Abstract
HIF-1α is an important transcriptional factor, which plays roles in cancer development and progression. But its regulation by miRNAs is not clear. Here, to investigate the regulation of HIF-1α by miRNAs, miRNAs were predicted and miR-526b-3p was verified as a regulator of HIF-1α in colon cancer cells. Using TaqMan RT-PCR analysis, we analyzed the expression of miR-526b-3p in tumor tissues and cell lines and found that miR-526b-3p was consistently under-expressed in cancer tissues and cell lines compared with their normal controls. When miR-526b-3p was induced into the colon cancer cells, cell proliferation, metastasis and glycolysis of colon cancer cells were suppressed. We also found that miR-526b-3p was down-regulated in metastatic colon cancer tissues and negatively with HIF-1α mRNA in colon cancer tissues. In a summary, miR-526b-3p plays as a tumor suppressor by down-regulation of HIF-1α expression in colon cancer and may be a new diagnosis or therapeutic target.
Keywords: miR-526b-3p, HIF-1α, colon cancer, glycolysis
Introduction
Colon cancer is a very common type of cancer. Its therapeutic methods usually include operation and chemotherapy. However, there are still many questions needing to resolve such as early diagnosis, drug resistance and metastasis. Hypoxia-inducible factors (HIFs), critical regulators in tumor hypoxia, are known to regulate multiple steps of tumorigenesis and are typically associated with changes in metabolism, angiogenesis, metastasis, drug resistance, and poor clinical outcomes [1,2]. HIFs consists of HIF-α and HIF-β subunits. HIF-1α and HIF-2α are the two isoforms of HIF-α family. In normoxia, α subunits of HIF are hydroxylated, recognized by the von Hippel-Lindau (pVHL), the substrate recognition component of an E3 ubiquitin ligase complex that targets HIF-α for proteasomal degradation. In hypoxia, HIF-α is stabilized by inhibiting prolyl hydroxylation, and in turn ubiquitin proteasomal degradation, making HIF-α capable of dimerizing with ARNT, binding to the hypoxia-responsive DNA element, and recruiting the transcription coactivator p300/CBP for the transcriptional activation of a host of hypoxia-responsive genes [3-5].
MicroRNAs (miRNAs) are small non-coding RNA molecules, which broadly express in various cells and regulate gene expression at the posttranscriptional level through interaction with the 3’-UTRs of target mRNAs [5-8]. There are reports showed that many miRNAs participate cell proliferation, development, differentiation, apoptosis, metabolism and metastasis of colon cancer. Indeed, recent studies show that aberrantly regulated miRNAs like miR-106b, miR-135, miR-21, miR-622 in colon cancers by targeting functional genes [9-12]. Studies show that miRNA expression is dysregulated in colon cancer.
In our study, miR-526b-3p was predicted as a regulator of HIF-1α. The expression of miR-526b-3p in colon cancer was examined in colon cancer tissue samples and cell lines. Role of miR-526b-3p in colon cancer cells were investigated, which included cell proliferation and metastasis. The study demonstrated that miR-526b-3p played as a suppressor in colon cancer by targeting HIF-1α.
Materials and methods
Tumor samples
Colon cancer and non-cancer samples were obtained from 86 patients diagnosed with colon cancer. The specimens of colon cancer were confirmed histologically. Adjacent noncancerous tissue samples were used as normal control tissues. The research protocol for the human tissue collection was approved by the review board of Liaoning cancer hospital & institute (Shenyang, China).
Cell culture
Human colon cancer cell lines CT26, SW116, LS174T, DLD-1, LOVO, SW620, HCT116, HT-29 and SW480 were obtained from China Center for Type Culture Collection (ATCC, Rockville, MD, USA) and Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Human benign colon cells NCM460 were obtained from ATCC. The cells were maintained in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum, 100 units/ml penicillin and streptomycin and maintained in cell culture flasks.
