Abstract
Introduction: Mosquito-borne viruses have a widespread distribution across the globe and are known to pose serious threats to human and animal health. The maintenance and dissemination of these viruses in nature are driven through horizontal and vertical transmission. In the temperate climate of northern Sweden, there is a dearth of knowledge on whether mosquito-borne viruses that occur are transmitted transovarially. To gain a better understanding of mosquito-borne virus circulation and maintenance, mosquito larvae were sampled in northern Sweden during the first and second year after a large outbreak of Ockelbo disease in 2013 caused by Sindbis virus (SINV).
Materials and Methods: A total of 3123 larvae were sampled during the summers of 2014 and 2015 at multiple sites in northern Sweden. The larvae were homogenized and screened for viruses using RT-PCR and sequencing. Species identification of selected larvae was performed by genetic barcoding targeting the cytochrome C oxidase subunit I gene.
Results and Discussion: SINV RNA was detected in mosquito larvae of three different species, Ochlerotatus (Oc.) communis, Oc. punctor, and Oc. diantaeus. Inkoo virus (INKV) RNA was detected in Oc. communis larvae. This finding suggested that these mosquitoes could support transovarial transmission of SINV and INKV. Detection of virus in mosquito larva may serve as an early warning for emerging arboviral diseases and add information to epidemiological investigations before, during, and after outbreaks. Furthermore, our results demonstrated the relevance of genetic barcoding as an attractive and effective method for mosquito larva typing. However, further mosquito transmission studies are needed to ascertain the possible role of different mosquito species and developmental stages in the transmission cycle of arboviruses.
Key Words: : Barcoding, Inkoo, Mosquito larvae, RNA, Sindbis, Virus
Introduction
Mosquito-borne viruses are globally spread and several members constitute a serious threat to human and animal health. To mitigate these infections, further knowledge about their distribution and circulation in nature is needed.
One of the most prevalent mosquito-borne viruses in Europe is Sindbis virus (SINV), family Togaviridae, genus Alphavirus (Hubalek et al. 2008). It causes polyarthritis, fever, and rash, mainly in northern Europe where the disease is named Ockelbo disease in Sweden, Pogosta disease in Finland, and Karelian fever in Russia (Brummer-Korvenkontio et al. 2002). Even though SINV is endemic in these areas, sudden outbreaks with large numbers of SINV cases are occurring (Lundström 1999, Brummer-Korvenkontio et al. 2002, Laine et al. 2004, Kurkela et al. 2008, Bergqvist et al. 2015). In Sweden, documented outbreaks of SINV disease have been reported in 1981/1982, 1988, 1995, 1997/1998, 2009, and 2013 (Lundström 1999, Bergqvist et al. 2015), and recently, more SINV infections are appearing in northern Sweden (Ahlm et al. 2014).
Another mosquito-borne virus known to circulate in northern Europe and Russia is Inkoo virus (INKV), family Bunyaviridae, genus Orthobunyavirus, and a member of the California serogroup (Brummer-Korvenkontio 1973, Niklasson and Vene 1996, L'vov et al. 1997, Putkuri et al. 2007, Hubalek 2008, Putkuri et al. 2014). Clinical manifestations of INKV in humans are mild and sometimes asymptomatic (Putkuri et al. 2007, 2014), but seroprevalence of up to 51% has been demonstrated in Finland (Putkuri et al. 2007) and 41% in northern Sweden (Evander et al. 2016).
The ecological transmission cycles of SINV and INKV have not been extensively studied. In northern Europe, SINV is detected in many species of birds (Lundstrom et al. 2001), as well as in the ornithophilic mosquitoes Culex (Cx.) pipiens, Cx. torrentium, Culiseta morsitans, and the bridge vector Aedes (Ae.) cinereus (Francy et al. 1989, Kurkela et al. 2008, Bergqvist et al. 2015). A mosquito-bird cycle has been suggested to be facilitated by migratory birds that carry SINV to new regions where local competent mosquito vectors establish the virus (Lundström and Pfeffer 2010). Regarding INKV, the virus has been isolated from Oc. communis mosquitoes in Finland, Sweden, and Russia and from Oc. hexodontus and Oc. punctor in Russia (Brummer-Korvenkontio and Saikku 1975, Francy et al. 1989, Mitchell et al. 1993), as well as from Lepus timidus (mountain hare), a possible vertebrate host (Hubalek et al. 2008, Putkuri et al. 2014).
