Abstract
Eight viruses are currently assigned to the family Filoviridae. Marburg virus, Sudan virus and, in particular, Ebola virus have received the most attention both by researchers and the public from 1967 to 2013. During this period, natural human filovirus disease outbreaks occurred sporadically in Equatorial Africa and, despite high case-fatality rates, never included more than several dozen to a few hundred infections per outbreak. Research emphasis shifted almost exclusively to Ebola virus in 2014, when this virus was identified as the cause of an outbreak that has thus far involved more than 28 646 people and caused more than 11 323 deaths in Western Africa. Consequently, major efforts are currently underway to develop licensed medical countermeasures against Ebola virus infection. However, the ecology of and mechanisms behind Ebola virus emergence are as little understood as they are for all other filoviruses. Consequently, the possibility of the future occurrence of a large disease outbreak caused by other less characterized filoviruses (i.e. Bundibugyo virus, Lloviu virus, Ravn virus, Reston virus and Taï Forest virus) is impossible to rule out. Yet, for many of these viruses, not even rudimentary research tools are available, let alone medical countermeasures. This review summarizes the current knowledge on these less well-characterized filoviruses.
Keywords: cuevavirus, Ebola, ebolavirus, Filoviridae, filovirus, marburgvirus
While Ebola virus dominates the headlines, other filoviruses remain largely uncharacterized but may pose equal risks to humans.
Graphical Abstract Figure.
While Ebola virus dominates the headlines, other filoviruses remain largely uncharacterized but may pose equal risks to humans.
INTRODUCTION
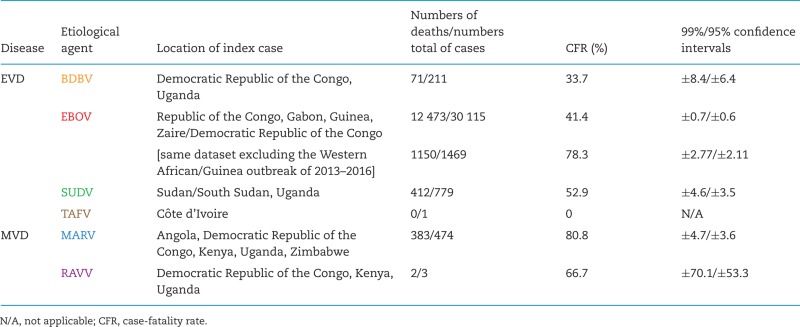
Viruses with single-stranded negative-sense RNA genomes can be assigned to three unofficial supergroups based on whether these genomes are multisegmented, circular or unsegmented, and on evolutionary relationships of the genome-encoded RNA-dependent RNA polymerases (Li et al. 2015). All of the unsegmented viruses are currently assigned to the rapidly expanding order Mononegavirales. The family Filoviridae is one of the eight mononegaviral families (Afonso et al. 2016). Filoviridae members are differentiated from other mononegaviruses not only by genomic sequence, but also by the formation of characteristically shaped, filamentous virions. In addition, filoviruses are also differentiated from other mononegaviruses by comparatively long (≈19 kb) genomes containing gene overlaps, unique transcriptional initiation and termination signals, and an open reading frame (ORF) encoding a unique structural protein without obvious homologs in other mononegaviruses (i.e. VP24) (Kuhn et al. 2010, 2011). Filoviruses are further assigned to seven species included in the three genera Cuevavirus, Ebolavirus and Marburgvirus (Table 1) based on individual molecular properties (Kuhn et al. 2010, 2011; Bao, Chetvernin and Tatusova 2012; Lauber and Gorbalenya 2012). Six of the currently eight recognized filoviruses are known human pathogens causing two diseases officially recognized by the World Health Organization: Ebola virus disease (EVD) and Marburg virus disease (MVD). The remaining two filoviruses are known or suspected animal pathogens (Table 2). In general, filoviruses can be considered exotic pathogens. Human and/or animal filovirus disease outbreaks are rare. Only 37 outbreaks have been recorded in the almost 50 years since the discovery of filoviruses, and of those, only 11 included more than 100 cases each (Table 3) (Kuhn 2015).
Table 1.
Official taxonomy of the family Filoviridae as of 2016 (Kuhn et al. 2010, 2011; Bukreyev et al. 2014).
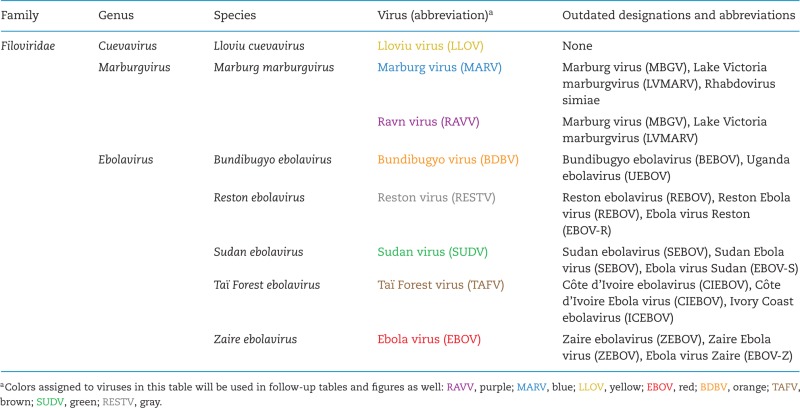 |
Table 2.
Official human filovirus disease classification and nomenclature as of 2016.
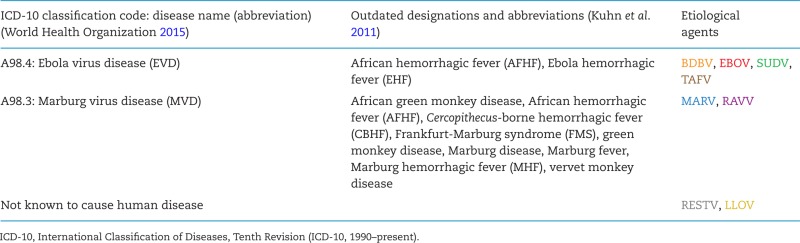 |
Table 3.
Cumulative human filovirus disease cases (updated from Kuhn 2015, as of March 27, 2016).
 |
Based on the publication records (Kuhn 2008), the filoviruses that are currently most characterized and understood are Ebola virus (EBOV), Sudan virus (SUDV) and Marburg virus (MARV). The filovirus best known and feared by the general public is EBOV. This notoriety is in part due to numerous popular science publications, fiction and movies (Semmler 1998; Kuhn 2008; Blakey et al. 2015) and in part because EBOV had caused the most recorded human filovirus infections of all filoviruses, i.e. 1101 until 2012 (Kuhn 2015). At the end of 2013, EBOV caused an unprecedented EVD outbreak in Western Africa, with thus far 28 646 human infections and 11 323 deaths (case-fatality rate [CFR] = 39.51%) (World Health Organization 2016). Unsurprisingly, this public-health emergency resulted in massively increased funding for basic and translational EBOV research, including the development and testing in clinical trials of potential medical countermeasures.
However, despite tremendous progress in understanding of filovirus infections in vitro and in vivo, where and how filoviruses are maintained in nature remain unclear. In addition, the circumstances or mechanisms that lead to the occasional filovirus emergence in human and other mammal populations are also unknown. To emphasize research more or less exclusively on EBOV, SUDV and MARV may quite literally be a fatal mistake (Anthony and Bradfute 2015; Kozak and Kobinger 2016). One cannot exclude the possibility of a future disease outbreak of the scope of the Western African outbreak caused by other filoviruses, including those viruses currently thought to be apathogenic for humans.
This review provides an overview of the current knowledge of these neglected filoviruses: Bundibugyo virus (BDBV), Lloviu virus (LLOV), Ravn virus (RAVV), Reston virus (RESTV) and Taï Forest virus (TAFV).
Bundibugyo virus
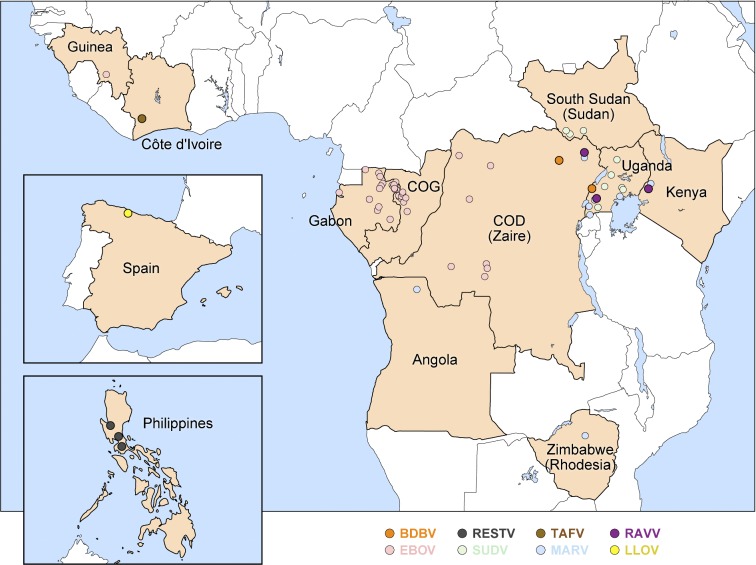
Bundibugyo virus (BDBV; pronounced ˌbʊndiː'bʊdʒɔː vɑɪrəs) was discovered in Eastern Africa during a human viral hemorrhagic fever (VHF) outbreak that probably began in or around August 2007 and lasted until January 2008. The outbreak affected the Bundibugyo and Kikyo townships in Bundibugyo District of Western Uganda Administrative Region, Uganda and ultimately amounted to 149 suspected cases and 37 deaths (CFR = 24.8%; alternative published statistics, based on different case definitions and index case identifications, are as follows: 116 cases and 39 deaths [33.6%]; and 131 cases and 42 deaths [32.1%]) (Towner et al. 2008; MacNeil et al. 2010; Wamala et al. 2010; Centers for Disease Control and Prevention 2015). A second EVD outbreak due to BDBV was recorded in August of 2012 around Isiro, Haut-Uele District, Province Orientale, in northeastern Democratic Republic of the Congo, roughly 400 km northwest from Bundibugyo (Fig. 1). When that outbreak was declared over in November 2012, 62 people had been infected and 34 had died (CFR = 54.8%; an alternative statistic, based on a different case definition, is 77 cases and 36 deaths [CFR = 46.8%]) (Albariño et al. 2013; Centers for Disease Control and Prevention 2015; Kratz et al. 2015). Therefore, the average CFR range of human BDBV infections (32.3%–41.1%; Table 3) is comparable to that of EBOV infections (≈41.4%). The often repeated notion that BDBV is ‘less virulent’ than EBOV in humans cannot be upheld based on available data.
Figure 1.
Geographical location of primary index cases causing disease outbreaks due to neglected filovirus infections. Countries with index cases are shown in light brown with outbreak locations marked as bold dots (disease outbreaks due to more prominent filoviruses, EBOV, SUDV or MARV, are indicated via faded dots for reference). Former country names are listed in parenthesis under present names. COD: Democratic Republic of the Congo; COG: Republic of the Congo. Adopted from (Kuhn 2015).
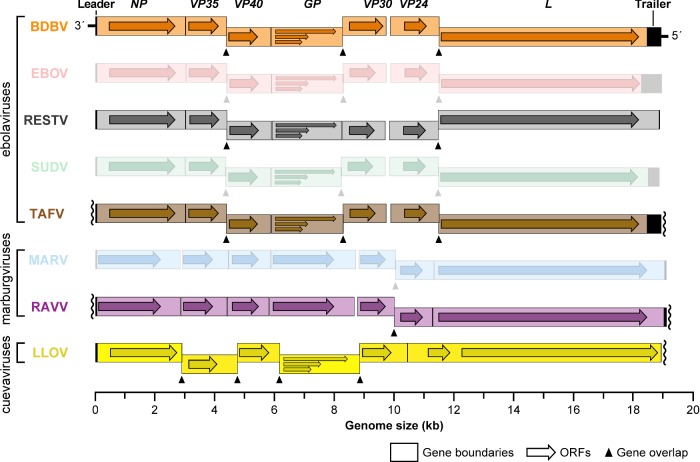
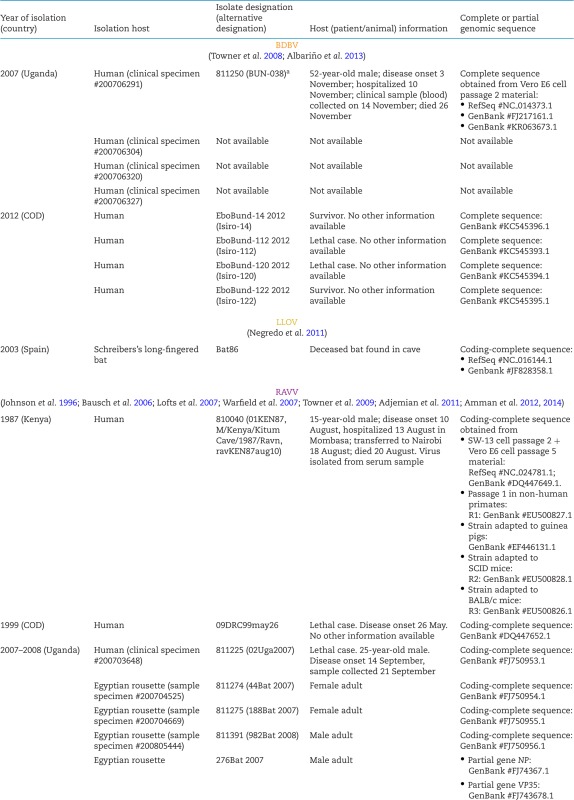
A number of BDBV isolates have been obtained (Table 4), and Ugandan Bundibugyo virus/H.sapiens-tc/UGA/2007/Butalya-811250 (BDBV/But-811250) was designated the type BDBV isolate (Kuhn et al. 2014). To date, all published in vitro and in vivo experiments involving BDBV have been performed exclusively with this isolate (Albariño et al. 2013). BDBV isolates from Uganda and Democratic Republic of the Congo share ≈98.6% genome identity. Genomic analyses revealed the BDBV genome organization to be identical to that of other ebolaviruses (Fig. 2) (Towner et al. 2008; Albariño et al. 2013).
