Abstract
Aim: Besides surgical treatment, systematic chemotherapy plays a crucial role in HCC treatment, especially for patients with advanced HCC. However, none of the single-drug-treatment strategies have shown significant survival benefit due to a high incidence rate of chemoresistance. This study was designed to observe the effect of small interfering of RNA (SiRNA) targeting multidrug resistance-related protein 1-4 (MRP1, MRP2, MRP3, and MRP4) in modulating drug resistance of HepG2/ADM and SMMC7721/ADM cells. Methods: HepG2/Adriamycin (ADM) and SMMC7721/ADM cell lines were developed by exposing parental cells to stepwise increasing concentrations of ADM. MTT assay was used to determine drug sensitivity and half inhibitory concentration (IC50) of drugs was calculated. Flow cytometry was employed to analyze cell cycle distribution. MRP1-4 mRNA expression levels were measured by quantitative real-time PCR (QRT-PCR). Expression of proteins was analyzed by Western blot. The growth curve was draw and the cell apoptosis was also observed. Animal experiment was used to compare the cell growth. Results: MTT assay showed that the values of IC50 and RI of HepG2/ADM and SMMC7721/ADM decreased after siRNA treatment in HepG2/ADM cells and SMMC7721/ADM cells. QRT-PCR analysis demonstrated the MRP1-4 mRNA expression decreased significantly in HepG2/ADM cells and SMMC7721/ADM cells after siRNA transfection. In addition, compared with parental cells, MRP1-4 protein expressions apparently decreased in SMMC7721/ADM and HepG2/ADM cells. Flow cytometry showed significantly elevated apoptosis rate following MRP1-4 siRNA transfection. Animal experiment suggested that silencing MRP1-4 gene in vivo inhibited tumor growth. Conclusion: Inhibition of MRP1-4 by small interfering RNA enhanced and selectively restored sensitivity of hepatoma cells to drugs. MRP1-4 siRNA might represent a new therapeutic option for HCC.
Keywords: Hepatocellular, multidrug resistance, MRP1-4, siRNA
Introduction
According to the latest statistics, there are about six hundred thousand new liver cancer cases worldwide each year, of which about 55% occur in our country. It has become a major killer that seriously threatens people’s health and lives [1]. Surgery and chemotherapy are the main treatments for liver cancer currently. However, liver cancer has low liver resection rate, low sensitivity to chemotherapy, and high recurrence rate, and its five year survival rate is only 14%-30% [2,3]. Although chemotherapy is an important means of treatment for liver cancer and there has been new chemotherapy drugs and launch, multidrug resistant (MDR) limits the application of liver cancer chemotherapy, and it is also a major cause of liver cancer recurrence and metastasis [4-6]. Multidrug resistance refers to that tumor cells produce drug resistance to a kind of anti-cancer drug, or different cross-resistance to anti-cancer drugs with the same structure, which is a major obstacle to cancer chemotherapy. Data shows that the incidence rate of MDR in primary liver cancer is 84.6%-100%, thus solving the multidrug resistance during chemotherapy is of great significance to the treatment of liver cancer [7-10]. In all resistance mechanisms, ABC transporter proteins combined with the nuclear membrane are considered to be the most important factor and are most studied by researchers, such as P-glycoprotein (P-gp), multidrug resistance associated protein (MRP), and breast cancer resistance protein [11-13].
RNA interference (RNAi) is a recently developed method for specific inhibition of gene expression, which refers to the degradation of double-stranded RNA (dsRNA) induced by homologous mRNA and blocking the corresponding gene expression, thus leading to specific post-transcriptional gene silencing (PTGS) [14]. Since RNAi has high sequence specificity, we can effectively and specifically block gene expression and it can be used as a simple and effective tool to replace the genetic knockout. Therefore, it was considered as one of the most important results of Science in 2001 [15-17]. Lentiviral vectors are capable of producing high-titer lentivirus with siRNA expression. It stably expresses siRNA in periodic and aperiodic cells, stem cells, fertilized ovum and differentiated progeny cells to achieve specific and stable gene silencing, which provides better tools for the study of gene functions and gene therapy [18-21]. All of the advantages of lentiviral vectors make them one of the best tools for investigating transgenosis and RNAi.
In this study, We will first import specific expression of siRNA gene fragment into drug-resistant cell line (HepG2/ADM, SMMC7721/ADM) models by lentiviral transfection techniques to downregulate or close MRP1, MRP2, MRP3 and MRP4 genes in multidrug-resistant hepatocellular carcinoma cell strains. Then we increase the intracellular drug concentration to promote the sensitivity of liver cancer cells to chemotherapy and improve the efficacy of chemotherapy for hepatocellular carcinoma.
Material and methods
Cell culture
Human HCC cell lines, HepG2 and SMMC7721, were purchased from Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Science, Chinese Academy of Sciences. HepG2 and SMMC7721 cells were cultured in DMEM medium containing 10% FBS, humidified incubator under 37°C, 5% CO2. Cells were added with different gradients concentration of doxorubicin (ADM) (0.01~2.0 μg/ml), each successive gradient sliding 0.1 μg/ml. The cells grown in the ADM with concentration stabilized at 2.0 μg/ml were named as HepG2/ADM and SMMC7721/ADM cells.