RNA Isolation and quantitative real-time-PCR
Total RNA from the various cells and tissues were extracted using RNeasy Mini Kit (QIAGEN, USA) according to the manufacturer’s instructions. The total RNA was treated with DNase I and purified using phenol-chloroform. The real-time RT-PCR for miRNA detection was performed using mirVana qRT-PCR miRNA detection kit. The real-time RT-PCR for HIF-1α detection was performed using SYBR Green One-step Real-Time RT-PCR Master Mixes (ThermoFisher). Reactions were carried out in Applied Biosystems 7500 Real time PCR system. The running conditions were according to the manuals.
Analysis of cell proliferation
Colon cancer cells HT-29 and SW480 were plated in 6-well plates and transfected with mimic miR-526b-3p or the control. Cell proliferation was assayed by CCK8 according to the instructions. The absorbance value was at 570 nm.
Invasion assay
Colon cancer cells HT-29 and SW480 were plated in 6-well plates and transfected with mimic miR-526b-3p or HIF-1α or the control. Cell invasion was evaluated using 24-transwell chambers with coating matrigel (8-μm pore size, Corning, NY). The invaded cells were fixed and stained with 0.1% crystal violet and counted under a microscope at least five randomly selected fields.
Luciferase reporter assay
The wild-typed HIF-1α 3’-UTR sequence was amplified from NCM460 cells by PCR. The mutated HIF-1α 3’-UTR was performed using QuickChange Site-Directed Mutagenesis Kit. The wide-type and mutated HIF-1α 3’-UTR were cloned into pMirTarget vector (OriGene). Colon cancer cells were co-transfected with HIF-1α 3’-UTR or miR-526b-3p mimics using Lipofectamine 2000 for 24 hour and then and performed for luciferase activity analysis using luciferase assay system (Promega).
Western blotting
Colon cancer cells HT-29 and SW480 were plated in 6-well plates and transfected with mimic miR-526b-3p or HIF-1α or the control. Total protein was extracted using RIPA buffer, qualified and then performed for SDS-PAGE. The protein on the gels was transferred to PVDF membranes. The blots were probed with the anti-HIF-1α, LDHA, PKM2, HK2, GLUT1 and GAPDH and detected using an ECL system (Pierce).
Statistical analysis
Data were analyzed using SPSS 15.0. All data were represented as mean ± SEM from triplicate experiments. Statistical analyses were performed using Student’s t-test, and a p < 0.05 value was considered significant.
Results
miR-526b-3p is identified as a potential regulator of HIF-1α in colon cancer cells
HIF-1α is an important transcriptional factor, which is induced by hypoxia or growth factors. To find the new miRNAs that regulates HIF-1α, miRNA target gene prediction software miRBase was used. The most potential miRNAs including the reported miRNAs were selected for real time RT-PCR, the data showed that the significant miRNAs were found in HT-29 colon cancer cells (Figure 1A). The significant miRNAs included miR-199a-5p, miR-199b-5p, miR-135b-5p, miR-135b-5p, miR-142-3p.2, miR-18b-5p, miR-18a-5p, miR-4735-3p, miR-20b-5p, miR-20a-5p, miR-106b-5p, miR-106a-5p, miR-93-5p, miR-17-5p, miR-338-3p, miR-481-3p, and miR-526b-3p (Figure 1A). The down-regulation of miR-526b-3p expression was verified in various colon cancer cells (Figure 1B). miR-526b-3p was selected for the following study.
Figure 1.