For both viruses, there is a possibility for transovarial transmission by the mosquito, but this has not been studied in northern Europe. As far as we know, only one study from Australia has suggested vertical transmission of SINV and the related Ross River virus, family Togaviridae, genus Alphavirus (Dhileepan et al. 1996). INKV has not been studied regarding vertical transmission, but the La Crosse virus (family Bunyaviridae, genus Orthobunyaviridae) in the California serogroup has been found in field collected larvae and mosquito eggs in North America (Reese et al. 2010).
In essence, it remains to be revealed if arboviruses are solely introduced into the mosquito population horizontally (e.g., from infected birds) or maintained in endemic regions through vertical infection of competent mosquito species. In 2013, an outbreak of Ockelbo disease was reported in northern Sweden (Bergqvist et al. 2015). This encouraged a screening of mosquitoes close to the locations where the highest number of human cases was reported. Subsequently, one new SINV strain (Lövånger; GenBank: KF737350.1) was detected, isolated, and fully sequenced (Bergqvist et al. 2015). Our aim was to further investigate the circulation and maintenance of SINV during postoutbreak seasons. In this study, mosquito larvae were collected in areas near the site where the SINV outbreak occurred and screened for the presence of arboviruses.
Materials and Methods
Study area and mosquito larvae collection
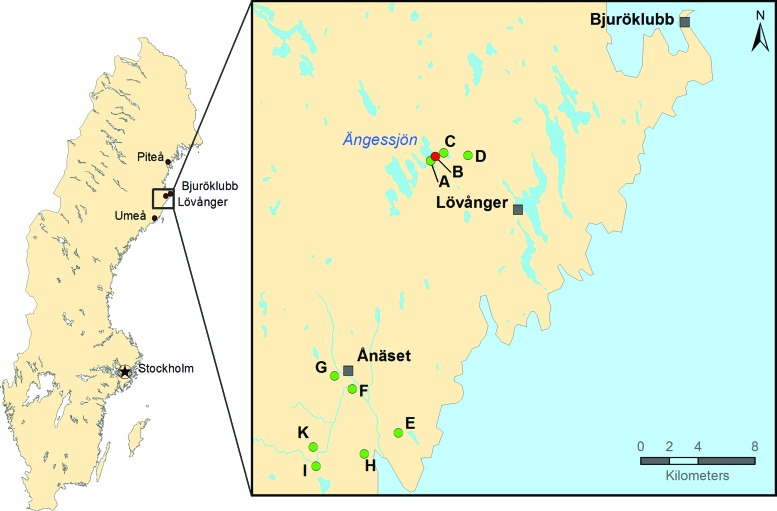
Mosquito larvae were sampled in May and June 2014 and June 2015, from two selected areas in Västerbotten County, northern Sweden. Within these areas, multiple water puddles in different types of environments were found (Fig. 1). The puddles were searched with a tuck and the collected larvae were placed in jars containing water from the source and immediately transported to the laboratory where single larva was placed in individual wells of 96-well plates containing sterile water and stored at −80°C until processing.
FIG. 1.
Mosquito larvae sampling sites. Map of the mosquito larvae sampling sites within Västerbotten County, Sweden. Sample site B (N64.40° E21.20°) is where Sindbis-positive mosquitoes were found in 2013. Sites A–D are in the vicinity of site B and E–K south of site B around N64.24° E21.03°. Color images available online at www.liebertpub.com/vbz
Homogenization and pooling of mosquito larvae
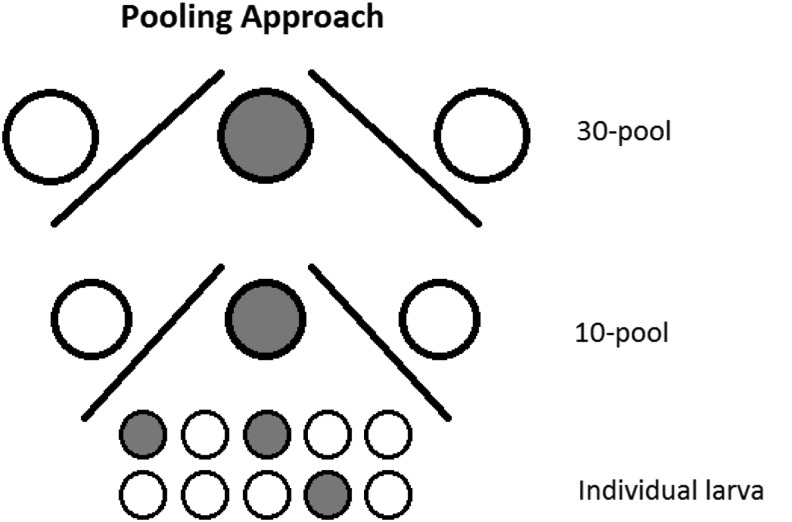
Larvae sampled in 2014 were homogenized individually in 2 mL round-bottomed microcentrifuge tubes containing five 2 mm stainless steel beads and 350 μL Glasgow medium (Gibco, Invitrogen). The homogenization was performed at 4°C with a Mixer Mill 400 (Retsch GmbH) for 20 s at a relative frequency of 30 cycles per second. Forty microliter from 10 individual larva homogenates was pooled and denoted 10× pools. Finally, 100 μL aliquots from three 10× pools were used to make 30× pools (Fig. 2).