SiRNA preparation and transfection
SiRNA was purchased from Thermo Fisher Scientific Inc. (Waltham, MA, USA). Target sequences were: GCUGGUAGCCCUAGUGUGU for MRP1, CCACCCUGCUGAUACAGUA for MRP2, GGUCAAGUGUUCUACAGAU for MRP3, and UCUGAAAGCUCCGGUAUUA for MRP4. Approximately 5 × 104 cells per well were seeded in a six-well plate 24 h prior to transfection. Cells were transfected with 25, 50 or 75 nM siRNA using Lipofectin2000 reagent (Invitrogen Corp., Madison, Carlsbad, CA.) following the manufacturer’s protocol. Cells were then incubated at 37°C in the presence of 5% CO2 for 24 h and the culture medium was replaced 48 h after transfection before cells were ready for assay of gene knockdown.
RNA extraction and quantitative real-time polymerase chain reactions (QRT-PCR)
The cell lines were seeded in six-well plates and cultured for 24 h before the cells were collected. Total cellular RNA was extracted from the four groups by application of Trizol. 2 μg RNA was taken to synthesis reverse transcription cDNA. The 18S RNA primers were used as an internal reference. The standard curve was drawn and the relative concentrations of MRP1, MRP2, MRP3, MRP4, and 18S RNA genes were got according to the standard curve plotted. DNASIS software was used for homology analysis of the sequencing results. CLUSTALW software was used on the arrangement of sequencing. MEGA software was used for the analysis of phylogenetic tree. Each group was repeated for 3 times, and the average value was got. Primer sequences are as follows: MRP1 forward, 5’-CTTCGCTGAGTTCCTGCGTA-3’, and reverse, 5’-GCTGAGCTGTCTCTGCAGTT-3’; MRP2 forward 5’-GAGCACCAGCAGCGATTTCT-3’, and reverse, 5’-AGCCAACAGTGTCCCCACTT-3’; MRP3 forward, 5’-ATCCTGGCGATCTACTTCCT-3’, and reverse, 5’-TACAGCTTCAGCACCTTGAT-3’; MRP4 forward, 5’-CCTTCTCAGAGTCTTCGGTTT-3’, and reverse, 5’-ACCTGAGCTGCAGTGTTTAGG-3’. QRT-PCR was performed at 95°C for 10 min, followed by 40 cycles at 95°C for 15 s, at 60°C for 30 s, and at 72°C for 30 s. Data were analyzed using the Sequence detector software (v1.9, Applied Biosystems). The mean Ct value for duplicate measurements was used to detect the expression of target gene with normalization to a housekeeping gene used as an internal control (18S rRNA) according to the 2-ΔCt formula.
MTT assay
The cell lines were inoculated in 96-well plates. Doxorubicin, 5-FU, vincristine or oxaliplatin with different concentration gradients were added respectively after 24 hours. The gradients concentration of doxorubicin were 0.8, 1.6, 3.2, 6.4 and 12.8 mg/L, 5-FU were 0.08, 0.40, 2.00, 10.00 and 50.00 mg/L, vincristine were 0.25, 0.5, 1.0, 2.0 and 4.0 mg/L, oxaliplatin were 5, 10, 20, 40 and 80 mg/L. Each concentration of the group was repeated for three times. Meanwhile, the set of wells without drugs were used as the blank group. Each group were detected by MTT at 12, 24, 36, 48 and 60 h. Poms software was applied to calculate half inhibitory concentration of each group (IC50) and resistance index was calculated (RI) = IC50 of resistance medicines cells/IC50 of blank cells.
Western blot analysis
Protein was collected from cultured HepG2, SMMC7721, HepG2/ADM and SMMC7721/ADM cells and the concentration was measured (protein assay dye, Bio-Rad). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal reference. Electrochemiluminescence (ECL) method was used to show protein bands, and gray Quantity One analysis software was applied for semi-quantitative analysis. Each group was repeated 3 times to get the mean value.
Flow cytometric analysis of cell apoptosis
The cell lines of HepG2/ADM and SMMC7721/ADM were treated with siRNA or NS, and then were collected respectively through trypsinization, washed with ice-cold PBS, centrifuged at 2000 g for 5 min at 4°C, and washed twice with ice-cold PBS. Samples were rehydrated with PBS and then incubated with propidium iodide and Annexin V-FITC for 15 min at room temperature. More cell apoptosis showed the effect of siRNA silencing resistant gene was remarkable, and vice versa showed resistance gene silencing was unsuccessful.
Animal experiment
Five-week-old male BALB/c mice were purchased from SHANGHAI SLAC LABORATORY ANIMAL CO. LTD. All nude mice were fed according to Chinese animal guidelines. In tumor formation experiments, siRNA was transfected with target gene fragment MRP1-4 in SMMC-7721/ADM. At 24 h after transfection, the tumor cells were resuspensed in 100 μL PBS and was then injected into the right shoulder area of mice. Tumor growth was detected, and the long diameter and the wide diameter of the tumor after cell inoculation were measured. Tumor volume is calculated according to the following formula: volume (mm3) = width2 (mm2) × length (mm)/2.
