Abstract
Wilson Disease (WD) is an inborn error of copper metabolism inherited in an autosomal recessive manner caused by the mutations in the P-type ATPase gene (ATP7B). In this study, we screen and detect the mutations of the ATP7B gene in unrelated Chinese WD patients. A total of 68 individuals from ten provinces of China with WD were recruited. Of them, 43 were males and 25 were females, and their onset ages were from 1 to 48 years with a median onset age of 22.2 years. All the exons and exon/intron boundaries of ATP7B gene of the patients were sequenced and aligned to the referred ATP7B gene sequence. The results suggested that 66 of the 68 patents carried with at least one mutation and 48 different mutations were identified including 34 missense, one synonymous, two nonsense, two splicing, and nine frameshift mutations (five insertion and four deletion). Among these mutations, c.2333G>T, c.2310C>G, c.2975C>T, and c.3443T>C were the most prevalent mutants and c.2310C>G always linked with c.2333G>T. The eighth, 11th, and 18th exons carried more mutations (6/48, 5/48, and 5/48, respectively) than others. After comparing with the mutations reported previously, 22 out of the 48 mutations were identified as novel mutations. A popular algorithm, Polyphen-2, was used to predict the effects of the amino-acid substitution due to the mutations on the structure and function of ATP7B function and the predicted results indicated that all the missense mutations were unfavorable except c.121A>G and c.748G>A. Phenotype/genotype correlation analysis suggested that the patients with c.2975C>T or c.3809A>G often presented WD features before 12 years old while the patients with c.3443T>C almost presented WD after 12 years old. This is the first time to identify the common mutations contributing to early onset age in Chinese WD patients. Our study will broaden our knowledge about ATP7B mutations in WD patients.
Keywords: ATP7B, Chinese, mutational analysis, Wilson disease
Introduction
Wilson Disease (WD, MIM#277900) is an inborn error of copper metabolism inherited in an autosomal recessive manner caused by the mutations in the P-type ATPase gene (ATP7B) [1]. It is firstly reported by Dr. Samuel Alexander Kinnier Wilson in the year of 1912 and characterized by excess accumulation of intracellular hepatic copper and hepatic and neurologic abnormalities [2]. The frequency of WD disease was 1/30000 to 1/100000 worldwide while the carrier frequency reaches a high value of 1/90 [3-5]. In addition, a higher incidence is observed in Han population in China [5-7]. The disease is diagnosed on the basis of typical symptoms and conventional biochemical indicators, which include low serum concentration of ceruloplasmin and elevated excretion of urinary copper [8]. The prognosis of the WD patients is very poor and an early diagnosis and therapy is verified to help the patients to prevent lifelong neurological disability and liver cirrhosis and improve life quality remarkably [9-11].
ATP7B gene is located at 13q14.3 (about 80 kb) and consisted of 21 exons and 20 introns encoding a protein containing 1465 amino acids [12]. It plays a great role in transforming apoceruloplasmin into ceruloplasmin and excreting copper into biliary canaliculi. Defects of ATP7B will reduce the blood ceruloplasmin and affect hepatocytes. And also many other organs including brain, eyes, and kidney will be involved [13,14].
Mutation screening has revealed a larger number of genetic mutations in ATP7B gene in WD patients. In this study, we performed a mutational analysis of ATP7B by exon deep sequencing in 68 subjects in China Han population with WD and identified 48 mutations, of which 22 mutations have never been reported previously.
Materials and methods
Subjects
A total of 68 individuals from ten provinces of China with WD were recruited in the present study. Of them, 43 were males and 25 were females with the mean age of 24.0 (5-61) years and mean onset age of 22.2 (1-48) years. All the patients were informed and signed for agreement. The diagnostics were performed according to the following criteria: (1) liver or brain failure symptoms; (2) presence of K-F ring in the cornea by slit-lamp examination; (3) reduced serum ceruloplasmin (<0.20 g/l) and/or elevated 24-hour urinary copper excretion (>1.6 μmol/24 h) and/or hepatic copper content >250 mg per g of dry weight [15].
DNA extraction and sequencing
The peripheral blood leukocyte of the patients was collected and genomic DNAs were extracted with QIAamp blood kits (Qiagen, Hilden, Gemany) based on the manufacture’s instructions. All of 21 exons of ATP7B and their ± 10 bp regions were analyzed by next sequencing.
Identification of mutations and prediction of deleterious variants
The sequencing results were aligned to referred ATP7B sequence (NM_000053.3) to figure out the mutations. The sequencing data from local 854 health controls were used to identify the polymorphisms. Allele and patient frequency of each mutation was also calculated. A novel mutant was identified when a genetic variant met all the following criteria: (1) no records in dbSNP database; (2) no reports in PubMed literatures. Then, in silico analysis tool PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/index.shtml) was used to predict the putative effects of each mutation on the structure and function of ATP7B protein.
Results
Clinical feathers and laboratory examinations of the included WD patients
We included 68 WD patients in the present study, of which 43 (63.2%) were males and 25 (36.8%) were females. Their onset age ranged from 1 to 48 years with the mean onset age of 22.2 years. The presence of K-F rings of 65 patients was examined and 50 (76.9%) of them were verified. Then serum ceruloplasmin and 24 h-urinary copper concentrations of the patients were determined. 67 of 68 patients (98.5%) owned low serum ceruloplasmin (<0.20 g/l) and 54 of 62 (87.1%) patients had high values of 24 h-urinary copper (>1.6 μmol/24 h). Further, the hepatic copper content of 19 patients was examined and that of 18 (94.7%) patients were >250 μg per g of dry weight. Based on the clinical analysis and laboratory examination, the phenotypes of the 68 WD patients were clarified into hepatic (32, 47.1%), hepatic and neurological (23, 33.8%), neurological (8, 11.8%), presymptomatic or no symptoms (5, 7.4%).
Identification of ATP7B genetic variants in Chinese patients with WD
The exons of ATP7B and 10 bps upstream and downstream of the exons of 68 Chinese WD patients were examined. All the sequencing results were aligned to the referred ATP7B gene sequence (NM_000053.3) and compared with the data from 854 health controls from China. Finally, 58 genetic variations including 48 mutations and eight SNPs were identified (Tables 1 and 3).
