Abstract
While it is evident that the metabolic machinery of stem cells should be fairly different from that of differentiated neurons, the basic energy production pathways in neural stem cells (NSCs) or in neurons are far from clear. Using the model of in vitro neuron production by NE-4C NSCs, this study focused on the metabolic changes taking place during the in vitro neuronal differentiation. O2 consumption, H+ production, and metabolic responses to single metabolites were measured in cultures of NSCs and in their neuronal derivatives, as well as in primary neuronal and astroglial cultures. In metabolite-free solutions, NSCs consumed little O2 and displayed a higher level of mitochondrial proton leak than neurons. In stem cells, glycolysis was the main source of energy for the survival of a 2.5-h period of metabolite deprivation. In contrast, stem cell-derived or primary neurons sustained a high-level oxidative phosphorylation during metabolite deprivation, indicating the consumption of own cellular material for energy production. The stem cells increased O2 consumption and mitochondrial ATP production in response to single metabolites (with the exception of glucose), showing rapid adaptation of the metabolic machinery to the available resources. In contrast, single metabolites did not increase the O2 consumption of neurons or astrocytes. In “starving” neurons, neither lactate nor pyruvate was utilized for mitochondrial ATP production. Gene expression studies also suggested that aerobic glycolysis and rapid metabolic adaptation characterize the NE-4C NSCs, while autophagy and alternative glucose utilization play important roles in the metabolism of stem cell-derived neurons.
Introduction
Neural stem cells (NSCs) are ideal tools for better understanding of neural cell differentiation, testing neuroactive drugs, modeling neural diseases, and elaborating effective cell therapies for the future. Despite world-wide research efforts, however, little is known on either inherent cellular or environmental conditions, which govern the formation of mature neural tissue-type cells from multipotent NSCs. Large body of data has demonstrated that NSCs and their differentiating progenies require a significantly different environment for survival and further differentiation, both in vitro and in vivo [1]. Besides the need for growth factors [2], adhesive surfaces [3], and cell activating stimuli [4], amendments of cellular metabolic machinery seem to play important roles in decision on propagation, differentiation, or death of neural progenitors during development, regeneration, or physiological neuron replacement [5,6].
The high O2 extraction by the brain and the almost 6 to 1 ratio of CO2 production from glucose led to the consent view that neural metabolism is based mainly on mitochondrial oxidative phosphorylation [7]. The importance of aerobic glycolysis in brain energy production, however, has been proved by multiple data [8] and was suggested to play important roles in fueling local activities of nerve terminals [9]. Astrocytes were shown to display a high glycolytic activity even in aerobic conditions and provide metabolic fuels, mainly glutamine and lactate, for neurons [10,11]. While important discussions have been focusing on the energy production and distribution of metabolites between astrocytes and neurons [12,13], little is known on the metabolic demands of other cellular components of the central neural tissue, including the diverse types of neural stem/progenitor cells [14].
Preceding the vascularization of the early embryonic brain primordia, NSCs exist in hypoxic conditions. Hypoxia characterizes the neurogenic zones in later phases of development and in the adult brain [15]. Our previous data [16] demonstrated that embryonic NSCs survive and proliferate under hypoxic conditions, but generation of neurons requires normoxic O2 supply, both in vitro and in vivo.
These data, together with available in vitro results on various neural stem-like cells [5,17–19], suggest that NSCs have mainly glycolysis-based metabolism, which shifts to oxidative metabolism during differentiation. Committed neural precursors become more sensitive to hypoxia, and maturing neurons die under hypoxic conditions. Depending on the level of differentiation, different stem cell populations display more or less developed mitochondria and utilize O2-dependent or O2-independent metabolic routes for energy production [20]. Besides HIF1α destabilization [15,21], the expression of several metabolic regulatory proteins was shown to change during neuronal differentiation [6], including the TSPO18 protein [22], a transmembrane protein associated with the mitochondrial permeability pore [23]. Available data suggest that the metabolite utilization and the routes of energy production change fundamentally with the advancement of cellular differentiation and maturation. While this view needs further support from solid experimental results, it raises a further question: whether metabolic changes represent adaptive cellular reactions to the environment, namely, to the gradually increasing O2 supply during in vivo tissue genesis or there are intrinsic metabolic changes characterizing the cellular differentiation.
Our recent studies have been addressed to reveal metabolic changes undertaken during in vitro neurogenesis by NSCs kept and differentiated in normoxic conditions. In vitro neuron formation by NE-4C embryonic mouse NSCs (ATCC-CRL-2925; [22,24–29]) provided a model to compare metabolic characteristics of stem cells and stem cell-derived neurons. NE-4C NSCs were cloned [30] from the anterior brain vesicles of 9-day-old p53−/− mouse embryos [31]. NE-4C cells proliferate continuously (with duplication time 16–20 h), display epithelial morphology and nestin immunoreactivity, and express Oct 4 and nanog stem cell genes. In response to treatment with 10−6 M all-trans retinoic acid (RA; 24–48 h), NE-4C cells generate neurons first (40%–50% of all cells on the 7th to 9th day of induction) and astrocytes later (from the 10th day of induction on; [24]). (More detailed description of induction, neuron and glia formation is presented in Supplementary Data 1; Supplementary Data are available online at www.liebertpub.com/scd)
Comparing the O2 consumption, H+ production, and metabolic responses to defined fuel molecules, important metabolic differences were revealed between NSCs and the differentiating neuronal progenies, and important similarities were found in the metabolic processes of stem cell-derived neurons and primary neurons isolated from the embryonic mouse forebrain.
Materials and Methods
Cell cultures
NE-4C ([30]; ATTC-CRL-2595) cells were maintained in minimum essential medium (MEM; Sigma), supplemented with 5% heat-inactivated fetal calf serum (FCS; PAA), 4 mM l-glutamine (Sigma), 40 μg/mL gentamicin (Chinoin), and 2.5 μg/mL amphotericin B (Sigma). The cells were seeded at 5 × 104 cells/cm2 density into poly-L-lysine-coated (PLL; Sigma) dishes and were let to grow to semiconfluence. Semiconfluent cultures were split by trypsinization (0.05% w/v trypsin with 1 mM EDTA in phosphate-buffered saline). The medium was changed three times a week.
Neuronal differentiation was induced by addition of 10−6 M all-trans retinoic acid (RA; Sigma) to confluent cultures of NE-4C cells. After 48 h, the media were changed to a RA- and serum-free neuronal medium: 50% Dulbecco's modified Eagle's medium (DMEM; Sigma), 50% F12 nutrient medium (Sigma), supplemented with 4 mM l-glutamine and 1% (v/v) insulin–transferrin–selenium (ITS; Gibco) and 1% (v/v) B27 (Gibco) neuronal supplements. Gentamycin (40 μg/mL) (Chinoin) and amphotericin B (2.5 μg/mL) (Sigma) were added as antibiotics. Differentiated NE-4C cells were used for the metabolic studies in the second week after the onset of induction.
