Abstract
The composition of the Gram-positive cell wall is typically described as containing peptidoglycan, proteins and essential secondary cell wall structures called teichoic acids, which comprise approximately half of the cell wall mass. The cell walls of many species within the genera Streptococcus, Enterococcus and Lactococcus contain large amounts of the sugar rhamnose, which is incorporated in cell wall-anchored polysaccharides (CWP) that possibly function as homologues of well-studied wall teichoic acids (WTA). The presence and chemical structure of many rhamnose-containing cell wall polysaccharides (RhaCWP) has sometimes been known for decades. In contrast to WTA, insight into the biosynthesis and functional role of RhaCWP has been lacking. Recent studies in human streptococcal and enterococcal pathogens have highlighted critical roles for these complex polysaccharides in bacterial cell wall architecture and pathogenesis. In this review, we provide an overview of the RhaCWP with regards to their biosynthesis, genetics and biological function in species most relevant to human health. We also briefly discuss how increased knowledge in this field can provide interesting leads for new therapeutic compounds and improve biotechnological applications.
Keywords: cell wall polysaccharide, rhamnose, pathogenesis, biosynthesis, glycobiology, Gram-positive bacteria
This review summarizes new insights into the genetics and function of rhamnose-containing cell wall polysaccharides expressed by lactic acid bacteria, which includes medically important pathogens, and discusses perspectives on possible future therapeutic and biotechnological applications.
INTRODUCTION
The composition of the bacterial cell wall is critical for fundamental features such as bacterial cell shape, protection from and interaction with the environment. Carbohydrates are the most abundant molecules in the Gram-positive cell wall with much of it incorporated in the thick layer of peptidoglycan (15–100 nm) that surrounds the cell membrane (Silhavy, Kahne and Walker 2010). Peptidoglycan is composed of alternating β-1,4-linked N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc) polysaccharide strands that are cross-linked by short peptides to form a three-dimensional network. Individual species tailor important physical properties of their peptidoglycan such as elasticity and porosity through the composition of the peptide bridge, the amount of crosslinking and chemical modifications of the composing glycan residues (Vollmer, Blanot and de Pedro 2008; Vollmer and Seligman 2010). Peptidoglycan also acts as a scaffold for other critical cell wall structures. For example, proteins containing LPXTG amino acid motifs are covalently attached to the peptidoglycan peptide bridge through the enzymatic action of sortase A (Hendrickx et al. 2011).
Cell wall polysaccharides (CWP), such as capsular polysaccharides and wall teichoic acids (WTA), are anchored to peptidoglycan GlcNAc or MurNAc, covering the bacterium with a layer of glycans that is directly exposed to the environment (Deng et al. 2000; Swoboda et al. 2010; Yother 2011). The predominance of capsulated species among bacterial pathogens instigated studies on the role of capsular polysaccharides in infectious disease pathogenesis. As a result, effective capsule polysaccharide conjugate vaccines were developed against various species including Neisseria meningitidis, Haemophilus influenzae type B and Streptococcus pneumoniae. For some human bacterial pathogens, such as Staphylococcus aureus and Streptococcus pyogenes, the role of polysaccharide capsule in pathogenesis is less pronounced (O'Riordan and Lee 2004; Flores et al. 2012). In addition, the discovery of host pattern-recognition receptors with specificity for carbohydrates as well as technological advances in the area of complex carbohydrate analysis and synthesis have fueled initiatives to investigate bacterial glycobiology more comprehensively by unraveling the structure, biosynthesis and functions of bacterial polysaccharide structures.
In this review, we focus on a subclass of secondary CWP that we refer to as rhamnose-containing CWP (RhaCWP). L-rhamnose is commonly found in bacteria but is not used or produced by humans (Maki and Renkonen 2004; Adibekian et al. 2011). Interestingly, L-rhamnose is often essential for bacterial virulence or even viability (Maki and Renkonen 2004), making its biosynthesis pathway an attractive therapeutic target. We will therefore review the current knowledge regarding L-rhamnose biosynthesis and functions in bacteria in more detail. Recent insights into the genetic basis and function of RhaCWP in two important human pathogens, S. pyogenes (Group A Streptococcus) and Streptococcus agalactiae (Group B Streptococcus), have emphasized the critical role of these molecules in cell wall biogenesis and pathogenesis (Caliot et al. 2012; van Sorge et al. 2014). This functional information combined with the localization and abundance of RhaCWP suggests that parallels can be drawn with WTA in other Gram-positive bacteria. WTA structure and function have been reviewed extensively (Weidenmaier and Peschel 2008; Swoboda et al. 2010; Brown, Santa Maria and Walker 2013). Therefore, we will only highlight specific parallels with WTA biology throughout this review. Finally, from their historic discovery (Lancefield 1933) and recent insight from bacterial genome sequences, it is apparent that RhaCWP are likely more widespread in Gram-positive cocci within the order Lactobacillales. We will provide an overview of their inferred distribution and review the literature for selected species with regard to structure, genetics, biosynthesis and function. We will end by discussing the potential therapeutic and biotechnological applications of this important class of CWP.
Non-classical CWP in Gram-positive cocci: a historical perspective
Most knowledge regarding the architecture and biology of the Gram-positive cell wall is derived from the model organisms Bacillus subtilis and S. aureus. For these and other species, WTA is a major cell wall component representing up to 60% of the total cell wall mass. WTA are anionic glycopolymers that are covalently attached to the peptidoglycan MurNAc residue (Swoboda et al. 2010). Most commonly, WTA are composed of a poly-ribitolphosphate (-RboP)- or poly-glycerolphosphate (-GroP-) backbone with modifications such as glycosylation and D-alanylation, the latter of which can neutralize the negative charge of the abundant phosphates in the WTA backbone (Brown, Santa Maria and Walker 2013). However, the exact chemical composition varies among and even within species (Neuhaus and Baddiley 2003; Weidenmaier and Peschel 2008; Winstel, Xia and Peschel 2014). It has long been recognized that not all Gram-positive bacteria incorporate polyRboP- or a polyGroP-based WTA into their cell wall. Instead, the cell walls of many species within the order Lactobacillales contain peptidoglycan-anchored polysaccharides that are characterized by the presence of L-rhamnose. Probably the first reports of RhaCWP in the genus Streptococcus date back to the late 1920s and early 1930s by Rebecca Lancefield (Lancefield 1928a,b, 1933). Her seminal work enabled the development of a new streptococcal classification system—in addition to hemolytic typing—based on the differential antigenic properties of streptococcal CWP, referred to as ‘C-substance’ or ‘Group Antigen’ (Lancefield 1933). The streptococcal serotyping scheme initially only discriminated Group A–E streptococci but was expanded to comprise as many as 20 serotypes called Groups A–V (excluding I and J) (Facklam 2002; Kohler 2007). The Lancefield typing system has been instrumental to link mild and severe human and animal diseases to specific bacterial groups within the genus Streptococcus, most notably S. pyogenes, causing more than 700 million infections resulting in over 500 000 deaths worldwide annually (Carapetis et al. 2005), and S. agalactiae, a pathogen affecting mainly neonates and the elderly (Phares et al. 2008; Edmond et al. 2012). Furthermore, the Lancefield typing scheme enabled the development of rapid diagnostic tests that aided clinically relevant ‘species identification’ (Lue, Howit and Ellner 1978). Increased recognition of additional strain characteristics, such as nutrient requirements and later the use of 16S rRNA for classification (and more recently whole genome sequencing), demonstrate that the Lancefield typing system cannot discriminate to the species level such that a single Lancefield Group represents one species. For example, the Group A antigen was long thought to be the exclusive molecular marker of S. pyogenes, yet Streptococcus castoreus is also noted to react with Group A antisera in commercial diagnostic kits (Lawson et al. 2005). Vice versa, a single species can express different Group antigens; strains of Streptococcus dysgalactiae subsp. equisimilis commonly express either the Group C or G antigen (Broyles et al. 2009; McMillan et al. 2010a; Takahashi, Ubukata and Watanabe 2011) and occasionally Group A carrying S. dysgalactiae subsp. equisimilis strains are identified (Tanaka et al. 2008; McMillan et al. 2010b). Consequently, the taxonomy and nomenclature of the genus Streptococcus has been reevaluated and reclassified over the years (Facklam 2002; Kohler 2007). This resulted in the split of the genus Streptococcus into three genera, i.e. Enterococcus, Lactococcus and Streptococcus (Schleifer et al. 1985), as well as subsequent description of several novel members within these genera. Two species covered in this review, Enterococcus faecalis and Lactococcus lactis, were formerly known as Streptococcus faecalis (Lancefield Group D) and Streptococcus lactis (Lancefield Group N). Today, more than 150 different species are known within these three genera, which remain classified within the order Lactobacillales (http://www.bacterio.net/-classifphyla.html#Firmicutes; Price et al. 2012).
Although the direct correlation between Lancefield serotyping and species clearly no longer persists, bioinformatic analyses of genome sequences indicates that many of the species within these three genera express RhaCWP (see section ‘Distribution of RhaCWP throughout bacteria’). We focus here on the species relevant to food production and human health and with experimental evidence for the presence of RhaCWP: Lancefield Groups A, B, C, E and G Streptococcus represented by S. pyogenes, S. agalactiae, Streptococcus equi subsp. zooepidemicus, Streptococcus mutans and S. dysgalactiae subsp. equisimilis, respectively, as well as E. faecalis and L. lactis.
Cell wall organization of RhaCWP
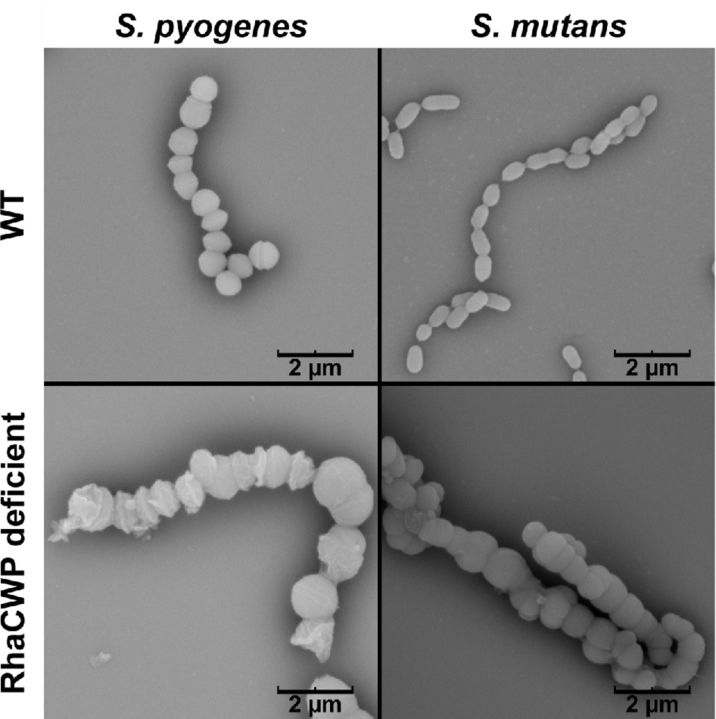
The RhaCWP is a major component in streptococcal species comprising about 40%–60% of the cell wall by weight (McCarty 1952; Krause and McCarty 1962a; Krause 1963; Doran and Mattingly 1982). Correspondingly, cell wall thickness is visually reduced by 40%–50% after chemical extraction of the Group-specific carbohydrate (Swanson and Gotschlich 1973; Wagner and Wagner 1978). Recent mutagenesis studies confirmed the structural importance of streptococcal group antigens; complete loss of RhaCWP expression resulted in severe growth and cell division abnormalities (Fig. 1; Tsukioka et al. 1997a; Caliot et al. 2012; van Sorge et al. 2014; van der Beek et al. 2015).
Figure 1.
Cell division and separation defects caused by RhaCWP deficiency in S. mutans and S. pyogenes. Representative scanning electron microscopy images of S. pyogenes and S. mutans wild-type (WT) strains and corresponding RhaCWP-deficient strains. In S. pyogenes loss of the GAC was enforced by inducible knockout of gacA (rmlD homologue) and in S. mutans, deletion of rmlD results in loss of RhaCWP (van der Beek et al. 2015). Scale bar is indicated in image.