FIG. 2.
Pooling approach. This figure illustrates how sampled larvae pools containing 30 and 10 mosquito larvae were screened by RT-PCR into individual PCR-positive larva. The filled circles illustrate PCR-positive samples. The same approach was used when screening 100× pools, 10× pools, and identification of individual virus-positive larva samples thereof.
The second set of larvae (those sampled in 2015) was processed in 350 μL Dulbecco's modified Eagle's medium (Invitrogen) and homogenized using a Mixer Mill 400 as above. Ten pools (10×) and 100 pools (100×) were generated by combining 30 μL from 10 homogenates and 30 μL from 10 (10×) pools. All larvae homogenates were stored at −80°C until analysis.
DNA and RNA extraction
To identify the larva species, DNA extraction for genetic barcoding of individual larvae was performed using the EZ1 Advanced Robotic Workstation and the EZ1 DNA Tissue Kit (Qiagen). Briefly, 100 μL of the homogenized samples were diluted with 100 μL of the DNA tissue buffer G2 from the kit before extraction. RNA extraction of the first set of larva homogenates was done with the QIAmp Viral RNA Mini Kit (Qiagen), according to the manufacturer's instructions. RNA extraction of the second set of larva homogenates was performed using the Viral NA Extraction Kit (DiaSorin) according the manufacturers' instructions (protocol: Total Viral NA 94).
PCR of the cytochrome C oxidase subunit I gene
The cytochrome C oxidase subunit I (COI) gene was amplified by PCR from DNA extracted from mosquito larva by a method previously developed for adult mosquitoes (Engdahl et al. 2014). Primers used were as follows: GB1310_29F, 5′-GAAGGAGTTTGATCAGGAATAGT-3′, GB1960-1936R, 5′-TCCTCCTCCAATAGGGTCAAAGAA-3′, LCO1490F, 5′-GGTCAACAAATCATAAAGATATTGG-3′ (Folmer et al. 1994), and TL2-N-3014R, 5′-TCCAATGCACTAATCTGC CATATTA-3′ (Simon et al. 1994, Tang et al. 2010). The PCR amplification was performed by StepOnePlus Real-time PCR System (Applied Biosystems) or with the KAPA SYBR FAST qPCR Kit (KAPA Biosystems). The preferred cycling conditions were as follows: initial denaturation at 95°C for 5 min followed by 40 cycles and 5 s at 95°C, 20 s at 57°C, and 15 s at 72°C. Positive and negative controls were included in each test. A melting curve was produced to evaluate the amplification. The PCR products were visualized on 1.5% agarose gels containing GelRed (Biotium, Inc.). PCR products were finally precipitated using ethanol, centrifuged, lyophilized, and sequenced (Eurofins Genomics). The obtained sequences were compared with available sequences in GenBank. Phylogenetic analysis was performed using Clustal Omega method, which uses Neighbor-joining tree without distance corrections (McWilliam et al. 2013).
Virus screening by RT-PCR
SINV screening of larva homogenates was performed by a probe-based qRT-PCR method using StepOnePlus Real-time PCR targeting the nonstructural gene (nsP1). Primers used were SINV-nsP1F, 5′-GGTTCCTACCACAGCGACGAT-3′, SINV-nsP1R, 5′-TGATACTGGTGCTCGGAAAACA-3′, and SINV-nsP1 probe, 5′-FAM-TTGGACATAGGCAGC GCA-3′ (Sane et al. 2012). The amplification was performed by qScript XLT One-step RT-qPCR ToughMix (Quanta Biosciences).
Larva homogenates were also screened using specific primers for Orthobunyavirus (Kuno et al. 1996), Flavivirus (Bryant et al. 2005), and Alphavirus (Eshoo et al. 2007). INKV PCR was performed using the Applied Biosystems® GeneAmp® PCR System 9700 (Applied Biosystems) with the SuperScript III One-step RT-PCR system and Platinum Taq DNA Polymerase (Invitrogen, Life Technologies). Specific primers used for INKV screening were as follows: INKV NS-Fw 5′-GAGAGTGGCAGGTGGAGATT-3′ and VIR2052R 5′-AAAGCCGGTGGATGGTAAGT-3′ (Huang et al. 1996, Vapalahti et al. 1996). Primers used to amplify overlapping fragments for sequencing of the S and M segment are given in Table 1. The complete open reading frames of the S and M segments of INKV strain Lövånger are stored in GenBank under the accession numbers KU302751 and KU681435.