Statistical analysis
Statistical data were analyzed using the SPSS 18.0 software (SPSS, Chicago, IL, USA). The rank sum test was used to examine the differential expression of the four multidrug resistance proteins in hepatoma cells. The results were analyzed using Student’s t test if two groups were compared and the Dunnett’s test if multiple groups were compared. If variances were inhomogeneous in the Student’s t test, the results were analyzed using the Welsh test. All data in this study were expressed as mean ± SEM. Results were considered statistically significant when P < 0.05.
Results
HepG2 and SMMC7721 resistant cell lines construction
In our study, each step of developing HepG2/ADM and SMMC7721/ADM cell lines took 10 months by gradually increasing concentrations of doxorubicin (ADM) in the DMEM medium. Each successive increasing 0.1 μg/ml concentration gradient from 0.01 to 2.0 ug/ml, and the cells induced in the concentration 2.0 μg/mL of ADM were called HepG2/ADM cells. When MTT assay was performed, we found that these cells were resistant not only to Adriamycin but also to multiple anticancer drugs including 5-FU, Vincristine, and Oxaliplatin. The lethal dose (IC50) and RI were significantly higher in HepG2/ADM and SMMC7721/ADM cells than in non-resistant parental cells (Table 1). MRP1-4 mRNA levels showed elevation in HepG2/ADM and SMMC7721/ADM cell lines tested by QRT-PCR assay.
Table 1.
Determination of IC50 and resistance index of different anticancer drugs (mean ± SD)
| A | ||||||
|
| ||||||
| IC50 | RI | IC50 | RI | |||
|
|
|
|||||
| SMMC7721 | SMMC7721/ADM | SMMC7721 | SMMC7721/ADM/MRP1 siRNA | |||
|
| ||||||
| Adriamycin (mg/L) | 0.0106±0.004 | 0.2657±0.003 | 25.43 | 0.0106±0.004 | 0.0608±0.033 | 5.73 |
| Fluorouracil (mg/L) | 0.1263±0.003 | 3.6129±0.045 | 28.60 | 0.1263±0.003 | 2.0031±0.004 | 15.86 |
| Vincristine (mg/L) | 0.0045±0.002 | 0.1763±0.008 | 39.17 | 0.0045±0.002 | 0.0112±0.002 | 2.49 |
| Oxaliplatin (mg/L) | 0.0063±0.004 | 0.1134±0.005 | 18.30 | 0.0063±0.004 | 0.0116±0.002 | 1.84 |
|
| ||||||
| B | ||||||
|
| ||||||
| IC50 | RI | IC50 | RI | |||
|
|
|
|||||
| SMMC7721 | SMMC7721/ADM | SMMC7721 | SMMC7721/ADM/MRP2 siRNA | |||
|
| ||||||
| Adriamycin (mg/L) | 0.0106±0.004 | 0.2657±0.003 | 25.43 | 0.0106±0.004 | 0.1023±0.006 | 9.65 |
| Fluorouracil (mg/L) | 0.1263±0.003 | 3.6129±0.045 | 28.60 | 0.1263±0.003 | 1.4417±0.023 | 11.41 |
| Vincristine (mg/L) | 0.0045±0.002 | 0.1763±0.008 | 39.17 | 0.0045±0.002 | 0.0452±0.010 | 10.04 |
| Oxaliplatin (mg/L) | 0.0063±0.004 | 0.1134±0.005 | 18.30 | 0.0063±0.004 | 0.0268±0.005 | 4.25 |
|
| ||||||
| C | ||||||
|
| ||||||
| IC50 | RI | IC50 | RI | |||
|
|
|
|||||
| SMMC7721 | SMMC7721/ADM | SMMC7721 | SMMC7721/ADM/MRP3 siRNA | |||
|
| ||||||
| Adriamycin (mg/L) | 0.0106±0.004 | 0.2657±0.003 | 25.43 | 0.0106±0.004 | 0.0521±0.009 | 4.92 |
