Abstract
Herein we present the tRNA core hypothesis, which emphasizes the central role of tRNAs molecules in the origin and evolution of fundamental biological processes. tRNAs gave origin to the first genes (mRNA) and the peptidyl transferase center (rRNA), proto-tRNAs were at the core of a proto-translation system, and the anticodon and operational codes then arose in tRNAs molecules. Metabolic pathways emerged from evolutionary pressures of the decoding systems. The transitions from the RNA world to the ribonucleoprotein world to modern biological systems were driven by three kinds of tRNAs transitions, to wit, tRNAs leading to both mRNA and rRNA.
Keywords: tRNA, origin of life, translation system, ribosome
1. Introduction
The formation of Earth was a violent cosmic event (circa 4.5 billion years ago), and shortly afterwards (circa 3.7 billions of years ago) particular conditions led to the emergence of a special state of matter that we call life. One of the main features of life is its capacity for self-generation, or self-reference, which is the property of biological systems to self-organize and to transmit genetic information with some modifications along generations (Darwinian evolution). At the core of this ability of self-referral, a primeval genetic code emerged for the translation of nucleic acids to proteins. Another vital property is the capacity of replication and chemical conversion of this information. Several experiments that simulate Earth’s primitive conditions have yielded the basic molecular components, which are still essential in the modern forms of life [1,2,3,4]. The discovery of catalytic activity by RNA molecules [5,6] opened a new way of thinking about the origins of biological systems. RNA molecules could simultaneously possess the system’s information and perform activities for their self-replication. This model of the origin of life is called the “RNA World Hypothesis” [7,8,9,10,11,12]. Another hypothesis for the origin of life is based on the abundance of amino acids and the diversity of functions performed by proteins. It is proposed that biological systems began essentially as a protein system which is dubbed the “Protein World” or “Protein First Model” [13,14,15]. A third model for the origin of life is based on the interactions between proteins and nucleic acids (RNA), which are the basis of the functioning of biological systems, and this model is called the “ribonucleoprotein world” [16,17]. Di Giulio proposed an early ribonucleoprotein world, based on the intrinsic relationship between nucleic acids and polypeptides observed in modern coenzymes. In his model, the ribonucleoproteins were RNAs covalently linked to polypeptides, such as peptidyl-tRNAs. The nonribosomal synthesis of peptides was conceived [18]. All these models look for answers to fundamental questions about the origin of life as: How did the first genes emerge? How was the information of these genes decoded? How did the metabolic systems arise?
This article presents a novel hypothesis for the transition from the RNA world to the ribonucleoprotein world, where proto-tRNAs molecules possessing similar folds to those observed in modern tRNAs guided the evolutionary process of the genetic code and the translation, and enabled its fixation.
The Hypothesis
The most plausible scenario of the origin of life is based on RNA molecules that exhibited simple catalytic functions. The tRNA molecules diversified novel structural conformations by the generation of new strands, and they formed new mini-helixes with catalytic function, i.e. ribozymes. With the stabilization of the catalytic reactions, these ribozymes began to participate in the first catalytic cycles. At this stage, the structural information emerged and was involved in the direct replication by complementarity between nucleotides. Amino acids in prebiotic conditions were abundant, but their incorporation to primitive forms of life should have been dependent on the chemical interaction with the ribozymes, which introduced a compositional bias induced by hydropathic correlations between amino acids and RNAs. Hydrophobic (hydrophilic) amino acids interacted with hydrophobic (hydrophilic) anticodons, leading to specific interactions, whereby a coding/decoding system of the biological information emerged [16]. At this point, the transition from the RNA world to a ribonucleoprotein world was not only possible but also favored. At the center of this transition, we have to explain how the translation system was organized. Nowadays the translation system is a complex system that involves a complex group of proteins that include ribosomal proteins, aminoacyl-tRNA synthetases, elongation and termination factors, recycling factory, among other proteins, and three kinds of RNA molecules, to wit, mRNA, rRNA, and tRNA. The mRNA contains the information codified in the DNA to synthesize a protein, the rRNA forms the ribosome where amino acids are joined by peptide bonds, and the tRNAs mediate the transmission of information codified in mRNA in the ribosome to accomplish the protein synthesis. The origin of protein biosynthesis machinery is still fertile ground in evolutionary biology and represents a critical evolutionary transition. Eigen (1971) suggested that RNA molecules had a limited capacity for replication without loss of information in an environment free of enzymes [19]. This phenomenon is known as the error catastrophe. The size of the nucleotide chain where biological information began to be conserved was present in relatively few nucleotides, around 70 nucleotides or less, like those of modern tRNAs [20]. It is possible that, in the beginning, tRNA-precursors were smaller than the modern tRNA. These initial molecules were similar to the building blocks of a proto-ribosome and functioned as a bonding apparatus in the RNA world before making peptide bonds [21,22,23,24]. These molecules had the ability to participate in the formation of new functional structures by joining two or more RNA molecules. Then, it was possible to increase the information stored in the RNA molecules, which allowed the appearance of the first genes. Thus, many combinations of proto-tRNAs worked as ribozymes, and their polymeric products gave origin to the peptidyl transferase center (PTC), a catalytic center capable of carrying out peptide bonds [21,25,26,27].