Table 1.
Information of ATP7B mutations and prediction of the functional effects of the mutations
| NT change | Location | Functional Region | AA change | Polyphen-2 | Mutation Type | Patient count | Patient Freq % | Allele count | Allele Freq % | Novelty | |
|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||
| Prediction | Score | ||||||||||
| c.121A>G | Exon 1 15021707309 | p.Asn41Asp | BENIGN | 0.008 | Missense | 1 | 1.47 | 1 | 0.74 | Novel | |
| c.748G>A | Exon 2 | p.Gly250Arg | BENIGN | 0.002 | Missense | 1 | 1.47 | 1 | 0.74 | Novel | |
| c.898_902 delAAGTA | Exon 2 | MBD3 | p.Lys300Ter | NA | NA | Nonsense | 1 | 1.47 | 1 | 0.74 | Novel |
| EX2 DEL | Exon 2 | NA | NA | Deletion | 2 | 2.94 | 2 | 1.47 | Novel | ||
| c.1708-1G>C | Intron 4 | NA | NA | Splicing | 2 | 2.94 | 2 | 1.47 | Novel | ||
| c.1745_1746 delTA | Exon 5 | MBD6 | NA | NA | Deletion | 1 | 1.47 | 1 | 0.74 | Novel | |
| c.1994T>G | Exon 7 | TMS1 | p.Met665Arg | PROBABLY DAMAGING | 0.995 | Missense | 1 | 1.47 | 1 | 0.74 | Novel |
| c.2012_2013 insATAT | Exon 7 | TMS1 | NA | NA | Insertion | 1 | 1.47 | 1 | 0.74 | Novel | |
| c.2128G>A | Exon 8 | TMS2 | p.Gly710Ser | PROBABLY DAMAGING | 1.000 | Missense | 1 | 1.47 | 1 | 0.74 | |
| c.2231C>T | Exon 8 | TMS3 | p.Ser744Phe | PROBABLY DAMAGING | 1.000 | Missense | 1 | 1.47 | 1 | 0.74 | |
| c.2304_2305 insC | Exon 8 | TMS4 | NA | NA | Insertion | 4 | 5.88 | 4 | 2.94 | Novel | |
| c.2308C>T | Exon 8 | TMS4 | p.Leu770Phe | PROBABLY DAMAGING | 1.000 | Missense | 1 | 1.47 | 1 | 0.74 | Novel |
| c.2310C>G | Exon 8 | TMS4 | p.Leu770Leu | NA | NA | Synonymous | 24 | 35.29 | 28 | 20.59 | |
| c.2333G>T | Exon 8 | TMS4 | p.Arg778Leu | PROBABLY DAMAGING | 1.000 | Missense | 31 | 45.59 | 35 | 25.74 | |
| c.2506G>A | Exon 10 | ATPase | p.Gly836Arg | PROBABLY DAMAGING | 1.000 | Missense | 1 | 1.47 | 1 | 0.74 | Novel |
| c.2549C>T | Exon 10 | ATPase | p.Thr850Ile | BENIGN | 0.037 | Missense | 1 | 1.47 | 1 | 0.74 | |
| c.2561A>G | Exon 10 | ATPase | p.Glu854Gly | PROBABLY DAMAGING | 1.000 | Missense | 1 | 1.47 | 1 | 0.74 | Novel |
| c.2593_2594 insGTCA | Exon 11 | ATPase | NA | NA | Insertion | 1 | 1.47 | 1 | 0.74 | Novel | |
| c.2605G>A | Exon 11 | ATPase | p.Gly869Arg | PROBABLY DAMAGING | 1.000 | Missense | 1 | 1.47 | 1 | 0.74 | |
| c.2620G>C | Exon 11 | ATPase | p.Ala874Pro | PROBABLY DAMAGING | 1.000 | Missense | 1 | 1.47 | 2 | 1.47 | |
| c.2621C>T | Exon 11 | ATPase | p.Ala874Val | PROBABLY DAMAGING | 1.000 | Missense | 6 | 8.82 | 6 | 4.41 | |
| c.2662A>C | Exon 11 | ATPase | p.Thr888Pro | PROBABLY DAMAGING | 0.998 | Missense | 3 | 4.41 | 3 | 2.21 | |
| c.2755C>G | Exon 12 | ATPase | p.Arg919Gly | POSSIBLY DAMAGING | 0.832 | Missense | 3 | 4.41 | 3 | 2.21 | |
| c.2804C>T | Exon 12 | TMS5 | p.Thr935Met | PROBABLY DAMAGING | 1.000 | Missense | 1 | 1.47 | 1 | 0.74 | |
| c.2828G>A | Exon 12 | p.Gly943Asp | PROBABLY DAMAGING | 1.000 | Missense | 1 | 1.47 | 1 | 0.74 | ||
| c.2924C>A | Exon 13 | TMS6 | p.Ser975Tyr | PROBABLY DAMAGING | 1.000 | Missense | 1 | 1.47 | 1 | 0.74 | |
| c.2975C>T | Exon 13 | TMS6 | p.Pro992Leu | PROBABLY DAMAGING | 1.000 | Missense | 10 | 14.71 | 11 | 8.09 | |
| c.3007G>A | Exon 13 | ATPase | p.Ala1003Thr | PROBABLY DAMAGING | 1.000 | Missense | 1 | 1.47 | 1 | 0.74 | |
| c.3056A>C | Exon 13 | p.His1019Pro | PROBABLY DAMAGING | 1.000 | Missense | 1 | 1.47 | 1 | 0.74 | Novel | |
| c.3061-3C>A | Intron 13 | NA | NA | Splicing | 1 | 1.47 | 1 | 0.74 | Novel | ||
| c.3140A>T | Exon 14 | p.Asp1047Val | POSSIBLY DAMAGING | 0.927 | Missense | 1 | 1.47 | 1 | 0.74 | ||
| c.3316G>A | Exon 15 | p.Val1106Ile | POSSIBLY DAMAGING | 0.863 | Missense | 5 | 7.35 | 5 | 3.68 | ||
| c.3377_3378 delAC | Exon 15 | NA | NA | Deletion | 2 | 2.94 | 2 | 1.47 | Novel | ||
| c.3443T>C | Exon 16 | ATP bind | p.Ile1148Thr | PROBABLY DAMAGING | 0.999 | Missense | 9 | 13.24 | 10 | 7.35 | |
| c.3445G>A | Exon 16 | ATP bind | p.Gly1149Arg | PROBABLY DAMAGING | 1.000 | Missense | 1 | 1.47 | 1 | 0.74 | |
| c.3451C>T | Exon 16 | ATP bind | p.Arg1151Cys | PROBABLY DAMAGING | 1.000 | Missense | 1 | 1.47 | 1 | 0.74 | |
