Abstract
Genome engineering has gone mainstream because of breakthroughs in defining and harnessing naturally occurring, customizable DNA recognition cursors (protein or RNA-guided). At present, most gene editing relies on these cursors to direct custom DNA endonucleases to a specific genomic sequence to induce a double-strand break. New tools for genome engineering are continuously being explored, and another advance in DNA targeting has recently been described. Argonaute isolated from Natronobacterium gregoryi (NgAgo) is an ssDNA-based cursor that thus far has no known limitations in sequence recognition, shows promise for high specificity, and for many applications may represent a potentially more accessible genome-editing system over prior tools as it requires only a single, 24-base, 5′ phosphorylated ssDNA for DNA targeting. Genome engineering is in a remarkable moment of unprecedented growth with exponential reduction in costs reminiscent of Moore's law in electronics. Many questions remain with regard to Argonaute utility in specific systems, but there is no doubt that genome engineering is expanding into new and exciting areas from synthetic biology to gene therapy.
Genome engineering requires precise and accessible gene-editing tools. At present, most gene editing relies on programmable DNA cursors (protein or RNA-guided) to localize a nuclease at a specific DNA location to induce a double-strand break (DSB) (for a comprehensive review, see ref.1). Edits in DNA are generated when a DSB is either erroneously repaired (i.e., generating insertions or deletions near the repair site) by nonhomologous end joining (NHEJ)2 or repaired using an exogenous DNA template via homology-directed repair (HDR).2 The most commonly used custom endonucleases include zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and the CRISPR-Cas9 (clustered regularly interspersed short palindromic repeats-CRISPR associated protein 9) system. Although ZFNs have been instrumental in pioneering genome-editing approaches, they are less commonly used today largely because of their substantial assembly cost (Fig. 1).3 Argonaute is a DNA-guided DNA cursor that represents an exciting new addition to the genome engineer's toolbox.4
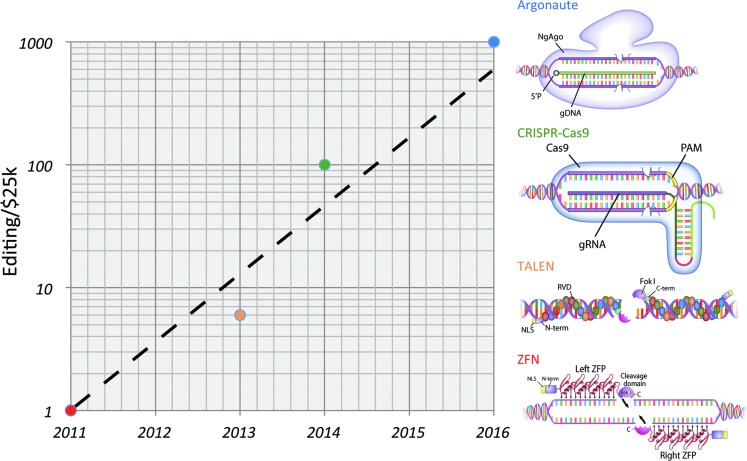
Figure 1.
Genomic Moore's law? The rate of gene editing as a function of fixed cost continues to rise at an exponential rate. Graphed are the initial commercial costs of purchasing custom DNA cursors (in units of US$25,000) for ZFNs, TALENs, CRISPR/Cas9, and Argonaute over time since 2011.
TALENs are composed of a protein-based TALE DNA cursor tethered to a FokI endonuclease domain. The TALE domains are programmable modular repeats of approximately 34 amino acids that encode, per repeat, two contiguous residues (repeat variable diresidues) for one-to-one DNA base recognition.5 Importantly, unlike zinc finger domains, TALE domains do not suffer from extensive context dependency and thus confer excellent targeting flexibility and efficacy. Furthermore, the catalytic activity of FokI is dependent on dimerization between two closely localized FokI domains, each tethered to unique TALE cursors. TALENs thus maintain a high targeting specificity by requiring two independent binding events at a predefined distance apart from one another. These attributes decrease off-target DSBs significantly. In addition, TALENs are substantially cheaper to make than ZFNs (Fig. 1). However, this protein-based tool for genome editing requires custom assembly of each TALE-based DNA cursor.