Eigen and Winkler-Oswatisch suggested an Ur-gene made of tRNA [28]. Bloch et al., comparing the tRNAs and 16S ribosomal RNA chain, suggested a common origin for these molecules [29,30]. Agmon showed that the modern tRNAs were built by the same building blocks as the modern, fully conserved PTC [23]. Tamura further analyzed the topological organization of the PTC of the large subunit of the ribosome and suggested that this region presents topological similarities with concatenated tRNAs [27]. Root-Bernstein and Root-Bernstein analyzed the origin of PTC and described its origin as a collection of tRNAs that could function as a primitive and functional genome [31]. Di Giulio suggested that the tRNAs molecules have a non-monophyletic origin and that, in the emergence of the biological system, the primitive genome could have been fragmented, and the trans-splicing events thus may have had an important function in the origin of the genes [32].
The hypothesis here proposed places the tRNA at the core of the origin of translation, i.e., tRNA molecules played a central role in the organization of the first codified biological system, established the information storage system, and participated in coding and decoding this information. They were the protagonists of the translation system via chemical interactions with amino acids, which are inherent to the transition between the RNA world to a ribonucleoprotein world.
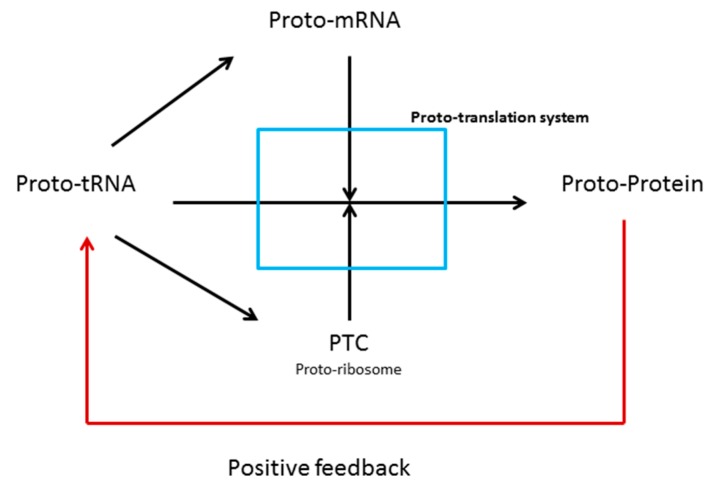
The present hypothesis contends that the tRNA-like molecules or proto-tRNAs (the 2D-cloverleaf tRNA canonical structure), through structural modification and concatenation, gave origin to the early genes, as well as to the PTC. In addition, we suggest that metabolic pathways resulted from evolutionary pressures of the decoding system. Hence, the first synthesized peptides by a proto-translation system could have been selected for binding and stabilization of the RNAs that acted as ribozymes and proto-genes [33]. In this manner, concatenated RNAs, which acted as a primitive genome, increased the amount of stored information, and, due to the increased size of RNAs strands, the new peptides emerged in a nascent ribonucleoprotein world. Thus, via positive feedback between peptides and RNA, the biological system developed robustness that was crucial for the establishment of the metabolic and informational processes, as we observe in a modern biological system. Figure 1 summarizes the functions of the tRNA-like molecules at the transition from the RNA world to the ribonucleoprotein world.
Figure 1.
Schematic model of the origin of the translation system originated from proto-tRNAs.
When proto-tRNAS joined in concatemers, they acquired new structural conformations, similar to the folds of both the proto-mRNA and proto-rRNA, which by interaction, were able to initiate translations into proteins. At this stage, positive feedback between proteins and RNAs was essential for further evolution.
