Abstract
The placenta is an organ that is formed transiently during pregnancy, and appropriate placental development is necessary for fetal survival and growth. Proper differentiation of the labyrinthine layer of the placenta is especially crucial, as it establishes the fetal–maternal interface that is involved in physiological exchange processes. Although previous studies have indicated the importance of inhibitor of differentiation/inhibitor of DNA binding-2 (Id2) helix-loop-helix transcriptional regulator in mediating cell differentiation, the ability of Id2 to regulate differentiation toward the labyrinthine (transport) lineage of the placenta has yet to be determined. In the current study, we have generated labyrinthine trophoblast progenitor cells with increased (SM10-Id2) or decreased (SM10-Id2-shRNA) Id2 expression and determined the effect on TGF-β-induced differentiation. Our Id2 overexpression and knockdown analyses indicate that Id2 mediates TGF-β-induced morphological differentiation of labyrinthine trophoblast cells, as Id2 overexpression prevents differentiation and Id2 knockdown results in differentiation. Thus, our data indicate that Id2 is an important molecular mediator of labyrinthine trophoblast differentiation. An understanding of the regulators of trophoblast progenitor differentiation toward the labyrinthine lineage may offer insights into events governing pregnancy-associated disorders, such as placental insufficiency, fetal growth restriction, and preeclampsia.
Introduction
The placenta is an organ that is only formed during pregnancy, and its proper development is essential for embryonic growth and fetal survival. The placenta is responsible for the transport of nutrients, gases, and wastes between the mother and the fetus [1–4]. Trophoblast cells that make up the placenta must properly differentiate into the appropriate cell types (lineages) to facilitate this transport [3–7]. Abnormal placental development has been proposed to lead to a reduction in placental function and subsequent pregnancy-associated disorders [1–8].
Numerous molecular events regulating placental development are conserved in both humans and mice. Both human and rodent placentas consist of analogous cell types that are involved in placental transport processes [3–11]. In rodents, the placenta comprises two zones: the junctional zone and the labyrinth. The placenta is further subdivided into three predominant cell lineages: labyrinthine cells, spongiotrophoblasts, and trophoblast giant cells [3,6,12–14]. Different trophoblast lineages are derived via differentiation of trophoblast progenitor cells that perform specialized functions during gestation [3–5,12]. The labyrinthine cells mediate the physiological fetal–maternal exchange processes, particularly gas, waste, and nutrients [3–5,9,10,12,15,16].
Transport across the labyrinthine layer of the placenta is the main means by which the fetus is able to obtain the appropriate nutrients for growth and development [5,12,14,17–20]. Thus, the differentiation of placental progenitors into labyrinthine cells is of critical importance for assuring fetal survival and well-being. In addition to offering insights into signals regulating differentiation of labyrinthine trophoblast cells, murine knockout and in vitro studies have been invaluable in advancing our understanding of genes that are required for proper placental development [5,6,15,16,21–25].
Morphological differentiation involves branching and fusion of labyrinthine trophoblast stem (TS) cells, leading to formation of a multinucleate exchange surface that is functionally suited for placental transport of nutrients, particularly glucose [3,6,12,26,27]. The molecular events governing labyrinthine trophoblast differentiation have been reported to involve alterations in the expression of several transcription factors, including Id2, Cdx2, and Gcm1 [21,28–36].
Several transcription factors belonging to the helix-loop-helix (HLH) family are essential for human and rodent placental development and for guiding trophoblast differentiation. Most HLH proteins possess a highly conserved basic region that allows DNA binding and transcriptional regulation. In the placenta, the basic-HLH (bHLH) transcription factors, Mash2, Hand1, and Stra13 direct trophoblast differentiation along the giant cell lineage [29,32,37–40]. In contrast, bHLH transcription factor Tfeb has been shown to be essential for the vascularization and functional differentiation of the labyrinthine trophoblast lineage of the placenta [41,42]. Additionally, spatiotemporal regulation of the expression of bHLH transcription factors and the ability of HLH inhibitor of differentiation/inhibitor of DNA binding (Id) proteins to modulate these proteins have been reported to be necessary for proper placental development and cell differentiation [29,20,43–46].
Id proteins are widely expressed throughout development and function in the determination of cell specification into specialized lineages [29,30,43,44,47–49]. Id proteins lack the basic DNA-binding region of bHLH transcription factors. Instead of binding DNA, Ids are capable of binding to bHLH proteins, thus inhibiting bHLH-induced transcription that is necessary for differentiation [29,48–50].
Four different Id isoforms (Id1–Id4) have been identified and Id1, Id2, and Id3 expression has been reported in the human and rodent placentas [28–30,43,48,49,51,52]. In particular, Id2 is known to be an important regulator of placental differentiation, since Id2 mRNA and protein are the highest in proliferative TS cells and decline during differentiation into lineage-specific trophoblast subtypes in both humans and rodents [16,30,53]. Additionally, previous studies in human and rodent cultures have indicated that sustained Id1 and Id2 expression can prevent differentiation into giant cells and extravillous trophoblasts [29,30,43].
Our previous studies have demonstrated a dramatic downregulation of Id2 expression during TGF-β-induced differentiation of the labyrinthine-specific trophoblast progenitor cell line, SM10 [15]. In the current study, we have generated SM10-Id2 clonal cell lines and determined the importance of Id2 in mediating TGF-β-induced morphological, functional, and molecular differentiation. We have furthermore analyzed the effects of RNAi-mediated Id2 knockdown on the differentiation process. Our results suggest that Id2 downregulation is necessary for labyrinthine trophoblast differentiation.
Materials and Methods
Materials
Cell culture media [RPMI1640/l-glutamine and Dulbecco's modified Eagle medium (DMEM)] was obtained from Mediatech. Antibiotic-antimycotic, trypsin-EDTA, sodium pyruvate, 100 bp DNA ladder, broad range protein marker, and dNTPs were obtained from Invitrogen. Human recombinant-fibroblast growth factor 4 (hrFGF4) and heparin were obtained from Sigma. 2β-mercaptoethanol and supersignal chemiluminescence reagent were purchased from Pierce. Reverse transcriptase was obtained from Stratagene, and random hexamers and Taq polymerase were from Promega. All reagents and primers for quantitative polymerase chain reaction (qPCR) were purchased from Ambion. Protease inhibitor cocktail tablets and RNase-free DNaseI were purchased from Roche. Rabbit anti-Id2 antibody (sc-489) was purchased from SantaCruz. Goat anti-mouse-IgG-HRP secondary antibody was obtained from BD Transduction Laboratories. Reverse transcription (RT)-PCR primers were purchased from Gibco. Hoechst dye (No. 33258) was obtained from Sigma, and Metafectene transfection reagent (T020) was from Biontex Laboratories. Vectashield mounting media was purchased from Vector Laboratories. Rhodamine-conjugated phalloidin was purchased from Molecular Probes. Polybrene was obtained from Millipore, and G418 was from Invivogen. 3[H]-2-deoxyglucose was purchased from MP Biochemicals. The shDNA sequences and the lentiviral infection system were purchased from Invitrogen. TaqMan® MGB probe was purchased from Applied Biosystems (4316034).
Cell culture and differentiation
Mouse SM10 cells were maintained in RPMI 1640/l-glutamine medium supplemented with 1 mM sodium pyruvate, 50 μM 2β-mercaptoethanol, 1% antibiotic-antimycotic, and 10% fetal bovine serum (FBS) [15,16,22,23]. Differentiation of SM10 cells was induced by treatment with 5 ng/mL of TGF-β in vehicle [4 mM HCl, 0.1% v/v bovine serum albumin in 1× phosphate-buffered saline (PBS)] for 72 h [15,53]. TS cells were propagated in RPMI 1640 containing 20% FBS, 100 μM 2β-mercaptoethanol, 1 mM sodium pyruvate, hrFGF4 (25 ng/mL), and heparin (1 μg/mL), as previously described [21]. Simultaneous differentiation of TS cells into three trophoblast subtypes was induced by withdrawal of hrFGF4 and heparin from the culture medium. All cells were maintained at sub-confluency at 37°C and 5% CO2. 293FT cells were maintained in DMEM/high glucose (Thermo Scientific; SH30027.01), 10% heat-inactivated FBS (Biowest; S01520), 1% antibiotic-antimycotic (Thermo Scientific; SV30079.01), 1 mM sodium pyruvate (Mediatech, Inc.; 25-000-CI), 2 mM glutaGRO Supplement (Mediatech, Inc.; 25-015-CI), 1 × NEAA Mixture (Lonza; 13-114E), and 500 μg/mL G418 (InvivoGen; ant-gn-1). Cos7 cells were cultured in DMEM/high glucose, 10% heat-inactivated FBS, and 1% antibiotic-antimycotic. Rcho-1 cells were cultured as previously described [54–56]. All cell lines were passaged at 80%–90% confluence.
Design of Id2 short hairpin sequences
All the Id2 siRNA and scrambled siRNA sequences were designed against the coding region of mouse Id2 gene (BC053699) using siRNA Wizard™ that was available online. Of the several recommended siRNA sequences, the sequences with the most favorable thermodynamic properties were chosen [57,58]. The siRNA sequences were converted into shRNA sequences using Block-it™ RNAi Designer with loop sequences 5′ TTCAAGAGA3′ or 5′CCACAC3′ [59,60]. Two previously published Id2 siRNA sequences were additionally converted into shDNA sequences [61].
Cloning of Id, GFP, and shDNA into lentiviral vector
The human Id2 cDNA was excised from pCDNA3-Id2, and the GFP cDNA was excised from pLv-CMV-GFP-V5 using BamHI and ApaI [45]. Restriction products were resolved by electrophoresis on 1% agarose gels and visualized by ethidium bromide staining. Appropriate restriction fragments were gel extracted and ligated as recommended by the manufacturer. Positive ligations were analyzed by restriction enzyme and western blotting analysis. Correct cloning of pLv-Id2 was further confirmed by sequencing using CMV-F and V5-R primers (Table 1). Id2 shDNA and scrambled shDNA sequences (Table 2) were cloned into the lentiviral expression plasmids using the BLOCK-iT Lentiviral RNAi Expression System. Briefly, shDNA sequences were annealed and cloned into linearized pENTR™/U6 vector as recommended by the manufacturer. Cloning products were analyzed by restriction enzyme analysis (BamH1, EcoRV), and positive clones were used for LR recombination into pLenti6/Block-it-DEST vector. Recombination reactions were performed as recommended by the supplier and confirmed by restriction enzyme analysis (BamHI, KpnI). Correct cloning into pLenti6/Block-it-DEST vector was further confirmed by sequencing using the U6-F and V5-R primers (Table 1).
Table 1.
