Abstract
Background:
Pancreatic ductal adenocarcinoma (PDAC) is frequently diagnosed at an advanced stage, leading to a poor prognosis. Therefore, interest in the development of non-invasive biomarkers for prognostic prediction has grown rapidly. Here, we assessed the clinical implications of v-Ki-ras2 kirsten rat sarcoma viral oncogene homolog (KRAS)-mutated circulating tumour DNA (ctDNA) as a useful surrogate biomarker in patients with resectable PDAC.
Methods:
We used droplet digital polymerase chain reaction to detect rare mutant tumour-derived KRAS genes in plasma cell-free DNA (cfDNA) as ctDNA. Samples were collected from 105 patients who underwent pancreatoduodenectomy for PDAC at a single institution. Overall survival (OS) was analysed according to the presence of ctDNA.
Results:
Among the 105 cases, ctDNA was detected in 33 (31%) plasma samples. The median OS durations were 13.6 months for patients with ctDNA (ctDNA+) and 27.6 months for patients without ctDNA. Patients who were ctDNA+ had a significantly poorer prognosis with respect to OS (P<0.0001).
Conclusions:
Our findings suggested that the presence of ctDNA in plasma samples could be an important and powerful predictor of poor survival in patients with PDAC. Accordingly, ctDNA detection might be a promising approach with respect to PDAC treatment.
Keywords: pancreatic cancer, cfDNA, ctDNA, KRAS, ddPCR, biomarker, prognosis, PDAC
Pancreatic ductal adenocarcinoma (PDAC) is a fatal disease that is often diagnosed at an advanced stage, leading to a poor prognosis (Poruk et al, 2013). This is partly attributed to the lack of suitable techniques for early detection and diagnosis. Hence, interest in the development of non-invasive biomarkers for early PDAC detection and prognostic prediction has grown rapidly.
Circulating cell-free DNA analysis, which is based on the concept of ‘liquid biopsy', was recently reported as a promising prognostic biomarker in patients with various types of cancer (Gormally et al, 2007; De Mattos-Arruda et al, 2011; Mead et al, 2011; Kim et al, 2014; Kato and Janku, 2015). Cell-free DNA is derived from somatic DNA that has been released into systemic circulation following cellular necrosis and apoptosis (Kamat et al, 2010). Similarly, the presence of tumour-derived cell-free DNA, known as circulating tumour DNA (ctDNA), has also been studied. (Diehl et al, 2008; Hashad et al, 2012; Nygaard et al, 2014). However, the practical and clinical feasibility of ctDNA has not yet been determined because of the lack of suitable techniques for quantification of rare target DNA. Quantitative real-time polymerase chain reaction (qPCR) is typically used for nucleic acid quantification; however, estimation via this method requires the use of external calibrators or normalisation to endogenous controls, which consequently limits the methodological sensitivity (Hindson et al, 2011). The recent introduction of droplet digital PCR (ddPCR), a novel next-generation PCR technique based on nanolitre-sized water-in-oil emulsion droplet technology, allows highly precise nucleic acid quantification without cumbersome processing, resulting in increased sensitivity and reproducibility relative to qPCR (Hindson et al, 2013). Therefore, the high detection capability of ddPCR may permit the elucidation of alternative biomarkers for PDAC.
A recent study confirmed the importance of mutations in various genes, including KRAS, TP53, SMAD4, and CDKN2A, in the pathogenesis of PDAC (Biankin et al, 2012). Among these genes, we assumed that KRAS was likely to be the best-characterised tumour-related gene because of following reasons. First, among all human malignancies, PDAC exhibits the highest frequency (75%–100%) of KRAS mutations (Smit et al, 1988; Grünewald et al, 1989; Tada et al, 1996). Second, in PDAC, the most frequent KRAS point mutations are located in two consecutive nucleotides in codon 12 (Almoguera et al, 1988; Bos, 1989). Third, alterations in this gene appear to occur at an early stage of pancreatic carcinogenesis (Uemura et al, 2003; Rhim et al, 2014). Therefore, KRAS-mutated ctDNA represents an important potential biomarker of PDAC.
Using ddPCR, we developed a high-precision method for ctDNA detection in patients with resectable PDAC, and conducted a survival analysis based on the KRAS status of ctDNA.
Materials and methods
Patients
Matched tumour and blood-derived samples were obtained from 105 patients with PDAC. All patients underwent pancreatoduodenectomy for PDAC at the Department of Surgery, Hiroshima University Hospital, between January 2007 and May 2013. Of 105 patients, 55 (52%) were men with a median age of 69 years (range: 43–88 years). Five (5%) and 100 (95%) patients underwent standard pancreatoduodenectomy and pylorus-preserving pancreatoduodenectomy, respectively. Fifty-two (50%) patients were diagnosed with borderline-resectable PDAC according to the 2014 National Comprehensive Cancer Network (NCCN) guidelines (Tempero et al, 2014). Eighteen (17%) patients harboured para-aortic lymph node metastases and were histopathologically diagnosed with stage IV disease after surgery. Eighty-six (82%) patients received adjuvant gemcitabine-based chemotherapy. At the time of the last follow-up, signs of disease progression were confirmed based on imaging findings. The demographic characteristics are summarised in Supplementary Table S1.
