min. The number of invaded cells were assessed using a light microscope (Olympus, Tokyo, Japan). All experiments were performed in triplicate.
Bioinformatics analysis of hsa-miR-599 target gene
The miRNA target computer-aided algorithms of TargetScan (http://www.targetscan.org/), PicTar (http://pictar.mdcberlin.de/), and miRanda (http://www.microrna.org) were used to predict the binding sites between hsa-miR-599 and MYC.
Luciferase reporter assay
The full length sequences of MYC 3’-UTR (WT) were amplified by PCR from cDNA and cloned at the Xhol and Notl sites into psiCHECK-2 luciferase vector (Promega, WI, USA). The primer sequences for the MYC 3’-UTR was 5’-GGAAAAGTAAGGAAAACGATTCCTT-3’ (forward) and 5’-AAGGAATCGTTTTCCTTACTTTTCC-3’ (reverse). The mutant construct of MYC 3’-UTR (MUT) was synthesized by life technology (Invitrogen, Carlsbad, CA, SUA). Co-transfection of WT and MUT reporter vectors and hsa-miR-599 mimics or NC into HepG2 and Bel7402 cells using Lipofectamine 2000. After 48 h of transfection, the relative luciferase activity was assessed by the Dual-Luciferase Reporter Assay system (Promega, WI, USA).
Western blot analysis
48 hours after transfection, HCC cells were extracted with RIPA buffer (Sigma, USA). The concentration of protein was quantified with BCA Protein Assay Kit (Millipore, USA). Then, samples were denatured at 105°C for 10~15 min and placed on ice for 5 min. Samples were loaded on 10% of sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and subsequently transferred to polyvinylidene difluoride membrane (PVDF) (Millipore, MA, USA). After that, the membranes were blocked for 2 h at 37°C and incubated with anti-MYC antibody overnight at 4°C. the membranes were incubated with appropriately secondary antibody after washing with TBST buffer for five times, and detected using commercial ECL kit (Millipore, MA, USA) according to the manufacturer’s instructions. The GAPDH protein was used as negative control.
Statistical analysis
Statistical analyses were performed with SPSS 18.0 software (SPSS, Inc., Chicago, IL, USA). Data are presented as the mean ± standard deviation (SD). Each experiment was repeated at least three times. Statistical analyses were performed with Student’s t-test or one-way ANOVA. Values of P<0.05 were considered to be statistically significant.
Results
Hsa-miR-599 is down-regulated in HCC tissues and cell lines
To explore the function and molecular mechanism of hsa-miR-599 in HCC development, we first detected the expression level of hsa-miR-599 in forty two paired HCC tissues and adjacent matched normal liver tissues by qRT-PCR. The results showed that hsa-miR-599 was down-regulated in HCC tissues compared with adjacent matched normal liver tissues (Figure 1A). Then, we assessed hsa-miR-599 expression in four human HCC cell lines (HepG2, Bel7402, SMMC7721 and Huh7) and a normal liver epithelial cell (L02). qRT-PCR data revealed that hsa-miR-599 expression was also markedly decreased in HCC cell lines as compared with L02 (Figure 1B). These results demonstrate hsa-miR-599 is down-regulated in HCC tissues and cells, which hints it may be a novel tumor suppressor in HCC.
Figure 1.
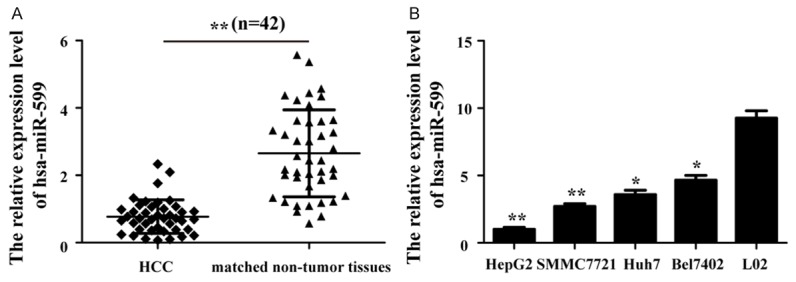
Hsa-miR-599 is down-regulated in HCC tissues and cell lines. U6 snRNA was used as a internal control. A. Comparing differences in the expression levels of hsa-miR-599 between HCC and matched non-tumor tissues. HCC: Hepatocellular carcinoma. B. qRT-PCR analysis detected the expression of hsa-miR-599 in a normal liver epithelial cell and HCC cells. qRT-PCR: real-time reverse-transcription PCR. *p<0.05; **p<0.01.