Table 1.
Sequencing Primers
| Name of primer | Primer sequence |
|---|---|
| INKVS1F | 5′-AGTAGTGTGCTCCACTGAATACATTTAA |
| INKVS600R | 5′-ACTGAGGATCCATCATACCATGCTT |
| INKVS400F | 5′-GATGACGATGAGTCCCAAAGAGAA |
| INKVS988R | 5′-GTAGTGTGCTCCACTGAATACATTTAA 3′ |
| INKVM1F | 5′-AGTAGTGTACTACCAAGCATAGAAAACGTTCA |
| INKVM1057R | 5′-TTCACCAATGACCATAGCACTAATAGGA |
| INKVM941F | 5′-CCTCAGAGCAGCCAGAGTTATGT |
| INKVM2049R | 5′-CGGGCCATACTTTCTGCAATATATGAA |
| INKVM1728F | 5′-CGAATATGCCTTCCTTACTAGATACTG |
| INKVM3086R | 5′-CCTATTACATCCATTAGGTAGTTGCCC |
| INKVM2957F | 5′-TGGAAGGTATTGAAACTGTTGAAGGGATA |
| INKVM4067R | 5′-TTGTGTTACAGATCTTAAATTTTAGAGACA |
| INKVM3545F | 5′-CTAGGTGTGTACTCCAAAGGATGTG |
| INKVM4507R | 5′-GTAGTGTGCTACCAAGTATATTTAAATGA |
The preferred cycling conditions were as follows: initial cDNA synthesis and denaturation at 55°C for 30 min and 94°C for 2 min, followed by 45 cycles of 94°C for 15 s, 60°C for 30 s, and 68°C for 1 min before a final extension at 68°C for 5 min. The PCR products were visualized on 1.2% agarose gels containing GelRed (Biotium, Inc.). PCR products were precipitated using ethanol, centrifuged, lyophilized, and sequenced (Eurofins Genomics).
Results
Species determination of mosquito larvae by molecular barcoding
Sampling of the first set of larvae captured in 2014 was carried out at 10 sites around Lövånger and Ånäset, Västerbotten, Sweden (Fig. 1). In total, 1258 mosquito larvae were collected with 672 originating from puddles (A–D) within the area where human cases of Ockelbo disease and SINV-infected mosquitoes were previously found (Bergqvist et al. 2015). Furthermore, 586 larvae were collected from the puddles (E–K), at an adjacent area, south of the village Ånäset. A second set of larvae captured in 2015 constituted in total 1865 larvae sampled at three locations close to puddle B.
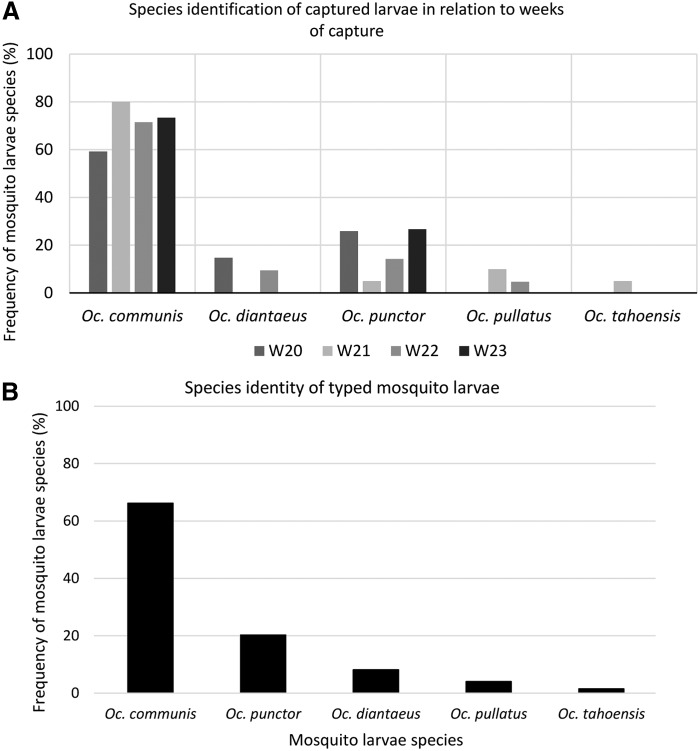
DNA was extracted from 76 random representatives of the first set of sampled larva, originating from all puddles and weeks of examination (Fig. 3A). They were sequenced and species identified by comparisons to sequence databases of adult mosquitoes followed by phylogenetic analysis for verification (data not shown). The typed larvae species are shown in (Fig. 3B). Oc. communis and Oc. punctor were the most abundant larvae species during the four consecutive weeks 20–23 in 2014 (Fig. 3). Interestingly, two of the larva samples displayed high sequence homology to multiple mosquito species in sequence databases, while the other 74 larvae show highest sequence identity only to one mosquito species.