Primary tumour samples
The samples used in this study were acquired and restricted to primary operable PDAC. After receiving ethical approval for the study, individual patients were recruited pre-operatively and provided consent through an approved process. Immediately following surgical extirpation, a special pathologist analysed the specimens macroscopically, and tumour samples were snap-frozen in liquid nitrogen. The remaining resected specimens underwent routine histopathological processing and examination. Only samples for which there was no doubt regarding the histopathological diagnosis of PDAC were included in the study. All samples were frozen at −80 °C, and genomic DNA was extracted from these cryopreserved samples using phenol-chloroform-isoamyl alcohol at a later date. Extracted DNA solutions were adjusted to approximately 100 ng μl−1 for use in ddPCR assays.
Plasma samples
A total of 125 plasma samples were collected; these included samples from 20 healthy volunteers (11 men and 9 women, ages 27–56 years with no evidence of malignancy). Whole blood samples were collected from patients with PDAC at the time of anaesthesia induction immediately before resection. Eight millilitres of whole blood were collected in EDTA-containing tubes and centrifuged at 3000 r.p.m. (1500 g) at room temperature for 10 min. Plasma samples were separated from the peripheral blood cells within 4 h after collection. Subsequently, cell-free DNA was extracted from 1 ml plasma and eluted in 100 μl elution buffer with a QIAamp circulating nucleic acid kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions (Dawson et al, 2013; Sun et al, 2014).
TaqMan assay for specific KRAS amplification
For ddPCR, we used a commercially available Prime PCR for ddPCR KRAS kit (Bio-Rad, Hercules, CA, USA), which contained sequence-specific forward and reverse primers with dual-labelled FAM- and HEX-labelled fluorescent TaqMan probes intended to conjugate the target and reference regions, respectively (Gao et al, 2015).
Early reports identified several point mutations in the KRAS oncogene, including Gly12Asp (G12D), Gly12Val (G12V), and Gly12Arg (G12R). Other types of KRAS mutations (e.g., codons 13 or 61) are rarely detected in PDAC (Chen et al, 2010; Takai et al, 2015). Accordingly, these three most frequent mutations in codon 12 of KRAS were amplified in each sample. Other types of KRAS mutations were not analysed because of the limited amounts of sample plasma. Samples lacking the above-mentioned types of mutant KRAS were designated as wild-type in this study (Kinugasa et al, 2015).
Droplet digital PCR (ddPCR)
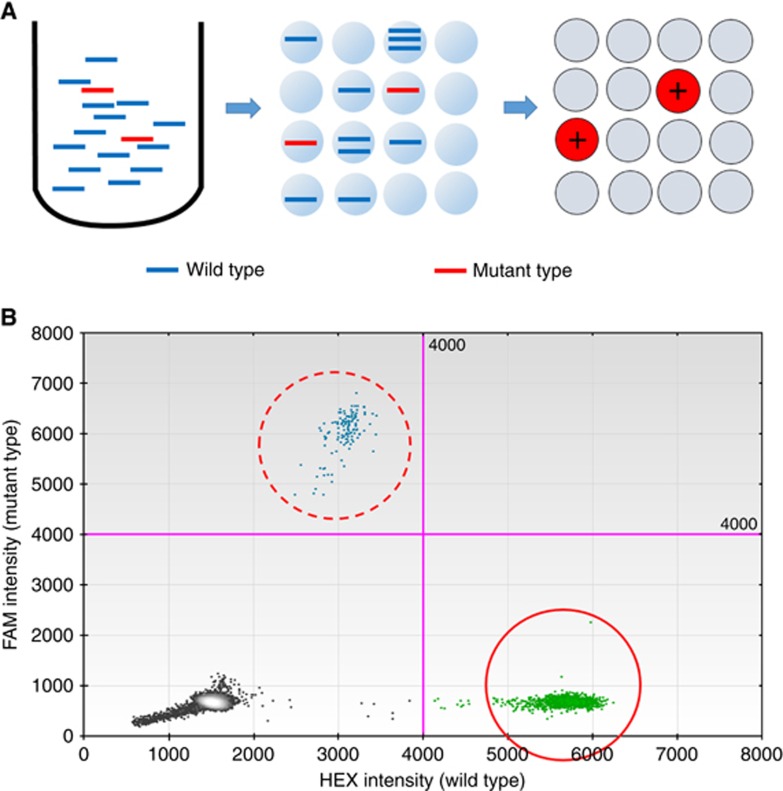
The KRAS mutation status of each sample was analysed using a Bio-Rad QX100 ddPCR system, based on nanolitre-sized water-in-oil emulsion droplet technology. In this method, target DNA molecules are uniformly distributed across thousands of emulsified droplets, after which PCR amplification is performed in each droplet. After amplification, reactions containing one or more target DNA molecules represent the positive end-point, whereas those without target DNA molecules represent the negative end-point (Figure 1A). The number of target DNA molecules (ctDNA concentration) present can be calculated from the fraction of positive end-point reactions using Poisson statistics (Hindson et al, 2011).
Figure 1.
