Abstract
Background:
Oral squamous cell carcinomas have high potential for locoregional invasion and nodal metastasis. Thus, early detection and elucidation of detailed molecular mechanisms of OSCCs are important. Roles of LEM domain containing 1 (LEMD1), a novel cancer–testis antigen, in OSCCs are unclear.
Methods:
We performed immunohistochemical analysis of LEMD1 in 289 OSCC patients and examined functions of LEMD1 in these carcinomas.
Results:
Immunohistochemical analysis showed that 101 patients were positive for LEMD1. LEM domain containing 1 expression levels in OSCCs significantly correlated with tumour progression (T factor and clinical stage), nodal metastasis, and poor prognosis. LEM domain containing 1 expression was an independent predictor of disease-free survival in OSCC patients. In OSCCs, LEMD1 knockdown suppressed cancer cell invasion. Moreover, downregulation of LEMD1 expression inhibited adhesion and transmigration of OSCCs and vascular or lymphatic vascular endothelial cells.
Conclusions:
Our findings suggest that LEMD1 is a novel tumour progressive factor and may be a useful diagnostic and therapeutic target in OSCCs.
Keywords: LEMD1, transendothelial migration, nodal metastasis, prognosis, oral cancer
Worldwide, head and neck cancer, including oral squamous cell carcinoma (OSCC), is the sixth most common malignancy, with 300 400 new cases diagnosed and 145 400 deaths every year (Torre et al, 2015). It is assumed that 8650 deaths due to OSCC will occur each year in the United States (Siegel et al, 2015), and the number of deaths due to OSCC per 100 000 individuals is 3.7 in Japan (Tanaka and Sobue, 2005). Despite advances in cancer diagnosis and therapy, the overall 5-year survival rate of OSCC patients has remained <50% over the past three decades, although over 80% of early-stage OSCCs can be cured by treatment, whereas >70% of advanced-stage cases cannot be cured (Sasahira et al, 2014b). Therefore, clarifying the detailed molecular mechanism of OSCCs is indispensable for their early detection and treatment.
Tumour formation, progression, and metastasis arise from the amplification of multiple oncogenes and inactivation of tumour suppressor genes. The molecular mechanisms of carcinogenesis, tumour invasion, and metastasis of OSCCs have been elucidated because of the recent advances in molecular biology; however, useful molecular biomarkers of OSCCs remain to be established. More detailed research is required to understand the biological and pathological mechanisms of OSCCs using molecular analysis.
LEM domain containing 1 (LEMD1) is cancer–testis antigen (CTA). Its overexpression has been found in prostate cancer and anaplastic large-cell lymphoma (ALCL) (Ghafouri-Fard et al, 2010; Matsuyama et al, 2011). Furthermore, results of gene expression profiling analysis showed that the expression levels of LEMD1 are elevated in colorectal carcinomas compared with those in their corresponding non-cancerous mucosa (Yuki et al, 2004). LEMD1 is a host gene of microRNA-135 (miR-135b), and its expression is promoted through the activation of signal transducer and activator of transcription 3 (Matsuyama et al, 2011). Furthermore, LEMD1 and miR-135b contribute to nucleophosmin-anaplastic lymphoma kinase-driven oncogenicity, IL-17 production, and the paracrine inflammatory response in ALCLs (Matsuyama et al, 2011). Although CTA expression is highly restricted to testicular germ cells and cancer cells (Balafoutas et al, 2013), the role of LEMD1 in OSCCs is unclear. In the present study, we examined the functional roles and expression of LEMD1 in OSCCs.
Materials and Methods
Surgical specimens
Formalin-fixed, paraffin-embedded samples originated from 289 cases with primary OSCC (158 men and 131 women; age range: 40–86 years, mean: 66.3 years) were used. The primary sites of OSCC were 150 tongue (51.9%), 62 gingiva (21.5%), 37 buccal mucosa (12.8%), 24 oral floor (8.3%), and 16 palate (5.5%). The study plan was performed according to the ethical standards presented in the Declaration of Helsinki, and was approved by the Medical Ethics Committee of Nara Medical University, Kashihara, Japan. All specimens were obtained from randomly selected patients attending Nara Medical University Hospital, Kashihara, Japan, without preoperative therapy. Tumour staging was performed according to the Union for International Cancer Control TNM classification system (seventh edition), and the histologic grade of OSCC was classified according to World Health Organisation criteria. Medical records and prognostic follow-up data were obtained from the patient database maintained by the hospital. The follow-up period was 126–1865 days (mean: 1378 days).