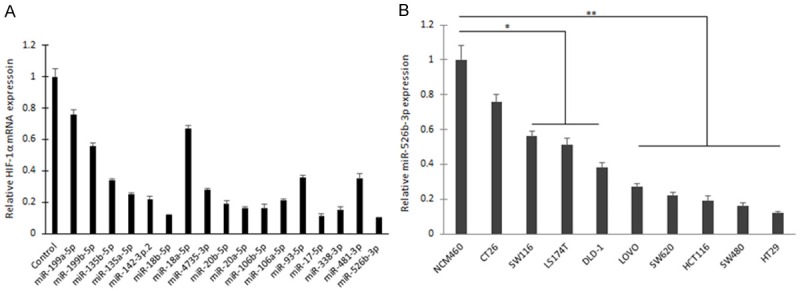
miR-526b-3p is identified as a potential regulator of HIF-1α in colon cancer cells. A. The expression miRNAs in HT-29 colon cancer cells that regulates HIF-1α. miRNA expression was examined by real time RT-PCR. B. miR-526b-3p was down-regulated in various colon cancer cells. *p < 0.05, **p < 0.01 vs control.
miR-526b-3p regulates HIF-1α expression in colon cancer cells
HIF-1α is a candidate target gene of miR-526b-3p using miRBase (Figure 2A). When HT-29 cells were co-transfected wide type of reporter plasmid carrying with 3’-UTR of HIF-1α or the mutated plasmid and miR-526b-3p, the dual reporter assays revealed that introduction of miR-526b-3p suppressed the activity of 3’-UTR of HIF-1α, but not mutated 3’-UTR of HIF-1α (Figure 2B). When the colon cancer cells including HT29, SW480 and HCT116 cells were transfected miR-526b-3p mimics, HIF-1α mRNA decreased significantly (Figure 2C). Because HIF-1α protein is sensitive to oxygen and easily degraded, so the colon cancers were transfected miR-526b-3p and then exposed to 1% oxygen for HIF-1α induction. The data from western blotting showed that HIF-1α protein was reduced in the cells with miR-526b-3p mimics transfection (Figure 2D). The results indicated that HIF-1α is a direct target of miR-526b-3p in colon cancer cells.
Figure 2.
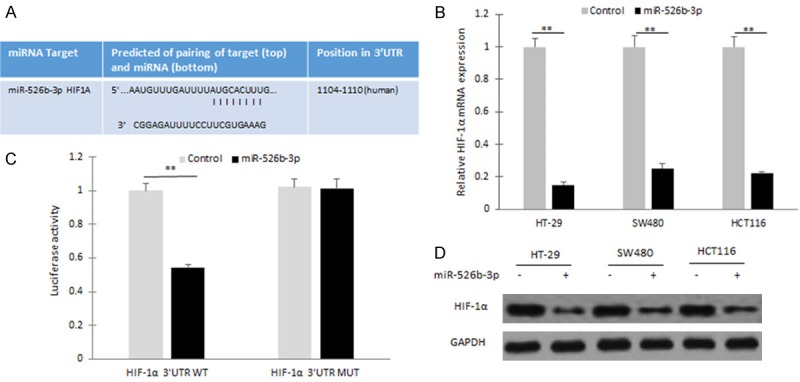
HIF-1α is a direct target of miR-526b-3p in colon cancer cells. A. Wild typed and mutant target sites for miR-526b-3p sequences in 3’UTR of HIF-1α were shown. B. Luciferase reporter assays were performed to verify the binding of miR-526b-3p in 3’-UTR of HIF-1α. WT: wild type; Mut: mutation. C. qRT-PCR assay was performed to detect the mRNA level of HIF-1α in HT29 cells treated with miR-526b-3p mimics. miR-526b-3p was measured by miRNA real-time RT-PCR in HT29, SW480 and HCT116 cells after miR-526b-3p mimic transfection. D. Western blotting analysis was used to measure HIF-1α protein in colon cancer cells exposing in hypoxia (1% O2) treated with miR-526b-3p mimics. *p < 0.05, **p < 0.01 vs control.
