Abstract
Transcription activator-like effectors (TALEs) are extremely effective, single-molecule DNA-targeting molecular cursors used for locus-specific genome science applications, including high-precision molecular medicine and other genome engineering applications. TALEs are used in genome engineering for locus-specific DNA editing and imaging, as artificial transcriptional activators and repressors, and for targeted epigenetic modification. TALEs as nucleases (TALENs) are effective editing tools and offer high binding specificity and fewer sequence constraints toward the targeted genome than other custom nuclease systems. One bottleneck of broader TALE use is reagent accessibility. For example, one commonly deployed method uses a multitube, 5-day assembly protocol. Here we describe FusX, a streamlined Golden Gate TALE assembly system that (1) is backward compatible with popular TALE backbones, (2) is functionalized as a single-tube 3-day TALE assembly process, (3) requires only commonly used basic molecular biology reagents, and (4) is cost-effective. More than 100 TALEN pairs have been successfully assembled using FusX, and 27 pairs were quantitatively tested in zebrafish, with each showing high somatic and germline activity. Furthermore, this assembly system is flexible and is compatible with standard molecular biology laboratory tools, but can be scaled with automated laboratory support. To demonstrate, we use a highly accessible and commercially available liquid-handling robot to rapidly and accurately assemble TALEs using the FusX TALE toolkit. Together, the FusX system accelerates TALE-based genomic science applications from basic science screening work for functional genomics testing and molecular medicine applications.
Introduction
Transcription activator-like effectors (TALEs) are extremely effective, single-molecule DNA-targeting molecular cursors used for locus-specific genome science applications, (for reviews see Campbell and colleagues1 and Scott and colleagues2). Diverse applications include locus-specific DNA editing, artificial transcriptional activators and repressors, locus-specific epigenetic modification, and live subcellular imaging. TALEs as nucleases (TALENs) are popular genome-editing tools that are extensively deployed in both in vitro cell systems and diverse model organisms.3–12 Native to the plant pathogen Xanthomonas, TALEs normally function as transcription factors.13,14 TALEs have been harnessed in molecular biology applications to target almost any DNA sequence and effect changes to nearby DNA and genes when fused to other proteins such as endonucleases or activator and repressor domains. The DNA recognition sequence of the TALE is typically a repetitive tract encoding 33–35 amino acid residues, with the last repetitive module being a partial or half (20-amino acid) length. Two variable, adjacent positions, positions 12 and 13, within this repetitive sequence (known as repeat-variable diresidues, or RVDs) are responsible for distinguishing between the four DNA nucleotides.15,16 Commonly used RVDs, each preferentially targeting a specific nucleotide, include the following: NI, NN = adenine; HD = cytosine; NK, NN, NH = guanine; NG = thymine. TALEs represent one of the most versatile programmable DNA-targeting cursors, with the widest potential programmability of any current widely used genome science tool.1,2 This wide targeting range for TALEs often enables single-base-level customization, a requirement for engineering personalized solutions for genome editing in molecular medicine. The main challenge to the broader use of TALEs has been reagent access, due in part to the labor- or capital-intensive standard manufacturing protocols.
TALEs are programmed to target DNA by fusing multiple RVD modules in a specific order according to the TALE cipher code. Diverse protocols have been developed to efficiently synthesize these repeat modules. The Golden Gate TALE system (Golden Gate TALEN and TAL effector kit 2.0, referred to herein as the GGT kit) is a popular assembly method.17 This is a three-step, 5-day Golden Gate process deployed in many laboratories thanks in part to its flexibility, low start-up cost, and requirement for small numbers of common molecular cloning reagents. Several high-throughput TALEN assembly methods18–25 offer unique advantages over the GGT kit and include features such as automation (reviewed in Campbell and colleagues1 and Scott and colleagues2), but each of these systems also comes with specific limitations. For example, the lowest supply cost assembly system reported to date uses custom-designed and capital-intensive research infrastructure.26 A TALE assembly platform that is rapid, flexible for use in many cloning scaffolds (such as common nuclease and nickase backbones), achievable with standard molecular biology laboratory tools, and scalable using modern laboratory automation tools would make TALEs a more accessible system for genome biology applications.