Table 4.
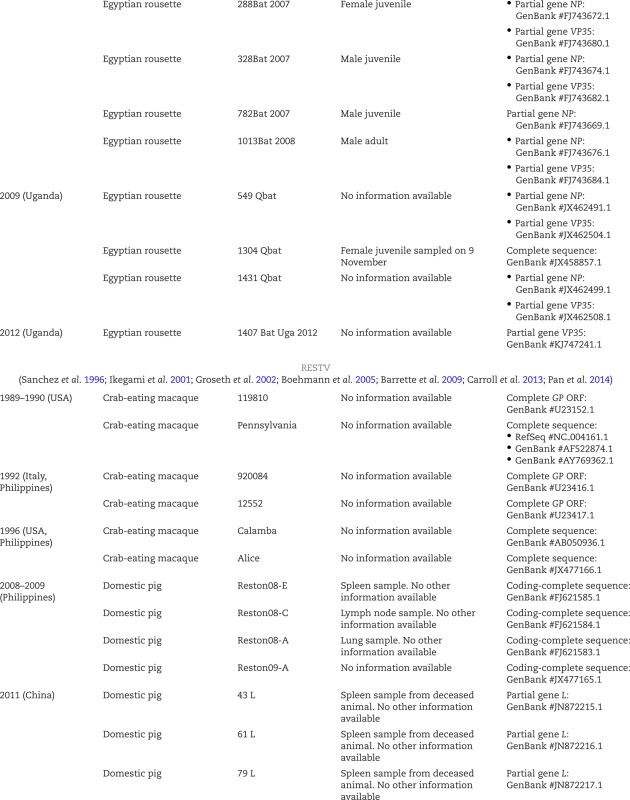
Neglected filovirus isolates and sequences.

|
Figure 2.
Organization of neglected filovirus genomes. Genes and open reading frames (ORFs) are shown as rectangles and horizontal arrows, respectively. Wavy lines indicate incomplete sequencing of 3′ leader or 5′ trailer sequences. Entries for prominent filoviruses, EBOV, SUDV or MARV, are muted.
Genomic analyses also indicated that each of the two outbreaks was in all likelihood caused through singular introductions into the human populations with subsequent human-to-human transmission (Albariño et al. 2013). However, how these introductions occurred and why no further BDBV introductions have occurred since the end of the second outbreak in 2012 remain unclear. The Bundibugyo District borders the Democratic Republic of the Congo. A large portion of the district is part of the Rwenzori Mountains, the Semliki National Park and Game Reserve with its associated wildlife, including non-human primates, and domestic or economical activities, such as cocoa farming, hunting and fishing (Wamala et al. 2010). These potential contacts with wildlife suggest that the BDBV introduction into the human population was a zoonotic event. However, the precise beginning of the 2007 Ugandan EVD outbreak and the identity and history of the human index case is unclear (Towner et al. 2008; MacNeil et al. 2010; Wamala et al. 2010). Likewise, no data have been published on the behavior and recent history of the suspected human index case of the 2012 EVD outbreak.
MVD outbreaks due to MARV and RAVV could be directly linked to infected frugivorous bats from which replicating virus isolates could be obtained (Towner et al. 2009; Amman et al. 2012; Wahl-Jensen et al. 2013). EVD outbreaks due to EBOV have been loosely associated with bats via anti-EBOV antibody or EBOV genome fragment detection in bat sera in the absence of virus isolation (Leroy et al. 2005; Wahl-Jensen et al. 2013). Likewise, massive ape (central chimpanzee and western lowland gorilla) population declines have been temporary and spatially associated with human EVD outbreaks, and while EBOV isolation was unsuccessful, EBOV genome fragments were detected in a low number of apes (Huijbregts et al. 2003; Walsh et al. 2003; Bermejo et al. 2006). However, neither bat nor ape associations could be established for BDBV thus far. Injection of BDBV into Egyptian rousettes (Rousettus aegyptiacus), the presumed natural bat reservoir of MARV and RAVV (Amman et al. 2012), did not result in replication (Jones et al. 2015). Therefore, the ecology of BDBV remains a mystery. Only one ecological survey for BDBV was reported. IgG antibodies against BDBV glycoprotein (GP1,2) antigen were detected by ELISA and western blot in 9/353 (2.6%) of orangutans (Pongo pygmaeus) sampled on Kalimantan Island, Indonesia, in 2006. BDBV-specific IgM could not be detected (Nidom et al. 2012). These results may suggest that the BDBV distribution is not restricted to Eastern Africa. However, caution is advised as the inter-ebolavirus and non-filovirus GP1,2 cross-reactivity of naturally occurring antibodies is undefined (antibodies to yet unknown filoviruses or unrelated antibodies may react with BDBV GP1,2). For instance, IgG and IgM antibodies from survivors of the 2007 BDBV/EVD outbreak strongly and weakly cross-react with other, non-BDBV, ebolavirus antigen preparations, respectively (Macneil, Reed and Rollin 2011). In the absence of BDBV genome detection by deep sequencing or BDBV isolation in cell culture, serological results at best hint toward the whereabouts of BDBV in nature. Thus, at the moment, future EVD outbreaks due to BDBV can neither be geographically nor temporally anticipated, let alone be prevented.
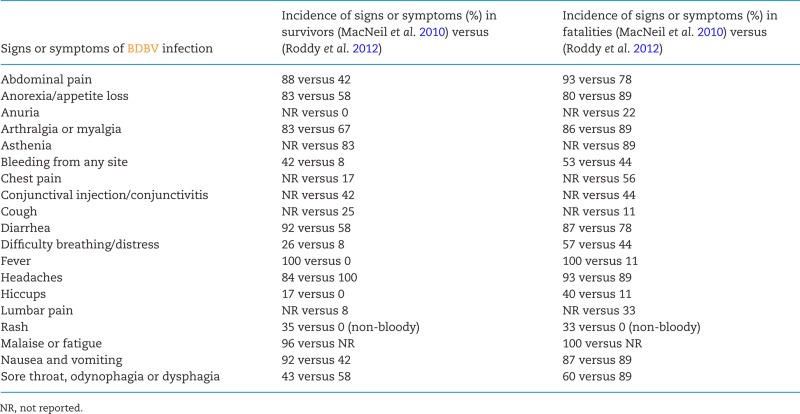
The clinical presentation of EVD due to BDBV infection in Uganda in 2007 is summarized in Table 5 (MacNeil et al. 2010; Roddy et al. 2012). Based on these very limited data and additional statistics published for the 2012 outbreak (Kratz et al. 2015), the overall distribution of clinical signs and symptoms of BDBV infections appears highly similar to that of EBOV, SUDV and MARV infections (not reviewed here, see Siegert et al. 1967; Egbring, Slenczka and Baltzer 1971; Isaäcson et al. 1978; Piot et al. 1978; Smith, Francis and Simpson 1978; Bwaka et al. 1999; Bausch et al. 2006; Barry et al. 2014; Maganga et al. 2014; Schieffelin et al. 2014; Bah et al. 2015; Dallatomasina et al. 2015; Lado et al. 2015; Qin et al. 2015; Yan et al. 2015). Consequently, BDBV infection cannot be diagnosed based on clinical observation alone. During the 2007 EVD/BDBV outbreak, the mean incubation period was 6.3 days (the longest measured incubation period was 25 days) (MacNeil et al. 2010, 2011) and, therefore, maybe somewhat shorter than that of EBOV infection [12.7 ± 4.3 days in a 1995 EVD/EBOV outbreak (Eichner, Dowell and Firese 2011) and 11.4 days in the 2013–2016 EVD/EBOV outbreak in Western Africa (World Health Organization Ebola Response Team 2014)]. Asthenia, diarrhea and headache were the most frequent symptoms/signs. Hemorrhagic signs were observed in approximately half of the cohort (MacNeil et al. 2010; Roddy et al. 2012), which is typical for all ebolavirus infections. As has been reported for EBOV, SUDV and MARV infections (Baltzer et al. 1979; Bwaka et al. 1999; Kibadi et al. 1999; Rowe et al. 1999; Qureshi et al. 2015), survivors of disease caused by BDBV may suffer of long-term sequelae years after convalescence. These sequelae include arthralgia, difficulty swallowing, hearing loss, ocular deficits (blurred vision and retro-orbital pain) and sleeplessness (Clark et al. 2015). Whether BDBV is able to persist in convalescent patients, as has been reported for EBOV and MARV (Martini and Schmidt 1968; Nikiforov et al. 1994; Rodriguez et al. 1999; Mate et al. 2015; Varkey et al. 2015), is unclear.
Table 5.
Comparison of clinical signs or symptoms of human BDBV infection during the 2007 Uganda EVD outbreak.
 |
On the molecular level, EVD due to BDBV appears to differ from other ebolavirus infections. Whereas strong inflammatory responses (‘cytokine storms’) have been reported during EBOV infections and were associated with concomitant multi-organ dysfunction syndrome (Baize et al. 1999, 2002; Villinger et al. 1999), lethal human BDBV infections were associated with low concentrations of proinflammatory cytokines (interleukin [IL]-1α, IL-1β, IL-6, tumor necrosis factor-α) and high concentrations of antiinflammatory IL-10 (Gupta et al. 2012). These results indirectly confirm in vitro experiments with BDBV-infected peripheral blood mononuclear cells (PBMCs). BDBV-infected cells produced 10–100-fold less progeny virions and reacted with 2–10-fold lower expression of tumor necrosis factor-α, monocyte chemoattractant protein-1, IL-1β and macrophage inflammatory protein 1-α compared to EBOV-infected control cells. Interestingly, in contrast to the in vivo data, IL-10 expression was also low (Gupta et al. 2010).
Thus, pathogenetic mechanisms or properties from relatively well-characterized filoviruses (EBOV, SUDV and MARV) cannot be generalized to other, less characterized filoviruses. Consequently, promising medical countermeasures against one filovirus should not be assumed automatically to be promising avenues for related filoviruses. This lack of translatability is all the more concerning in regard to BDBV, as the virus itself is completely uncharacterized. With the exception of genomic sequence determination and functional deductions one can possibly make from alignment with other ebolavirus sequences, only three reports have been published addressing BDBV molecular biology directly. BDBV uses Niemann-Pick C1 (NPC1) as a cell-entry receptor (Ng et al. 2015); cell entry is dependent on cathepsin B in vitro (Misasi et al. 2012); and BDBV Δ-peptide has no effect on MARV replication (Radoshitzky et al. 2011).
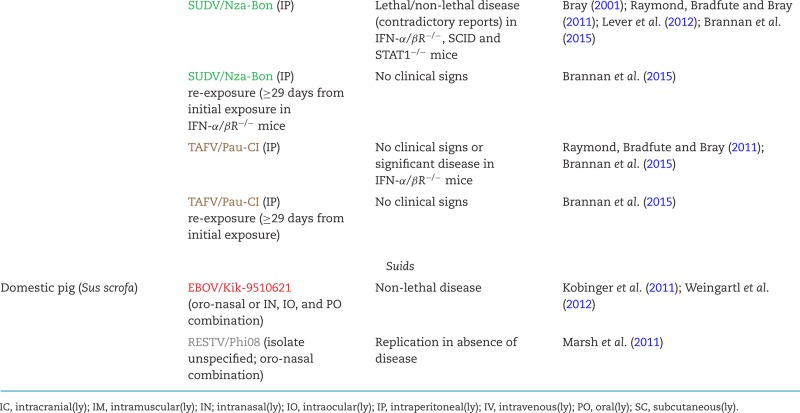
The pathology of human BDBV infection remains unknown as neither autopsies nor biopsies were performed during the two disease outbreaks. Until future outbreaks might occur, the delineation of BDBV pathogenesis is solely dependent on animal models. Whereas numerous animal models are available for EBOV, SUDV and MARV infection (Table 6), the only established animal model for BDBV infection is based on intramuscular inoculation of crab-eating macaques (Macaca fascicularis). The conditions to achieve 100% fatality in this model have yet to be determined (typically, 25%–34% of infected animals survive) (Hensley et al. 2010; Falzarano et al. 2011; Mire et al. 2013). No detailed clinical or pathological descriptions of BDBV infection in crab-eating macaques are available. The establishment of a mouse model of BDBV infection was attempted using type 1 interferon α/β receptor knockout (IFN-α/βR−/−) mice (Table 6). When exposed to BDBV, these mice did not develop signs of clinical disease, weight loss or lethality (Brannan et al. 2015). However, in some of BDBV-infected mice, transient viremia and increased tissue titers (spleen and liver) were noted. Results from serial sampling experiments indicated hepatic disseminated intravascular coagulation (DIC). Splenic lymphoid hyperplasia occurred concurrently with mild lymphoid depletion, and immunohistochemical analysis detected viral antigen in red pulp macrophage-like cells. Platelets were transiently decreased, and white blood cells and absolute lymphocytes were transiently increased.
Table 6.
Overview of animal models for filovirus infections.



|
The absence of true rodent models for BDBV infection excludes the possibility of high-throughput in vivo drug screening and initial candidate countermeasure evaluation. The only partially lethal non-human primate model requires countermeasure evaluation to be performed with large animal numbers to achieve statistical significance of outcomes, which is both ethically problematic and in most cases prohibited by cost and logistics in limited biosafety level-4 space. Despite these obstacles, the BDBV crab-eating macaque model has been used for limited evaluation of candidate medical countermeasure platforms that are under advanced development for the prevention or treatment of EVD due to EBOV infection. At least two candidate vaccine platforms (i.e. DNA prime/recombinant adenovirus boost, recombinant vesicular stomatitis Indiana virus) appear promising against BDBV (Table 7) (Hensley et al. 2010; Falzarano et al. 2011; Mire et al. 2013). Theoretically, administration of a pan-filovirus vaccine comprised of mosaic ebolavirus proteins could elicit antibody responses to a number of ebolaviruses, including BDBV (Fenimore et al. 2012), but such a vaccine has not been tested in animals with BDBV infection.
Table 7.
Overview of candidate vaccine development against neglected filoviruses.