2. Experimental Section
tRNA and the First Genes
The tRNA sequences were obtained from the tRNA database (http://trnadb.bioinf.uni-leipzig.de) and correspond to 361 organisms distributed in the three domains of life. The RNY code was proposed as a primeval genetic code by Eigen and Schuster (1978). This code displays the property of base complementarity with respect to plus and minus strands, and it minimizes errors in the replication and translation processes [34]. Hence, we reconstructed the ancestor sequences for each type of tRNA with RNY anticodons (Supplementary Information 1). The total of 600 sequences for AlatRNA, 313 for AsntRNA, 267 for AsptRNA, 608 for GlytRNA, 326 for IletRNA, 855 for SertRNA, 634 for ThrtRNA, and 538 for ValtRNA were analyzed. Model Tests was performed for each group of tRNAs to choose the best evolutionary model, which pointed to Kimura 2 parameters. To reconstruct the ancestral sequences, a phylogenetic tree was generated using the maximum-likelihood method. In order to achieve statistical significance, bootstrapping with 1000 replicates was carried out. These evaluations were performed with the MEGA5 program [35]. From the ancestor sequences for each tRNAs with anticodons of the RNY type (Ile, Asn, Vau, Asp, Gly, Ser, Thr, and Ala), an initial set of tRNAs that composed the progenotes was proposed [34]. Sequences were constructed from all possible combinations (3 × 3) of those elements that take place without the presence of the same tRNA in two positions in the final sequence. A search for similar proteins from the combined ancestral tRNA sequences in NCBI with the BLASTX algorithm [36] in the database UniProtKB/Swiss-Prot (swissprot) was performed. All sequences that had similarities in the database were analyzed (Supplementary Information 2) and ranked according to their functional category with the support of the phylogenetic classification of proteins tool encoded in complete genomes by COG (Clusters of Orthologous Groups of proteins) [37].
3. Results and Discussion
3.1. tRNAs and the Origin of Genes
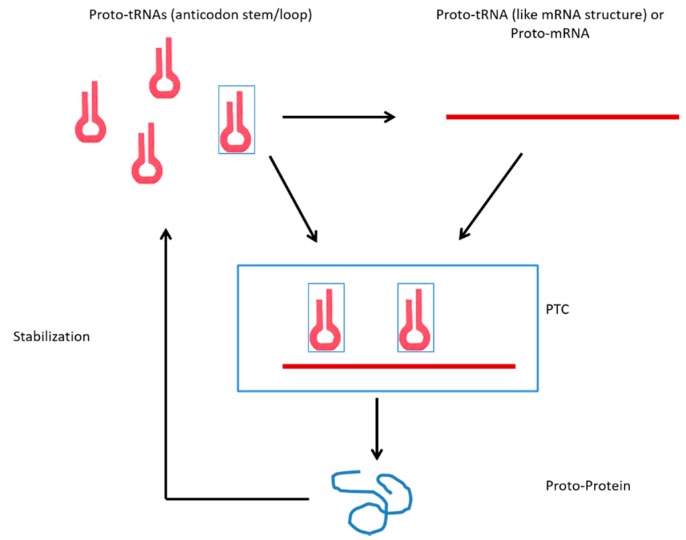
In this hypothesis, the first genes derived from tRNA by structural changes (tRNA-like-mRNA structure) enabled other tRNAs (cloverleaf tRNA canonical structure) to bind this sequence by the loop of the proto-anticodon. Szathmáry proposed that, in the origin of the biological systems, amino acids worked as cofactors binding to ribozymes that had the primordial function of aminoacylation and, from this interaction, the biological relation between amino acids and specific oligonucleotides emerged. Thereby, this interaction was important for the emergence of the genetic code [39]. Da Silva suggested that the interaction between amino acids and nucleic acids, as riboswitches, was important for the stabilization of the RNA structure and relevant for its catalytic activity as a ribozyme [40]. Di Giulio proposed that the primordial mRNA originated by interactions of peptidated-RNAs (aminoacylated), which had the aspect of hairpin-like structures [41]. In the present proposal, amino acids and small peptides worked like riboswitches or cofactors to stabilize the alternative conformations of the proto-tRNAs. Thus, the binding of two or more tRNAs, which showed distinct structural organization (tRNA-like mRNA and cloverleaf tRNA canonical structure), allowed the stability of these molecules to increase (Figure 2).
Figure 2.
Schematic model of the origin of the translation system by the change of the tertiary structure of proto-tRNAs (anticodon stem/loop) and the mechanisms of interactions between the alternative structural states.