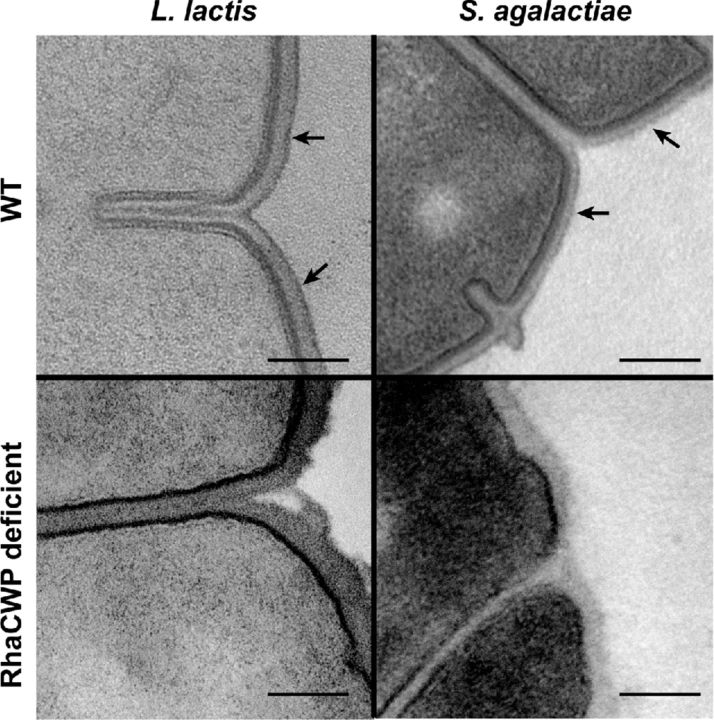
RhaCWP are localized on the outermost surface of the cell wall but are likely also intercalated within the mesh-like structure of the peptidoglycan layer since antibodies directed against these structures bind to both sides of isolated cell walls (Swanson and Gotschlich 1973; Wagner and Wagner 1978; Wagner et al. 1980). Group-specific antigens can also be isolated at high yield in the growth medium (Carey et al. 1980; De Cueninck, Shockman and Swenson 1982; Doran and Mattingly 1982), possibly as a result of cell wall catabolism during growth. Compared to the cell walls of S. aureus and B. subtilis, streptococcal species appear to lack expression of polyol-based WTA and, typically, lack orthologues of the critical WTA biosynthesis enzymes TagB, TagD and TagF (Sutcliffe, Black and Harrington 2008) (and unpublished observations). In contrast to streptococci, homologues of WTA biosynthesis genes are found in lactococcal genomes. Consequently, L. lactis strains likely express both polyol-based WTA and RhaCWP as part of their cell wall (Chapot-Chartier and Kulakauskas 2014). E. faecalis expresses the most extensive surface glycome including WTA, LTA, capsular polysaccharide and a RhaCWP called Enterococcal polysaccharide antigen (Epa) (Hancock and Gilmore 2002; Teng et al. 2009; Thurlow, Thomas and Hancock 2009; Theilacker et al. 2012). In contrast to RhaCWP in streptococci and L. lactis, Epa appears to be buried in the cell wall precluding interaction with the immune system, at least under laboratory conditions (Hancock and Gilmore 2002). It should however be noted that sera from patients do contain Epa-specific antibodies (Xu, Murray and Weinstock 1998; Xu et al. 2000; Teng et al. 2002). Also, the presence of Epa is visible under transmission electron microscopy (TEM) as a separate electron dense outer layer (Rigottier-Gois et al. 2015), similar to the RhaCWP layer observed in L. lactis and S. agalactiae (Chapot-Chartier et al. 2010; Caliot et al. 2012) (Fig. 2). In early studies, this electron dense outer layer was known as the outer lamina and was initially described as microcapsule (Baker and Kasper 1976). Recently, the term pellicle was coined (Chapot-Chartier et al. 2010) and is preferred since the term does not inherently imply that this layer is either ‘outer’ or of a specific composition (such as capsule). Interestingly, the pellicle is not as impenetrable as TEM images suggest. In S. agalactiae, topographic imaging and atomic force microscopy-based single-molecule mapping on live bacteria revealed that peptidoglycan strands can still be probed in the presence of the GBC (Beaussart et al. 2014). However, the pellicle does shield peptidoglycan to some extent, since loss of GBC expression through genetic manipulation renders S. agalactiae extremely sensitive to the activity of peptidoglycan-cleaving mutanolysin (Caliot et al. 2012). Similarly, atomic force microscopy studies on pellicle-deficient L. lactis uncovers peptidoglycan periodic bands orientated parallel to the short axis of the cell (Andre et al. 2010).
Figure 2.
Visualization of the pellicle by transmission electron microscopy. Lactococcus lactis and S. agalactiae transmission electron microscope images contrasted with heavy metal staining method. In wild-type (WT) strains the pellicle is visible and indicated by arrows. Loss of RhaCWP expression, due to of genetic mutation, result in loss of pellicle expression. The scale bar represents 0.1 μm.
Chemical structure of RhaCWP
Studies focusing on the chemical composition of Lancefield Group antigens have demonstrated that rhamnose is the major constituent, along with variable combinations and linkages of Glc, GlcNAc, Gal, GalNAc and phosphate (Pritchard et al. 1981). Elucidation of Group-specific RhaCWP structures in individual streptococcal species provided structural evidence for the discriminating capacity of the Lancefield typing scheme (Fig. 3A). The serological distinction between Group A and Group C Streptococcus is explained by expression of a terminal β-linked GlcNAc side chain in the Group A Carbohydrate (GAC) versus a (GalNAc)2 side chain in the Group C Carbohydrate (GCC), respectively (McCarty 1956; Krause and McCarty 1962a; Coligan, Kindt and Krause 1978) (Fig. 3A). However, biochemical and immunological characterization of CWP isolated from so-called Group A- and Group C-variant strains also noted that the GAC and GCC are structurally related. These variant strains lost Lancefield serum reactivity and displayed a variant Group antigen comprised of unsubstituted rhamnan with only trace amount of the N-acetylated sugars (McCarty 1956; Krause and McCarty 1962b). Occurrence of such variant strains appears to be a rare event; one Group C-variant strain was isolated as a resistant clone after exposure to virulent Group C bacteriophages (Krause 1963), whereas Group A-variant strains appear after multiple passages through an unnatural host, such as mice, but have never been isolated from humans (McCarty and Lancefield 1955). Whether strains are able to vary expression, composition or length of RhaCWP during natural infection is currently unclear. Possibly such variants may be missed in routine diagnostics screening. Alternatively, loss of specific RhaCWP epitopes or loss of the complete structure would severely hamper the ability of the bacterium to colonize or infect the host, allowing rapid eradication by the host immune system as will be described in more detail below.
Figure 3.
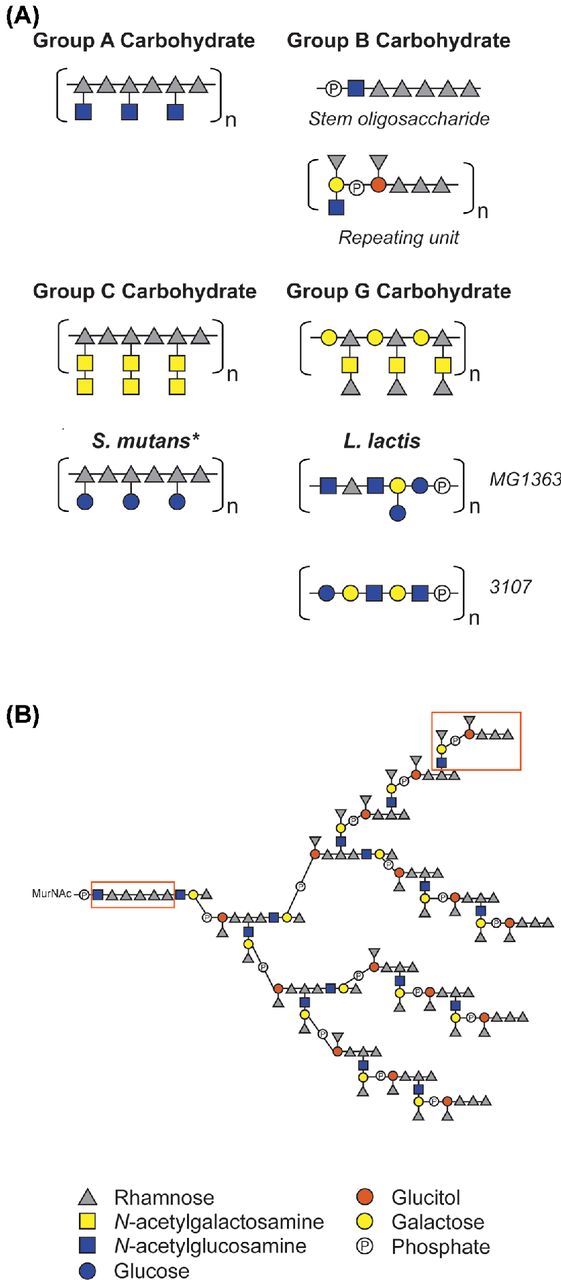
Schematic representation of known RhaCWP structures. (A) Schematic representation of RhaCWP core structures in different streptococcal species and L. lactis. GAC structure (McCarty and Lancefield 1955; Coligan, Kindt and Krause 1978; Pritchard et al. 1982; Huang, Rama Krishna and Pritchard 1986); GBC structure (Pritchard, Gray and Dillon 1984; Michon et al. 1987; 1988); GCC structure (Krause and McCarty 1962a; Coligan, Kindt and Krause 1978); GGC structure (Pritchard et al. 1988); S. mutans serotypes c, e, f, k (Pritchard and Furner 1985; Pritchard et al. 1986, 1987; Nakano and Ooshima 2009); L lactis (Chapot-Chartier et al. 2010; Ainsworth et al. 2014; Chapot-Chartier and Kulakauskas 2014). *For S. mutans the glucose side chain can either be absent (serotype k) or be linked to the rhamnan backbone in α-1,2 configuration (serotype c), β-1,2 configuration (serotype e), or α-1,3 configuration (serotype f). It must be noted that the (distribution of) length(s) of the RhaCWP have not been experimentally determined. RhaCWP are likely covalently attached to peptidoglycan MurNAc. (B) Full structure of GBC as described by (Pritchard, Gray and Dillon 1984; Michon et al. 1987, 1988) with newly recognized structural elements highlighted in boxes. Because both the repeating unit and the rhamnan stem have a basal GlcNAc moiety, we hypothesize that the synthesis of each building block is initiated separately on the undecaprenyl lipid carrier by GbcO. It is recognized that either incomplete substitution (at branch points located on the penultimate rhamnose of the repeating unit) or further extension could create a more heterogeneous final structure than presented here. Phosphate groups are involved in phosphodiester bonds linking oligosaccharides into polysaccharides.
For the Group B (GBC) and G carbohydrate (GGC), rhamnose is the major antigenic determinant (Curtis and Krause 1964a,b). Species carrying these structures are serologically discriminated based on the presence of either a single rhamnose in GGC versus triterminal rhamnose in GBC (Curtis and Krause 1964a) (Fig. 3A and B). Among the streptococcal group antigens, the GBC is unique since it forms a multiantenna branching structure and is negatively charged, due to the presence of phosphodiester bonds that link different GBC repeat units (Fig. 3B). Similarly, the L. lactis RhaCWP contains phosphodiester bonds that link hexasaccharide or pentasaccharide repeating units (Fig. 3A) (Chapot-Chartier et al. 2010; Ainsworth et al. 2014). Unfortunately, the structure of E. faecalis Epa has not yet been elucidated but is composed of the monosaccharides glucose, rhamnose, GlcNAc, GalNAc, galactose and probably phosphate (Hancock and Gilmore 2002; Teng et al. 2009). The presence of phosphates in RhaCWP of S. agalactiae, L. lactis and possibly E. faecalis likely suggests that the functions of these structures more closely resemble functions exerted by WTA in other Gram-positive bacteria (Michon et al. 1987, 1988, 1991; Sutcliffe 2008).
General aspects of RhaCWP genetics and biosynthesis
CWP can be structurally highly complex due the number and variation in their monosaccharide components, diverse linkage types and chemical substitutions such as (de)acetylation and hydroxylation. Their biosynthesis requires the coordinated action of glycosyltransferases (enzymes that link monosaccharides), transporters and metabolic enzymes for the production of nucleotide-sugar precursors. Often, genes encoding proteins required for glycoconjugate biosynthesis are clustered on the chromosome. Their identification allows subsequent structure–function studies that help to understand the role of glycosylation in bacterial (infection) biology.
It is surprising that despite the historical and medical importance of the Lancefield Group antigens, no studies were undertaken to decipher the genetic basis of their biosynthesis. By contrast, capsule synthesis was quickly recognized as a major virulence factor in many bacterial species and has consequently been the subject of many genetic, biochemical and functional studies (Llull, Lopez and Garcia 2001; Cress et al. 2014). Initial predictions for the genetic basis of Lancefield Group carbohydrate biosynthesis were postulated upon availability of the first streptococcal genome sequences (Ferretti et al. 2001; Glaser et al. 2002; Holden et al. 2009; Shimomura et al. 2011). For S. agalactiae, the availability of the GBC structure and genome sequence enabled a comprehensive and detailed in silico analysis that linked protein-encoding genes to the different glycosidic linkages in the GBC structure (Sutcliffe, Black and Harrington 2008). That study demonstrated that most, but not all, of the enzymatic activities for synthesis and transport of the mature GBC molecule are present in the predicted 15 kB GBC gene cluster (Sutcliffe, Black and Harrington 2008); proteins required for lipid carrier activation and peptidoglycan anchoring of GBC seemed to be encoded elsewhere on the genome (Sutcliffe, Black and Harrington 2008). As will be discussed below, this split in gene organization, correlating with different biosynthetic steps, appears to be a common feature of the streptococcal and lactoccocal RhaCWP biosynthesis pathway. In addition, the biosynthesis of RhaCWP requires the production of appropriate nucleotide sugars precursors. We will only cover dTDP-L-rhamnose biosynthesis here given its characteristic presence in RhaCWP, as well as the possible therapeutic implications of this pathway. We will then discuss the genetic organization and putative biosynthesis pathway of RhaCWP.
L-rhamnose biosynthesis
Incorporation of L-rhamnose into polysaccharide structures in both Gram-positive and Gram-negative bacteria requires the formation of the nucleotide sugar precursor dTDP-L-rhamnose. dTDP-L-rhamnose is produced from glucose-1-phophate through a conserved four-step enzymatic reaction that has been characterized both biochemically and structurally (Fig. 4) (Giraud and Naismith 2000; Dong et al. 2003a). In the first step of the pathway, RmlA, a glucose-1-phosphate thymidyltransferase, converts glucose-1-phosphate into dTDP-glucose (Blankenfeldt et al. 2000), which is subsequently oxidized and dehydrated to form dTDP-4-keto-6-deoxy-D-glucose by the dTDP-D-glucose 4,6-dehydratase RmlB (Beis et al. 2003). RmlC catalyzes an unusual double epimerization reaction (Giraud et al. 2000; Dong et al. 2003b; 2007), the product of which is finally reduced by RmlD, a dTDP-4-dehydrorhamnosereductase, to form dTDP-L-rhamnose (Blankenfeldt et al. 2002; van der Beek et al. 2015). Initial structure elucidation of the Rml enzymes from different species, including Pseudomonas aeruginosa (RmlA), Salmonella enterica (RmlC and RmlD) and Streptococcus suis (RmlB and RmlC), demonstrated that rhamnose biosynthesis enzymes require dimerization or even dimerization of dimers to catalyze the respective enzymatic reactions (Blankenfeldt et al. 2000; Giraud et al. 2000; Blankenfeldt et al. 2002; Beis et al. 2003; Dong et al. 2007). However, recent structural and biochemical characterization of the S. pyogenes RmlD homologue provided surprising insight that, in this species, RmlD is active as a monomer (van der Beek et al. 2015). Subsequent comprehensive bioinformatics analysis of 213 putative RmlD sequences indicates that the monomeric form of RmlD is more widespread throughout the bacterial kingdom compared to the originally described RmlD from Salmonella (Blankenfeldt et al. 2002; van der Beek et al. 2015). The benefit of either structure to the enzymatic reaction is currently unknown.