Primary neuronal cultures were prepared from embryonic (E15–E16) mouse (CD1) forebrains according to the animal experimentation license 22.1/354/3/2011 (expiry date 2016 July) issued by the Animal Welfare Directorate of National Food Chain Safety Office (NFCSO). Cell suspensions were obtained by mechanical dissociation in MEM containing 10% FCS, over a nylon mesh with a pore diameter of 40–42 μm [32]. The cells were seeded at 105 cells/cm2 density onto PLL-coated dishes for molecular studies, onto PLL-coated coverslips for immunocytochemical investigations, or onto PLL-coated 96-well plates for metabolic assays. The cultures were maintained in MEM supplemented with 10% heat-inactivated FCS, 4 mM l-glutamine (Sigma), 40 μg/mL gentamicin (Chinoin), and 2.5 μg/mL amphotericin B (Sigma). The first medium exchange was carried out 24 h after plating, and then, the medium was changed twice a week. In the second week after plating, the medium was changed to serum-free neuronal medium. The cultures were used for metabolic assays on the 11th–13th days after seeding.
Astrocyte cultures were prepared from late fetal or perinatal CD1 mouse forebrains [33]. The cells were seeded in 10% FCS MEM on PLL-coated surfaces, at a density of 105 cells/cm2. The cells were let to grow in serum-containing medium with two changes of medium a week. The cultures were split maximum three times before plating into PLL-coated 96-well plates for metabolic studies.
The cells were maintained in water-saturated air atmosphere containing 5% CO2 at 37°C.
Viability assays
MTT (3-[4,5-dimethylthiazol-2yl]-2-5-diphenyl-tetrazolium bromide; Sigma) reduction test [34] was used to check the viability of cells in metabolite-free (starving) conditions. Cells were seeded onto PLL-coated 96-well plates, at a density of 2 × 104 cells/well. NE-4C stem cells were assayed right after firm attachment (6–8 h after plating). NE-4C-derived neurons were assayed on the 8th to 10th day after the induction of neuronal differentiation. For the assays, the medium was changed to artificial cerebrospinal fluid (ACSF) (145 mM NaCl, 3 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 10 mM HEPES; pH = 7.2; 50 μL/well) on half of the cultures. In the other wells, the cells were maintained in the normal medium. After a 3-h incubation in a CO2 incubator (37°C, 21% v/v O2, 5% CO2), 10 μL MTT was added to each well to reach a final MTT concentration of 0.208 mg/mL. After 1.5 h, the formazan precipitate and the cellular material were dissolved in acidified (0.08 M HCl) isopropanol and measured at 570 nm measuring and 690 nm reference wavelengths (Wellcome MR5000 plate reader). Cell viability was calculated by relating the absorbance values of each well to the mean of optical densities measured in normal medium on the same plate (n = 5–8). Relative viability was calculated as the percentage of the mean OD value in normal medium. The assays were repeated three times. Significance was calculated by t-test (see statistical analysis section).
Assays on O2 consumption and extracellular pH changes
The Seahorse XF96 Extracellular Flux Analyzer (Seahorse Bioscience) was used for in situ measurement of O2 consumption rate (OCR) and the rate of the extracellular pH drift (ECAR) in a 2.28 μL fluid volume above the cells.
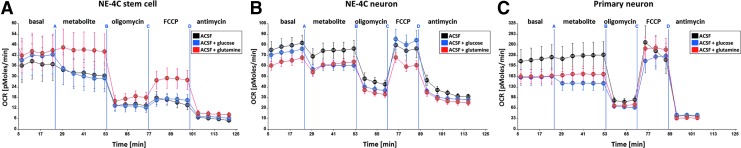
NE-4C cells were seeded onto 96-well Seahorse plates (1.5–2.0 × 104cells/well) coated with PLL. Before the assays, the cultures were washed with metabolite-free ACSF to remove metabolic components of the maintaining media. The cells were incubated in 180 μL ACSF for 1.5 h in the incubator of the instrument to equilibrate the gas concentrations. The oxygen content and acidity (pH) of the extracellular medium were measured in parallel in 96 wells. The OCR and ECAR data were calculated by the instrument from the changes of the O2 and H+ concentrations in a small fluid volume above the cells. The OCR and ECAR were first recorded in ACSF (five cycles of 2–3 min measuring and 3-min mixing; see Supplementary Data 2), and the measured changes were averaged in 5 data points (Fig. 1) and used as a baseline (basal OCR and ECAR). Single metabolites dissolved in ACSF were then added through the injection port to reach the following final concentrations in the culture fluids: 5 mM D-glucose (Sigma), 5 mM Na-lactate (Sigma), 5 mM D,L-β-hydroxybutyrate (Sigma), 2.5 mM glutamine (Merck), or 5 mM pyruvate (Sigma). On each plate, 6–12 microcultures received the same metabolite or were left in ACSF (as starving control). In the presence of metabolites, O2 and H+ concentrations were further measured with interruptions of mixing (Supplementary Data 2), and the data were averaged into five time points (Fig. 1). At the end of metabolite testing, mitochondrial inhibitors (2 μM oligomycin; Sigma), 0.4 μM FCCP (fluoro 3-carbonyl cyanide-methoxyphenyl hydrazone; Sigma), or 100 μM DNP (2,4-dinitrophenol; Sigma), finally 1 μM antimycin (Sigma), in the case of DNP antimycin with 1 μM rotenone (Sigma), were injected and further data points were measured with each. Cells responding accurately to the above drugs possessed functional mitochondria and thus were regarded healthy.
FIG. 1.
Representative graphs of the oxygen consumption (OCR; O2 pMoles/min) by NE-4C stem cells (A), NE-4C-derived neurons (B), and primary forebrain neurons (C) in starvation (in metabolite-free ACSF; basal), in response to addition of 5 mM glucose or 2.5 mM glutamine or just fuel-free ACSF (metabolites), then oligomycin, FCCP, and antimycin successively. Averages and standard errors of mean of n ≥ 7 cultures are shown. ACSF, artificial cerebrospinal fluid; FCCP, fluoro 3-carbonyl cyanide-methoxyphenyl hydrazone.
The data were plotted as OCR (oxygen consumption rate: pMoles O2 consumption/min) and ECAR (extracellular acidification rate mpH/min) as a function of time (Fig. 1). For comparing the reactions of cells in different wells (not always containing the same amount of cells), OCR and ECAR values in the presence of metabolites/inhibitors were related to the OCR and ECAR values recorded in the nontreated state of the same well (basal values: 100%) and were plotted as relative OCR% and ECAR% values. The presented data were obtained from n ≥ 3 independent series of assays.
The data sets were processed with MATLAB. Statistical analysis was performed with R statistical programing [35].
Quantitative polymerase chain reaction
Total RNAs were isolated from confluent (60-mm-diameter dishes) cultures with the RNeasy Mini Kit (Qiagen) according to the manufacturer's protocol. DNA contamination was eliminated by RNase-Free DNase Set (Qiagen). For reverse transcription, 2 μg amounts of total RNA were used with the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) according to the manufacturer's instructions, with 0.5 μL RNase inhibitor. The 20 μL reaction mixture was incubated for 10 min at 25°C (annealing), for 10 min at 37°C (transcription), and for 3 min at 72°C (inactivation). The reaction product cDNA was diluted with equal volume of water and its 2 μL was added to 8 μL mixtures of Master mix (Roche R480 LightCycler probes master lc480), with 1 μL primers (Table 1; final concentration 5 pM) and 0.5 μL EvaGreen diluted 20x in water (Biotium). The qPCR sequences were as follows: 95°C for 10 min, 40 repetitions of 95°C for 15 s, 60°C for 30 s. The reactions were run in StepOnePlus (Applied Biosystems) Real-Time PCR apparate. Primer efficiencies determined by 5-point standard curves in duplicate assays on dilution series of 1 to 5 for each cDNA, ranged between 87% and 106% (Table 1). The results were analyzed according to the ΔΔ CT method [36] with StepOne v2.3 software (Applied Biosystems).