The tRNA molecules with the proto-tRNA (anticodon stem/loop) could interact with the amino acids (as cofactor or riboswitches), which were present in the prebiotic environment; hence, the binding between tRNA and amino acids was established. The tRNAs binding to amino acids could interact with other tRNAs in open conformation (tRNA-like mRNA structure). This interaction stabilized the complex cloverleaf tRNA canonical structure-tRNA-like mRNA structure. In addition, this interaction allowed the assembly of the binary complex proto-tRNA/mRNA, to be closer to the tRNAs charged with amino acids, which, after their stabilization, enabled the interaction with the proto-rRNA (PTC), forming the ternary complex proto-tRNA/mRNA/rRNA (PTC) and thus stabilizing the primitive protein synthesis system. Figure 2 portrays the mechanism of interaction between tRNAs that gave rise to the primitive protein synthesis system. It is suggested that the first peptides that were fixed in the biological system had the ability to bind and stabilize tRNAs in different structural conformations, which in turn increased the stability and efficiency of the process, thereby establishing the first positive feedback between peptides and nucleic acids [42,43,44]. With an increased stability of the primitive protein synthesis system, its efficiency increased, and the rate of consumption of amino acids involved in proto-proteins increased. At this stage, the biological system had a new selective pressure because the prebiotic substrates decreased, and the replacement of essential components for maintenance of the system was required (Figure 3).
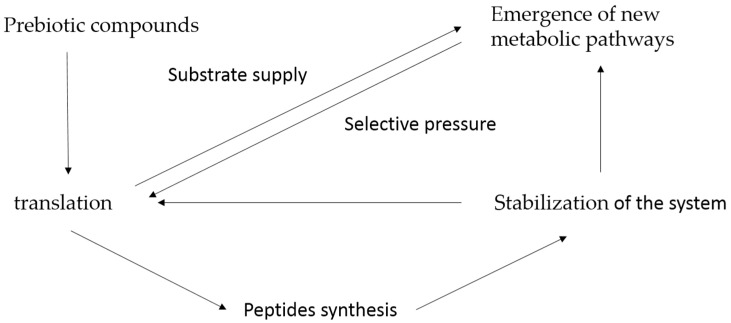
Figure 3.
Sequential stages in the emergence of the translation system and the initial selective pressure that originated the first metabolic pathways.
Thus, the primordial metabolic pathways of amino acid synthesis started its organization, as well as the primitive metabolic pathways of nucleotides catalyzed by enzymes. Keller et al. demonstrated that the main reactions of glycolysis and pentose phosphate pathways were plausible in a prebiotic condition [45]. In modern metabolic pathways, those of the glycolysis and pentose phosphate act as a distribution center for precursors of amino acids and nucleotides biosynthesis pathways. Thus, these primordial routes that emerged in an environment without proteins were gradually replaced by primordial enzyme-catalyzed versions after the emergence of the primordial translation system of peptides, which increased the efficiency of this synthesis process, and amino acids were supplied for the translation system in formation. Thus, the translation system became the attractor, the state where the system channeled the production of amino acids, and therefore was the first selective force to organize the metabolic pathways that we observe in modern biological systems.
Table 1 shows the results obtained by the translation of ancestral tRNAs, indicating which motifs had similarities with current proteins. We observe that most of the proteins that had similarities with proto-genes, constructed with ancestral sequences of tRNAs, are involved in the translation process. This implies that they have the capability of binding to RNA. We also note a similarity with the enzyme RNA-polymerase directed by RNA, which replicates RNA molecules. Another fact that stands out is the similarity of the proto-genes with proteins involved in the metabolism of molecules with three carbon, which are present at the core of the glycolytic pathway, which turns out to be a distribution center of molecules to various pathways in modern cells. Among these pathways, we have amino acid synthesis, nucleotide synthesis, lipid synthesis, and energy generation.
Table 1.
Enzymes and processes that matched between ancestor tRNA and the modern proteins. The original BLAST results can be found in the Supplementary Information 2.