Overview of droplet digital PCR assay.(A) Schematic representation of the droplet digital PCR (ddPCR) assay, which is based on nanolitre-sized water-in-oil emulsion droplet technology. In this assay, target DNA molecules are uniformly distributed across thousands of emulsified droplets, after which PCR amplification is performed in each partitioned droplet. After amplification, reactions containing one or more target DNA molecules represent the positive end-point, whereas those without target DNA molecules represent the negative end-point. The number of target DNA molecules present can be calculated from the fraction of positive end-point reactions using Poisson statistics. (B) Two-dimensional histogram of ddPCR assay for KRAS amplification. FAM (blue) and HEX (green) fluorescence levels were plotted for each droplet. Clusters in the upper and right halves of the plot (dashed circle and solid circle) represent the positive mutant and wild-type KRAS end-point results, respectively.
The reaction mixture comprised 10 μl of 2 × Bio-Rad ddPCR Supermix, 1 μl of each reference and variant 20 × Bio-Rad Prime PCR for ddPCR KRAS, and 10 μl of template DNA in a final volume of 22 μl. The mixtures were loaded into a droplet generator cartridge with 45 μl of droplet generation oil. After placing the cartridge in the Bio-Rad QX100 droplet generator, approximately 15 000–20 000 droplets per well were generated, transferred to a 96-well reaction plate, heat-sealed with a foil seal for 5 s, and subjected to thermocycling in a Veriti 96-well thermal cycler (Thermo Fisher Scientific, Waltham, MA, USA) under the following cycling conditions: 95 °C for 10 min; 45 cycles of denaturation at 95 °C for 30 s and extension at 55 °C for 60 s; and a final 10-min extension step at 95 °C. After cycling, the droplets were analysed immediately using a Bio-Rad QX100 droplet reader. Finally, Bio-Rad QuantaSoft analysis software was used to calculate the number of target DNA molecules from the fraction of positive end-point reactions that were plotted in a two-dimensional image (Figure 1B). Eight replicates of the same reaction were run per plasma sample to increase the reproducibility and counteract the high possibility of false-negative results because of very low levels of target DNA molecules.
Statistical analysis
Patients with PDAC were subjected to survival analyses according to the following 11 variables: age, gender, pathological differentiation, pT factor (defined by the Union for International Cancer Control (UICC)), lymph node metastasis, UICC final stage, residual tumour, pre-operative carcinoembryonic antigen value, carbohydrate antigen 19-9 (CA19-9) value, KRAS status of primary tumour, and presence of ctDNA. Samples were classified into two groups: ctDNA-positive (ctDNA+) and ctDNA-negative (ctDNA−). Statistical analyses were performed using the Mann–Whitney U test, chi-squared test, Fisher's exact test, or Spearman correlation coefficient where appropriate. Overall survival (OS) was estimated using the Kaplan–Meier method, and differences were evaluated using the log-rank test. Variables with P values less than 0.05 according to log-rank tests were retained in the multivariate Cox proportional hazards regression model. All statistical analyses were performed using JMP software, version 11.
Ethical approval to conduct human research
Approval for this study, including approval for the retrospective analysis of collected samples, was granted by the applicable Institutional Review Boards at Hiroshima University Hospital. All patients provided written informed consent.
Results
Limit of detection and threshold baseline for a positive result
To evaluate the high capability of ddPCR, we performed an initial study using serial dilutions of a positive control and wild-type plasma samples from healthy controls. We found that our assay could detect a mutation prevalence of 0.01–0.1%, which corresponded to 1 mutant copy per background of 1000–10 000 wild-type copies, and we established the threshold baseline for a positive result for each types of KRAS mutation (Supplementary Figure S1).
Patient characteristics
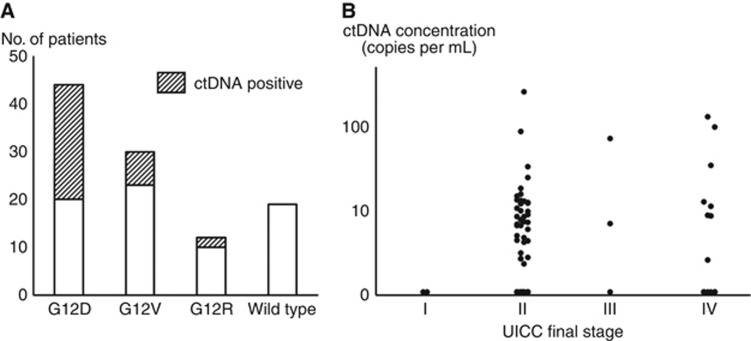
A total of 105 patients with PDAC were evaluated in this study. KRAS mutations were identified in 86 (82%) of the 105 primary tumour specimens. The frequencies of the G12D, G12V, G12R, and wild-type KRAS alleles were 44 (42%), 30 (29%), 12 (11%), and 19 (18%) of 105 samples, respectively. KRAS-mutated ctDNA was detected in 33 (31%) of the 105 matched plasma samples at a mean ctDNA concentration of 10.1 copies per ml (range: 2.4–255 copies per ml). There was a median of 2805 copies per ml of wild-type KRAS fragments (range: 1212–34 320 copies per ml) in patients with PDAC, and the percentage of mutant KRAS fragments in the ctDNA+ samples ranged from 0.05% to 9.5%. The frequencies of the G12D, G12V, and G12R KRAS subtypes were 24 (73%), 7 (21%), and 2 (6%) out of 33 ctDNA-positive samples, respectively (Figure 2A).