Reverse Transcription-Polymerase Chain Reaction Primer Sequences Used in This Study
| Gene | PCR primer sequences | Tm (°C) | Size (bp) | Reference | |
|---|---|---|---|---|---|
| Id1 | F5′tggacgaacagcaggtgaacg3′ | R5′gcactgatctcgccgttcagg3′ | 58 | 243 | 59 |
| Id2 | F5′tctgagcttatgtcgaatgatagc3′ | R5′cacagcattcagtaggctcgtgtc3′ | 62 | 497 | 12,60 |
| Id3 | F5′tgctacgaggcggtgtgctg3′ | R5′agtgagctcagctgtctggatcgg3′ | 58 | 287 | 59 |
| Id4 | F5′gcgatatgaacgactgctac3′ | R5′tcaccctgcttgttcacggc3′ | 58 | 270 | 59 |
| β-Actin | F5′atcgtgggccgccctaggca3′ | R5′tggccttagggttcagagggg3′ | 55–65 | 243 | 12,61 |
| U6 | F5′ggactatcatatcgttaccg | 62 | * | C | |
| CMV | F5′cgcaaatgggcggtaggcgtg | 62 | * | C | |
| V5 | R5′accgaggagagggttagggat | 62 | * | C | |
Forward (F) and reverse (R) RT-PCR primer sequences, amplification temperature (Tm°C), expected size of the amplification product, and the reference for the primer sequence sources are shown. Genes for RT-PCR amplification primers are bolded. Genes for PCR amplification from genomic DNA are in italics. The size of the amplification product may vary, depending on the size of the insert, and these primers were additionally used to confirm correct cloning of Id2 and short hairpin constructs into the lentiviral vectors (*). The primer sequence was obtained from Invitrogen (C).
RT-PCR, reverse transcription-polymerase chain reaction.
Table 2.
Short Hairpin Sequences Against Id2 RNA Used in This Study
| SiRNA | Sense sequences | Loop | Antisense sequence | Targets |
|---|---|---|---|---|
| Id2-96 | T5′gatgagtctgctctacaaca | ttcaagaga | tgttgtagagcagactcatc3′ | 96–115 |
| B5′gatgagtctgctctacaaca | tctcttgaa | tgttgtagagcagactcatc3′ | ||
| Id2-369 | T5′gcttatgtcgaatgatagca | ttcaagaga | tgctatcattcgacataagc3′ | 369–388 |
| B5′gcttatgtcgaatgatagca | tctcttgaa | tgctatcattcgacataagc3′ | ||
| Id2-96s | T5′gtcgataccttagactcaga | ttcaagaga | tctgagtctaaggtatcgac3′ | Scrambled |
| B5′gtcgataccttagactcaga | tctcttgaa | tctgagtctaaggtatcgac3′ | ||
| Id2-369s | T5′gatttcggatcagaacatgt | ttcaagaga | acatgttctgatccgaaatc3′ | Scrambled |
| B5′gatttcggatcagaacatgt | tctcttgaa | acatgttctgatccgaaatc3′ |
The number after Id2 indicates the first coding region nucleotide recognized by the siRNA. Sense and antisense DNA sequences for the top (T) strand and the bottom (B) strand oligos, loop regions, and complete Id2 coding region targeted by the siRNA are shown. The Id2 shDNA sequences against the coding region of the mouse Id2 gene used in this study were generated using the siRNA Wizard™ and Block-iT™ RNAi designers and confirmed by NCBI nucleotide Blast.
Viral production and transductions
The viral supernatant was produced as recommended by the supplier. Briefly, 6 × 106 293FT cells were plated 24 h before transfection. The cells were transfected with the recommended amount of ViraPower mix and 10 μg of lentiviral expression vector using Metafectene transfection reagent. Sixteen hours later, the transfection media were removed and cells were provided fresh media. The virus containing media was collected at 60 h and applied to cells with 12 μg/mL of polybrene. Infections were allowed to proceed for 12 h, after which the cells were supplied with fresh media. All infections were performed using fresh virus on SM10 cells seeded the previous day.
Establishing Id2 and GFP expressing SM10 stable cell lines
Cells were plated at 3.3 × 103 cells/mL in 60 mm dishes. Twenty-four hours post-seeding, SM10 cells were infected with freshly isolated pLv-Id2 or pLv-GFP virus in media containing 12 μg/mL polybrene. After selection in blasticidin (8 μg/mL), Id2 and GFP stable clones were established by limiting dilution of 0.5 cell/well. Positive clones were analyzed by PCR amplification of genomic DNA using CMV-F and V5-R primers flanking the Id2 and GFP genes and by western blotting analysis of Id2 (Table 1 and Fig. 3).
FIG. 3.
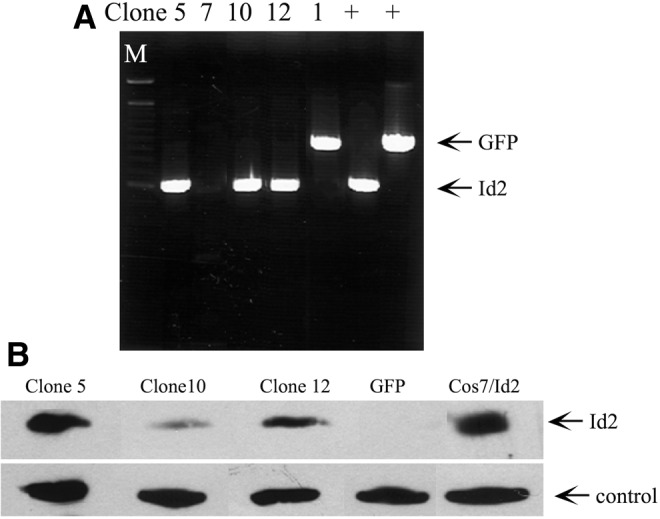
Generation of SM10-Id2 and SM10-GFP stable cell lines. (A) SM10 cells were transduced with lentiviral-Id2 or lentiviral-GFP, and stable cell lines were generated by limiting dilution. Total genomic DNA was extracted, PCR amplified using primers flanking the gene, and resolved on a 1% agarose gel. Genomic integration of Id2 in clones 5, 10, 12, and GFP in clone 1 is shown. One hundred basepair DNA marker (M) and amplification from lentiviral plasmids (+) were utilized in estimating the sizes of PCR amplification products and for positive controls, respectively. (B) Western blotting analysis of Id2 in the SM10-GFP and SM10-Id2 clones. Two hundred fifty micrograms of whole cell lysate was resolved on an 11% SDS-PAGE, transferred to PVDF membrane, and analyzed for Id2 protein levels with Id2 antibody. Cos7 cells transiently expressing lentiviral Id2 were used as a positive control. A cross-reactive band served as a fortuitous loading control. PVDF, polyvinylidene difluoride; SDS-PAGE, sodium dodecyl sulphate-polyacrylamide gel electrophoresis.
Reverse-transcription-PCR
Cells were harvested by trypsinization and centrifuged at 2,000 rpm for 5 min. The media were aspirated, and total RNA was extracted using the RNeasy Kit (Qiagen) according to the manufacturer's instructions. To remove any contaminating DNA, the samples were treated with RNase-free DNaseI for 30 min at 37°C. For RT-PCR analysis, 5 μg of total RNA was reverse transcribed using random primers and PCR was amplified for 40 cycles at appropriate temperatures for each primer set (Table 1) [62–64]. Briefly, PCR was performed using 2.5 U of Taq DNA polymerase, 1× PCR buffer without MgCl2, 1.875 mM MgCl2, 200 μM dNTPs, and 200 nM reverse and forward primers. Co-amplification of β-actin in each sample was used for semiquantitative normalization. After the initial denaturation at 94°C for 5 min, all PCRs were performed under the following conditions: 95°C denaturation for 1 min, specific primer annealing temperature (Table 1) for 1 min, and primer extension at 72°C for 1 min for a total of 40 cycles. The PCR products were resolved by electrophoresis on 1% agarose gels and visualized by ethidium bromide staining.
Real-time qPCR
Total RNA was collected and processed as described earlier. RNA (2 μg) was reverse transcribed using random primers, and samples were diluted 1:2. Real-time qPCR was performed using an ABI Prism 7700 thermal cycler (Applied Biosystems). Id2 and β-actin amplification was detected using a sequence-specific TaqMan MGB probe and Id2 or β-actin-specific primers. Briefly, each reaction contained 900 nM antisense primers, 900 nM sense primers, a 250 nM probe, 1 × Taqman Universal Master Mix (10 μL), and cDNA sample (1 μL) in a final reaction volume of 20 μL. Samples from three independent experiments were analyzed in triplicate. All samples were run simultaneously on 96-well reaction plates as follows: PCR amplification included an initial phase of 10 min at 95°C, then 40 cycles of 15 s at 95°C, and 1 min at 60°C. Expression levels of Id2 were determined by normalization to β-actin (endogenous control) in each sample using the comparative CT method 2−ΔΔCt [53,65]. All real-time qRT-PCR experiments included a no-template (H2O) control. To quantify decreased Id2 expression, we analyzed changes in Id2 expression by qPCR. Undifferentiated TS cells were used as a positive control for semiquantitative RT-PCR and qPCR analyses.
Western blotting and immunostaining
Western blotting was performed as previously described using rabbit polyclonal anti-Id2 antibody at 1/500 overnight at 4°C overnight in blocking buffer [5% (w/v) non-fat milk, pH 7.4 in 1× PBS containing 0.05% Tween-20 (PBST)] [54,66]. The membrane was washed with PBST, probed with HRP-conjugated goat anti-rabbit IgG secondary antibody at 1/10,000 for 1 h at room temperature, and visualized using Enhanced Chemi-Luminiscence reagent according to the manufacturer's instructions. For immunostaining, the SM10 cells were seeded at 3 × 104 cells/mL in 35 mm culture dishes. Twenty-four hours post-seeding, the cells were infected and subsequently treated with TGF-β or vehicle for 72 h. Cells were rinsed and fixed with 3.7% paraformaldehyde in 1× PBS. Fixation was quenched in 10 mM glycine in 1× PBS for 10 min, and cells were rinsed with 1× PBS. Cells were permeabilized and blocked in buffer containing 3% goat serum, 250 mM KCl, 20 mM HEPES pH7.8, 0.1% glycine, and 0.5% Triton X-100 in 1× PBS for 30 min. The cells were incubated with anti-Id2 antibody at 1/200 overnight at 4°C in blocking buffer; rinsed and probed with Alexfluor 594 F(ab′)2 fragmented goat anti-rabbit IgG antibody at 1/1,000 for 1.5 h; and incubated with 1 μg/mL Hoechst dye for 1 min to stain the cell nuclei. The coverslips were mounted using Vectashield mounting media, and images were visualized by epifluorescence microscopy and MetaMorph software.
Luciferase reporter transactivation
SM10 cells were plated at 5 × 104 cells/mL in a six-well plate. Twenty-four hours post-seeding, the cells were transfected for 24 h with 1 μg E7-TK-luc and 0.1 μg pRLSV40 using Metafectene transfection reagent, as previously described [53,67,68]. The cells were placed in fresh media and 5 ng/mL TGF-β was added for 72 h, after which the cells were rinsed in 1× PBS and analyzed using the Dual Luciferase Reporter system according to the manufacturer's recommendation. All E7-TK-Luc values were normalized to pRLSV40.