| Protein | Process | Protein | Process |
|---|---|---|---|
| Elongation factor 1–alfa RNA transport | Translation | Putative ribose/galactose/methyl galactose import | Glycolysis/glycogenesis |
| Elongation factor 4–mRNA translation assisting | Translation | Glycerate kinase | Glycolysis/glycogenesis |
| tRNA uridine 5-carboxymethylaminomethyl modification | Translation | Triosephosphate isomerase | Glycolysis/glycogenesis |
| 60S ribosomal protein L3 | Translation | Beta-glucosidase A | Glycolysis/glycogenesis |
| 60S ribosomal protein L7a | Translation | Glucose-6-phosphate-1-dehydrogenase | Glycolysis/glycogenesis |
| 60S ribosomal protein L27a-1 | Translation | Glucose 6 phosphate isomerase | Glycolysis/glycogenesis |
| 60S ribosomal protein L27a-3 | Translation | Phosphoglycerate kinase | Glycolysis/glycogenesis |
| 60S ribosomal protein L27a-2/4 | Translation | Glycerol-3-phosphate dehydrogenase | Glycolysis/glycogenesis |
| 50S ribosomal protein L10 | Translation | Transketolase | Glycolysis/glycogenesis |
| Methionyl–tRNA formyl transferase | Translation | α-galactosidase | Glycolysis/glycogenesis |
| Lysys-tRNA synthetase | Translation | Diaminopimelate epimerase | Amino acids pathways |
| Asparaginyl-tRNA synthetase | Translation | l-asparaginase | Amino acids pathways |
| Glutamyl-tRNA synthetase | Translation | ATP phosphoribosyl transferase | Amino acids pathways |
| Leucyl-tRNA synthetase | Translation | Histidinol phosphate aminotransfarase | Amino acids pathways |
| Valyl-tRNA synthetase | Translation | 4-aminobutyrate aminotransferase | Amino acids pathways |
| Phenylalanine-tRNA synthetase | Translation | Ornithine carboxylase antizyme | Amino acids pathways |
| RNA methyltransferase—tRNA modification | Translation | N-acetyl-γ-glutamyl-phosphate redutase | Amino acids pathways |
| DNA-direct RNA polymerase or RNA-direct RNA polymerase | Transcription | Homoserine kinase | Amino acids pathways |
| Thymidylate kinase | Nucleotides pathways | Aromatic amino acid aminotransferase | Amino acids pathways |
| Cytidine deaminase | Nucleotides pathways | Ornithine carbomyltransferase | Amino acids pathways |
| Uridylate kinase | Nucleotides pathways | Tryptophan synthase α-chain | Amino acids pathways |
| Orotidine 5-phosphate decarboxilase | Nucleotides pathways | Fatty acid synthase | Lipids pathways |
| Dihydroorotate dehydrogenase | Nucleotides pathways | CoA mutase | Lipids pathways |
| Phosphoribosyl formyl glycinamidine cyclo-ligase | Nucleotides pathways | Phosphate acyltransferase | Lipids pathways |
| Phosphoribosyl glycinamidine synthase | Nucleotides pathways | Lycopene cyclase | Lipids pathways |
| 3-β-hydroxisteroid dehydrogenase | Lipids pathways |
Conclusions are consistent with the ones obtained by Delaye et al., who reconstructed the probable metabolic pathways of LUCA via methods based in homology [46]. In this work, a likely proteome for this biological entity is suggested. Thus, the results suggest that the tRNAs may have originated the early genes, which encoded proteins involved in the binding to RNA, and proteins involved in the pathways for the replacement of prebiotic components in peptides synthesis. The results also suggest that the system of encoding/decoding of biological information arose early in the organization of biological systems, where the same tRNAs participated in the information storage. The incorporation of new metabolic pathways may have been selected from their chemical collaboration between the replacement of organic compounds and the process of peptide synthesis. In this context, the modern biological system was initially established as a ribonucleoprotein system, where the nascent proteins could bind to tRNAs and stabilize them. This stabilization enhanced the fidelity of peptides synthesis, which conferred strength to the nascent biological system.
3.2. tRNAs and the Origin of Ribosomes
The emergence of the first genes, as well as the need to decode the information contained in these genes, must have been a major challenge in the origin of biological systems. Bloch et al. suggested, when comparing the sequences of tRNAs and 16S ribosomal RNA, that these molecules had a common origin [29,30]. Tamura, in a comparative structural study between the PTC and tRNAs, suggested that the former arose by the concatenation of the latter [27]. Davidovich et al. analyzed the possibility of small RNAs to acquire a similar structure to the PTC and noted that RNA with structural organization of the type stem-elbow-stem, when joined, had comparable structural organization with the PTC of the ribosome [47]. Root-Bernstein and Root-Bernstein suggested that the ribosome contained information of the tRNAs that functioned as the primordial genome [31]. In the search for evidence that might suggest an origin of the PTC from tRNA-like molecules, ancestral sequences of tRNAs were obtained [38]. From individual alignments of tRNAs with the PTC of Thermus thermophilus, it was possible to construct a concatamer of tRNAs and compare its similarity to the PTC as a whole. The ancestral sequences of tRNAs used for the construction of the concatamer and its comparison with the PTC were LeutRNA-SertRNA-HistRNA-ProtRNA-TyrtRNA-Phe-tRNA-GlntRNA-GlytRNA-LystRNA. The alignment showed an identity of 50.53% between the sequence constructed with ancestral tRNAs and the PTC from T. thermophiles. We have previously published the results of an alignment of tRNA ancestral sequences with the PTC in [38].