Figure 4.
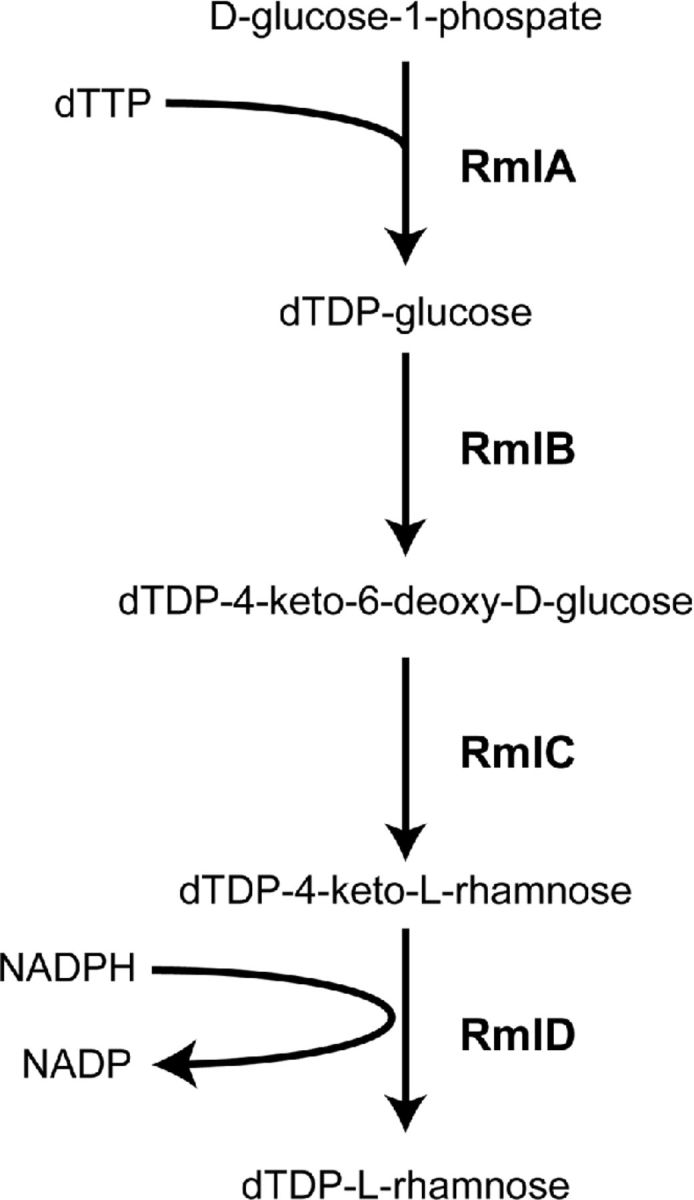
L-rhamnose biosynthesis pathway. Four-step catalytic reaction resulting in dTDP-L-rhamnose production from D-glucose-1-phosphate.
In many bacteria, such as in E. faecalis (Xu, Murray and Weinstock 1998), Shigella flexneri (Macpherson, Manning and Morona 1994), L. lactis (Dupont et al. 2004), S. pneumoniae (Bentley et al. 2006) and S. enterica (Jiang et al. 1991), the RmlA-D proteins are encoded by a single genetic locus, rmlABCD. The rml genes are likely transcribed as an operon, although experimental evidence for that is currently lacking. In contrast, in some bacteria the rml genes display a split architecture, often clustering rmlABC, but excluding rmlD. This is for example the case in Mycobacterium tuberculosis (Ma et al. 2001) and several streptococcal species (Tsukioka et al. 1997a,b; van der Beek et al. 2015). The evolutionary origin or functional benefit of a split versus clustered gene architecture is remains to be determined.
The contribution of Rml enzymes to L-rhamnose biosynthesis has been demonstrated in several species through a genetics approach, i.e. mutation of any of the rml genes results in loss of L-rhamnose in the bacterial cell walls (Tsukioka et al. 1997a,b; Rahim et al. 2000; Carvalho et al. 2015; van der Beek et al. 2015). For other species, loss of L-rhamnose in the cell wall was not confirmed by cell wall composition analysis upon mutation of rml genes (Chiang and Mekalanos 1999; Xu et al. 2000). Importantly, disruption of dTDP-L-rhamnose biosynthesis severely attenuates bacterial fitness and/or virulence (Tsukioka et al. 1997a; Chiang and Mekalanos 1999; Rahim et al. 2000; Xu et al. 2000; Carvalho et al. 2015; van der Beek et al. 2015). The position or percentage of incorporated L-rhamnose likely dictates whether rml genes are essential. For example, in M. tuberculosis, L-rhamnose covalently links arabinogalactan to peptidoglycan, which is critical for the overall architecture of the Mycobacterial cell wall, making L-rhamnose biosynthesis essential (McNeil, Daffe and Brennan 1990; Ma, Pan and McNeil 2002). Similarly in S. pyogenes, L-rhamnose is incorporated in the GAC that comprises half of the cell wall mass (McCarty 1952), rendering depletion lethal (Le Breton et al. 2015; van der Beek et al. 2015). In the case of uropathogenic Escherichia coli and P. aeruginosa, mutation of rmlD results in loss of O-antigen expression but leaves the lipopolysaccharide core and lipid A structure intact, yielding viable bacteria (Burns and Hull 1998; Rahim et al. 2000). However, loss of rhamnose does come at a certain cost, since rhamnose-deficient E. coli are extremely sensitized to serum-mediated killing (Burns and Hull 1998).
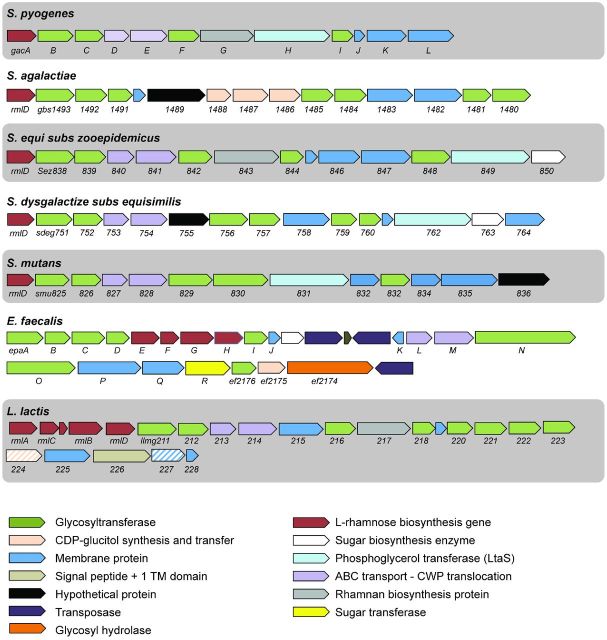
Analysis of its genomic location in members of the family Streptococcaceae and in E. faecalis reveal that rmlD is systematically associated with a large cluster of genes encoding glycosyltransferases, polysaccharide transport systems, sugar biosynthesis enzymes and genes of unknown functions (Fig. 5). Experimental evidence in selected streptococcal species, L. lactis and E. faecalis confirms that rmlD-associated loci participate in the biosynthesis of RhaCWP as will be discussed in the next section.
Figure 5.
Comparison of the RhaCWP biosynthesis gene clusters in streptococcal species, L. lactis and E. faecalis. The RhaCWP biosynthesis genes are often located in rmlD-associated gene clusters. The loci vary between 14 and 26 kB in size encoding between 12 and 25 genes with annotated functions such as glycosyltransferases, polysaccharide biosynthesis proteins, rhamnose biosynthesis proteins (Rml proteins) and putative transport molecules. Each function is indicated by a different color. A representative gene cluster of a single species is presented and abbreviated gene annotations are indicated below. Arrows are drawn to scale with gene size. The RhaCWP gene clusters of the following strains are shown: S. pyogenes strain M5005 spy0602 – spy0613; S. agalactiae NEM316 gbs1480 – gbs1494; S. equi subs equisimilis MGCS10565 Sez837 – Sez850; S. dysgalactiae sub equisimilis GGS_124 SDEG750 – SDEG764; S. mutans UA159 SMU.824 – SMU.836; E. faecalis V583 EF2198 – EF2174; L. lactis MG1363 llmg0206 – llmg0228. TM, transmembrane
Biosynthesis of RhaCWP
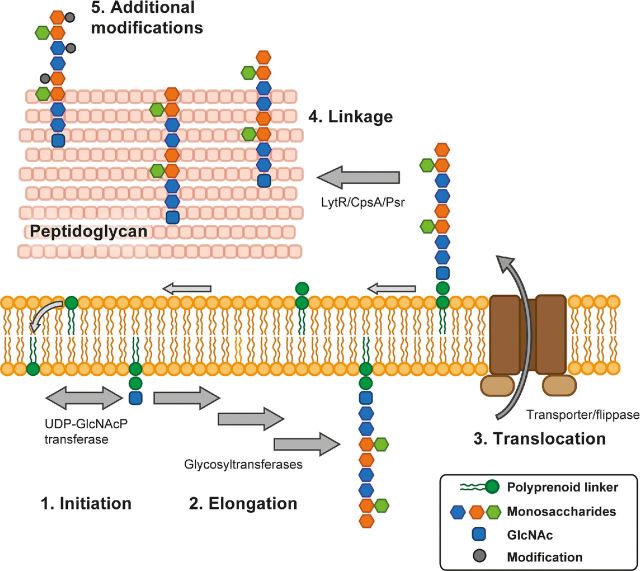
Biosynthesis of glycosylated surface structures in bacteria displays some common themes, despite the considerable diversity in the chemical composition of glycosylated structures (reviewed in Tytgat and Lebeer 2014). In general, glycoconjugate biosynthesis proceeds as follows (Fig. 6); (1) initiation of biosynthesis through activation of a lipid carrier, often undecaprenylphosphate, on the cytoplasmic side of the membrane, (2) elongation of the polysaccharide (building block) on the lipid carrier by sequential addition of activated sugar precursors, (3) translocation of lipid-linked precursors, either repeating units or the complete glycoconjugate, across the membrane by ABC transporters or ‘flippases’ (Lazarevic and Karamata 1995), (4) linkage of the glycoconjugate to peptidoglycan or protein (Kawai et al. 2011; Chan et al. 2013) and (5) additional modifications to the glycoconjugate that can occur after cell wall anchoring. In some cases, the mature glycoconjugate is formed by polymerization of translocated repeating units between steps (3) and (4) by a dedicated polymerase. Since undecaprenylphosphate serves as a common scaffold to build structurally diverse glycoconjugates including capsules, peptidoglycan, lipopolysaccharides and protein-modifying glycans, the availability of ‘free’ undecaprenylphosphate is essential for bacterial survival (Hartley and Imperiali 2012).
Figure 6.
General steps in glycoconjugate biosynthesis. Despite glycoconjugate diversity, bacterial glycoconjugate biosynthesis is quite conserved and proceeds in five basic steps: (1) initiation of biosynthesis through activation of a lipid carrier on the cytoplasmic side of the membrane, (2) elongation by sequential addition of activated monosaccharides by glycosyltransferases to form the polysaccharide on the polyprenoid linker, (3) translocation of lipid-linked precursors across the membrane by ABC transporters or ‘flippases’, (4) linkage of the glycoconjugate to peptidoglycan by LytR/CpsA/Psr proteins, and (5) additional modifications to the glycoconjugate that can occur after anchoring to the cell wall, for example alanylation of wall teichoic acid. GlcNAc, N-acetylglucosamine. UDP-GlcNAcP transferase, UDP-GlcNAc:lipid phosphate transferase
In accordance with the general steps of glycoconjugate biosynthesis (Fig. 6), biosynthesis of RhaCWP is likely initiated on the inside of the cytoplasmic membrane on an undecaprenylphosphate lipid carrier, before the structure is transported across the membrane and attached to peptidoglycan. Indeed, most enzymes involved in RhaCWP biosynthesis are predicted to be intracellular or incorporated in the membrane. Furthermore, lysed protoplasts of S. pyogenes incorporate 14C-labeled dTDP-rhamnose into a polysaccharide structure, presumably the GAC (Zeleznick et al. 1963). The proteins required for elongation and translocation of RhaCWP appear to be encoded by rmlD-associated gene clusters (Fig. 5). Within each cluster, we find genes encoding glycosyltransferases, putative transport systems (either an ABC transport system or putative flippase), sugar modifying enzymes for the production of specific sugar precursors, and hypothetical proteins which can be unique for the species (Sutcliffe, Black and Harrington 2008). Enzymes for initiation (Fig. 6; step 1) and peptidoglycan anchoring after transport (Fig. 6; step 4) are encoded elsewhere on the genome in most species. It is beyond the scope of this review to discuss the putative role of each gene in detail; instead, we aim to provide genetic and biochemical insights concerning the proposed steps of the RhaCWP biosynthesis pathway.
Lipid carrier activation by UDP-GlcNAc:lipid phosphate transferases
The available structures of RhaCWP as well as evidence for their covalent linkage to peptidoglycan MurNAc (Heymann, Manniello and Barkulis 1967; Deng et al. 2000) implies that activation of the lipid carrier undecaprenylphosphate by the transfer of GlcNAc by a UDP-GlcNAc:lipid phosphate transferase is the first enzymatic step of RhaCWP biosynthesis. This step is similar to the initiating reaction for WTA biosynthesis in B. subtilis and S. aureus, which is catalyzed by integral membrane proteins called TarO/TagO (Swoboda et al. 2010). A gene encoding a TarO/TagO homologue is not present in the rmlD-associated gene clusters in streptococci and L. lactis (Fig. 5), but can be readily identified elsewhere in the genome through homology searches. Atypically in E. faecalis, the first gene of the putative RhaCWP gene cluster, epaA, may encode the required UDP-GlcNAc:lipid phosphate transferase (Fig. 5). Experimental proof for a role of these enzymes in the biosynthesis of RhaCWP has been provided for S. pyogenes GacO, S. agalactiae GbcO, S. mutans RgpG and E. faecalis EpaA through genetic mutation, bacterial complementation studies or pharmacological inhibition of the transferase by the compound tunicamycin (Yamashita et al. 1999; Teng et al. 2009; Campbell et al. 2011; Caliot et al. 2012; van Sorge et al. 2014). Indeed, interference with expression or enzymatic function of these transferases depleted the cell wall of L-rhamnose, attenuated bacterial growth and induced aberrant morphology and cell division resulting in an increased chain length (Yamashita et al. 1999; Teng et al. 2009; Caliot et al. 2012; van Sorge et al. 2014). Correspondingly, the S. agalactiae gbcO mutant lost expression of the pellicle (Fig. 2) (Caliot et al. 2012), confirming previous observations in L. lactis that this outer layer correlates with the presence of RhaCWP (Fig. 2) (Chapot-Chartier et al. 2010). For streptococci, this phenotype mimics the defects observed in rml mutants, which also lose expression of RhaCWP and display attenuated growth, aberrant morphology and growth in long chains (Fig. 1) (Tsukioka et al. 1997a; van der Beek et al. 2015).