Over-expression of hsa-miR-599 inhibits cells proliferation in HCC
qRT-PCR analysis was conducted to measure the expression level of hsa-miR-599 in HepG2 and Bel7402 cells after transfection with hsa-miR-599 mimics or NC. We found that hsa-miR-599 expression was significantly increased in HepG2 and Bel7402 cells treated with hsa-miR-599 mimics, as compared with NC group (Figure 2A). The result showed hsa-miR-599 mimics could enhance hsa-miR-599 expression in HCC cells, which laid a foundation for further experiments.
Figure 2.
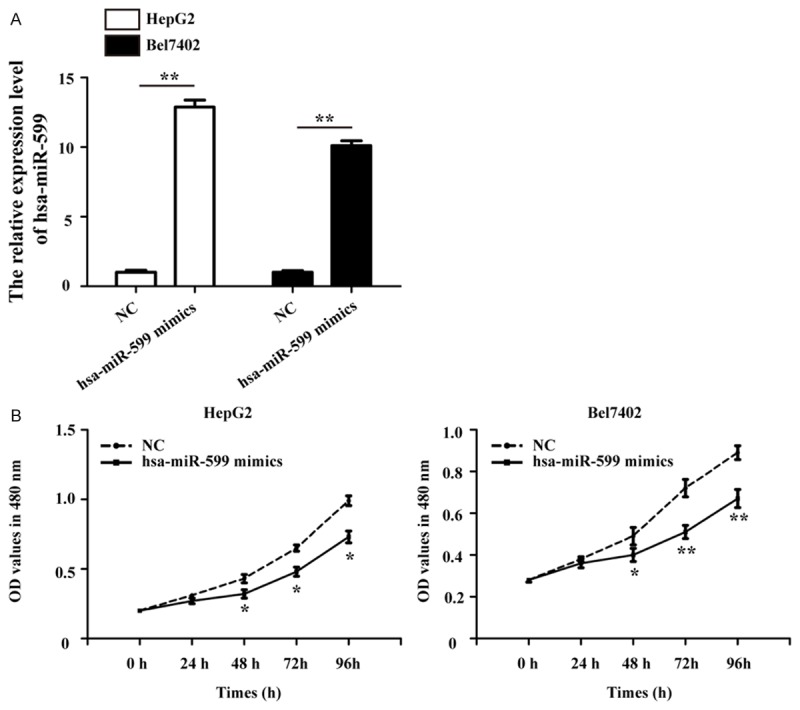
Over-expression of hsa-miR-599 inhibits cells proliferation in HCC. A. Transfection of hsa-miR-599 mimics to HepG2 and Bel7402 cells increased the expression level of hsa-miR-599 detected by qRT-PCR. B. The MTT assay showed significantly cells proliferation inhibition in hsa-miR-599 mimics group compared with NC group from HepG2 and Bel7402 cells. NC: negative control. *p<0.05; **p<0.01.
Furthermore, the MTT assay showed that the growth rates of HepG2 and Bel7402 cells was attenuated after transfected with hsa-miR-599 mimics than the cells treated with NC (Figure 2B). These results demonstrate that hsa-miR-599 inhibits the proliferation potential of HCC cells.
Over-expression of hsa-miR-599 hinders cells migration and invasion in HCC
The roles of hsa-miR-599 in the regulation of HCC cells migration and invasion were further explored by using migration scratch and transwell assay, respectively. As shown in Figure 3A, the hsa-miR-599-overexpressed HepG2 and Bel7402 cells showed reduced migration ability compared to the NC group, which demonstrates that hsa-miR-599 has inhibitory effect on HCC cells migration in vitro.