Table 1.
Primers Used
| Gene | Forward | Reverse | Efficiency (%) |
|---|---|---|---|
| hprt | CACAGGACTAGAACACCTGC | GCTGGTGAAAAGGACCTCT | 106.298 |
| glut1 (slc2a1) | TGCCCAGGTGTTTGGCTTAG | TCCCTCGAAGCTTCTTCAGC | 94.470 |
| glut3 (slc2a3) | AACACTTGCTGCCGAGAACA | GATGGGGTCACCTTCGTTGTC | 97.303 |
| pdk4 | CCAACCCTACGGATCCTAACC | GGCATTTTCTGAACCAAAGTCC | 88.043 |
| atg12 | TGGCCTCGGAACAGTTGTTTAT | GGAAGGGGCAAAGGACTGAT | 94.727 |
| tfam | TCCCCTCGTCTATCAGTCTTGT | CCACAGGGCTGCAATTTTCC | 102.946 |
| oct4 (pou5f1) | GAGGCTACAGGGACACCTTTC | GTGCCAAAGTGGGGACCT | 86.627 |
| ngn2 (neurog2) | ACATCTGGAGCCGCGTAG | CAGCAGCATCAGTACCTCCTC | 93.215 |
| math2 (neurod6) | CGACACTCAGCCTGAAAAGA | CAAACTTTCTGCACATCTGGG | 96.756 |
HPRT was used as endogen reference for each amplification. The amplified amount of each sequence was normalized to that of HPRT from the same cell source. The different preparations were compared by relating the amplified product/HPRT ratio to the same ratio obtained from NE-4C neurons ( = 1). Data are presented as median and MAD (Median Absolute Deviation) values (n ≥ 3).
Immunocytochemistry
NE-4C cells were seeded onto 24-well plates (2-4 × 105 cells/cm2) coated with PLL and were maintained as nondifferentiated stem cells or were induced for neural differentiation and used on the 6th–12th day of induction. Before the assays, the media were changed to ACSF, and the cells were incubated in external metabolite-free conditions for 3.5 h. The cells were fixed with 4% w/v paraformaldehyde (TAAB) in PBS for 20 min at room temperature, washed with PBS, and then permeabilized with 0.1% Triton-X 100 (Reanal) in PBS. For blocking nonspecific antibody binding, the cells were incubated with 2% BSA (bovine serum albumin; Sigma) in PBS for 1 h. Primary antibodies, anti-β-III-tubulin mouse IgG (Sigma) (for NE-4C neurons) and anti-nestin mouse IgG (Chemicon) (for NE-4C stem cells), were diluted 1:1,000 with BSA-PBS and added to the cells for overnight at 4°C. As a secondary antibody, anti-mouse IgG-Alexa488 (Invitrogen) was diluted 1:1,000 and added to the cells for 1 h at room temperature. After washing, the stained preparations were mounted with Mowiol (Calbiochem) containing 10 μg/mL bisbenzimide (Hoechst 33258 nuclear stain; Sigma). The preparations were examined with the Zeiss Axiovert 200 M Microscope equipped with AxioVision 4.8 software (Carl Zeiss).
Statistical analysis
Statistical analysis was performed with R statistical programing [35]. Results are presented as mean ± SEM; in case of qPCR, results are shown as median ± MAD.
For viability, OCR%, ECAR%, and maximal respiration data, t-test was performed between two experimental groups.
For calculations of proton leak significance, one-way ANOVA and Tukey HSD tests were performed.
Statistical relevance of qPCR data was analyzed by the Kruskal–Wallis rank-sum test and Dunn's test (of multiple comparisons using rank sums).
For t-tests and ANOVA, data were first checked for normality with q-q plot and homogeneity of variances with Bartlett test. The graphs were plotted with ggplot2 package. In each case, P < 0.05 (*), P < 0.01 (**), and P < 0.001 (***) were considered statistically significant.
Results
To investigate the metabolic changes during in vitro neuron formation, metabolic characteristics of noninduced NE-4C NSCs were compared to those of retinoic acid-induced neuronal NE-4C progenies. The rate of neuron formation was checked by immunocytochemical assays and by qPCR verification of the expression of proneural (ngn2) and neuronal (math2) genes, as well as the repression of oct4 stemness genes (Supplementary Data 1). Primary neuronal cultures prepared from E15-16 embryonic mouse forebrain [32] and cultures of astrocytes [33] were also investigated for comparison.
The basal oxygen consumption, the extracellular acidification, and the effects of selected single fuel molecules (glucose, lactate, β-OH-butyrate, glutamine, and pyruvate) on OCR and ECAR were determined by instrumental cell metabolism assays using Seahorse Cell Analyzer XF96 [37] (Supplementary Data 2) (Fig. 1).
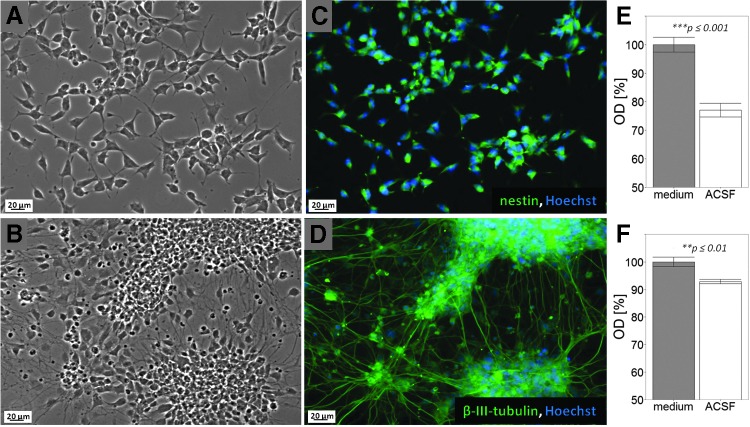
Before the assays, the cells were washed with ACSF and maintained in ACSF without any metabolic fuel compounds. In response to starvation for a period as long as 3.5 h, morphological changes were not detected in any of the cultures by light microscopy (Fig. 2A–D).
FIG. 2.
NE-4C neural stem cells (A, C) and NE-4C-derived neurons (B, D) on the 6th day after induction with retinoic acid did not show morphological damages in response to 3.5-h starvation. For immunocytochemical identification, NE-4C stem cells were stained for nestin (green on C), and NE-4C-derived neurons were visualized by staining for β-III-tubulin (green on D). The nuclei were stained with Hoechst (blue). (E, F) MTT reduction test indicated higher sensitivity to starvation (in metabolite-free ACSF) of NE-4C stem cells (E) than differentiating NE-4C neuronal derivatives (F). Averages and standard errors of mean (n ≥ 10) are shown.
