Abstract
MicroRNAs (miRNAs) are a class of small, non-coding RNAs that play a crucial role in tumor procession. It has been demonstrated that miR-449a expression was downregulated and served as tumor suppressor in many types of tumor. However, the biological function and molecular mechanism of miR-449a in hepatocellular carcinoma (HCC) still remains largely unknown. Therefore, the aims of this study were to investigate biological role and molecular mechanism of miR-449a in HCC by a serial of molecular experiments. Here, we demonstrated that miR-449a expression was downregulated in HCC tissues and cell lines compared with the adjacent nontumor tissues and normal hepatic cell line. Ectopic expression of miR-449a suppressed HCC cell proliferation, colony formation, migration and invasion. Moreover, A Disintegrin And Metalloproteinases 10 (ADAM10) was identified as a direct target gene of miR-449a in HCC cell. ADAM10 expression was upregulated in HCC tissues and cell lines, and was negatively correlated with the expression level of miR-449a in HCC tissues. Interesting, overexpression of ADAM10 attenuated the inhibition effect of miR-449a-mediated HCC cell proliferation, colony formation, migration and invasion. These results suggested that miR-449a might function as a tumor suppressor miRNA, at least in part, through regulating ADAM10 expression in HCC.
Keywords: Hepatocellular carcinoma, miR-449a, ADAM10, proliferation, invasion
Introduction
Hepatocellular carcinoma (HCC) is one of the most common human cancers in the world, which ranks as the fifth most common malignancy and the second leading cause of cancer death worldwide, resulting in about 745,000 HCC-related deaths annually [1]. Despite recent advances in therapeutic strategies such as surgery resection, liver transplantation, radiotherapy, and adjuvant chemotherapy, longtime survival of advance HCC patients remains unsatisfactory mainly due to its high recurrent and metastatic rate [2,3]. Therefore, it is crucial to understand the molecular mechanism by which regulate the growth and metastasis of HCC for exploring the novel biomarkers and therapy agent in HCC patients.
MicroRNAs (miRNAs) are a class of small (18-25 nucleotides), endogenously expressed, well-conserved noncoding RNA molecules that regulate gene expression post-transcriptionally by binding to the 3’-untranslated region (3’UTR) of target mRNAs, leading to the degradation of the mRNA or translational inhibition of functional proteins [4-6]. Given that more than 50% of miRNAs are located in cancer-associated genomic regions or in fragile sites, miRNAs may involve in regulation of cancer initiation and progression [7]. Emerging evidence has demonstrated that miRNAs contributes to development and progression of human cancers including HCC by promoting oncogene expression or by inhibiting tumor suppressor genes [8,9].
miR-449a, located in the first intron of CDC20B on chromosome 5q11, has been reported to be downregulated and function as a tumor suppressor by regulating cell proliferation, cycle procession, apoptosis and migration and invasion in several types of human cancers [10-14]. Recently studies has been demonstrated that miR-449a expression was downregulated, suppressed the epithelial-mesenchymal transition and metastasis of HCC by targeting FOS and Met [15], as well as induced HCC cell apoptosis by targeting Calpain6 and POU2F1 [16]. However, the detail biological function, and underlying molecular mechanism of miR-449a in HCC has not been total elucidated. Therefore, the goal of this study was to explore the role and underlying mechanism of miR-449a in HCC. We demonstrated that miR-449a expression was downregulated in the HCC tissues and cell lines, and overexpression of miR-449a in HCC cells suppressed cell proliferation, colony formation, migration and invasion by targeting A Disintegrin and Metalloproteinases (ADAM 10). These results suggested that miR-449a may serve as a promising target for HCC.
Materials and methods
Tissue samples and cell culture
Forty pair surgical specimens of HCC tissues and their adjacent non-cancerous tissues (more than 5 cm from the cancer tissue) were obtained from patients with HCC who underwent surgery between July 2010 and September 2014 at Department of Hepatobiliary and Pancreatic Surgery, the First Hospital, Jilin University (Changchun, China). All tissue samples were immediately frozen in liquid nitrogen after resection, and stored at -80°C until RNA extraction. Informed consent was obtained from each patient or family. This study protocol was approved by the ethics committee of the First Hospital of Jilin University.
HCC cell lines (SMMC-7721, Hep3B, HepG2, Huh-7) and normal hepatic cell line, HL-7702 were brought from Institute of Cell Biology of Chinese Academy of Science (Shanghai, China), and cultured in Dulbecco’s modified Eagle’s medium (DMEM, Hyclone) supplemented with 10% (v/v) fetal bovine serum (FBS, HyClone, USA), 2 mmol/L glutamine, 100 units/mL penicillin or 100 μg/ml streptomycin, and incubated at 37°C in a humidified incubator with an atmosphere of 5% CO2.
Cell transfection
miR-448 mimic and corresponding negative control (miR-NC) were synthesized by RiboBio (Guangzhou, China). ADAM10 overexpression plasmid was purchased from Genechem (Shanghai, China). Cell transfections were performed using Lipofectamine2000 kit (Invitrogen, Carlsbad, CA, USA) following to the manufacturer’s instructions.