Figure 2.
The results of the ctDNA detection.(A) Frequency of KRAS mutations in all primary tumour specimens and plasma samples. (B) Scattergram of ctDNA concentrations in all patients subdivided according to the Union for International Cancer Control (UICC) classification.
The KRAS status concordance of the tumour specimens and matched plasma samples was 100% (33/33). Two or more KRAS mutations did not co-exist in the same sample. Comparisons of the clinicopathological features between the ctDNA+ and ctDNA− groups are summarised in Supplementary Table S2. No significant differences in the demographic characteristics and tumour burden were associated with ctDNA+ results, except for adjuvant chemotherapy. No distinct increase in ctDNA concentration was observed as the disease progressed (Figure 2B).
Survival analysis
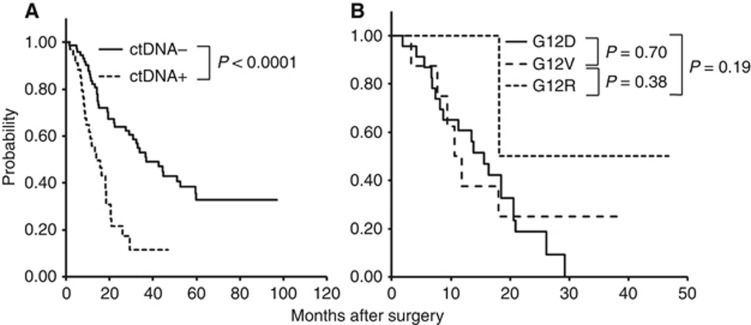
The overall follow-up period ranged from 14 to 96 months, with an average of 54 months. There was no significant difference in OS (P=0.18) between patients with wild-type KRAS tumours (n=19) and those with mutant KRAS (n=86; Supplementary Figure S2a). Similarly, no differences in median OS were observed according to KRAS mutation subtypes (Supplementary Figure S2b). In contrast, the presence of ctDNA in plasma samples was significantly associated with a poor prognosis in both disease-free survival and OS analyses. The median disease-free survival times were 6.1 months in patients who were ctDNA+ vs 16.1 months in patients who were ctDNA−, and the median OS times were 13.6 months in patients who were ctDNA+ vs 27.6 months in patients who were ctDNA− (P<0.001 and P<0.0001, respectively). However, there were no differences in OS according to KRAS mutation subtype in ctDNA. As shown in Figure 3A and B, we performed Kaplan–Meier analyses of OS according to patient stratification based on their classification into the ctDNA+ or ctDNA− group followed by KRAS mutation subtypes of ctDNA.
Figure 3.
Overall survival curves according to the (A) presence of ctDNA and (B) KRAS mutation subtypes of ctDNA.Overall survival did not differ significantly according to the KRAS mutation subtypes of ctDNA. In contrast, significant differences in OS were observed according the categorisation of patients into ctDNA+ and ctDNA− groups.
In addition, univariate OS analysis of 11 independent demographic and clinicopathological variables, including the presence of ctDNA was conducted. In these univariate OS analyses, four variables (lymph node metastasis, UICC final stage, CA19-9 value, and presence of ctDNA) were identified as prognostic factors significantly associated with OS. These four variables were retained in a multivariate Cox proportional hazards regression model, through which lymph node metastasis (hazard ratio=2.2, 95% confidence interval: 1.1–4.2, P=0.023), UICC final stage (hazard ratio=2.0, 95% confidence interval: 1.1–3.5, P=0.016), and the presence of ctDNA (hazard ratio=3.2, 95% confidence interval: 1.8–5.4, P<0.001) were identified as independent factors associated with poor prognosis. However, a high level of CA19-9 (hazard ratio=1.2, 95% confidence interval: 0.7–2.0, P=0.57) did not affect long-term survival (Table 1).
Table 1. Univariate and multivariate analyses of OS among patients with PDAC.