At the end of a 3.5-h starvation period in fuel-free ACSF, the MTT reduction (viability) test [34] indicated about 20%–25% viability reduction of NE-4C stem cells, while less than 10% reduction was detected in the cultures of differentiating NE-4C-derived neurons (Fig. 2E, F). The data demonstrated that while the cells survived a 3.5-h starvation, stem cells displayed enhanced sensitivity to the lack of metabolic fuel molecules.
For checking mitochondrial integrity and getting further parameters on the mitochondrial activity, oligomycin (an inhibitor of mitochondrial ATP synthase), FCCP (disruptor of mitochondrial proton gradient), and antimycin (an inhibitor of the electron transport in complex III) were added for 30 min successively, during continuous OCR/ECAR determinations. Metabolic changes were calculated from microcultures, which responded to the inhibitors (Supplementary Data 2).
In line with previous data [5,16,18], stem cells displayed the lowest oxygen consumption rate in the metabolite-free ACSF, as it was determined in absolute pMoles/min/15,000 cell values (48.17 ± 3.02; n = 28). Differentiating NE-4C neurons displayed a significantly higher oxygen consumption (101.49 ± 10.1; n = 23), but did not reach the level of neuron-enriched primary cultures (204.67 ± 9.79; n = 46). The OCR of primary astrocytes (113.61 ± 3.62; n = 39) stayed behind that of primary neurons corresponding to the high aerobic glycolytic activity of astrocytes [11]. While the data indicated a raise of O2 consumption with cellular differentiation, the absolute OCR values could not be directly compared between the different cultures due to the hardly controllable variations of cell numbers in the different types of cultures. The differentiation-dependent increase of mitochondrial activity, however, was clearly demonstrated by the maximal mitochondrial respiration values [38] (Fig. 3A). Maximal mitochondrial respiration was calculated for each microculture by subtracting nonmitochondrial O2 consumption (OCR in antimycin block) from the OCR measured in response to the mitochondrial uncoupling agent, FCCP, in the same culture. The maturation of the mitochondrial activity was also shown by the decrease in proton leak ([38]; [17,18]) with cell development; after blocking the mitochondrial ATP synthase with oligomycin, the O2 consumption remained relatively high in stem cells in comparison to more differentiated progenies (Fig. 3B). In line with the O2 consumption data, the expression of the mitochondrial transcription factor A (TFAM; [39]) was significantly lower in NE-4C stem cell cultures than in more differentiated cultures, suggesting differentiation-dependent changes in mitochondrial maturation.
FIG. 3.
(A) Maximum mitochondrial respiration of NE-4C stem cells and differentiating NE-4C neurons was determined as the OCR of FCCP-treated cells in the percentage of OCR in starvation (100%). The columns represent averages and standard errors of means (n ≥ 8). (B) The proton leak of NE-4C stem cells, differentiating NE-4C neurons, primary neurons, and astrocytes was determined as the percentage of OCR remaining after blocking the mitochondrial ATP-synthase with oligomycin and subtracting the nonmitochondrial O2 consumption. The columns represent averages and standard errors of means (n ≥ 23). (C) The relative expression (RQ) of mitochondrial transcription factor A (tfam) in the investigated cells (n = 3) is shown in relation to the tfam/hprt ratio (1.00) in NE-4C neurons. OCR, oxygen consumption rate.
After the determination of basal OCR and ECAR values in metabolite-free ACSF, a single metabolite (5 mM final concentration of glucose, lactate, β-OH-butyrate, pyruvate, or 2.5 mM final concentration of glutamine) was added to the cultures or the cells were maintained for a further 30 min in fuel-free conditions (starving controls).
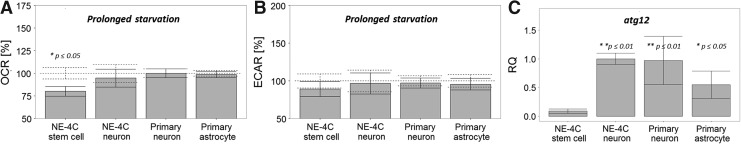
The prolonged (+ 30 min) starvation reduced further the O2 consumption of NSCs, while did not cause changes in the OCR of differentiating NE-4C neurons, primary neurons, and astrocytes (Fig. 4A). The extracellular proton production (Fig. 4B) indicated a moderate drop in the glycolytic activity of all cell types with most marked (about 10%) changes in NE-4C NSCs. The nonchanging viability (Fig. 2), and the nonreduced O2 consumption and glycolytic activity of differentiated neural cells suggested that more differentiated cells utilize their own cellular material for preserving the energy production in starvation. In line with these findings, an enhanced expression of autophagy-related gene 12 (ATG12; [40]) was found in NE-4C neuronal progenies, primary neurons, and astrocytes in comparison to stem cells by qPCR (Fig. 4C), suggesting some higher role of autophagy in the preservation of metabolic integrity of differentiated cells.
FIG. 4.
Prolonged (+ 30 min) starvation reduced the OCR only in neural stem cells (A) (the dashed line indicates the basal OCR for each cell type; 100%) and decreased slightly the extracellular acidification rate (an indicator of glycolytic activity) (B) (dashed line indicates the basal ECAR for each cell type; 100%). The columns represent averages and standard errors of means (n ≥ 23). (C) The relative expression (RQ) of autophagy-related gene 12 (atg12) is shown in relation to the atg12/hprt ratio (1.00) in NE-4C neurons (n = 3). ECAR, extracellular acidification rate.
When 5 mM glucose was added to starving cells, the O2 consumption decreased and the extracellular acidification increased markedly in all cultures. The highest OCR reduction and ECAR increase was found in the cultures of NSCs, indicating a high rate of glucose utilization through aerobic glycolysis in these cells (Fig. 5).
FIG. 5.
Effect of glucose addition on the O2 consumption (OCR) (A) and the extracellular acidification (ECAR) (B) by starving cells. The basal OCR (100%) values and their standard errors of mean are shown as dashed lines (n ≥ 23). The relative expression (RQ) of glucose transporters, Glut1 (C) and Glut3 (D) (glut1/HPRT and glut3/HPRT), by NE-4C neural stem cells is shown in relation to the RQ values of glut1/HPRT and glut3/HPRT in NE-4C-derived neuronal cultures (RQ = 1.00), respectively (n = 3).
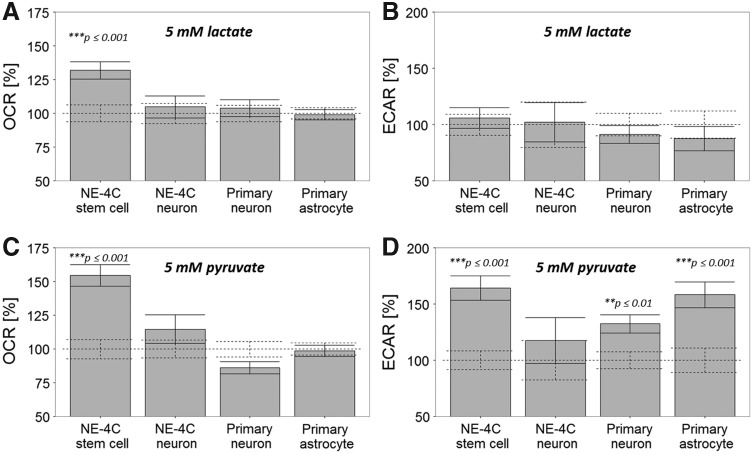
The relatively moderate decrease in O2 consumption and the only about 1.5-times increase in the acidification by differentiating NE-4C neuronal progenitors raised the question whether glucose is available for these cells. qPCR studies demonstrated that the expression of glucose transporters, both the universal Glut1 and the neuronally abundant Glut3 [41], increased with neuronal differentiation (Fig. 5C,D). While glucose transporters were not checked at the protein level, the developmental increase in the expression of glucose transporters suggested that glucose might be even more available for differentiating neurons than for stem cells. As a second step, the effects of glycolytic end-products were investigated by measuring the O2 consumption after adding 5 mM pyruvate or lactate to the cells (Fig. 6). Addition of lactate and pyruvate increased the O2 consumption only in NE-4C stem cells. NE-4C-derived or primary neurons apparently did not respond to lactate with enhanced O2 uptake. Rather surprisingly, pyruvate caused a mild decrease in the O2 consumption (Fig. 6C) of primary neurons.