Glycosyltransferases required for RhaCWP biosynthesis
Following lipid carrier activation by UDP-GlcNAc:lipid phosphate transferases, biosynthesis of the actual RhaCWP structure occurs through the step-wise addition of monosaccharides by specific glycosyltransferases. In most cases, each glycosidic bond requires a dedicated enzyme. Such knowledge informs detailed structure–function studies of RhaCWP as will be discussed below.
The Lancefield GAC, GCC and S. mutans RhaCWP contain a backbone composed of α-1,2-/α-1,3-linked polyrhamnose (Fig. 3A). The first seven genes of the respective RhaCWP gene clusters contain a high sequence identity, suggesting that these genes are required to construct the identical rhamnan backbone (Fig. 5). Indeed, heterologous expression of these seven genes of S. mutans in E. coli results in the production of α-1,2-/α-1,3-linked polyrhamnose (Shibata et al. 2002). Subsequently, Shibata et al. identified that three glycosyltransferases encoded in this partial cluster, RgpA, RgpB and RgpF, are required for rhamnan biosynthesis, with RgpA adding the first rhamnose to the undecaprenylphosphate-GlcNAc lipid carrier (Shibata et al. 2002). Homologous glycosyltransferases in S. pyogenes and S. zooepidemicus likely catalyze a similar reaction. Decoration of the rhamnan backbone with specific side chains produces the discriminating epitopes of the GAC, GCC and RhaCWP of S. mutans (Fig. 3A). The remaining glycosyltransferases in the respective gene clusters are likely implicated in these reactions. For S. mutans, RgpE and RgpI add α1,2-linked glucose to RhaCWP (Yamashita et al. 1998; Ozaki et al. 2002), whereas GacI in S. pyogenes likely adds the characteristic β-linked GlcNAc side chain since mutation of gacI results in loss of side chain expression (van Sorge et al. 2014). For construction of the GCC side chain, a disaccharide GalNAc (Fig. 3A), glycosyltransferases encoded by Sez_0844 and Sez_0848 likely play a crucial role. Similar to the GAC, GCC and RhaCWP of S. mutans, the Lancefield GGC is also reported to be a linear structure (Pritchard et al. 1988), albeit with a different disaccharide backbone (Fig. 3A). No studies have experimentally addressed the contribution of specific glycosyltransferases to the biosynthesis of the GGC, but the increased number of glycosidic bonds compared to the GAC, GCC and S. mutans RhaCWP, suggest that the activity of all seven glycosyltransferases encoded in the gene cluster (Fig. 5) are required for its biosynthesis.
The situation is more complex in the case of the pellicle-forming GBC and RhaCWP of L. lactis strains. The GBC was previously postulated to be composed of four modular subunits (Sutcliffe 2008), which are ultimately linked together via phosphodiester bonds to form a multiantenna branching structure (Fig. 3B). Despite its structural complexity, it was hypothesized that a minimum of 11 glycosyltransferases would theoretically be required to compose all glycosidic linkages in the GBC, seven of which are encoded within the GBC gene cluster (Sutcliffe, Black and Harrington 2008). However, upon reexamination of the GBC structure (Michon et al. 1987, 1988, 1991), we have recognized the presence of two key structural elements, both beginning with GlcNAc: a stem oligosaccharide and a repeating unit (Fig. 3A, B). Following transfer of GlcNAc to the undecaprenylphosphate lipid carrier by the GbcO UDP-GlcNAc:lipid phosphate transferase, cytoplasmic synthesis of both of these subunits could be achieved by the glycosyltransferases encoded in the GBC gene cluster (unpublished observations). Identification of this repeat unit allows for a revised model of GBC assembly (see below) and solves the mystery of the two ‘missing’ GlcNAc transferases (Sutcliffe, Black and Harrington 2008).
In contrast to the invariable structure of the GAC, GBC, GCC and GGC in the respective streptococci, L. lactis strains can express structurally diverse CWP structures, correlating to diversity in the responsible gene cluster (Chapot-Chartier et al. 2010; Mahony et al. 2013; Ainsworth et al. 2014; Farenc et al. 2014). Rhamnose is not incorporated into the CWP of every strain (Ainsworth et al. 2014). Bioinformatics predict the presence of eight glycosyltransferases in the L. lactis pellicle gene cluster but for none of them a contribution or role has been experimentally addressed. Currently, the structure of the E. faecalis Epa awaits elucidation, making it difficult to speculate about the specific glycosyltransferases involved. However, genetic disruption of epaB and epaN, which encode putative glycosyltransferases (Fig. 5), changes immunoreactivity of Epa, indicating that these enzymes are involved in Epa biosynthesis (Xu, Murray and Weinstock 1998; Teng et al. 2002, 2009). Moreover, the Epa polysaccharide extracted from the ΔepaB mutant was incapable of incorporating rhamnose and instead included mannose, suggesting that epaB encodes a rhamnosyltransferase (Teng et al. 2009).
RhaCWP translocation and incorporation into the cell wall
Transport of big sugar complexes across cell membranes is energetically unfavorable and requires one of three mechanisms, i.e. synthase-dependent transporters, ABC transporters, or so-called ‘flippases’ (Cuthbertson, Kos and Whitfield 2010). The synthase-dependent pathway is least well defined and is involved in the formation of polysaccharides such as chitin, cellulose, hyaluronan and poly-N-GlcNAc. It was recently confirmed that a single protein, the synthase, executes both polymerization and export of the growing polysaccharide chain (Morgan, Strumillo and Zimmer 2013). ABC transporters translocate longer glycan chains and can be composed of a single protein (for example PglK; Perez et al. 2015) or of a two-protein complex consisting of a permease protein and an ATP-binding protein (Cuthbertson, Kos and Whitfield 2010). Finally, classical Wzx flippase systems generally transport oligosaccharide repeat units that are polymerized on the extracytoplasmic face of the membrane by a Wzy-like polymerase to complete the mature polysaccharide (Islam and Lam 2013). Interestingly, genes consistent with either the Wzx flippase and/or the ABC transport system seem to be present in all RhaCWP gene clusters (Fig. 5). Currently, the only experimental evidence supporting a role for the cognate ABC transport system is for S. mutans (Shibata et al. 2002). After heterologous expression of the first seven (rmlD-rgpABCDEF) genes in E. coli and subsequent disruption of the ABC transporter-encoding genes (rgpCD), the bacteria were unable to produce rhamnan (Shibata et al. 2002). This transport mechanism would resemble WTA translocation, which involves the TagG/H ABC transport system (Lazarevic and Karamata 1995).
The translocation of the GBC structure remains particularly enigmatic, given that assembly of a fully branched polymer before translocation would present a major challenge (Sutcliffe, Black and Harrington 2008). In contrast to the other RhaCWP gene clusters, the GBC gene cluster only contains a putative Wzx flippase gene (gbs1482) as well as two integral membrane proteins (encoded by gbs1483 and gbs1490) that may act as accessory proteins (Sutcliffe, Black and Harrington 2008). Recognition that the GBC is composed of two structural elements (stem oligosaccharide and a repeat unit, Fig. 3B) instead of the previously predicted four elements, now allows for a revised route of synthesis. Flipping of these two building blocks (by GBS1482 and, presumptively, a second flippase) could allow assembly of the mature GBC on the extracytoplasmic face of the membrane. The integral membrane proteins (GBS1484, GBS1489 or GBS1490) can be proposed as the second flippase and/or assembly proteins. Assembly of repeating units requires formation of linkages between the GlcNAc and C3 of a terminal rhamnose in the growing chain, whilst the branch points are formed by repeating unit linkage to the C4 of the penultimate rhamnose. An attractive aspect of this revised proposed biosynthesis is that it removes the need for as yet unidentified cytoplasmic GlcNAc transferases. In addition, it would allow flipping of smaller units instead of the fully branched GBC, which is likely energetically more favorable. Thus, complete synthesis, translocation and assembly of the GBC can be predicted to occur through the action of GbcO and the proteins encoded in GBC gene cluster. Experimental evidence to support these hypotheses should now be sought.
Detailed biochemical analysis has demonstrated that the GAC and GBC are attached to the MurNAc moiety of peptidoglycan, similar to WTA in other Gram-positive bacteria (Swoboda et al. 2010). GAC is presumably connected to MurNAc through a phosphate containing bridge composed of one or more units of glycerol (Heymann, Manniello and Barkulis 1967), which would concur with the linkage of WTA to peptidoglycan in S. aureus (Swoboda et al. 2010). The transfer of nascent RhaCWP from the flipped lipid carrier onto the peptidoglycan is likely catalyzed by a member of the LytR-CpsA-Psr family. These proteins were initially identified to catalyze the attachment of WTA to peptidoglycan in B. subtilis (Kawai et al. 2011). This family of proteins is widespread throughout Gram-positive bacteria, with at least two family members present in the genomes of streptococci up to five in E. faecalis (Hubscher et al. 2008). The LytR-CpsA-Psr proteins appear to be highly redundant since lack of peptidoglycan-attached glycopolymers, such as capsule and WTA, only becomes apparent upon genetic mutation of all encoded enzymes (Kawai et al. 2011; Eberhardt et al. 2012; Chan et al. 2013, 2014). The LytR-CpsA-Psr phosphotransferases typically hydrolyze the phosphodiester linkage between the lipid-carrier and the first GlcNAc at the stem base of the polysaccharide (i.e. they hydrolyze the linkage created by the UDP-GlcNAc:lipid phosphate transferase such as GacO and GbcO) and attach the polymers to peptidoglycan via a phosphate ester linkage (Kawai et al. 2011; Eberhardt et al. 2012; Chan et al. 2013). Overall, it seems likely that LytR-CpsA-Psr proteins are involved in anchoring RhaCWP to the cell wall, but experimental proof is currently lacking.
Distribution of RhaCWP throughout bacteria: identification of additional RhaCWP gene clusters
As mentioned above, the Lancefield typing scheme is unable to discriminate bacteria up to the species level. The availability of genome sequences, as well as knowledge regarding RhaCWP gene clusters, provides an opportunity to gain insight into the distribution and potential structure of RhaCWP among Gram-positive bacteria. For example, S. castoreus was noted to react with Group A-specific antisera and indeed its draft genome contains a biosynthetic locus syntenous with that of the GAC of S. pyogenes (Table S1, Supporting Information). However, the presence of an additional glycosyltransferase compared to the GAC gene cluster in S. pyogenes suggests some fine structural variation. Likewise, the GBC has been reported to be expressed by different streptococcal species most notably Streptococcus porcinus, Streptococcus pseudoporcinus, Streptococcus troglodytidis and Streptococcus plurextorum. Correspondingly, the genomes of S. pseudoporcinus and S. porcinus contain a fully syntenous GBC biosynthetic gene cluster except for the lack of a gbs1485 orthologue (unpublished observations). Presumably, the expressed structures only lack one of the monosaccharide rhamnose side-branches present in the GBC repeating unit (Fig. 3B) (Sutcliffe, Black and Harrington 2008), but can still make the linear trirhamnosyl immunodominant epitope with a terminal rhamnose that is detected by Group B-specific serotyping (Curtis and Krause 1964b). In contrast, Streptococcus thoraltensis contains a GBC-variant gene cluster that lacks orthologues of two of the predicted rhamnosyltransferases, gbs1481 and gbs1485, present in S. agalactiae (Table S2, Supporting Information) and this species is non-groupable by Lancefield serotyping assays. Instead of gbs1481 and gbs1485, the S. thoraltensis locus harbors two additional glycosyltransferases absent in the otherwise syntenous locus of S. agalactiae (Table S2, Supporting Information). Absence of a gbs1481 orthologue is likely responsible for abrogated cross-reactivity in the Group B antigen serotyping tests due to the loss of the terminal rhamnose from the dominant trirhamnosyl epitope. This analysis therefore suggests that S. thoraltensis is capable of synthesizing a RhaCWP that is a structural variant of the GBC. Increased availability of genome sequences for streptococci and related species will help identify additional rmlD-linked gene clusters for RhaCWP biosynthesis that are consistent with either known Lancefield serotyping reactions (as exemplified here by S. castoreus) or from which variant or novel RhaCWP structures can be predicted.
Physiological role of RhaCWP
Since their identification and structural characterization from the 1930s onwards, the biological roles of the Lancefield Group antigens or other RhaCWP have received little attention. It was long thought that Group-specific antigens were only of structural importance (McCarty 1952). Indeed, complete loss of RhaCWP expression through genetic mutation results in severe growth and cell division abnormalities (Fig. 1) and can be essential under competing conditions (McCarty 1952; Tsukioka et al. 1997a; Caliot et al. 2012; van der Beek et al. 2015). Caliot et al. (2012) targeted the UDP-GlcNAc:lipid phosphate transferase gbcO (gbs0136) of S. agalactiae to initiate functional studies on the GBC. The resulting GBC-negative S. agalactiae strain was devoid of cell wall rhamnose and phosphate and lost expression of the pellicle structure (Caliot et al. 2012) (Fig. 2). The loss of GBC was associated with major morphological and cell growth defects, including mislocated septa and defects in cell division and separation, which resulted in the formation of very long chains (Caliot et al. 2012). This phenotype corresponds to previous descriptions of a stable opacity variant of S. agalactiae that had lost GBC expression and displayed growth and morphological defects (Pincus et al. 1992, 1993). For this spontaneous opaque S. agalactiae mutant strain the underlying genetic defect has never been clarified. For the gbcO mutant strain, the defects result from reduced levels of highly cross-linked peptidoglycan and mislocalization of the important peptidoglycan hydrolase PcsB (Caliot et al. 2012), a protein required for streptococci cell wall separation (Reinscheid et al. 2003; Sham et al. 2011). The latter observation is reminiscent of studies in S. aureus, where the preferential localization of the major autolysin Atl is lost in absence of WTA (Schlag et al. 2010). Collectively, these observations support a role of GBC in cell wall homeostasis of S. agalactiae. In line with observations in the S. agalactiae gbcO mutant (Caliot et al. 2012), pharmacological inhibition of the S. pyogenes enzyme GacO (encoded by M5005_Spy0240) by tunicamycin resulted in depletion of GAC from the cell wall, increased mutanolysin susceptibility and increased chain length as a result of cell separation defects (van Sorge et al. 2014). However, interpretations from these studies may be obscured by the interconnection between biosynthesis of Group-specific antigens and that of other cell wall glycopolymers, including peptidoglycan, since most glycoconjugates use undecaprenylphosphate as a carrier for biosynthesis. Coordinated regulation between biosynthesis of different glycoconjugates is further supported by the observation that the S. agalactiae gbcO mutant increases capsule production suggesting a coordinated regulation between the two glycoconjugates (Beaussart et al. 2014). Knowledge regarding the interrelatedness of glycoconjugate biosynthesis pathways is relevant for the design of new antibiotics since interference of such connected pathways may have synergistic effects as recently demonstrated for WTA and peptidoglycan biosynthesis (Sewell and Brown 2014).