|
Survival |
Univariate analysis |
Multivariate analysis |
||||
|---|---|---|---|---|---|---|
| Prognostic factors | No. of patients | Median (months) | 2-year (%) | P-value | HR (95% CI) | P-value |
|
Age | ||||||
| <Median (69) | 52 | 20.6 | 49 | |||
| ⩾Median | 53 | 18.9 | 51 | 0.62 | ||
|
Gender | ||||||
| Male | 55 | 19.4 | 43 | |||
| Female | 50 | 20 | 58 | 0.09 | ||
|
Pathological differentiation | ||||||
| Well/moderate | 86 | 20.1 | 52 | |||
| Poorly | 19 | 14.3 | 40 | 0.45 | ||
|
UICC pT factor | ||||||
| T1/T2 | 3 | 46.8 | 100 | |||
| T3/T4 | 102 | 19.4 | 49 | 0.08 | ||
|
Lymph node metastasis | ||||||
| No | 29 | 38.3 | 40 | |||
| Yes | 76 | 16.6 | 78 | <0.001 | 2.2 (1.1–4.2) | 0.023 |
|
UICC final stage | ||||||
| IA/IB/IIA/IIB | 84 | 22.6 | 60 | |||
| III/IV | 21 | 14.1 | 14.3 | <0.001 | 2.0 (1.1–3.5) | 0.016 |
|
Residual tumour | ||||||
| R0 | 46 | 24.5 | 57 | |||
| R1 | 59 | 12.5 | 37 | 0.07 | ||
|
KRAS status of primary tumour | ||||||
| Wild-type | 19 | 31 | 72 | |||
| Mutant | 86 | 18.4 | 46 | 0.18 | ||
|
Pre-operative CEA value | ||||||
| <5.0 ng ml−1 | 74 | 21.5 | 47 | |||
| ⩾5.0 ng ml−1 | 31 | 14.5 | 32 | 0.14 | ||
|
Pre-operative CA19-9 value | ||||||
| <90 U ml−1 | 44 | 25 | 57 | |||
| ⩾90 U ml−1 | 61 | 19.4 | 41 | 0.023 | 1.2 (0.7–2.0) | 0.57 |
|
Presence of ctDNA | ||||||
| Negative | 72 | 27.6 | 53 | |||
| Positive | 33 | 13.6 | 21 | <0.0001 | 3.2 (1.8–5.4) | <0.001 |
Abbreviations: CA19-9=carbohydrate antigen 19-9; CEA=carcinoembryonic antigen; CI=confidence interval; ctDNA=circulating tumour DNA; HR=hazard ratio; KRAS=v-Ki-ras2 kirsten rat sarcoma viral oncogene homolog; OS=overall survival; PDAC=pancreatic ductal adenocarcinoma; UICC=Union for International Cancer Control.
In addition, subgroup analyses of OS according to resectability as defined by the 2014 NCCN guidelines were performed. Our study population included 53 (50%) patients with resectable cancers and KRAS-mutated ctDNA was detected in 14 (26%) patients of them. The presence of ctDNA had a significant impact on OS among patients with resectable PDAC (P<0.001; Supplementary Figure S3).
Discussion
Pancreatic ductal adenocarcinoma is one of the most fatal diseases and has an extremely poor prognosis, which may be associated with the lack of effective screening modalities. Accordingly, a very precise predictive surrogate marker is urgently needed to clarify the pre-operative treatment strategies for this lethal disease.
In a previous study of PDAC, CA19-9 was the biomarker most commonly used as a diagnostic aid for treatment monitoring and survival prediction (Ferrone et al, 2006). However, elevated CA19-9 levels are also observed in many benign conditions, such as liver disease, cholangitis, and pancreatitis, and only applicable in patients with the Lewis-positive blood group (Ballehaninna and Chamberlain, 2012).
The KRAS status has also been reported as a useful prognostic biomarker in patients with PDAC. Some reports have described significant differences in OS between patients with wild-type KRAS-bearing primary tumours and those with mutant KRAS-bearing tumours (Tada et al, 1991). Relationships between codon 12 KRAS mutation subtypes (e.g., G12D, G12R) and shorter survival durations have also been reported (Kawesha et al, 2000; Ogura et al, 2013). These previous studies suggested the clinical feasibility of the KRAS mutation status of primary tumour specimens as prognostic biomarkers in patients with PDAC. However, other studies have reported conflicting results (Jimeno et al, 2008; Boeck et al, 2013; Kinugasa et al, 2015), and our study observed no significant differences among patients with tumours expressing wild-type and mutant KRAS (Supplementary Figure S2a and b). The clinical implications of these factors remain controversial and more prospective studies are needed.
In contrast, our data demonstrated that circulating KRAS-mutated ctDNA was associated with significantly poorer survival in both univariate and multivariate analyses, and the presence of ctDNA was the only prognostic factor pre-operatively available in this study. In addition, among patients with resectable cancer (defined by the NCCN guidelines), ctDNA+ patients showed significantly poorer prognosis in OS. This finding suggested that tumour-derived ctDNA disseminated into systemic circulation in some cases in which cancer was diagnosed at an earlier stage via pre-operative imaging. Positivity for ctDNA may correlate with micrometastases that cannot be found in diagnostic imaging. Similar considerations have also been described by other researchers. Takai et al (2015) reported that nine patients with resectable PDAC exhibiting detectable levels of ctDNA relapsed because of formation of metastatic tumours within a short period of time, and had a very poor prognosis. Accordingly, patients who are positive for ctDNA should be considered for indications of any pre-operative options, such as neoadjuvant chemotherapy, even if radiographic evidence of disease progression is absent.
No distinct correlation between ctDNA concentrations and stage of disease was observed in this study (Figure 2B). However, it is notable that no patients with greater than 15 copies per ml of ctDNA survived beyond 2 years (n=12, data not shown). In addition, we experienced a unique case of advanced PDAC with multiple organ metastases; this patient, who had a KRAS-mutated ctDNA concentration exceeding 5500 copies per ml, died within 1 month after blood collection (Supplementary Figure S4). These results suggested the potential for a clearer association between a high ctDNA concentration and poorer prognosis in future larger-scale studies. In support of this potential finding, Bettegowda et al (2014) also reported a steady decrease in the survival rate as the ctDNA concentration increased.