|
Neither post-exposure prophylactics nor antivirals have yet been identified, let alone been tested, against BDBV infections in vivo. Consequently, treatment of human BDBV infections is limited to general supportive measures applied to other ebolavirus infections, such as nutritional supplementation, oral or intravenous fluid rehydration and medication against anxiety (e.g. diazepam), secondary infections (antibiotics, antimalarials, antimycotics), dyspepsia (e.g. cimetidine, ranitidine, omeprazole), nausea/vomiting (e.g., metaclopramide) and pain (e.g. acetominophen, morphine) (Roddy et al. 2012).
Diagnostic tests for BDBV infection are currently limited to various pan-filovirus reverse transcriptase-polymerase chain reaction (RT-PCR) or quantitative reverse transcriptase-PCR (qRT-PCR) protocols (Lu et al. 2015) and BDBV-specific sequence capture probes for next-generation sequencing (Koehler et al. 2014). A handful of BDBV-specific monoclonal or polyclonal murine, rabbit or crab-eating macaque antibodies (Ou et al. 2011; Holtsberg et al. 2015; Keck et al. 2015; Wang et al. 2015), BDBV cross-reactive murine antibodies (Fusco et al. 2015; Wang et al. 2015; Furuyama et al. 2016), BDBV cross-reactive murine single-chain variable domain fragments (scFv) and thermostable single-domain nurse shark antibodies (IgNAR V) raised against inactivated EBOV particles (Goodchild et al. 2011) are available, but have not yet been tested in diagnostic serological assays.
Lloviu virus
Lloviu virus (LLOV; pronounced j'ɔːvju vɑɪrəs) was discovered in 2002 in Cueva del Lloviu in Spain. Next-generation sequencing of tissues collected from several of hundreds of deceased (insectivorous) Schreibers's long-fingered bats (Miniopterus schreibersii) within the cave revealed infection with this novel filovirus (Fig. 1) (Negredo et al. 2011). Therefore, LLOV is only the third (LLOV, MARV and RAVV) of the eight known filoviruses to unambiguously infect bats. However, analysis of available data does not permit drawing conclusions on whether LLOV caused the bats’ deaths. Similar to MARV- and RAVV-infected bats, LLOV-infected bats could have been subclinically and persistently infected with LLOV and died of other causes. As LLOV has not yet been rediscovered, Schreiber's long-fingered bats may not be the natural host reservoir of LLOV.
A single coding-complete genomic sequence of LLOV, Lloviu virus/M.schreibersii-wt/ESP/2003/Asturias-Bat86 (LLOV/Ast-Bat86; Table 4) was assembled from one of the Spanish samples (Negredo et al. 2011) and was designated the type LLOV ‘isolate’ (Kuhn et al. 2014). The LLOV genome is highly reminiscent in organization of other filovirus genomes containing the same overall linear ORF arrangement (Fig. 2). However, LLOV appears to express seven structural proteins (nucleoprotein [NP], VP35, VP40, GP, VP30, VP24, L) from six genes rather than the filovirus-typical seven genes (Negredo et al. 2011). All attempts failed to isolate this virus in culture (Negredo et al. 2011), and all sample material was depleted. Because the 5′ genomic terminus of the virus could not be sequenced, it is unclear whether a functional LLOV genome could be synthesized for in vitro rescue. Consequently, only four studies are published targeting LLOV specifically. These studies relied on recombinantly expressed LLOV proteins (Maruyama et al. 2014; Feagins and Basler 2015) or on virus surrogate systems such as vesiculoviral pseudotypes (Maruyama et al. 2014), retroviral pseudotypes (Ng et al. 2014) or recombinant vesiculoviruses (Ng et al. 2014, 2015) to study parts of the presumed LLOV replication cycle. Results indicate that the LLOV surface GP1,2 mediates LLOV cell entry by binding to the universal endosomal filovirus receptor NPC1 (Ng et al. 2014, 2015) in a pH-, cathepsin L-dependent (not cathepsin B-dependent) manner reminiscent of EBOV (Maruyama et al. 2014; Ng et al. 2014). As shown for other filoviruses, co-expression of LLOV GP1,2 and matrix protein VP40 results in the formation of filamentous filovirion-like particles (Maruyama et al. 2014). In terms of interferon response inhibitory functions, LLOV proteins VP35, VP40 and VP24 are functional analogs of EBOV rather than MARV (Feagins and Basler 2015). Finally, LLOV Δ-peptide inhibits cell transduction of retroviral particles pseudotyped with MARV GP1,2 just like EBOV and MARV Δ-peptides (Ng et al. 2014).
The potential of LLOV to infect humans is unknown. Because of the absence of a replicating LLOV isolate, no animal model of LLOV infection is available. Consequently, possible persistence of LLOV in animals, pathogenicity/virulence, pathogenesis or potential countermeasures against infection are not known. Specific diagnostic tests for LLOV infection have not yet been reported. However, a recently established, novel system using recombinant EBOV expressing LLOV GP1,2 instead of EBOV GP1,2 aided in the identification of LLOV GP1,2-specific antibodies that could possibly be used in diagnostic assays such as immunofluorescent assay (IFA), enzyme-linked immunosorbent assay (ELISA) or western blot, assuming that LLOV GP1,2 is presented correctly on the recombinant EBOV/LLOV chimeric virion (Ilinykh et al. 2016).
Ravn virus
Ravn virus (RAVV; pronounced rævn vɑɪrəs) was discovered in 1987 and has been reencountered in 1999, 2007 and 2009 (Fig. 1). Thus far, only three human infections, two of them lethal, have been recorded. These cases result in a mean CFR of 66.7% (higher than that of any ebolavirus infection), but the low case number results in extremely high confidence intervals (Table 3). Of note, several human RAVV infections may have been overlooked during the relatively extensive MVD outbreak in the Democratic Republic of Congo in 1998–2000 (Bausch et al. 2006).
In addition to the scientific literature (Tukei 1988; Johnson et al. 1996), the initial 1987 infection of a 15-year-old Danish boy with RAVV has been vividly described in a popular science book using the synonym ‘Peter Cardinal’ (Preston 1994). The boy had lived with his expatriate sister and parents in Kisumu District of the now dissolved Nyanza Province in Kenya. The boy fell sick on 10 August 1987, with symptoms and clinical signs indistinguishable from MVD. Despite extensive treatment and life-sustaining measures started at Aga Khan Hospital in Mombasa on 13 August and continued at Nairobi Hospital on 18 August, the boy died on 20 August (Tukei 1988; Preston 1994; Johnson et al. 1996). Where and under which circumstances the boy became infected with RAVV are unclear. Epizootiological studies focused primarily on Kitum Cave in Kenya's Mt. Elgon National Park, which the family had visited on 2 August, because this cave was also loosely associated with a fatal MARV infection of a French engineer in 1980 (Smith et al. 1982; Preston 1994). Being large enough to give shelter to animals including bats, large felids and elephants, Kitum Cave was thought to be a possible hotspot for zoonotic viral spillover. However, even after examination of several thousand collected samples and sentinel animals from the cave, no filovirus was found (Tukei 1988; Preston 1994).
The second case of human RAVV infection was recorded in 1999 in Durba, Haut-Uele District, Province Orientale, Democratic Republic of the Congo, during a much larger outbreak of MVD due to MARV infection (1998–2000) including 153 infections and 128 deaths. All these human MARV infections and the single RAVV infection were traced back to an illegal underground gold mine. The original source of the RAVV and MARV infections is unclear. However, the outbreak came to an abrupt end when the gold mine was flooded, indicating a marburgvirus (MARV and RAVV) host reservoir inside of the cave and, therefore, zoonotic transmission (Bausch et al. 2006).
Between June and July 2007, three people developed MVD due to MARV infection in Ibanda District, Western Region, Uganda, after entering and working in the local Kitaka Cave (Adjemian et al. 2011). This lead ore mine was closed and secured by a guard who developed MVD in mid-September due to RAVV infection (Towner et al. 2009; Adjemian et al. 2011).
A total of three human RAVV isolates have been sequenced, one from each outbreak (Table 4). The coding-complete sequence of the isolate obtained from the 1987 case, Ravn virus/H.sapiens-tc/KEN/1987/Kitum Cave-810040 (RAVV/KiC-810040), was designated the type RAVV isolate (Kuhn et al. 2014). RAVV/KiC-810040 is also the isolate that has been used for virtually all laboratory studies with RAVV. The RAVV genome is identical in organization to the MARV genome (Fig. 2) (Johnson et al. 1996). Molecular-biological characterization studies focusing on RAVV have been extremely rare. In one study, RAVV VP40 was found to be a functional analog to MARV VP40, inhibiting the host cell's interferon type I and II responses (Valmas and Basler 2011).
Among the neglected filoviruses, RAVV's ecology is the least shrouded in mystery. RAVV (and MARV) genome-related nucleic acids fragments were detected in Egyptian rousettes (R. aegyptiacus) sampled in Kitaka Cave in 2007–2008 and 2012. Several replicating isolates of RAVV (and MARV) could be isolated from Kitaka Cave Egyptian rousettes sampled in 2007–2008 and from Egyptian rousettes sampled from 2008 and 2009 in Python Cave, a tourist site in Uganda's Queen Elizabeth National Park less than 30 miles away from Kitaka Cave (Table 4) (Towner et al. 2009; Amman et al. 2012, 2014). These findings indicate that Egyptian rousettes are reservoir hosts for marburgviruses. The circumstances under which extremely rare marburgvirus transmission occurs to humans has not been defined, and explanations are not available for the lack of marburgviruses in Egyptian rousette populations elsewhere in Africa (Wahl-Jensen et al. 2013).
The clinical presentation of human RAVV infection has only been described for the Danish boy who died in 1987 (Preston 1994; Johnson et al. 1996). As the source of infection remains unknown, so is the incubation time. The boy first developed headache and fever, accompanied by malaise, anorexia and vomiting. The disease then worsened, resulting in bloody diarrhea and hypotension, ecchymoses, leukocytosis (20 000/mm3) and thrombocytopenia (26 000/mm3). Deterioration was characterized by high fever (40°C), disseminated Pseudomonas aeruginosa superinfection, continuously falling blood pressure reaching unrecordable values, delirium, cyanosis, blood coagulation abnormalities resembling DIC, and increases in serum potassium and urea levels. The boy then died because of cardiac arrest due to shock despite all efforts, including administration of antibiotics, steroids, heparin, fresh plasma and blood, and dialysis. Autopsy revealed massive petechial and purpuric hemorrhages in the skin, conjunctivae and gastrointestinal mucosa; hemorrhages in the lungs, tracheobronchal tree, epicardium, renal cortices and bladder (but not in spleen, pancreas, adrenals or in the acutely congested liver); and retroperitoneal edema; and pleural, pericardial and peritoneal effusions (Preston 1994; Johnson et al. 1996).
RAVV is the best researched neglected filoviruses in regard to medical countermeasures. Immunocompetent and immunodeficient mouse models and guinea pig models of RAVV infection have been established for high-throughput medical countermeasure evaluation, and immunopathological studies and uniformly lethal crab-eating macaque and rhesus monkey models are available for pathogenesis studies and specific countermeasure evaluation (Table 6). RAVV was long seen as an outlier ‘subtype’ or ‘strain’ of MARV. Consequently, many experiments aiming at identification of medical countermeasures against MARV infection included RAVV. This inclusion explains why several promising candidate vaccines for prevention (adenovirus, DNA, virion-like particles [VLP], vesicular stomatitis Indiana virus [VSIV]-based, inactivated and subunit; Table 7) and candidate therapeutic treatments (antisense/interfering RNAs and small molecules; Table 8) of RAVV infection have been studied. However, candidate therapeutic treatments have not been evaluated in non-human primates.
Table 8.
Overview of candidate peri-exposure treatment against infections with neglected filoviruses.
 |
Reston virus
Reston virus (RESTV; pronounced ‘rɛstən vɑɪrəs) was discovered during a highly lethal VHF epizootic that occurred almost simultaneously in Virgina, Pennsylvania and Texas in the USA in 1989–1990 among captive crab-eating macaques (Geisbert and Jahrling 1990; Jahrling et al. 1990; Miranda and Miranda 2011). The first affected US location was Hazleton Research Products’ Primate Quarantine Unit in Reston, Virginia, for which RESTV was named, and this virus became a household name through the vivid descriptions of the epizootic in Richard Preston's popular science book The Hot Zone (Preston 1994). The affected macaques of all US locations, several of them also co-infected with a yet-to-be-identified simian arterivirus (suspected to be simian hemorrhagic fever virus), had been imported from a Ferlite Scientific Research non-human primate export facility in Calamba, Luzon and Philippines (Fig. 1). Follow-up studies indeed found RESTV circulating in that facility (Hayes et al. 1992; Miranda et al. 1999), suggesting that RESTV may be an Asian filovirus or that the virus had been imported to the Philippines from another location. Since then, RESTV epizootics were recorded three more times. In 1992 and 1996, RESTV once again killed numerous crab-eating macaques in Hazleton non-human primate facilities in Siena, Italy and Alice, Texas, respectively, after being imported from the same Philippine facility in Calamba that was implicated in the 1989 epizootic (Centers for Disease Control and Prevention 1996; Ciorba et al. 1997; Miranda et al. 1999, 2002; Rollin et al. 1999; Miranda and Miranda 2011). Depopulation of the facility terminated RESTV circulation (Miranda et al. 2000). From 2008 to 2009, RESTV was repeatedly isolated in the Philippines from several of hundreds of captive domestic pigs (Sus scrofa) dying of a respiratory and abortion disease and co-infected with a porcine arterivirus (porcine reproductive and respiratory disease syndrome virus) and/or circoviruses (Barrette et al. 2009). Whether RESTV caused or contributed to the observed clinical signs in the affected pigs remains unclear.
Only a few RESTV isolates have been obtained (Table 4). Reston virus/M.fascicularis-tc/USA/1989/Philippines89-Pennsylvania (RESTV/Phi89-Pen) was designated the type RESTV isolate (Kuhn et al. 2014).