RhaCWP as phage receptors
In addition to their significant role in cell wall architecture, it is appreciated that RhaCWP are important phage receptors for many species. This again highlights a parallel with WTA in other Gram-positive bacteria where WTA is critical to phage-mediated horizontal gene transfer (Baptista, Santos and Sao-Jose 2008; Brown et al. 2012; Winstel et al. 2014). For several streptococci, the specificity of phage adsorption correlates to the side chain anchored to the rhamnan backbone. Indeed, the (GalNAc)2 side chain of the GCC serves as an attachment site for Group C1 bacteriophage in Group C Streptococcus (Krause 1957; Fischetti and Zabriskie 1968). Correspondingly, a Group C-variant strain, which completely lacks the immunodominant (GalNAc)2 epitope, was isolated from Group C Streptococcus that survived exposure to Group C1 lytic phages (Krause 1963). Also for S. pyogenes, the GAC-specific GlcNAc terminal moiety appears to be involved in both lytic and temperate phage adsorption although additional unidentified cell wall factors are also involved (Fischetti and Zabriskie 1968). Finally, specific phages recognize the α-1,2-linked glucose side chain of serotype c S. mutans strains (Shibata, Yamashita and van der Ploeg 2009). For L. lactis, selection of phage resistant strains from a random insertional mutagenesis library originally identified the presence and genetic locus of the RhaCWP (Dupont et al. 2004). Moreover, the precise structure of the RhaCWP dictates bacteriophage sensitivity (Dupont et al. 2004; Mahony et al. 2013; Ainsworth et al. 2014). Finally, structural changes in E. faecalis Epa through genetic manipulation greatly affect phage sensitivity despite similar adsorption levels (Teng et al. 2009). Phage dynamics within the bacterial population will impact fitness and may also be important for horizontal gene transfer, even across long phylogenetic distances (Winstel et al. 2013), affecting virulence or antibiotic resistance of pathogens. Thus the interactions of phages with RhaCWP are likely to impact on bacterial population structure. In the case of L. lactis, knowledge regarding the molecular mechanisms of phage adsorption and infection may benefit the food industry.
Role of RhaCWP in virulence
The localization of RhaCWP at the host–pathogen interface suggests that their biological function might be broader than a role in cell wall biogenesis. Similarly for WTA, evidence is accumulating for its role in virulence by increasing adherence and immune evasion (Carvalho et al. 2015; Winstel et al. 2015). Recent studies in S. pyogenes and E. faecalis now highlight that subtle modifications to the RhaCWP structure, which do not impact bacteria growth, can significantly impact virulence (Xu et al. 2000; Teng et al. 2009; van Sorge et al. 2014). For E. faecalis, disruption of epaB, epaE, epaM and epaN, which may completely eliminate Epa expression or only modify its structure, all caused significant attenuation in a mouse peritonitis model (Xu et al. 2000; Teng et al. 2009). Reduced virulence correlated with increased phagocytic uptake and clearance by neutrophils (Teng et al. 2002). Similarly in L. lactis, loss of the pellicle results in 10-fold more efficient uptake by macrophage cell lines compared to wild-type bacteria (Chapot-Chartier et al. 2010). These results therefore indicate that the pellicle can exert an anti-phagocytic effect both for L. lactis and E. faecalis. For S. pyogenes, structure–function studies focused on the role of the GAC GlcNAc side chain, which was selectively removed through genetic mutation of the glycosyltransferase-encoding gene gacI (van Sorge et al. 2014). The gacI mutant bacteria still expressed the rhamnan backbone but did not display apparent cell wall abnormalities. However, bacteria were increasingly susceptible to innate immune clearance by neutrophils and antimicrobial components (van Sorge et al. 2014). Moreover, virulence of this genetically engineered Group A-variant strain was significantly attenuated in two animal models (van Sorge et al. 2014). Again, this indicates that specific epitopes of RhaCWP increase bacterial immune resistance, although the mechanism has not been well defined. In addition to increased immune resistance, RhaCWP may modulate host immune responses by targeting specific lectin receptors (carbohydrate-recognizing pattern-recognition receptors) (Sancho and Reis e Sousa 2012). Human lectins regulate fundamental immunological processes but also directly engage microbial carbohydrates, linking pathogen recognition to appropriate immune responses. Although this often promotes bacterial clearance, lectin targeting can also promote bacterial survival by skewing immune reponses (van Vliet et al. 2009). Research in this area should provide insight whether the virulence-promoting effect of RhaCWP occurs through interaction with lectin receptors. Alternatively, decoration of the rhamnan backbone or rhamnose moieties by common sugars such as GlcNAc, glucose and GalNAc may be a strategy for microorganisms to avoid immune recognition. This is relevant since the absence of rhamnose in humans makes rhamnose an attractive pattern-associated molecular pattern (PAMP). Indeed, in fish and invertebrates, rhamnose is targeted by the innate immune system through germ-line encoded pattern-recognition receptors called rhamnose-binding lectins (Ogawa et al. 2011; Ng et al. 2014). Interestingly, rhamnose-binding lectins agglutinate both Gram-positive and Gram-negative bacteria through interaction with glycan structures such as LPS or lipoteichoic acid in the bacterial cell wall (Tateno et al. 2002; Cammarata et al. 2014; Ng et al. 2014). Rhamnose-binding lectins are also involved in inflammatory responses through the induction of cytokines (Watanabe et al. 2009). Humans lectins with specificity for rhamnose have not been identified yet, but their existence might be anticipated given the estimated presence of over 150 glycan-binding proteins in humans, many with uncharacterized ligand specificity (Zelensky and Gready 2005; Drickamer 2014). Overall, the role of RhaCWP in pathogenesis and cell physiology is just starting to be appreciated. Further investigations are likely to unravel new bacterial immune evasion strategies but may also contribute to new insights into innate immune responses and recognition.
Rhamnose containing capsules in Gram-positive bacteria
Rhamnose is not just incorporated in RhaCWP, but is also present in capsular polysaccharides. We consider this structure distinct from RhaCWP given their localization in the cell wall; RhaCWP are interpolated within the peptidoglycan wall layer, whereas capsular polysaccharides are typically the outermost layers of the cell envelope. Nevertheless, it is worth noting that several clinically relevant Gram-positive bacteria synthesize rhamnose-containing capsules. The significance of the S. pneumoniae capsule as a key virulence factor has been established since the 1928 landmark ‘Griffiths experiment’ and the capsular polysaccharides are major protective antigens utilized in current vaccine formulations (Geno et al. 2015). Of the 41 (out of 46) serogroups of pneumococcal capsule for which carbohydrate composition or structural information is available, 17 of these (∼40%) contain rhamnose, including those of clinically significant serogroups such as 6, 19 and 23. The capsular biosynthetic loci for these serogroups all contain rmlABCD genes (Bentley et al. 2006). The presence of these genes in the biosynthetic loci for which capsular polysaccharide structures are not yet available or may be incomplete (e.g. serogroups 21, 40 and 48) suggests additional structures also contain rhamnose. Thus, rhamnose biology is likely of significance in much of the S. pneumoniae population and at least nine of the serotype antigens included in the current 23-valent vaccin are rhamnose-containing polysaccharides (Geno et al. 2015).
In addition to S. pneumoniae, S. agalactiae strains belonging to serotype VIII contain rhamnose within the polysaccharide repeating unit (Kogan et al. 1996; Cieslewicz et al. 2005). This serotype remains relatively rare globally but has been reported to be of significance in some population groups, notably in Japan and the Pacific (Lachenauer et al. 1999; Edmond et al. 2012).
Therapeutic and technological applications
Increased knowledge regarding the biosynthesis and function of RhaCWP could aid the development of new antimicrobial agents but may also have applications in metabolic engineering to optimize food production (Chapot-Chartier 2014) or glycoconjugate production for medical purposes (Jaffe et al. 2014). Clearly, the L-rhamnose biosynthesis pathway holds promise for antimicrobial drug targeting, given the loss of virulence and/or viability upon rhamnose depletion in a wide range of Gram-positive and Gram-negative bacteria. More importantly, lack of L-rhamnose in humans should preclude off-target effects lowering risks of unwanted side effects. Thus far, several inhibitors screens for Rml enzymes have been initiated, resulting in only one RmlA inhibitor targeting P. aeruginosa with some marginal activity against M. tuberculosis (Alphey et al. 2013). In addition to targeting rhamnose, inhibition of other steps in the RhaCWP biosynthesis pathway could also be of interest. Although this may not immediately kill the bacterium, it may act as anti-virulence agent, increasing susceptibility to host defense mechanisms such as phagocytic clearance (Nizet 2015). A similar strategy is currently exploited for S. aureus, where inhibition of the UDP-GlcNAc:lipid phosphate transferase TarO is not detrimental to the bacterium, but render the bacterium avirulent (Sewell and Brown 2014). An additional effect of TarO inhibition is the re-sensitization of resistant bacteria to β-lactam antibiotics due to an interaction between the WTA and peptidoglycan biosynthesis pathways in S. aureus (Campbell et al. 2011). Similar synergy may occur between conventional antibiotics and inhibitors of the RhaCWP pathway.
RhaCWP are also attractive vaccine candidates due to their conserved and constant expression in species of medical importance, such as S. pyogenes and S. agalactiae. Indeed, different strategies are currently explored to develop protective vaccines against these streptococcal species (Dale et al. 2013; Steer, Dale and Carapetis 2013; Nuccitelli, Rinaudo and Maione 2015). For both pathogens, much research has focused on type-specific vaccine strategies, i.e. a capsule-conjugate vaccine for S. agalactiae (Johri et al. 2006; Nuccitelli, Rinaudo and Maione 2015) and a multivalent M-protein vaccine for S. pyogenes (Dale et al. 2013; Steer, Dale and Carapetis 2013). A more elegant and globally effective approach would employ a vaccine antigen that is universally expressed on all strains in an invariant manner. Indeed, the GBC is immunogenic in rabbits but antibodies raised are not protective in a newborn mouse model (Marques et al. 1994), likely due to shielding of the GBC by the polysaccharide capsule. More encouraging are the results with regard to the GAC as a universal S. pyogenes vaccine antigen. Conjugate vaccines of either isolated or synthetic GAC protect mice from subsequent infection after active and passive immunization (Sabharwal et al. 2006; Kabanova et al. 2010). However, there is controversy with regards to safety in the use of the native GAC for vaccine purposes, since several groups have indicated a role for anti-GlcNAc antibodies in the pathogenesis of rheumatic fever (Goldstein et al. 1968; Ayoub and Dudding 1970; Kirvan et al. 2003). Recent elucidation of the molecular pathway for GlcNAc side chain formation allows for the development of an alternate vaccine antigen consisting of the polyrhamnose backbone of the GAC (van Sorge et al. 2014). Anti-rhamnan antibodies raised against the GlcNAc-deficient GAC enhanced phagocytic killing of multiple M-serotypes in vitro and protected mice from lethal challenge with wild-type S. pyogenes through passive immunization (van Sorge et al. 2014). Of interest is the observation that some streptococci also release RhaCWP into their surroundings. Although it is unclear whether this involves an active mechanism of release, it possibly has implications for the efficiency of vaccine strategies. Overall, further exploration on the application of RhaCWP for vaccination purposes is warranted.
Finally, dissection of the RhaCWP biosynthesis pathway could benefit food production, most notably the dairy industry that uses lactic acid bacteria for food fermentations (Chapot-Chartier 2014). Phage infection of these fermenting cultures results in product variations but can also lead to huge economic losses (Samson and Moineau 2013). The selection or engineering of phage-resistant strains is therefore of considerable interest. The role of RhaCWP in phage–host interaction has opened up new possibilities for the development of bacteriophage insensitive mutants for food production purposes. In addition to food preparation, polysaccharides often have cosmetic, pharmaceutical and biomedical applications. Elucidation of specific transferase activities could be used towards metabolic engineering of new materials or compounds with interesting biological or physical properties. In particular, the incorporation of rare sugars such as rhamnose and uronic acids is a rather unexplored area but may be of interest to different areas including the biomedical field (Roca et al. 2015).
CONCLUDING REMARKS
Despite their long history in streptococcal diagnostics, investigations on the biological roles and possible applications of RhaCWP have lagged behind. Genome sequencing has initiated genetic studies to elucidate structure–function relationships of RhaCWP, highlighting their critical importance in proper cell wall architecture and pathogenesis. Their indispensable nature identifies the RhaCWP biosynthesis pathway as an attractive therapeutic target for antimicrobial drug development. Spin offs will likely find applications in the area of metabolic engineering for food production and biomedical purposes.
Supplementary Material
Acknowledgments
NvS would like to thank Samantha van der Beek for providing scanning electron microscopy pictures of S. pyogenes and S. mutans and for construction of Figs 1 and 6. M-Y M. would like to thank S. Kulakauskas for providing transmission electron micrographs of L. lactis.
FUNDING
This work was supported by the [VIDI grant and ASPASIA grant ] to NvS.