FIG. 6.
Effects of lactate (A, B) and pyruvate (C, D) on the O2 consumption (OCR) of cells in relation to basal OCR (100%; dashed lines) (A, C), and on the extracellular acidification in relation to basal ECAR (100%; dashed lines) (B, D). Averages and standard errors of mean are shown (n ≥ 23).
Pyruvate significantly contributed to the acidification of the extracellular fluid of stem cells and astrocytes (Fig. 6D), presumably due to CO2 generation by pyruvate dehydrogenase. Rather unexpectedly, however, pyruvate while apparently did not enhance the mitochondrial energy production of neurons, did not increase markedly the acidity of their fluid environment, either.
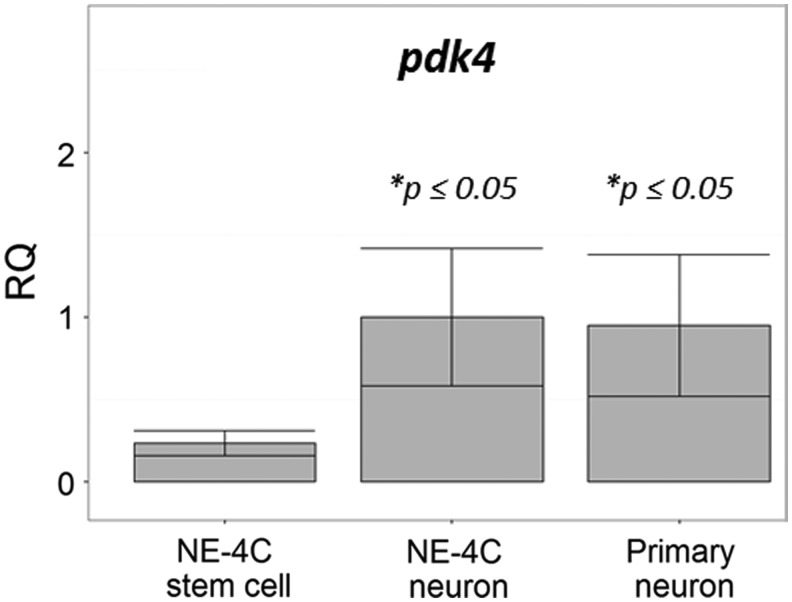
The data suggested that while stem cells utilize glycolytic end-products for mitochondrial energy production, neurons, at least in starvation, seem to utilize glucose through some different pathways. The pyruvate–dehydrogenase complex (PDC) is a key point to direct pyruvate to the citrate cycle and so to fuel mitochondrial oxidative phosphorylation [42,43]. Pyruvate dehydrogenase E1 enzyme (PDH) is inactivated by phosphorylation by pyruvate dehydrogenase kinases (PDKs). PDK4 was reported to be present in the rodent brain [44]. qPCR studies on the expression of pdk4 in NE-4C NSCs and in NE-4C-derived neurons (Fig. 7) showed an increase with development, suggesting a differentiation-dependent capacity to regulate the entry of pyruvate into the citrate cycle.
FIG. 7.
The relative expression (RQ) of pyruvate dehydrogenase kinase 4 (pdk4) in NE-4C neural stem cells and in NE-4C-derived and primary neuronal cultures shown in relation to the pdk4/hprt ratio (1.00) in NE-4C neurons (n = 3).
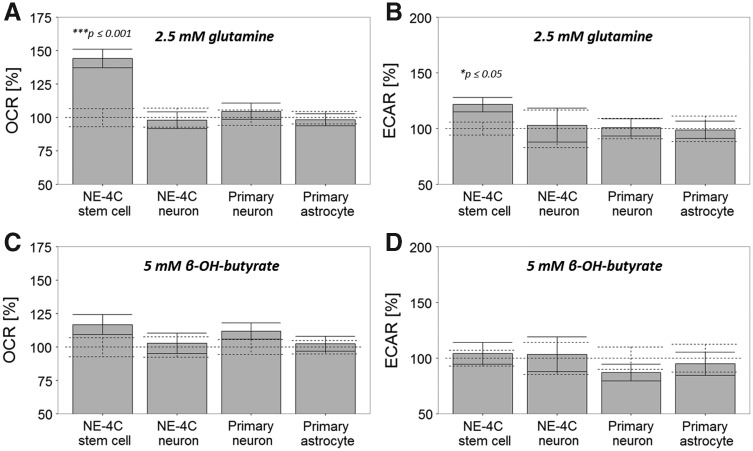
If glycolysis is not the main input for mitochondrial energy production in starving neurons, what metabolic pathway might be responsible for the high neuronal OCR even in starving conditions? As potential fuel compounds, β-OH-butyrate (5 mM) and glutamine (2.5 mM) were investigated (Fig. 8).
FIG. 8.
Effects of glutamine (A, B) and β-OH-butyrate (C, D) on the O2 consumption (A, C) and extracellular acidification (B, D) of the cells. The basal OCR and ECAR with the standard errors of mean are shown as dashed lines (100%) for each type of cells. Averages and standard errors of mean of n ≥ 23 cultures are shown.
The studies demonstrated that NE-4C NSCs utilized both β-OH-butyrate and glutamine in mitochondrial oxidation, while none of these substrates evoked significant increase in neuronal O2 consumption.
Summarizing, our data showed that NSCs and their neuronal derivatives display different metabolite utilization under starvation. Stem cells display low mitochondrial activity and require low O2 supply. However, if metabolites such as lactate, pyruvate, β−OH-butyrate, and glutamine are available, stem cells use them for mitochondrial energy production. Glucose, if added as a single metabolic fuel, is not utilized for mitochondrial ATP production by either stem cells or neurons and astrocytes. For metabolite-deprived neurons, glycolytic end-products, pyruvate, or lactate did not serve as fuels for mitochondrial ATP production. Addition of a single metabolite did not activate the mitochondrial energy production of neurons. Under the applied conditions, the high O2 consumption of starving neurons seemed to be fueled by cellular self-material.