The present study included only data from patients who had undergone pancreatic resection and therefore provided important, novel findings. However, this study was limited by its retrospective nature, and the collection of blood samples only at a single pre-operative time point. A comparison of pre-operative and post-operative ctDNA concentrations would be informative, as changes in ctDNA concentration after treatment may provide important information regarding the therapeutic effect, and management strategies for PDAC (Diaz and Bardelli, 2014; Sausen et al, 2015). Therefore, additional prospective studies will be needed, and are currently underway.
In conclusion, this exploratory study revealed the utility of KRAS-mutated ctDNA as a prognostic biomarker in patients with PDAC. Robust technologies, such as ddPCR, could therefore contribute to the establishment of new therapeutic strategies for PDAC.
Acknowledgments
We thank the staff of the Analysis Center of Life Science, Hiroshima University for their valuable cooperation in this study. We would also like to thank Enago for English language review.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
Supplementary Material
References
- Almoguera C, Shibata D, Forrester K, Martin J, Arnheim N, Perucho M (1988) Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell 53: 549–554. [DOI] [PubMed] [Google Scholar]
- Ballehaninna UK, Chamberlain RS (2012) The clinical utility of serum CA 19-9 in the diagnosis, prognosis and management of pancreatic adenocarcinoma: an evidence based appraisal. J Gastrointest Oncol 3: 1051–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM, Antonarakis ES, Azad NS, Bardelli A, Brem H, Cameron JL, Lee CC, Fecher LA, Gallia GL, Gibbs P, Le D, Giuntoli RL, Goggins M, Hogarty MD, Holdhoff M, Hong SM, Jiao Y, Juhl HH, Kim JJ, Siravegna G, Laheru DA, Lauricella C, Lim M, Lipson EJ, Marie SK, Netto GJ, Oliner KS, Olivi A, Olsson L, Riggins GJ, Sartore-Bianchi A, Schmidt K, Shih LM, Oba-Shinjo SM, Siena S, Theodorescu D, Tie J, Harkins TT, Veronese S, Wang TL, Weingart JD, Wolfgang CL, Wood LD, Xing D, Hruban RH, Wu J, Allen PJ, Schmidt CM, Choti MA, Velculescu VE, Kinzler KW, Vogelstein B, Papadopoulos N, Diaz Jr LA (2014) Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 6: 224ra24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biankin AV, Waddell N, Kassahn KS, Gingras MC, Muthuswamy LB, Johns AL, Miller DK, Wilson PJ, Patch AM, Wu J, Chang DK, Cowley MJ, Gardiner BB, Song S, Harliwong I, Idrisoglu S, Nourse C, Nourbakhsh E, Manning S, Wani S, Gongora M, Pajic M, Scarlett CJ, Gill AJ, Pinho AV, Rooman I, Anderson M, Holmes O, Leonard C, Taylor D, Wood S, Xu Q, Nones K, Fink JL, Christ A, Bruxner T, Cloonan N, Kolle G, Newell F, Pinese M, Mead RS, Humphris JL, Kaplan W, Jones MD, Colvin EK, Nagrial AM, Humphrey ES, Chou A, Chin VT, Chantrill LA, Mawson A, Samra JS, Kench JG, Lovell JA, Daly RJ, Merrett ND, Toon C, Epari K, Nguyen NQ, Barbour A, Zeps N. Australian Pancreatic Cancer Genome Initiative, Kakkar N, Zhao F, Wu YQ, Wang M, Muzny DM, Fisher WE, Brunicardi FC, Hodges SE, Reid JG, Drummond J, Chang K, Han Y, Lewis LR, Dinh H, Buhay CJ, Beck T, Timms L, Sam M, Begley K, Brown A, Pai D, Panchal A, Buchner N, De Borja R, Denroche RE, Yung CK, Serra S, Onetto N, Mukhopadhyay D, Tsao MS, Shaw PA, Petersen GM, Gallinger S, Hruban RH, Maitra A, Iacobuzio-Donahue CA, Schulick RD, Wolfgang CL, Morgan RA, Lawlor RT, Capelli P, Corbo V, Scardoni M, Tortora G, Tempero MA, Mann KM, Jenkins NA, Perez-Mancera PA, Adams DJ, Largaespada DA, Wessels LF, Rust AG, Stein LD, Tuveson DA, Copeland NG, Musgrove EA, Scarpa A, Eshleman JR, Hudson TJ, Sutherland RL, Wheeler DA, Pearson JV, McPherson JD, Gibbs RA, Grimmond SM (2012) Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 491: 399–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeck S, Jung A, Laubender RP, Neumann J, Egg R, Goritschan C, Ormanns S, Haas M, Modest DP, Kirchner T, Heinemann V (2013) KRAS mutation status is not predictive for objective response to anti-EGFR treatment with erlotinib in patients with advanced pancreatic cancer. J Gastroenterol 48: 544–548. [DOI] [PubMed] [Google Scholar]
- Bos JL (1989) Ras oncogenes in human cancer: a review. Cancer Res 49: 4682–4689. [PubMed] [Google Scholar]