The GoldyTALEN backbone (RCIscript-GoldyTALEN) coupled with 15 RVDs (14.5 TALE repeats) was selected as the TALE assembly platform for development as this system is highly mutagenic in diverse systems and technical improvements would thus be of broad potential value to many different researchers.7,9,10,27 Herein we describe a single-tube modified assembly method, FusX, that is fully backward compatible with the GGT kit with a 3-day optimal completion time. Hundreds of TALE domains have been successfully assembled using this protocol to date, with more than 90% of these TALEs successfully assembled in the first pass. Twenty-seven have been quantitatively tested for activity using the zebrafish gene targeting system, each showing high somatic and germline activity, with many TALENs showing biallelic somatic targeting rates. We show that the FusX system can be readily adapted using a laboratory automation tool, the OpenTrons liquid-handling robot, enabling the rapid scaling of this protocol from in silico TALEN design tools. Once established, the FusX system is comparable in assembly time and reagent cost to other custom genomic tools, while offering high activity and unparalleled specificity and genomic targeting design flexibility.
Materials and Methods
Generating the FusX TALE assembly system
Modification of tandem TALE intermediate vectors: pFusX1–4
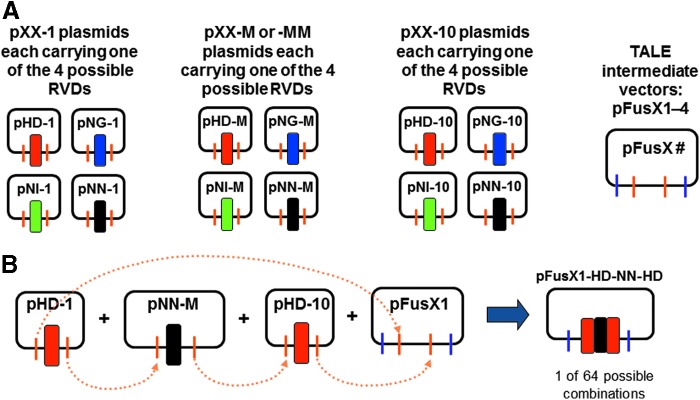
No modification was required for pFusX1, which is the same as pFus_A30A in the GGT kit.17 To generate pFusX2, pFusX3, and pFusX4, modifications to pFusA_30B from the GGT kit17 were made via removal of a small fragment, using different restriction endonuclease combinations, and replaced with modified fragments. For pFusX2, the replacement fragment was produced by self-annealing oligonucleotides; for pFusX3 and pFusX4, the fragment was generated by PCR (Table 1) and these new intermediate vectors will generate specific four-base overhangs on BsmBI digestion (Table 2). All four pFusX# vectors will be available from Addgene (Cambridge, MA: addgene.org).
Table 1.
Modification of tandem transcription activator-like effector intermediate vectors
| Vector | Backbone | Insert type | Subcloning enzymes | Sequence of short oligos or PCR primers |
|---|---|---|---|---|
| pFusX1* | pFus_A30A | N/A | N/A | N/A |
| pFusX2 | pFus_A30B | Short oligos | AgeI/AatII | pFusX2-S: |
| CCGGTGGTCTCTGGCGGCAAGCAAGCGCTCGAAACGGTGCAGCGGCTGTTGCCGGTGCTGTGCCAGGACCATGGCGAGACGT | ||||
| pFusX2-AS: CTCGCCATGGTCCTGGCACAGCACCGGCAACAGCCGCTGCACCGTTTCGAGCGCTTGCTTGCCGCCAGAGACCA | ||||
| pFusX3 | pFus_A30B | PCR | AflII/AatII | pFusX3-F: ATTCTTAAGCGTCTCCATGGCCTGACCCCG |
| pFusX3-R: ATTGACGTCTCGCAGGCCATGGTCCTGG | ||||
| pFusX4 | pFus_A30B | PCR | AflII/AatII | pFusX4-F: GTTCTTAAGCGTCTCCCCTGACCCCG |
| pFusX4-R: GTTGACGTCTCGGCCATGGTCCTGG |
Shown are restriction enzymes and sequences of short oligos or primers used in modification of pFus_A30B into pFusX2, pFusX3, and pFusX4. For pFusX2, annealed reverse complementary short oligos are used directly as the insert. For pFusX3 and pFusX4, PCR products from primers with pFus_A30B as template are used as inserts. No modification is needed for pFusX1, which is identical to pFus_A30B.
N/A, not applicable.
Table 2.