Real-time quantitative polymerase chain reaction (RT-qPCR)
Total RNA from cell lines and tissues was extracted with TRIzol® reagent (Invitrogen) according to the manufacturer’s instructions, and the concentration of total RNA was quantified using a NanoDrop 2000 (Thermo Fisher Scientific, Waltham, MA, USA). For quantify miR-449a, cDNA was synthesized with the TaqMan MicroRNA reverse transcription kit (Applied Biosystems, Foster City, CA, USA), and then was quantified using the QuantMir RT Kit (System Biosciences, Mountain View, CA, USA) under ABI 7900 Sequence Detection System (Applied Biosystems) with miR-449a und U6 primers (Applied Biosystems). For detection ADAM10 expression, RNA was reverse-transcribed into cDNA by a Primescript™ RT reagent kit (Takara, Dalian, China), then was quantified using Real-time PCR Mixture Reagent (Takara) under ABI 7900 Sequence Detection System (Applied Biosystems) with primes of ADAM10 and β-action as described previously [17]. The relative miR-449a expression was normalized to U6 and the expression of ADAM10 was normalized to β-action by using 2-ΔΔCt method.
Cell proliferation and colony formation assay
Cell proliferation was determined using the Cell Counting Kit-8 kit (CCK-8 kit, Dojindo, Japan). In briefly, transfected cells were seeded into 96-well culture plates and cultured at DMEM medium supplemented with 10% (v/v) FBS for 24 h-72 h. At the indicated time points (24 h, 48 h and 72 h), 10 μl CCK-8 reagent (DoJinDo, Japan) was added to each well, followed by measuring the absorbance at 450 nm on an enzyme immunoassay analyzer (Bio-Rad, USA).
For the colony formation assay, transfected cells were resuspended and seeded onto 6-well plates at a density of 1000 cells/well and maintained in DMEM containing 10% FBS for 2 weeks. Colonies were fixed with methanol and stained with 0.1% crystal violet for 15 min, and then were counted by light microscope (Olympus, Tokyo, Japan). The percentage colony formation was calculated by adjusting control to 100%.
Wound healing and invasion assays
Cell migration was assessed by measuring the movement of cells into a scraped. In briefly, 1 × 105 transfected cells were plated in 12-well plates in DMEM containing 10% FBS. After 24 h, a scratch was made through the confluent cell monolayer using a 200-μl pipette tube, and the spread of wound closure was observed after 24 h and photographed under a microscope (Olympus, Tokyo, Japan).
For invasion assays, 1 × 105 transfected cells in serum-free medium were plated in the upper of the membranes coated with Matrigel (BD Biosciences, Becton Dickson Labware, Flanklin Lakes, NJ, USA), and 600 μl of DMEM medium containing 10% FBS were added to the lower chamber to serve as chemoattractant. After 48 hours, the noninvading cells were gently removed with a cotton swab, while invasive cells located on the lower side of the chamber were fixed with 4% paraformaldehyde, stained with 0.1% crystal violet, air dried, photographed, and quantified by counting them in randomly five fields by a light microscope (Olympus).
Dual-luciferase reporter assay
The wild type and mutant 3’untranslated region (3’UTR) of ADAM10 were synthesized from Genechem (Shanghai, China) and subcloned into psicheck-2 vector (Promega, Madison, WI, USA) downstream of the firefly luciferase coding region, named as Wt-ADAM10-3’UTR and Mut-ADAM10-3’UTR. For the luciferase reporter assay, HepG2 cells were cultured in the 96-well plate at 50% confluence before transfection. Cells were tranfected with Wt/Mut ADAM10-3’UTR report plasmid, renilla, and miR-449a mimic or miR-NC using Lipofectamine 2000 kit (Invitrogen) according to the manufacturer’s information. After 48 h transection, luciferase activities were determined using a dual-luciferase reporter analysis System (Promega, USA). The relative luciferase activity was normalized to renilla luciferase activity.
Western blot analysis
The cultured cells or tissues were harvested and then homogenized in a lysis buffer (Tris-HCl 50 mmol/L, EDTA 5 mmol/L, NaCl 150 mmol/L, sodium deoxycholate 1%, Na3VO4 500 μmol/L, Triton X-100 0.5%, AEBSF 10 μmol/L, NaF 10 mmol/L) on ice for 30 min, then cell lysates were collected by centrifugation at 10,000 g for 15 min. The concentrations of protein were determined using the Bradford reagent kit (Sigma, German). Equal amounts of protein (20 μg/lane) from the cell lysates were separated on a 10% SDS-polyacrylamide gel (SDS-PAGE) and transferred onto polyvinylidene difluoride (PVDF) membranes (Bio-Rad, Richmond, CA, USA). The protein in the PVDF membrane was blocked with 5% nonfat skim milk in PBS plus 0.1% Tween-20 (PBST) for 2 h at room temperature. Then the membranes were incubated overnight at 4°C with antibodies against ADAM10 (1:1000; Santa Cruz Biotechnology Inc., California, USA) and β-actin (1:5000; Santa Cruz Biotechnology Inc). The membranes were incubated with secondary horseradish peroxidase (HRP)-conjugated anti-mouse antibodies (1:10000; Santa Cruz Biotechnology) at room temperature for 2 h. The protein bland was visualized using a chemiluminescent detection system (ECL, Thermo Scientific, Rockford, IL, USA).