| Fluorouracil (mg/L) | 0.1263±0.003 | 3.6129±0.045 | 28.60 | 0.1263±0.003 | 1.2740±0.018 | 10.09 |
| Vincristine (mg/L) | 0.0045±0.002 | 0.1763±0.008 | 39.17 | 0.0045±0.002 | 0.0146±0.002 | 3.24 |
| Oxaliplatin (mg/L) | 0.0063±0.004 | 0.1134±0.005 | 18.30 | 0.0063±0.004 | 0.0311±0.002 | 4.94 |
|
| ||||||
| D | ||||||
|
| ||||||
| IC50 | RI | IC50 | RI | |||
|
|
|
|||||
| SMMC7721 | SMMC7721/ADM | SMMC7721 | SMMC7721/ADM/MRP4 siRNA | |||
|
| ||||||
| Adriamycin (mg/L) | 0.0106±0.004 | 0.2657±0.003 | 25.43 | 0.0106±0.004 | 0.1052±0.004 | 9.92 |
| Fluorouracil (mg/L) | 0.1263±0.003 | 3.6129±0.045 | 28.60 | 0.1263±0.003 | 2.9576±0.006 | 23.41 |
| Vincristine (mg/L) | 0.0045±0.002 | 0.1763±0.008 | 39.17 | 0.0045±0.002 | 0.0532±0.001 | 11.82 |
| Oxaliplatin (mg/L) | 0.0063±0.004 | 0.1134±0.005 | 18.30 | 0.0063±0.004 | 0.0071±0.002 | 1.13 |
Silencing effects of siRNA on RNA transcription
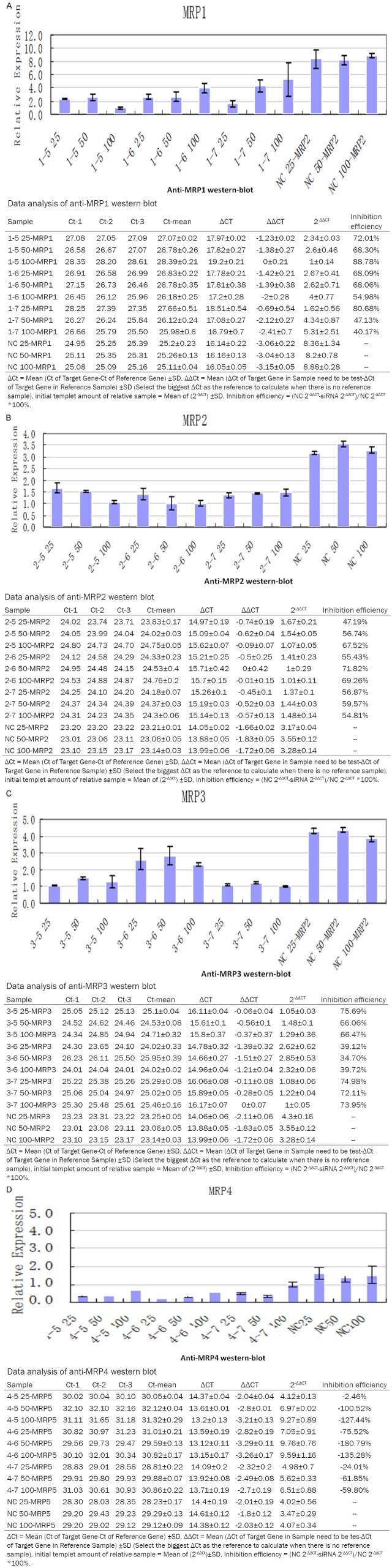
QRT-PCR was performed to detect the alteration in mRNA levels of MRP1-4 genes after the cells were treated with siRNA. mRNA level increased significantly when multidrug resistance was developed. However, when HepG2/ADM and SMMC7721/ADM cell lines were treated with siRNA, the levels of mRNA decreased to almost the same level as non-resistant cells. Quantities of RNA in each lane were normalized by 18s RNA expression (Figure 1).
Figure 1.
mRNA expression after siRNA treatment in MDR cells. mRNA levels were measured by QRT-PCR. Results were normalized by 18 sRNA mRNA expression and compared with the levels in parental cells (n = 3). Statistical analyses comparing MDR cells with parental cells were performed using Student’s t-test. P < 0.05 vs parental cells (data not shown). ΔCt = gene Ct valu-normalized gene Ct value; ΔΔCt = MRP1-4 genes ΔCt = valu-normalized genes ΔCt value; when the PCR efficacy is approaching 100%, relative value = 2-ΔΔCt.
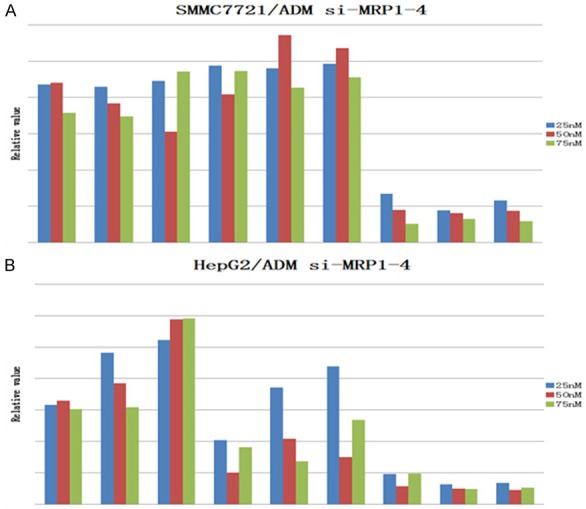
MRP1-4 protein expression after siRNA transfection
Western blot analysis was performed to assess the effects of siRNA on protein expression. Higher levels of MRP1-4 expression were detected in HepG2/ADM and SMMC7721/ADM cells. However, when these cells were treated with siRNA, the protein levels significantly decreased (Figure 2).
Figure 2.
Expression of MRP1-4 in MDR cells.
siRNA sensitizes ADM cell lines to chemotherapy drugs
To assess whether siRNA-directed MRP1-4 suppression sensitizes MDR cancer cells to cytotoxic agents, we compared the drug sensitivity of the siRNA-treated to that of the mock-treated MDR cells using MTT assay. As shown in Table 2, the sensitivity of the MDR cells to doxorubicin, 5-fluorouracil, vinblastine and Oxaliplatin increased significantly after the introduction of targeted siRNA (p < 0.05).
Table 2.