3[H]-2-deoxyglucose uptake
Glucose uptake was assayed as previously described [15,53,69]. Briefly, the cells were plated at 2 × 104 cells/mL in 24-well plates. Twenty-four hours post-seeding, the cells were differentiated for 72 h. The media were then aspirated, and the cells were washed three times with pre-warmed 1× PBS. The cells were pulsed for 10 min with 1 μCi of 3[H]-2-deoxyglucose in transport buffer (25 mM HEPES, 0.8 mM MgSO4, 140 mM NaCl, 5.4 mM KCl, 1.8 mM CaCl2) at 37°C. Glucose uptake was quenched by washing cells with ice-cold transport buffer. The cells were detached by adding 600 μL of 0.03% sodium dodecyl sulfate in H2O, and a 400 μL aliquot was analyzed for radioactivity by liquid scintillation counting. Cells were counted by trypan blue exclusion using the Beckman Coulter cell analyzer and Vi-Cell 1.01 software. Uptake values were normalized to cell counts. For the RNAi analyses, the cells were infected with 0.5 mL virus containing media and briefly selected (3 days) in media containing blasticidin (8 μg/mL).
Phalloidin staining
SM10 cells were seeded at 3 × 104 cells/mL in 35 mm culture dishes. Twenty-four hours post-seeding, the cells were infected and subsequently treated with 5 ng/mL TGF-β or vehicle. The cells were processed as described earlier for immunostaining. Cells were incubated with 6.6 μM rhodamine-conjugated phalloidin in blocking buffer for 1 h, rinsed in 1× PBS, and incubated with 1 μg/mL Hoechst dye for 1 min to stain the cell nuclei. Coverslips were mounted using Vectashield mounting media, and images were visualized by epifluorescence microscopy and MetaMorph software.
Statistical analysis
Experiments were independently replicated at least three times with similar results. Quantitative data from experiments are represented as the average ± SEM. Statistical analysis was performed by the Statistical Consulting Center at Wright State University by using one sample t-test for glucose uptake experiments and two-sample t-test for qPCR analyses. P values for 95% confidence interval were calculated. P < 0.025 was considered significant and is denoted by *.
Results
Id2 is the only Id family member that exhibits decreased RNA expression on differentiation of the labyrinthine trophoblast progenitor cell line, SM10
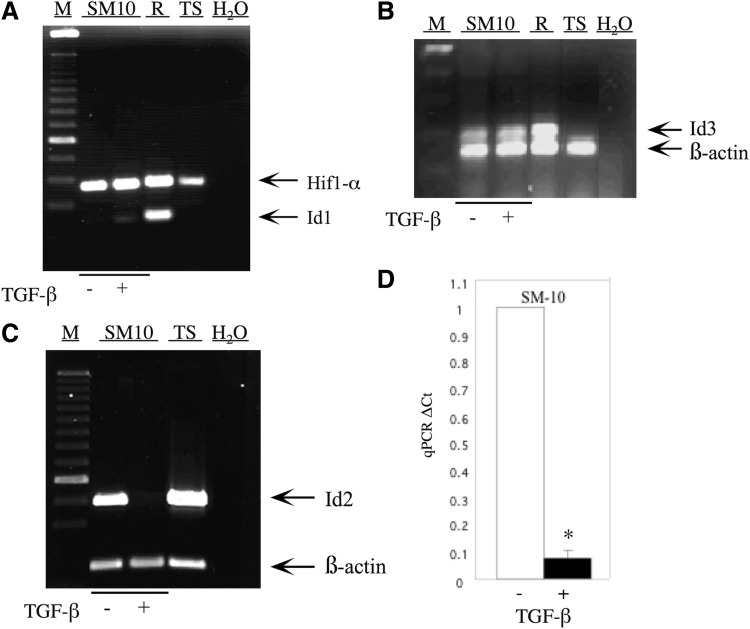
Previous reports have shown expression of several Id isoforms in rodent and human trophoblast cells [28–30,43,52]. We have previously demonstrated that the expression of Id2 is dramatically reduced during differentiation of labyrinthine-specific, trophoblast progenitor SM10 cells [15]. To determine the presence of other Id isoforms in SM10 cells and their response to TGF-β or vehicle treatment, the expression of Id1–Id4 in SM10 cells was analyzed by RT-PCR and qPCR. Expression of Id1 was minimally detectable in undifferentiated and differentiated SM10 cells (Fig. 1A). Rcho-1 and TS cells served as controls and were analyzed for Id1 expression, and only the Rcho-1 cells expressed Id1. In contrast to Id1, Id2 expression was readily detectable in vehicle-treated SM10 cells and dramatically downregulated in response to TGF-β, as previously described (Fig. 1C). Quantification of Id2 expression by real-time qPCR analysis indicated a significant 13-fold reduction in Id2 levels in the SM10 cell line in response to TGF-β (Fig. 1D). Low levels of Id3 expression were detected in the SM10 cell line; however, addition of TGF-β did not change the level of expression (Fig. 1B). Similar to Id1, Id3 could also be detected in Rcho-1 cells that were used as a control, but expression was absent in TS cells (Fig. 1B). Expression of Id4 could not be detected in TGF-β or vehicle-treated SM10 cells (data not shown).
FIG. 1.
Id2 expression is inhibited on differentiation of the labyrinthine trophoblast progenitor cell line, SM10. Total RNA from TGF-β or vehicle-treated, SM10 cells was RT-PCR amplified with appropriate primers (Table 1) and resolved on a 1% agarose gel. Expression of Id1 (A), Id2 (C), and Id3 (B) in TGF-β or vehicle-treated SM10 cells is shown. Undifferentiated Rcho-1 (R) and TS cells (TS) were used as positive controls; 100 bp DNA marker (M) was utilized in estimating the sizes of PCR amplification products. Co-amplification of β-actin was used for semiquantitative normalization. Due to similarity in size, Hif1-α was co-amplified for semiquantitative normalization of Id1 samples (A). H2O was substituted for reverse-transcribed RNA as a control to indicate no genomic contamination (A–C). TGF-β-induced changes in Id2 expression were quantified by real-time qPCR (D). Expression of Id2 levels was determined by normalization to β-actin (endogenous control) in each sample by using the comparative CT method with the TaqMan MGB probe for qPCR with a fold change = 2−ΔΔCT method. Error bars represent standard deviation from the mean *P < 0.0005. RT-PCR, reverse transcription-polymerase chain reaction; qPCR, quantitative PCR; TS, trophoblast stem.
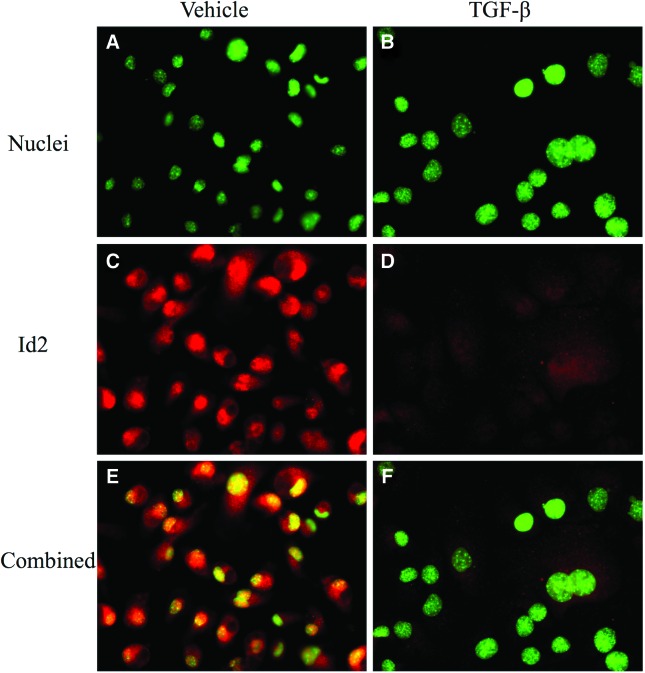
Id2 protein is decreased in differentiated SM10 cells
In addition to mRNA downregulation during trophoblast progenitor cell differentiation, Id2 protein levels during differentiation were also investigated. To analyze changes in Id2 protein levels during differentiation of progenitors into labyrinthine cells, Id2 protein levels were investigated via immunofluorescent staining of TGF-β and vehicle-treated SM10 cells. Id2 protein was readily detected in vehicle-treated SM10 cells (Fig. 2C); however, its presence was nearly abolished in response to TGF-β treatment (Fig. 2D). The color combination of Id2 immunofluorescent staining and nuclear images (Fig. 2E, F) additionally indicated increased nuclear concentration of Id2 in the undifferentiated SM10 cells (Fig. 2E).
FIG. 2.
Id2 protein is decreased in differentiated SM10 cells. SM10 cells were treated for 72 h with TGF-β or vehicle. The cells were paraformaldehyde fixed and probed with anti-Id2 primary and Alexa Fluor 594–conjugated secondary antibodies. The cell nuclei were stained with Hoeschst dye. Epifluorescence microscopy analysis of Id2 immunoreactivity (C, D) and nuclei (A, B) is shown. Combined images were generated using the MetaMorph software (E, F). Color images available online at www.liebertpub.com/scd
Id2 overexpression prevents TGF-β-induced downregulation of Id2 RNA, protein, and activity in clonal SM10-Id2
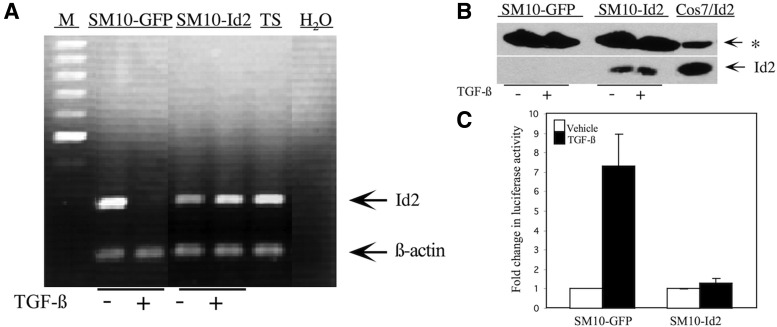
The dramatic downregulation of Id2 mRNA and protein levels in response to TGF-β suggested its involvement in directing labyrinthine trophoblast differentiation. To determine whether Id2 is required for TGF-β-induced differentiation, Id2 and GFP expressing SM10 cell lines were generated. Stable clones (5, 10, and 12) were confirmed by PCR analysis of the genomically integrated Id2 gene using CMV-F and V5-R primers flanking the cDNA sequence (Fig. 3A and Table 1) and by western blot analysis (Fig. 3B). No endogenous Id2 protein could be detected by western blot analysis, as indicated in the GFP control (Fig. 3B). Id2-transfected Cos7 cells served as a positive control (Cos7/Id2). The inability to detect endogenous Id2 in GFP clones is most likely due to antibody sensitivity, as transfected Id2 was readily detectable (Fig. 3B), which is consistent with the manufacturer's specifications.
Id2 mRNA and protein levels in TGF-β differentiated or undifferentiated, vehicle-treated SM10-Id2 and SM10-GFP clones were additionally analyzed for Id2 expression (Figs. 4 and 5). In contrast to SM10-GFP clones, SM10-Id2 clones maintained Id2 mRNA expression after addition of TGF-β, as shown by RT-PCR (Fig. 4A). Western blotting analysis further confirmed that Id2 protein levels were maintained in SM10-Id2 expressing cells (Fig. 4B). Cos7 cells transiently expressing lentiviral Id2 served as a positive control (Fig. 4B). To further evaluate Id2 function, we analyzed E-box transactivation luciferase assays in Id2-SM10 clones using the E7-TK-luciferase reporter, which contains seven E-box sequences in its promoter (Fig. 4C) [70]. Id2 is known to bind to bHLH transcription factors and to inhibit bHLH transcription factor-induced expression of genes containing E-box elements [49,50]. Therefore, a reduced level of Id2 would lead to an increase in E-box luciferase activation. In SM10-GFP clones, in which TGF-β inhibits Id2, we see a corresponding 7.5-fold increase in E-box luciferase activity (Fig. 4A, C). In SM10 cells stably expressing Id2, however, Id2 overexpression abolishes TGF-β-induced transactivation of E7-Tk-luciferase reporter activity (Fig. 4A, C). This result is consistent with the fact that maintenance of Id2 expression, even under differentiating conditions, prevents E-box activation.