Immunohistochemistry
Consecutive 3-μm sections were cut from each block and immunohistochemistry was performed and antigen retrieval by microwaving in citrate buffer at 95 °C for 30 min. After endogenous peroxidase blocking with 3% H2O2-methanol, specimens were incubated in a 10% skim milk solution (Morinaga Milk, Tokyo, Japan) for 20 min to block non-specific antibody reactions. Anti-LEMD1 antibody (Abcam, Cambridge, UK; polyclonal, dilution at 1 : 50) was applied as the primary antibody for 2 h, followed by incubation with the secondary antibody peroxidase-conjugated anti-mouse or rabbit (Medical & Biological Laboratories, Nagoya, Japan; dilution at 1 : 200) for 30 min at room temperature. The specimens were colour-developed with diaminobenzidine solution (Dako, Carpinteria, CA, USA) and specimens were counterstained with Meyer's hematoxylin (Sigma-Aldrich Corporation, St Louis, MO, USA). Immunoreactivity was classified according to Allred's score (AS) (Allred et al, 1998). We divided immunoreactivity into three criteria based on AS according to our prior report (Kurihara et al, 2013): negative, AS of 0; LEMD1 low, AS of 2∼4; LEMD1 high, AS of 5∼8.
Cell culture
The human OSCC lines HSC2 and KON were obtained from the Health Science Research Resources Bank, National Institute of Biomedical Innovation, Osaka, Japan. HSC2 cells have low metastatic potential, whereas KON cells have metastatic ability (Momose et al, 1989; Kurihara et al, 2013). Total RNA from the normal tongue was purchased from Biochain Institute (Newark, CA, USA) and used as a control. Cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Wako Pure Chemical Industries, Osaka, Japan) supplemented with 10% fetal bovine serum (Nichirei Biosciences, Tokyo, Japan) in 5% CO2 in air at 37 °C.
RNA extraction and quantitative reverse-transcription polymerase chain reaction
Total RNA was extracted using a TRIzol reagent (Invitrogen, Carlsbad, CA, USA), and total RNA (1 μg) was synthesised using a ReverTra Ace qPCR RT Kit (Toyobo, Osaka, Japan). Quantitative reverse-transcription polymerase chain reaction was performed on a StepOnePlus Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) using TaqMan Fast Universal PCR Master Mix (Applied Biosystems), and analysed using the relative standard curve quantification method. The PCR conditions used were selected according to the manufacturer's manual and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA was amplified as an internal control. TaqMan Gene Expression Assays of LEMD1 (ID: Hs01077215_m1) and GAPDH (ID: Hs03929097_g1) were purchased from Applied Biosystems. All PCRs were performed in triplicate.
Immunoblotting
Whole-cell lysate was obtained using M-PER mammalian protein extraction reagent (Thermo Fisher Scientific, Rockford, IL, USA) according to the manufacturer's protocol, and 50 μg of the lysate was subjected to immunoblotting in 12.5% SDS–PAGE, followed by electrotransfer to polyvinylidene fluoride membranes (Thermo Fisher Scientific). The membranes were incubated with anti-LEMD1 antibody (Abcam) and then with peroxidase-conjugated IgG (Medical & Biological Laboratories). The immune complex was visualised by ECL western blotting detection system (GE Healthcare, Amersham place, UK). Anti-GAPDH antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA, clone V-18) was used as an internal control.
Transient transfection
Silencer Select RNAi, a short interfering RNA (siRNA) for LEMD1 (ID: s41117), was purchased from Ambion. AllStars Negative Control siRNA (Qiagen, Venlo, Limburg, The Netherlands) was used as a control. Further, LEMD1 cDNA was amplified by PCR and sub-cloned into pcDNA3.1 (Invitrogen). In total, 20 nM of siRNA and LEMD1-pcDNA3.1 and control vector were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's recommendations.
Cell growth and apoptosis assays
Cells were seeded at density of 2000 cells per well in 96-well tissue culture plates, and incubated for 48 h at 37 °C. Cell growth was assessed using a Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan), and apoptotic cells were detected using the APOPercentage Apoptosis Assay (Biocolor, Carrickfergus, County Antrim, UK). Absorbance at 450 nm (for the measurement of cell growth) and 550 nm (for the measurement of apoptosis) was recorded in each well using a Multiskan GO Microplate Spectrophotometer (Thermo Fischer Scientific, Waltham, MA, USA). All experiments were performed in triplicate.
Cell invasion assay
In vitro invasion assay was performed using BD BioCoat Cell Culture Inserts coated with Type IV Collagen (BD Biosciences, Bedford, MA, USA) as described previously (Kurihara et al, 2013). In brief, cells were suspended in 500 μl of DMEM and placed in the insert. After incubation for 48 h at 37 °C, the filters were dyed with a Diff-Quick Staining Kit (Siemens Healthcare Diagnostics, Newark, DE, USA). The stained cells were counted in whole inserts at × 100 magnification. Each experiment was repeated at least three times.