Figure 3.
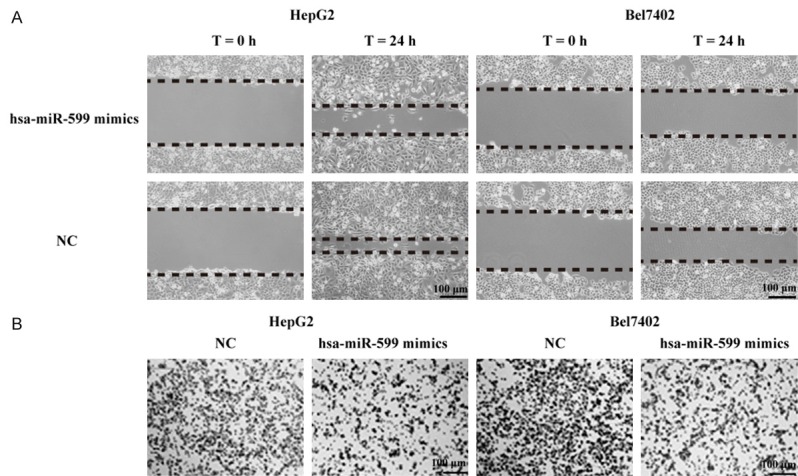
Over-expression of hsa-miR-599 hinders cells migration and invasion in HCC. A. The inhibitory effect of hsa-miR-599 toward the migration of HepG2 and Bel7402 cells. B. The repressive effect of hsa-miR-599 toward the invasion of HepG2 and Bel7402 cells. Bar = 100 µm.
We further surveyed the function of hsa-miR-599 in the regulation of cells invasion in HCC. As shown in Figure 3B, compared to NC group, HCC cells transfected with hsa-miR-599 mimics significantly decreased the number of HepG2 and Bel7402 cells invading the chamber membrane with matrigel. Our results indicate that hsa-miR-599 acts as a tumor suppressor miRNA and contributes to inhibition of migration and invasion in HCC cells.
MYC is a direct target gene of hsa-miR-599 in HCC cells
Previous studies demonstrated that miRNAs executed posttranscriptional regulation by directly binding to the 3’-UTR of their downstream genes. Bioinformatics analysis was performed in order to identify the potential targeted gene of hsa-miR-599. The complementary sequences between hsa-miR-599 and MYC 3’-UTR was presented in Figure 4A. As shown in Figure 4B, the expression levels of MYC mRNA and protein were decreased after treated with hsa-miR-599 mimics for 48 h than treated with NC in HepG2 and Bel7402 cells. Moreover, to verify whether MYC is a direct target of hsa-miR-599 in HCC, dual-luciferase reporter assay was performed after co-transfection of WT and MUT reporter vectors and hsa-miR-599 mimics or NC into HepG2 cell. The results showed that hsa-miR-599 markedly down-regulated the relatively luciferase activity of the WT vector, while the relatively luciferase expression of MUT vector was no significantly difference (Figure 4C). These results suggest that the bind sites in the MYC 3’-UTR was exactly re-gulated by hsa-miR-599 in HCC cells.
Figure 4.