FIG. 4.
Id2 is expressed and transcriptionally active in clonal SM10-Id2 with TGF-β or vehicle control treatment. (A) Total RNA from TGF-β or vehicle-treated clonal SM10-GFP and SM10-Id2 cells was RT-PCR amplified with appropriate primers (Table 1) and resolved on a 1% agarose gel. Expression of Id2 in the SM10-GFP and SM10-Id2 cells is shown. Undifferentiated TS cells (TS) were used as a positive control. One hundred basepair DNA marker (M) was utilized in estimating the sizes of PCR amplification products. Co-amplification of β-actin was used for semiquantitative normalization (A). H2O was substituted for reverse-transcribed RNA as a control to indicate no genomic contamination. (B) Western blotting analysis of Id2 in the TGF-β or vehicle-treated clonal SM10-GFP and SM10-Id2 cells. Two hundred fifty micrograms of cell lysate was resolved on an 11% SDS-PAGE, transferred to PVDF membrane, and analyzed for the presence of Id2 protein with Id2 antibody. Cos7 cells transiently expressing lentiviral Id2 (Cos7/Id2) were used as a positive control. A cross-reactive band served as a fortuitous loading control (*). (C) E-box luciferase reporter activity in SM10-GFP and SM10-Id2 cells treated with TGF-β or vehicle control. Cells were transiently transfected with pE7-TK-luc and pRLSV40 using Metafectene. Twenty-four hours post-transfection, media were changed and cells were treated with TGF-β (5 ng/mL) or vehicle for 72 h. Luciferase activity was analyzed using the dual luciferase assay system. The pE7-TK-luc transactivation values were normalized to the constitutively active reporter (pRLSV40). The experiment was repeated at least three independent times for each clone with similar results. Error bars represent standard deviation from the mean.
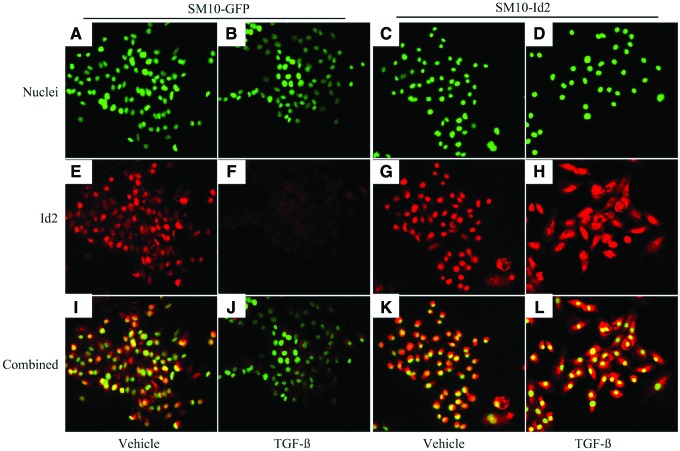
FIG. 5.
Id2 overexpression prevents TGF-β-induced downregulation of Id2 protein in clonal SM10-Id2. Epifluorescence microscopic analysis of Id2 protein immunoreactivity in SM10-GFP and SM10-Id2 cells treated with TGF-β or vehicle control. Id2 expression was analyzed using anti-Id2 primary antibody and Alexa Fluor 594–conjugated secondary antibody. Cell nuclei were stained with Hoechst dye before imaging, and nuclei are indicated in green (A–D). Id2 immunoreactivity is indicated in red (E–H). Combined images were generated using the MetaMorph software (I–L). Color images available online at www.liebertpub.com/scd
In addition, we examined Id2 protein in our stable clones by immunofluorescent staining. TGF-β and vehicle-treated SM10-GFP and SM10-Id2 cells were stained for nuclei (Fig. 5A–D) or Id2 (Fig. 5E–H). Additionally, color-combined images were examined to assess nuclear localization of Id2 (Fig. 5I–L). As expected for SM10 cells stably expressing GFP, Id2 was downregulated (Fig. 5E, F). Id2 localization, in SM10-GFP cells, was similar to that seen in nontransduced SM10 cells (Fig. 2C, D). Our immunofluorescent staining confirmed that Id2 protein levels were maintained in SM10-Id2 cells, even in the presence of TGF-β (Fig. 5H, L).
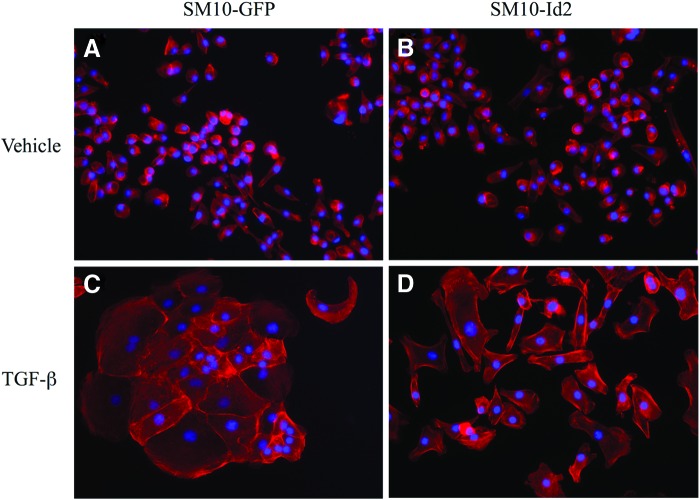
Id2 overexpression inhibits TGF-β-induced morphological differentiation
Previous studies have demonstrated that differentiation of labyrinthine trophoblast cells involves morphological changes, such as cell aggregation and colony formation [15,53]. To investigate the effects of Id2 overexpression on TGF-β-induced morphological differentiation, we analyzed cell morphology by using rhodamine-conjugated phalloidin and nuclear staining of TGF-β or vehicle-treated SM10-GFP and SM10-Id2 cells (Fig. 6). No morphological differentiation was detected in cells exposed to the vehicle (Fig. 6A, B). SM10-Id2 cells showed inhibition of morphological differentiation when exposed to TGF-β (Fig. 6D), as compared with SM10-GFP cells exposed to TGF-β (Fig. 6C). Instead of aggregating and forming colonies, the cells retained a more progenitor-like, single-cell morphology (Fig. 6D). Formation of multinucleated colonies in TGF-β-treated SM10-GFP, which is indicative of differentiation, was clearly evident (Fig. 6C); whereas this response was almost completely inhibited by Id2 overexpression (Fig. 6D).
FIG. 6.
Id2 overexpression inhibits TGF-β-induced morphological differentiation. Epifluorescence microscopy images of rhodamine-conjugated, phalloidin-stained clonal SM10-GFP (A, C) and SM10-Id2 (B, D) cells after 72 h of vehicle or TGF-β treatment. The actin cytoskeleton (red) of SM10 cells was stained with rhodamine-conjugated phalloidin, and nuclei (blue) were stained with Hoechst dye. Images were combined using the MetaMorph software. Color images available online at www.liebertpub.com/scd
Id2 overexpression inhibits TGF-β-induced SM10 functional and molecular differentiation
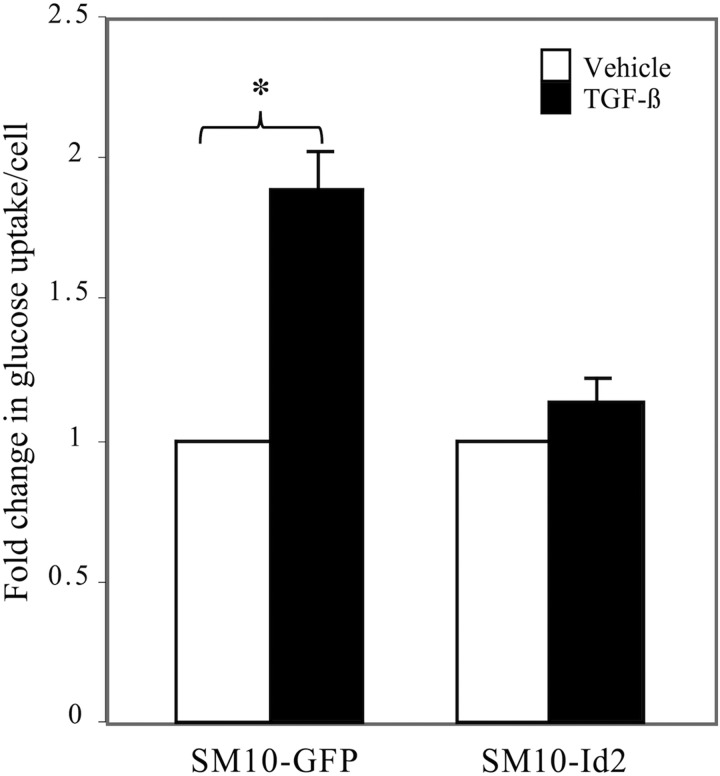
Functional and molecular differentiation accompanies the morphological differentiation of labyrinthine trophoblast cells. One of the main functions of the labyrinthine layer of the placenta is the physiological exchange of nutrients between the mother and the fetus [4,5,12,14,17–20]. Thus, the effects of Id2 overexpression on functional differentiation were examined by analyzing glucose uptake in TGF-β or vehicle-treated SM10-GFP and SM10-Id2 cells. TGF-β treatment resulted in a significantly induced twofold increase in glucose uptake into SM10-GFP cells; however, this upregulation was abolished by the overexpression of Id2 in the SM10-Id2 cells, which is suggestive of inhibition of differentiation (Fig. 7).
FIG. 7.
Id2 overexpression prevents the TGF-β-induced increase in glucose uptake. After 72 h of TGF-β or vehicle treatment, clonal SM10-GFP and SM10-Id2 cells were pulsed with 1 μCi/mL of 2-deoxyglucose for 10 min and uptake was measured by liquid scintillation counting. Cells were counted by trypan blue exclusion, and glucose uptake values were normalized to cell number. Fold change in glucose uptake is indicated as a ratio of normalized TGF-β-treated values to normalized vehicle control-treated values. Each experiment was conducted at least three independent times. Error bars represent standard deviations from the mean *P < 0.01.
Likewise, TGF-β-induced molecular differentiation was inhibited in Id2 overexpressing SM10 cells, as the expression of labyrinthine TS cell marker, Cdx2, was maintained in TGF-β-treated SM10-Id2 cells (Fig. 8A) [47,71,72]. In addition, TGF-β treatment induced the expression of a marker of differentiating labyrinthine trophoblasts, Gcm1, in SM10-GFP cells but did not induce Gcm-1 in SM10-Id2 cells, further supporting the maintenance of a progenitor cell-like state of SM10-Id2 cells (Fig. 8B) [35,73,74].