FIG. 3.
(A) Species identification of captured larvae in relation to weeks of capture. The results are shown in percentage of specific species collected in 2014 in relation to the total number of barcoded mosquitoes at weeks (W) 20–23. (B) Species identity. Species identity of 76 genetically typed mosquito larvae collected in 2014 after direct sequence comparison and species verification by phylogeny toward previously published sequences.
Screening for SINV RNA in mosquito larvae
When samples of mosquito larvae from the 2014 collection were analyzed for the presence of SINV RNA, surprisingly 29 out of 41, 30× pools showed a positive signal for SINV RNA (cycle threshold [CT] values below 40). PCR positive 30× pools were found at 9 of the 10 investigated sites. Furthermore, when ten 10× pools of the 29 positive 30× pools were screened for SINV RNA, 56 larva samples showed presence of SINV RNA. No difference was noticed between samples from the two collection areas during week 20 (2014). From the 56 positives, a selection of 40 individual larvae, based on CT-values, was investigated further. Interestingly, there was a tendency that the level of virus RNA decreased with time of harvest (between weeks 20 and 23). Of the 40 tested single larva samples, nine showed CT-values between 31 and 35. Six SINV-positive mosquito larvae were successfully species determined by genetic barcoding, and three COI sequences corresponded to Oc. communis, two to Oc. punctor, and one mosquito larva was identified as Oc. diantaeus. However, efforts to infect cells or sequence larger parts of the SINV genome were not successful with these samples.
Screening for INKV RNA in mosquito larvae
The finding of SINV RNA in the larvae collection from 2014 encouraged us to analyze a second set of larvae sampled in 2015 with primers for several Arboviruses. Interestingly, one 100× pool sample (S3) became PCR positive when using Orthobunyavirus primers and it was confirmed to be INKV when using INKV-specific PCR primers.
Further screening of 10× pools of the positive 100× pool was performed and the results indicated that seven out of the ten 10× pools were positive. Two of the seven positive 10× pools were then selected as representatives of all the positive 10× pools. Furthermore, when individual larva homogenates of the two selected 10× pools were then analyzed, seven homogenates (39%) were found to be positive for INKV and five larvae were species identified by sequencing the COI gene. All five mosquito larvae that contained INKV RNA were Oc. communis. This would then suggest 1.5% positives out of all collected larvae. Even though large parts of the INKV genome were amplified and sequenced, efforts to infect cells with these samples were not successful.
Discussion
Mosquito-borne virus transmission involves the presence of competent vectors, which have the capacity to acquire, replicate, and transmit the virus to susceptible hosts (Schneider and Higgs 2008, Dubrulle et al. 2009). In the hosts, the virus is amplified to sufficient levels leading to viremia that facilitates dissemination of the virus by mosquitoes through blood meals to susceptible hosts (Weaver 2005).
In this study, possible vertical transmission was addressed by analyzing samples of mosquito larvae for mosquito-borne viruses within a previous SINV outbreak area (Bergqvist et al. 2015). In northern Sweden, SINV cases occur during late summer, before a long and cold winter. In the early summer of 2014, 1 year after the outbreak, we detected SINV RNA in mosquito larvae, indicating vertical viral transmission. Interestingly, another prevalent mosquito-borne virus, INKV, was also detected in collected larvae during 2015. Our results suggest that SINV and INKV may hibernate in mosquito eggs and amplify during development. Unfortunately, the samples were not in condition to allow isolation of infectious virus.
Maintenance and dissemination of mosquito-borne virus in nature are mainly through horizontal transmission between susceptible vertebrate hosts and mosquitoes or through vertical transmission when the virus is passed from the infected adult mosquitoes to their offspring. Only a few studies have experimentally demonstrated transovarial transfer of viruses. One example is Rift Valley fever virus that has been shown to persist in mosquito Aedes eggs (Linthicum et al. 1985, Balkhy and Memish 2003). It has also been demonstrated that dengue virus-infected Ae. aegypti eggs may produce infected adult mosquitoes (Khin and Than 1983). Arboviruses have also been found in field-collected mosquito larva that has been reared to adult stage (Dhileepan et al. 1996, Reese et al. 2010).