Interaction assays of OSCCs and endothelial cells
Primary human umbilical vein endothelial cells (HUVEC) and primary human dermal lymphatic microvascular endothelial cells (HDLMVEC) were purchased from Cell Applications (San Diego, CA, USA) to examine interaction of OSCCs and endothelial cells and maintained in Endothelial growth media (Cell Applications) and Microvacular endothelial growth media (Cell Applications) under the conditions of 5% CO2 in air at 37 °C, respectively. Reciprocal action of OSCCs and endothelial cells were tested using CytoSelect Tumor-Endothelium Adhesion Assay (Cell Biolabs, San Diego, CA, USA) and CytoSelect Tumor Transendothelial Migration Assay systems (Cell Biolabs) as we described previously (Sasahira et al, 2014a). In brief, added the fluorescent labelled cancer cells suspension in monolayer culture of endothelial cells and lysed in lysis buffer. Adhesion or migration cells were measured by Multiskan GO Microplate Spectrophotometer (Thermo Fischer Scientific) at 480 nm/520 nm. We performed at least three times of experiments.
Statistical analysis
Statistical analysis was performed using the χ2-test, Fisher's exact test, one-factor analysis of variance, the Student's t-test, Welch's t-test, and the Mann–Whitney U-test. Disease-free survival was analysed using the Kaplan–Meier method, and the differences between groups were calculated using the log-rank test. Univariate and multivariate analyses were performed using the Cox proportional hazards model. All statistical analyses were conducted using JMP8 (SAS Institute, Cary, NC, USA).
Results
Expression of LEMD1 in human OSCC specimens
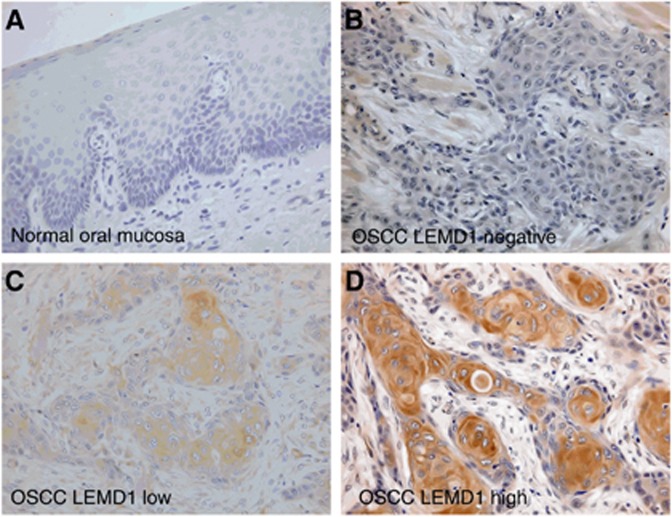
We examined the localisation of LEMD1 in 289 OSCC patients by immunohistochemistry. The immunostaining of LEMD1 was negative or very weak in the normal oral epithelium (Figure 1A). Cytoplasmic staining for LEMD1 was found in 35% (101/289) of OSCCs; these 101 patients included 55 with low LEMD1 and 46 with high LEMD1 expression (Figure 1B–D). Relationships between LEMD1 expression and clinicopathological parameters are summarised in Table 1. Immunoreactivity to LEMD1 was strongly associated with tumour progression (T factor) (P<0.0001). LEMD1 expression was 10.2% (12/118) among Tis/T1 to T2 patients, whereas it was 52.1% (89/171) among T3 and T4 patients. A significant association was also observed between LEMD1 expression and clinical stage (P<0.0001). Among 186 patients, 94 (50.5%) with LEMD1 positivity were at stages III and IV, whereas only 7 of 103 patients (6.8%) expressing LEMD1 were at stages I and II. Among patients with high LEMD1 expression, 72.1% (44/61) and 75.4% (46/61) were at T4 and stage IV, respectively. Furthermore, LEMD1 overexpression was more common in patients with nodal metastasis (46/70, 65.7%) than in those without it (55/219, 25.1% P<0.0001). No significant relationship was observed between LEMD1 expression levels and other clinicopathological factors.
Figure 1.
Expression of LEMD1 in OSCC patients.(A–D) Immunoreactivity to LEMD1 in normal oral mucosa and OSCCs. (A) Weak or no expression of LEMD1 was found in normal oral mucosa. (B) This OSCC patient showed negative LEMD1 expression. (C and D) Cytoplasmic expression of LEMD1 was observed in OSCCs. Patients with low (C) and high LEMD1 expression (D). Original magnification was × 400. HE: hematoxylin and eosin.
Table 1. Association between LEMD1 expression and clinicopathological parameters in OSCCs.