The CRISPR-Cas9 platform utilizes a single guide RNA (sgRNA) as a cursor, and is the leading RNA cursor-based genome-editing tool. In these systems, the Cas9 protein is programmed through an sgRNA to mediate base-to-base genomic DNA recognition, followed by the triggering of the innate endonuclease activity of Cas9.6 The genome engineering community has developed a particular excitement for the CRISPR-Cas9 system in part because of their substantially lower cost and consequently generally higher accessibility compared with ZFNs or TALENs (Fig. 1). However, the CRISPR-Cas9 system is not without limitations. For example, the most commonly used Cas9 protein (isolated from Streptococcus pyogenes) requires a three-nucleotide protospacer adjacent motif (PAM) sequence—NGG—at the 3′ end of potential target sequences.7 In addition and for reasons yet unknown, not all sgRNAs can initiate effective DNA cutting, and the relatively large size of sgRNAs makes their direct synthesis a technical challenge. As a result, sgRNAs are typically generated as a synthetic message in vitro or expressed from a DNA construct in a cell. After the identification of two unique catalytic domains of Cas9 (each involved in cutting one strand of DNA), mutagenized Cas9 proteins—Cas9 nickases and dead Cas9—have been widely adopted as RNA-based DNA cursors by genome engineers for broad applications.8,9
Recently, the first DNA-based genome engineering cursors were described from the Argonaute gene family. Argonaute (Ago) proteins are well known for their key role in the RNA interference pathway in eukaryotes, where members of this protein family utilize RNA guides to bind and cleave complementary mRNA (see ref.10 for review). Intriguingly, early reports showed that prokaryotic Ago proteins could use DNA guides to target and cleave double-strand DNA, although only at supraphysiological temperatures.11,12 This high thermal requirement reduced their general utility.
However, one Ago protein isolated from Natronobacterium gregoryi (NgAgo) has just been shown to work at physiological temperatures for effective DNA cursor-based targeting and genome editing in vitro and in human cells (Fig. 2).4 NgAgo is unique in several ways: (1) the only known requirement for this protein-based DNA cursor is a 5′-phosphorylated single-strand DNA guide with a functional length of about 24 bases, (2) much like CRISPR-Cas9, it is a two-component system requiring only the gDNA and NgAgo protein, capable of inducing DSBs in human cells, and (3) NgAgo is about two-thirds the size of S. pyogenes Cas9, making it potentially easier to deploy using delivery tools such as viral vectors. Furthermore, NgAgo does not have any noted sequence targeting limitations, such as the obligate PAM site for CRISPR-Cas9. Furthermore, the initial specificity screens indicate a promisingly low tolerance for base-pair mismatches at a rate substantially higher than a standard CRISPR-Cas9 system, suggesting low inherent off-targeting issues. Together with its simplicity and its potential for continued cost reductions for genome-editing applications (i.e., only needing a synthetic oligonucleotide DNA as a guide instead of RNA; Fig. 1), NgAgo is an attractive addition to modern genome engineering science. Although the utility of NgAgo in vivo is to be validated, it is foreseeable for this tool to become rapidly adapted to many biological systems.
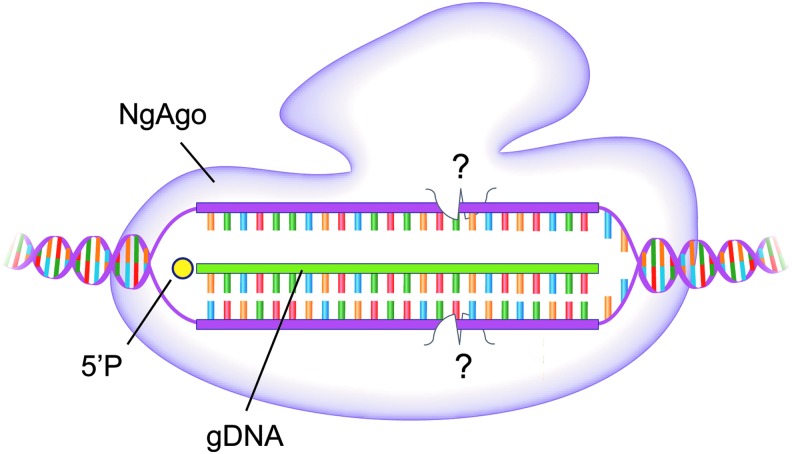
Figure 2.
Argonaute genome editor. Cartoon of the Argonaute protein isolated from Natronobacterium gregoryi (NgAgo) drawn in complex with its ssDNA cursor and its double-stranded DNA target. Specificity is achieved by a DNA:DNA complex. The Ago protein induces double-stranded breaks at yet-unclear positions (shown as question marks) and using an uncertain enzymatic mechanism.4
The precise mechanism of DNA-guided DNA cleavage by NgAgo remains unknown at present. However, there could be substantial similarity to other proteins in the PIWI superfamily. Once the correct target is identified through Watson–Crick base pairing, subsequent cleavage results in a double-strand break (Fig. 2).11 Intriguingly, NgAgo appears to be more efficient at inducing DSBs when guide DNA loading takes place inside of live cells and remains faithful to the first guide DNA that it is loaded with.4 As such, the kinetics, utility, and practical limitations of gDNA loading will be important considerations for the use of NgAgo in genome-editing applications.