FIG. 8.
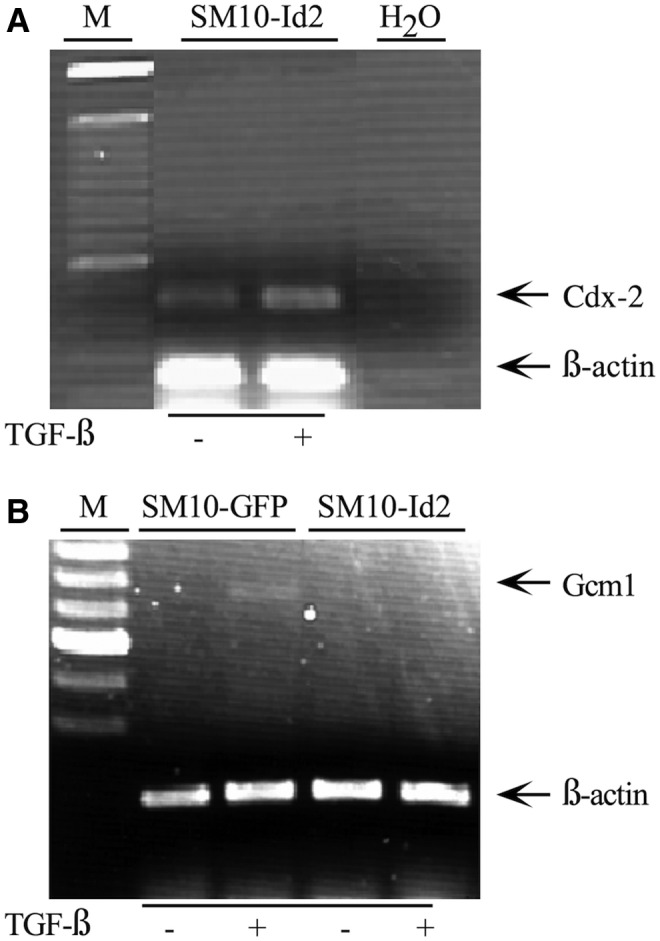
Id2 overexpression results in sustained expression of stem cell marker Cdx2 and repression of labyrinthine differentation marker Gcm1 SM10 cells. Total RNA from subconfluent SM10-GFP and SM10-Id2 cells was RT-PCR amplified with appropriate primers and resolved on a 1% agarose gel. Expression of stem cell-specific marker (Cdx2) (A), and marker of differentiated labyrinthine trophoblasts (Gcm1) (B) in TGF-β or vehicle-treated cells is shown. One hundred basepair DNA marker (M) was utilized in estimating the sizes of PCR amplification products. Co-amplification of β-actin was used for semiquantitative normalization. H2O was substituted for reverse-transcribed RNA as a control to indicate no genomic amplification.
Knockdown of Id2 in SM10 cells results in labyrinthine trophoblast differentiation
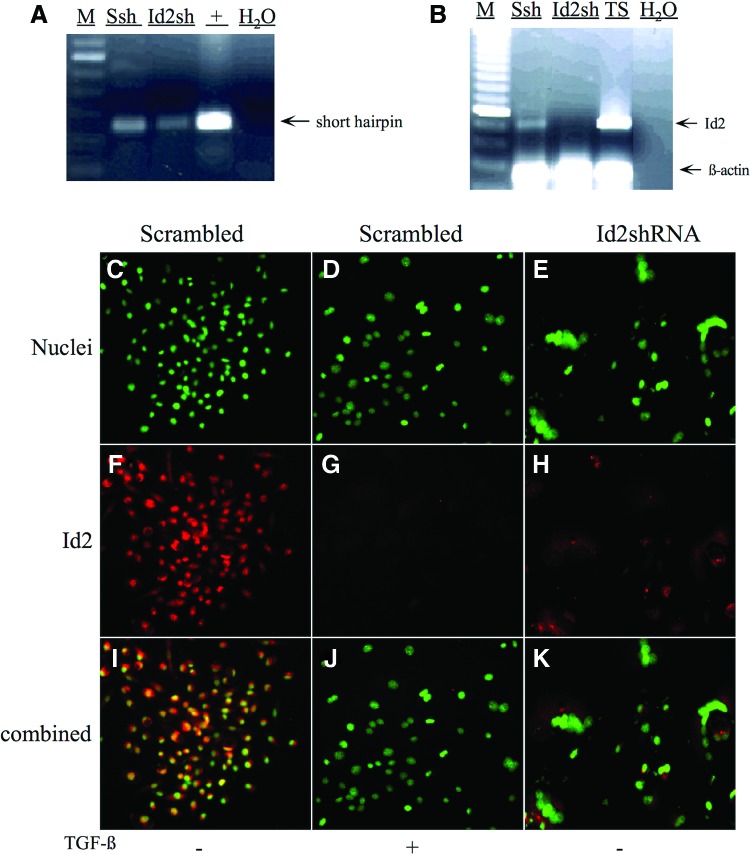
Even though overexpression analyses suggested that Id2 is a mediator of TGF-β-induced labyrinthine trophoblast differentiation, Id2 knockdown experiments were additionally designed to confirm that inhibition of endogenous Id2 is necessary for labyrinthine trophoblast differentiation. Several sequences targeting the Id2 coding region were designed. The two most effective Id2-shRNA sequences and their scrambled control sequences were generated and cloned into lentiviral vectors (Table 2). Although both shRNAs displayed similar results, the most effective Id2-shRNA (Id2-369) and its control-scrambled shRNA (Id2-369s) were used in subsequent experiments (Table 2). Genomic integration of the Id2-shRNA (Id2sh) and control scrambled Id2-shRNA (Ssh) sequences into the SM10 cells was confirmed by PCR amplification of genomic DNA of infected SM10 cells using primers flanking the exogenous insert (Fig. 9A and Table 1). Knockdown of Id2 RNA in SM10 cells was confirmed by PCR using primers specific to endogenous Id2 (Fig. 9B and Table 1). Additionally, RNA collected from TS cells and PCR amplified using primers specific to Id2 were used as positive controls (Fig. 9B). Expression of Id2 is abolished by the expression of Id2-shRNA, but not by the expression of scrambled Id2-shRNA (Fig. 9B).
FIG. 9.
Generation of Id2 knockdown and scrambled control shRNA SM10 cells. (A) SM10 cells were transduced with lentiviral scrambled Id2-shRNA (Ssh) or Id2-shRNA targeted against Id2 (Id2sh). Five days post-transduction, total genomic DNA was extracted and PCR amplified using primers flanking the shRNA sequence. One hundred basepair DNA marker (M) and amplification from lentiviral plasmids (+) were utilized in estimating the sizes of PCR products and as a positive control, respectively. H2O was substituted for genomic DNA to confirm absence of contamination (H2O). (B) SM10 cells were transduced with lentiviral scrambled Id2-shRNA (Ssh) or Id2-shRNA (Id2sh). Three days post-transduction, cells were placed under blasticidin (8 μg/mL) selection for 5 days. Total RNA was RT-PCR amplified with appropriate Id2-specific primers and resolved on a 1% agarose gel. Expression of Id2 in lentivirally transduced cells is shown. Undifferentiated TS (TS) cells were used as a positive control, and 100 bp DNA marker (M) was utilized in estimating the sizes of PCR amplification products. Co-amplification of β-actin was used for semiquantitative normalization. H2O was substituted for reverse-transcribed RNA as a control to indicate no genomic contamination. (C–K) Epifluorescence microscopic analysis of SM10 cells transduced with lentiviral scrambled Id2-shRNA (Scrambled) or Id2-shRNA (Id2-shRNA). Three days post-transduction, infected cells were selected in blasticidin (8 μg/mL) and treated with 5 ng/mL TGF-β (+) or vehicle (−) for 72 h. Id2 expression was analyzed using anti-Id2 primary antibody and Alexa Fluor 594–conjugated secondary antibody. Cell nuclei were stained with Hoechst dye before imaging and are indicated in green (C–E). Id2 immunoreactivity is indicated in red (F–H). Combined images were generated using the MetaMorph software (I–K). Color images available online at www.liebertpub.com/scd
In addition to PCR amplification, immunofluorescent staining for Id2 and nuclear staining was performed to assess abundance and location of Id2 protein in TGF-β or vehicle control-treated SM10 cells expressing scrambled Id2-shRNA or vehicle-treated Id2-shRNA SM10 cells (Fig. 9C–K). It should be noted that repeated attempts to propagate a stable cell line expressing Id2-shRNA were unsuccessful due to the induction of continual terminal differentiation. As expected for SM10 cells with scrambled control Id2-shRNA (Scrambled), Id2 localization was similar to the localization seen in nontransduced SM10 cells (Fig. 9F, G and Fig. 2C, D). The immunofluorescent staining also confirmed that Id2 is reduced in vehicle-treated Id2-shRNA expressing SM10 cells (Id2-shRNA) (Fig. 9H) compared with the vehicle-treated scrambled Id2-shRNA expressing SM10 cells (Scrambled) (Fig. 9F). Interestingly, this reduction in Id2 protein, in the Id2-shRNA expressing SM10 cells (Fig. 9H), is similar to that of the TGF-β-treated scrambled Id2-shRNA expressing SM10 cells (Fig. 9G).
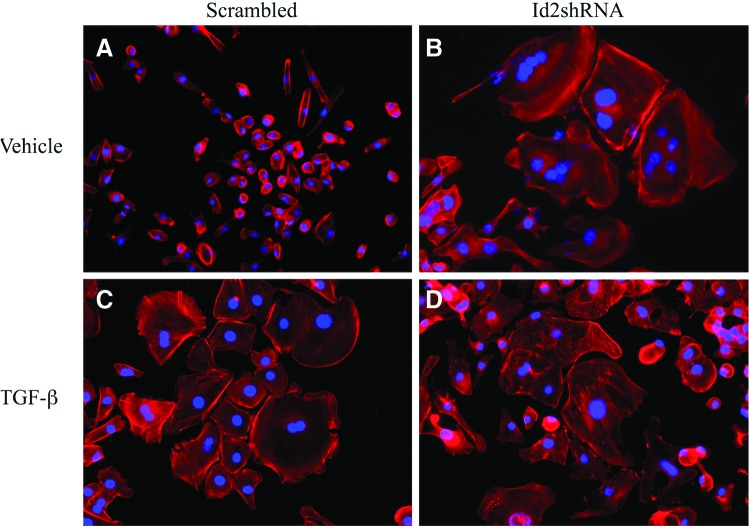
Our Id2 overexpression studies suggested that Id2 may mediate events associated with TGF-β-induced changes in labyrinthine trophoblast cell morphology and that Id2 knockdown promotes labyrinthine trophoblast differentiation. Analysis of cell morphology using rhodamine-conjugated phalloidin and nuclear staining showed that SM10 cells infected with scrambled Id2-shRNA maintained a progenitor-like, single-cell morphology (Fig. 10A) and differentiated as expected when treated with TGF-β (Fig. 10C). In Id2-shRNA expressing SM10 cells, in the absence of TGF-β, the formation of multinucleate cells could be observed 3–4 days post-infection in vehicle-treated controls and this response was maintained up to 10 days, as observed by rhodamine-conjugated phalloidin and nuclear staining (Fig. 10B). Thus, reduced Id2 expression resulted in morphological differentiation of SM10 cells (Fig. 10B). Addition of TGF-β to Id2-shRNA expressing SM10 cells did not have any further differentiating effect on the morphology of Id2-shRNA expressing SM10 cells (Fig. 10D).