Statistical analysis
All data are presented as mean ± SD (standard deviation) from at least three independent experiments with similar results. Statistical analyses were undertaken using SPSS version 19.0 software (SPSS, Chicago, IL, USA). The differences between groups were analyzed using Student’s t-test or one-way ANOVA. The relationship between ADAM10 and miR-449a expressions was analyzed with Pearson’s correlation. A value of P<0.05 was taken as an indication of statistical significance.
Results
miR-449a expression is downregulated in HCC tissues and cell lines
We firstly measured the expression of miR-449a in the 40 pairs of HCC tissues and adjacent nontumor tissues by quantitative RT-PCR. As shown in Figure 1A, miR-449a was significantly downregulated in HCC tissues compared with adjacent nontumor tissues (P<0.01). Furthermore, we also investigated the expression of miR-449a in four HCC cell lines (SMMC-7721, Hep3B, HepG2 and Huh-7) and normal hepatic cell line HL-7702 by qRT-PCR. Compared with normal hepatic cell line HL-7702, miR-449a expression was significantly decreased in four HCC cell lines (Figure 1B). HepG2 displayed a lowest expression level of miR-449a among four cell lines (Figure 1B), thus, it was selected as a model for below study.
Figure 1.
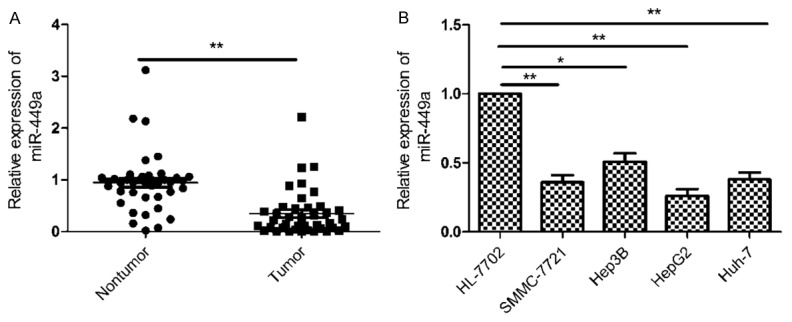
miR-449a expression was downregulated in HCC tissues and cell lines. A. The expression of miR-449a was measured in 40 pairs HCC tissues and adjacent nontumor tissues by qRT-PCR. **P<0.01 versus nontumor tissues. B. The expression of miR-449a was measured in four HCC cell lines (SMMC-7721, Hep3B, HepG2, Huh-7) and normal hepatic cell line HL-7702 by qRT-PCR. *P<0.05, **P<0.01 versus HL-7702.
miR-449a inhibits cell proliferation and colony formation in HCC cells
To assess the function of miR-449a in HCC procession, miR-449a mimic and negative controls (miR-NC) were transfected into HepG2 cells, respectively. miR-449a expression was upregulated in the HepG2 cells after transfected with miR-449a mimic (Figure 2A), suggesting that the efficiency of miR-449a mimic was high. We evaluated the effect of miR-449a on cell proliferation using CCK8 assay. As shown in Figure 2B, restoration of miR-449a significantly inhibited cell proliferation in HepG2 cells (P<0.01). Consistent with this result, miR-449a overexpression significantly inhibited colony formation of HepG2 cells (Figure 2C).
Figure 2.
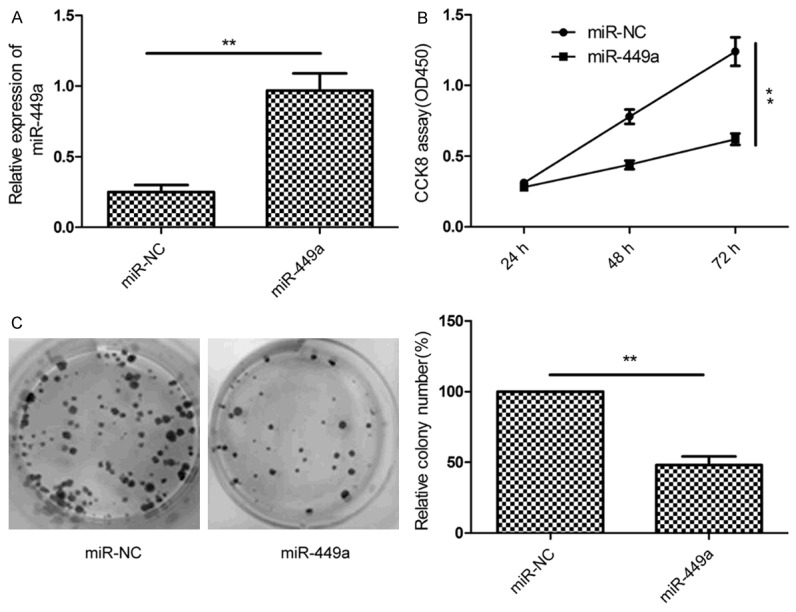
miR-449a inhibits cell proliferation and colony formation in HCC cells. A. The expression of miR-449a was measured in HepG2 cell after tranfected with miR-449a mimic or miR-NC using qRT-PCR. B. Cell proliferation was determined in HepG2 cell after tranfected with miR-449a mimic or miR-NC using CCK8 assay. C. Colony formation was determined in HepG2 cell after tranfected with miR-449a mimic or miR-NC. *P<0.05, **P<0.01 versus miR-NC.