Many interesting functional and mechanistic validation experiments remain to be completed to better understand the NgAgo system for use in downstream applications in both basic and clinical research. These include (1) examining its utility in vivo, including flies, worms, zebrafish, and rodents; (2) identifying functional domain(s) for the cleavage of targeted and nontargeted DNA strands to cause a DSB; (3) determining any sequence requirements or biases for optimal DNA cursor binding; (4) defining the degree of off-target effects by unbiased approaches, especially relative to other modern custom restriction enzyme systems; and (5) developing effective ways to generate 5′ phosphorylated ssDNA guides when long-term endonuclease activity is desired. Despite these unknowns, this exciting genome engineering tool represents a new critical data point for cost reduction in genome editing (Fig. 1) and potentially offers substantively improved practicality and accessibility compared with other genome engineering tools we have at our disposal.
Conclusions
The field of genome engineering has seen an unprecedented rate of growth in the last half decade. The key has been the systematic development of customizable DNA cursor systems. In addition to the diversity of science and applications now possible, the accessibility and cost has shown remarkable exponential growth (Fig. 1). The door to this new era of genome engineering was opened with the introduction of ZFNs that enabled the first semimodular targeting of custom endonucleases to genomic sequences of interest. However, accessibility and utility were very limited when ZFNs first became available as their commercial production cost 25,000 U.S. dollars, plus taking months for synthesis and validation. A newer protein-based system, TALENs, represented a substantial reduction in both reagent cost and time to deployment. The RNA-guided CRISPR-Cas9 toolkits reduced this cost further, empowering about 100 times more genome-editing science for the same cost as a single ZFN. NgAgo represents another potential data point on this exponential curve (Fig. 1). Indeed, the cost–efficiency analysis for the synthesis of custom restriction enzymes seems overly similar to Moore's law in electronics, which has accurately predicted exponential growth of the processing power of computers for the last five decades. The comparison between computer programming and DNA editing costs likely represents just the start of the potential for continued exponential growth in the field of genome engineering in the 21st century.
Acknowledgments
This work was supported by National Institutes of Health Grants GM63904 and P30DK084567 to S.C.E., NIH Training Grant UL1 TR000135 to J.M.C., and the Mayo Foundation.
Author Disclosure
The authors declare no competing financial interests.
References
- 1.Peng Y, Clark KJ, Campbell JM, et al. Making designer mutants in model organisms. Development 2014;141:4042–4054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Govindan G, Ramalingam S. Programmable site-specific nucleases for targeted genome engineering in higher eukaryotes. J Cell Physiol 2016. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 3.Cornu TI, Thibodeau-Beganny S, Guhl E, et al. DNA-binding specificity is a major determinant of the activity and toxicity of zinc-finger nucleases. Mol Ther 2008;16:352–358 [DOI] [PubMed] [Google Scholar]
- 4.Gao F, Shen XZ, Jiang F, et al. DNA-guided genome editing using the Natronobacterium gregoryi Argonaute. Nat Biotechnol 2016. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 5.Bogdanove AJ, Voytas DF. TAL effectors: Customizable proteins for DNA targeting. Science 2011;333:1843–1846 [DOI] [PubMed] [Google Scholar]
- 6.Barrangou R, Fremaux C, Deveau H, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science 2007;315:1709–1712 [DOI] [PubMed] [Google Scholar]
- 7.Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 2009;155:733–740 [DOI] [PubMed] [Google Scholar]
- 8.Guilinger JP, Thompson DB, Liu DR. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat Biotechnol 2014;32:577–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ran FA, Hsu PD, Lin CY, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 2013;154:1380–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Swarts DC, Makarova K, Wang Y, et al. The evolutionary journey of Argonaute proteins. Nat Struct Mol Biol 2014;21:743–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Swarts DC, Hegge JW, Hinojo I, et al. Argonaute of the archaeon Pyrococcus furiosus is a DNA-guided nuclease that targets cognate DNA. Nucleic Acids Res 2015;43:5120–5129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Swarts DC, Jore MM, Westra ER, et al. DNA-guided DNA interference by a prokaryotic Argonaute. Nature 2014;507:258–261 [DOI] [PMC free article] [PubMed] [Google Scholar]