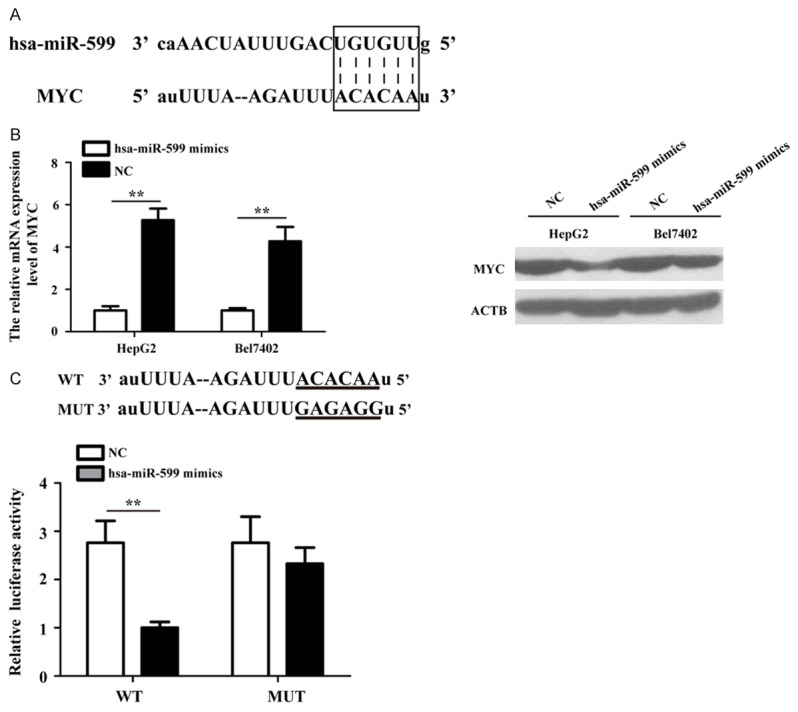
MYC is a direct target gene of hsa-miR-599 in HCC cells. A. Sequence alignment of hsa-miR-599 and MYC 3’-UTR using miRanda algorithm. B. The qRT-PCR and Western blot analyses detection of MYC expression in HepG2 and Bel7402 cells transfected with hsa-miR-599 mimics or NC. C. The HepG2 cell were co-transfected with hsa-miR-599 mimics or NC and a luciferase reporter plasmid containing a fragment of the MYC 3’-UTR harboring either the hsa-miR-599 binding site (WT) or the mutant (MUT). **p<0.01.
MYC is up-regulated in HCC tissues and cell lines
The expressions levels of MYC mRNA and protein were performed by qRT-PCR and Western blot analyses, respectively. We observed that both the mRNA and protein expression levels of MYC were significantly enhanced in HCC tissues, compared with adjacent matched normal liver tissues (Figure 5A). Consistent with our expectation, MYC in all four HCC cells were also observably up-regulated than L-02 cell (Figure 5B).
Figure 5.
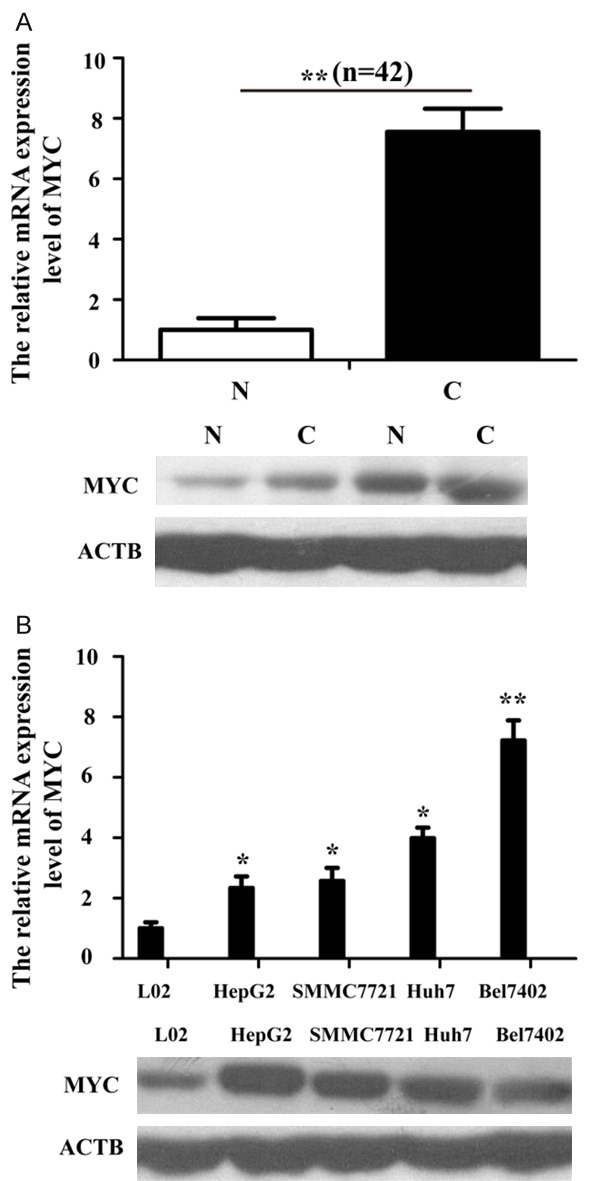
MYC is up-regulated in HCC tissues and cell lines. A. The mRNA and protein expression of MYC in HCC tissues. N: Normal tissues; C: HCC tissues. B. qPT-PCR and Western blot were used to detect MYC expression in four HCC cells (HepG2, Bel7402, SMMC7721 and Huh7) and a normal liver epithelial cell (L-02). The data was normalized with ACTB by the 2-ΔΔCt method. *p<0.05; **p<0.01.