miR-449a inhibits cell migration and invasion in HCC cells
To investigate the effect of miR-449a on metastasis ability of HCC cells, migration and invasion were determined in Hep2 cells transfected with miR-449a or miR-NC by using wound healing and transwell invasion assays, respectively. The result of wound-healing showed that miR-449a overexpression caused a suppression of cell migration in the HepG2 cells (Figure 3A). Transwell invasion assay demonstrated that miR-449a markedly reduced invasiveness of HepG2 cells (Figure 3B).
Figure 3.
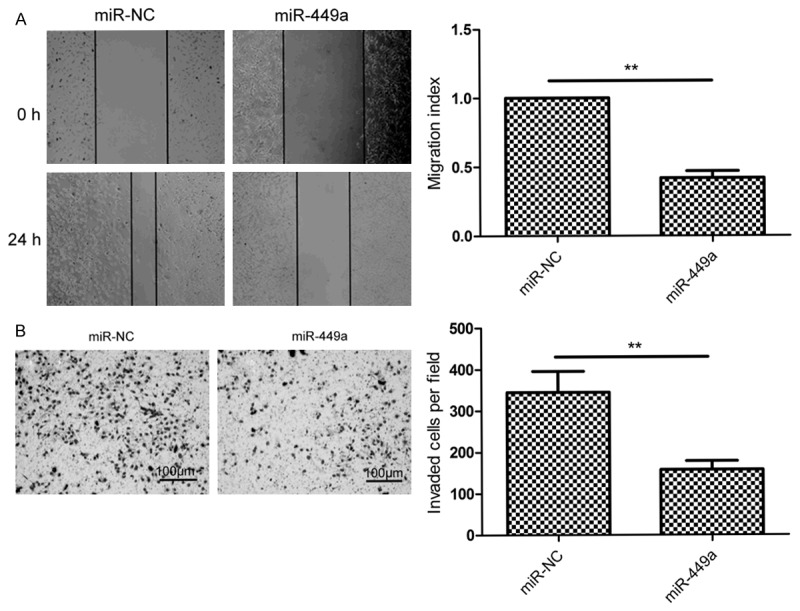
miR-449a inhibits cell migration and invasion in HCC cells. A. Cell migration was measured in HepG2 cell after tranfected with miR-449a mimic or miR-NC using wound healing assay. B. Cell invasion was determined in HepG2 cell after tranfected with miR-449a mimic or miR-NC using transwell invasion assay. *P<0.05, **P<0.01 versus miR-NC.
miR-449a directly targets ADAM10 3’-UTR
To explore the mechanism underlying inhibitory effects of miR-449 on growth and metastasis of HCC cells, putative miR-449a targets were predicted using target prediction algorithm (miRanda and TargetScan7.0). Our analysis revealed that ADAM10 was a potential target of miR-449a since the 3’-UTR of ADAM10 mRNA contains a complementary site for the seed region of miR-449a at position 287-293 (Figure 4A). To verify whether or not ADAM10 is a direct target of miR-449a, luciferase reporter assay was performed in HepG2 cells. As shown in Figure 4B, overexpression of miR-449a caused a significant decrease in luciferase activity in HepG2 cells transfected with the reporter plasmid with Wt-ADAM10-3’UTR, but not reporter plasmid with Mut-ADAM10-3’UTR. In addition, endogenous ADAM10 expression in HepG2 cells transfected with miR-449a mimic was examined. The results showed that ADAM10 expression on mRNA level and protein level was decreased in HepG2 cells transfected with miR-449a (Figure 4C and 4D). These results suggest that miR-449a directly targets ADAM10 3’-UTR, and inhibits its expression.
Figure 4.