Infection of domestic piglets with RESTV in the laboratory only resulted in viremia in the absence of disease signs, indicating that pigs or other suids may be able to maintain subclinical infections and, therefore, serve as RESTV reservoir hosts (Marsh et al. 2011). However, no evidence has been obtained thus far supporting RESTV infection in wild suids. RESTV is widely assumed to be apathogenic for humans. This assumption is based on the absence of recorded clinically overt human infections despite numerous possibilities for non-human primate-to-human or pig-to-human transmission during the various epizootics and on the detection of anti-RESTV antibodies in a few clinically healthy individuals that were exposed to RESTV (Center for Disease Control 1990a,b; Miranda et al. 1991; World Health Organization 2009; Miranda and Miranda 2011). Consequently, ecological studies focusing on RESTV have been sparse as the virus is frequently not considered as an immediate threat to humans as are most other filoviruses. Anti-RESTV antibodies were detected in pteropodid bats sampled in Africa (Ogawa et al. 2015), Bangladesh (Olival et al. 2013), China (Yuan et al. 2012) and the Philippines (Taniguchi et al. 2011; Jayme et al. 2015), and in orangutans in Indonesia (Niikura et al. 2001). Short RESTV NP gene-like fragments (519 bp) were amplified from samples taken from bats trapped in the Philippines (Jayme et al. 2015), and short RESTV L gene-like fragments could be detected by PCR in samples taken from domestic pigs in China (Pan et al. 2014). However, neither replicating RESTV isolates could be isolated from any sample nor could complete or coding-complete genomes be assembled to prove actual infection of these animals. Injection of RESTV into Egyptian rousettes did not result in virus replication (Jones et al. 2015). Consequently, Asian endemicity of RESTV remains a hypothesis at this time, and epizootics due to RESTV infection can neither be geographically nor temporally anticipated.
Clinical descriptions of RESTV infections are limited. Due to the co-infection with simian arteriviruses (non-human primate epizootics) or porcine arteriviruses and circoviruses (pig epizootic), clinical descriptions of the disease outbreaks (Dalgard et al. 1992; Geisbert et al. 1992; Hutchinson et al. 2001; Ikegami et al. 2002a) have to be viewed with extreme caution. Which virus caused which clinical sign and how the various co-infecting viruses interfered with each other remain unclear. Therefore, our understanding of the clinical presentation of RESTV infection relies on results of very few published experimental laboratory infections (Table 6). Initial experiments suggested that RESTV infection in crab-eating macaques presents clinically and pathologically similar to EBOV and SUDV infections, but progresses more slowly (death within 8–14 days after infection compared to 8 days, respectively), is less lethal (<100%), and results in lower viremia. Interestingly, whereas EBOV is known to also cause lethal infection in grivets (Chlorocebus aethiops), RESTV and SUDV do not (Fisher-Hoch et al. 1992; Jahrling et al. 1996b).
On the molecular-biological level, RESTV is arguably the best characterized of the neglected filoviruses, but very few studies have been published overall. The genome structure of RESTV is highly similar to other ebolaviruses (Ikegami et al. 2001; Groseth et al. 2002) (Fig. 2). Crystal structures have been determined for parts or the entire polypeptide chain of RESTV NP (Baker et al. 2016), VP35 (Leung et al. 2010), VP30 (Preston 1994) and VP24 (Reid et al. 2007; Zhang et al. 2012). In comparison to EBOV and MARV, RESTV was found to be less able to inhibit cellular type I IFN responses (Kash et al. 2006; Zhang et al. 2012). Just like EBOV, RESTV suppresses the type I interferon response through VP35 and VP24, and the type II interferon response through VP24 (Kash et al. 2006; Zhang et al. 2012). RESTV uses NPC1 for cell entry (Ng et al. 2015) and is dependent on cathepsin B (but not cathepsin L) for cell entry in vitro (Misasi et al. 2012). Akin to LLOV, RESTV Δ-peptide has little effect on MARV replication (Radoshitzky et al. 2011).
A RESTV minigenome system and RESTV reverse genetics have been used to study the RESTV lifecycle (Boehmann et al. 2005; Groseth et al. 2005). No reports on medical countermeasures against RESTV infection are available, most likely due to the perception that RESTV is not an imminent threat to humans. Diagnosis of RESTV infection is possible via pan-filovirus RT-PCR or qRT-PCR protocols (Lu et al. 2015) and RESTV-specific sequence capture probes for next-generation sequencing (Koehler et al. 2014). Several serological or nucleic-acid based RESTV-specific diagnostic systems have been described (Kalter et al. 1995; Ksiazek et al. 1999; Niikura et al. 2001; Ikegami et al. 2002b, 2003a,b; Ou et al. 2011). RESTV-specific monoclonal or polyclonal murine, rabbit or crab-eating macaque antibodies (Ou et al. 2011; Holtsberg et al. 2015; Keck et al. 2015; Wang et al. 2015), RESTV cross-reactive murine antibodies (Fusco et al. 2015; Wang et al. 2015; Furuyama et al. 2016), RESTV cross-reactive murine scFvs and IgNAR Vs raised against inactivated EBOV particles (Goodchild et al. 2011) are available.
Taï Forest virus
Like LLOV, Taï Forest virus (TAFV; pronounced tɑː'iː 'fɔːrɨst vɑɪrəs) is a filovirus that thus far has only been encountered once (Table 3). TAFV was discovered in 1994 through the infection of a Swiss ethologist during a necropsy she performed with two colleagues on a western chimpanzee (Pan troglodytes verus) in Taï National Park, western Côte d'Ivoire (Fig. 1) (Le Guenno et al. 1995; le Guenno, Formenty and Boesch 1999). The ape belonged to a troop under observation since 1979 whose numbers had been reduced by at least two episodes of epizootic hemorrhagic fever in November 1992 (8 deaths) and November 1994 (12 deaths). The 34-year-old ethologist, who performed the necropsy on 16 November 1994 to shed light on the etiological cause of these episodes, developed a febrile disease 8 days later. On 26 November, she was hospitalized in Abidjan, and on 1 December she was transported to a hospital in Basel, Switzerland, where she recovered. Filovirus infection was confirmed by electron-microscopic and serological methods (ELISA and IFA) and by virus isolation in tissue culture (Le Guenno et al. 1995). Molecular characterization identified TAFV as a distinct filovirus most closely related to BDBV with a genome organization similar to all ebolaviruses (Fig. 2), an ebolavirus that only was encountered in Eastern Africa ≈4200 km away (Towner et al. 2008). Taï Forest virus/H.sapiens-tc/CIV/1994/Pauléoula-CI (TAFV/Pau-CI) was designated the type (and only) isolate of TAFV (Table 4) (Kuhn et al. 2014).
Histopathological examination of tissues from the western chimpanzee necropsied by the ethologist strongly indicated that the ethologist acquired TAFV from this animal. Lesions resembled those found in macaques experimentally infected with EBOV: multifocal necroses infiltrated with inflammatory cells, Kupffer cell hyperplasia in the liver, diffuse fibrinoid and hemorrhagic necrosis in the splenic red pulp and lymphoid depletion in all lymphatic tissues. More importantly, macrophages in various affected tissues reacted with TAFV-specific and TAFV-cross-reactive antibodies. However, typical hemorrhagic, thrombotic or vascular lesions of EBOV infection were not present in the ape (Wyers et al. 1999), and ultimate confirmation of TAFV infection in chimpanzees by either virus isolation or next-generation sequencing is lacking. Likewise, how the chimpanzees may have become infected is unclear. Whereas MARV and RAVV subclinically infect bats in nature, and data for EBOV–bat associations are suggestive (Wahl-Jensen et al. 2013), TAFV does not replicate in Egyptian rousette bats (Jones et al. 2015). No clues have been found in regard to TAFV ecology other than possibly TAFV-specific antibodies in Indonesian orangutans (Nidom et al. 2012).
Because of the recording of only a single case, the clinical presentation of EVD due to TAFV infection is unclear. The TAFV-infected ethologist developed fever, headaches, myalgia, chills, cough, abdominal pain and nausea accompanied with acute non-bloody diarrhea and vomiting, a generalized maculopapular rash and hematuria. Clinical tests revealed proteinuria, marked liver enzymes (aspartate aminotransferase, alanine aminotransferase) and lactate dehydrogenase elevations, thrombocytopenia, lymphopenia and neutrophilia, and suggested DIC. The woman was released on day 15 after disease onset and recovered without sequelae other than temporary hair loss (Le Guenno et al. 1995; le Guenno, Formenty and Boesch 1999; Formenty et al. 1999). Thus, the disease resembled non-lethal EVD caused by BDBV (Table 5) and other ebolaviruses and MVD. The CFR of TAFV infection cannot be determined from a single survivor. The molecular and immunological responses to human TAFV infection have not been studied in detail. Consequently, neither cytokine response nor other biomarker data are available. Since the patient fortunately survived and no biopsies had been performed, there are no human pathological data.
Establishment of experimental disease caused by TAFV is currently only possible using a partially lethal crab-eating macaque model (Table 6), thereby development of medical countermeasures is challenging. Consequently, results from only a single study on a candidate vaccine for the prevention of TAFV are ambiguous (Table 7), and no TAFV-specific antivirals have been brought forward. Molecular-biological studies on TAFV are close to absent with the exception of the determination of the crystal structure of the TAFV NP C-terminal domain (Baker et al. 2016) and the characterization of TAFV GP1,2-mediated cell-entry (NPC1 and cathepsin B) requirements (Misasi et al. 2012; Ng et al. 2015). Minigenomes and reverse genetics systems have yet to be established for TAFV. TAFV detection is possible via pan-filovirus RT-PCR (Lu et al. 2015) and TAFV-specific sequence capture probes for next-generation sequencing (Koehler et al. 2014). Very few TAFV-specific antibodies have been described (Ou et al. 2011; Furuyama et al. 2016), including ebolavirus cross-reactive llama single-domain antibodies (Sherwood and Hayhurst 2013). TAFV-specific serological diagnostic assays have not yet been described.
Other filoviruses
The geographically broad distribution of filoviruses (western to eastern Equatorial Africa, Philippines and Spain) suggests that many filoviruses remain to be discovered. Supporting this hypothesis, He et al. (2015) recently described the amplification of filovirus NP-, VP35- and L-like nucleic acids from (frugivorous) Leschenault's rousette bats (Rousettus leschenaultii) captured in Yúnnán Province, China. Whereas most fragments were too short (129–354 bp) to unambiguously assign them to the family Filoviridae, two fragments from the same bat (‘Bt-DH04‘) could be extended to 2750 and 2682-bp length, respectively. The first fragment aligned with the 3′ end of a filoviral NP and almost an entire filoviral VP35-like gene; the second covered a large portion of a filoviral L-like gene. Phylogenetically, these fragments represent a novel clade of filoviruses basal to ebolaviruses and in between ebolaviruses and marburgviruses/cuevaviruses.
CONCLUSIONS
The ecology of filoviruses needs to be defined, and the natural host reservoirs of cuevaviruses and ebolaviruses are still unclear (Wahl-Jensen et al. 2013). The discovery of Egyptian rousettes as natural host reservoirs of marburgviruses in Ugandan caves (Towner et al. 2009) was a major step forward in filovirology, but questions remain as to why MARV and RAVV cannot be found in other Egyptian rousette populations in and outside of Africa. Because of these uncertainties, how and under which circumstances filovirus host-human transmission occurs are unclear. The geographic distribution of filoviruses (i.e. their endemicity) remains undefined despite several ecological filovirus niche modeling studies (Peterson, Bauer and Mills 2004; Peterson et al. 2006; Pigott et al. 2014, 2015). Finally, the discovery of LLOV in Spain (Negredo et al. 2011) and the detection of filovirus-like entities in Chinese bat populations (He et al. 2015) indicate that the family Filoviridae is undersampled. Its members are probably much more diverse and distributed than previously thought.
Because of this lack of ecological knowledge, prediction of when and where human and/or animal filovirus disease outbreaks may occur is impossible. Since knowledge on neglected filoviruses (BDBV, LLOV, RAVV, RESTV and TAFV) is extremely limited, one cannot exclude the possibility of future large EVD or MVD outbreaks caused by these viruses. The recent EVD/EBOV outbreak in Western Africa, involving 28 646 cases and 11 323 deaths, demonstrates for the first time that EVD outbreaks do not necessarily remain geographically confined or involve only dozens to several hundreds of cases (Table 3). Yet, a disease outbreak of the magnitude of that EVD outbreak caused by a neglected filovirus may prove even more disastrous to Africa or the world. Among the already very low global number of research institutes that are permitted to perform biosafety level 4 research on filoviruses, few even have access to neglected filoviruses. Reagents and assays for neglected filoviruses are not available or extremely limited, especially for commercial products, which primarily fulfill the need for research on EBOV, SUDV and MARV research. The only commerical reagents for neglected filoviruses include: irradiated BDBV, RAVV, RESTV and TAFV or isolated genomic RNA; recombinant complete, partial or tagged GP1,2 (BDBV, RESTV, TAFV), VP40 (TAFV), VP24 (RESTV); polyclonal rabbit anti-GP1,2 antibodies (BDBV, RESTV, TAFV); and monoclonal antibodies against VP40 (BDBV); commercial ELISA systems for the detection of circulating guinea pig, human, non-human primate and pig antibodies (RESTV) and circulating GP1,2 antigen (BDBV, RESTV, TAFV); and qPCR systems for RESTV and TAFV (all based on catalog searches of Alpha Diagnostics, BEI Resources, Integrated BioTherapeutics, Genesig and Sino Biological; Anthony and Bradfute 2015). Animal models for neglected filovirus infections are either absent, not 100% lethal, or not established in non-human primates (Table 6). Consequently, there are few specific candidate vaccines and almost no specific therapeutics in the pipeline to prevent or treat neglected filovirus infections (Tables 7 and 8).
Therefore, we appeal to the international filovirus research community, and even more so to the funders of filovirus (currently almost exclusively EBOV) research and development activities, to create and maintain a global, coordinated and highly collaborative program to prospectively create basic reagents, assays, methodologies, databases, animal models and medical countermeasure platforms that include neglected filoviruses on a routine basis.
FUNDING
This work was supported in part through Battelle Memorial Institute's prime contract with the US National Institute of Allergy and Infectious Diseases (NIAID) under Contract No. HHSN272200700016I. LB, JCJ and JW performed this work as employees of Battelle Memorial Institute. A subcontractor to Battelle Memorial Institute who performed this work is: JHK, an employee of Tunnell Government Services, Inc. RB is supported by the Hartmut Hoffmann-Berling International Graduate School of Molecular and Cellular Biology (HBIGS). The content of this publication does not necessarily reflect the views or policies of the US Department of Health and Human Services, or the institutions and companies affiliated with the authors.