In this article, we showed for the first time that SINV and INKV could be detected in mosquito larvae in northern Europe, indicating that these viruses could be maintained in the environment through vertical transmission. However, even though SINV was detected in a large proportion of the larvae during 2014, no larvae were positive in 2015, suggesting that horizontal transfer is an important contributor. INKV is a more prevalent infection than SINV in the population in this region (Ahlm et al. 2014, Evander et al. 2016) and it seems likely that vertical transmission in mosquitoes of INKV is more common, although it needs to be further studied. As far as we know, SINV was detected for the first time in three Ochlerotatus species, which merit further investigation in field studies and laboratory experiments. INKV was detected in a mosquito species previously known to harbor INKV (Francy et al. 1989).
Using a molecular barcoding method developed for adult mosquitoes (Engdahl et al. 2014), we have successfully typed mosquito larvae, circumventing the need for rearing the larvae to adult mosquitoes for species determination. Five different mosquito species were identified and the results clearly indicated that the distribution of mosquito species was similar, but differed greatly in numbers between the different collection sites.
Three of the mosquito species (Oc. diantaeus, Oc. communis, and Oc. punctor) have previously been found in the county of Västerbotten, while the remaining two mosquito species (Oc. pullatus and Oc. tahoensis) have not been detected in Västerbotten before (Lundström et al. 2013), although Oc. pullatus has been found in several places in Sweden (Lundström et al. 2013). To our understanding, Oc. tahoensis has not been found in Sweden before. However, there is an uncertainty whether it is a subspecies to Oc. communis or a separate species (Shultz et al. 1993).
It is also evident that demographic, climatic, and societal changes have a major impact on the ecology of arboviruses in general. Some of the factors include population growth, urbanization, ecotourism, human encroachment on natural disease foci, alterations in land use, lack of efficient vector control strategies, and climate change among other factors (Gubler 2002). Weather and climate factors in endemic areas have been shown to affect the probability of SINV outbreaks (Uejio et al. 2012, Jalava et al. 2013). For instance, snow in late spring combined with warm temperatures during the late summer has been suggested to precede a higher incidence of SINV infections in Finland (Jalava et al. 2013).
Conclusions
From our understanding, this is the first report on species identification by DNA barcoding of mosquito larvae. The detection of SINV and INKV RNA in mosquito larvae indicated a likelihood of the viruses to be vertically transmitted. These findings imply that these viruses may hibernate in mosquito eggs and continue to be infective after hatching into the larval stage.
Acknowledgments
The authors acknowledge the following: Emma Åhlén, Joakim Bergqvist, Alexandra Andersson, and Maj Bylund for their technical support; Per Wikström for the graphics; and Kristoffer Ahlm for sampling mosquito larvae.
Financial Support
The study was supported by grants from the Medical Faculty of Umeå University, the County Councils of Norrbotten and Västerbotten, and the Swedish Research Council Formas (grant no. 221-2014-1556).
Author Contributions
C.A., M.E., and G.B. conceptualized the idea and initiated the study. O.T., O.W.L., and I.S. collected the larva, prepared RNA samples, did PCR analysis, prepared samples for sequencing, and contributed to the writing. J.N. participated in the experimental design, preparation of samples, and contributed to the writing. P.V.S. collected and analyzed weather data. C.E. contributed to the experimental design. C.A. and M.E. organized the trapping and contributed to writing and proofreading. G.B. participated in the experimental design, coordinated this study, and drafted the article. All the authors read and approved the final article.
Author Disclosure Statement
No competing financial interests exist.