Low expression of miR-526b-3p is positively correlated with clinincal features of colon cancer
miR-526b-3p expression levels in colon cancer tissues were examined by qRT-PCR analyses and significantly lower in 86 human Colon cancer tissues than their normal adjacent Colon tissues (Figure 3A). Furthermore, miR-526b-3p expression levels were compared between metastatic tissues and non-metastatic colon ones and the former was greatly lower than the latter (Figure 3B). Further analysis showed that miR-526b-3p expression was negatively associated with the histological grade of colon cancer (Figure 3C). The results demonstrated that lack of miR-526b-3p expression in colon cancer is positively related to the advanced stage, metastasis, and poor prognosis.
Figure 3.
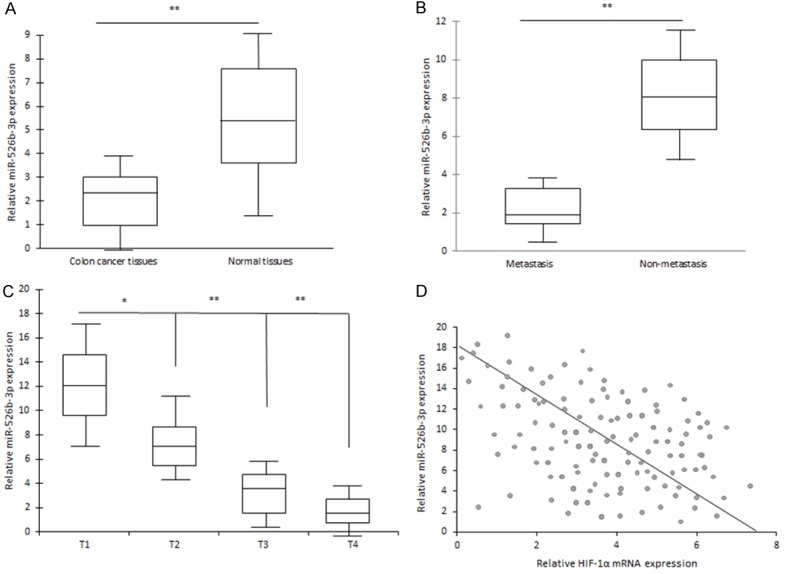
Low expression of miR-526b-3p is positively correlated with clinic features of colon cancer. A. miR-526b-3p was down-regulated in various colon cancer tissues. B. miR-526b-3p was down-regulated in metastatic colon cancer tissues. C. miR-526b-3p was related to colon cancer features in clinic. D. HIF-1α mRNA expression was negatively with miR-526b-3p expression in colon cancer tissues. *p < 0.05, **p < 0.01 vs control.
HIF-1α was found to be negatively related to miR-526b-3p expression in colon cancer tissues (Figure 3D).
miR-526b-3p suppresses colon cancer cell proliferation and invasion by down-regulation of HIF-1α
HIF-1α was identified as a target gene of miR-526b-3p, but the functional relationship between miR-526b-3p and HIF-1α is not known. HT29 cells were transfected with both HIF-1α/ΔODD and miR-526b-3p, and then cell proliferation was assayed by CCK8. It was shown that cells with HIF-1α overexpression grew faster, and miR-526b-3p inhibited HIF-1α induced cell growing (Figure 4A and 4B). Moreover, upon transfection with the HIF-1α/ΔODD construct, the suppression of miR-526b-3p mediated cell invasion (Figure 4C and 4D) in HT29 and SW480 cells was also abolished. Taken together, the results demonstrated that the repression of cell progression by miR-526b-3p was typically a consequence of decreased HIF-1α expression in colon cancer cells.
Figure 4.