Conflict of interest.None declared.
REFERENCES
- Adibekian A, Stallforth P, Hecht M-L, et al. Comparative bioinformatics analysis of the mammalian and bacterial glycomes. Chem Sci. 2011;2:337–44. [Google Scholar]
- Ainsworth S, Sadovskaya I, Vinogradov E, et al. Differences in lactococcal cell wall polysaccharide structure are major determining factors in bacteriophage sensitivity. mBio. 2014;5:e00880–14. doi: 10.1128/mBio.00880-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alphey MS, Pirrie L, Torrie LS, et al. Allosteric competitive inhibitors of the glucose-1-phosphate thymidylyltransferase (RmlA) from Pseudomonas aeruginosa. ACS Chem Biol. 2013;8:387–96. doi: 10.1021/cb300426u. [DOI] [PubMed] [Google Scholar]
- Andre G, Kulakauskas S, Chapot-Chartier MP, et al. Imaging the nanoscale organization of peptidoglycan in living Lactococcus lactis cells. Nat Commun. 2010;1:27. doi: 10.1038/ncomms1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayoub EM, Dudding BA. Streptococcal Group A carbohydrate antibody in rheumatic and nonrheumatic bacterial endocarditis. J Lab Clin Med. 1970;76:322–32. [PubMed] [Google Scholar]
- Baker CJ, Kasper DL. Microcapsule of type III strains of Group B Streptococcus: production and morphology. Infect Immun. 1976;13:189–94. doi: 10.1128/iai.13.1.189-194.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baptista C, Santos MA, Sao-Jose C. Phage SPP1 reversible adsorption to Bacillus subtilis cell wall teichoic acids accelerates virus recognition of membrane receptor YueB. J Bacteriol. 2008;190:4989–96. doi: 10.1128/JB.00349-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaussart A, Pechoux C, Trieu-Cuot P, et al. Molecular mapping of the cell wall polysaccharides of the human pathogen Streptococcus agalactiae. Nanoscale. 2014;6:14820–7. doi: 10.1039/c4nr05280c. [DOI] [PubMed] [Google Scholar]
- Beis K, Allard ST, Hegeman AD, et al. The structure of NADH in the enzyme dTDP-d-glucose dehydratase (RmlB) J Am Chem Soc. 2003;125:11872–8. doi: 10.1021/ja035796r. [DOI] [PubMed] [Google Scholar]
- Bentley SD, Aanensen DM, Mavroidi A, et al. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet. 2006;2:e31. doi: 10.1371/journal.pgen.0020031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenfeldt W, Asuncion M, Lam JS, et al. The structural basis of the catalytic mechanism and regulation of glucose-1-phosphate thymidylyltransferase (RmlA) EMBO J. 2000;19:6652–63. doi: 10.1093/emboj/19.24.6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenfeldt W, Kerr ID, Giraud MF, et al. Variation on a theme of SDR. dTDP-6-deoxy-L- lyxo-4-hexulose reductase (RmlD) shows a new Mg2+-dependent dimerization mode. Structure. 2002;10:773–86. doi: 10.1016/s0969-2126(02)00770-0. [DOI] [PubMed] [Google Scholar]
- Brown S, Santa Maria JP, Jr, Walker S. Wall teichoic acids of Gram-positive bacteria. Annu Rev Microbiol. 2013;67:313–36. doi: 10.1146/annurev-micro-092412-155620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S, Xia G, Luhachack LG, et al. Methicillin resistance in Staphylococcus aureus requires glycosylated wall teichoic acids. P Natl Acad Sci USA. 2012;109:18909–14. doi: 10.1073/pnas.1209126109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broyles LN, Van Beneden C, Beall B, et al. Population-based study of invasive disease due to beta-hemolytic streptococci of groups other than A and B. Clin Infect Dis. 2009;48:706–12. doi: 10.1086/597035. [DOI] [PubMed] [Google Scholar]
- Burns SM, Hull SI. Comparison of loss of serum resistance by defined lipopolysaccharide mutants and an acapsular mutant of uropathogenic Escherichia coli O75:K5. Infect Immun. 1998;66:4244–53. doi: 10.1128/iai.66.9.4244-4253.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caliot E, Dramsi S, Chapot-Chartier MP, et al. Role of the Group B antigen of Streptococcus agalactiae: a peptidoglycan-anchored polysaccharide involved in cell wall biogenesis. PLoS Pathog. 2012;8:e1002756. doi: 10.1371/journal.ppat.1002756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammarata M, Parisi MG, Benenati G, et al. A rhamnose-binding lectin from sea bass (Dicentrarchus labrax) plasma agglutinates and opsonizes pathogenic bacteria. Dev Comp Immunol. 2014;44:332–40. doi: 10.1016/j.dci.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell J, Singh AK, Santa Maria JP, Jr, et al. Synthetic lethal compound combinations reveal a fundamental connection between wall teichoic acid and peptidoglycan biosyntheses in Staphylococcus aureus. ACS Chem Biol. 2011;6:106–16. doi: 10.1021/cb100269f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carapetis JR, Steer AC, Mulholland EK, et al. The global burden of Group A streptococcal diseases. Lancet infect Dis. 2005;5:685–94. doi: 10.1016/S1473-3099(05)70267-X. [DOI] [PubMed] [Google Scholar]
- Carey RB, Eisenstein TK, Shockman GD, et al. Soluble group- and type-specific antigens from type III Group B Streptococcus. Infect Immun. 1980;28:195–203. doi: 10.1128/iai.28.1.195-203.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho F, Atilano ML, Pombinho R, et al. L-Rhamnosylation of Listeria monocytogenes wall teichoic acids promotes resistance to antimicrobial peptides by delaying interaction with the membrane. PLoS Pathog. 2015;11:e1004919. doi: 10.1371/journal.ppat.1004919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YG, Frankel MB, Dengler V, et al. Staphylococcus aureus mutants lacking the LytR-CpsA-Psr family of enzymes release cell wall teichoic acids into the extracellular medium. J Bacteriol. 2013;195:4650–9. doi: 10.1128/JB.00544-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YG, Kim HK, Schneewind O, et al. The capsular polysaccharide of Staphylococcus aureus is attached to peptidoglycan by the LytR-CpsA-Psr (LCP) family of enzymes. J Biol Chem. 2014;289:15680–90. doi: 10.1074/jbc.M114.567669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapot-Chartier MP. Interactions of the cell-wall glycopolymers of lactic acid bacteria with their bacteriophages. Front Microbiol. 2014;5:236. doi: 10.3389/fmicb.2014.00236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapot-Chartier MP, Kulakauskas S. Cell wall structure and function in lactic acid bacteria. Microb Cell Fact. 2014;13(Suppl 1):S9. doi: 10.1186/1475-2859-13-S1-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapot-Chartier MP, Vinogradov E, Sadovskaya I, et al. Cell surface of Lactococcus lactis is covered by a protective polysaccharide pellicle. J Biol Chem. 2010;285:10464–71. doi: 10.1074/jbc.M109.082958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang SL, Mekalanos JJ. rfb mutations in Vibrio cholerae do not affect surface production of toxin-coregulated pili but still inhibit intestinal colonization. Infect Immun. 1999;67:976–80. doi: 10.1128/iai.67.2.976-980.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cieslewicz MJ, Chaffin D, Glusman G, et al. Structural and genetic diversity of Group B Streptococcus capsular polysaccharides. Infect Immun. 2005;73:3096–103. doi: 10.1128/IAI.73.5.3096-3103.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coligan JE, Kindt TJ, Krause RM. Structure of the streptococcal Groups A, A-variant and C carbohydrates. Immunochemistry. 1978;15:755–60. doi: 10.1016/0161-5890(78)90105-0. [DOI] [PubMed] [Google Scholar]
- Cress BF, Englaender JA, He W, et al. Masquerading microbial pathogens: capsular polysaccharides mimic host-tissue molecules. FEMS Microbiol Rev. 2014;38:660–97. doi: 10.1111/1574-6976.12056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis SN, Krause RM. Antigenic Relationships between Groups B and G Streptococci. J Exp Med. 1964a;120:629–37. doi: 10.1084/jem.120.4.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis SN, Krause RM. Immunochemical studies on the specific carbohydrate of Group G streptococci. J Exp Med. 1964b;119:997–1003. doi: 10.1084/jem.119.6.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuthbertson L, Kos V, Whitfield C. ABC transporters involved in export of cell surface glycoconjugates. Microbiol Mol Biol Rev. 2010;74:341–62. doi: 10.1128/MMBR.00009-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale JB, Fischetti VA, Carapetis JR, et al. Group A streptococcal vaccines: paving a path for accelerated development. Vaccine. 2013;31(Suppl 2):B216–22. doi: 10.1016/j.vaccine.2012.09.045. [DOI] [PubMed] [Google Scholar]
- De Cueninck BJ, Shockman GD, Swenson RM. Group B, type III streptococcal cell wall: composition and structural aspects revealed through endo-N-acetylmuramidase-catalyzed hydrolysis. Infect Immun. 1982;35:572–81. doi: 10.1128/iai.35.2.572-581.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Kasper DL, Krick TP, et al. Characterization of the linkage between the type III capsular polysaccharide and the bacterial cell wall of Group B Streptococcus. J Biol Chem. 2000;275:7497–504. doi: 10.1074/jbc.275.11.7497. [DOI] [PubMed] [Google Scholar]
- Dong C, Beis K, Giraud MF, et al. A structural perspective on the enzymes that convert dTDP-d-glucose into dTDP-l-rhamnose. Biochem Soc Trans. 2003a;31:532–6. doi: 10.1042/bst0310532. [DOI] [PubMed] [Google Scholar]
- Dong C, Major LL, Allen A, et al. High-resolution structures of RmlC from Streptococcus suis in complex with substrate analogs locate the active site of this class of enzyme. Structure. 2003b;11:715–23. doi: 10.1016/s0969-2126(03)00098-4. [DOI] [PubMed] [Google Scholar]
- Dong C, Major LL, Srikannathasan V, et al. RmlC, a C3′ and C5′ carbohydrate epimerase, appears to operate via an intermediate with an unusual twist boat conformation. J Mol Biol. 2007;365:146–59. doi: 10.1016/j.jmb.2006.09.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doran TI, Mattingly SJ. Association of type- and group-specific antigens with the cell wall of serotype III Group B Streptococcus. Infect Immun. 1982;36:1115–22. doi: 10.1128/iai.36.3.1115-1122.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drickamer K. 2014 http://www.imperial.ac.uk/research/animallectins/ctld/mammals/humandata%20updated.html (3 March 2016, date last accessed) [Google Scholar]
- Dupont K, Janzen T, Vogensen FK, et al. Identification of Lactococcus lactis genes required for bacteriophage adsorption. Appl Environ Microb. 2004;70:5825–32. doi: 10.1128/AEM.70.10.5825-5832.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhardt A, Hoyland CN, Vollmer D, et al. Attachment of capsular polysaccharide to the cell wall in Streptococcus pneumoniae. Microb Drug Resist. 2012;18:240–55. doi: 10.1089/mdr.2011.0232. [DOI] [PubMed] [Google Scholar]
- Edmond KM, Kortsalioudaki C, Scott S, et al. Group B streptococcal disease in infants aged younger than 3 months: systematic review and meta-analysis. Lancet. 2012;379:547–56. doi: 10.1016/S0140-6736(11)61651-6. [DOI] [PubMed] [Google Scholar]
- Facklam R. What happened to the streptococci: overview of taxonomic and nomenclature changes. Clin Microbiol Rev. 2002;15:613–30. doi: 10.1128/CMR.15.4.613-630.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farenc C, Spinelli S, Vinogradov E, et al. Molecular insights on the recognition of a Lactococcus lactis cell wall pellicle by the phage 1358 receptor binding protein. J Virol. 2014;88:7005–15. doi: 10.1128/JVI.00739-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti JJ, McShan WM, Ajdic D, et al. Complete genome sequence of an M1 strain of Streptococcus pyogenes. P Natl Acad Sci USA. 2001;98:4658–63. doi: 10.1073/pnas.071559398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischetti VA, Zabriskie JB. Studies on streptococcal bacteriophages. II. Adsorption studies on Group A and Group C streptococcal bacteriophages. J Exp Med. 1968;127:489–505. doi: 10.1084/jem.127.3.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores AR, Jewell BE, Fittipaldi N, et al. Human disease isolates of serotype M4 and M22 Group A Streptococcus lack genes required for hyaluronic acid capsule biosynthesis. mBio. 2012;3:e00413–2. doi: 10.1128/mBio.00413-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geno KA, Gilbert GL, Song JY, et al. Pneumococcal capsules and their types: past, present, and future. Clin Microbiol Rev. 2015;28:871–99. doi: 10.1128/CMR.00024-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud MF, Leonard GA, Field RA, et al. RmlC, the third enzyme of dTDP-L-rhamnose pathway, is a new class of epimerase. Nat Struct Biol. 2000;7:398–402. doi: 10.1038/75178. [DOI] [PubMed] [Google Scholar]
- Giraud MF, Naismith JH. The rhamnose pathway. Curr Opin Struct Biol. 2000;10:687–96. doi: 10.1016/s0959-440x(00)00145-7. [DOI] [PubMed] [Google Scholar]
- Glaser P, Rusniok C, Buchrieser C, et al. Genome sequence of Streptococcus agalactiae, a pathogen causing invasive neonatal disease. Mol Microbiol. 2002;45:1499–513. doi: 10.1046/j.1365-2958.2002.03126.x. [DOI] [PubMed] [Google Scholar]
- Goldstein I, Rebeyrotte P, Parlebas J, et al. Isolation from heart valves of glycopeptides which share immunological properties with Streptococcus haemolyticus Group A polysaccharides. Nature. 1968;219:866–8. doi: 10.1038/219866a0. [DOI] [PubMed] [Google Scholar]