| c.3452G>A | Exon 16 | ATP bind | p.Arg1151His | PROBABLY DAMAGING | 1.000 | Missense | 1 | 1.47 | 1 | 0.74 | |
| c.3584C>T | Exon 17 | HAD | p.Ala1195Val | PROBABLY DAMAGING | 1.000 | Missense | 1 | 1.47 | 1 | 0.74 | Novel |
| c.3677C>T | Exon 17 | HAD | p.Thr1226Ile | PROBABLY DAMAGING | 0.999 | Missense | 1 | 1.47 | 1 | 0.74 | Novel |
| c.3679G>C | Exon 17 | HAD | p.Ala1227Pro | PROBABLY DAMAGING | 1.000 | Missense | 1 | 1.47 | 1 | 0.74 | Novel |
| c.3700delG | Exon 18 | HAD | NA | NA | Deletion | 1 | 1.47 | 1 | 0.74 | Novel | |
| c.3809A>G | Exon 18 | HAD | p.Asn1270Ser | PROBABLY DAMAGING | 1.000 | Missense | 4 | 5.88 | 4 | 2.94 | |
| c.3843_3844 insT | Exon 18 | HAD | NA | NA | Insertion | 1 | 1.47 | 1 | 0.74 | Novel | |
| c.3884C>T | Exon 18 | p.Ala1295Val | PROBABLY DAMAGING | 1.000 | Missense | 1 | 1.47 | 1 | 0.74 | ||
| c.3901_3902 insA | Exon 18 | NA | NA | Insertion | 1 | 1.47 | 1 | 0.74 | Novel | ||
| c.3960G>C | Exon 19 | p.Arg1320Ser | PROBABLY DAMAGING | 1.000 | Missense | 1 | 1.47 | 1 | 0.74 | ||
| c.3982G>A | Exon 19 | TMS7 | p.Ala1328Thr | PROBABLY DAMAGING | 1.000 | Missense | 1 | 1.47 | 1 | 0.74 | |
| c.4114C>T | Exon 20 | p.Gln1372Ter | NA | NA | Nonsense | 1 | 1.47 | 1 | 0.74 | ||
Abbreviations: AA, amino acid; ATPase, copper-(or silver)-translocating P-type ATPase segment; ATP bind, ATP binding segment; HAD, haloacid dehalogenase-like hydrolases segment; MBD, mental-binding domain; NT, nucleotide; TMS, transmembrane segment.
Table 3.
Information of ATP7B SNPs
| SNP | RS-ID | dbSNP freq | Hapmap freq | 1000 G freq | Patient freq | Control freq |
|---|---|---|---|---|---|---|
| p.Lys832Arg | rs1061472 | 0.5 | 0.507 | 0.4753 | 0.6119 | 0.4940 |
| p.Val1297Ile | rs148399850 | 0.012 | 0 | 0.0101 | 0.0448 | 0.0386 |
| p.Ser406Ala | rs1801243 | 0.489 | 0.482 | 0.4414 | 0.6716 | 0.5024 |
| p.Val456Leu | rs1801244 | 0.486 | 0.416 | 0.4423 | 0.6418 | 0.4952 |
| p.Ala1003Ala | rs1801247 | 0.055 | 0 | 0.0568 | 0.0149 | 0.0193 |
| p.Val1140Ala | rs1801249 | 0.498 | 0.5 | 0.4652 | 0.5970 | 0.4964 |
| c.3093+6C>T | rs2282057 | 0.5 | 0 | 0.4753 | 0.5970 | 0.4976 |
| p.Arg952Lys | rs7322774 | 0.499 | 0.504 | 0.4725 | 0.5970 | 0.4964 |
Among the 48 different mutations, 34 were missense, one was synonymous, two were nonsense, two were splicing, and nine were frameshift (five insertion and four deletion). The mutations were distributed in exon 1, 2, 5, 7, 8, 10-19, and 20 and intron 4 and 13. Exon 8 (6/48) was showed to more frequent to suffer the mutations, followed by exon 11 (5/48), exon 18 (5/48), exon 13 (4/48), and exon 16 (4/48) suggesting these exons might be more susceptible to Wilson disease in Chinese Han population, consisted with the previous studies [6,16,17]. 11 mutations were identified to locate at the transmembrane segments encoding region, nine at copper-(or silver)-translocating P-type ATPase segment region, six at haloacid dehalogenase-like hydrolases segment region, two at mental-binding region, and five at the ATP binding region. Then the allele and patient frequencies of every mutation were calculated. The top four allele frequencies were 35/136 (c.2333G>T), 28/136 (c.2310C>G), 11/136 (c.2975C>T), and 10/136 (c.3443T>C). And their corresponding patient frequencies were 31/68, 24/68, 10/68, and 9/68. After comparing with the previous reports, 22 out of the 48 mutations were identified to be novel and they were made up of all the splicing (2) and frameshift (9) mutations, one nonsense mutation, and 10 missense mutations.
Predicting the functional effects of mutations
We applied a popular algorithm, PolyPhen-2, to predict the effects of the mutations on ATP7B function. Polyphen-2 can predict the possible impact of an amino-acid substitution on the structure and function of a human protein by using straightforward physical and comparative considerations [17]. The predict results indicated that all the missense mutations were unfavorable except c.121A>G and c.748G>A, both of which were novel mutations (Table 1). And also two nonsense mutations, c.898_902 delAAGTA and c.4114C>T identified in our study should be most deleterious.