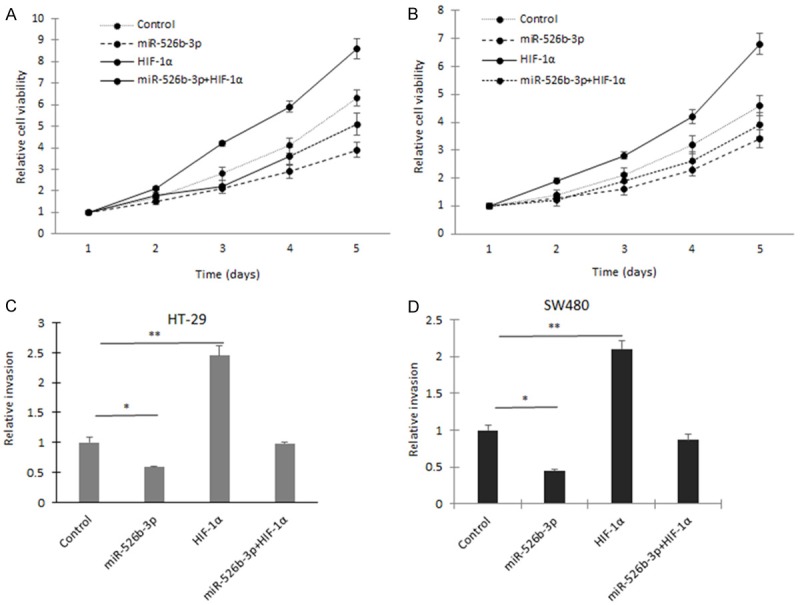
miR-526b-3p suppresses colon cancer cell proliferation and invasion by down-regulation of HIF-1α. A. Cell grow was assayed in HT-29 cells with miR-526b-3p or HIF-1α transfection. B. Cell grow was assayed in SW480 cells with miR-526b-3p or HIF-1α transfection. C. Cell migration was assayed in HT-29 and SW480 cells with miR-526b-3p or HIF-1α transfection. D. Cell invasion was assayed in HT-29 and SW480 cells with miR-526b-3p or HIF-1α transfection. *p < 0.05, **p < 0.01 vs control.
miR-526b-3p regulates colon cancer cell glycolysis through HIF-1α
HIF-1α could regulate cancer cell metabolism by its targets genes such as CA9, LDHA. To investigate whether miR-526b-3p involves in colon cancer cell metabolism, HT-29 and SW480 cells were transfected miR-526b-3p and HIF-1α/ΔODD, and glycolysis rate and lactate were examined. It was showed that cells with miR-526b-3p overexpression had low rate of glycolysis than in the cells with HIF-1α overexpression and miR-526b-3p could inhibit the glycolysis rate of the cells induced by HIF-1α (Figure 5A and 5B). Lactate production was also inhibited in HT-29 and SW480 cells with miR-526b-3p overexpression and miR-526b-3p could inhibit the lactate production of the cells induced by HIF-1α (Figure 5C and 5D). The changes of glycolysis was related to up-regulation of glycolysis associated gene expression (Figure 5E). LDHA, HK2 and GLUT1 were suppressed in the cells with miR-526b-3p restoration.
Figure 5.
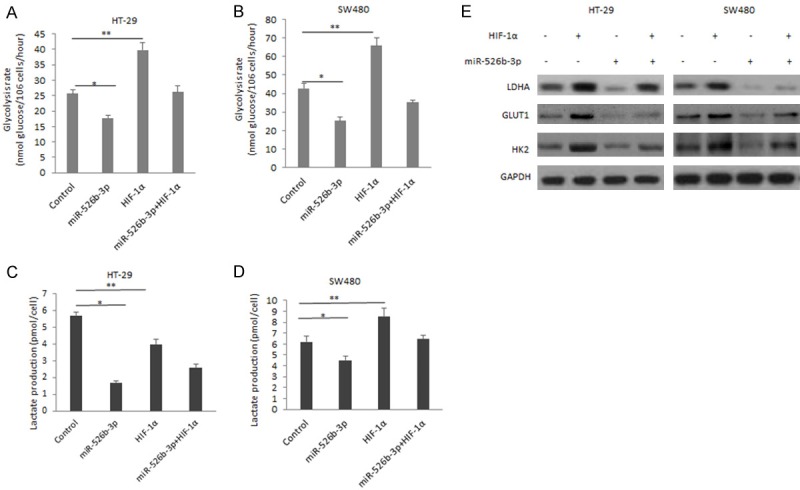
miR-526b-3p regulates colon cancer cell glycolysis through HIF-1α. A. Glycolysis rate in HT-29 cells co-transfected miR-526b-3p or HIF-1α/ΔODD. B. Glycolysis rate in SW480 cells co-transfected miR-526b-3p and HIF-1α/ΔODD. C. Lactate production in HT-29 cells co-transfected miR-526b-3p and HIF-1α/ΔODD. D. Lactate production in SW480 cells co-transfected miR-526b-3p and HIF-1α/ΔODD. E. The changes of glycolysis associated protein in cells transfected miR-526b-3p or HIF-1α/ΔODD. *p < 0.05, **p < 0.01 vs control.