- Hancock LE, Gilmore MS. The capsular polysaccharide of Enterococcus faecalis and its relationship to other polysaccharides in the cell wall. P Natl Acad Sci USA. 2002;99:1574–9. doi: 10.1073/pnas.032448299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley MD, Imperiali B. At the membrane frontier: a prospectus on the remarkable evolutionary conservation of polyprenols and polyprenyl-phosphates. Arch Biochem Biophys. 2012;517:83–97. doi: 10.1016/j.abb.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickx AP, Budzik JM, Oh SY, et al. Architects at the bacterial surface - sortases and the assembly of pili with isopeptide bonds. Nat Rev Microbiol. 2011;9:166–76. doi: 10.1038/nrmicro2520. [DOI] [PubMed] [Google Scholar]
- Heymann H, Manniello JM, Barkulis SS. Structure of streptococcal cell walls. V. Phosphate esters in the walls of Group A Streptococcus pyogenes. Biochem Biophys Res Commun. 1967;26:486–91. doi: 10.1016/0006-291x(67)90574-8. [DOI] [PubMed] [Google Scholar]
- Holden MT, Heather Z, Paillot R, et al. Genomic evidence for the evolution of Streptococcus equi: host restriction, increased virulence, and genetic exchange with human pathogens. PLoS Pathog. 2009;5:e1000346. doi: 10.1371/journal.ppat.1000346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DH, Rama Krishna N, Pritchard DG. Characterization of the group A streptococcal polysaccharide by two-dimensional 1H-nuclear-magnetic-resonance spectroscopy. Carbohydr Res. 1986;155:193–9. doi: 10.1016/s0008-6215(00)90145-9. [DOI] [PubMed] [Google Scholar]
- Hubscher J, Luthy L, Berger-Bachi B, et al. Phylogenetic distribution and membrane topology of the LytR-CpsA-Psr protein family. BMC Genomics. 2008;9:617. doi: 10.1186/1471-2164-9-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam ST, Lam JS. Wzx flippase-mediated membrane translocation of sugar polymer precursors in bacteria. Environ Microbiol. 2013;15:1001–15. doi: 10.1111/j.1462-2920.2012.02890.x. [DOI] [PubMed] [Google Scholar]
- Jaffe SR, Strutton B, Levarski Z, et al. Escherichia coli as a glycoprotein production host: recent developments and challenges. Curr Opin Biotechnol. 2014;30:205–10. doi: 10.1016/j.copbio.2014.07.006. [DOI] [PubMed] [Google Scholar]
- Jiang XM, Neal B, Santiago F, et al. Structure and sequence of the rfb (O antigen) gene cluster of Salmonella serovar typhimurium (strain LT2) Mol Microbiol. 1991;5:695–713. doi: 10.1111/j.1365-2958.1991.tb00741.x. [DOI] [PubMed] [Google Scholar]
- Johri AK, Paoletti LC, Glaser P, et al. Group B Streptococcus: global incidence and vaccine development. Nat Rev Microbiol. 2006;4:932–42. doi: 10.1038/nrmicro1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabanova A, Margarit I, Berti F, et al. Evaluation of a Group A Streptococcus synthetic oligosaccharide as vaccine candidate. Vaccine. 2010;29:104–14. doi: 10.1016/j.vaccine.2010.09.018. [DOI] [PubMed] [Google Scholar]
- Kawai Y, Marles-Wright J, Cleverley RM, et al. A widespread family of bacterial cell wall assembly proteins. EMBO J. 2011;30:4931–41. doi: 10.1038/emboj.2011.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirvan CA, Swedo SE, Heuser JS, et al. Mimicry and autoantibody-mediated neuronal cell signaling in Sydenham chorea. Nat Med. 2003;9:914–20. doi: 10.1038/nm892. [DOI] [PubMed] [Google Scholar]
- Kogan G, Uhrin D, Brisson JR, et al. Structural and immunochemical characterization of the type VIII Group B Streptococcus capsular polysaccharide. J Biol Chem. 1996;271:8786–90. doi: 10.1074/jbc.271.15.8786. [DOI] [PubMed] [Google Scholar]
- Kohler W. The present state of species within the genera Streptococcus and Enterococcus. Int J Med Microbiol. 2007;297:133–50. doi: 10.1016/j.ijmm.2006.11.008. [DOI] [PubMed] [Google Scholar]
- Krause RM. Studies on bacteriophages of hemolytic streptococci. I. Factors influencing the interaction of phage and susceptible host cell. J Exp Med. 1957;106:365–84. doi: 10.1084/jem.106.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause RM. Symposium on relationship of structure of microorganisms to their Immunological properties. IV. Antigenic and biochemical composition of hemolytic streptococcal cell walls. Bacteriol Rev. 1963;27:369–80. doi: 10.1128/br.27.4.369-380.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause RM, McCarty M. Studies on the chemical structure of the streptococcal cell wall. II. The composition of Group C cell walls and chemical basis for serologic specificity of the carbohydrate moiety. J Exp Med. 1962a;115:49–62. doi: 10.1084/jem.115.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause RM, McCarty M. Variation in the group-specific carbohydrate of Group C hemolytic Streptococci. J Exp Med. 1962b;116:131–40. doi: 10.1084/jem.116.2.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachenauer CS, Kasper DL, Shimada J, et al. Serotypes VI and VIII predominate among Group B streptococci isolated from pregnant Japanese women. J Infect Dis. 1999;179:1030–3. doi: 10.1086/314666. [DOI] [PubMed] [Google Scholar]
- Lancefield RC. The antigenic complex of Streptococcus Haemolyticus: I. Demonstration of a type-specific substance in extracts of Streptococcus Haemolyticus. J Exp Med. 1928a;47:91–103. doi: 10.1084/jem.47.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancefield RC. The antigenic complex of Streptococcus Haemolyticus: III. Chemical and immunological properties of the species-specific substance. J Exp Med. 1928b;47:481–91. doi: 10.1084/jem.47.3.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancefield RC. A serological differentiation of human and other groups of hemolytic streptococci. J Exp Med. 1933;57:571–95. doi: 10.1084/jem.57.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson PA, Foster G, Falsen E, et al. Streptococcus castoreus sp. nov., isolated from a beaver (Castor fiber) Int J Syst Evol Micr. 2005;55:843–6. doi: 10.1099/ijs.0.63433-0. [DOI] [PubMed] [Google Scholar]
- Lazarevic V, Karamata D. The tagGH operon of Bacillus subtilis 168 encodes a two-component ABC transporter involved in the metabolism of two wall teichoic acids. Mol Microbiol. 1995;16:345–55. doi: 10.1111/j.1365-2958.1995.tb02306.x. [DOI] [PubMed] [Google Scholar]
- Le Breton Y, Belew AT, Valdes KM, et al. Essential genes in the core genome of the human pathogen Streptococcus pyogenes. Sci Rep. 2015;5:9838. doi: 10.1038/srep09838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llull D, Lopez R, Garcia E. Genetic bases and medical relevance of capsular polysaccharide biosynthesis in pathogenic streptococci. Curr Mol Med. 2001;1:475–91. doi: 10.2174/1566524013363618. [DOI] [PubMed] [Google Scholar]
- Lue YA, Howit IP, Ellner PD. Rapid grouping of beta-hemolytic streptococci by latex agglutination. J Clin Microbiol. 1978;8:326–8. doi: 10.1128/jcm.8.3.326-328.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Pan F, McNeil M. Formation of dTDP-rhamnose is essential for growth of mycobacteria. J Bacteriol. 2002;184:3392–5. doi: 10.1128/JB.184.12.3392-3395.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Stern RJ, Scherman MS, et al. Drug targeting Mycobacterium tuberculosis cell wall synthesis: genetics of dTDP-rhamnose synthetic enzymes and development of a microtiter plate-based screen for inhibitors of conversion of dTDP-glucose to dTDP-rhamnose. Antimicrob Agents Chemother. 2001;45:1407–16. doi: 10.1128/AAC.45.5.1407-1416.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty M. The lysis of Group A hemolytic streptococci by extracellular enzymes of Streptomyces albus. II. Nature of the cellular substrate attacked by the lytic enzymes. J Exp Med. 1952;96:569–80. doi: 10.1084/jem.96.6.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty M. Variation in the Group-specific carbohydrate of Group A streptococci. II. Studies on the chemical basis for serological specificity of the carbohydrates. J Exp Med. 1956;104:629–43. doi: 10.1084/jem.104.5.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty M, Lancefield RC. Variation in the group-specific carbohydrate of Group A streptococci. I. Immunochemical studies on the carbohydrates of variant strains. J Exp Med. 1955;102:11–28. doi: 10.1084/jem.102.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan DJ, Bessen DE, Pinho M, et al. Population genetics of Streptococcus dysgalactiae subspecies equisimilis reveals widely dispersed clones and extensive recombination. PLoS One. 2010a;5:e11741. doi: 10.1371/journal.pone.0011741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan DJ, Vu T, Bramhachari PV, et al. Molecular markers for discriminating Streptococcus pyogenes and S. dysgalactiae subspecies equisimilis. Eur J Clin Microbiol Inf Dis. 2010b;29:585–9. doi: 10.1007/s10096-010-0899-x. [DOI] [PubMed] [Google Scholar]
- McNeil M, Daffe M, Brennan PJ. Evidence for the nature of the link between the arabinogalactan and peptidoglycan of mycobacterial cell walls. J Biol Chem. 1990;265:18200–6. [PubMed] [Google Scholar]
- Macpherson DF, Manning PA, Morona R. Characterization of the dTDP-rhamnose biosynthetic genes encoded in the rfb locus of Shigella flexneri. Mol Microbiol. 1994;11:281–92. doi: 10.1111/j.1365-2958.1994.tb00308.x. [DOI] [PubMed] [Google Scholar]
- Mahony J, Kot W, Murphy J, et al. Investigation of the relationship between lactococcal host cell wall polysaccharide genotype and 936 phage receptor binding protein phylogeny. Appl Environ Microb. 2013;79:4385–92. doi: 10.1128/AEM.00653-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki M, Renkonen R. Biosynthesis of 6-deoxyhexose glycans in bacteria. Glycobiology. 2004;14:1R–15R. doi: 10.1093/glycob/cwh040. [DOI] [PubMed] [Google Scholar]
- Marques MB, Kasper DL, Shroff A, et al. Functional activity of antibodies to the Group B polysaccharide of Group B streptococci elicited by a polysaccharide-protein conjugate vaccine. Infect Immun. 1994;62:1593–9. doi: 10.1128/iai.62.5.1593-1599.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michon F, Brisson JR, Dell A, et al. Multiantennary group-specific polysaccharide of Group B Streptococcus. Biochemistry. 1988;27:5341–51. doi: 10.1021/bi00414a059. [DOI] [PubMed] [Google Scholar]
- Michon F, Chalifour R, Feldman R, et al. The alpha-L-(1–-2)-trirhamnopyranoside epitope on the group-specific polysaccharide of Group B streptococci. Infect Immun. 1991;59:1690–6. doi: 10.1128/iai.59.5.1690-1696.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michon F, Katzenellenbogen E, Kasper DL, et al. Structure of the complex group-specific polysaccharide of Group B Streptococcus. Biochemistry. 1987;26:476–86. doi: 10.1021/bi00376a020. [DOI] [PubMed] [Google Scholar]
- Morgan JL, Strumillo J, Zimmer J. Crystallographic snapshot of cellulose synthesis and membrane translocation. Nature. 2013;493:181–6. doi: 10.1038/nature11744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano K, Ooshima T. Serotype classification of Streptococcus mutans and its detection outside the oral cavity. Future Microbiol. 2009;4:891–902. doi: 10.2217/fmb.09.64. [DOI] [PubMed] [Google Scholar]
- Neuhaus FC, Baddiley J. A continuum of anionic charge: structures and functions of D-alanyl-teichoic acids in gram-positive bacteria. Microbiol Mol Biol Rev. 2003;67:686–723. doi: 10.1128/MMBR.67.4.686-723.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SK, Huang YT, Lee YC, et al. A recombinant horseshoe crab plasma lectin recognizes specific pathogen-associated molecular patterns of bacteria through rhamnose. PLoS One. 2014;9:e115296. doi: 10.1371/journal.pone.0115296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nizet V. Stopping superbugs, maintaining the microbiota. Sci Transl Med. 2015;7:295ed298. doi: 10.1126/scitranslmed.aab2373. [DOI] [PubMed] [Google Scholar]
- Nuccitelli A, Rinaudo CD, Maione D. Group B Streptococcus vaccine: state of the art. Ther Adv Vaccines. 2015;3:76–90. doi: 10.1177/2051013615579869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T, Watanabe M, Naganuma T, et al. Diversified carbohydrate-binding lectins from marine resources. J Amino Acids. 2011;2011:838914. doi: 10.4061/2011/838914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Riordan K, Lee JC. Staphylococcus aureus capsular polysaccharides. Clin Microbiol Rev. 2004;17:218–34. doi: 10.1128/CMR.17.1.218-234.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki K, Shibata Y, Yamashita Y, et al. A novel mechanism for glucose side-chain formation in rhamnose-glucose polysaccharide synthesis. FEBS Lett. 2002;532:159–63. doi: 10.1016/s0014-5793(02)03661-x. [DOI] [PubMed] [Google Scholar]
- Perez C, Gerber S, Boilevin J, et al. Structure and mechanism of an active lipid-linked oligosaccharide flippase. Nature. 2015;524:433–8. doi: 10.1038/nature14953. [DOI] [PubMed] [Google Scholar]
- Phares CR, Lynfield R, Farley MM, et al. Epidemiology of invasive Group B streptococcal disease in the United States, 1999–2005. JAMA. 2008;299:2056–65. doi: 10.1001/jama.299.17.2056. [DOI] [PubMed] [Google Scholar]
- Pincus SH, Cole RL, Kamanga-Sollo E, et al. Interaction of Group B streptococcal opacity variants with the host defense system. Infect Immun. 1993;61:3761–8. doi: 10.1128/iai.61.9.3761-3768.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pincus SH, Cole RL, Wessels MR, et al. Group B streptococcal opacity variants. J Bacteriol. 1992;174:3739–49. doi: 10.1128/jb.174.11.3739-3749.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price CE, Zeyniyev A, Kuipers OP, et al. From meadows to milk to mucosa - adaptation of Streptococcus and Lactococcus species to their nutritional environments. FEMS Microbiol Rev. 2012;36:949–71. doi: 10.1111/j.1574-6976.2011.00323.x. [DOI] [PubMed] [Google Scholar]