Discussion
The aim of the presented studies was to reveal whether alterations in the metabolite utilization may reason the differences known in the O2 requirement of stem cells and neurons [5,16–19]. Comparing the metabolic characteristics of stem cells to features of neurons derived from them, offered a way to reduce cellular heterogeneity between the cell preparations to be compared. The model, however, suffered some drawbacks:
(i) The NE-4C NSC line was isolated from p53-deficient mice [31] and got presumably immortalized just by the lack of functional p53 protein. The tumor suppressor p53 protein, besides regulating cell cycle progression, DNA repair, and apoptosis [45], is known to interfere with molecular networks that can sense and regulate the cellular energy production (for review: [46]). The p53-deficient mice, in contrast, develop normally, and a higher tumor incidence appears only in aged populations of such mice [31]. In case of NE-4C cells, the lack of functional p53 does not prevent the exit from the cell cycle upon induction with retinoic acid, and to produce well-developed postmitotic neuronal phenotypes in vitro [22,30] or neurons integrating into the embryonic forebrain [47]. Because of the metabolic regulatory effects of p53, however, wild-type neurons and astrocytes had to be included in the studies addressed to cellular metabolism. NE-4C neurons and wild-type primary neurons responded similarly to metabolite deprivation and to the addition of various fuel-molecules. The data demonstrated that neuronal reactions to the applied metabolic challenges were fairly different from the responses of stem cells, regardless of the presence or absence of p53.
(ii) While the cultures of NE-4C stem cells were composed of phenotypically identical cells, the retinoic acid-induced cultures contained persisting stem cells and progenies in various stages of differentiation, besides of about 40%–50% of NE-4C neurons. The cellular heterogeneity caused variations in the detected cellular responses and resulted in rather large standard error of mean values. Cellular heterogeneity had to be taken into account in case of primary neuronal cultures as well. Primary neuronal cultures, even if enriched in neurons, always contain glial cells, and this condition cannot be neglected if neuronal metabolic characteristics are investigated.
In the presented study, primary neuronal cultures were included to investigate whether the developmental changes during the neuron formation from NE-4C stem cells result in a similar metabolic phenotype as primary neuronal cultures of the mouse forebrain. Embryonic (E15-16) forebrain-derived and NE-4C-derived neuronal cultures share a number of structural, cytochemical, and functional characteristics. Briefly, both NE-4C and primary embryonic forebrain cultures contain a substrate-attached, non-neuronal cell layer, and neurons develop on the top of these basal cells [30,32]. The basal cell layer is composed by GFAP-positive astroglial cells in primary cultures, while a more scattered, GFAP-negative basal cell layer is formed under NE-4C neurons. NE-4C-derived GFAP-positive cells appear in this layer from the 10th day of induction [24]. Nondifferentiated stem/progenitor cells in different stages of cell commitment persist in both types of neural cultures. In both types of cultures, neurons with long branching processes, MAP2 and β-III-tubulin, synaptophysin and synapsin immunoreactivity, and with recordable spike potentials give the 40%–50% of the total cells by the end of the second week after plating (primary cultures) or retinoid induction (NE-4C). Both neuronal cultures contain GABAergic and less glutamatergic neurons; monoaminergic cells do not develop. The neuronal circuit activity had been characterized in primary neuronal cultures [48], while only single-cell activities were investigated in NE-4C-derived neuronal cultures [27]. Both cultures display spontaneous neuronal activities, which fluctuate in time. Cells in different stages of neural development are expected to display different ion-redistribution patterns in response to intracellular processes and to bioelectric activities in their environment (Kőhidi et al., manuscript in preparation). As changing bioelectric activity would cause alterations in the energy homeostasis, differentiation-dependent changes in the metabolism will reflect the different excitability of developing NSCs, as well. To reduce the fluctuations caused by different excitability, the metabolic activity of stem cells and their more differentiated progenies were compared under strictly identical conditions.
While the drawbacks of the models resulted in large statistical deviations, the data outlined some marked cell-type-related metabolic features.
In accord with results of other laboratories [5,17–19], the stem cells consumed less O2, and produced less mitochondrial ATP than neurons. The data showed that NE-4C stem cells could gain enough energy from glycolytic sources for the survival of a 2.5-h period of metabolite deprivation. In response to the addition of single metabolites (with the exception of glucose), however, the mitochondrial ATP production increased, indicating a rapid adaptation of the metabolic machinery to the available resources. In contrast, astrocytes and neurons, both NE-4C-derived and primary, hardly responded to starvation and sustained a rather high-level of oxidative phosphorylation during metabolite deprivation. The observation suggests that more differentiated cells can sustain their energy status by mobilizing internal sources for a relatively long time. Rather surprisingly, addition of single metabolites to neurons or astrocytes did not result in an immediate increase of the O2 consumption and in an importantly increased mitochondrial ATP production. Starving neurons did not respond with enhanced O2 consumption either to lactate or to pyruvate, indicating that these glycolytic end-products were not conducted toward the tricarboxylic acid (TCA) cycle and were not utilized for mitochondrial ATP production.
While these data seem to contradict to the “astrocyte-neuron lactate shuttle” model [11], one should keep in mind that (i) starvation might be a condition fairly different from the physiological or neuronally activated states and (ii) the uptake of lactate and pyruvate might change importantly during neuronal differentiation.
Monocarboxylic acid transporters (MCT1-4) responsible for the transmembrane transport of lactate/pyruvate show cell-type-specific distribution [49]. While astrocytes carry MCT4 all over the cell membranes, the neurons, and only excitatory neurons, express MCT2 at differentiated postsynaptic sites. As the high-affinity but low-capacity MCT2 transporter molecules are localized in these highly specialized areas of well-differentiated glutamatergic neurons, it was not surprising that we could not demonstrate measurable expression either in the embryo-derived (11–13 days in vitro old) primary neuronal cultures or in NE-4C-derived neurons on the second week after induction (data not shown). NSCs, on the contrary, display a number of astrocytic features and can gain energy from both lactate and pyruvate; thus, they necessarily do express MCT1 and/or MCT4, similarly to a number of tumor stem cells [50].
The transcription of PDK4 was reported to increase in response to starvation [44]. This kinase, in contrast, was found to be highly expressed by both primary and NE-4C-derived neurons, while almost absent in stem cells. The pdk upregulation at the transcriptional level, however, does not mean necessarily that the PDC is inactivated. Without data on the protein level and the enzymatic activity of PDKs and PDC, only hypotheses can be formulated: enhanced expression of pdk4 might result in the uncoupling of glycolysis from the TCA cycle and might be responsible for the enhanced rate of aerobic glycolysis [51].
Aerobic glycolysis was clearly detected in all investigated cells in response to glucose addition. In response to glucose, all cells dropped O2 consumption, and mitochondrial ATP production did not increase in any of the cells. Interestingly, the extracellular acidification grew enormously in cultures of NE-4C stem cells, while significant, less extreme increase was found in more differentiated cultures. The moderate ECAR raised the question: what might be the fate of glucose in neurons. One possible assumption is that glucose is guided to the pentose-phosphate pathway. Utilization of glucose through the pentose-phosphate pathway can provide nucleotides for DNA repair of postmitotic and/or starvation-stressed cells and can protect against oxidative stress through increased NADPH production [52]. These later mechanisms would highly help the survival of finally differentiated neurons throughout their extremely long life span.