Over-expression of MYC partly reverses HCC cells proliferation suppressed by hsa-miR-599
To confirm whether hsa-miR-599 could exert its tumor suppressive role through MYC, MYC was ectopic expressed by a MYC over-expression plasmid (pCMV-HA-MYC). The over-expression effect was assessed by qRT-PCR and Western blot analyses. As shown in Figure 6A, the mRNA and protein expression levels of MYC were significantly increased after over-expressed MYC gene by pCMV-HA-MYC-transfected HepG2 and Bel7402 cells. Furthermore, the MTT assay showed that the over-expression of MYC could attenuate the repression of HCC cells growth by hsa-miR-599 (Figure 6B).
Figure 6.
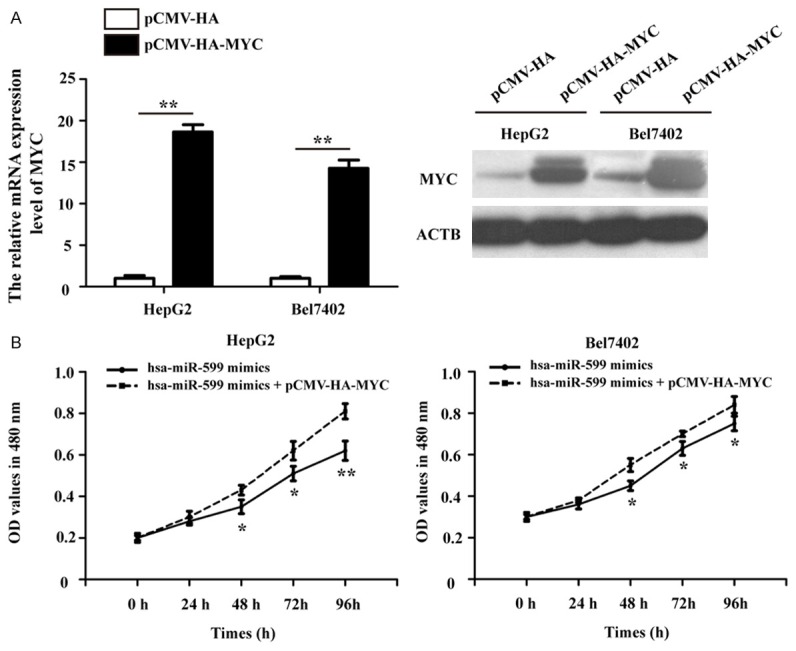
Over-expression of MYC partly reverses HCC cells proliferation suppressed by hsa-miR-599. A. The relative MYC mRNA and protein expression level were significantly increased in pCMV-HA-MYC-transfected HepG2 and Bel7402 cells. B. Ectopic expression MYC attenuated the repression of cells growth by hsa-miR-599. *p<0.05; **p<0.01.
Over-expression of MYC partly reverses the effect of hsa-miR-599 on cells migration and invasion in HCC
Using migration assay, we found the inhibition of HepG2 and Bel7402 cells migration by hsa-miR-599 was partly reversed by the over-expression of MYC (Figure 7A). Also, restoration of MYC abrogated the hsa-miR-599-restrained cells invasion (Figure 7B). Take together, hsa-miR-599 functions as a tumor suppressor inhibits HCC cells proliferation, migration and invasion by directly targeting MYC.
Figure 7.