- Pritchard DG, Coligan JE, Geckle JM, et al. High-resolution 1H- and 13C-n.m.r. spectra of the Group A-variant streptococcal polysaccharide. Carbohydr Res. 1982;110:315–9. doi: 10.1016/0008-6215(82)84013-5. [DOI] [PubMed] [Google Scholar]
- Pritchard DG, Coligan JE, Speed SE, et al. Carbohydrate fingerprints of streptococcal cells. J Clin Microbiol. 1981;13:89–92. doi: 10.1128/jcm.13.1.89-92.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard DG, Furner RL. Structure of the Group-specific polysaccharide of group E Streptococcus. Carbohydr Res. 1985;144:289–96. doi: 10.1016/s0008-6215(00)90676-1. [DOI] [PubMed] [Google Scholar]
- Pritchard DG, Gray BM, Dillon HC, Jr. Characterization of the Group-specific polysaccharide of Group B Streptococcus. Arch Biochem Biophys. 1984;235:385–92. doi: 10.1016/0003-9861(84)90211-x. [DOI] [PubMed] [Google Scholar]
- Pritchard DG, Gregory RL, Michalek SM, et al. Characterization of the serotype e polysaccharide antigen of Streptococcus mutans. Mol Immunol. 1986;23:141–5. doi: 10.1016/0161-5890(86)90035-0. [DOI] [PubMed] [Google Scholar]
- Pritchard DG, Michalek SM, McGhee JR, et al. Structure of the serotype f polysaccharide antigen of Streptococcus mutans. Carbohydr Res. 1987;166:123–31. doi: 10.1016/0008-6215(87)80049-6. [DOI] [PubMed] [Google Scholar]
- Pritchard DG, Rener BP, Furner RL, et al. Structure of the Group G streptococcal polysaccharide. Carbohydr Res. 1988;173:255–62. doi: 10.1016/s0008-6215(00)90821-8. [DOI] [PubMed] [Google Scholar]
- Rahim R, Burrows LL, Monteiro MA, et al. Involvement of the rml locus in core oligosaccharide and O polysaccharide assembly in Pseudomonas aeruginosa. Microbiology. 2000;146(Pt 11):2803–14. doi: 10.1099/00221287-146-11-2803. [DOI] [PubMed] [Google Scholar]
- Reinscheid DJ, Ehlert K, Chhatwal GS, et al. Functional analysis of a PcsB-deficient mutant of Group B Streptococcus. FEMS Microbiol Lett. 2003;221:73–79. doi: 10.1016/S0378-1097(03)00167-8. [DOI] [PubMed] [Google Scholar]
- Rigottier-Gois L, Madec C, Navickas A, et al. The surface rhamnopolysaccharide epa of Enterococcus faecalis is a key determinant of intestinal colonization. J Infect Dis. 2015;211:62–71. doi: 10.1093/infdis/jiu402. [DOI] [PubMed] [Google Scholar]
- Roca C, Alves VD, Freitas F, et al. Exopolysaccharides enriched in rare sugars: bacterial sources, production, and applications. Front Microbiol. 2015;6:288. doi: 10.3389/fmicb.2015.00288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabharwal H, Michon F, Nelson D, et al. Group A Streptococcus (GAS) carbohydrate as an immunogen for protection against GAS infection. J Infect Dis. 2006;193:129–35. doi: 10.1086/498618. [DOI] [PubMed] [Google Scholar]
- Samson JE, Moineau S. Bacteriophages in food fermentations: new frontiers in a continuous arms race. Annu Rev Food Sci Technol. 2013;4:347–68. doi: 10.1146/annurev-food-030212-182541. [DOI] [PubMed] [Google Scholar]
- Sancho D, Reis e Sousa C. Signaling by myeloid C-type lectin receptors in immunity and homeostasis. Annu Rev Immunol. 2012;30:491–529. doi: 10.1146/annurev-immunol-031210-101352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlag M, Biswas R, Krismer B, et al. Role of staphylococcal wall teichoic acid in targeting the major autolysin Atl. Mol Microbiol. 2010;75:864–73. doi: 10.1111/j.1365-2958.2009.07007.x. [DOI] [PubMed] [Google Scholar]
- Schleifer KH, Kraus J, Dvorak C, et al. Transfer of Streptococcus lactis and related streptococci to the genus Lactococcus gen. nov. Syst Appl Microbiol. 1985;6:183–95. [Google Scholar]
- Sewell EW, Brown ED. Taking aim at wall teichoic acid synthesis: new biology and new leads for antibiotics. J Antibiot. 2014;67:43–51. doi: 10.1038/ja.2013.100. [DOI] [PubMed] [Google Scholar]
- Sham LT, Barendt SM, Kopecky KE, et al. Essential PcsB putative peptidoglycan hydrolase interacts with the essential FtsXSpn cell division protein in Streptococcus pneumoniae D39. P Natl Acad Sci USA. 2011;108:E1061–9. doi: 10.1073/pnas.1108323108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y, Yamashita Y, Ozaki K, et al. Expression and characterization of streptococcal rgp genes required for rhamnan synthesis in Escherichia coli. Infect Immun. 2002;70:2891–8. doi: 10.1128/IAI.70.6.2891-2898.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y, Yamashita Y, van der Ploeg JR. The serotype-specific glucose side chain of rhamnose-glucose polysaccharides is essential for adsorption of bacteriophage M102 to Streptococcus mutans. FEMS Microbiol Lett. 2009;294:68–73. doi: 10.1111/j.1574-6968.2009.01546.x. [DOI] [PubMed] [Google Scholar]
- Shimomura Y, Okumura K, Murayama SY, et al. Complete genome sequencing and analysis of a Lancefield Group G Streptococcus dysgalactiae subsp. equisimilis strain causing streptococcal toxic shock syndrome (STSS) BMC Genomics. 2011;12:17. doi: 10.1186/1471-2164-12-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silhavy TJ, Kahne D, Walker S. The bacterial cell envelope. Cold Spring Harb Perspect Biol. 2010;2:a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steer AC, Dale JB, Carapetis JR. Progress toward a global Group A streptococcal vaccine. Pediatr Infect Dis J. 2013;32:180–2. doi: 10.1097/INF.0b013e318281da11. [DOI] [PubMed] [Google Scholar]
- Sutcliffe IC, Black GW, Harrington DJ. Bioinformatic insights into the biosynthesis of the Group B carbohydrate in Streptococcus agalactiae. Microbiology. 2008;154:1354–63. doi: 10.1099/mic.0.2007/014522-0. [DOI] [PubMed] [Google Scholar]
- Swanson J, Gotschlich EC. Electron microscopic studies on streptococci. II. Group A carbohydrate. J Exp Med. 1973;138:245–58. doi: 10.1084/jem.138.1.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swoboda JG, Campbell J, Meredith TC, et al. Wall teichoic acid function, biosynthesis, and inhibition. Chembiochem. 2010;11:35–45. doi: 10.1002/cbic.200900557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Ubukata K, Watanabe H. Invasive infection caused by Streptococcus dysgalactiae subsp. equisimilis: characteristics of strains and clinical features. J Infect Chemother. 2011;17:1–10. doi: 10.1007/s10156-010-0084-2. [DOI] [PubMed] [Google Scholar]
- Tanaka D, Isobe J, Watahiki M, et al. Genetic features of clinical isolates of Streptococcus dysgalactiae subsp. equisimilis possessing Lancefield's Group A antigen. J Clin Microbiol. 2008;46:1526–9. doi: 10.1128/JCM.02188-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateno H, Ogawa T, Muramoto K, et al. Rhamnose-binding lectins from steelhead trout (Oncorhynchus mykiss) eggs recognize bacterial lipopolysaccharides and lipoteichoic acid. Biosci Biotech Bioch. 2002;66:604–12. doi: 10.1271/bbb.66.604. [DOI] [PubMed] [Google Scholar]
- Teng F, Jacques-Palaz KD, Weinstock GM, et al. Evidence that the enterococcal polysaccharide antigen gene (epa) cluster is widespread in Enterococcus faecalis and influences resistance to phagocytic killing of E. faecalis. Infect Immun. 2002;70:2010–5. doi: 10.1128/IAI.70.4.2010-2015.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng F, Singh KV, Bourgogne A, et al. Further characterization of the epa gene cluster and Epa polysaccharides of Enterococcus faecalis. Infect Immun. 2009;77:3759–67. doi: 10.1128/IAI.00149-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theilacker C, Holst O, Lindner B, et al. The structure of the wall teichoic acid isolated from Enterococcus faecalis strain 12030. Carbohydr Res. 2012;354:106–9. doi: 10.1016/j.carres.2012.03.031. [DOI] [PubMed] [Google Scholar]
- Thurlow LR, Thomas VC, Hancock LE. Capsular polysaccharide production in Enterococcus faecalis and contribution of CpsF to capsule serospecificity. J Bacteriol. 2009;191:6203–10. doi: 10.1128/JB.00592-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukioka Y, Yamashita Y, Nakano Y, et al. Identification of a fourth gene involved in dTDP-rhamnose synthesis in Streptococcus mutans. J Bacteriol. 1997a;179:4411–4. doi: 10.1128/jb.179.13.4411-4414.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukioka Y, Yamashita Y, Oho T, et al. Biological function of the dTDP-rhamnose synthesis pathway in Streptococcus mutans. J Bacteriol. 1997b;179:1126–34. doi: 10.1128/jb.179.4.1126-1134.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tytgat HL, Lebeer S. The sweet tooth of bacteria: common themes in bacterial glycoconjugates. Microbiol Mol. 2014;78:372–417. doi: 10.1128/MMBR.00007-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Beek SL, Le Breton Y, Ferenbach AT, et al. GacA is essential for Group A Streptococcus and defines a new class of monomeric dTDP-4-dehydrorhamnose reductases (RmlD) Mol Microbiol. 2015;98:946–62. doi: 10.1111/mmi.13169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Sorge NM, Cole JN, Kuipers K, et al. The classical lancefield antigen of Group A Streptococcus is a virulence determinant with implications for vaccine design. Cell Host Microbe. 2014;15:729–40. doi: 10.1016/j.chom.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vliet SJ, Steeghs L, Bruijns SC, et al. Variation of Neisseria gonorrhoeae lipooligosaccharide directs dendritic cell-induced T helper responses. PLoS Pathog. 2009;5:e1000625. doi: 10.1371/journal.ppat.1000625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer W, Blanot D, de Pedro MA. Peptidoglycan structure and architecture. FEMS Microbiol Rev. 2008;32:149–67. doi: 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- Vollmer W, Seligman SJ. Architecture of peptidoglycan: more data and more models. Trends Microbiol. 2010;18:59–66. doi: 10.1016/j.tim.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Wagner B, Wagner M, Kubin V, et al. Immunoelectron microscopic study of the location of Group-specific and protein type-specific antigens of Group B streptococci. J Gen Microbiol. 1980;118:95–105. doi: 10.1099/00221287-118-1-95. [DOI] [PubMed] [Google Scholar]
- Wagner M, Wagner B. An electron microscopic study of the location of peptidoglycan in Group A and C streptococcal cell walls. J Gen Microbiol. 1978;108:283–94. doi: 10.1099/00221287-108-2-283. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Tateno H, Nakamura-Tsuruta S, et al. The function of rhamnose-binding lectin in innate immunity by restricted binding to Gb3. Dev Comp Immunol. 2009;33:187–97. doi: 10.1016/j.dci.2008.08.008. [DOI] [PubMed] [Google Scholar]
- Weidenmaier C, Peschel A. Teichoic acids and related cell-wall glycopolymers in Gram-positive physiology and host interactions. Nat Rev Microbiol. 2008;6:276–87. doi: 10.1038/nrmicro1861. [DOI] [PubMed] [Google Scholar]
- Winstel V, Kuhner P, Salomon F, et al. Wall Teichoic Acid Glycosylation Governs Staphylococcus aureus Nasal Colonization. mBio. 2015;6 doi: 10.1128/mBio.00632-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winstel V, Liang C, Sanchez-Carballo P, et al. Wall teichoic acid structure governs horizontal gene transfer between major bacterial pathogens. Nat Commun. 2013;4:2345. doi: 10.1038/ncomms3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winstel V, Sanchez-Carballo P, Holst O, et al. Biosynthesis of the unique wall teichoic acid of Staphylococcus aureus lineage ST395. mBio. 2014;5:e00869. doi: 10.1128/mBio.00869-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winstel V, Xia G, Peschel A. Pathways and roles of wall teichoic acid glycosylation in Staphylococcus aureus. Int J Med Microbiol. 2014;304:215–21. doi: 10.1016/j.ijmm.2013.10.009. [DOI] [PubMed] [Google Scholar]
- Xu Y, Murray BE, Weinstock GM. A cluster of genes involved in polysaccharide biosynthesis from Enterococcus faecalis OG1RF. Infect Immun. 1998;66:4313–23. doi: 10.1128/iai.66.9.4313-4323.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Singh KV, Qin X, et al. Analysis of a gene cluster of Enterococcus faecalis involved in polysaccharide biosynthesis. Infect Immun. 2000;68:815–23. doi: 10.1128/iai.68.2.815-823.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita Y, Shibata Y, Nakano Y, et al. A novel gene required for rhamnose-glucose polysaccharide synthesis in Streptococcus mutans. J Bacteriol. 1999;181:6556–9. doi: 10.1128/jb.181.20.6556-6559.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita Y, Tsukioka Y, Tomihisa K, et al. Genes involved in cell wall localization and side chain formation of rhamnose-glucose polysaccharide in Streptococcus mutans. J Bacteriol. 1998;180:5803–7. doi: 10.1128/jb.180.21.5803-5807.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yother J. Capsules of Streptococcus pneumoniae and other bacteria: paradigms for polysaccharide biosynthesis and regulation. Annu Rev Microbiol. 2011;65:563–81. doi: 10.1146/annurev.micro.62.081307.162944. [DOI] [PubMed] [Google Scholar]
- Zelensky AN, Gready JE. The C-type lectin-like domain superfamily. FEBS J. 2005;272:6179–217. doi: 10.1111/j.1742-4658.2005.05031.x. [DOI] [PubMed] [Google Scholar]
- Zeleznick LD, Boltralik JJ, Barkulis SS, et al. Biosynthesis of streptococcal cell walls: a rhamnose polysaccharide. Science. 1963;140:400–1. doi: 10.1126/science.140.3565.400. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.