Discussion
Increasing number of studies has demonstrated that miRNAs may serve as novel prognostic markers for prediction of chemotherapeutic responses and prognosis in patients with colon cancer. In the current study, we show that miR-526b-3p can regulate HIF-1α in colon cancer cell lines. We revealed an inverse correlation between miR-526b-3p and HIF-1α in a panel of colon cancer cell lines as compared to a normal colon epithelial cell line. miR-526b-3p restoration in colon cancer cell lines, reduces HIF-1α mRNA and protein levels, and inhibits cell growth, colony formation and metastasis. In conclusion, our data suggest that miR-526b-3p acts as a tumor suppressive miRNA and when down-regulated promotes tumorigenesis through the up regulation of HIF-1α.
There are several miRNAs targeting HIF-1α and playing roles in tumor and other diseases. For example, Xu et al reported that miR-338-3p is involved in sensitivity to sorafenib in hepatocarcinoma cells via targeting HIF-1α [13]. Krutilina reported that miR-18a inhibits lung metastasis in breast cancer by down-regulation of HIF-1α [14]. Except in cancer, regulation of HIF-1α by miRNAs is also reported. MiR-210 could regulates TH17 cell differentiation by targeting HIF-1α expression [15]. In our study, we found a new regulator of HIF-1α, miR-526b-3p. Firstly, it was verified that HIF-1α is indeed a direct target gene of miR-526b-3p in colon cancer cells. Further research demonstrated that HIF-1α mRNA and protein were negatively regulated by miR-526b-3p. The cellular function assays indicated that miR-526b-3p could inhibit colon cancer cell proliferation, metastasis and glycolysis by targeting HIF-1α.
Overall, some of the important tumor-suppressive mechanism of miR-526b-3p through the negative regulation of HIF-1α was investigated. miR-526b-3p may be a miRNA-based molecular therapy candidate of colon cancer. Our findings provide insight into the mechanism of dysregulation of colon cancer stem cells and eventually colorectal cancer initiation and progression. By understanding the molecular mechanisms of colorectal cancer biology, there are more methods or targets of colon cancer therapy be developed.
Acknowledgements
Supported by science and technology key project of Liaoning Province, China (NO. 2012225016).
Disclosure of conflict of interest
None.