The results of our experiments, however, did not answer the question: what type of molecules can fuel the sustained oxidative phosphorylation of starving neurons. PDKs are regarded as important regulators of the glucose catabolism versus lipid oxidation [53], and utilization of ketone bodies and unsaturated fatty acids was suggested to attenuate sustained neuronal burst activities [54]. To understand the rather controversial roles of ketone bodies and free fatty acids in the energy production of neurons [55], a ketone body, β-OH-butyrate, was also added to the cells. NE-4C-derived or primary neurons, however, failed to show any measurable changes in O2 consumption in response to β-OH-butyrate. Further studies are in progress to find nutrients that may serve as metabolic fuel molecules for neurons in starvation or under other stress situations.
Supplementary Material
Acknowledgments
The authors are indebted to Mrs. Katalin Gaál for the excellent technical assistance and to Dr. Tünde Kovács, Dr. Zsuzsanna Környei, and Mr. Gergő Horváth for their valuable help in experiments and evaluations.
The work was supported by the National Science Foundation (OTKA) grant: K 106191 (to E.M.) and NK 81983 (to V.A.-V.), by Richter Gedeon Pharmaceutical, Inc. Tematic Grant 4700148815 (to L.T. and E.M.), by Richter Gedeon Centenary Fund (to A.J.), and by the Hungarian Brain Research Program (KTIA_13_NAP-A-III/6) (to V.A.-V.).
Author Disclosure Statement
The authors declare that there is no conflict of interest with any commercial enterprise or financing body.
References
- 1.Gage FH. and Temple S. (2013). Neural Stem Cells: generating and Regenerating the Brain. Neuron 80:588–601 [DOI] [PubMed] [Google Scholar]
- 2.Ramasamy S, Narayanan G, Sankaran S, Yu YH. and Ahmed S. (2013). Neural stem cell survival factors. Arch Biochem Biophys 534:71–87 [DOI] [PubMed] [Google Scholar]
- 3.Faissner A. and Reinhard J. (2015). The extracellular matrix compartment of neural stem and glial progenitor cells: extracellular Matrix of Stem/Progenitor Cells. Glia 63:1330–1349 [DOI] [PubMed] [Google Scholar]
- 4.Spitzer NC. (2006). Electrical activity in early neuronal development. Nature 444:707–712 [DOI] [PubMed] [Google Scholar]
- 5.Folmes CDL, Dzeja PP, Nelson TJ. and Terzic A. (2012). Metabolic plasticity in stem cell homeostasis and differentiation. Cell Stem Cell 11:596–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rafalski VA. and Brunet A. (2011). Energy metabolism in adult neural stem cell fate. Prog Neurobiol 93:182–203 [DOI] [PubMed] [Google Scholar]
- 7.Kety SS. (1957). The general metabolism of the brain in vivo. In: The Metabolism of the Nervous System, Richter D., ed. Pergamon Press, London, pp 221–237 [Google Scholar]
- 8.Vaishnavi SN, Vlassenko AG, Rundle MM, Snyder AZ, Mintun MA. and Raichle ME. (2010). Regional aerobic glycolysis in the human brain. Proc Natl Acad Sci USA 107:17757–17762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pellerin L. and Magistretti PJ. (1996). Excitatory amino acids stimulate aerobic glycolysis in astrocytes via an activation of the Na+/K+ ATPase. Dev Neurosci 18:336–342 [DOI] [PubMed] [Google Scholar]
- 10.Bak LK, Schousboe A. and Waagepetersen HS. (2006). The glutamate/GABA-glutamine cycle: aspects of transport, neurotransmitter homeostasis and ammonia transfer. J Neurochem 98:641–653 [DOI] [PubMed] [Google Scholar]
- 11.Bélanger M, Allaman I. and Magistretti PJ. (2011). Brain energy metabolism: focus on astrocyte-neuron metabolic cooperation. Cell Metab 14:724–738 [DOI] [PubMed] [Google Scholar]
- 12.Dienel GA. (2012). Brain lactate metabolism: the discoveries and the controversies. J Cereb Blood Flow Metab 32:1107–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hertz L, Peng L. and Dienel GA. (2007). Energy metabolism in astrocytes: high rate of oxidative metabolism and spatiotemporal dependence on glycolysis/glycogenolysis. J Cereb Blood Flow Metab 27:219–249 [DOI] [PubMed] [Google Scholar]
- 14.Madarász E. (2013). Diversity of neural stem/progenitor populations: varieties by age, regional origin and environment. In: Neural Stem Cells—New Perspectives, Bonfanti L, ed. InTech; Rijeka, Croatia [Google Scholar]
- 15.Lange C, Turrero Garcia M, Decimo I, Bifari F, Eelen G, Quaegebeur A, Boon R, Zhao H, Boeckx B, et al. (2016). Relief of hypoxia by angiogenesis promotes neural stem cell differentiation by targeting glycolysis. EMBO J 35:924–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zádori A, Ágoston VA, Demeter K, Hádinger N, Várady L, Kőhídi T, Gőbl A, Nagy Z. and Madarász E. (2011). Survival and differentiation of neuroectodermal cells with stem cell properties at different oxygen levels. Exp Neurol 227:136–148 [DOI] [PubMed] [Google Scholar]
- 17.Schneider L, Giordano S, Zelickson BR, Johnson MS, Benavides GA, Ouyang X, Fineberg N, Darley-Usmar VM. and Zhang J. (2011). Differentiation of SH-SY5Y cells to a neuronal phenotype changes cellular bioenergetics and the response to oxidative stress. Free Radic Biol Med 51:2007–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xun Z, Lee D-Y, Lim J, Canaria CA, Barnebey A, Yanonne SM. and McMurray CT. (2012). Retinoic acid-induced differentiation increases the rate of oxygen consumption and enhances the spare respiratory capacity of mitochondria in SH-SY5Y cells. Mech Ageing Dev 133:176–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang J, Nuebel E, Daley GQ, Koehler CM. and Teitell MA. (2012). Metabolic Regulation in Pluripotent Stem Cells during Reprogramming and Self-Renewal. Cell Stem Cell 11:589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi HW, Kim JH, Chung MK, Hong YJ, Jang HS, Seo BJ, Jung TH, Kim JS, Chung HM, et al. (2015). Mitochondrial and metabolic remodeling during reprogramming and differentiation of the reprogrammed cells. Stem Cells Dev 24:1366–1373 [DOI] [PubMed] [Google Scholar]
- 21.Panchision DM. (2009). The role of oxygen in regulating neural stem cells in development and disease. J Cell Physiol 220:562–568 [DOI] [PubMed] [Google Scholar]
- 22.Varga B, Markó K, Hádinger N, Jelitai M, Demeter K, Tihanyi K, Vas Á. and Madarász E. (2009). Translocator protein (TSPO 18 kDa) is expressed by neural stem and neuronal precursor cells. Neurosci Lett 462:257–262 [DOI] [PubMed] [Google Scholar]
- 23.Veenman L, Shandalov Y. and Gavish M. (2008). VDAC activation by the 18 kDa translocator protein (TSPO), implications for apoptosis. J Bioenerg Biomembr 40:199–205 [DOI] [PubMed] [Google Scholar]