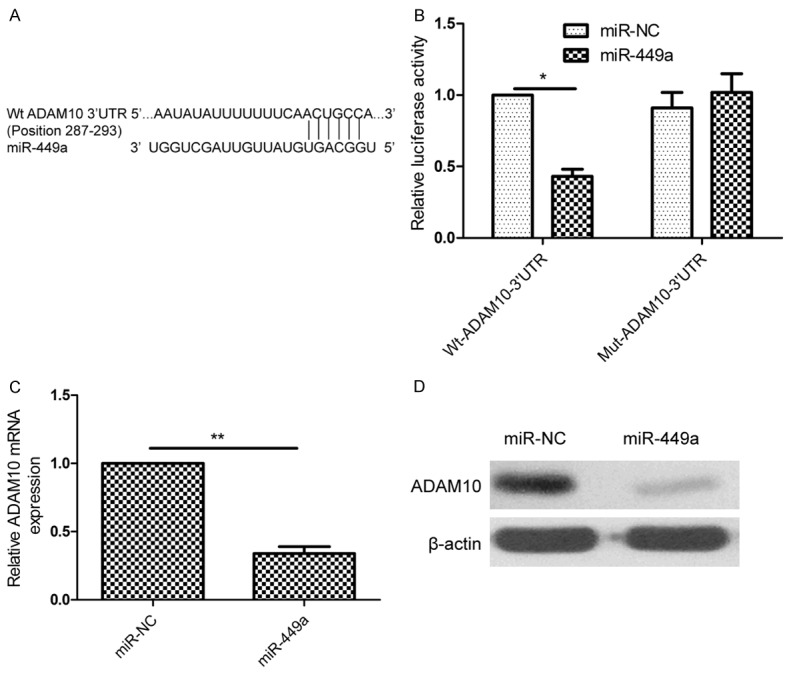
miR-449a directly targets ADAM10 3’-UTR. (A) Schematic illustration of the predicted miR-449a-binding sites in ADAM10 3’-UTR. (B) Luciferase activity was measured in HepG2 cells after co-transfected with miR-449a mimic or miR-NC and Wt or Mut ADAM10 3’UTR report plasmid. Wt: wide-type; Mut: mutant-type. (C and D) ADAM10 expression on mRNA level (C) and protein level (D) was measured in HepG2 cell after tranfected with miR-449a mimic or miR-NC by qRT-PCR and western blot, respectively. β-actin was used as a control. *P<0.05, **P<0.01 versus miR-NC.
miR-449a expression is negatively associated with ADAM10 in HCC tissues
To investigate the relationship between miR-449a and ADAM10 in HCC, ADAM10 expression on mRNA level and protein level was detected by qRT-PCR and Western blot, respectively. ADAM10 expression on mRNA level and protein level was increased in HCC tissues compared with adjacent nontumor tissues (Figure 5A and 5B). Through two-tailed Pearson’s correlation analysis, we found that ADAM10 mRNA expression was inversely correlated with miR-449a expression (Figure 5C; r=-0.655, P<0.001). In addition, we also found that found that ADAM10 protein expression was significantly increased in four HCC cell lines (SMMC-7721, Hep3B, HepG2, Huh-7) compared with normal hepatic cell line HL-7702 (Figure 5D).
Figure 5.
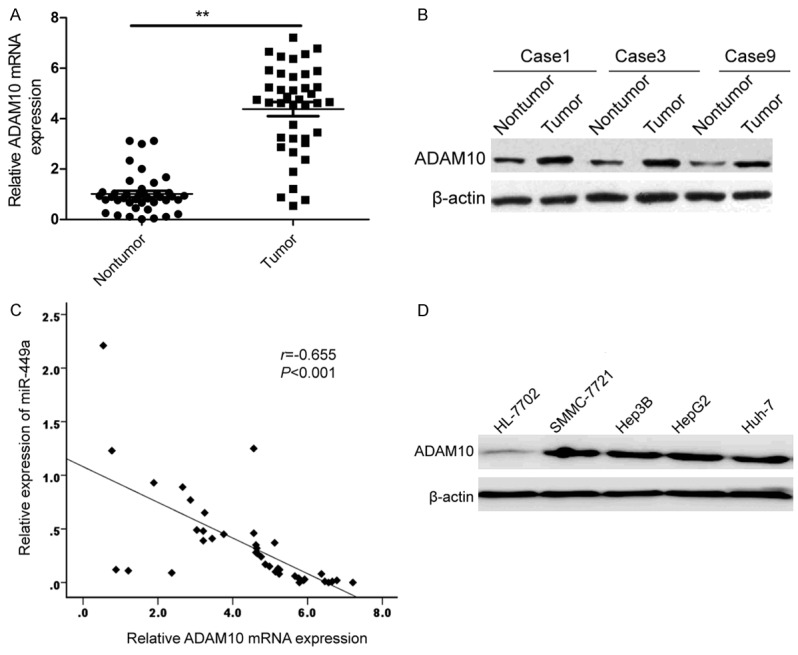
miR-449a expression is negatively associated with ADAM10 in HCC tissues. A. The mRNA expression of ADAM10 was measured in 40 pairs HCC tissues and adjacent nontumor tissues by qRT-PCR. **P<0.01 versus nontumor tissues. B. ADAM10 protein was measured in HCC tissues and corresponding nontumor tissues by western blot. β-actin was used as a control. C. The negative correlation between ADAM10 mRNA expression and miR-449a expression was determined by Pearson’s correlation. D. The ADAM10 protein expression was measured in four HCC cell lines (SMMC-7721, Hep3B, HepG2, Huh-7) and normal hepatic cell line HL-7702 by western blot. β-actin was used as a control.