References
- 1.Xie L, Xue X, Taylor M, Ramakrishnan SK, Nagaoka K, Hao C, Gonzalez FJ, Shah YM. Hypoxia-inducible factor/MAZ-dependent induction of caveolin-1 regulates colon permeability through suppression of occludin, leading to hypoxia-induced inflammation. Mol Cell Biol. 2014;34:3013–3023. doi: 10.1128/MCB.00324-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shay JE, Imtiyaz HZ, Sivanand S, Durham AC, Skuli N, Hsu S, Mucaj V, Eisinger-Mathason TS, Krock BL, Giannoukos DN, Simon MC. Inhibition of hypoxia-inducible factors limits tumor progression in a mouse model of colorectal cancer. Carcinogenesis. 2014;35:1067–1077. doi: 10.1093/carcin/bgu004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jung KH, Lee JH, Thien Quach CH, Paik JY, Oh H, Park JW, Lee EJ, Moon SH, Lee KH. Resveratrol suppresses cancer cell glucose uptake by targeting reactive oxygen species-mediated hypoxia-inducible factor-1α activation. J Nucl Med. 2013;54:2161–2167. doi: 10.2967/jnumed.112.115436. [DOI] [PubMed] [Google Scholar]
- 4.Zhdanov AV, Dmitriev RI, Papkovsky DB. Bafilomycin A1 activates HIF-dependent signalling in human colon cancer cells via mitochondrial uncoupling. Biosci Rep. 2012;32:587–595. doi: 10.1042/BSR20120085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tokarz P, Blasiak J. The role of microRNA in metastatic colorectal cancer and its significance in cancer prognosis and treatment. Acta Biochim Pol. 2012;59:467–474. [PubMed] [Google Scholar]
- 6.Rossi S, Di Narzo AF, Mestdagh P, Jacobs B, Bosman FT, Gustavsson B, Majoie B, Roth A, Vandesompele J, Rigoutsos I, Delorenzi M, Tejpar S. microRNAs in colon cancer: a roadmap for discovery. FEBS Lett. 2012;586:3000–3007. doi: 10.1016/j.febslet.2012.07.048. [DOI] [PubMed] [Google Scholar]
- 7.Schetter AJ, Harris CC. Alterations of microRNAs contribute to colon carcinogenesis. Semin Oncol. 2011;38:734–742. doi: 10.1053/j.seminoncol.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu WK, Law PT, Lee CW, Cho CH, Fan D, Wu K, Yu J, Sung JJ. MicroRNA in colorectal cancer: from benchtop to bedside. Carcinogenesis. 2011;32:247–253. doi: 10.1093/carcin/bgq243. [DOI] [PubMed] [Google Scholar]
- 9.Schlörmann W, Naumann S, Renner C, Glei M. Influence of miRNA-106b and miRNA-135a on butyrate-regulated expression of p21 and Cyclin D2 in human colon adenoma cells. Genes Nutr. 2015;10:50. doi: 10.1007/s12263-015-0500-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fang Y, Sun B, Li Z, Chen Z, Xiang J. MiR-622 inhibited colorectal cancer occurrence and metastasis by suppressing K-Ras. Mol Carcinog. 2015 doi: 10.1002/mc.22380. [DOI] [PubMed] [Google Scholar]
- 11.Yu Y, Nangia-Makker P, Farhana L, G Rajendra S, Levi E, Majumdar AP. miR-21 and miR-145 cooperation in regulation of colon cancer stem cells. Mol Cancer. 2015;14:98. doi: 10.1186/s12943-015-0372-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones MF, Hara T, Francis P, Li XL, Bilke S, Zhu Y, Pineda M, Subramanian M, Bodmer WF, Lal A. The CDX1-microRNA-215 axis regulates colorectal cancer stem cell differentiation. Proc Natl Acad Sci U S A. 2015;112:E1550–1558. doi: 10.1073/pnas.1503370112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu H, Zhao L, Fang Q, Sun J, Zhang S, Zhan C, Liu S, Zhang Y. MiR-338-3p inhibits hepatocarcinoma cells and sensitizes these cells to sorafenib by targeting hypoxia-induced factor 1α. PLoS One. 2014;9:e115565. doi: 10.1371/journal.pone.0115565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krutilina R, Sun W, Sethuraman A, Brown M, Seagroves TN, Pfeffer LM, Ignatova T, Fan M. MicroRNA-18a inhibits hypoxia-inducible factor 1α activity and lung metastasis in basal breast cancers. Breast Cancer Res. 2014;16:R78. doi: 10.1186/bcr3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang H, Flach H, Onizawa M, Wei L, McManus MT, Weiss A. Negative regulation of Hif1a expression and TH17 differentiation by the hypoxia-regulated microRNA miR-210. Nat Immunol. 2014;15:393–401. doi: 10.1038/ni.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]