Identification of the linker mutations and identification of the SNPs
As shown in Table 2, the paired emerging of the mutations with an allele count of no less than two was analyzed to investigate the potential linkage between mutations. The resulted demonstrated that a pair of mutations, c.2310C>G and c.2333G>T, was most closely correlated. Then the SNPs of the patients were analyzed, as shown in Table 3, a total of eight SNPs were also identified and were compared with the frequencies in NCBI dsSNPs, Hapmap, 1000 genomes, and health controls.
Table 2.
Linkage analysis of the mutations
| EX2 Del | 2 | ||||||||||||
| c.1708-1G>C | 0 | 2 | |||||||||||
| c.2304_2305 insC | 0 | 0 | 4 | ||||||||||
| c.2310C>G | 0 | 1 | 1 | 28 | |||||||||
| c.2333G>T | 1 | 1 | 1 | 28 | 35 | ||||||||
| c.2621C>T | 0 | 0 | 0 | 1 | 3 | 6 | |||||||
| c.2662A>C | 0 | 0 | 0 | 0 | 0 | 0 | 3 | ||||||
| c.2755C>G | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 3 | |||||
| c.2975C>T | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 10 | ||||
| c.3316G>A | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 5 | |||
| c.3377_3378 delAC | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 2 | ||
| c.3443T>C | 0 | 0 | 1 | 2 | 2 | 0 | 2 | 0 | 0 | 2 | 0 | 9 | |
| c.3809A>G | 0 | 0 | 1 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 |
| EX2 Del | c.1708-1G>C | c.2304_2305 insC | c.2310C>G | c.2333G>T | c.2621C>T | c.2662A>C | c.2755C>G | c.2975C>T | c.3316G>A | c.3377_3378 delAC | c.3443T>C | c.3809A>G |
Correlation between genotype and phenotype of each WD patient
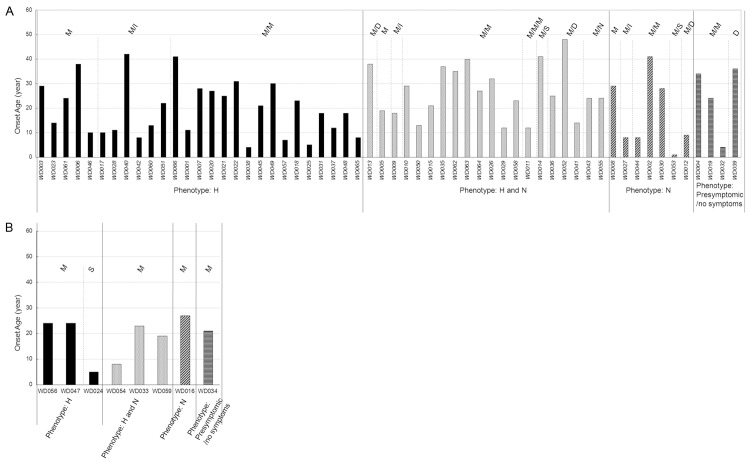
66 out of the 68 WD patients (97.1%) harbored at least one mutation. As shown in Table 4, 19 out of the 68 patients (24.6%) harbored three mutations, 38 patients (60.6%) harbored two mutations, and 9 patients (14.8%) harbored one mutation. The c.2333G>T linked with c.2310C>G mutations exist in all patients who harbored three mutations except one and exist in 21 patients who had two mutations. Eight patients were found to harbor homozygous mutations. Four of the homozygous mutations were c.2333G>T and c.2310C>G homozygote. The genotype of each patient and their corresponding detailed phenotypes were also showed in Table 4. Furthermore, to investigate the correlation between genotype and phenotype of the patients, the synonymous mutation c.2310C>G were excluded and we established a genotype/phenotype matrix by plotting the onset ages of each patient on the ATP7B mutation background and grouping by phenotypes. The ATP7B mutation backgrounds of individual patient were classified by mutation type and numbers, such as Missense (M), Missense/Missense (M/M), Missense/Splicing (M/S), and Missense/Insertion (M/I). As shown in Figure 1A, for the patients with heterozygous mutations, the mutation types and distribution were different in different phenotype groups. The top three mutation types were M/M (15/27), M/I (6/27) in H group and M/M (11/20), M/D (3/20), and M/N (2/20) in H and N group. And only the number of M/M was over two in N group and Presymptomic/no symptoms group due to the limited amount of patients. For the 8 patients with homozygous mutations, 7 patients carried with only unique missense mutation while one patient carried with unique splicing mutation (Figure 1B). The patient frequency with onset age ≤ 12 years old in H group and N group were bigger than that in H and N group and Presymptomic/no symptoms group in regardless of the heterozygous and homozygous conditions although the difference was not significant (Figure 1). And the mutation c.2333G>T were most prevalent in four groups while the other top prevalent mutations were not consistent. c.2975C>T was prevalent in H group, c.2621C>T was prevalent in H and N group, and c.3443T>C was prevalent in N group. The different mutation types and distribution in four groups might result in the difference of onset age.
Table 4.