Determination of IC50 and resistance index of different anticancer drugs (mean ± SD)
| A | ||||||
|
| ||||||
| IC50 | RI | IC50 | RI | |||
|
|
|
|||||
| HepG2 | HepG2/ADM | HepG2 | HepG2/ADM/MRP1 siRNA | |||
|
| ||||||
| Adriamycin (mg/L) | 0.0187±0.004 | 0.2754±0.010 | 14.73 | 0.0187±0.004 | 0.0596±0.006 | 3.19 |
| Fluorouracil (mg/L) | 0.1319±0.002 | 3.5388±0.016 | 26.83 | 0.1319±0.002 | 2.0103±0.016 | 15.24 |
| Vincristine (mg/L) | 0.0057±0.007 | 0.1821±0.019 | 31.95 | 0.0057±0.007 | 0.0236±0.003 | 4.14 |
| Oxaliplatin (mg/L) | 0.0088±0.003 | 0.1956±0.008 | 22.23 | 0.0088±0.003 | 0.0128±0.002 | 1.46 |
|
| ||||||
| B | ||||||
|
| ||||||
| IC50 | RI | IC50 | RI | |||
|
|
|
|||||
| HepG2 | HepG2/ADM | HepG2 | HepG2/ADM/MRP2 siRNA | |||
|
| ||||||
| Adriamycin (mg/L) | 0.0187±0.004 | 0.2754±0.010 | 14.73 | 0.0187±0.004 | 0.1832±0.007 | 9.80 |
| Fluorouracil (mg/L) | 0.1319±0.002 | 3.5388±0.016 | 26.83 | 0.1319±0.002 | 1.7210±0.019 | 13.04 |
| Vincristine (mg/L) | 0.0057±0.007 | 0.1821±0.019 | 31.95 | 0.0057±0.007 | 0.0307±0.005 | 5.39 |
| Oxaliplatin (mg/L) | 0.0088±0.003 | 0.1956±0.008 | 22.23 | 0.0088±0.003 | 0.0236±0.008 | 2.68 |
|
| ||||||
| C | ||||||
|
| ||||||
| IC50 | RI | IC50 | RI | |||
|
|
|
|||||
| HepG2 | HepG2/ADM | HepG2 | HepG2/ADM/MRP3 siRNA | |||
|
| ||||||
| Adriamycin (mg/L) | 0.0187±0.004 | 0.2754±0.010 | 14.73 | 0.0187±0.004 | 0.0724±0.010 | 3.87 |
| Fluorouracil (mg/L) | 0.1319±0.002 | 3.5388±0.016 | 26.83 | 0.1319±0.002 | 1.2260±0.013 | 9.29 |
| Vincristine (mg/L) | 0.0057±0.007 | 0.1821±0.019 | 31.95 | 0.0057±0.007 | 0.0354±0.009 | 6.21 |
| Oxaliplatin (mg/L) | 0.0088±0.003 | 0.1956±0.008 | 22.23 | 0.0088±0.003 | 0.0291±0.006 | 3.31 |
|
| ||||||
| D | ||||||
|
| ||||||
| IC50 | RI | IC50 | RI | |||
|
|
|
|||||
| HepG2 | HepG2/ADM | HepG2 | HepG2/ADM/MRP4 siRNA | |||
|
| ||||||
| Adriamycin (mg/L) | 0.0187±0.004 | 0.2754±0.010 | 14.73 | 0.0187±0.004 | 0.1106±0.013 | 5.91 |
| Fluorouracil (mg/L) | 0.1319±0.002 | 3.5388±0.016 | 26.83 | 0.1319±0.002 | 2.4238±0.022 | 18.38 |
| Vincristine (mg/L) | 0.0057±0.007 | 0.1821±0.019 | 31.95 | 0.0057±0.007 | 0.0621±0.004 | 10.89 |
| Oxaliplatin (mg/L) | 0.0088±0.003 | 0.1956±0.008 | 22.23 | 0.0088±0.003 | 0.0349±0.005 | 3.97 |
MRP1-4 silencing induces apoptosis
To explore whether MRP1-4 silencing could induce apoptosis, we analyzed apoptosis in ADM cells by flow cytometry. The average rates of apoptosis in SMMC7721/ADM and HepG2/ADM with siRNA were 8.50% and 8.11%, respectively (Figure 3). These figures were significantly higher than those with NS (0.14% and 0.11%, p < 0.05), indicating that MRP1-4 expression could effectively protect SMMC7721/ADM and HepG2/ADM cells from apoptosis.
Figure 3.
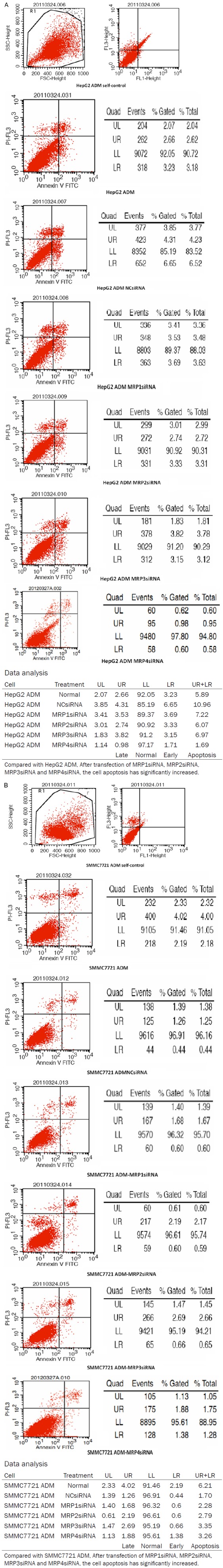
Apoptosis of MDR cells with or without siRNA.