|
LEMD1 | ||||
|---|---|---|---|---|
| Parameters | Negative | LEMD1 low | LEMD1 high | P-value |
|
Gender | ||||
| Male | 104 | 18 | 36 | |
| Female | 84 | 22 | 25 | 0.3667 |
|
Age | ||||
| ⩽65 | 82 | 22 | 26 | |
| >65 | 106 | 18 | 35 | 0.3866 |
|
Site | ||||
| Tongue | 100 | 25 | 25 | |
| Gingiva | 35 | 6 | 21 | |
| Other | 53 | 9 | 15 | 0.0582 |
|
Histological differentiation | ||||
| Well | 80 | 13 | 33 | |
| Moderately | 85 | 23 | 21 | |
| Poorly | 23 | 4 | 7 | 0.2157 |
|
T classification | ||||
| Tis, T1 | 48 | 4 | 0 | |
| T2 | 58 | 3 | 5 | |
| T3 | 44 | 13 | 12 | |
| T4 | 38 | 20 | 44 | <0.0001 |
|
Clinical stage | ||||
| I | 47 | 4 | 0 | |
| II | 49 | 3 | 0 | |
| III | 52 | 11 | 15 | |
| IV | 40 | 22 | 46 | <0.0001 |
|
Nodal metastasis | ||||
| Negative | 164 | 27 | 28 | |
| Positive | 24 | 13 | 33 | <0.0001 |
Relationship between LEMD1 expression and each parameters were calculated by χ2-test or Fisher's exact test. T classification and clinical stage were classified according to the TNM classification.
Relationship between LEMD1 expression and OSCC prognosis
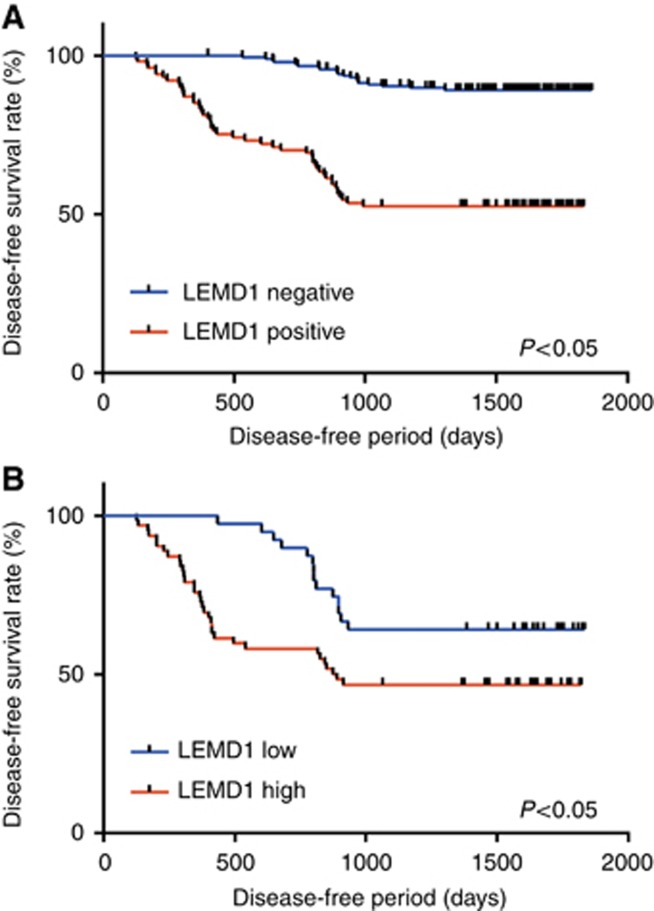
Local and nodal recurrence arose in 68 of the 289 patients, and the disease-free survival was calculated using the Kaplan–Meier method. The disease-free survival among all LEMD1-positive patients was significantly worse than that among LEMD1-negative patients (P<0.0001) (Figure 2A). With regard to LEMD1 expression, patients with high LEMD1 expression also showed a significantly worse prognosis than those with low LEMD1 expression (P=0.0095) (Figure 2B).
Figure 2.
Disease-free survival curves in OSCC patients.Disease-free survival curves of OSCCs were calculated by the Kaplan–Meier method. (A) LEMD1 expression was strongly associated with poor survival (P<0.0001). (B) In LEMD1-positive patients, those with high LEMD1 expression had a significantly worse prognosis than the negative patients (P=0.0002).
We then performed univariate and multivariate Cox proportional hazards analyses. The univariate analysis indicated that clinical stage (P=0.0445), nodal metastasis (P<0.0001), and LEMD1 expression (P<0.0001) were associated with a poor outcome in OSCC patients. In the multivariate analysis, nodal metastasis (P=0.0003), and LEMD1 expression (P<0.0001) were independent predictors of disease-free survival in OSCCs (Table 2).
Table 2. Univariate and multivariate analysis of disease-free survival.