These findings, along with the results of other groups [26,30], suggest that the tRNA-like molecules originated the catalytic portion of the ribosome and assisted in the decoding of the information contained in the first genes that in turn could have originated from the tRNA molecules [38]. Thus, our hypothesis provides novel insight on the origin of the ribonucleoprotein world, where the tRNAs originated the peptidyl transferase center and the first genes, important elements in the origin of the translation system.
4. Conclusions
The present hypothesis suggests that tRNA molecules organized the essence of the biological system, giving rise to the first genes and the catalytic center of the ribosome, the PTC, thereby orchestrating the origin of the primitive protein synthesis system. The first synthesized proteins or peptides could bind to the RNA apparatus that produced them and stabilize the other tRNA-like molecules in its specific functional conformations, as well as act as a primitive genome, thus increasing the robustness of the biological system in formation. The data obtained from the analysis of the reconstruction of the ancestral sequences of the tRNAs showed evidence of the cardinal involvement of the tRNAs in the origin of the early genes. These genes encoded proteins which were involved in protein synthesis and the metabolism of molecules with 3 carbons of the glycolytic pathway. The latter is part of the distribution and supply of compounds for various other metabolic pathways. The data also show that tRNA-like molecules may have led to the assembly of the catalytic center of the ribosome. From the data, we suggest that the tRNAs were the key molecules in the organization and evolution of the biological systems, initiating the feedback relationships between proteins and nucleic acids observed today in modern organisms. Thus, we propose a model about the transitions from the RNA world to ribonucleoprotein world to the origin of the modern biological systems, where the system encoding/decoding emerged as a natural pressure for the maintenance of the primitive translation system.
Acknowledgments
We would like to thank Gustavo Caetano-Anollés and Ada Yonath for their comments and critical review. Marco V. Jose was financially supported by PAPIIT-IN224015, UNAM, México.
Supplementary Materials
The following are available online at http://www.mdpi.com/2075-1729/6/2/15/s1.
Author Contributions
Sávio Torres de Farias, Thais Gaudencio do Rego and Marco V. Jose conceived and designed the experiments. Sávio Torres de Farias and Thais Gaudencio do Rego performed the experiments. Sávio Torres de Farias, Thais Gaudencio do Rego and Marco V. Jose analyzed the data and wrote the article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Miller S.L. A production of amino acids under possible primitive earth conditions. Science. 1953;117:528–529. doi: 10.1126/science.117.3046.528. [DOI] [PubMed] [Google Scholar]
- 2.Parker E.T., Cleaves H.J., Dworkin J.P., Glavin D.P., Callahan M., Aubrey A., Lazcano A., Bada J.L. Primordial synthesis of amines and amino acids in a 1958 Miller H2S-rich spark discharge experiment. Proc. Natl. Acad. Sci. USA. 2011;108:5526–5531. doi: 10.1073/pnas.1019191108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parker E.T., Zhou M., Burton A.S., Glavin D.P., Dworkin J.P., Krishnamurthy R., Fernández F.M., Bada J.L. A plausible simultaneous synthesis of amino acids and simple peptides on the primordial Earth. Angew. Chem. Int. Ed. Engl. 2014;53:8132–8136. doi: 10.1002/anie.201403683. [DOI] [PubMed] [Google Scholar]
- 4.Bada J.L. New insights into prebiotic chemistry from Stanley Miller’s spark discharge experiments. Chem. Soc. Rev. 2013;42:2186–2196. doi: 10.1039/c3cs35433d. [DOI] [PubMed] [Google Scholar]
- 5.Kruger K., Grabowski P.J., Zaug A.J., Sands J., Gottschling D.E., Cech T.R. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell. 1982;31:147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- 6.Guerrier-Takada C., Gardiner K., Marsh T., Pace N., Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- 7.Poole A.M., Jeffares D.C., Penny D. The path from the RNA world. J. Mol. Evol. 1998;46:1–17. doi: 10.1007/PL00006275. [DOI] [PubMed] [Google Scholar]