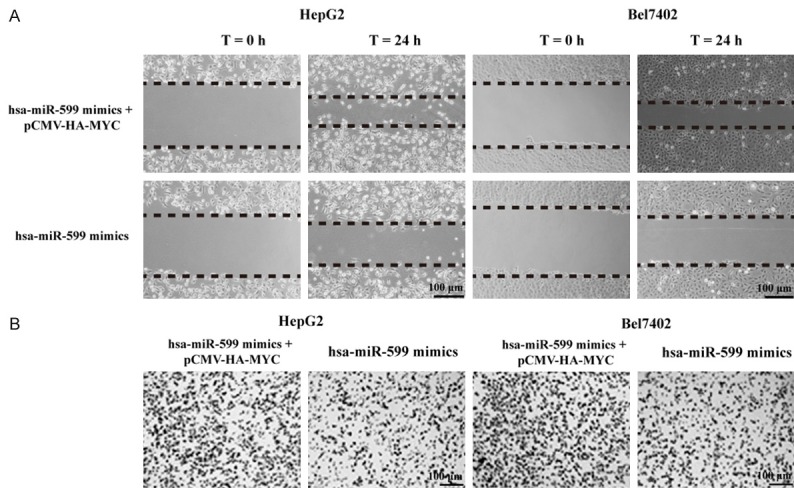
Over-expression of MYC partly reverses the effect of hsa-miR-599 on cells migration and invasion in HCC. A. Over-expression of MYC could reverse the inhibition of HepG2 and Bel7402 cells migration by hsa-miR-599. B. Restoration of MYC abrogated the hsa-miR-599-restrained cell invasion.
Discussion
HCC is especially health problems in China, where the overall incidence of HCC is much higher than others Asian countries. HCC is a highly malignancy and poor prognosis, which is characterized with high potential for vascular invasion [19]. It is essential to comprehend the exact molecular mechanisms underlying HCC tumorigenesis to further improve overall survival for HCC [20].
Increasing evidences have reported that miRNAs could regulated hepatocarcinogenesis-related genes expression, which prompted a new insight in the progression and development of HCC [21]. Numerous miRNAs have demonstrated that played crucial roles in modifying HCC cells proliferation, metastasis, apoptosis and neoplastic transformation [22]. The hsa-miR-599 is located at 8q22.2 on human genome. A previous miRNAs expression profiles reports have showed that hsa-miR-599 was don-regulated in HCC [13,14]. As a novel miRNA, the function and molecular mechanism of hsa-miR-599 in HCC has not been reported.
In the present study, we analyzed the detailed expression and function of hsa-miR-599 in HCC development. Consistent with our speculation, the results indicated that the expression of hsa-miR-599 was significantly down-regulated in HCC tissues and cell lines, compared to adjacent liver normal tissues and normal liver epithelial cell. The subsequently experiments showed that hsa-miR-599 significantly inhibited HCC cells proliferation and metastasis. MYC was identified as an important target gene, which was negative regulated by hsa-miR-599 in HCC cells. Furthermore, the dual-luciferase reporter assays indicated that hsa-miR-599 could directly bind the complementary region of MYC mRNA 3’-UTR. Our findings suggest that hsa-miR-599 has a tumor-suppressor role in HCC development.
MYC is a key basic helix-loop-helix leucine zipper transcription factor that functions as an crucial regulator of several cellular processes, including growth, proliferation and migration in different cells [23-25]. In addition, the oncogene of MYC is up-regulated in many types of human tumors and plays an important role in cell invasion, migration, autophagy and angiogenesis [26,27]. Moreover, the over-expression MYC is considered to be a new potential or independent predictor of poor prognosis for clinical patients in multiple types of cancers [28]. In our study, we identified MYC as a directly and functional target of hsa-miR-599 in HCC. We found that MYC was up-regulated in HCC tissues and cells, and hsa-miR-599-inhibited cells proliferation, migration and invasion were partly reversed by over-expression of MYC. Taken together, these results demonstrate a functional correlation between hsa-miR-599 and MYC, and confirm that hsa-miR-599 acts as an anti-metastatic miRNA in HCC cells by targeting MYC.