MRP1-4 silencing suppresses tumor growth in vivo
After inoculated to mice for 14 days, the tumor was enucleated to calculate tumor growth. Compared with SMMC-7721/ADM, tumor growth significantly reduced after silencing MRP1-4 gene. The results suggested that silencing MRP1-4 gene in vivo could inhibit tumor growth (Figure 4).
Figure 4.
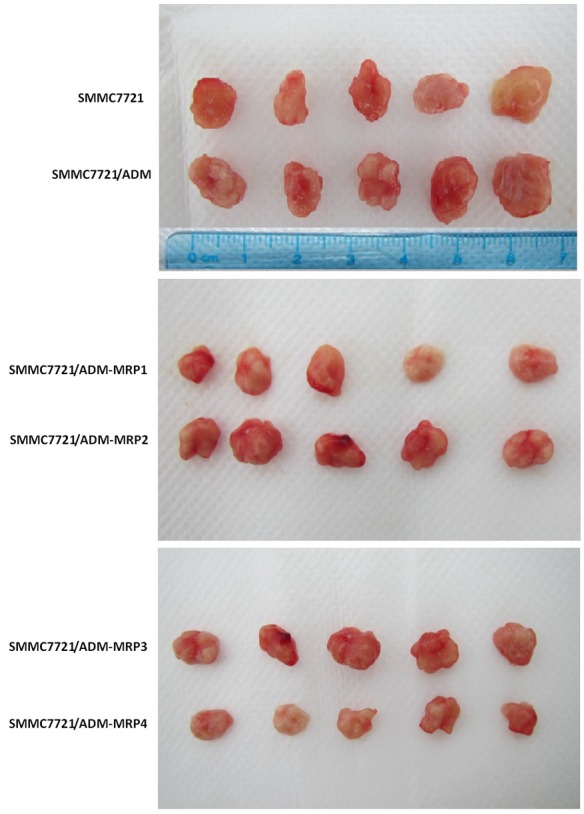
Animal experiment Tumor cells were injected to mice (four per group) on the right side scapular area after resuspended with 100 μL PBS, detect the growth of the tumor cells after inoculating for 14 days and measure the major axis diameter and width. Tumor volume was calculated according to the following formula: volume (mm3) = width2 (mm2) × length (mm)/2.
Discussion
Human hepatocellular carcinoma is one of the most common cancers in males and females over the world. How-ever, no chemotherapeutic agents have been found to provide a clinically effective treatment [1]. Studies showed that MRPs expressed in HCC may confer HCC resistance to chemotherapeutic drugs. Some reports suggested that MRPs are better candidates to mediate chemo-resistance in HCC than MDR1. However, since their expression in mature hepatocytes is negligible, some researchers have ruled out the involvement of MRPs in the MDR phenotype of HCC [22-24]. However, other researchers demonstrated that MRPs expression is at significant levels in HCCs and suggested that MRPs expression is closely related to MDR in HCC [25-27]. MRPs play a major role in chemotherapy failure. In this study, we screened a variety of anticancer drugs or cytotoxic agents, including natural products to detect the impact of MRPs on the multidrug resistance phenotype, using two human hepatocellular carcinoma-derived cell lines as an in vitro model.
As shown in Table 1D, the IC50 of anticancer drugs to the MDR cell subclone was much higher than that to parental cells, suggesting that the acquired MDR of HepG2/ADM and SMMC7721/ADM was reliable. We also found clear differences by western blot between MDR cells and the parental cell lines. Higher levels of MRP1-4 expression were detected in MDR cell lines, indicating that MRP1-4 over-expression certainly contributed to the MDR cells [8] .
Sara Vander B [28] et al declared a diffuse protein expression of MRP1-4 compared with negative hepatocytic expression observed in normal (surrounding) hepatocytes. In addition, MRP1-4 expression was high in poorly differentiated HCCs, large tumors (> 7 cm) and microvascular invasive tumors. These results corresponded to our findings that plenty of MRP1-4 mRNA and protein expressed in MDR cells. It may also explain the confusion that some reports deemed high P-gp expression was seen along with low MRP1-4 expression in HCC cell lines. We could detect high levels of MRP1-4 mRNA and protein expression. For the MDR cell models (as we established) tolerated the stress of anti-cancer drug for ages, it was more closed to advanced HCCs.
Since Elbashir et al. [29] have reported that RNA interference could be triggered in mammalian cells by introduction of 21-nucleotide siRNA, siRNA has been shown to be an effective approach for silencing gene expression and has been applied to inhibit oncogene in hepatoma. We now demonstrate that introduction of siRNA duplex decreased MRP1-4 expression (Figure 1), induced SMMC7721/ADM and HepG2/ADM cells apoptosis to drugs (Figure 3), and restored drug sensitivity (Table 1D) in human MDR cancer cells. We also found that the modulation of MDR results from the siRNA-directed degradation of MRP1-4 mRNA (Figure 1).