Conflict of interest. None declared.
REFERENCES
- Adjemian J, Farnon EC, Tschioko F, et al. Outbreak of Marburg hemorrhagic fever among miners in Kamwenge and Ibanda Districts, Uganda, 2007. J Infect Dis. 2011;204(Suppl 3):S796–9. doi: 10.1093/infdis/jir312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonso CL, Amarasinghe GK, Bányai K, et al. Taxonomy of the order Mononegavirales: update 2016. Arch Virol. 2016 doi: 10.1007/s00705-016-2880-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albariño CG, Shoemaker T, Khristova ML, et al. Genomic analysis of filoviruses associated with four viral hemorrhagic fever outbreaks in Uganda and the Democratic Republic of the Congo in 2012. Virology. 2013;442:97–100. doi: 10.1016/j.virol.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves DA, Glynn AR, Steele KE, et al. Aerosol exposure to the Angola strain of Marburg virus causes lethal viral hemorrhagic fever in cynomolgus macaques. Vet Pathol. 2010;47:831–51. doi: 10.1177/0300985810378597. [DOI] [PubMed] [Google Scholar]
- Amman BR, Carroll SA, Reed ZD, et al. Seasonal pulses of Marburg virus circulation in juvenile Rousettus aegyptiacus bats coincide with periods of increased risk of human infection. PLoS Pathog. 2012;8:e1002877. doi: 10.1371/journal.ppat.1002877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amman BR, Jones ME, Sealy TK, et al. Oral shedding of Marburg virus in experimentally infected Egyptian fruit bats (Rousettus aegyptiacus) J Wildlife Dis. 2015;51:113–24. doi: 10.7589/2014-08-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amman BR, Nyakarahuka L, McElroy AK, et al. Marburgvirus resurgence in Kitaka mine bat population after extermination attempts, Uganda. Emerg Infect Dis. 2014;20:1761–4. doi: 10.3201/eid2010.140696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony SM, Bradfute SB. Filoviruses: one of these things is (not) like the other. Viruses. 2015;7:5172–90. doi: 10.3390/v7102867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bah EI, Lamah MC, Fletcher T, et al. Clinical presentation of patients with Ebola virus disease in Conakry, Guinea. New Engl J Med. 2015;372:40–7. doi: 10.1056/NEJMoa1411249. [DOI] [PubMed] [Google Scholar]
- Baize S, Leroy EM, Georges AJ, et al. Inflammatory responses in Ebola virus-infected patients. Clin Exp Immunol. 2002;128:163–8. doi: 10.1046/j.1365-2249.2002.01800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baize S, Leroy EM, Georges-Courbot MC, et al. Defective humoral responses and extensive intravascular apoptosis are associated with fatal outcome in Ebola virus-infected patients. Nat Med. 1999;5:423–6. doi: 10.1038/7422. [DOI] [PubMed] [Google Scholar]
- Baker LE, Ellena JF, Handing KB, et al. Molecular architecture of the nucleoprotein C-terminal domain from the Ebola and Marburg viruses. Acta Crystallogr D. 2016;72:49–58. doi: 10.1107/S2059798315021439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltzer G, Slenczka W, Stöppler L, et al. Marburg-Virus-Krankheit. Verlaufsbeobachtungen über 12 Jahre (1967–1979) [Marburg virus disease. Long-term observations over 12 years (1967–1979)] In: Schlegel B, editor. Verhandlungen der Deutschen Gesellschaft für Innere Medizin [Proceedings of the German Society of Internal Medicine] Munich, Germany: J. F. Bergmann; 1979. pp. 1203–6. [German] [Google Scholar]
- Bao Y, Chetvernin V, Tatusova T. PAirwise Sequence Comparison (PASC) and its application in the classification of filoviruses. Viruses. 2012;4:1318–27. doi: 10.3390/v4081318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrette RW, Metwally SA, Rowland JM, et al. Discovery of swine as a host for the Reston ebolavirus. Science. 2009;325:204–6. doi: 10.1126/science.1172705. [DOI] [PubMed] [Google Scholar]
- Barry M, Traore FA, Sako FB, et al. Ebola outbreak in Conakry, Guinea: epidemiological, clinical, and outcome features. Med Mal Infect. 2014;44:491–4. doi: 10.1016/j.medmal.2014.09.009. [DOI] [PubMed] [Google Scholar]
- Baskerville A, Bowen ET, Platt GS, et al. The pathology of experimental Ebola virus infection in monkeys. J Pathol. 1978;125:131–8. doi: 10.1002/path.1711250303. [DOI] [PubMed] [Google Scholar]
- Bausch DG, Nichol ST, Muyembe-Tamfum JJ, et al. Marburg hemorrhagic fever associated with multiple genetic lineages of virus. New Engl J Med. 2006;355:909–19. doi: 10.1056/NEJMoa051465. [DOI] [PubMed] [Google Scholar]
- Bazhutin NB, Belanov EF, Spiridonov VA, et al. The effect of the methods for producing an experimental Marburg virus infection on the characteristics of the course of the disease in green monkeys. Vop Virusol. 1992;37:153–6. [Russian] [PubMed] [Google Scholar]
- Bechtelsheimer H, Korb G, Gedigk P. Die ‘Marburg-Virus’-Hepatitis - Untersuchungen bei Menschen und Meerschweinchen [The ‘Marburg-virus’-hepatitis. Studies in man and guinea pigs] Virchows Arch A. 1970;351:273–90. [German] [PubMed] [Google Scholar]
- Bermejo M, Rodríguez-Teijeiro JD, Illera G, et al. Ebola outbreak killed 5000 gorillas. Science. 2006;314:1564. doi: 10.1126/science.1133105. [DOI] [PubMed] [Google Scholar]
- Blakey SM, Reuman L, Jacoby RJ, et al. Tracing ‘Fearbola’: psychological predictors of anxious responding to the threat of Ebola. Cognitive Ther Res. 2015;39:816–25. doi: 10.1007/s10608-015-9701-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehmann Y, Enterlein S, Randolf A, et al. A reconstituted replication and transcription system for Ebola virus Reston and comparison with Ebola virus Zaire. Virology. 2005;332:406–17. doi: 10.1016/j.virol.2004.11.018. [DOI] [PubMed] [Google Scholar]
- Brannan JM, Froude JW, Prugar LI, et al. Interferon alpha/beta receptor-deficient mice as a model for Ebola virus disease. J Infect Dis. 2015;212(Suppl 2):S282–94. doi: 10.1093/infdis/jiv215. [DOI] [PubMed] [Google Scholar]
- Bray M. The role of the Type I interferon response in the resistance of mice to filovirus infection. J Gen Virol. 2001;82:1365–73. doi: 10.1099/0022-1317-82-6-1365. [DOI] [PubMed] [Google Scholar]
- Bray M, Davis K, Geisbert T, et al. A mouse model for evaluation of prophylaxis and therapy of Ebola hemorrhagic fever. J Infect Dis. 1998;178:651–61. doi: 10.1086/515386. [DOI] [PubMed] [Google Scholar]
- Bray M, Hatfill S, Hensley L, et al. Haematological, biochemical and coagulation changes in mice, guinea-pigs and monkeys infected with a mouse-adapted variant of Ebola Zaire virus. J Comp Pathol. 2001;125:243–53. doi: 10.1053/jcpa.2001.0503. [DOI] [PubMed] [Google Scholar]
- Bukreyev AA, Chandran K, Dolnik O, et al. Discussions and decisions of the 2012–2014 International Committee on Taxonomy of Viruses (ICTV) Filoviridae Study Group, January 2012-June 2013. Arch Virol. 2014;159:821–30. doi: 10.1007/s00705-013-1846-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bwaka MA, Bonnet MJ, Calain P, et al. Ebola hemorrhagic fever in Kikwit, Democratic Republic of the Congo: clinical observations in 103 patients. J Infect Dis. 1999;179(Suppl 1):S1–7. doi: 10.1086/514308. [DOI] [PubMed] [Google Scholar]
- Carrion R, Jr, Ro Y, Hoosien K, et al. A small nonhuman primate model for filovirus-induced disease. Virology. 2011;420:117–24. doi: 10.1016/j.virol.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SA, Towner JS, Sealy TK, et al. Molecular evolution of viruses of the family Filoviridae based on 97 whole-genome sequences. J Virol. 2013;87:2608–16. doi: 10.1128/JVI.03118-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Center for Disease Control. Update: filovirus infection in animal handlers. MMWR Morb Mortal Wkly Rep. 1990a;39:221. [PubMed] [Google Scholar]
- Center for Disease Control. Update: filovirus infections among persons with occupational exposure to nonhuman primates. MMWR Morb Mortal Wkly Rep. 1990b;39:266–7. 273. [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. Ebola-Reston virus infection among quarantined nonhuman primates - Texas, 1996. MMWR Morb Mort Wkly Rep. 1996;45:314–6. [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. Ebola Outbreaks 2000–2014. 2015. http://www.cdc.gov/vhf/ebola/outbreaks/history/summaries.html (5 April 2016, date last accessed) [Google Scholar]
- Ciorba A, Matteucci G, Perini L, et al. Infezione da virus Ebola nella scimmia: Reperti clinici et anatomoistopatologici. Osservati nel corso del primo episidio verificatosi in Europa [Ebola virus infections of monkeys. Report of the clinical and anatomic-histopathologic observations in the course of the first verified outbreak in Europe] Veterinaria (Cremona) 1997;11:109–12. [Italian] [Google Scholar]
- Clark DV, Kibuuka H, Millard M, et al. Long-term sequelae after Ebola virus disease in Bundibugyo, Uganda: a retrospective cohort study. Lancet Infect Dis. 2015;15:905–12. doi: 10.1016/S1473-3099(15)70152-0. [DOI] [PubMed] [Google Scholar]
- Connolly BM, Steele KE, Davis KJ, et al. Pathogenesis of experimental Ebola virus infection in guinea pigs. J Infect Dis. 1999;179(Suppl 1):S203–17. doi: 10.1086/514305. [DOI] [PubMed] [Google Scholar]
- Cross RW, Fenton KA, Geisbert JB, et al. Comparison of the pathogenesis of the Angola and Ravn Strains of Marburg virus in the outbred guinea pig model. J Infect Dis. 2015;212(Suppl 2):S258–70. doi: 10.1093/infdis/jiv182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daddario-DiCaprio KM, Geisbert TW, Geisbert JB, et al. Cross-protection against Marburg virus strains by using a live, attenuated recombinant vaccine. J Virol. 2006a;80:9659–66. doi: 10.1128/JVI.00959-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daddario-DiCaprio KM, Geisbert TW, Ströher U, et al. Postexposure protection against Marburg haemorrhagic fever with recombinant vesicular stomatitis virus vectors in non-human primates: an efficacy assessment. Lancet. 2006b;367:1399–404. doi: 10.1016/S0140-6736(06)68546-2. [DOI] [PubMed] [Google Scholar]
- Dalgard DW, Hardy RJ, Pearson SL, et al. Combined simian hemorrhagic fever and Ebola virus infection in cynomolgus monkeys. Lab Anim Sci. 1992;42:152–7. [PubMed] [Google Scholar]
- Dallatomasina S, Crestani R, Sylvester Squire J, et al. Ebola outbreak in rural West Africa: epidemiology, clinical features and outcomes. Trop Med Int Health. 2015;20:448–54. doi: 10.1111/tmi.12454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E, Munster VJ, Metwally SA, et al. Assessment of rodents as animal models for Reston ebolavirus. J Infect Dis. 2011;204(Suppl 3):S968–72. doi: 10.1093/infdis/jir330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebihara H, Zivcec M, Gardner D, et al. A Syrian golden hamster model recapitulating Ebola hemorrhagic fever. J Infect Dis. 2013;207:306–18. doi: 10.1093/infdis/jis626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egbring R, Slenczka W, Baltzer G. Clinical syndrome. In: Martini G, Siegert R, editors. Marburg Virus Disease. New York: Springer; 1971. pp. 41–9. [Google Scholar]
- Eichner M, Dowell SF, Firese N. Incubation period of Ebola hemorrhagic virus subtype Zaire. Osong Public Health Res Perspect. 2011;2:3–7. doi: 10.1016/j.phrp.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis DS, Bowen ET, Simpson DI, et al. Ebola virus: a comparison, at ultrastructural level, of the behaviour of the Sudan and Zaire strains in monkeys. Brit J Exp Pathol. 1978;59:584–93. [PMC free article] [PubMed] [Google Scholar]
- Falzarano D, Feldmann F, Grolla A, et al. Single immunization with a monovalent vesicular stomatitis virus-based vaccine protects nonhuman primates against heterologous challenge with Bundibugyo ebolavirus. J Infect Dis. 2011;204(Suppl 3):S1082–9. doi: 10.1093/infdis/jir350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feagins AR, Basler CF. Lloviu virus VP24 and VP35 proteins function as innate immune antagonists in human and bat cells. Virology. 2015;485:145–52. doi: 10.1016/j.virol.2015.07.010. [DOI] [PubMed] [Google Scholar]
- Fenimore PW, Muhammad MA, Fischer WM, et al. Designing and testing broadly-protective filoviral vaccines optimized for cytotoxic T-lymphocyte epitope coverage. PLoS One. 2012;7:e44769. doi: 10.1371/journal.pone.0044769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher-Hoch SP, Brammer TL, Trappier SG, et al. Pathogenic potential of filoviruses: role of geographic origin of primate host and virus strain. J Infect Dis. 1992;166:753–63. doi: 10.1093/infdis/166.4.753. [DOI] [PubMed] [Google Scholar]
- Fisher-Hoch SP, Platt GS, Neild GH, et al. Pathophysiology of shock and hemorrhage in a fulminating viral infection (Ebola) J Infect Dis. 1985;152:887–94. doi: 10.1093/infdis/152.5.887. [DOI] [PubMed] [Google Scholar]
- Formenty P, Hatz C, Le Guenno B, et al. Human infection due to Ebola virus, subtype Côte d'Ivoire: clinical and biologic presentation. J Infect Dis. 1999;179(Suppl 1):S48–53. doi: 10.1086/514285. [DOI] [PubMed] [Google Scholar]