FusX system libraries required for constructing transcription activator-like effectors with 14.5, 15.5, and 16.5 repeats
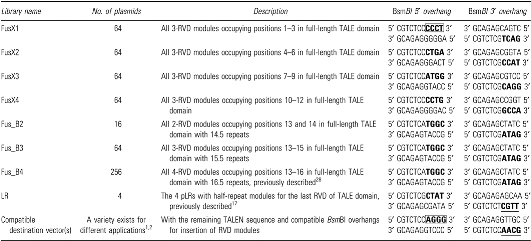 |
Shown is detailed information including library names, plasmid quantities, description, and 5′ and 3′ overhang sequences (in boldface) produced by BsmBI digestion of the nine plasmid libraries required for making TALEs with 14.5 to 16.5 repeats. Compatible overhangs of destination vector with pFusX1 and pLR are boxed and underlined, respectively.
pLR, last half-repeat plasmid; RVD, repeat-variable diresidue; TALE, transcription activator-like effector; TALEN, TALE as nuclease.
Synthesis of “middle” TALE repeat monomers: pXX-M and pXX-MM
To synthesize the middle TALE repeat monomers (M), sense and antisense short oligonucleotides were designed for each RVD to produce inserts with specific 5′ and 3′ BsaI overhangs (Table 3). For simplicity, the designation XX is used to refer to any of the four RVD monomers. Another set of oligonucleotides was designed for each middle RVD monomer with silent mutations introduced into a region of the consensus repeat sequence. These modified-middle repeats (MM) were only introduced into the FusX3 intermediate library to create a unique priming site in the full-length TALE domain. All oligonucleotides were self-annealed and TA cloned into pSC-A, using a StrataClone PCR cloning kit (Agilent Technologies, Santa Clara, CA). All plasmids carrying middle (pSC-A-XX-M) and modified-middle (pSC-A-XX-MM) monomers will be available from Addgene.
Table 3.
Components used to build the FusX system: FusX# and Fus_B# libraries
 |
Detailed information including plasmid names, plasmid quantities, description, and 5′ and 3′ overhang sequences (in boldface) produced by BsaI digestion of all the component plasmids used to synthesize FusX system libraries are listed. XX represents any of the four RVDs (HD, NG, NI, NN), and compatible overhangs with intermediate vectors are highlighted in the same styles.
RVD, repeat-variable diresidue.
Synthesis of TALE intermediate libraries
All TALE intermediate libraries (Table 2) were synthesized through mixed first Golden Gate (GG-1) reactions.28 For libraries FusX1, FusX2, and FusX4, all 12 pXX-1, pXX-M, and pXX-10 were combined with a single pFusX# vector. For FusX3, pXX-MM was used instead of pXX-M, and for Fus_B2 and Fus_B3 that are used to add the penultimate RVDs to an assembly (Fig. 1), pXX-1 and pXX-2 or pXX-1, pXX-2, and pXX-3 were used, respectively. Briefly, for each reaction, 25 ng of each plasmid (for trimer libraries, 13 plasmids; for dimer libraries, 9 plasmids) (Table 3) was combined with 10 U of BsaI (New England BioLabs, Ipswich, MA) and 400 U of T4 DNA ligase (New England BioLabs) in 1× T4 DNA ligase buffer (reaction volume, 20 μl). Three digest–ligation cycles were performed (10-min digest at 37°C; 15-min ligation at 16°C), followed by two 5-min inactivation steps at 50°C and 80°C. To each reaction 0.5 μl of ATP (25 mM) and 5 U of plasmid-safe DNase (Epicentre, Madison, WI) were added and incubated at 37°C for 1 hr, followed by inactivation at 70°C for 30 min. Chemically competent DH5α cells were transformed with 2–5 μl of the reaction and screened via blue/white selection on Luria broth (LB) agar + carbenicillin (100 μg/ml) +5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) + isopropyl β-d-1-thiogalactopyranoside (IPTG). Sequencing screens of 144 colonies per mixed reaction typically yielded 90% of all 64 possible clones in each trimer library. All remaining clones in each library were individually synthesized according to the same protocol. All libraries will be made available through Addgene, and the Fus_B4 library28 is currently available from Addgene (Ekker Laboratory TALEN accessory pack, cat. no. 1000000038).
Figure 1.