This report demonstrated the feasibility of using siRNA to specifically and effectively modulate MDR. MRP-targeted siRNA inhibits the expression of MRP1-4 RNA and MRP1-4 protein with minimum effect on 18s RNA and β-actin expression in comparison with mock treatment (Figures 1, 2); GAPD siRNA decreased GAPD expression but had no effect on the expression of MRP1-4 (Figure 1). These data suggested that silencing of MRP1-4 expression mediated by siRNA is specific.
Furthermore, SMMC7721/ADM and HepG2/ADM cell lines were selected by prolonged exposure to doxorubicin, while additional mechanisms of drug resistance are known to exist. The drug resistance of these cells was significantly restored after inhibition of MRP1-4, indicating that over-expression of MRP1-4 also contributes to MDR of HepG2/ADM and SMMC7721/ADM cells. In animal experiment, we detected tumor growth and enucleated the tumor from the mice after inoculating for 14 days. Compared with SMMC-7721/ADM, tumor growth significantly reduced after silencing MRP1-4 gene. These results suggested that silencing MRP1-4 gene in vivo experiments can inhibit tumor growth (Figure 4).
In summary, in this study, we successfully established SMMC-7721/ADM and HepG2/ADM multidrug-resistant HCC cell subclones. We demonstrated that the MDR cells were associated with the over-expression of MRP1-4. Our study reveled the effect of silencing MRP1-4 gene in reversing MDR. Therefore, the introduction of MRP1-4 siRNA may hold promise for the treatment of drug-resistant cancer.
Acknowledgements
This study was supported by Guangdong province science and technology plan projects No. 2011B031800296. We gratefully acknowledge the assistance of the Department of Hepatobiliary Surgery for their help in collecting medical records. In addition, we express our thanks to all the participants in the study, without whom the study would not have been possible.
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Bray F, Center MM. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Okuda K. Hepatocellular carcinoma. J Hepatol. 2000;32:225–237. doi: 10.1016/s0168-8278(00)80428-6. [DOI] [PubMed] [Google Scholar]
- 3.Avila MA, Berasain C, Sangro B, Prieto J. New therapies for hepatocellular carcinoma. Oncogene. 2006;25:3866–3884. doi: 10.1038/sj.onc.1209550. [DOI] [PubMed] [Google Scholar]
- 4.Slot AJ, Molinski SV, Cole SP. Mammalian multidrug resistance proteins (MRPs) Essays Biochem. 2011;50:179–207. doi: 10.1042/bse0500179. [DOI] [PubMed] [Google Scholar]
- 5.Wakamatsu T, Nakahashi Y, Hachimine D, Seki T, Okazaki K. The combination of glycyrrhizin and lamivudine can reverse the cisplatin resistance in hepatocellular carcinoma cells through inhibition of multidrug resistance-associated proteins. Int J Oncol. 2007;31:1465–1472. [PubMed] [Google Scholar]
- 6.Perez-Tomas R. Multidrug resistance: retrospect and prospects in anti-cancer drug treatment. Curr Med Chem. 2006;13:1859–1876. doi: 10.2174/092986706777585077. [DOI] [PubMed] [Google Scholar]
- 7.Folmer Y, Schneider M, Blum HE, Hafkemeyer P. Reversal of drug resistance of hepatocellular carcinoma cells by adenoviral delivery of anti-ABCC2 antisense constructs. Cancer Gene Ther. 2007;14:875–884. doi: 10.1038/sj.cgt.7701082. [DOI] [PubMed] [Google Scholar]
- 8.Modok S, Mellor HR, Callaghan R. Modulation of multidrug resistance efflux pump activity to overcome chemoresistance in cancer. Curr Opin Pharmacol. 2006;6:350–354. doi: 10.1016/j.coph.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 9.Rigalli JP, Ciriaci N, Arias A, Ceballos MP, Villanueva SS, Luquita MG, Mottino AD, Ghanem CI, Catania VA, Ruiz ML. Regulation of multidrug resistance proteins by genistein in a hepatocarcinoma cell line: impact on sorafenib cytotoxicity. PLoS One. 2015;10:e0119502. doi: 10.1371/journal.pone.0119502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang PP, Xu DJ, Huang C, Wang WP, Xu WK. Astragaloside IV reduces the expression level of P-glycoprotein in multidrug-resistant human hepatic cancer cell lines. Mol Med Rep. 2014;9:2131–2137. doi: 10.3892/mmr.2014.2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang CF, Wang YQ, Huang FZ, Nie WP, Liu XY, Jiang XZ. Association between reversal of multidrug resistance by methyl jasmonate and P-glycoprotein ATPase activity in hepatocellular carcinoma. J Int Med Res. 2013;41:964–974. doi: 10.1177/0300060513483401. [DOI] [PubMed] [Google Scholar]