FIG. 10.
Knockdown of Id2 in SM10 cells results in labyrinthine trophoblast differentiation. SM10 cells were transduced with lentiviral scrambled Id2-shRNA (Scrambled) or Id2-shRNA (Id2-shRNA). Three days post-transduction, infected cells were enriched under (8 μg/mL) blasticidin selection and treated with 5 ng/mL TGF-β or vehicle for 72 h. Epifluorescence microscopy images of rhodamine-conjugated, phalloidin-stained SM10 cells transduced with scrambled Id2-shRNA (Scrambled) (A, C) or Id2-shRNA (Id2sh) (B, D) and treated with TGF-β (C, D) or vehicle (A, B) are shown. The actin cytoskeleton (red) of SM10 cells was stained with rhodamine-conjugated phalloidin, and nuclei (blue) were stained with Hoechst dye. Images were combined using the MetaMorph software. Color images available online at www.liebertpub.com/scd
Discussion
Appropriate placental development is essential for fetal survival and a healthy pregnancy. This process is highly regulated and involves differentiation of placental trophoblast cells into lineage-specific subtypes that fulfill several roles as the interface between the mother and the fetus. The proper development of the placental labyrinth is required for establishing the fetal–maternal interface needed for nutrient and waste exchange. Although several studies using knockout mice have been conducted and have been useful for understanding the molecular events of trophoblast differentiation, a detailed examination of the molecular pathways governing placental development has been hindered by the lack of suitable cell culture systems to model labyrinthine trophoblast differentiation [6,25,43].
We have previously shown that the trophoblast progenitor cell line SM10 expresses several labyrinthine-specific lineage markers (Esx1, Tfeb, and Tec) and undergoes TGF-β-induced differentiation [15,16]. Notably, this differentiation was associated with downregulation of Id2 expression [15]. In this investigation, the expression of Id isoforms in the SM10 labyrinthine progenitor cell line and the effect of Id2 overexpression and knockdown on TGF-β-induced differentiation were examined. The expression of Id proteins is essential for normal development and differentiation of several cell types [28,51,75–80]. Id isoforms are also expressed in the placenta, and this suggests their importance in directing trophoblast self-renewal and differentiation [15,29–32,43,76]. Even though mice that are deficient in a single Id isoform are viable, double deficiency of Id1, Id2, and/or Id3 isoforms leads to embryonic lethality, implying functional redundancy of expression [48,49,81–87]. Unfortunately, studies have not yet been conducted on investigating the possible placenta defects in these knockout mice [48,49,81–87].
Of the four Ids characterized, expression of Id1, Id2, and Id3 has been shown in human and rodent placentas [28–30,43,48,49,51,52]. The labyrinthine trophoblast progenitor cell line, SM10, was shown to express Id isoforms. However, expression of Id1 and Id3 is constitutive and not responsive to treatment with TGF-β; whereas the expression of Id2 is readily detectable in undifferentiated SM10 cells, but it is drastically reduced on treatment with TGF-β and subsequent differentiation. The lack of Id4 expression in the SM10 cells confirms previously reported observations in placental cells [30,51]. In contrast to previous studies done in placental giant cells that showed a decrease of both Id1 and Id2 on differentiation, only the expression of Id2 mRNA and protein was shown to be downregulated during labyrinthine trophoblast differentiation in the current study and suggests that Id2 is a major mediator of labyrinthine trophoblast differentiation [15,29].
Although Id2 downregulation has been reported during trophoblast differentiation into invasive and transport subtypes, triggers inducing lineage-specific differentiation appear complex and suggest multifaceted control of Id2 expression [15,29,30,32]. Differentiation into the transport lineage by TGF-β has been reported in both humans and rodents [13,15,26,88,89]. Even though the intracellular mediators guiding trophoblast differentiation and Id2 downregulation in response to TGF-β are still being investigated, studies in other epithelial cell models have reported TGF-β-induced downregulation of Id1, Id2, and Id3 [61,90–92]. Additionally, TGF-β-induced Id2 downregulation has been shown to depend on Smad4, suggesting the possibility of direct regulation of Id2 expression by the TGF-β-activated Smad-signaling pathway [61]. In addition, Sp1 transcription factor binding sites that are capable of interacting with Smad transcriptional complexes have been identified in the promoter of the Id2 gene and may be involved in Smad-dependent regulation of Id2 expression [61,93–96].
No previous investigations have addressed the effects of Id isoforms, specifically on the labyrinthine trophoblast differentiation. Our studies indicate that overexpression and knockdown of Id2 in the labyrinthine-specific trophoblast cell line SM10 alters TGF-β induced morphological, functional, and molecular differentiation, which is suggestive of its importance in differentiation into the transport lineage. Indeed, Id2 overexpression inhibited TGF-β-induced morphological differentiation and maintained cells in a progenitor-like phenotype. In contrast, morphological differentiation (cell aggregation, colony formation, inability to proliferate) was clearly induced when the Id2 gene expression was knocked down. Considering the changes in phalloidin staining patterns in Id2 overexpressing and Id2 knockdown SM10 cells, Id2 may be involved in regulating the function of cytoskeletal proteins that have been implicated in altering trophoblast cell morphology, including β-catenin and connexins [47,79,97].
In addition to morphological differentiation, functional differentiation was inhibited in Id2 overexpressing SM10 cells, as demonstrated by a lack of increase in glucose uptake on treatment with TGF-β. Three glucose transporters, Glut1, Glut3, and Glut4, have been shown to be present in the placenta [53,98,99]. TGF-β has previously been shown to induce glucose uptake in several models via upregulation of Glut1 [100–103]. In contrast, Glut3 expression appears to be TGF-β insensitive [103]. Further studies on the effects of TGF-β and Id2 on Glut expression and function are warranted.
In summary, we have demonstrated that Id2 is one of the primary mediators regulating labyrinthine trophoblast progenitor cell differentiation. We have further demonstrated that SM10 cells can be easily transduced to express genes of interest using lentiviral constructs to study development of labyrinthine trophoblasts and can lead to the elucidation of events underlying development. An understanding of the basic molecular pathways of trophoblast differentiation offers valuable insights into the mechanisms that govern placental development. Lineage-specific trophoblast cell lines can also help identify possible subtype-specific placental abnormalities that may lead to pre-eclampsia, fetal growth restriction, and placental insufficiency.
Acknowledgments
Plasmid pcDNA-Id2 was a generous gift of Dr. John D. Norton of the University of Essex, Colchester, the United Kingdom. The lentiviral plasmid, pLv-CMV-GFP-V5, was kindly provided by Dr. Steven Berberich of the Wright State University, Dayton, Ohio. Mouse SM10 cells were kindly provided by Dr. Joan S. Hunt of the Kansas University Medical Center, Kansas City, Kansas. TGF-β2 was a kind gift of Dr. Steven Ledbetter of Genzyme, Inc., Boston, Massachusetts. TS3.5 cells (TS cells) were generously provided by Dr. Janet Rossant, The Hospital for Sick Children, Toronto, Canada. The pE7-TK-luc construct was kindly provided by Dr. Yoshifumi Yokota, the University of Fukui, Fukui, Japan. The authors would like to thank Amy Gultice for her valuable assistance and input with the TS and Rcho cells. This work was supported in part by grants from the Wright State University Research Incentive Program, The Ohio Board of Reagents (T.L.B.), the Biomedical Sciences Ph.D. Program (K.S., R.E.A.), the Wright State University Graduate Council Scholarship (R.E.A.), The Wright State University Endowment for Research on Pregnancy Associated Disorders (www.wright.edu/give/pregnancyassociateddisorders), and The National Institutes of Health NICHD-R01 HD059969-(T.L.B.).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Jansson T, Aye ILMH. and Goberdhan DCI. (2012). The emerging role of mTORC1 signaling in placental nutrient-sensing. Placenta 33:e23–e29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lager S. and Powell TL. (2012). Regulation of nutrient transport across the placenta. J Pregnancy 2012:Article ID 179827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cross JC. (2000). Genetic insights into trophoblast differentiation and placental morphogenesis. Semin Cell Dev Biol 11:105–113 [DOI] [PubMed] [Google Scholar]
- 4.Cross JC, Baczyk D, Dobric N, Hemberger M, Hughes M, Simmons DG, Yamamoto H. and Kingdom JC. (2003). Genes, development and evolution of the placenta. Placenta 24:123–130 [DOI] [PubMed] [Google Scholar]
- 5.Hemberger M. and Cross JC. (2001). Genes governing placental development. Trends Endocrinol Metab 12:162–168 [DOI] [PubMed] [Google Scholar]
- 6.Rossant J. and Cross JC. (2001). Placental development: lessons from mouse mutants. Nat Rev Genet 2:538–548 [DOI] [PubMed] [Google Scholar]
- 7.Kam EPY, Gardner L, Loke YW. and King A. (1999). The role of trophoblast in the physiological change in decidual spiral arteries. Hum Reprod 14:2131–2138 [DOI] [PubMed] [Google Scholar]
- 8.Cross JC. (2006). Placental function in development and disease. Reprod Fertil Dev 18:71–76 [DOI] [PubMed] [Google Scholar]
- 9.Georgiades P, Ferguson-Smith AC. and Burton GJ. (2002). Comparative developmental anatomy of the murine and human definitive placentae. Placenta 23:3–19 [DOI] [PubMed] [Google Scholar]
- 10.Malassiné A, Frendo JL. and Evain-Brion D. (2003). A comparison of placental development and endocrine functions between the human and mouse model. Hum Reprod Update 9:531–539 [DOI] [PubMed] [Google Scholar]
- 11.Adamson SL, Lu Y, Whiteley KJ, Holmyard D, Hemberger M, Pfarrer C. and Cross JC. (2002). Interactions between trophoblast cells and the maternal and fetal circulation in the mouse placenta. Dev Biol 250:358–373 [DOI] [PubMed] [Google Scholar]