- Chen H, Tu H, Meng ZQ, Chen Z, Wang P, Liu LM (2010) K-ras mutational status predicts poor prognosis in unresectable pancreatic cancer. Eur J Surg Oncol 36: 657–662. [DOI] [PubMed] [Google Scholar]
- Dawson SJ, Tsui DW, Murtaza M, Biggs H, Rueda OM, Chin SF, Dunning MJ, Gale D, Forshew T, Mahler-Araujo B, Rajan S, Humphray S, Becq J, Halsall D, Wallis M, Bentley D, Caldas C, Rosenfeld N (2013) Analysis of circulating tumour DNA to monitor metastatic breast cancer. N Engl J Med 368: 1199–1209. [DOI] [PubMed] [Google Scholar]
- De Mattos-Arruda L, Olmos D, Tabernero J (2011) Prognostic and predictive roles for circulating biomarkers in gastrointestinal cancer. Future Oncol 7: 1385–1397. [DOI] [PubMed] [Google Scholar]
- Diaz LA Jr, Bardelli A (2014) Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol 32: 579–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, Thornton K, Agrawal N, Sokoll L, Szabo SA, Kinzler KW, Vogelstein B, Diaz Jr LA (2008) Circulating mutant DNA to assess tumor dynamics. Nat Med 14: 985–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrone CR, Finkelstein DM, Thayer SP, Muzikansky A, Fernandez-delCastillo C, Warshaw AL (2006) Perioperative CA19-9 levels can predict stage and survival in patients with resectable pancreatic adenocarcinoma. J Clin Oncol 24: 2897–2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S, Zheng C, Chang G, Liu W, Kou X, Tan K, Tao L, Xu K, Wang H, Cai J, Tian J, Gao S (2015) Unique features of mutations revealed by sequentially reprogrammed induced pluripotent stem cells. Nat Commun 6: 6318. [DOI] [PubMed] [Google Scholar]
- Gormally E, Caboux E, Vineis P, Hainaut P (2007) Circulating free DNA in plasma or serum as biomarker of carcinogenesis: practical aspects and biological significance. Mutat Res 635: 105–117. [DOI] [PubMed] [Google Scholar]
- Grünewald K, Lyons J, Fröhlich A, Feichtinger H, Weger RA, Schwab G, Janssen JW, Bartram CR (1989) High frequency of Ki-ras codon 12 mutations in pancreatic adenocarcinomas. Int J Cancer 43: 1037–1041. [DOI] [PubMed] [Google Scholar]
- Hashad D, Sorour A, Ghazal A, Talaat I (2012) Free circulating tumor DNA as a diagnostic marker for breast cancer. J Clin Lab Anal 26: 467–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, Bright IJ, Lucero MY, Hiddessen AL, Legler TC, Kitano TK, Hodel MR, Petersen JF, Wyatt PW, Steenblock ER, Shah PH, Bousse LJ, Troup CB, Mellen JC, Wittmann DK, Erndt NG, Cauley TH, Koehler RT, So AP, Dube S, Rose KA, Montesclaros L, Wang S, Stumbo DP, Hodges SP, Romine S, Milanovich FP, White HE, Regan JF, Karlin-Neumann GA, Hindson CM, Saxonov S, Colston BW (2011) High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem 83: 8604–8610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindson CM, Chevillet JR, Briggs HA, Gallichotte EN, Ruf IK, Hindson BJ, Vessella RL, Tewari M (2013) Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat Methods 10: 1003–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimeno A, Tan AC, Coffa J, Rajeshkumar NV, Kulesza P, Rubio-Viqueira B, Wheelhouse J, Diosdado B, Messersmith WA, Iacobuzio-Donahue C, Maitra A, Varella-Garcia M, Hirsch FR, Meijer GA, Hidalgo M (2008) Coordinated epidermal growth factor receptor pathway gene overexpression predicts epidermal growth factor receptor inhibitor sensitivity in pancreatic cancer. Cancer Res 68: 2841–2849. [DOI] [PubMed] [Google Scholar]
- Kamat AA, Baldwin M, Urbauer D, Dang D, Han LY, Godwin A, Karlan BY, Simpson JL, Gershenson DM, Coleman RL, Bischoff FZ, Sood AK (2010) Plasma cell-free DNA in ovarian cancer: an independent prognostic biomarker. Cancer 116: 1918–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawesha A, Ghaneh P, Andrén-Sandberg A, Ograed D, Skar R, Dawiskiba S, Evans JD, Campbell F, Lemoine N, Neoptolemos JP (2000) K-ras oncogene subtype mutations are associated with survival but not expression of p53, p16(INK4A), p21(WAF-1), cyclin D1, erbB-2 and erbB-3 in resected pancreatic ductal adenocarcinoma. Int J Cancer 89: 469–474. [DOI] [PubMed] [Google Scholar]
- Kato S, Janku F (2015) Cell-free DNA as a novel marker in cancer therapy. Biomark Med 9: 703–712. [DOI] [PubMed] [Google Scholar]