- Fritz EA, Geisbert JB, Geisbert TW, et al. Cellular immune response to Marburg virus infection in cynomolgus macaques. Viral Immunol. 2008;21:355–63. doi: 10.1089/vim.2008.0023. [DOI] [PubMed] [Google Scholar]
- Furuyama W, Marzi A, Nanbo A, et al. Discovery of an antibody for pan-ebolavirus therapy. Sci Rep. 2016;6:20514. doi: 10.1038/srep20514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusco ML, Hashiguchi T, Cassan R, et al. Protective mAbs and cross-reactive mAbs raised by immunization with engineered Marburg virus GPs. PLoS Pathog. 2015;11:e1005016. doi: 10.1371/journal.ppat.1005016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisbert TW, Daddario-DiCaprio KM, Geisbert JB, et al. Marburg virus Angola infection of rhesus macaques: pathogenesis and treatment with recombinant nematode anticoagulant protein c2. J Infect Dis. 2007;196(Suppl 2):S372–81. doi: 10.1086/520608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisbert TW, Geisbert JB, Leung A, et al. Single-injection vaccine protects nonhuman primates against infection with Marburg virus and three species of Ebola virus. J Virol. 2009;83:7296–304. doi: 10.1128/JVI.00561-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisbert TW, Hensley LE, Jahrling PB, et al. Treatment of Ebola virus infection with a recombinant inhibitor of factor VIIa/tissue factor: a study in rhesus monkeys. Lancet. 2003a;362:1953–8. doi: 10.1016/S0140-6736(03)15012-X. [DOI] [PubMed] [Google Scholar]
- Geisbert TW, Hensley LE, Kagan E, et al. Postexposure protection of guinea pigs against a lethal Ebola virus challenge is conferred by RNA interference. J Infect Dis. 2006;193:1650–7. doi: 10.1086/504267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisbert TW, Hensley LE, Larsen T, et al. Pathogenesis of Ebola hemorrhagic fever in cynomolgus macaques: evidence that dendritic cells are early and sustained targets of infection. Am J Pathol. 2003b;163:2347–70. doi: 10.1016/S0002-9440(10)63591-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisbert TW, Jahrling PB. Use of immunoelectron microscopy to show Ebola virus during the 1989 United States epizootic. J Clin Pathol. 1990;43:813–6. doi: 10.1136/jcp.43.10.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisbert TW, Jahrling PB, Hanes MA, et al. Association of Ebola-related Reston virus particles and antigen with tissue lesions of monkeys imported to the United States. J Comp Pathol. 1992;106:137–52. doi: 10.1016/0021-9975(92)90043-t. [DOI] [PubMed] [Google Scholar]
- Goodchild SA, Dooley H, Schoepp RJ, et al. Isolation and characterisation of Ebolavirus-specific recombinant antibody fragments from murine and shark immune libraries. Mol Immunol. 2011;48:2027–37. doi: 10.1016/j.molimm.2011.06.437. [DOI] [PubMed] [Google Scholar]
- Grant-Klein RJ, Van Deusen NM, Badger CV, et al. A multiagent filovirus DNA vaccine delivered by intramuscular electroporation completely protects mice from ebola and Marburg virus challenge. Hum Vaccin Immunother. 2012;8:1703–6. doi: 10.4161/hv.21873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groseth A, Feldmann H, Theriault S, et al. RNA polymerase I-driven minigenome system for Ebola viruses. J Virol. 2005;79:4425–33. doi: 10.1128/JVI.79.7.4425-4433.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groseth A, Ströher U, Theriault S, et al. Molecular characterization of an isolate of the 1989/90 epizootic of Ebola virus Reston among macaques imported into the United States. Virus Res. 2002;87:155–63. doi: 10.1016/s0168-1702(02)00087-4. [DOI] [PubMed] [Google Scholar]
- Gupta M, Goldsmith CS, Metcalfe MG, et al. Reduced virus replication, proinflammatory cytokine production, and delayed macrophage cell death in human PBMCs infected with the newly discovered Bundibugyo ebolavirus relative to Zaire ebolavirus. Virology. 2010;402:203–8. doi: 10.1016/j.virol.2010.03.024. [DOI] [PubMed] [Google Scholar]
- Gupta M, MacNeil A, Reed ZD, et al. Serology and cytokine profiles in patients infected with the newly discovered Bundibugyo ebolavirus. Virology. 2012;423:119–24. doi: 10.1016/j.virol.2011.11.027. [DOI] [PubMed] [Google Scholar]
- Gupta M, Mahanty S, Bray M, et al. Passive transfer of antibodies protects immunocompetent and immunodeficient mice against lethal Ebola virus infection without complete inhibition of viral replication. J Virol. 2001;75:4649–54. doi: 10.1128/JVI.75.10.4649-4654.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes CG, Burans JP, Ksiazek TG, et al. Outbreak of fatal illness among captive macaques in the Philippines caused by an Ebola-related filovirus. Am J Trop Med Hyg. 1992;46:664–71. doi: 10.4269/ajtmh.1992.46.664. [DOI] [PubMed] [Google Scholar]
- He B, Feng Y, Zhang H, et al. Filovirus RNA in fruit bats, China. Emerg Infect Dis. 2015;21:1675–7. doi: 10.3201/eid2109.150260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensley LE, Alves DA, Geisbert JB, et al. Pathogenesis of Marburg hemorrhagic fever in cynomolgus macaques. J Infect Dis. 2011;204(Suppl 3):S1021–31. doi: 10.1093/infdis/jir339. [DOI] [PubMed] [Google Scholar]
- Hensley LE, Mulangu S, Asiedu C, et al. Demonstration of cross-protective vaccine immunity against an emerging pathogenic ebolavirus species. PLoS Pathog. 2010;6:e1000904. doi: 10.1371/journal.ppat.1000904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hevey M, Negley D, Geisbert J, et al. Antigenicity and vaccine potential of Marburg virus glycoprotein expressed by baculovirus recombinants. Virology. 1997;239:206–16. doi: 10.1006/viro.1997.8883. [DOI] [PubMed] [Google Scholar]
- Holtsberg FW, Shulenin S, Vu H, et al. Pan-ebolavirus and pan-filovirus mouse monoclonal antibodies: protection against Ebola and Sudan viruses. J Virol. 2015;90:266–78. doi: 10.1128/JVI.02171-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijbregts B, De Wachter P, Obiang LSN, et al. Ebola and the decline of gorilla Gorilla gorilla and chimpanzee Pan troglodytes populations in Minkebe Forest, north-eastern Gabon. Oryx. 2003;37:437–43. [Google Scholar]
- Hutchinson KL, Villinger F, Miranda ME, et al. Multiplex analysis of cytokines in the blood of cynomolgus macaques naturally infected with Ebola virus (Reston serotype) J Med Virol. 2001;65:561–6. [PubMed] [Google Scholar]
- Ikegami T, Calaor AB, Miranda ME, et al. Genome structure of Ebola virus subtype Reston: differences among Ebola subtypes. Arch Virol. 2001;146:2021–7. doi: 10.1007/s007050170049. [DOI] [PubMed] [Google Scholar]
- Ikegami T, Miranda ME, Calaor AB, et al. Histopathology of natural Ebola virus subtype Reston infection in cynomolgus macaques during the Philippine outbreak in 1996. Exp Anim. 2002a;51:447–55. doi: 10.1538/expanim.51.447. [DOI] [PubMed] [Google Scholar]
- Ikegami T, Niikura M, Saijo M, et al. Antigen capture enzyme-linked immunosorbent assay for specific detection of Reston Ebola virus nucleoprotein. Clin Diagn Lab Immunol. 2003a;10:552–7. doi: 10.1128/CDLI.10.4.552-557.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami T, Saijo M, Niikura M, et al. Development of an immunofluorescence method for the detection of antibodies to Ebola virus subtype Reston by the use of recombinant nucleoprotein-expressing HeLa Cells. Microbiol Immunol. 2002b;46:633–8. doi: 10.1111/j.1348-0421.2002.tb02745.x. [DOI] [PubMed] [Google Scholar]
- Ikegami T, Saijo M, Niikura M, et al. Immunoglobulin G enzyme-linked immunosorbent assay using truncated nucleoproteins of Reston Ebola virus. Epidemiol Infect. 2003b;130:533–9. [PMC free article] [PubMed] [Google Scholar]
- Ilinykh PA, Shen X, Flyak AI, et al. Chimeric filoviruses for identification and characterization of monoclonal antibodies. J Virol. 2016;90:3890–901. doi: 10.1128/JVI.00101-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaäcson M, Sureau P, Courteille G, et al. Clinical aspects of Ebola virus disease at the Ngaliema Hospital, Kinshasa, Zaire, 1976. In: Pattyn SR, editor. Ebola Virus Haemorrhagic Fever. Amsterdam, The Netherlands: Elsevier/North-Holland Biomedical Press; 1978. pp. 15–20. [Google Scholar]
- Iversen PL, Warren TK, Wells JB, et al. Discovery and early development of AVI-7537 and AVI-7288 for the treatment of Ebola virus and Marburg virus infections. Viruses. 2012;4:2806–30. doi: 10.3390/v4112806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaax N, Jahrling P, Geisbert T, et al. Transmission of Ebola virus (Zaire strain) to uninfected control monkeys in a biocontainment laboratory. Lancet. 1995;346:1669–71. doi: 10.1016/s0140-6736(95)92841-3. [DOI] [PubMed] [Google Scholar]
- Jahrling PB, Geisbert J, Swearengen JR, et al. Passive immunization of Ebola virus-infected cynomolgus monkeys with immunoglobulin from hyperimmune horses. Arch Virol Suppl. 1996a;11:135–40. doi: 10.1007/978-3-7091-7482-1_12. [DOI] [PubMed] [Google Scholar]
- Jahrling PB, Geisbert TW, Dalgard DW, et al. Preliminary report: isolation of Ebola virus from monkeys imported to USA. Lancet. 1990;335:502–5. doi: 10.1016/0140-6736(90)90737-p. [DOI] [PubMed] [Google Scholar]
- Jahrling PB, Geisbert TW, Jaax NK, et al. Experimental infection of cynomolgus macaques with Ebola-Reston filoviruses from the 1989–1990 U.S. epizootic. In: Schwarz TF, Siegl G, editors. Imported Virus Infections. Vienna, Austria: Springer; 1996b. pp. 115–34. Vol. 11. [DOI] [PubMed] [Google Scholar]
- Jayme SI, Field HE, de Jong C, et al. Molecular evidence of Ebola Reston virus infection in Philippine bats. Virol J. 2015;12:107. doi: 10.1186/s12985-015-0331-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E, Jaax N, White J, et al. Lethal experimental infections of rhesus monkeys by aerosolized Ebola virus. Int J Exp Pathol. 1995;76:227–36. [PMC free article] [PubMed] [Google Scholar]
- Johnson ED, Johnson BK, Silverstein D, et al. Characterization of a new Marburg virus isolate from a 1987 fatal case in Kenya. In: Schwarz TF, Siegl G, editors. Imported Virus Infections. Vienna, Austria: Springer; 1996. pp. 101–14. Vol. 11. [DOI] [PubMed] [Google Scholar]
- Jones ME, Schuh AJ, Amman BR, et al. Experimental inoculation of Egyptian rousette bats (Rousettus aegyptiacus) with viruses of the Ebolavirus and Marburgvirus genera. Viruses. 2015;7:3420–42. doi: 10.3390/v7072779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalter SS, Heberling RL, Barry JD, et al. Detection of Ebola-Reston (Filoviridae) virus antibody by dot-immunobinding assay. Lab Anim Sci. 1995;45:523–5. [PubMed] [Google Scholar]
- Kash JC, Mühlberger E, Carter V, et al. Global suppression of the host antiviral response by ebola- and marburgviruses: increased antagonism of the Type I interferon response is associated with enhanced virulence. J Virol. 2006;80:3009–20. doi: 10.1128/JVI.80.6.3009-3020.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keck ZY, Enterlein SG, Howell KA, et al. Macaque monoclonal antibodies targeting novel conserved epitopes within filovirus glycoprotein. J Virol. 2015;90:279–91. doi: 10.1128/JVI.02172-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kibadi K, Mupapa K, Kuvula K, et al. Late ophthalmologic manifestations in survivors of the 1995 Ebola virus epidemic in Kikwit, Democratic Republic of the Congo. J Infect Dis. 1999;179(Suppl 1):S13–4. doi: 10.1086/514288. [DOI] [PubMed] [Google Scholar]
- Kobinger GP, Leung A, Neufeld J, et al. Replication, pathogenicity, shedding, and transmission of Zaire ebolavirus in pigs. J Infect Dis. 2011;204:200–8. doi: 10.1093/infdis/jir077. [DOI] [PubMed] [Google Scholar]
- Koehler JW, Hall AT, Rolfe PA, et al. Development and evaluation of a panel of filovirus sequence capture probes for pathogen detection by next-generation sequencing. PLoS One. 2014;9:e107007. doi: 10.1371/journal.pone.0107007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korb G, Slenczka W, Bechtelsheimer H, et al. Die ‘Marburg-Virus’-Hepatitis im Tierexperiment - Versuche an Meerschweinchen [The ‘Marburg virus’ hepatitis in the animal model - experiments with guinea pigs] Virchows Arch A. 1971;353:169–84. [German] [PubMed] [Google Scholar]
- Kozak RA, Kobinger GP. Vaccines against ‘the other’ Ebolavirus species. Expert Rev Vaccines. 2016:1–8. doi: 10.1586/14760584.2016.1170597. [DOI] [PubMed] [Google Scholar]
- Kratz T, Roddy P, Tshomba Oloma A, et al. Ebola virus disease outbreak in Isiro, Democratic Republic of the Congo, 2012: signs and symptoms, management and outcomes. PLoS One. 2015;10:e0129333. doi: 10.1371/journal.pone.0129333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek TG, West CP, Rollin PE, et al. ELISA for the detection of antibodies to Ebola viruses. J Infect Dis. 1999;179(Suppl 1):S192–8. doi: 10.1086/514313. [DOI] [PubMed] [Google Scholar]
- Kuhn JH. Filoviruses. A compendium of 40 years of epidemiological, clinical, and laboratory studies. Arch Virol Suppl. 2008;20:13–360. [PubMed] [Google Scholar]