- 12.Cross JC. (2005). How to make a placenta: mechanisms of trophoblast cell differentiation in mice–a review. Placenta 26 Supplement:S3–S9 [DOI] [PubMed] [Google Scholar]
- 13.Rama S, Suresh Y. and Rao AJ. (2003). TGF β1 induces multiple independent signals to regulate human trophoblastic differentiation: mechanistic insights. Mol Cell Endocrinol 206:123–136 [DOI] [PubMed] [Google Scholar]
- 14.Watson ED. and Cross JC. (2005). Development of structures and transport functions in the mouse placenta. Physiology 20:180–193 [DOI] [PubMed] [Google Scholar]
- 15.Selesniemi K, Reedy M, Gultice A, Guilbert LJ. and Brown TL. (2005). Transforming growth factor-beta induces differentiation of the labyrinthine trophoblast stem cell line SM10. Stem Cells Dev 14:697–711 [DOI] [PubMed] [Google Scholar]
- 16.Selesniemi KL, Reedy MA, Gultice AD. and Brown TL. (2005). Identification of committed placental stem cell lines for studies of differentiation. Stem Cells Dev 14:535–547 [DOI] [PubMed] [Google Scholar]
- 17.Cross JC, Simmons DG. and Watson ED. (2003). Chorioallantoic morphogenesis and formation of the placental villous tree. Ann N Y Acad Sci 995:84–93 [DOI] [PubMed] [Google Scholar]
- 18.Knipp GT, Audus KL. and Soares MJ. (1999). Nutrient transport across the placenta. Adv Drug Deliv Rev 38:41–58 [DOI] [PubMed] [Google Scholar]
- 19.Bell AW, Hay WW., Jr. and Ehrhardt RA. (1999). Placental transport of nutrients and its implications for fetal growth. J Reprod Fertil Suppl 54:401–410 [PubMed] [Google Scholar]
- 20.Takata K. and Hirano H. (1997). Mechanism of glucose transport across the human and rat placental barrier: a review. Microsc Res Tech 38:145–152 [DOI] [PubMed] [Google Scholar]
- 21.Tanaka S, Kunath T, Hadjantonakis A-K, Nagy A. and Rossant J. (1998). Promotion of trophoblast stem cell proliferation by FGF4. Science 282:2072–2075 [DOI] [PubMed] [Google Scholar]
- 22.Sharma RK. (1998). Mouse trophoblastic cell lines: I—relationship between invasive potential and TGF-beta 1. In Vivo 12:431–440 [PubMed] [Google Scholar]
- 23.Sharma RK. (1998). Mouse trophoblastic cell lines: II—relationship between invasive potential and proteases. In Vivo 12:209–217 [PubMed] [Google Scholar]
- 24.Hess KA, Waltz SE, Chan EL. and Degen SJF. (2003). Receptor tyrosine kinase ron is expressed in mouse reproductive tissues during embryo implantation and is important in trophoblast cell function. Biol Reprod 68:1267–1275 [DOI] [PubMed] [Google Scholar]
- 25.Du X, Dong Y, Shi H, Li J, Kong S, Shi D, Sun LV, Xu T, Deng K. and Tao W. (2014). Mst1 and Mst2 are essential regulators of trophoblast differentiation and placenta morphogenesis. PLoS One 9:e90701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rama S, Suresh Y. and Rao AJ. (2001). Regulation of telomerase during human placental differentiation: a role for TGFβ1. Mol Cell Endocrinol 182:233–248 [DOI] [PubMed] [Google Scholar]
- 27.Red-Horse K, Zhou Y, Genbacev O, Prakobphol A, Foulk R, McMaster M. and Fisher SJ. (2004). Trophoblast differentiation during embryo implantation and formation of the maternal-fetal interface. J Clin Invest 114:744–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jen Y, Manova K. and Benezra R. (1997). Each member of the Id gene family exhibits a unique expression pattern in mouse gastrulation and neurogenesis. Dev Dyn 208:92–106 [DOI] [PubMed] [Google Scholar]
- 29.Cross JC, Flannery ML, Blanar MA, Steingrimsson E, Jenkins NA, Copeland NG, Rutter WJ. and Werb Z. (1995). Hxt encodes a basic helix-loop-helix transcription factor that regulates trophoblast cell development. Development 121:2513–2523 [DOI] [PubMed] [Google Scholar]
- 30.Janatpour MJ, McMaster MT, Genbacev O, Zhou Y, Dong J, Cross JC, Israel MA. and Fisher SJ. (2000). Id-2 regulates critical aspects of human cytotrophoblast differentiation, invasion and migration. Development 127:549–558 [DOI] [PubMed] [Google Scholar]
- 31.Evans SM. and O'Brien TX. (1993). Expression of the helix-loop-helix factor Id during mouse embryonic development. Dev Biol 159:485–499 [DOI] [PubMed] [Google Scholar]
- 32.Jiang B, Kamat A. and Mendelson CR. (2000). Hypoxia prevents induction of aromatase expression in human trophoblast cells in culture: potential inhibitory role of the hypoxia-inducible transcription factor Mash-2 (mammalian achaete-scute homologous protein-2). Mol Endocrinol 14:1661–1673 [DOI] [PubMed] [Google Scholar]
- 33.Strumpf D, Mao C-A, Yamanaka Y, Ralston A, Chawengsaksophak K, Beck F. and Rossant J. (2005). Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development 132:2093–2102 [DOI] [PubMed] [Google Scholar]
- 34.Hay DC, Sutherland L, Clark J. and Burdon T. (2004). Oct-4 knockdown induces similar patterns of endoderm and trophoblast differentiation markers in human and mouse embryonic stem cells. Stem Cells 22:225–235 [DOI] [PubMed] [Google Scholar]
- 35.Anson-Cartwright L, Dawson K, Holmyard D, Fisher SJ, Lazzarini RA. and Cross JC. (2000). The glial cells missing-1 protein is essential for branching morphogenesis in the chorioallantoic placenta. Nat Genet 25:311–314 [DOI] [PubMed] [Google Scholar]
- 36.Yu C, Shen K, Lin M, Chen P, Lin C, Chang G-D. and Chen H. (2002). GCMa regulates the syncytin-mediated trophoblastic fusion. J Biol Chem 277:50062–50068 [DOI] [PubMed] [Google Scholar]
- 37.Knofler M, Meinhardt G, Vasicek R, Husslein P. and Egarter C. (1998). Molecular cloning of the human Hand1 gene/cDNA and its tissue-restricted expression in cytotrophoblastic cells and heart. Gene 224:77–86 [DOI] [PubMed] [Google Scholar]
- 38.Guillemot F, Nagy A, Auerbach A, Rossant J. and Joyner AL. (1994). Essential role of Mash-2 in extraembryonic development. Nature 371:333–336 [DOI] [PubMed] [Google Scholar]
- 39.Scott IC, Anson-Cartwright L, Riley P, Reda D. and Cross JC. (2000). The HAND1 basic helix-loop-helix transcription factor regulates trophoblast differentiation via multiple mechanisms. Mol Cell Biol 20:530–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hughes M, Dobric N, Scott IC, Su L, Starovic M, St-Pierre B, Egan SE, Kingdom JCP. and Cross JC. (2004). The Hand1, Stra13 and Gcm1 transcription factors override FGF signaling to promote terminal differentiation of trophoblast stem cells. Dev Biol 271:26–37 [DOI] [PubMed] [Google Scholar]
- 41.Steingrimsson E, Tessarollo L, Reid SW, Jenkins NA. and Copeland NG. (1998). The bHLH-Zip transcription factor Tfeb is essential for placental vascularization. Development 125:4607–4616 [DOI] [PubMed] [Google Scholar]
- 42.Kuiper RP, Schepens M, Thijssen J, Schoenmakers EFPM. and van Kessel AG. (2004). Regulation of the MiTF/TFE bHLH-LZ transcription factors through restricted spatial expression and alternative splicing of functional domains. Nucleic Acids Res 32:2315–2322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Townley-Tilson WHD, Wu Y, Ferguson JE. and Patterson C. (2014). The ubiquitin ligase ASB4 promotes trophoblast differentiation through the degradation of ID2. PLoS One 9:e89451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lasorella A, Noseda M, Beyna M. and Iavarone A. (2000). Id2 is a retinoblastoma protein target and mediates signalling by Myc oncoproteins. Nature 407:592–598 [DOI] [PubMed] [Google Scholar]
- 45.Norton JD. and Atherton GT. (1998). Coupling of cell growth control and apoptosis functions of Id proteins. Mol Cell Biol 18:2371–2381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ruzinova MB. and Benezra R. (2003). Id proteins in development, cell cycle and cancer. Trends Cell Biol 13:410–418 [DOI] [PubMed] [Google Scholar]
- 47.Liu Y-P, Burleigh D, Durning M, Hudson L, Chiu I-M. and Golos TG. (2004). Id2 is a primary partner for the E2-2 basic helix-loop-helix transcription factor in the human placenta. Mol Cell Endocrinol 222:83–92 [DOI] [PubMed] [Google Scholar]
- 48.Lasorella A, Benezra R. and Iavarone A. (2014). The ID proteins: master regulators of cancer stem cells and tumour aggressiveness. Nat Rev Cancer 14:77–91 [DOI] [PubMed] [Google Scholar]
- 49.Wang L-H. and Baker NE. (2015). E proteins and ID proteins: helix-loop-helix partners in development and disease. Dev Cell 35:269–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Benezra R, Davis RL, Lockshon D, Turner DL. and Weintraub H. (1990). The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell 61:49–59 [DOI] [PubMed] [Google Scholar]
- 51.Riechmann V, van Crüchten I. and Sablitzky F. (1994). The expression pattern of Id4, a novel dominant negative helix-loop-helix protein, is distinct from Id1, 1d2 and Id3. Nucleic Acids Res 22:749–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xue WC, Feng HC, Chan KYK, Chiu PM, Ngan HYS, Khoo US, Tsao SW, Chan KW. and Cheung ANY. (2005). Id helix-loop-helix proteins are differentially expressed in gestational trophoblastic disease. Histopathology 47:303–309 [DOI] [PubMed] [Google Scholar]
- 53.Carey EA, Albers RE, Doliboa SR, Hughes M, Wyatt CN, Natale DR. and Brown TL. (2014). AMPK knockdown in placental trophoblast cells results in altered morphology and function. Stem Cells Dev 23:2921–2930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gultice AD, Kulkarni-Datar K. and Brown TL. (2009). Hypoxia-inducible factor 1alpha (HIF1A) mediates distinct steps of rat trophoblast differentiation in gradient oxygen. Biol Reprod 80:184–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gultice AD, Selesniemi KL. and Brown TL. (2006). Hypoxia inhibits differentiation of lineage-specific Rcho-1 trophoblast giant cells. Biol Reprod 74:1041–1050 [DOI] [PubMed] [Google Scholar]
- 56.Hamlin GP, Lu XJ, Roby KF. and Soares MJ. (1994). Recapitulation of the pathway for trophoblast giant cell differentiation in vitro: stage-specific expression of members of the prolactin gene family. Endocrinology 134:2390–2396 [DOI] [PubMed] [Google Scholar]
- 57.Khvorova A, Reynolds A. and Jayasena SD. (2003). Functional siRNAs and miRNAs Exhibit Strand Bias. Cell 115:209–216 [DOI] [PubMed] [Google Scholar]
- 58.Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N. and Zamore PD. (2003). Asymmetry in the assembly of the RNAi enzyme complex. Cell 115:199–208 [DOI] [PubMed] [Google Scholar]
- 59.Brummelkamp TR, Bernards R. and Agami R. (2002). A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550–553 [DOI] [PubMed] [Google Scholar]