|
Univariate analysis |
Multivariate analysis |
|||||
|---|---|---|---|---|---|---|
| Parameters | HR | 95% CI | P-value | HR | 95% CI | P-value |
|
Gender | ||||||
| Male | 1.00 | |||||
| Female | 1.0937 | 0.6784–1.7635 | 0.7119 | |||
|
Age | ||||||
| ⩽65 | 1.00 | |||||
| >65 | 0.8033 | 0.4982–1.2951 | 0.3669 | |||
|
Site | ||||||
| Tongue | 1.00 | |||||
| Other | 0.7345 | 0.4489–1.1852 | 0.2072 | |||
|
Histology | ||||||
| Well | 1.00 | |||||
| Mod, Poor | 1.0658 | 0.6618–1.7319 | 0.7942 | |||
|
T factor | ||||||
| Tis-3 | 1.00 | |||||
| T4 | 1.5113 | 0.9270–2.4362 | 0.0967 | |||
|
Clinical stage | ||||||
| I–III | 1.00 | 1.00 | ||||
| IV | 1.6414 | 1.0126–2.6430 | 0.0445 | 0.6289 | 0.3679–1.0759 | 0.0902 |
|
Nodal metastasis | ||||||
| Negative | 1.00 | 1.00 | ||||
| Positive | 3.7439 | 2.3122–6.03511 | <0.0001 | 2.5698 | 1.5535–4.2419 | 0.0003 |
|
LEMD1 | ||||||
| Negative | 1.00 | 1.00 | ||||
| Low, high | 6.1300 | 3.6904–10.5703 | <0.0001 | 5.8863 | 3.2915–10.7371 | <0.0001 |
Univariate and multivariate analysis was calculated by means of Cox proportional hazard model. HR and 95% CI mean hazard ratio and 95% confidence intervals, respectively.
In vitro analysis of LEMD1 in OSCCs
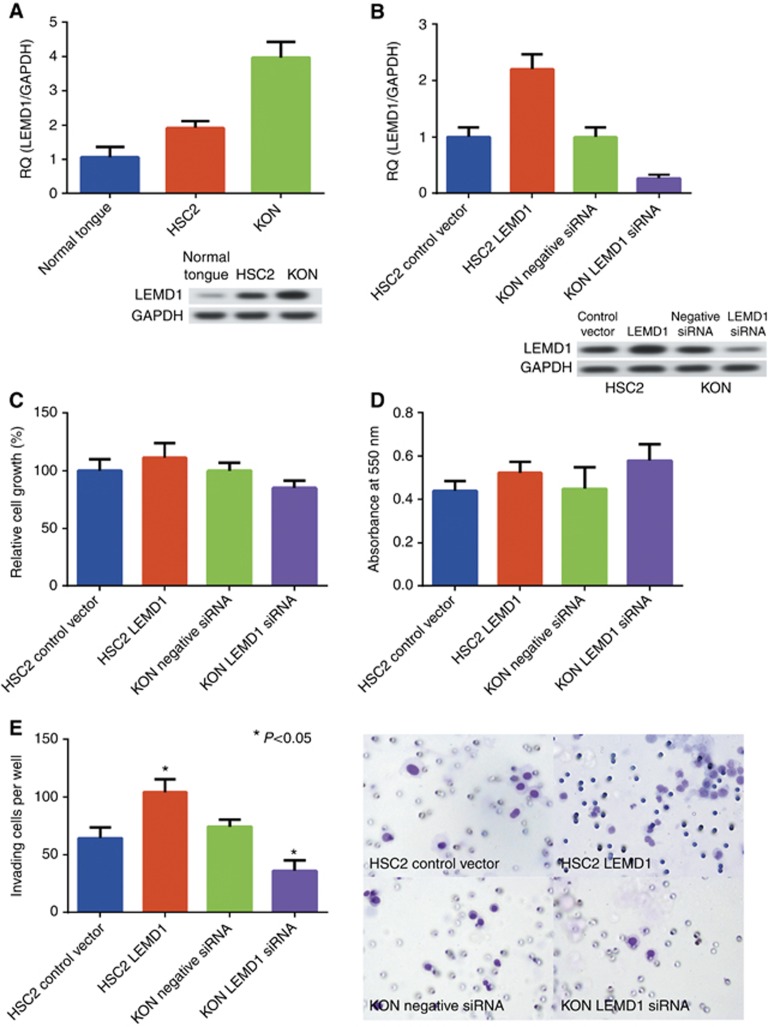
Subsequently, we performed an in vitro analysis using OSCCs. The expression levels of LEMD1 in HSC2 and KON cells were higher than those in normal tongue mucosa (Figure 3A, P<0.01). KON cells with metastatic potential showed higher expression of LEMD1, whereas HSC2 cells that lacked the ability to invade and metastasise showed lower expression (Figure 3A, P<0.05). To examine the effects of LEMD1 in OSCCs, we performed LEMD1 knockdown or upregulation treatment (Figure 3B). In OSCCs treated with LEMD1 knockdown or overexpression, cell growth and apoptotic ability were unchanged compared with those in cells treated with negative siRNA or control vector (Figure 3C and D). However, the number of invading OSCCs on LEMD1 knockdown or overexpression was significantly lower or higher, respectively, than that of cells with the control treatment (Figure 3E).
Figure 3.
Function of LEMD1 in OSCCs.(A) Expression of LEMD1 in OSCCs. (B) Expression of LEMD1 in OSCCs transfected with an LEMD1 short interfering RNA (siRNA) or cDNA. (C and D) Effects of inhibition or upregulation of LEMD1 expression on growth (C) and apoptosis (D) in OSCCs. (E) Changes in invasive ability upon treatment with LEMD1 knockdown or overexpression in OSCCs. Error bar, standard deviation (s.d.). RQ: relative quantification.