Phenotypes and genotypes of the WD patients
| No. | Sex | Onset age | NT change | Mutation type | Phenotype | Presense of K-F ring | Serum ceruloplamin (g/L) | 24h-urinary copper (μmol) | Liver copper content (μg/g) |
|---|---|---|---|---|---|---|---|---|---|
| WD001 | female | 11 | c.2975C>T; c.2561A>G | heterozygous | H | + | 0.07 | 8.63 | 1,434.00 |
| WD002 | male | 41 | c.3818C>A; c.3316G>A | heterozygous | N | + | 0.09 | 5.98 | NA |
| WD003 | male | 29 | c.2975C>T | heterozygous | H | - | 0.14 | 0.53 | 348.00 |
| WD004 | male | 34 | c.2333G>T; c.3982G>A | heterozygous | presymptomic or no symptoms | + | 0.09 | 5.44 | 400.00 |
| WD005 | female | 19 | c.3901_3902 insA; c.2333G>T; c.2310C>G | heterozygous | H and N | + | 0.02 | 4.88 | NA |
| WD006 | male | 38 | c.2333G>T; c.2310C>G | heterozygous | H | - | 0.09 | 2.45 | 1,504.00 |
| WD007 | female | 28 | c.2975C>T; c.2755C>G | heterozygous | H | + | 0.11 | 5.70 | NA |
| WD008 | male | 29 | c.2333G>T; c.2310C>G | heterozygous | N | + | 0.03 | 4.50 | NA |
| WD009 | male | 18 | c.3443T>C; c.2662A>C | heterozygous | H and N | + | 0.03 | 3.28 | NA |
| WD010 | female | 29 | c.2333G>T; c.2549C>T | heterozygous | H and N | + | 0.14 | 2.33 | NA |
| WD011 | female | 12 | c.2975C>T; c.3445G>A; c.748G>A | heterozygous | H and N | + | 0.04 | 3.09 | 1,331.00 |
| WD012 | male | 9 | c.3443T>C; c.1745_1746 delTA | heterozygous | N | + | 0.03 | 2.50 | 295.80 |
| WD013 | male | 38 | c.2621C>T | heterozygous | H and N | + | 0.03 | 6.15 | NA |
| WD014 | male | 41 | c.1708-1G>C; c.3960G>C | heterozygous | H and N | + | 0.15 | 2.33 | NA |
| WD015 | male | 21 | c.2333G>T; c.3451C>T | heterozygous | H and N | - | 0.02 | 2.72 | 764.00 |
| WD016 | male | 27 | c.2620G>C | homozygous | N | + | 0.02 | 9.92 | NA |
| WD017 | male | 10 | c.2975C>T; c.3843_3844 insT | heterozygous | H | + | 0.12 | 5.17 | NA |
| WD018 | female | 23 | c.2333G>T; c.2506G>A; c.2310C>G | heterozygous | H | + | 0.02 | 3.53 | 1,109.00 |
| WD019 | male | 24 | c.3443T>C; c.3316G>A | heterozygous | presymptomic or no symptoms | - | 0.06 | 1.47 | 1,110.70 |
| WD020 | female | 27 | c.3443T>C; c.3316G>A | heterozygous | H | - | 0.07 | 4.11 | 956.00 |
| WD021 | female | 25 | c.3809A>G; c.2804C>T | heterozygous | H | + | 0.38 | 5.13 | NA |
| WD022 | male | 31 | c.2333G>T; c.2310C>G; c.2231C>T | heterozygous | H | + | 0.08 | 59.18 | NA |
| WD023 | female | 14 | c.3884C>T | heterozygous | H | + | 0.05 | 4.73 | NA |
| WD024 | female | 5 | c.3061-3C>A | homozygous | H | + | 0.03 | 2.34 | NA |
| WD025 | female | 5 | c.2333G>T; c.3679G>C; c.2310C>G | heterozygous | H | - | 0.02 | 1.25 | NA |
| WD026 | female | 32 | c.2333G>T; c.2310C>G; c.2621C>T | heterozygous | H and N | + | 0.03 | 5.86 | NA |
| WD027 | male | 8 | c.2304_2305 insC; c.3809A>G | heterozygous | N | + | 0.04 | 1.92 | 421.00 |
| WD028 | male | 11 | c.2975C>T; c.2304_2305 insC | heterozygous | H | + | 0.07 | 4.70 | NA |
| WD029 | male | 12 | c.2333G>T; c.2924C>A; c.2310C>G | heterozygous | H and N | + | 0.07 | 5.53 | NA |
| WD030 | male | 28 | c.2333G>T; c.3443T>C; c.2310C>G | heterozygous | N | + | 0.03 | 1.28 | NA |
| WD031 | male | 18 | c.2333G>T; c.2310C>G; c.121A>G | heterozygous | H | - | 0.06 | 2.13 | 950.00 |
| WD032 | male | 4 | c.3809A>G; c.2333G>T; c.2310C>G | heterozygous | presymptomic or no symptoms | + | 0.04 | 5.95 | 395.00 |
| WD033 | female | 23 | c.2333G>T; c.2310C>G | homozygous | H and N | + | 0.04 | 1.42 | 1,161.00 |
| WD034 | male | 21 | c.2333G>T; c.2310C>G | homozygous | presymptomic or no symptoms | + | 0.03 | 6.55 | NA |
| WD035 | female | 37 | c.3140A>T; c.2333G>T | heterozygous | H and N | + | 0.03 | 3.15 | NA |
| WD036 | male | 25 | c.3700 delG; c.1994T>G | heterozygous | H and N | + | 0.02 | NA | NA |
| WD037 | female | 12 | c.3584C>T; c.2333G>T; c.2310C>G | heterozygous | H | + | 0.20 | 20.51 | NA |
| WD038 | male | 4 | c.2975C>T; c.2662A>C | heterozygous | H | - | 0.03 | 1.31 | NA |
| WD039 | male | 36 | c.3377_3378 delAC | heterozygous | presymptomic or no symptoms | NA | 0.19 | NA | NA |
| WD040 | male | 42 | c.2593_2594insGTCA; c.3316G>A | heterozygous | H | + | 0.03 | 1.97 | NA |
| WD041 | female | 14 | EX2 DEL; c.3056A>C | heterozygous | H and N | + | 0.05 | 55.08 | NA |
| WD042 | female | 8 | c.2333G>T; c.2012_2013 insATAT | heterozygous | H | + | 0.04 | 4.19 | NA |
| WD043 | male | 24 | c.898_902 delAAGTA; c.2308C>T | heterozygous | H and N | + | 0.09 | 12.19 | NA |
| WD044 | male | 8 | c.2975C>T; c.2828G>A | heterozygous | N | + | 0.05 | NA | NA |
| WD045 | male | 21 | c.3007G>A; c.3677C>T | heterozygous | H | - | 0.12 | 2.53 | 1,080.00 |