- 12.Korita PV, Wakai T, Shirai Y, Matsuda Y, Sakata J, Takamura M, Yano M, Sanpei A, Aoyagi Y, Hatakeyama K, Ajioka Y. Multidrug resistance-associated protein 2 determines the efficacy of cisplatin in patients with hepatocellular carcinoma. Oncol Rep. 2010;23:965–972. doi: 10.3892/or_00000721. [DOI] [PubMed] [Google Scholar]
- 13.Zhao J, Yu BY, Wang DY, Yang JE. Promoter polymorphism of MRP1 associated with reduced survival in hepatocellular carcinoma. World J Gastroenterol. 2010;16:6104–6110. doi: 10.3748/wjg.v16.i48.6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ye L, Zhao X, Lu J, Qian G, Zheng JC, Ge S. Knockdown of TIGAR by RNA interference induces apoptosis and autophagy in HepG2 hepatocellular carcinoma cells. Biochem Biophys Res Commun. 2013;437:300–306. doi: 10.1016/j.bbrc.2013.06.072. [DOI] [PubMed] [Google Scholar]
- 15.Zeng XC, Zhang T, Huang DH, Wang GY, Chen W, Li H, Zhang J, Fang TL, Zhang Q, Chen GH. RNA interfering targeting human leukocyte antigen-G enhanced immune surveillance mediated by the natural killer cells on hepatocellular carcinoma. Ann Clin Lab Sci. 2013;43:135–144. [PubMed] [Google Scholar]
- 16.Yan LL, Huang YJ, Yi X, Yan XM, Cai Y, He Q, Han ZJ. Effects of silencing S100A8 and S100A9 with small interfering RNA on the migration of CNE1 nasopharyngeal carcinoma cells. Oncol Lett. 2015;9:2534–2540. doi: 10.3892/ol.2015.3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xie J, Lei P, Hu Y. Small interfering RNA-induced inhibition of epithelial cell transforming sequence 2 suppresses the proliferation, migration and invasion of osteosarcoma cells. Exp Ther Med. 2015;9:1881–1886. doi: 10.3892/etm.2015.2306. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Di Cresce C, Figueredo R, Rytelewski M, Maleki Vareki S, Way C, Ferguson PJ, Vincent MD, Koropatnick J. siRNA knockdown of mitochondrial thymidine kinase 2 (TK2) sensitizes human tumor cells to gemcitabine. Oncotarget. 2015;6:22397–22409. doi: 10.18632/oncotarget.4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu S, Yin X, Spollen W, Zhang N, Xu D, Schoelz J, Bilyeu K, Zhang ZJ. Analysis of the siRNA-Mediated Gene Silencing Process Targeting Three Homologous Genes Controlling Soybean Seed Oil Quality. PLoS One. 2015;10:e0129010. doi: 10.1371/journal.pone.0129010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Montazami N, Kheir Andish M, Majidi J, Yousefi M, Yousefi B, Mohamadnejad L, Shanebandi D, Estiar MA, Khaze V, Mansoori B, Baghbani E, Baradaran B. siRNA-mediated silencing of MDR1 reverses the resistance to oxaliplatin in SW480/OxR colon cancer cells. Cell Mol Biol. 2015;61:98–103. [PubMed] [Google Scholar]
- 21.Li T, Niu L, Li M, Liu Y, Xu Z, Gao X, Liu D. Effects of small interfering RNA-mediated downregulation of the Krüppel-like factor 4 gene on collagen metabolism in human hepatic stellate cells. Mol Med Rep. 2015;12:3972–3978. doi: 10.3892/mmr.2015.3848. [DOI] [PubMed] [Google Scholar]
- 22.Fallica B, Makin G, Zaman MH. Bioengineering approaches to study multidrug resistance in tumor cells. Integr Biol (Camb) 2011;3:529–539. doi: 10.1039/c0ib00142b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perez-Tomas R. Multidrug resistance: retrospect and prospects in anti-cancer drug treatment. Curr Med Chem. 2006;13:1859–1876. doi: 10.2174/092986706777585077. [DOI] [PubMed] [Google Scholar]
- 24.Modok S, Mellor HR, Callaghan R. Modulation of multidrug resistance efflux pump activity to overcome chemoresistance in cancer. Curr Opin Pharmacol. 2006;6:350–354. doi: 10.1016/j.coph.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 25.Lage H. ABC-transporters: implications on drug resistance from microorganisms to human cancers. Int J Antimicrob Agents. 2003;22:188–199. doi: 10.1016/s0924-8579(03)00203-6. [DOI] [PubMed] [Google Scholar]
- 26.Leslie EM, RG Deeley, SP Cole. Toxicological relevance of the multidrug resistance protein 1, MRP1 (ABCC1) and related transporters. Toxicology. 2001;167:3–23. doi: 10.1016/s0300-483x(01)00454-1. [DOI] [PubMed] [Google Scholar]
- 27.Stavrovskaya AA. Cellular mechanisms of multidrug resistance of tumor cells. Biochemistry (Mosc) 2000;65:95–106. [PubMed] [Google Scholar]
- 28.Vander Borght S, Komuta M, Libbrecht L, Katoonizadeh A, Aerts R, Dymarkowski S, Verslype C, Nevens F, Roskams T. Expression of multidrug resistance-associated protein 1 in hepatocellular carcinoma is associated with a more aggressive tumor phenotype and may reflect a progenitor cell origin. Liver Int. 2008;28:1370–1380. doi: 10.1111/j.1478-3231.2008.01889.x. [DOI] [PubMed] [Google Scholar]
- 29.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]