- 60.Tangeman L, Wyatt CN. and Brown TL. (2012). Knockdown of AMP-activated protein kinase alpha 1 and alpha 2 catalytic subunits. J RNAi Gene Silencing 8:470–478 [PMC free article] [PubMed] [Google Scholar]
- 61.Kowanetz M, Valcourt U, Bergström R, Heldin C-H. and Moustakas A. (2004). Id2 and Id3 define the potency of cell proliferation and differentiation responses to transforming growth factor β and bone morphogenetic protein. Mol Cell Biol 24:4241–4254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chaudhary J, Johnson J, Kim G. and Skinner MK. (2001). Hormonal regulation and differential actions of the helix-loop-helix transcriptional inhibitors of differentiation (Id1, Id2, Id3, and Id4) in Sertoli cells. Endocrinology 142:1727–1736 [DOI] [PubMed] [Google Scholar]
- 63.Ikawa T, Fujimoto S, Kawamoto H, Katsura Y. and Yokota Y. (2001). Commitment to natural killer cells requires the helix–loop–helix inhibitor Id2. Proc Natl Acad Sci U S A 98:5164–5169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dai G, Imagawa W, Liu B, Szpirer C, Levan G, Kwok SC. and Soares MJ. (1996). Rcho-1 trophoblast cell placental lactogens: complementary deoxyribonucleic acids, heterologous expression, and biological activities. Endocrinology 137:5020–5027 [DOI] [PubMed] [Google Scholar]
- 65.Livak KJ. and Schmittgen TD. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 66.Caserta TM, Smith AN, Gultice AD, Reedy MA. and Brown TL. (2003). Q-VD-OPh, a broad spectrum caspase inhibitor with potent antiapoptotic properties. Apoptosis 8:345–352 [DOI] [PubMed] [Google Scholar]
- 67.Doran DM, Kulkarni-Datar K, Cool DR. and Brown TL. (2011). Hypoxia activates constitutive luciferase reporter constructs. Biochimie 93:361–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Selesniemi K. and Brown TL. (2004). Efficiency of metafectene transfection in a murine placental trophoblast cell line. Available at: www.biontex.com/navagation/applicationnotes/Brown2.pdf
- 69.Ogura K, Sakata M, Yamaguchi M, Kurachi H. and Murata Y. (1999). High concentration of glucose decreases glucose transporter-1 expression in mouse placenta in vitro and in vivo. J Endocrinol 160:443–452 [DOI] [PubMed] [Google Scholar]
- 70.Kurooka H. and Yokota Y. (2005). Nucleo-cytoplasmic shuttling of Id2, a negative regulator of basic helix-loop-helix transcription factors. J Biol Chem 280:4313–4320 [DOI] [PubMed] [Google Scholar]
- 71.Beck F, Erler T, Russell A. and James R. (1995). Expression of Cdx-2 in the mouse embryo and placenta: possible role in patterning of the extra-embryonic membranes. Dev Dyn 204:219–227 [DOI] [PubMed] [Google Scholar]
- 72.Chawengsaksophak K, de Graaff W, Rossant J, Deschamps J. and Beck F. (2004). Cdx2 is essential for axial elongation in mouse development. Proc Natl Acad Sci U S A 101:7641–7645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hashemolhosseini S, Hadjihannas M, Stolt CC, Haas CS, Amann K. and Wegner M. (2002). Restricted expression of mouse GCMa/Gcm1 in kidney and thymus. Mech Dev 118:175–178 [DOI] [PubMed] [Google Scholar]
- 74.Basyuk E, Cross JC, Corbin J, Nakayama H, Hunter P, Nait-Oumesmar B. and Lazzarini RA. (1999). Murine Gcm1 gene is expressed in a subset of placental trophoblast cells. Dev Dyn 214:303–311 [DOI] [PubMed] [Google Scholar]
- 75.Awonuga AO, Zhong W, Abdallah ME, Slater JA, Zhou SC, Xie YF, Puscheck EE. and Rappolee DA. (2011). Eomesodermin, HAND1, and CSH1 proteins are induced by cellular stress in a stress-activated protein kinase-dependent manner. Mol Reprod Dev 78:519–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Puscheck EE, Awonuga AO, Yang Y, Jiang Z. and Rappolee DA. (2015). Molecular biology of the stress response in the early embryo and its stem cells. In: Cell Signaling During Mammalian Early Embryo Development: Leese HJ, Brison DR, eds. Springer, New York, pp. 77–128 [DOI] [PubMed] [Google Scholar]
- 77.Xie Y, Abdallah ME, Awonuga AO, Slater JA, Puscheck EE. and Rappolee DA. (2010). Benzo(a)pyrene causes PRKAA1/2-dependent ID2 loss in trophoblast stem cells. Mol Reprod Dev 77:533–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhong W, Xie Y, Abdallah M, Awonuga AO, Slater JA, L Sipahi L, Puscheck EE. and Rappolee DA. (2010). Cellular stress causes reversible, PRKAA1/2-, and proteasome-dependent ID2 protein loss in trophoblast stem cells. Reproduction 140:921–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kibschull M, Colaco K, Matysiak-Zablocki E, Winterhager E. and Lye SJ. (2014). Connexin31.1 (Gjb5) deficiency blocks trophoblast stem cell differentiation and delays placental development. Stem Cells Dev 23:2649–2660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ling F, Kang B. and Sun X-H. (2014). Chapter five—Id proteins: small molecules, mighty regulators. In: Current Topics in Developmental Biology. Reshma T, ed. Academic Press, pp. 189–216 [DOI] [PubMed] [Google Scholar]
- 81.Lyden D, Young AZ, Zagzag D, Yan W, Gerald W, O'Reilly R, Bader BL, Hynes RO, Zhuang Y, Manova K. and Benezra R. (1999). Id1 and Id3 are required for neurogenesis, angiogenesis and vascularization of tumour xenografts. Nature 401:670–677 [DOI] [PubMed] [Google Scholar]
- 82.Bedford L, Walker R, Kondo T, van Crüchten I, King ER. and Sablitzky F. (2005). Id4 is required for the correct timing of neural differentiation. Dev Biol 280:386–395 [DOI] [PubMed] [Google Scholar]
- 83.Mori S, Nishikawa S-I. and Yokota Y. (2000). Lactation defect in mice lacking the helix–loop–helix inhibitor Id2. EMBO J 19:5772–5781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pan L, Sato S, Frederick JP, Sun X-H. and Zhuang Y. (1999). Impaired immune responses and B-cell proliferation in mice lacking the Id3 gene. Mol Cell Biol 19:5969–5980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Satyanarayana A, Klarmann KD, Gavrilova O. and Keller JR. (2012). Ablation of the transcriptional regulator Id1 enhances energy expenditure, increases insulin sensitivity, and protects against age and diet induced insulin resistance, and hepatosteatosis. FASEB J 26:309–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yan W, Young AZ, Soares VC, Kelley R, Benezra R. and Zhuang Y. (1997). High incidence of T-cell tumors in E2A-null mice and E2A/Id1 double-knockout mice. Mol Cell Biol 17:7317–7327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yokota Y, Mansouri A, Mori S, Sugawara S, Adachi S, Nishikawa S-I. and Gruss P. (1999). Development of peripheral lymphoid organs and natural killer cells depends on the helix-loop-helix inhibitor Id2. Nature 397:702–706 [DOI] [PubMed] [Google Scholar]
- 88.Graham CH, Lysiak JJ, McCrae KR. and Lala PK. (1992). Localization of transforming growth factor-beta at the human fetal-maternal interface: role in trophoblast growth and differentiation. Biol Reprod 46:561–572 [DOI] [PubMed] [Google Scholar]
- 89.Rama S, Petrusz P. and Rao AJ. (2004). Hormonal regulation of human trophoblast differentiation: a possible role for 17β-estradiol and GnRH. Mol Cell Endocrinol 218:79–94 [DOI] [PubMed] [Google Scholar]
- 90.Ling MT, Wang X, Tsao SW. and Wong YC. (2002). Down-regulation of Id-1 expression is associated with TGFβ1-induced growth arrest in prostate epithelial cells. Biochim Biophys Acta 1570:145–152 [DOI] [PubMed] [Google Scholar]
- 91.Siegel PM, Shu W. and Massagué J. (2003). Mad upregulation and Id2 repression accompany transforming growth factor (TGF)-β-mediated epithelial cell growth suppression. J Biol Chem 278:35444–35450 [DOI] [PubMed] [Google Scholar]
- 92.Xie L, Law BK, Aakre ME, Edgerton M, Shyr Y, Bhowmick NA. and Moses HL. (2003). Transforming growth factor beta-regulated gene expression in a mouse mammary gland epithelial cell line. Breast Cancer Res 5:R187–R198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kruger O, Plum A, Kim JS, Winterhager E, Maxeiner S, Hallas G, Kirchhoff S, Traub O, Lamers WH. and Willecke K. (2000). Defective vascular development in connexin 45-deficient mice. Development 127:4179–4193 [DOI] [PubMed] [Google Scholar]
- 94.Li X-J, Hata K. and Mizuguchi J. (2005). Engagement of membrane immunoglobulin enhances Id3 promoter activity in WEHI-231 B lymphoma cells. Acta Pharmacol Sin 26:486–491 [DOI] [PubMed] [Google Scholar]
- 95.Mantani A, Hernandez M-C, Kuo W-L. and Israel MA. (1998). The mouse Id2 and Id4 genes: structural organization and chromosomal localization. Gene 222:229–235 [DOI] [PubMed] [Google Scholar]
- 96.Pagliuca A, Cannada-Bartoli P. and Lania L. (1998). A role for Sp and helix-loop-helix transcription factors in the regulation of the human Id4 gene promoter activity. J Biol Chem 273:7668–7674 [DOI] [PubMed] [Google Scholar]
- 97.Rockman SP, Currie SA, Ciavarella M, Vincan E, Dow C, Thomas RJS. and Phillips WA. (2001). Id2 is a target of the β-catenin/T cell factor pathway in colon carcinoma. J Biol Chem 276:45113–45119 [DOI] [PubMed] [Google Scholar]
- 98.Boileau P, Mrejen C, Girard J. and Hauguel-de Mouzon S. (1995). Overexpression of GLUT3 placental glucose transporter in diabetic rats. J Clin Invest 96:309–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ericsson A, HamarK B, Powell TL. and Jansson T. (2005). Glucose transporter isoform 4 is expressed in the syncytiotrophoblast of first trimester human placenta. Hum Reprod 20:521–530 [DOI] [PubMed] [Google Scholar]
- 100.Inoki K, Haneda M, Maeda S, Koya D. and Kikkawa R. (1999). TGF-beta 1 stimulates glucose uptake by enhancing GLUT1 expression in mesangial cells. Kidney Int 55:1704–1712 [DOI] [PubMed] [Google Scholar]
- 101.Kitagawa T, Masumi A. and Akamatsu Y. (1991). Transforming growth factor-beta 1 stimulates glucose uptake and the expression of glucose transporter mRNA in quiescent Swiss mouse 3T3 cells. J Biol Chem 266:18066–18071 [PubMed] [Google Scholar]
- 102.Okamoto Y, Sakata M, Yamamoto T, Nishio Y, Adachi K, Ogura K, Yamaguchi M, Takeda T, Tasaka K. and Murata Y. (2001). Involvement of nuclear transcription factor Sp1 in regulating glucose transporter-1 gene expression during rat trophoblast differentiation. Biochem Biophy Res Commun 288:940–948 [DOI] [PubMed] [Google Scholar]
- 103.Phillips T, Ferraz I, Bell S, Clegg PD, Carter SD. and Mobasheri A. (2005). Differential regulation of the GLUT1 and GLUT3 glucose transporters by growth factors and pro-inflammatory cytokines in equine articular chondrocytes. Vet J 169:216–222 [DOI] [PubMed] [Google Scholar]