References
- Ahlm C, Eliasson M, Vapalahti O, Evander M. Seroprevalence of sindbis virus and associated risk factors in northern Sweden. Epidemiol Infect 2014; 142:1559–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balkhy HH, Memish ZA. Rift Valley fever: an uninvited zoonosis in the Arabian peninsula. Int J Antimicrob Agents 2003; 21:153–157 [DOI] [PubMed] [Google Scholar]
- Bergqvist J, Forsman O, Larsson P, Naslund J, et al. Detection and isolation of sindbis virus from mosquitoes captured during an outbreak in Sweden, 2013. Vector Borne Zoonotic Dis 2015; 15:133–140 [DOI] [PubMed] [Google Scholar]
- Brummer-Korvenkontio M. Arboviruses in Finland. V. Serological survey of antibodies against Inkoo virus (California group) in human, cow, reindeer, and wildlife sera. Am J Trop Med Hyg 1973; 22:654–661 [DOI] [PubMed] [Google Scholar]
- Brummer-Korvenkontio M, Saikku P. Mosquito-borne viruses in Finland. Med Biol 1975; 53:279–281 [PubMed] [Google Scholar]
- Brummer-Korvenkontio M, Vapalahti O, Kuusisto P, Saikku P, et al. Epidemiology of Sindbis virus infections in Finland 1981–1996: possible factors explaining a peculiar disease pattern. Epidemiol Infect 2002; 129:335–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant JE, Crabtree MB, Nam VS, Yen NT, et al. Isolation of arboviruses from mosquitoes collected in northern Vietnam. Am J Trop Med Hyg 2005; 73:470–473 [PubMed] [Google Scholar]
- Dhileepan K, Azuolas JK, Gibson CA. Evidence of vertical transmission of Ross River and Sindbis viruses (Togaviridae: Alphavirus) by mosquitoes (Diptera: Culicidae) in southeastern Australia. J Med Entomol 1996; 33:180–182 [DOI] [PubMed] [Google Scholar]
- Dubrulle M, Mousson L, Moutailler S, Vazeille M, et al. Chikungunya virus and Aedes mosquitoes: saliva is infectious as soon as two days after oral infection. PLoS One 2009; 4:e5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engdahl C, Larsson P, Naslund J, Bravo M, et al. Identification of Swedish mosquitoes based on molecular barcoding of the COI gene and SNP analysis. Mol Ecol Resour 2014; 14:478–488 [DOI] [PubMed] [Google Scholar]
- Eshoo MW, Whitehouse CA, Zoll ST, Massire C, et al. Direct broad-range detection of alphaviruses in mosquito extracts. Virology 2007; 368:286–295 [DOI] [PubMed] [Google Scholar]
- Evander M, Putkuri N, Eliasson M, Lwande OW, et al. Seroprevalence and risk factors of Inkoo Virus in Northern Sweden. Am J Trop Med Hyg [Epub ahead of print]; DOI: 10.4269/ajtmh.15-0270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folmer O, Black M, Hoeh W, Lutz R, et al. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol 1994; 3:294–299 [PubMed] [Google Scholar]
- Francy DB, Jaenson T, Lundström JO, Schildt E-B, et al. Ecologic studies of mosquitoes and birds as hosts of Ockelbo virus in Sweden and isolation of Inkoo and Batai viruses from mosquitoes. Am J Trop Med Hyg 1989; 41:355–363 [PubMed] [Google Scholar]
- Gubler DJ. The global emergence/resurgence of arboviral diseases as public health problems. Arch Med Res 2002; 33:330–342 [DOI] [PubMed] [Google Scholar]
- Huang C, Shope RE, Spargo B, Campbell WP. The S RNA genomic sequences of Inkoo, San Angelo, Serra do Navio, South River and Tahyna bunyaviruses. J Gen Virol 1996; 77:1761–1768 [DOI] [PubMed] [Google Scholar]
- Hubalek Z. Mosquito-borne viruses in Europe. Parasitol Res 2008; 103 Suppl 1:S29–S43 [DOI] [PubMed] [Google Scholar]
- Hubalek Z, Halouzka J, Juricova Z, Sikutova S, et al. Serologic survey of birds for West Nile flavivirus in southern Moravia (Czech Republic). Vector Borne Zoonotic Dis 2008; 8:659–666 [DOI] [PubMed] [Google Scholar]
- Jalava K, Sane J, Ollgren J, Ruuhela R, et al. Climatic, ecological and socioeconomic factors as predictors of Sindbis virus infections in Finland. Epidemiol Infect 2013; 141:1857–1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khin MM, Than KA. Transovarial transmission of dengue 2 virus by Aedes aegypti in nature. Am J Trop Med Hyg 1983; 32:590–594 [DOI] [PubMed] [Google Scholar]
- Kuno G, Mitchell CJ, Chang GJ, Smith GC. Detecting bunyaviruses of the Bunyamwera and California serogroups by a PCR technique. J Clin Microbiol 1996; 34:1184–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurkela S, Helve T, Vaheri A, Vapalahti O. Arthritis and arthralgia three years after Sindbis virus infection: clinical follow-up of a cohort of 49 patients. Scand J Infect Dis 2008; 40:167–173 [DOI] [PubMed] [Google Scholar]