In conclusion, the current study provides first directly evidences that hsa-miR-599 is significantly lower-expressed in HCC specimens and cells, and appears to function as a tumor suppressor in HCC cells through the regulation of MYC oncogene expression. The identification of novel miRNA-mediated tumor-suppressor pathways may provide new proof on potential therapeutic targets in HCC treatment.
Disclosure of conflict of interest
None.
References
- 1.Qasim W, Brunetto M, Gehring AJ, Xue SA, Schurich A, Khakpoor A, Zhan H, Ciccorossi P, Gilmour K, Cavallone D, Moriconi F, Farzhenah F, Mazzoni A, Chan L, Morris E, Thrasher A, Maini MK, Bonino F, Stauss H, Bertoletti A. Immunotherapy of HCC metastases with autologous T cell receptor redirected T cells, targeting HBsAg in a liver transplant patient. J Hepatol. 2015;62:486–491. doi: 10.1016/j.jhep.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 2.Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245–1255. doi: 10.1016/S0140-6736(11)61347-0. [DOI] [PubMed] [Google Scholar]
- 3.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 4.Minguez B, Lachenmayer A. Diagnostic and prognostic molecular markers in hepatocellular carcinoma. Dis Markers. 2011;31:181–190. doi: 10.3233/DMA-2011-0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyault S, Rickman DS, de Reynies A, Balabaud C, Rebouissou S, Jeannot E, Herault A, Saric J, Belghiti J, Franco D, Bioulac-Sage P, Laurent-Puig P, Zucman-Rossi J. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology. 2007;45:42–52. doi: 10.1002/hep.21467. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 8.Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 9.Sun K, Lai EC. Adult-specific functions of animal microRNAs. Nat Rev Genet. 2013;14:535–548. doi: 10.1038/nrg3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu L, Liu R, Zhang W, Qian S, Wang JH. MicroRNA-205 regulates ubiquitin specific peptidase 7 protein expression in hepatocellular carcinoma cells. Mol Med Rep. 2015;12:4652–4656. doi: 10.3892/mmr.2015.3998. [DOI] [PubMed] [Google Scholar]
- 11.Bai J, Zhang Z, Li X, Liu H. MicroRNA-365 inhibits growth, invasion and metastasis of malignant melanoma by targeting NRP1 expression. Int J Clin Exp Pathol. 2015;8:4913–4922. [PMC free article] [PubMed] [Google Scholar]
- 12.Yang X, Ye J, Yan H, Tang Z, Shen J, Zhang J, Yang L. MiR-491 attenuates cancer stem cells-like properties of hepatocellular carcinoma by inhibition of GIT-1/NF-kappaB-mediated EMT. Tumour Biol. 2016;37:201–9. doi: 10.1007/s13277-015-3687-5. [DOI] [PubMed] [Google Scholar]
- 13.Wang Y, Gao Y, Shi W, Zhai D, Rao Q, Jia X, Liu J, Jiao X, Du Z. Profiles of differential expression of circulating microRNAs in hepatitis B virus-positive small hepatocellular carcinoma. Cancer Biomark. 2015;15:171–180. doi: 10.3233/CBM-140451. [DOI] [PubMed] [Google Scholar]
- 14.Murakami Y, Tanahashi T, Okada R, Toyoda H, Kumada T, Enomoto M, Tamori A, Kawada N, Taguchi YH, Azuma T. Comparison of hepatocellular carcinoma miRNA expression profiling as evaluated by next generation sequencing and microarray. PLoS One. 2014;9:e106314. doi: 10.1371/journal.pone.0106314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang H, Sheng C, Yin Y, Wen S, Yang G, Cheng Z, Zhu Q. PABPC1 interacts with AGO2 and is responsible for the microRNA mediated gene silencing in high grade hepatocellular carcinoma. Cancer Lett. 2015;367:49–57. doi: 10.1016/j.canlet.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 16.Chen S, Li F, Chai H, Tao X, Wang H, Ji A. miR-502 inhibits cell proliferation and tumor growth in hepatocellular carcinoma through suppressing phosphoinositide 3-kinase catalytic subunit gamma. Biochem Biophys Res Commun. 2015;464:500–5. doi: 10.1016/j.bbrc.2015.06.168. [DOI] [PubMed] [Google Scholar]