- Kuhn JH. Ebolavirus and marburgvirus infections. In: Kasper DL, Fauci AS, Hauser SL, et al., editors. Harrison's Principles of Internal Medicine. 19th edn. Columbus: McGraw-Hill Education; 2015. pp. 1323–9. Vol. 2. [Google Scholar]
- Kuhn JH, Andersen KG, Bao Y, et al. Filovirus RefSeq entries: evaluation and selection of filovirus type variants, type sequences, and names. Viruses. 2014;6:3663–82. doi: 10.3390/v6093663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn JH, Becker S, Ebihara H, et al. Proposal for a revised taxonomy of the family Filoviridae: classification, names of taxa and viruses, and virus abbreviations. Arch Virol. 2010;155:2083–103. doi: 10.1007/s00705-010-0814-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn JH, Becker S, Ebihara H, et al. Family Filoviridae. In: King AMQ, Adams MJ, Carstens EB, et al., editors. Virus Taxonomy—Ninth Report of the International Committee on Taxonomy of Viruses. London: Elsevier/Academic Press; 2011. pp. 665–71. [Google Scholar]
- Lado M, Walker NF, Baker P, et al. Clinical features of patients isolated for suspected Ebola virus disease at Connaught Hospital, Freetown, Sierra Leone: a retrospective cohort study. Lancet Infect Dis. 2015;15:1024–33. doi: 10.1016/S1473-3099(15)00137-1. [DOI] [PubMed] [Google Scholar]
- Lauber C, Gorbalenya AE. Genetics-based classification of filoviruses calls for expanded sampling of genomic sequences. Viruses. 2012;4:1425–37. doi: 10.3390/v4091425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- le Guenno B, Formenty P, Boesch C. Ebola virus outbreak in the Ivory Coast and Liberia, 1994–1995. In: Klenk H-D, editor. Marburg and Ebola Viruses. Berlin: Springer; 1999. pp. 77–84. Vol. 235. [DOI] [PubMed] [Google Scholar]
- Le Guenno B, Formenty P, Wyers M, et al. Isolation and partial characterisation of a new strain of Ebola virus. Lancet. 1995;345:1271–4. doi: 10.1016/s0140-6736(95)90925-7. [DOI] [PubMed] [Google Scholar]
- Leroy EM, Kumulungui B, Pourrut X, et al. Fruit bats as reservoirs of Ebola virus. Nature. 2005;438:575–6. doi: 10.1038/438575a. [DOI] [PubMed] [Google Scholar]
- Leung DW, Shabman RS, Farahbakhsh M, et al. Structural and functional characterization of Reston Ebola virus VP35 interferon inhibitory domain. J Mol Biol. 2010;399:347–57. doi: 10.1016/j.jmb.2010.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lever MS, Piercy TJ, Steward JA, et al. Lethality and pathogenesis of airborne infection with filoviruses in A129 α/β -/- interferon receptor-deficient mice. J Med Microbiol. 2012;61:8–15. doi: 10.1099/jmm.0.036210-0. [DOI] [PubMed] [Google Scholar]
- Li CX, Shi M, Tian JH, et al. Unprecedented genomic diversity of RNA viruses in arthropods reveals the ancestry of negative-sense RNA viruses. Elife. 2015;4:e05378. doi: 10.7554/eLife.05378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lofts LL, Ibrahim MS, Negley DL, et al. Genomic differences between guinea pig lethal and nonlethal Marburg virus variants. J Infect Dis. 2007;196(Suppl 2):S305–12. doi: 10.1086/520585. [DOI] [PubMed] [Google Scholar]
- Lu G, Zhang J, Zhang C, et al. One-step reverse-transcription FRET-PCR for differential detection of five ebolavirus species. PLoS One. 2015;10:e0126281. doi: 10.1371/journal.pone.0126281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNeil A, Farnon EC, Morgan OW, et al. Filovirus outbreak detection and surveillance: lessons from Bundibugyo. J Infect Dis. 2011;204(Suppl 3):S761–7. doi: 10.1093/infdis/jir294. [DOI] [PubMed] [Google Scholar]
- MacNeil A, Farnon EC, Wamala J, et al. Proportion of deaths and clinical features in Bundibugyo Ebola virus infection, Uganda. Emerg Infect Dis. 2010;16:1969–72. doi: 10.3201/eid1612.100627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macneil A, Reed Z, Rollin PE. Serologic cross-reactivity of human IgM and IgG antibodies to five species of Ebola virus. PLoS Neglect Trop D. 2011;5:e1175. doi: 10.1371/journal.pntd.0001175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maganga GD, Kapetshi J, Berthet N, et al. Ebola virus disease in the Democratic Republic of Congo. New Engl J Med. 2014;371:2083–91. doi: 10.1056/NEJMoa1411099. [DOI] [PubMed] [Google Scholar]
- Marsh GA, Haining J, Robinson R, et al. Ebola Reston virus infection of pigs: clinical significance and transmission potential. J Infect Dis. 2011;204(Suppl 3):S804–9. doi: 10.1093/infdis/jir300. [DOI] [PubMed] [Google Scholar]
- Martini GA, Schmidt HA. Spermatogene Übertragung des ‘Virus Marburg’ (Erreger der ‘Marburger Affenkrankheit’) [Spermatogenic transmission of the ‘Marburg virus’. (Causes of ‘Marburg simian disease’)] Klin Wochenschr. 1968;46:398–400. doi: 10.1007/BF01734141. [DOI] [PubMed] [Google Scholar]
- Maruyama J, Miyamoto H, Kajihara M, et al. Characterization of the envelope glycoprotein of a novel filovirus, Lloviu virus. J Virol. 2014;88:99–109. doi: 10.1128/JVI.02265-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzi A, Ebihara H, Callison J, et al. Vesicular stomatitis virus-based Ebola vaccines with improved cross-protective efficacy. J Infect Dis. 2011;204(Suppl 3):S1066–74. doi: 10.1093/infdis/jir348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mate SE, Kugelman JR, Nyenswah TG, et al. Molecular evidence of sexual transmission of Ebola virus. New Engl J Med. 2015;373:2448–54. doi: 10.1056/NEJMoa1509773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhailov VV, Borisevich IV, Chernikova NK, et al. The evaluation in hamadryas baboons of the possibility for the specific prevention of Ebola fever. Vop Virusol. 1994;39:82–4. [Russian] [PubMed] [Google Scholar]
- Miranda ME, Ksiazek TG, Retuya TJ, et al. Epidemiology of Ebola (subtype Reston) virus in the Philippines, 1996. J Infect Dis. 1999;179(Suppl 1):S115–9. doi: 10.1086/514314. [DOI] [PubMed] [Google Scholar]
- Miranda ME, Miranda NL. Reston ebolavirus in humans and animals in the Philippines: a review. J Infect Dis. 2011;204(Suppl 3):S757–60. doi: 10.1093/infdis/jir296. [DOI] [PubMed] [Google Scholar]
- Miranda MEG, Calaor AB, Cho F, et al. Updates on Ebola-Reston virus research activities on long-tailed macaques in the Philippines. Sylvatrop (Laguna) 2000;10:40–3. [Google Scholar]
- Miranda MEG, White ME, Dayrit MM, et al. Seroepidemiological study of filovirus related to Ebola in the Philippines. Lancet. 1991;337:425–6. doi: 10.1016/0140-6736(91)91199-5. [DOI] [PubMed] [Google Scholar]
- Miranda MEG, Yoshikawa Y, Manalo DL, et al. Chronological and spatial Analysis of the 1996 Ebola Reston virus outbreak in a monkey breeding facility in the Philippines. Exp Anim. 2002;51:173–9. doi: 10.1538/expanim.51.173. [DOI] [PubMed] [Google Scholar]
- Mire CE, Geisbert JB, Marzi A, et al. Vesicular stomatitis virus-based vaccines protect nonhuman primates against Bundibugyo ebolavirus. PLoS Neglect Trop D. 2013;7:e2600. doi: 10.1371/journal.pntd.0002600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misasi J, Chandran K, Yang JY, et al. Filoviruses require endosomal cysteine proteases for entry but exhibit distinct protease preferences. J Virol. 2012;86:3284–92. doi: 10.1128/JVI.06346-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negredo A, Palacios G, Vázquez-Morón S, et al. Discovery of an ebolavirus-like filovirus in Europe. PLoS Pathog. 2011;7:e1002304. doi: 10.1371/journal.ppat.1002304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng M, Ndungo E, Jangra RK, et al. Cell entry by a novel European filovirus requires host endosomal cysteine proteases and Niemann-Pick C1. Virology. 2014;468-470C:637–46. doi: 10.1016/j.virol.2014.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng M, Ndungo E, Kaczmarek ME, et al. Filovirus receptor NPC1 contributes to species-specific patterns of ebolavirus susceptibility in bats. Elife. 2015;4:e11785. doi: 10.7554/eLife.11785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nidom CA, Nakayama E, Nidom RV, et al. Serological evidence of Ebola virus infection in Indonesian orangutans. PLoS One. 2012;7:e40740. doi: 10.1371/journal.pone.0040740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niikura M, Ikegami T, Saijo M, et al. Detection of Ebola viral antigen by enzyme-linked immunosorbent assay using a novel monoclonal antibody to nucleoprotein. J Clin Microbiol. 2001;39:3267–71. doi: 10.1128/JCM.39.9.3267-3271.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikiforov VV, Turovskii YI, Kalinin PP, et al. A case of a laboratory infection with Marburg fever. Zh Mikrob Epid Immun. 1994:104–6. [Russian] [PubMed] [Google Scholar]
- Ogawa H, Miyamoto H, Nakayama E, et al. Seroepidemiological prevalence of multiple species of filoviruses in fruit bats (Eidolon helvum) migrating in Africa. J Infect Dis. 2015;212(Suppl 2):S101–8. doi: 10.1093/infdis/jiv063. [DOI] [PubMed] [Google Scholar]
- Olival KJ, Islam A, Yu M, et al. Ebola virus antibodies in fruit bats, Bangladesh. Emerg Infect Dis. 2013;19:270–3. doi: 10.3201/eid1902.120524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou W, Delisle J, Konduru K, et al. Development and characterization of rabbit and mouse antibodies against ebolavirus envelope glycoproteins. J Virol Methods. 2011;174:99–109. doi: 10.1016/j.jviromet.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Zhang W, Cui L, et al. Reston virus in domestic pigs in China. Arch Virol. 2014;159:1129–32. doi: 10.1007/s00705-012-1477-6. [DOI] [PubMed] [Google Scholar]
- Panchal RG, Reid SP, Tran JP, et al. Identification of an antioxidant small-molecule with broad-spectrum antiviral activity. Antivir Res. 2012;93:23–9. doi: 10.1016/j.antiviral.2011.10.011. [DOI] [PubMed] [Google Scholar]
- Peterson AT, Bauer JT, Mills JN. Ecologic and geographic distribution of filovirus disease. Emerg Infect Dis. 2004;10:40–7. doi: 10.3201/eid1001.030125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson AT, Lash RR, Carroll DS, et al. Geographic potential for outbreaks of Marburg hemorrhagic fever. Am J Trop Med Hyg. 2006;75:9–15. doi: 10.4269/ajtmh.2006.75.1.0750009. [DOI] [PubMed] [Google Scholar]
- Pigott DM, Golding N, Mylne A, et al. Mapping the zoonotic niche of Ebola virus disease in Africa. Elife. 2014;3:e04395. doi: 10.7554/eLife.04395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigott DM, Golding N, Mylne A, et al. Mapping the zoonotic niche of Marburg virus disease in Africa. T Roy Soc Trop Med H. 2015;109:366–78. doi: 10.1093/trstmh/trv024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piot P, Sureau P, Breman G, et al. Clinical aspects of Ebola virus infection in Yambuku area, Zaire, 1976. In: Pattyn SR, editor. Ebola Virus Haemorrhagic Fever. Amsterdam: Elsevier/North-Holland Biomedical Press; 1978. pp. 7–14. [Google Scholar]
- Preston R. The Hot Zone. A Terrifying New Story. New York: Random House; 1994. [Google Scholar]
- Qin E, Bi J, Zhao M, et al. Clinical features of patients With Ebola virus disease in Sierra Leone. Clin Infect Dis. 2015;61:491–5. doi: 10.1093/cid/civ319. [DOI] [PubMed] [Google Scholar]
- Qiu X, Wong G, Audet J, et al. Establishment and characterization of a lethal mouse model for the Angola strain of Marburg virus. J Virol. 2014;88:12703–14. doi: 10.1128/JVI.01643-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi AI, Chughtai M, Loua TO, et al. Study of Ebola virus disease survivors in Guinea. Clin Infect Dis. 2015;61:1035–42. doi: 10.1093/cid/civ453. [DOI] [PubMed] [Google Scholar]
- Radoshitzky SR, Warfield KL, Chi X, et al. Ebolavirus delta-peptide immunoadhesins inhibit marburgvirus and ebolavirus cell entry. J Virol. 2011;85:8502–13. doi: 10.1128/JVI.02600-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen AL, Okumura A, Ferris MT, et al. Host genetic diversity enables Ebola hemorrhagic fever pathogenesis and resistance. Science. 2014;346:987–91. doi: 10.1126/science.1259595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond J, Bradfute S, Bray M. Filovirus infection of STAT-1 knockout mice. J Infect Dis. 2011;204(Suppl 3):S986–90. doi: 10.1093/infdis/jir335. [DOI] [PubMed] [Google Scholar]
- Reed DS, Lackemeyer MG, Garza NL, et al. Aerosol exposure to Zaire ebolavirus in three nonhuman primate species: differences in disease course and clinical pathology. Microbes Infect. 2011;13:930–6. doi: 10.1016/j.micinf.2011.05.002. [DOI] [PubMed] [Google Scholar]
- Reid SP, Valmas C, Martinez O, et al. Ebola virus VP24 proteins inhibit the interaction of NPI-1 subfamily karyopherin alpha proteins with activated STAT1. J Virol. 2007;81:13469–77. doi: 10.1128/JVI.01097-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riemenschneider J, Garrison A, Geisbert J, et al. Comparison of individual and combination DNA vaccines for B. anthracis, Ebola virus, Marburg virus and Venezuelan equine encephalitis virus. Vaccine. 2003;21:4071–80. doi: 10.1016/s0264-410x(03)00362-1. [DOI] [PubMed] [Google Scholar]