- L'vov SD, Gromashevskii VL, Morozova TN, Aristova VA, et al. [Distribution of viruses from the Californian encephalitis serogroup (Bunyaviridae, Bunyavirus) in the northern expanses of Russia]. Vopr Virusol 1997; 42:229–235 (Article in Russian). [PubMed] [Google Scholar]
- Laine M, Luukkainen R, Toivanen A. Sindbis viruses and other alphaviruses as cause of human arthritic disease. J Intern Med 2004; 256:457–471 [DOI] [PubMed] [Google Scholar]
- Linthicum KJ, Davies FG, Kairo A, Bailey CL. Rift Valley fever virus (family Bunyaviridae, genus Phlebovirus). Isolations from Diptera collected during an inter-epizootic period in Kenya. J Hyg (Lond) 1985; 95:197–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundstrom JO, Lindstrom KM, Olsen B, Dufva R, et al. Prevalence of sindbis virus neutralizing antibodies among Swedish passerines indicates that thrushes are the main amplifying hosts. J Med Entomol 2001; 38:289–297 [DOI] [PubMed] [Google Scholar]
- Lundström JO. Mosquito-borne viruses in western Europe: a review. J Vector Ecol 1999; 24:1–39 [PubMed] [Google Scholar]
- Lundström JO, Pfeffer M. Phylogeographic structure and evolutionary history of Sindbis virus. Vector Borne Zoonotic Dis 2010; 10:889–907 [DOI] [PubMed] [Google Scholar]
- Lundström JO, Schäfer ML, Hesson JC, Blomgren E, et al. The geographic distribution of mosquito species in Sweden. J Eur Mosq Control Assoc 2013; 31:21–35 [Google Scholar]
- McWilliam H, Li W, Uludag M, Squizzato S, et al. Analysis tool web services from the EMBL-EBI. Nucleic Acids Res 2013; 41:W597–W600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell CJ, Lvov SD, Savage HM, Calisher CH, et al. Vector and host relationships of California serogroup viruses in western Siberia. Am J Trop Med Hyg 1993; 49:53–62 [DOI] [PubMed] [Google Scholar]
- Niklasson B, Vene S. Vector-borne viral diseases in Sweden—a short review. Arch Virol Suppl 1996; 11:49–55 [DOI] [PubMed] [Google Scholar]
- Putkuri N, Kurkela S, Levanov L, Huhtamo E, et al. Isolation and characterization of a California encephalitis serogroup orthobunyavirus from Finnish mosquitoes. Infect Genet Evol 2014; 22:164–173 [DOI] [PubMed] [Google Scholar]
- Putkuri N, Vaheri A, Vapalahti O. Prevalence and protein specificity of human antibodies to Inkoo virus infection. Clin Vaccine Immunol 2007; 14:1555–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese SM, Mossel EC, Beaty MK, Beck ET, et al. Identification of super-infected Aedes triseriatus mosquitoes collected as eggs from the field and partial characterization of the infecting La Crosse viruses. Virol J 2010; 7:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sane J, Kurkela S, Levanov L, Nikkari S, et al. Development and evaluation of a real-time RT-PCR assay for Sindbis virus detection. J Virol Methods 2012; 179:185–188 [DOI] [PubMed] [Google Scholar]
- Schneider BS, Higgs S. The enhancement of arbovirus transmission and disease by mosquito saliva is associated with modulation of the host immune response. Trans R Soc Trop Med Hyg 2008; 102:400–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shultz SJ, Eldridge FB. Biogeography of the Aedes (ochlerotatus) communis species complex (diftera: culicidae) in the western United States. Mosq Syst 1993; 25:170–176 [Google Scholar]
- Simon C, Frati F, Beckenbach A, Crespi B, et al. Evolution, weighting, and phylogenetic utility of mitochondrial gene-sequences and a compilation of conserved polymerase chain-reaction primers. Ann Entomol Soc Am 1994; 87:651–701 [Google Scholar]
- Tang KL, Agnew MK, Hirt MV, Sado T, et al. Systematics of the subfamily Danioninae (Teleostei: Cypriniformes: Cyprinidae). Mol Phylogenet Evol 2010; 57:189–214 [DOI] [PubMed] [Google Scholar]
- Uejio CK, Kemp A. Comrie AC. Climatic controls on West Nile virus and Sindbis virus transmission and outbreaks in South Africa. Vector Borne Zoonotic Dis 2012; 12:117–125 [DOI] [PubMed] [Google Scholar]
- Vapalahti O, Plyusnin A, Cheng Y, Manni T, et al. Inkoo and Tahyna, the European California serogroup bunyaviruses: sequence and phylogeny of the S RNA segment. J Gen Virol 1996; 77:1769–1774 [DOI] [PubMed] [Google Scholar]
- Weaver SC. Host range, amplification and arboviral disease emergence. Arch Virol Suppl 2005; 33–44 [DOI] [PubMed] [Google Scholar]