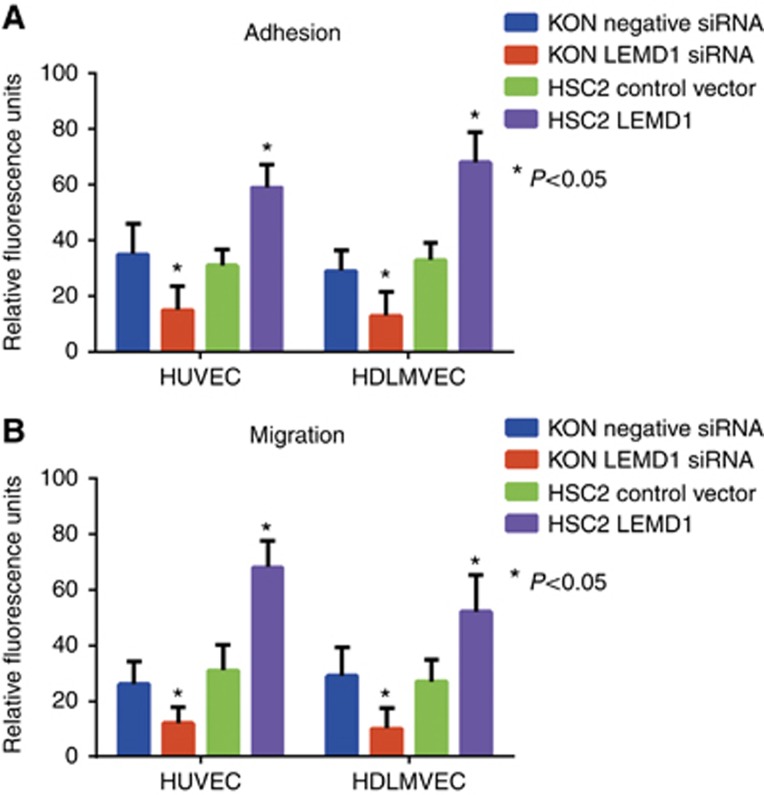
Finally, we investigated the influence of LEMD1 on the interaction between OSCCs and endothelial cells (Figure 4). KON cells with LEMD1 knockdown showed a decreased ability to adhere to primary HUVEC and HDLMVEC (Figure 4A). Moreover, the transmigration ability of KON cells to endothelial cells was inhibited by LEMD1 siRNA treatment (Figure 4B). Opposite results were obtained using HSC2 cells with LEMD1 cDNA transfection (Figure 4B). These results suggest that LEMD1 is a novel tumour-promoting factor in OSCCs.
Figure 4.
Interaction of OSCCs and endothelial cells.(A) A change in the ability to adhere was found in LEMD1 siRNA or cDNA-treated OSCCs and HUVECs or HDLMVECs. (B) Migration ability of OSCCs to HUVECs or HDLMVECs was effected by LEMD1 knockdown or overexpression. Error bar, s.d.
Discussion
Tumour biomarkers are classified into a number of different markers: screening or early diagnostic markers, staging markers, prognostic markers, and predictive and monitoring markers (Biomarkers Definitions Working G, 2001). It has been reported that tumour biomarkers must meet the following criteria: (1) the change can be objectively quantified; (2) the change must be measureable in small samples; (3) the biomarker must be altered in cancer, but not in normal, tissues; and (4) the biomarker must be detected at an early stage of cancer development (Wu et al, 2010). Although a large number of studies have been performed, no practical and beneficial tumour biomarkers of OSCC have yet been established.
In the present study, LEMD1 was identified as a novel metastasis-associated factor for OSCCs. The upregulation of LEMD1 is found in colorectal cancer, prostate cancer, and ALCL (Yuki et al, 2004; Ghafouri-Fard et al, 2010; Matsuyama et al, 2011). However, the biological functions and roles of LEMD1 in tumour cells are still incompletely understood. In this research, we determined that LEMD1 accelerates the invasion of OSCCs. Moreover, LEMD1 promoted the adhesion and transmigration of OSCCs and vascular or lymphatic vascular endothelial cells. In an expression analysis using immunohistochemistry, the expression levels of LEMD1 were closely associated with tumour progression (local progression and clinical stage), nodal metastasis, and worse prognosis in OSCCs. In univariate and multivariate analyses, LEMD1 expression was an independent poor prognostic factor in OSCCs. LEMD1 may thus be a useful target for the diagnosis and treatment of OSCCs. More detailed research is needed to explore the possibility of using LEMD1 as a tumour biomarker in OSCCs.