- Roddy P, Howard N, Van Kerkhove MD, et al. Clinical manifestations and case management of Ebola haemorrhagic fever caused by a newly identified virus strain, Bundibugyo, Uganda, 2007–2008. PLoS One. 2012;7:e52986. doi: 10.1371/journal.pone.0052986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez LL, de Roo A, Guimard Y, et al. Persistence and genetic stability of Ebola Virus during the outbreak in Kikwit, Democratic Republic of the Congo, 1995. J Infect Dis. 1999;179(Suppl. 1):S170–S6. doi: 10.1086/514291. [DOI] [PubMed] [Google Scholar]
- Rollin PE, Williams RJ, Bressler DS, et al. Ebola (subtype Reston) virus among quarantined nonhuman primates recently imported from the Philippines to the United States. J Infect Dis. 1999;179(Suppl 1):S108–14. doi: 10.1086/514303. [DOI] [PubMed] [Google Scholar]
- Rowe AK, Bertolli J, Khan AS, et al. Clinical, virologic, and immunologic follow-up of convalescent Ebola hemorrhagic fever patients and their household contacts, Kikwit, Democratic Republic of the Congo. J Infect Dis. 1999;179(Suppl 1):S28–35. doi: 10.1086/514318. [DOI] [PubMed] [Google Scholar]
- Ryabchikova E, Kolesnikova L, Smolina M, et al. Ebola virus infection in guinea pigs: presumable role of granulomatous inflammation in pathogenesis. Arch Virol. 1996a;141:909–21. doi: 10.1007/BF01718165. [DOI] [PubMed] [Google Scholar]
- Ryabchikova E, Price BBS. Ebola and Marburg Viruses: A View of Infection Using Electron Microscopy. Columbus: Battelle Press; 2004. [Google Scholar]
- Ryabchikova E, Strelets L, Kolesnikova L, et al. Respiratory Marburg virus infection in guinea pigs. Arch Virol. 1996b;141:2177–90. doi: 10.1007/BF01718224. [DOI] [PubMed] [Google Scholar]
- Ryabchikova EI, Kolesnikova LV, Luchko SV. An analysis of features of pathogenesis in two animal models of Ebola virus infection. J Infect Dis. 1999;179(Suppl 1):S199–202. doi: 10.1086/514293. [DOI] [PubMed] [Google Scholar]
- Sanchez A, Trappier SG, Mahy BW, et al. The virion glycoproteins of Ebola viruses are encoded in two reading frames and are expressed through transcriptional editing. P Natl Acad Sci USA. 1996;93:3602–7. doi: 10.1073/pnas.93.8.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schieffelin JS, Shaffer JG, Goba A, et al. Clinical illness and outcomes in patients with Ebola in Sierra Leone. New Engl J Med. 2014;371:2092–100. doi: 10.1056/NEJMoa1411680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semmler IA. Ebola goes pop: the filovirus from literature into film. Lit Med. 1998;17:149–74. doi: 10.1353/lm.1998.0006. [DOI] [PubMed] [Google Scholar]
- Sherwood LJ, Hayhurst A. Ebolavirus nucleoprotein C-termini potently attract single domain antibodies enabling monoclonal affinity reagent sandwich assay (MARSA) formulation. PLoS One. 2013;8:e61232. doi: 10.1371/journal.pone.0061232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegert R, Shu HL, Slenczka W, et al. Zur Ätiologie einer unbekannten, von Affen ausgegangenen menschlichen Infektionskrankheit [On the etiology of an unknown human infection originating from monkeys] Dtsch Med Wochenschr. 1967;92:2341–3. doi: 10.1055/s-0028-1106144. [German] [DOI] [PubMed] [Google Scholar]
- Simpson DI. Marburg agent disease: in monkeys. T Roy Soc Trop Med H. 1969a;63:303–9. doi: 10.1016/0035-9203(69)90002-9. [DOI] [PubMed] [Google Scholar]
- Simpson DIH. Marburg virus disease: experimental infection in monkeys. In: Perkins FT, O'Donoghue PN, Beveridge WIB, et al., editors. Laboratory Animal Handbooks. London: London Laboratory Animals, Ltd.; 1969b. pp. 149–54. Vol. 4. [Google Scholar]
- Simpson DIH. Vervet monkey disease—transmission to the hamster. Brit J Exp Pathol. 1969c;50:389–92. [PMC free article] [PubMed] [Google Scholar]
- Smith DH, Francis F, Simpson DIH. African haemorrhagic fever in the southern Sudan, 1976: the clinical manifestations. In: Pattyn SR, editor. Ebola Virus Haemorrhagic Fever. Amsterdam: Elsevier/North-Holland Biomedical Press; 1978. pp. 21–6. [Google Scholar]
- Smith DH, Johnson BK, Isaacson M, et al. Marburg-virus disease in Kenya. Lancet. 1982;1:816–20. doi: 10.1016/s0140-6736(82)91871-2. [DOI] [PubMed] [Google Scholar]
- Smither SJ, Nelson M, Eastaugh L, et al. Experimental respiratory Marburg virus haemorrhagic fever infection in the common marmoset (Callithrix jacchus) Int J Exp Pathol. 2013;94:156–68. doi: 10.1111/iep.12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smither SJ, Nelson M, Eastaugh L, et al. Experimental respiratory infection of marmosets (Callithrix jacchus) with Ebola virus Kikwit. J Infect Dis. 2015;212(Suppl 2):S336–45. doi: 10.1093/infdis/jiv371. [DOI] [PubMed] [Google Scholar]
- Subbotina E, Dadaeva A, Kachko A, et al. Genetic factors of Ebola virus virulence in guinea pigs. Virus Res. 2010;153:121–33. doi: 10.1016/j.virusres.2010.07.015. [DOI] [PubMed] [Google Scholar]
- Sullivan NJ, Sanchez A, Rollin PE, et al. Development of a preventive vaccine for Ebola virus infection in primates. Nature. 2000;408:605–9. doi: 10.1038/35046108. [DOI] [PubMed] [Google Scholar]
- Swenson DL, Wang D, Luo M, et al. Vaccine to confer to nonhuman primates complete protection against multistrain Ebola and Marburg virus infections. Clin Vaccine Immunol. 2008a;15:460–7. doi: 10.1128/CVI.00431-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swenson DL, Warfield KL, Larsen T, et al. Monovalent virus-like particle vaccine protects guinea pigs and nonhuman primates against infection with multiple Marburg viruses. Expert Rev Vaccines. 2008b;7:417–29. doi: 10.1586/14760584.7.4.417. [DOI] [PubMed] [Google Scholar]
- Taniguchi S, Watanabe S, Masangkay JS, et al. Reston Ebolavirus antibodies in bats, the Philippines. Emerg Infect Dis. 2011;17:1559–60. doi: 10.3201/eid1708.101693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towner JS, Amman BR, Sealy TK, et al. Isolation of genetically diverse Marburg viruses from Egyptian fruit bats. PLoS Pathog. 2009;5:e1000536. doi: 10.1371/journal.ppat.1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towner JS, Sealy TK, Khristova ML, et al. Newly discovered Ebola virus associated with hemorrhagic fever outbreak in Uganda. PLoS Pathog. 2008;4:e1000212. doi: 10.1371/journal.ppat.1000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tukei PM. US Army Medical Research and Development Command; Fort Detrick, Frederick, Maryland, USA: Virus Research Centre, Kenya Medical Research Institute; Nairobi, Kenya: 1988. Epidemiology and epizootiological investigations of hemorrhagic fever viruses in Kenya. Annual Report (May 30, 1988) and. [Google Scholar]
- Twenhafel NA, Shaia CI, Bunton TE, et al. Experimental aerosolized guinea pig-adapted Zaire ebolavirus (variant: Mayinga) causes lethal pneumonia in guinea pigs. Vet Pathol. 2015;52:21–5. doi: 10.1177/0300985814535612. [DOI] [PubMed] [Google Scholar]
- Ursic-Bedoya R, Mire CE, Robbins M, et al. Protection against lethal Marburg virus infection mediated by lipid encapsulated small interfering RNA. J Infect Dis. 2014;209:562–70. doi: 10.1093/infdis/jit465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valmas C, Basler CF. Marburg virus VP40 antagonizes interferon signaling in a species-specific manner. J Virol. 2011;85:4309–17. doi: 10.1128/JVI.02575-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varkey JB, Shantha JG, Crozier I, et al. Persistence of Ebola virus in ocular fluid during convalescence. New Engl J Med. 2015;372:2423–7. doi: 10.1056/NEJMoa1500306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villinger F, Rollin PE, Brar SS, et al. Markedly elevated levels of interferon (IFN)-gamma, IFN-alpha, interleukin (IL)-2, IL-10, and tumor necrosis factor-alpha associated with fatal Ebola virus infection. J Infect Dis. 1999;179(Suppl 1):S188–91. doi: 10.1086/514283. [DOI] [PubMed] [Google Scholar]
- Wahl-Jensen V, Radoshitzky SR, de Kok-Mercado F, et al. Role of rodents and bats in human viral hemorrhagic fevers. In: Singh SK, Ruzek D, editors. Viral Hemorrhagic Fevers. Boca Raton: Taylor & Francis/CRC Press; 2013. pp. 99–127. [Google Scholar]
- Walsh PD, Abernethy KA, Bermejo M, et al. Catastrophic ape decline in western equatorial Africa. Nature. 2003;422:611–4. doi: 10.1038/nature01566. [DOI] [PubMed] [Google Scholar]
- Wamala JF, Lukwago L, Malimbo M, et al. Ebola hemorrhagic fever associated with novel virus strain, Uganda, 2007–2008. Emerg Infect Dis. 2010;16:1087–92. doi: 10.3201/eid1607.091525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Kluwe CA, Lungu OI, et al. Facile discovery of a diverse panel of anti-Ebola virus antibodies by immune repertoire mining. Sci Rep. 2015;5:13926. doi: 10.1038/srep13926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Hevey M, Juompan LY, et al. Complex adenovirus-vectored vaccine protects guinea pigs from three strains of Marburg virus challenges. Virology. 2006;353:324–32. doi: 10.1016/j.virol.2006.05.033. [DOI] [PubMed] [Google Scholar]
- Warfield KL, Alves DA, Bradfute SB, et al. Development of a model for marburgvirus based on severe-combined immunodeficiency mice. Virol J. 2007;4:108. doi: 10.1186/1743-422X-4-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warfield KL, Bradfute SB, Wells J, et al. Development and characterization of a mouse model for Marburg hemorrhagic fever. J Virol. 2009;83:6404–15. doi: 10.1128/JVI.00126-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warfield KL, Dye JM, Wells JB, et al. Homologous and heterologous protection of nonhuman primates by Ebola and Sudan virus-like particles. PLoS One. 2015;10:e0118881. doi: 10.1371/journal.pone.0118881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren TK, Warfield KL, Wells J, et al. Antiviral activity of a small-molecule inhibitor of filovirus infection. Antimicrob Agents Ch. 2010;54:2152–9. doi: 10.1128/AAC.01315-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren TK, Wells J, Panchal RG, et al. Protection against filovirus diseases by a novel broad-spectrum nucleoside analogue BCX4430. Nature. 2014;508:402–5. doi: 10.1038/nature13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingartl HM, Embury-Hyatt C, Nfon C, et al. Transmission of Ebola virus from pigs to non-human primates. Sci Rep. 2012;2:811. doi: 10.1038/srep00811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. Ebola reston in pigs and humans, Philippines. Wkly Epidemiol Rec. 2009;84:49–50. [PubMed] [Google Scholar]
- World Health Organization. International Statistical Classification of Diseases and Related Health Problems, 10th Revision (ICD-10) 2015. http://apps.who.int/classifications/icd10/browse/2015/en (5 April 2016, date last accessed) [Google Scholar]
- World Health Organization. Ebola Situation Report – 30 March 2016. 2016. http://apps.who.int/ebola/current-situation/ebola-situation-report-30-march-2016 (5 April 2016, date last accessed) [Google Scholar]
- World Health Organization Ebola Response Team. Ebola virus disease in West Africa—the first 9 months of the epidemic and forward projections. New Engl J Med. 2014;371:1481–95. doi: 10.1056/NEJMoa1411100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyers M, Formenty P, Cherel Y, et al. Histopathological and immunohistochemical studies of lesions associated with Ebola virus in a naturally infected chimpanzee. J Infect Dis. 1999;179(Suppl 1):S54–9. doi: 10.1086/514300. [DOI] [PubMed] [Google Scholar]
- Yan T, Mu J, Qin E, et al. Clinical characteristics of 154 patients suspected of having Ebola virus disease in the Ebola holding center of Jui Government Hospital in Sierra Leone during the 2014 Ebola outbreak. Eur J Clin Microbiol. 2015;34:2089–95. doi: 10.1007/s10096-015-2457-z. [DOI] [PubMed] [Google Scholar]
- Yuan J, Zhang Y, Li J, et al. Serological evidence of ebolavirus infection in bats, China. Virol J. 2012;9:236. doi: 10.1186/1743-422X-9-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang AP, Bornholdt ZA, Liu T, et al. The Ebola virus interferon antagonist VP24 directly binds STAT1 and has a novel, pyramidal fold. PLoS Pathog. 2012;8:e1002550. doi: 10.1371/journal.ppat.1002550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlotnik I, Simpson DI. The pathology of experimental vervet monkey disease in hamsters. Brit J Exp Pathol. 1969;50:393–9. [PMC free article] [PubMed] [Google Scholar]
- Zumbrun EE, Abdeltawab NF, Bloomfield HA, et al. Development of a murine model for aerosolized ebolavirus infection using a panel of recombinant inbred mice. Viruses. 2012a;4:3468–93. doi: 10.3390/v4123468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumbrun EE, Bloomfield HA, Dye JM, et al. A characterization of aerosolized Sudan virus infection in African green monkeys, cynomolgus macaques, and rhesus macaques. Viruses. 2012b;4:2115–36. doi: 10.3390/v4102115. [DOI] [PMC free article] [PubMed] [Google Scholar]