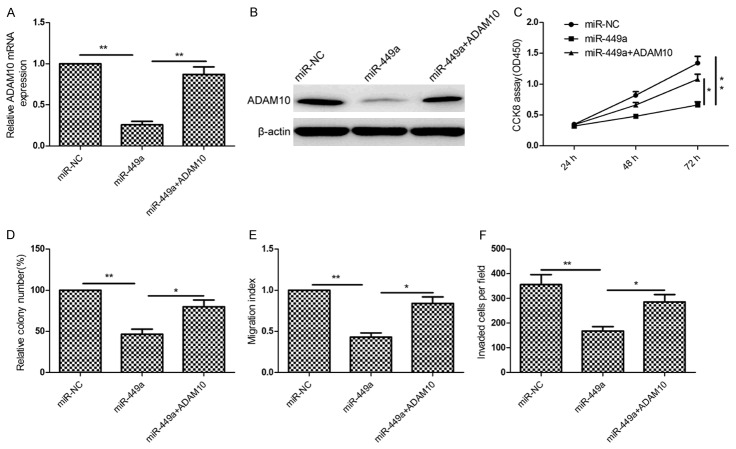
ADAM10 rescued the suppressive effect of miR-449a-mediated HCC on cell proliferation, colon formation, and invasion
To determine whether or not ADAM10 is involved in miR-449a-inhibition effect on cell proliferation, colon formation, and invasion, ADAM10 overexpression plasmid was transfected into the HepG2 cells with high levels of miR-449a or miR-NC to counteract the function of miR-449a. qRT-PCR and western blot assays demonstrated that miR-449a overexpression obviously ADAM10 expression on mRNA level and protein level in HepG2 cells, while overexpression ADAM10 plasmid abolished the inhibition expression of ADAM10 caused by the miR-449a mimic (Figure 6A and 6B). In addition, we found that overexpression of ADAM10 can rescue the suppressive effect on proliferation (Figure 6C), colony formation (Figure 6D), migration (Figure 6E) and invasion (Figure 6F) in HepG2 cells caused by miR-449a overexpression. These results suggested that miR-449a exerted its suppressive roles in HCC cells, at least in part, by repressing ADAM10.
Figure 6.
ADAM10 rescued the suppressive effect of miR-449a-mediated HCC on cell proliferation, colon formation, and invasion. (A and B) ADAM10 expression on mRNA level (A) and protein level (B) was measured in HepG2 cells transfected with miR-449a with/without ADAM10 overexpression plasmid. β-actin was used as an internal control. (C-F) Cell proliferation (C), colon formation (D), migration (E) and invasion (F) were determined in HepG2 cells transfected with miR-449a with/without ADAM10 overexpression plasmid. *P<0.05, **P<0.01 versus miR-449a.
Discussion
Accumulating evidence revealed that altered expression of miRNAs contribute to initiation and progression of HCC by promoting oncogene expression or by inhibiting tumor suppressor genes [8,9]. For instance, miR-222 could significantly promote cell proliferation, migration and invasion, and decrease cell apoptosis, as well as enhance the resistance of HCC cells to sorafenib at least in part through activating PI3K/AKT signaling pathway [18]. miR-140-5p overexpression suppressed HCC growth and metastasis by regulating TGFB receptor 1 (TGFBR1) and fibroblast growth factor 9 (FGF9) [19]. miR-154 overexpression in HepG2 cells could inhibit cell proliferation, migration and invasion, and induced apoptosis and cell arrest at G1 phase in vitro, as well as suppress tumor growth in nude mice model by repressing ZEB2 [20]. Here, we showed that miR-449a expression was downregulated in the HCC tissues and cell lines compared with the adjacent nontumor tissues and normal hepatic cell line; which is consistent with the previous reports that miR-449a expression was suppressed in HCC tissue and cell lines [15,16]. In addition, our study also showed that ectopic expression of miR-449a suppressed HCC cell proliferation, colony formation, migration and invasion by repressing ADAM10. These data suggested that miR-449a played important roles in HCC procession.
miR-449a is a member miR-449 family cluster, located in the first intron of CDC20B on chromosome 5q11, which is a susceptibility locus in a variety of malignancies, including HCC [15]. It has been reported that miR-449a expression is downregulated, and functioned as a tumor suppressor miRNA in majority maglignancies, such as prostate cancer [10], nasopharyngeal carcinoma [11], gastric cancer [12], non-small cell lung cancer [13], glioblastoma [21], endometrial cancer [22]. On the contrary, in breast cancer, miR-449a was found to be upregulated, and promote breast cancer cell proliferation, clonogenic survival, migration, and invasion by targeting Cysteine-Rich Protein 2 [23]. In the present study, our results showed that miR-449a expression was downregulated in the HCC tissues and cell lines, and that restoration of miR-449a suppressed cell proliferation, colony formation, migration and invasion of HCC, which is consistent with the previous reports that miR-449a expression was suppressed HCC growth and metastasis, and induced cell apoptosis [15,16]. Our results together with previous results suggest that miR-449a function as tumor suppressor in HCC.