| WD046 | male | 10 | c.2333G>T; c.2310 C>G | heterozygous | H | - | 0.02 | 3.92 | 1,030.30 |
| WD047 | female | 24 | c.2333G>T; c.2310C>G | homozygous | H | + | 0.02 | 2.08 | NA |
| WD048 | male | 18 | c2333G>T; c3452G>A; c.2310C>G | heterozygous | H | - | 0.02 | 4.10 | NA |
| WD049 | male | 30 | c.2975C>T; c.2605G>A | heterozygous | H | - | 0.06 | 2.81 | 908.00 |
| WD050 | male | 13 | c.3443T>C; c.2662A>C | heterozygous | H and N | + | 0.12 | 8.61 | NA |
| WD051 | male | 22 | c.2333G>T; c.2304_2305 insC; c.2310C>G | heterozygous | H | + | 0.03 | 5.39 | NA |
| WD052 | female | 48 | c.2621C>T; c.3377_3378 delAC | heterozygous | H and N | + | NA | 14.45 | NA |
| WD053 | male | 1 | c.2333G>T; c.1708-1G>C; c.2310C>G | heterozygous | N | + | 0.05 | NA | 111.00 |
| WD054 | female | 8 | c.2975C>T | homozygous | H and N | + | 0.09 | 8.95 | NA |
| WD055 | male | 24 | c.4114C>T; c.2128G>A | heterozygous | H and N | + | 0.03 | 5.26 | NA |
| WD056 | female | 24 | c.3443T>C | homozygous | H | + | 0.03 | 5.47 | NA |
| WD057 | male | 7 | c.2333G>T; c.3809A>G | heterozygous | H | + | 0.04 | 7.08 | NA |
| WD058 | female | 23 | c.2333G>T; c.3443T>C; c.2310C>G | heterozygous | H and N | + | 0.07 | 5.41 | 1,540.00 |
| WD059 | male | 19 | c.2333G>T; c.2310 C>G | homozygous | H and N | + | 0.04 | 5.39 | NA |
| WD060 | male | 13 | c.3443T>C; c.2304_2305 insC | heterozygous | H | + | 0.03 | 5.47 | NA |
| WD061 | male | 24 | c.3316G>A | heterozygous | H | - | 0.07 | 13.92 | NA |
| WD062 | male | 35 | c.2333G>T; c.2310C>G; c.2621C>T | heterozygous | H and N | + | 0.02 | 14.26 | NA |
| WD063 | female | 40 | c.2333G>T; c.2310C>G; c.2621C>T | heterozygous | H and N | - | 0.03 | NA | NA |
| WD064 | male | 27 | c.2755C>G; c.2333G>T; c.2310C>G | heterozygous | H and N | NA | 0.05 | NA | NA |
| WD065 | male | 8 | c.2333G>T; EX2 DEL | heterozygous | H | + | 0.02 | 33.69 | NA |
| WD066 | female | 41 | c.2621C>T; c.2755C>G | heterozygous | H | NA | 0.07 | 1.95 | NA |
| WD067 | male | 34 | - | - | H | - | 0.12 | 0.13 | NA |
| WD068 | female | 42 | - | - | H | + | 0.17 | 0.03 | NA |
Abbreviations: H, hepatic; H and N, hepatic and neurological; N, neurological.
Figure 1.
The Genotype-phenotype matrix for WD patients. A. The patients with heterozygous mutations. B. The patients with homozygous mutations. Abbreviations: M, Missense; I, Insertion; S, Splicing; D, Deletion.
To further study the correlation between onset age and ATP7B mutation background, the mutations in patients with onset age less than 12 years were analyzed. As shown in Table 5, a total of 19 mutations were identified and most the mutations located in TMS and HAD regions. Out of the mutations, 13 were missense mutations, three were insertion mutations, two were splicing mutations, and two were deletion mutation. The patients with c.2975C>T or c.3809A>G mostly presented WD features before 12 years old while the patients with c.3443T>C almost presented WD features after 12 years old. On the other hand, the patients with c.2333G>T, the prevalent mutations in Chinese WD patients, might have no little contribution on the onset age that the patients frequency with c.2333G>T with onset age ≤ 12 years old (9/31) was similar with that in total patients (19/66).
Table 5.
Information of ATP7B mutations in WD patients with onset age less than 12 years old
| Mutation | Location | Function region | Mutation type | No. of Total patient with the mutation | No. of ≤ 12 years patient with the mutation |
|---|---|---|---|---|---|
| c.1708-1G>C | Intron 4 | Splicing | 2 | 1 | |
| c.1745_1746 delTA | Exon 5 | MBD6 | Deletion | 1 | 1 |
| c.2012_2013insATAT | Exon 7 | TMS1 | Insertion | 1 | 1 |
| c.2304_2305 insC | Exon 8 | TMS4 | Insertion | 4 | 2 |
| c.2333G>T | Exon 8 | TMS4 | Missense | 31 | 9 |
| c.2561A>G | Exon 10 | ATPase | Missense | 1 | 1 |
| c.2662A>C | Exon 11 | ATPase | Missense | 3 | 1 |
| c.2828G>A | Exon 12 | Missense | 1 | 1 | |
| c.2924C>A | Exon 13 | TMS6 | Missense | 1 | 1 |
| c.2975C>T | Exon 13 | TMS6 | Missense | 10 | 7 |
| c.3061-3C>A | Intron 13 | Splicing | 1 | 1 | |
| c.3443T>C | Exon 16 | ATP bind | Missense | 9 | 1 |
| c.3445G>A | Exon 16 | ATP bind | Missense | 1 | 1 |
| c.3584C>T | Exon 17 | HAD | Missense | 1 | 1 |
| c.3679G>C | Exon 17 | HAD | Missense | 1 | 1 |
| c.3809A>G | Exon 18 | HAD | Missense | 4 | 3 |
| c.3843_3844insT | Exon 18 | HAD | Insertion | 1 | 1 |
| c.748G>A | Exon 2 | Missense | 1 | 1 | |
| EX2 DEL | Exon 2 | Deletion | 2 | 1 |
Discussion
In the present study, we performed mutational analysis of 68 WD patients from China. Finally, 48 mutations were identified, of which 22 mutations were novel. And the rate of mutation detection was 97.1% (66/68).