- 8.Jeffares D.C., Poole A.M., Penny D. Relics from the RNA world. J. Mol. Evol. 1998;46:18–36. doi: 10.1007/PL00006280. [DOI] [PubMed] [Google Scholar]
- 9.Dworkin J.P., Lazcano A., Miller S.L. The roads to and from the RNA world. J. Theor. Biol. 2003;222:127–134. doi: 10.1016/S0022-5193(03)00020-1. [DOI] [PubMed] [Google Scholar]
- 10.Lazcano A. The biochemical roots of the RNA world: From zymonucleic acid to ribozymes. Hist. Philos. Life Sci. 2012;34:407–423. [PubMed] [Google Scholar]
- 11.Reyes-Prieto F., Hernández-Morales R., Jácome R., Becerra A., Lazcano A. Coenzymes, viruses and the RNA world. Biochimie. 2012;94:1467–1473. doi: 10.1016/j.biochi.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 12.Neveu M., Kim H.J., Benner S.A. The “strong” RNA world hypothesis: Fifty years old. Astrobiology. 2013;13:391–403. doi: 10.1089/ast.2012.0868. [DOI] [PubMed] [Google Scholar]
- 13.Ikehara K. Possible steps to the emergence of life: the [GADV] protein world hypothesis. Chem. Rec. 2005;5:107–118. doi: 10.1002/tcr.20037. [DOI] [PubMed] [Google Scholar]
- 14.Caetano-Anollés D., Kim K.M., Mittenthal J.E., Caetano-Anollés G. Proteome evolution and the metabolic origins of translation and cellular life. J. Mol. Evol. 2011;72:14–33. doi: 10.1007/s00239-010-9400-9. [DOI] [PubMed] [Google Scholar]
- 15.Caetano-Anollés G., Wang M., Caetano-Anollés D. Structural phylogenomics retrodicts the origin of the genetic code and uncovers the evolutionary impact of protein flexibility. PLoS ONE. 2013;8:15. doi: 10.1371/journal.pone.0072225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farias S.T., Moreira C.H., Guimarães R.C. Structure of the genetic code suggested by the hydropathy correlation between anticodons and amino acid residues. Orig. Life Evol. Biosph. 2007;37:83–103. doi: 10.1007/s11084-006-9008-7. [DOI] [PubMed] [Google Scholar]
- 17.Guimarães R.C. Metabolic basis for the self-referential genetic code. Orig. Life Evol. Biosph. 2011;41:357–371. doi: 10.1007/s11084-010-9226-x. [DOI] [PubMed] [Google Scholar]
- 18.Di Giulio M. On the RNA world: evidence in favor of an early ribonucleopeptide world. J. Mol Evol. 1997;45:571–578. doi: 10.1007/PL00006261. [DOI] [PubMed] [Google Scholar]
- 19.Eigen M. Self-organization of matter and the evolution of biological macromolecules. Naturwissenschaften. 1971;58:465–523. doi: 10.1007/BF00623322. [DOI] [PubMed] [Google Scholar]
- 20.Kreysing M., Keil L., Lanzmich S., Braun D. Heat flux across an open pore enables the continuous replication and selection of oligonucleotides towards increasing length. Nat. Chem. 2015;7:203–208. doi: 10.1038/nchem.2155. [DOI] [PubMed] [Google Scholar]
- 21.Agmon I., Bashan A., Zarivach R., Yonath A. Symmetry at the active site of the ribosome: Structural and functional implications. Biol. Chem. 2005;386:833–844. doi: 10.1515/BC.2005.098. [DOI] [PubMed] [Google Scholar]
- 22.Agmon I., Bashan A., Yonath A. On ribosome conservation and evolution. Isr. J. Ecol. Evol. 2006;52:359–374. doi: 10.1560/IJEE_52_3-4_359. [DOI] [Google Scholar]
- 23.Agmon I. The dimeric proto-ribosome: Structural details and possible implications on the origin of life. Int. J. Mol. Sci. 2009;30:2921–2934. doi: 10.3390/ijms10072921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Belousoff M.J., Davidovich C., Bashan A., Yonath A. On the development towards the modern world: A plausible role of uncoded peptides in the RNA world. Orig. Life Evol. Biosph. 2010;40:415–419. [Google Scholar]
- 25.Bashan A., Yonath A. Ribosome crystallography: Catalysis and evolution of peptide-bond formation, nascent chain elongation and its co-translational folding. Biochem. Soc. Trans. 2005;33:488–492. doi: 10.1042/BST0330488. [DOI] [PubMed] [Google Scholar]
- 26.Bashan A., Belousoff M.J., Davidovich C., Yonath A. Linking the RNA world to modern life: The proto-ribosome conception. Orig. Life Evol. Biosph. 2010;40:425–429. [Google Scholar]