LEMD1 belongs to the CTA family (Yuki et al, 2004; Matsuyama et al, 2011; Gjerstorff et al, 2015). In normal cells, the expression of CTAs is highly restricted to testicular germ cells, but these antigens are often expressed in various types of cancer (Balafoutas et al, 2013). One of the representative CTAs is melanoma-associated antigen-A, the expression of which is observed in various malignancies including OSCCs (Ries et al, 2008; Schultz-Thater et al, 2011; Lian et al, 2012). Recently, it has been suggested that oncogenic CTAs in malignancies are useful targets of immunotherapy through the activation of cytotoxic T lymphocytes (Gjerstorff et al, 2015). The normalisation of LEMD1 may be useful for the activation of the host immune function, but further in vivo and in vitro studies are required to clarify the possibility of LEMD1-targeted immunotherapy.
The following multiple steps are required for cancer cell invasion and metastasis: disruption of the basement membrane, decreased attachment and segregation of cancer cells, acquisition of cancer cell proliferation and stromal intrusion, vascular infiltration, intravascular migration, transmigration out of the vessels, and cancer cell growth in the metastatic organ (Chen et al, 2013; Sasahira et al, 2014a, 2014b). The initial arrest and attachment of tumour cells to the vascular endothelium are pivotal steps in the tumour metastatic cascade. Tumour cell adhesion to the endothelium and transendothelial migration are impacted by endothelial activation and rely on the expression of specific cell surface molecules such as E-selectin, vascular cell adhesion molecule-1, the Ras homologue (Rho) family, and Rho-associated protein kinase signaling (Tremblay et al, 2006; Ferjancic et al, 2013; Brown et al, 2014). In addition, several growth factors, such as vascular endothelial growth factor and platelet-derived growth factor, can trigger tumour angiogenesis and lymphangiogenesis (Holopainen et al, 2011). We previously revealed that the melanoma inhibitory activity (MIA) gene family members including MIA, MIA2, and transport and Golgi organisation protein 1 promote neoplastic angiogenesis and lymphangiogenesis in OSCCs (Sasahira et al, 2008, 2010; Kurihara et al, 2013; Sasahira et al, 2014a). Nevertheless, the role of transendothelial migration was still unclear in OSCCs. Our results suggest that LEMD1 is a novel tumour-promoting factor that acts by inducing transendothelial migration. However, no studies have identified the detailed angiogenic and/or lymphangiogenic role of LEMD1 in cancer; thus, further studies are required to clarify the underlying mechanism.
In conclusion, the present study demonstrates that LEMD1 is associated with tumour progression, nodal metastasis, and poor prognosis in OSCCs. In addition, LEMD1 facilitates cancer cell invasion and promotes the interaction of OSCCs and endothelial cells. However, there is a need to clarify the mechanism by which LEMD1 acts as an oncogene in OSCCs in detail. Although adequate in vivo and in vitro and large-scale clinical studies are needed, our results indicate that LEMD1 is a relevant target for cancer treatment and diagnosis.
Acknowledgments
This work was supported in part by Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science, Japan.
The authors declare no conflict of interest.
Footnotes
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
References
- Allred DC, Harvey JM, Berardo M, Clark GM (1998) Prognostic and predictive factors in breast cancer by immunohistochemical analysis. Mod Pathol 11(2): 155–168. [PubMed] [Google Scholar]
- Balafoutas D, zur Hausen A, Mayer S, Hirschfeld M, Jaeger M, Denschlag D, Gitsch G, Jungbluth A, Stickeler E (2013) Cancer testis antigens and NY-BR-1 expression in primary breast cancer: prognostic and therapeutic implications. BMC Cancer 13: 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biomarkers Definitions Working G (2001) Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin Pharmacol Ther 69(3): 89–95. [DOI] [PubMed] [Google Scholar]
- Brown M, Roulson JA, Hart CA, Tawadros T, Clarke NW (2014) Arachidonic acid induction of Rho-mediated transendothelial migration in prostate cancer. Br J Cancer 110(8): 2099–2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SF, Lin YS, Jao SW, Chang YC, Liu CL, Lin YJ, Nieh S (2013) Pulmonary Adenocarcinoma in malignant pleural effusion enriches cancer stem cell properties during metastatic cascade. PLoS One 8(5): e54659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferjancic S, Gil-Bernabe AM, Hill SA, Allen PD, Richardson P, Sparey T, Savory E, McGuffog J, Muschel RJ (2013) VCAM-1 and VAP-1 recruit myeloid cells that promote pulmonary metastasis in mice. Blood 121(16): 3289–3297. [DOI] [PubMed] [Google Scholar]