Up to now, more than 500 mutations have been reported in many different populations and the frequency and type of mutations are closely related with the geographic areas and ethnicities. For example, the most prevalent mutation in Europe is H1069Q missense mutation of exon 14, which is associated with neurological manifestation with allele frequency of 30%-65% of allele [18-20]. c.441_427del is present in 61.5% of patients in Sardinia [21], mutation M645R is frequent in Spain with a high value over 55% [22], and R778L in exon 8 associating with hepatic presentation is most frequent in China Han population with an allele frequency of 12-39% [6,23-25]. In this study, the allele frequency of R778L is 25.7% and the mutation is present in 45.6% patients. And we did not found the mutations H1069Q, c.441_427del, and M645R. Another most prevalent mutation in the study was p.P992L with an allele frequency of 8.1%, similar with the result in a previous north China WD patient cohort study (6.1%) [17]. Consistent with the results in other studies performed in Chinese population, we also found that the mutation p.L770L, was completely linked with the most prevalent mutation p.R778L [6,17]. Although p.L770L was a synonymous mutation, it was rare in the normal population (only five of the 854 healthy controls in the present study were identified to harbor this mutation) while appeared frequently in WD patients (24/68) and seemed to be a polymorphism in European population. All the exons and exon/intron boundaries of ATP7B gene were analyzed in our study and the rate of mutation detection is 97.1%, more than that in the previous reports. It also means that almost all the WD patients own pathological mutations in exons and exon/intro boundaries of ATP7B and tell us that a deep sequencing of the ATP7B exons and flanking regions other than the whole length will be economical and relative accuracy to help diagnose WD. The failure of detecting any mutations in the two patient might be because that some unknown mutations have not been detected due to the they may locate on the outside of the exons and flanking regions, such as the promoter, introns or other DNA control regions [26,27].
Although there are a number of studies analyzing the mutations of ATP7B in WD patients around the world, the detailed and specific correlation between phenotypes and mutation in WD patients is not well known. In the present study, we established a phenotype/genotype matrix by plotting onset age on ATP7B mutation types according to phenotype groups and we found the mutation types and distribution in different groups were different. Further analysis suggested that the mutations c.2975C>T or c.3809A>G contributed to early onset age in WD patients. This is the first time to identify the common mutations in the Chinese WD patients with an early onset age of less than 12 years old. A further multicenter study with a larger scale of WD patients needs to be performed to clarify the conclusions.
Conclusions
In this study, we perform mutational analysis of the ATP7B gene in 68 unrelated Chinese WD patients. The results suggested that mutations are present in 66 of the 68 patents and a total of 48 different mutations are identified, of which 22 mutations have never been reported previously. Among these mutations, c.2333G>T, c.2310C>G, c.2975C>T, and c.3443T>C are the most prevalent mutants. Phenotype/genotype correlation analysis suggests that the patients with c.2975C>T or c.3809A>G often present WD features before 12 years old while the patients with c.3443T>C almost present WD after 12 years old. Our study will broaden our knowledge about ATP7B mutations and their associations with phenotypes in WD patients in China.
Acknowledgements
This study was supported by the grant from Science and Technology Agency of Jilin Province (No. 20160101137JC).
Disclosure of conflict of interest
None.
References
- 1.Thomas GR, Forbes JR, Roberts EA, Walshe JM, Cox DW. The Wilson disease gene: spectrum of mutations and their consequences. Nat Genet. 1995;9:210–217. doi: 10.1038/ng0295-210. [DOI] [PubMed] [Google Scholar]
- 2.Compston A. Progressive lenticular degeneration: a familial nervous disease associated with cirrhosis of the liver, by S. A. Kinnier Wilson, (From the National Hospital, and the Laboratory of the National Hospital, Queen Square, London) Brain 1912: 34; 295-509. Brain. 2009;132:1997–2001. doi: 10.1093/brain/awp193. [DOI] [PubMed] [Google Scholar]
- 3.Ala A, Walker AP, Ashkan K, Dooley JS, Schilsky ML. Wilson’s disease. Lancet. 2007;369:397–408. doi: 10.1016/S0140-6736(07)60196-2. [DOI] [PubMed] [Google Scholar]
- 4.Gupta A, Aikath D, Neogi R, Datta S, Basu K, Maity B, Trivedi R, Ray J, Das SK, Gangopadhyay PK, Ray K. Molecular pathogenesis of Wilson disease: haplotype analysis, detection of prevalent mutations and genotype-phenotype correlation in Indian patients. Hum Genet. 2005;118:49–57. doi: 10.1007/s00439-005-0007-y. [DOI] [PubMed] [Google Scholar]
- 5.Ye S, Gong L, Shui QX, Zhou LF. Wilson disease: identification of two novel mutations and clinical correlation in Eastern Chinese patients. World J Gastroenterol. 2007;13:5147–5150. doi: 10.3748/wjg.v13.i38.5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mak CM, Lam CW, Tam S, Lai CL, Chan LY, Fan ST, Lau YL, Lai ST, Yuen P, Hui J, Fu CC, Wong KS, Mak WL, Tze K, Tong SF, Lau A, Leung N, Hui A, Cheung KM, Ko CH, Chan YK, Ma O, Chau TN, Chiu A, Chan YW. Mutational analysis of 65 Wilson disease patients in Hong Kong Chinese: identification of 17 novel mutations and its genetic heterogeneity. J Hum Genet. 2008;53:55–63. doi: 10.1007/s10038-007-0218-2. [DOI] [PubMed] [Google Scholar]