- Kim K, Shin DG, Park MK, Baik SH, Kim TH, Kim S, Lee S (2014) Circulating cell-free DNA as a promising biomarker in patients with gastric cancer: diagnostic validity and significant reduction of cfDNA after surgical resection. Ann Surg Treat Res 86: 136–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinugasa H, Nouso K, Miyahara K, Morimoto Y, Dohi C, Tsutsumi K, Kato H, Matsubara T, Okada H, Yamamoto K (2015) Detection of K-ras gene mutation by liquid biopsy in patients with pancreatic cancer. Cancer 121: 2271–2280. [DOI] [PubMed] [Google Scholar]
- Mead R, Duku M, Bhandari P, Cree IA (2011) Circulating tumour markers can define patients with normal colons, benign polyps, and cancers. Br J Cancer 105: 239–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nygaard AD, Holdgaard PC, Spindler KL, Pallisgaard N, Jakobsen A (2014) The correlation between cell-free DNA and tumor burden was estimated by PET/CT in patients with advanced NSCLC. Br J Cancer 110: 363–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura T, Yamao K, Hara K, Mizuno N, Hijioka S, Imaoka H, Sawaki A, Niwa Y, Tajika M, Kondo S, Tanaka T, Shimizu Y, Bhatia V, Higuchi K, Hosoda W, Yatabe Y (2013) Prognostic value of K-ras mutation status and subtypes in endoscopic ultrasound-guided fine-needle aspiration specimens from patients with unresectable pancreatic cancer. J Gastroenterol 48: 640–646. [DOI] [PubMed] [Google Scholar]
- Poruk KE, Firpo MA, Adler DG, Mulvihill SJ (2013) Screening for pancreatic cancer: why, how, and who? Ann Surg 257: 17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhim AD, Thege FI, Santana SM, Lannin TB, Saha TN, Tsai S, Maggs LR, Kochman ML, Ginsberg GG, Lieb JG, Chandrasekhara V, Drebin JA, Ahmad N, Yang YX, Kirby BJ, Stanger BZ (2014) Detection of circulating pancreas epithelial cells in patients with pancreatic cystic lesions. Gastroenterology 146: 647–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sausen M, Phallen J, Adleff V, Jones S, Leary RJ, Barrett MT, Anagnostou V, Parpart-Li S, Murphy D, Kay LiQ, Hruban CA, Scharpf R, White JR, O'Dwyer PJ, Allen PJ, Eshleman JR, Thompson CB, Klimstra DS, Linehan DC, Maitra A, Hruban RH, Diaz LA Jr, Von Hoff DD, Johansen JS, Drebin JA, Velculescu VE (2015) Clinical implications of genomic alterations in the tumour and circulation of pancreatic cancer patients. Nat Commun 6: 7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit VT, Boot AJ, Smits AM, Fleuren GJ, Cornelisse CJ, Bos JL (1988) KRAS codon 12 mutations occur very frequently in pancreatic adenocarcinomas. Nucleic Acids Res 16: 7773–7782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B, Tao L, Zheng YL (2014) Simultaneous quantification of alternatively spliced transcripts in a single droplet digital PCR reaction. Biotechniques 56: 319–325. [DOI] [PubMed] [Google Scholar]
- Tada M, Omata M, Ohto M (1991) Clinical application of ras gene mutation for diagnosis of pancreatic adenocarcinoma. Gastroenterology 100: 233–238. [DOI] [PubMed] [Google Scholar]
- Tada M, Ohashi M, Shiratori Y, Okudaira T, Komatsu Y, Kawabe T, Yoshida H, Machinami R, Kishi K, Omata M (1996) Analysis of K-ras gene mutation in hyperplastic duct cells of the pancreas without pancreatic disease. Gastroenterology 110: 227–231. [DOI] [PubMed] [Google Scholar]
- Takai E, Totoki Y, Nakamura H, Morizane C, Nara S, Hama N, Suzuki M, Furukawa E, Kato M, Hayashi H, Kohno T, Ueno H, Shimada K, Okusaka T, Nakagama H, Shibata T, Yachida S (2015) Clinical utility of circulating tumor DNA for molecular assessment in pancreatic cancer. Sci Rep 5: 18425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tempero MA, Malafa MP, Behrman SW, Benson AB 3rd, Casper ES, Chiorean EG, Chung V, Cohen SJ, Czito B, Engebretson A, Feng M, Hawkins WG, Herman J, Hoffman JP, Ko A, Komanduri S, Koong A, Lowy AM, Ma WW, Merchant NB, Mulvihill SJ, Muscarella P 2nd, Nakakura EK, Obando J, Pitman MB, Reddy S, Sasson AR, Thayer SP, Weekes CD, Wolff RA, Wolpin BM, Burns JL, Freedman-Cass DA (2014) Pancreatic adenocarcinoma, version 2.2014: featured updates to the NCCN guidelines. J Natl Compr Canc Netw 12: 1083–1093. [DOI] [PubMed] [Google Scholar]
- Uemura K, Hiyama E, Murakami Y, Kanehiro T, Ohge H, Sueda T, Yokoyama T (2003) Comparative analysis of K-ras point mutation, telomerase activity, and p53 overexpression in pancreatic tumours. Oncol Rep 10: 277–283. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.