A disintegrin and metalloproteinase 10 (ADAM10) is a member of the ADAM family which involved in multiple cellular processes such as cell proliferation, differentiation, migration, and invasion [24,25]. Growing evidence demonstrated that ADAM10 plays important roles in cancer initiation and progression [26,27]. It has been showed that ADAM10 expression was significantly up-regulated in HCC tissues, and its expression was significantly associated with higher tumor grade, tumor size and metastasis [28]. Recently studies showed that ADAM10 could promote the cell invasion and cell migration in HepG2 cells [29], and that downregulation of ADAM10 by siRNA could inhibit HCC cell migration and invasion [17,30], increase sorafenib antitumor activity of liver cancer [31]. These results suggest that ADAM10 serve as oncogene in HCC. Here, we identified ADAM10 as a direct target gene of miR-449a in HCC cell. ADAM10 expression was upregulated in HCC tissues and cell lines, and its mRNA expression level was negatively correlated with the expression level of miR-449a in the HCC tissues. ADAM10 overexpression rescued the effect of miR-449a-mediated HCC on cell proliferation, colony formation, migration and invasion. These results suggested that miR-449a exerts a tumor suppressive role in HCC partly through targeting ADAM10 expression. Previous studies has been demonstrated that miR-449a inhibited HCC growth and metastasis of HCC by targeting FOS and Met [15], as well as induced cell apoptosis by targeting Calpain6 and POU2F1 [16]. These results together with our results imply that miR-449a exert its suppressive role in HCC by targeting multiple genes.
In conclusion, the present study showed that miR-449a expression was downregulated in HCC tissues and cell lines, and that miR-449a functioned as a tumor suppressor miRNA by inhibiting cell proliferation, colony formation, migration, and invasion of HCC via partially repressing ADAM10 expression. These results suggested that miR-449a might be employed as a novel and effective therapeutic target for HCC in future.
Disclosure of conflict of interest
None.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Mazzola A, Costantino A, Petta S, Bartolotta TV, Raineri M, Sacco R, Brancatelli G, Camma C, Cabibbo G. Recurrence of hepatocellular carcinoma after liver transplantation: an update. Future Oncol. 2015;11:2923–2936. doi: 10.2217/fon.15.239. [DOI] [PubMed] [Google Scholar]
- 3.Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245–1255. doi: 10.1016/S0140-6736(11)61347-0. [DOI] [PubMed] [Google Scholar]
- 4.Valinezhad Orang A, Safaralizadeh R, Kazemzadeh-Bavili M. Mechanisms of miRNA-Mediated Gene Regulation from Common Downregulation to mRNA-Specific Upregulation. Int J Genomics. 2014;2014:970607. doi: 10.1155/2014/970607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 6.Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2006;94:776–780. doi: 10.1038/sj.bjc.6603023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mao B, Wang G. MicroRNAs involved with hepatocellular carcinoma (Review) Oncol Rep. 2015;34:2811–2820. doi: 10.3892/or.2015.4275. [DOI] [PubMed] [Google Scholar]
- 9.Giordano S, Columbano A. MicroRNAs: new tools for diagnosis, prognosis, and therapy in hepatocellular carcinoma? Hepatology. 2013;57:840–847. doi: 10.1002/hep.26095. [DOI] [PubMed] [Google Scholar]
- 10.Kumar P, Sharad S, Petrovics G, Mohamed A, Dobi A, Sreenath TL, Srivastava S, Biswas R. Loss of miR-449a in ERG-associated prostate cancer promotes the invasive phenotype by inducing SIRT1. Oncotarget. 2016 doi: 10.18632/oncotarget.8061. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niu M, Gao D, Wen Q, Wei P, Pan S, Shuai C, Ma H, Xiang J, Li Z, Fan S, Li G, Peng S. MiR-29c regulates the expression of miR-34c and miR-449a by targeting DNA methyltransferase 3a and 3b in nasopharyngeal carcinoma. BMC Cancer. 2016;16:218. doi: 10.1186/s12885-016-2253-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi J, Liu Y, Liu J, Zhou J. Hsa-miR-449a genetic variant is associated with risk of gastric cancer in a Chinese population. Int J Clin Exp Pathol. 2015;8:13387–13392. [PMC free article] [PubMed] [Google Scholar]
- 13.You J, Zhang Y, Li Y, Fang N, Liu B, Zu L, Zhou Q. MiR-449a suppresses cell invasion by inhibiting MAP2K1 in non-small cell lung cancer. Am J Cancer Res. 2015;5:2730–2744. [PMC free article] [PubMed] [Google Scholar]