- 27.Tamura K. Ribosome evolution: Emergence of peptide synthesis machinery. J. Biosci. 2011;36:921–928. doi: 10.1007/s12038-011-9158-2. [DOI] [PubMed] [Google Scholar]
- 28.Eigen M., Winkler-Oswatitsch R. Transfer-RNA, an early gene? Naturwissenschaften. 1981;68:282–292. doi: 10.1007/BF01047470. [DOI] [PubMed] [Google Scholar]
- 29.Bloch D., McArthur B., Widdowson R., Spector D., Guimaraes R.C., Smith J. tRNA-rRNA sequence homologies: A model for the origin of a common ancestral molecule, and prospects for its reconstruction. Orig. Life. 1984;14:571–578. doi: 10.1007/BF00933706. [DOI] [PubMed] [Google Scholar]
- 30.Bloch D.P., McArthur B., Guimarães R.C., Smith J., Staves M.P. tRNA-rRNA sequence matches from inter- and intraspecies comparisons suggest common origins for the two RNAs. Braz. J. Med. Biol. Res. 1989;22:931–944. [PubMed] [Google Scholar]
- 31.Root-Bernstein M., Root-Bernstein R. The ribosome as a missing link in the evolution of life. J. Theor. Biol. 2015;367:130–158. doi: 10.1016/j.jtbi.2014.11.025. [DOI] [PubMed] [Google Scholar]
- 32.Di Giulio M. The non-monophyletic origin of the tRNA molecule and the origin of genes only after the evolutionary stage of the last universal common ancestor (LUCA) J. Theor Biol. 2006;240:343–352. doi: 10.1016/j.jtbi.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 33.Krupkin M., Matzov D., Tang H., Metz M., Kalaora R., Belousoff M.J., Zimmerman E., Bashan A., Yonath A. A vestige of a prebiotic bonding machine is functioning within the contemporary ribosome. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2011;366:2972–2978. doi: 10.1098/rstb.2011.0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eigen M., Schuster P. The hypercycle. A principle of natural self-organization. Part C: The realistic hypercycle. Naturwissenschaften. 1978;65:341–369. doi: 10.1007/BF00439699. [DOI] [PubMed] [Google Scholar]
- 35.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 37.Tatusov R.L., Fedorova N.D., Jackson J.D., Jacobs A.R., Kiryutin B., Koonin E.V., Krylov D.M., Mazumder R., Mekhedov S.L., Nikolskaya A.N., et al. The COG database: An updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Farias S.T., Rêgo T.G., José M.V. Origin and evolution of the Peptidyl Transferase Center from proto-tRNAs. FEBS Open Bio. 2014;4:175–178. doi: 10.1016/j.fob.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Szathmáry E. The origin of the genetic code: amino acids as cofactors in an RNA world. Trends. Genet. 1999;15:223–229. doi: 10.1016/S0168-9525(99)01730-8. [DOI] [PubMed] [Google Scholar]
- 40.Da Silva J.A. From the RNA world to the RNA/protein world: contribution of some riboswitch-binding species? J. Theor. Biol. 2015;7:197–201. doi: 10.1016/j.jtbi.2014.12.013. [DOI] [PubMed] [Google Scholar]
- 41.Di Giulio M. A model for the origin of the first mRNAs. J Mol Evol. 2015;81:10–17. doi: 10.1007/s00239-015-9691-y. [DOI] [PubMed] [Google Scholar]
- 42.Noller H.F. The driving force for molecular evolution of translation. RNA. 2004;10:1833–1837. doi: 10.1261/rna.7142404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Noller H.F. Evolution of protein synthesis from an RNA world. Cold Spring Harb. Perspect. Biol. 2012;4 doi: 10.1101/cshperspect.a003681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Szostak J. The eightfold path to non-enzymatic RNA replication. J. Syst. Chem. 2012;3 doi: 10.1186/1759-2208-3-2. [DOI] [Google Scholar]
- 45.Keller M.A., Turchyn A.V., Ralser M. Non-enzymatic glycolysis and pentose phosphate pathway-like reactions in a plausible Archean ocean. Mol. Syst. Biol. 2014;10:725. doi: 10.1002/msb.20145228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Delaye L., Becerra A., Lazcano A. The last common ancestor: What’s in a name? Orig. Life Evol. Biosph. 2005;35:537–554. doi: 10.1007/s11084-005-5760-3. [DOI] [PubMed] [Google Scholar]
- 47.Davidovich C., Belousoff M., Bashan A., Yonath A. The evolving ribosome: From non-coded peptide bond formation to sophisticated translation machinery. Res. Microbiol. 2009;160:487–492. doi: 10.1016/j.resmic.2009.07.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.