- 24.Hádinger N, Varga BV, Berzsenyi S, Környei Z, Madarász E. and Herberth B. (2009). Astroglia genesis in vitro: distinct effects of retinoic acid in different phases of neural stem cell differentiaion. Int J Dev Neurosci 27:365–375 [DOI] [PubMed] [Google Scholar]
- 25.Jelitai M, Schlett K, Varju P, Eisel U. and Madarász E. (2002). Regulated appearance of NMDA receptor subunits and channel functions duringin vitro neuronal differentiation. J Neurobiol 51:54–65 [DOI] [PubMed] [Google Scholar]
- 26.Jelitai M, Anděrová M, Markó K, Kékesi K, Koncz P, Syková E. and Madarász E. (2004). Role of ß-aminobutyric acid in early neuronal development: studies with an embryonic neuroectodermal stem cell clone. J Neurosci Res 76:801–811 [DOI] [PubMed] [Google Scholar]
- 27.Jelitai M, Anderová M, Chvátal A. and Madarász E. (2007). Electrophysiological characterization of neural stem/progenitor cells during in vitro differentiation: study with an immortalized neuroectodermal cell line. J Neurosci Res 85:1606–1617 [DOI] [PubMed] [Google Scholar]
- 28.Schlett K, Herberth B. and Madarász E. (1997). In Vitro pattern formation during neurogenesis in neuroectodermal progenitor cells immortalized by p53-deficiency. Int J Dev Neurosci 15:795–804 [DOI] [PubMed] [Google Scholar]
- 29.Tarnok K, Pataki A, Kovacs J, Schlett K. and Madarasz E. (2002). Stage-dependent effects of cell-to-cell connections on in vitro induced neurogenesis. Eur J Cell Biol 81:403–412 [DOI] [PubMed] [Google Scholar]
- 30.Schlett K. and Madarász E. (1997). Retinoic acid induced neural differentiation in a neuroectodermal cell line immortalized by p53 deficiency. J Neurosci Res 47:405–415 [PubMed] [Google Scholar]
- 31.Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CAJ, Butel JS. and Bradley A. (1992). Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 356:215–221 [DOI] [PubMed] [Google Scholar]
- 32.Madarasz E, Kiss J. and Bartok I. (1984). Cell production and morphological pattern formation in primary brain cell cultures. I. Pattern formation within the basal layer(s). Brain Res 304:339–349 [DOI] [PubMed] [Google Scholar]
- 33.Kornyei Z, Czirók A, Vicsek T. and Madarász E. (2000). Proliferative and migratory responses of astrocytes to in vitro injury. J Neurosci Res 61:421–429 [DOI] [PubMed] [Google Scholar]
- 34.Mosmann T. (1983). Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods 65:55–63 [DOI] [PubMed] [Google Scholar]
- 35.Core Team R. (2015). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 36.Livak KJ. and Schmittgen TD. (2001). Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 37.Wu M, Neilson A, Swift AL, Moran R, Tamagnine J, Parslow D, Armistead S, Lemire K, Orrell J, et al. (2006). Multiparameter metabolic analysis reveals a close link between attenuated mitochondrial bioenergetic function and enhanced glycolysis dependency in human tumor cells. AJP Cell Physiol 292:C125–C136 [DOI] [PubMed] [Google Scholar]
- 38.Seahorse Bioscience. (2015). Seahorse XFe Brochure: www.seahorsebio.com
- 39.Uchiumi T. and Kang D. (2012). The role of TFAM-associated proteins in mitochondrial RNA metabolism. Biochim Biophys Acta 1820:565–570 [DOI] [PubMed] [Google Scholar]
- 40.Rogov V, Dötsch V, Johansen T. and Kirkin V. (2014). Interactions between Autophagy Receptors and Ubiquitin-like Proteins Form the Molecular Basis for Selective Autophagy. Mol Cell 53:167–178 [DOI] [PubMed] [Google Scholar]
- 41.Duelli R. and Kuschinsky W. (2001). Brain glucose transporters: relationship to local energy demand. Physiology 16:71–76 [DOI] [PubMed] [Google Scholar]
- 42.Jha MK, Jeon S. and Suk K. (2012). Pyruvate Dehydrogenase Kinases in the nervous system: their principal functions in Neuronal-glial metabolic interaction and Neuro-metabolic disorders. Curr Neuropharmacol 10:393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sugden MC. and Holness MJ. (2003). Recent advances in mechanisms regulating glucose oxidation at the level of the pyruvate dehydrogenase complex by PDKs. Am J Physiol Endocrinol Metab 284:E855–E862 [DOI] [PubMed] [Google Scholar]
- 44.Wu P, Blair PV, Sato J, Jaskiewicz J, Popov KM. and Harris RA. (2000). Starvation Increases the Amount of Pyruvate Dehydrogenase Kinase in Several Mammalian Tissues. Arch Biochem Biophys 381:1–7 [DOI] [PubMed] [Google Scholar]
- 45.Levine AJ, Momand J. and Finlay CA. (1991). The p53 tumour suppressor gene. Nature 351:453–456 [DOI] [PubMed] [Google Scholar]
- 46.Berkers CR, Maddocks ODK, Cheung EC, Mor I. and Vousden KH. (2013). Metabolic Regulation by p53 Family Members. Cell Metab 18:617–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Demeter K, Herberth B, Duda E, Domonkos A, Jaffredo T, Herman JP. and Madarász E. (2004). Fate of cloned embryonic neuroectodermal cells implanted into the adult, newborn and embryonic forebrain. Exp Neurol 188:254–267 [DOI] [PubMed] [Google Scholar]
- 48.Janossy V, Toth A, Bodocs L, Imrik P, Madarasz E. and Gyevai A. (1990). Multielectrode culture chamber: a device for long-term recording of bioelectric activities in vitro. Acta Biol Hung 41:309–320 [PubMed] [Google Scholar]
- 49.Bergersen LH. (2007). Is lactate food for neurons? Comparison of monocarboxylate transporter subtypes in brain and muscle. Neuroscience 145:11–19 [DOI] [PubMed] [Google Scholar]
- 50.Grotius J, Dittfeld C, Huether M, Mueller-Klieser W, Baumann M. and Kunz-Schughart LA. (2009). Impact of exogenous lactate on survival and radioresponse of carcinoma cells in vitro. Int J Radiat Biol 85:989–1001 [DOI] [PubMed] [Google Scholar]
- 51.Vander Heiden MG, Cantley LC. and Thompson CB. (2009). Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324:1029–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bouzier-Sore A-K. and Bolaños JP. (2015). Uncertainties in pentose-phosphate pathway flux assessment underestimate its contribution to neuronal glucose consumption: relevance for neurodegeneration and aging. Front Aging Neurosci 7:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rardin MJ, Wiley SE, Naviaux RK, Murphy AN. and Dixon JE. (2009). Monitoring phosphorylation of the pyruvate dehydrogenase complex. Anal Biochem 389:157–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.John Freeman, Veggiotti Pierangelo, Lanzi Giovanni, Tagliabue Anna. and Perucca Emilio. (2006). The Ketogenic diet: from molecular mechanisms to clinical effects. Epilepsy Res 68:145–180 [DOI] [PubMed] [Google Scholar]
- 55.Schönfeld P. and Reiser G. (2013). Why does brain metabolism not favor burning of fatty acids to provide energy?-Reflections on disadvantages of the use of free fatty acids as fuel for brain. J Cereb Blood Flow Metab 33:1493–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.