- 17.Zhang W, Liu K, Liu S, Ji B, Wang Y, Liu Y. MicroRNA-133a functions as a tumor suppressor by targeting IGF-1R in hepatocellular carcinoma. Tumour Biol. 2015;36:9779–88. doi: 10.1007/s13277-015-3749-8. [DOI] [PubMed] [Google Scholar]
- 18.Arocho A, Chen B, Ladanyi M, Pan Q. Validation of the 2-DeltaDeltaCt calculation as an alternate method of data analysis for quantitative PCR of BCR-ABL P210 transcripts. Diagn Mol Pathol. 2006;15:56–61. doi: 10.1097/00019606-200603000-00009. [DOI] [PubMed] [Google Scholar]
- 19.Bolos D, Finn RS. Systemic therapy in HCC: lessons from brivanib. J Hepatol. 2014;61:947–950. doi: 10.1016/j.jhep.2014.06.019. [DOI] [PubMed] [Google Scholar]
- 20.Oishi N, Yamashita T, Kaneko S. Molecular biology of liver cancer stem cells. Liver Cancer. 2014;3:71–84. doi: 10.1159/000343863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang X, Jia Z. Construction of HCC-targeting artificial miRNAs using natural miRNA precursors. Exp Ther Med. 2013;6:209–215. doi: 10.3892/etm.2013.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gramantieri L, Fornari F, Callegari E, Sabbioni S, Lanza G, Croce CM, Bolondi L, Negrini M. MicroRNA involvement in hepatocellular carcinoma. J Cell Mol Med. 2008;12:2189–2204. doi: 10.1111/j.1582-4934.2008.00533.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y, Liu H, Lai C, Du X, Su Z, Gao S. The Lin28/let-7a/c-Myc pathway plays a role in non-muscle invasive bladder cancer. Cell Tissue Res. 2013;354:533–541. doi: 10.1007/s00441-013-1715-6. [DOI] [PubMed] [Google Scholar]
- 24.Akinyeke T, Matsumura S, Wang X, Wu Y, Schalfer ED, Saxena A, Yan W, Logan SK, Li X. Metformin targets c-MYC oncogene to prevent prostate cancer. Carcinogenesis. 2013;34:2823–2832. doi: 10.1093/carcin/bgt307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dueck AC, Reinholz MM, Geiger XJ, Tenner K, Ballman K, Jenkins RB, Riehle D, Chen B, McCullough AE, Davidson NE, Martino S, Sledge GW, Kaufman PA, Kutteh LA, Gralow J, Harris LN, Ingle JN, Lingle WL, Perez EA. Impact of c-MYC protein expression on outcome of patients with early-stage HER2+ breast cancer treated with adjuvant trastuzumab NCCTG (alliance) N9831. Clin Cancer Res. 2013;19:5798–5807. doi: 10.1158/1078-0432.CCR-13-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li K, Chen MK, Situ J, Huang WT, Su ZL, He D, Gao X. Role of co-expression of c-Myc, EZH2 and p27 in prognosis of prostate cancer patients after surgery. Chin Med J (Engl) 2013;126:82–87. [PubMed] [Google Scholar]
- 27.Liu Z, Jiang Y, Hou Y, Hu Y, Cao X, Tao Y, Xu C, Liu S, Wang S, Wang L, Shi Y, Siebenlist U, Zhang X. The IkappaB family member Bcl-3 stabilizes c-Myc in colorectal cancer. J Mol Cell Biol. 2013;5:280–282. doi: 10.1093/jmcb/mjt020. [DOI] [PubMed] [Google Scholar]
- 28.Pello OM, Andres V. Role of c-MYC in tumor-associated macrophages and cancer progression. Oncoimmunology. 2013;2:e22984. doi: 10.4161/onci.22984. [DOI] [PMC free article] [PubMed] [Google Scholar]