- 7.Wu ZY, Zhao GX, Chen WJ, Wang N, Wan B, Lin MT, Murong SX, Yu L. Mutation analysis of 218 Chinese patients with Wilson disease revealed no correlation between the canine copper toxicosis gene MURR1 and Wilson disease. J Mol Med (Berl) 2006;84:438–442. doi: 10.1007/s00109-005-0036-y. [DOI] [PubMed] [Google Scholar]
- 8.Sternlieb I. Perspectives on Wilson’s disease. Hepatology. 1990;12:1234–1239. doi: 10.1002/hep.1840120526. [DOI] [PubMed] [Google Scholar]
- 9.Gitlin JD. Wilson disease. Gastroenterology. 2003;125:1868–1877. doi: 10.1053/j.gastro.2003.05.010. [DOI] [PubMed] [Google Scholar]
- 10.Rosencrantz R, Schilsky M. Wilson disease: pathogenesis and clinical considerations in diagnosis and treatment. Semin Liver Dis. 2011;31:245–259. doi: 10.1055/s-0031-1286056. [DOI] [PubMed] [Google Scholar]
- 11.Huster D. Wilson disease. Best Pract Res Clin Gastroenterol. 2010;24:531–539. doi: 10.1016/j.bpg.2010.07.014. [DOI] [PubMed] [Google Scholar]
- 12.Tanzi RE, Petrukhin K, Chernov I, Pellequer JL, Wasco W, Ross B, Romano DM, Parano E, Pavone L, Brzustowicz LM, et al. The Wilson disease gene is a copper transporting ATPase with homology to the Menkes disease gene. Nat Genet. 1993;5:344–350. doi: 10.1038/ng1293-344. [DOI] [PubMed] [Google Scholar]
- 13.Menkes JH. Menkes disease and Wilson disease: two sides of the same copper coin. Part I: Menkes disease. Eur J Paediatr Neurol. 1999;3:147–158. doi: 10.1016/s1090-3798(99)90048-x. [DOI] [PubMed] [Google Scholar]
- 14.Menkes JH. Menkes disease and Wilson disease: two sides of the same copper coin. Part II: Wilson disease. Eur J Paediatr Neurol. 1999;3:245–253. doi: 10.1016/s1090-3798(99)90979-0. [DOI] [PubMed] [Google Scholar]
- 15.Roberts EA, Schilsky ML American Association for Study of Liver Diseases (AASLD) Diagnosis and treatment of Wilson disease: an update. Hepatology. 2008;47:2089–2111. doi: 10.1002/hep.22261. [DOI] [PubMed] [Google Scholar]
- 16.Gu YH, Kodama H, Du SL, Gu QJ, Sun HJ, Ushijima H. Mutation spectrum and polymorphisms in ATP7B identified on direct sequencing of all exons in Chinese Han and Hui ethnic patients with Wilson’s disease. Clin Genet. 2003;64:479–484. doi: 10.1046/j.1399-0004.2003.00179.x. [DOI] [PubMed] [Google Scholar]
- 17.Li K, Zhang WM, Lin S, Wen L, Wang ZF, Xie D, Wei M, Qiu ZQ, Dai Y, Lin MC, Kung HF, Yao FX. Mutational analysis of ATP7B in north Chinese patients with Wilson disease. J Hum Genet. 2013;58:67–72. doi: 10.1038/jhg.2012.134. [DOI] [PubMed] [Google Scholar]
- 18.Caca K, Ferenci P, Kuhn HJ, Polli C, Willgerodt H, Kunath B, Hermann W, Mossner J, Berr F. High prevalence of the H1069Q mutation in East German patients with Wilson disease: rapid detection of mutations by limited sequencing and phenotype-genotype analysis. J Hepatol. 2001;35:575–581. doi: 10.1016/s0168-8278(01)00219-7. [DOI] [PubMed] [Google Scholar]
- 19.Firneisz G, Lakatos PL, Szalay F, Polli C, Glant TT, Ferenci P. Common mutations of ATP7B in Wilson disease patients from Hungary. Am J Med Genet. 2002;108:23–28. doi: 10.1002/ajmg.10220. [DOI] [PubMed] [Google Scholar]
- 20.Maier-Dobersberger T, Ferenci P, Polli C, Balac P, Dienes HP, Kaserer K, Datz C, Vogel W, Gangl A. Detection of the His1069Gln mutation in Wilson disease by rapid polymerase chain reaction. Ann Intern Med. 1997;127:21–26. doi: 10.7326/0003-4819-127-1-199707010-00004. [DOI] [PubMed] [Google Scholar]
- 21.Loudianos G, Dessi V, Lovicu M, Angius A, Figus A, Lilliu F, De Virgiliis S, Nurchi AM, Deplano A, Moi P, Pirastu M, Cao A. Molecular characterization of wilson disease in the Sardinian population--evidence of a founder effect. Hum Mutat. 1999;14:294–303. doi: 10.1002/(SICI)1098-1004(199910)14:4<294::AID-HUMU4>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 22.Margarit E, Bach V, Gomez D, Bruguera M, Jara P, Queralt R, Ballesta F. Mutation analysis of Wilson disease in the Spanish population -- identification of a prevalent substitution and eight novel mutations in the ATP7B gene. Clin Genet. 2005;68:61–68. doi: 10.1111/j.1399-0004.2005.00439.x. [DOI] [PubMed] [Google Scholar]
- 23.Chuang LM, Wu HP, Jang MH, Wang TR, Sue WC, Lin BJ, Cox DW, Tai TY. High frequency of two mutations in codon 778 in exon 8 of the ATP7B gene in Taiwanese families with Wilson disease. J Med Genet. 1996;33:521–523. doi: 10.1136/jmg.33.6.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nanji MS, Nguyen VT, Kawasoe JH, Inui K, Endo F, Nakajima T, Anezaki T, Cox DW. Haplotype and mutation analysis in Japanese patients with Wilson disease. Am J Hum Genet. 1997;60:1423–1429. doi: 10.1086/515459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park S, Park JY, Kim GH, Choi JH, Kim KM, Kim JB, Yoo HW. Identification of novel ATP7B gene mutations and their functional roles in Korean patients with Wilson disease. Hum Mutat. 2007;28:1108–1113. doi: 10.1002/humu.20574. [DOI] [PubMed] [Google Scholar]
- 26.Al Jumah M, Majumdar R, Al Rajeh S, Awada A, Al Zaben A, Al Traif I, Al Jumah AR, Rehana Z. A clinical and genetic study of 56 Saudi Wilson disease patients: identification of Saudi-specific mutations. Eur J Neurol. 2004;11:121–124. doi: 10.1046/j.1351-5101.2003.00729.x. [DOI] [PubMed] [Google Scholar]
- 27.Wang LH, Huang YQ, Shang X, Su QX, Xiong F, Yu QY, Lin HP, Wei ZS, Hong MF, Xu XM. Mutation analysis of 73 southern Chinese Wilson’s disease patients: identification of 10 novel mutations and its clinical correlation. J Hum Genet. 2011;56:660–665. doi: 10.1038/jhg.2011.76. [DOI] [PubMed] [Google Scholar]