- Ghafouri-Fard S, Ousati Ashtiani Z, Sabah Golian B, Hasheminasab SM, Modarressi MH (2010) Expression of two testis-specific genes, SPATA19 and LEMD1, in prostate cancer. Arch Med Res 41(3): 195–200. [DOI] [PubMed] [Google Scholar]
- Gjerstorff MF, Andersen MH, Ditzel HJ (2015) Oncogenic cancer/testis antigens: prime candidates for immunotherapy. Oncotarget 6(18): 15772–15787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holopainen T, Bry M, Alitalo K, Saaristo A (2011) Perspectives on lymphangiogenesis and angiogenesis in cancer. J Surg Oncol 103(6): 484–488. [DOI] [PubMed] [Google Scholar]
- Kurihara M, Kirita T, Sasahira T, Ohmori H, Matsushima S, Yamamoto K, Bosserhoff AK, Kuniyasu H (2013) Protumoral roles of melanoma inhibitory activity 2 in oral squamous cell carcinoma. Br J Cancer 108(7): 1460–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian Y, Sang M, Ding C, Zhou X, Fan X, Xu Y, Lu W, Shan B (2012) Expressions of MAGE-A10 and MAGE-A11 in breast cancers and their prognostic significance: a retrospective clinical study. J Cancer Res Clin Oncol 138(3): 519–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama H, Suzuki HI, Nishimori H, Noguchi M, Yao T, Komatsu N, Mano H, Sugimoto K, Miyazono K (2011) miR-135b mediates NPM-ALK-driven oncogenicity and renders IL-17-producing immunophenotype to anaplastic large cell lymphoma. Blood 118(26): 6881–6892. [DOI] [PubMed] [Google Scholar]
- Momose F, Araida T, Negishi A, Ichijo H, Shioda S, Sasaki S (1989) Variant sublines with different metastatic potentials selected in nude mice from human oral squamous cell carcinomas. J Oral Pathol Med 18(7): 391–395. [DOI] [PubMed] [Google Scholar]
- Ries J, Vairaktaris E, Mollaoglu N, Wiltfang J, Neukam FW, Nkenke E (2008) Expression of melanoma-associated antigens in oral squamous cell carcinoma. J Oral Pathol Med 37(2): 88–93. [DOI] [PubMed] [Google Scholar]
- Sasahira T, Kirita T, Kurihara M, Yamamoto K, Bhawal UK, Bosserhoff AK, Kuniyasu H (2010) MIA-dependent angiogenesis and lymphangiogenesis are closely associated with progression, nodal metastasis and poor prognosis in tongue squamous cell carcinoma. Eur J Cancer 46(12): 2285–2294. [DOI] [PubMed] [Google Scholar]
- Sasahira T, Kirita T, Oue N, Bhawal UK, Yamamoto K, Fujii K, Ohmori H, Luo Y, Yasui W, Bosserhoff AK, Kuniyasu H (2008) High mobility group box-1-inducible melanoma inhibitory activity is associated with nodal metastasis and lymphangiogenesis in oral squamous cell carcinoma. Cancer Sci 99(9): 1806–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasahira T, Kirita T, Yamamoto K, Ueda N, Kurihara M, Matsushima S, Bhawal UK, Bosserhoff AK, Kuniyasu H (2014. a) Transport and Golgi organisation protein 1 is a novel tumour progressive factor in oral squamous cell carcinoma. Eur J Cancer 50(12): 2142–2151. [DOI] [PubMed] [Google Scholar]
- Sasahira T, Kurihara M, Yamamoto K, Ueda N, Nakashima C, Matsushima S, Bhawal UK, Kirita T, Kuniyasu H (2014. b) HuD promotes progression of oral squamous cell carcinoma. Pathobiology 81(4): 206–214. [DOI] [PubMed] [Google Scholar]
- Schultz-Thater E, Piscuoglio S, Iezzi G, Le Magnen C, Zajac P, Carafa V, Terracciano L, Tornillo L, Spagnoli GC (2011) MAGE-A10 is a nuclear protein frequently expressed in high percentages of tumor cells in lung, skin and urothelial malignancies. Int J Cancer 129(5): 1137–1148. [DOI] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A (2015) Cancer statistics, 2015. CA Cancer J Clin 65(1): 5–29. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Sobue T (2005) Comparison of oral and pharyngeal cancer mortality in five countries: France, Italy, Japan, UK and USA from the WHO Mortality Database (1960-2000). Jpn J Clin Oncol 35(8): 488–491. [DOI] [PubMed] [Google Scholar]
- Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A (2015) Global cancer statistics, 2012. CA Cancer J Clin 65(2): 87–108. [DOI] [PubMed] [Google Scholar]
- Tremblay PL, Auger FA, Huot J (2006) Regulation of transendothelial migration of colon cancer cells by E-selectin-mediated activation of p38 and ERK MAP kinases. Oncogene 25(50): 6563–6573. [DOI] [PubMed] [Google Scholar]
- Wu JY, Yi C, Chung HR, Wang DJ, Chang WC, Lee SY, Lin CT, Yang YC, Yang WC (2010) Potential biomarkers in saliva for oral squamous cell carcinoma. Oral Oncol 46(4): 226–231. [DOI] [PubMed] [Google Scholar]
- Yuki D, Lin YM, Fujii Y, Nakamura Y, Furukawa Y (2004) Isolation of LEM domain-containing 1, a novel testis-specific gene expressed in colorectal cancers. Oncol Rep 12(2): 275–280. [PubMed] [Google Scholar]