- 14.Li Q, Peng J, Li X, Leng A, Liu T. miR-449a targets Flot2 and inhibits gastric cancer invasion by inhibiting TGF-beta-mediated EMT. Diagn Pathol. 2015;10:202. doi: 10.1186/s13000-015-0435-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen SP, Liu BX, Xu J, Pei XF, Liao YJ, Yuan F, Zheng F. MiR-449a suppresses the epithelial-mesenchymal transition and metastasis of hepatocellular carcinoma by multiple targets. BMC Cancer. 2015;15:706. doi: 10.1186/s12885-015-1738-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Y, Wang Y, Sun X, Mei C, Wang L, Li Z, Zha X. miR-449a promotes liver cancer cell apoptosis by downregulation of Calpain6 and POU2F1. Oncotarget. 2016;7:13491–501. doi: 10.18632/oncotarget.4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu S, Zhang W, Liu K, Ji B, Wang G. Silencing ADAM10 inhibits the in vitro and in vivo growth of hepatocellular carcinoma cancer cells. Mol Med Rep. 2015;11:597–602. doi: 10.3892/mmr.2014.2652. [DOI] [PubMed] [Google Scholar]
- 18.Liu K, Liu S, Zhang W, Ji B, Wang Y, Liu Y. miR222 regulates sorafenib resistance and enhance tumorigenicity in hepatocellular carcinoma. Int J Oncol. 2014;45:1537–1546. doi: 10.3892/ijo.2014.2577. [DOI] [PubMed] [Google Scholar]
- 19.Yang H, Fang F, Chang R, Yang L. MicroRNA-140-5p suppresses tumor growth and metastasis by targeting transforming growth factor beta receptor 1 and fibroblast growth factor 9 in hepatocellular carcinoma. Hepatology. 2013;58:205–217. doi: 10.1002/hep.26315. [DOI] [PubMed] [Google Scholar]
- 20.Pang X, Huang K, Zhang Q, Zhang Y, Niu J. miR-154 targeting ZEB2 in hepatocellular carcinoma functions as a potential tumor suppressor. Oncol Rep. 2015;34:3272–3279. doi: 10.3892/or.2015.4321. [DOI] [PubMed] [Google Scholar]
- 21.Yao Y, Ma J, Xue Y, Wang P, Li Z, Li Z, Hu Y, Shang X, Liu Y. MiR-449a exerts tumor-suppressive functions in human glioblastoma by targeting Myc-associated zinc-finger protein. Mol Oncol. 2015;9:640–656. doi: 10.1016/j.molonc.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ye W, Xue J, Zhang Q, Li F, Zhang W, Chen H, Huang Y, Zheng F. MiR-449a functions as a tumor suppressor in endometrial cancer by targeting CDC25A. Oncol Rep. 2014;32:1193–1199. doi: 10.3892/or.2014.3303. [DOI] [PubMed] [Google Scholar]
- 23.Shi W, Bruce J, Lee M, Yue S, Rowe M, Pintilie M, Kogo R, Bissey PA, Fyles A, Yip KW, Liu FF. MiR-449a promotes breast cancer progression by targeting CRIP2. Oncotarget. 2016 doi: 10.18632/oncotarget.7753. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klein T, Bischoff R. Active metalloproteases of the A Disintegrin and Metalloprotease (ADAM) family: biological function and structure. J Proteome Res. 2011;10:17–33. doi: 10.1021/pr100556z. [DOI] [PubMed] [Google Scholar]
- 25.Seals DF, Courtneidge SA. The ADAMs family of metalloproteases: multidomain proteins with multiple functions. Genes Dev. 2003;17:7–30. doi: 10.1101/gad.1039703. [DOI] [PubMed] [Google Scholar]
- 26.Mochizuki S, Okada Y. ADAMs in cancer cell proliferation and progression. Cancer Sci. 2007;98:621–628. doi: 10.1111/j.1349-7006.2007.00434.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crawford HC, Dempsey PJ, Brown G, Adam L, Moss ML. ADAM10 as a therapeutic target for cancer and inflammation. Curr Pharm Des. 2009;15:2288–2299. doi: 10.2174/138161209788682442. [DOI] [PubMed] [Google Scholar]
- 28.Zhang W, Liu S, Liu K, Wang Y, Ji B, Zhang X, Liu Y. A disintegrin and metalloprotease (ADAM)10 is highly expressed in hepatocellular carcinoma and is associated with tumour progression. J Int Med Res. 2014;42:611–618. doi: 10.1177/0300060513505500. [DOI] [PubMed] [Google Scholar]
- 29.Yuan S, Lei S, Wu S. ADAM10 is overexpressed in human hepatocellular carcinoma and contributes to the proliferation, invasion and migration of HepG2 cells. Oncol Rep. 2013;30:1715–1722. doi: 10.3892/or.2013.2650. [DOI] [PubMed] [Google Scholar]
- 30.Liu S, Zhang W, Liu K, Wang Y, Ji B, Liu Y. Synergistic effects of co-expression plasmidbased ADAM10-specific siRNA and GRIM-19 on hepatocellular carcinoma in vitro and in vivo. Oncol Rep. 2014;32:2501–2510. doi: 10.3892/or.2014.3503. [DOI] [PubMed] [Google Scholar]
- 31.Zhang W, Liu S, Liu K, Ji B, Wang Y, Liu Y. Knockout of ADAM10 enhances sorafenib antitumor activity of hepatocellular carcinoma in vitro and in vivo. Oncol Rep. 2014;32:1913–1922. doi: 10.3892/or.2014.3418. [DOI] [PubMed] [Google Scholar]