Construction of the FusX1–4 libraries. (A) Component plasmids used to construct the pFusX1–4 libraries. pXX-1 and pXX-10 are single-RVD (repeat-variable diresidue) encoding plasmids from the original Golden Gate system (2.0)16; pXX-M and -MM are new, single RVD modules with designated sequence and BsaI overhangs for ligation in between pXX-1 and pXX-10 to form 3-mer intermediates in pFusX1–4 libraries. pXX-MM includes extra silent mutations and is used only to construct the pFusX3 library, providing a specific primer-binding site for sequencing of long TALE (transcription activator-like effector) domain. “XX” represents any of the four RVD modules: HD, NG, NI, NN. (B) Schematic diagram showing sequential ligation of single RVD component plasmids into the four intermediate vectors: pFusX1, pFusX2, pFusX3, pFusX4. Dotted arrows indicate ligation at compatible overhangs generated by BsaI.
Testing and using the FusX system
TALEN design
TALEN pairs FLT3 P1, P2, and P3 targeting zebrafish flt3 were described previously.28 All other TALENs assembled via the FusX method were designed with Mojo Hand software (see www.talendesign.org)29 applying the default parameters: an upstream T nucleotide to the TALE domain (consensus sequence, Ts.*e); minimal binding length of 15 bp; maximal binding length of 17 (14.5–16.5 RVDs). Spacer minima and maxima were set for 14–17 bp. For every target locus, one to three TALEN pairs were saved with a unique restriction site in the middle of the spacer region for a restriction fragment length polymorphism (RFLP) activity assay. Each locus was screened via PCR from pooled genomic DNA (minimum, 10 individuals) to confirm efficient amplification and restriction endonuclease digestion while confirming the target sequence before committing to a TALEN pair. Newly designed TALENs were assembled into the RCIscript-GoldyTALEN mRNA transcription vector, following parameters described below. Assemblies were also performed with a mammalian expression Golden Gate compatible backbone, pC-GoldyTALEN, to ascertain the versatility of this assembly method. These GoldyTALEN receiver vectors are available through Addgene (RCIscript-GoldyTALEN, cat. no. 38142; pC-GoldyTALEN, cat. no. 38143).
Three-day TALEN assembly using the FusX system
All required components for FusX TALEN assembly are reported in Table 2. Day 1: Seven-plasmid assembly similar to second Golden Gate (GG-2) reaction. For each TALEN repeat array, 50 ng of each component plasmid was combined with the RCIscript-GoldyTALEN in a 10-μl reaction volume. Reactions were digested with 5 U of BsmBI, in 1× NEBuffer 3.1 (New England BioLabs) for 30 min at 55°C, and then cooled to 4°C. To each reaction, 1.5 μl of 10× T4 DNA ligase reaction buffer, 400 U of T4 DNA ligase (New England BioLabs), 5 U of Esp3I (Thermo Fisher Scientific, Waltham, MA), and water were added to a final volume of 15 μl. Reactions were incubated in a thermocycler (37°C, 5 min; 16°C, 10 min) for five or six cycles (37°C 15 min, 80°C 5 min, and 4°C hold). To each reaction, 0.5 μl of ATP (25 mM) and 5 U of plasmid-safe DNase (Epicentre) were added and incubated at 37°C for 1 hr, followed by inactivation at 70°C for 30 min. Chemically competent DH5α cells (minimal efficiency, 107) were transformed with 2–5 μl of the reaction and screened via blue/white selection on LB-agar + carbenicillin + X-Gal + IPTG. Day 2: Four to eight white colonies were screened by colony PCR using primers TAL_F1 (5′ ttggcgtcggcaaacagtgg 3′) and TAL-R2 (5′ ggcgacgaggtggtcgttgg 3′).17 Note that for Golden Gate receiver vectors other than RCIscript-GoldyTALEN, colony PCR conditions may need to be further optimized to predict positive clones. For each TALEN, one or two PCR-positive colonies were selected and cultured in LB + carbenicillin overnight. Day 3: Plasmids were purified from overnight cultures and double-digested with AatII and StuI (New England BioLabs) to confirm the presence and size of TALE domains. Each domain was sequenced from the forward and reverse direction using primers TAL_F1 and TAL_R2 or using SEQ-TAL-F2 (5′ accgcaatggaggcagtg 3′) and SEQ-TAL-R3 (5′ cgtgcggcaatcccttt 3′). If middle RVDs could not be confirmed, sequencing primer RVD-MM-F (5′ ctcacacccgatcaggtc 3′) was used to prime off the eighth RVD in the array.
Testing TALEN somatic activities in vivo
RCIscript-GoldyTALEN constructs were linearized by SacI digestion: 5 μg of DNA, 10 U of SacI (New England BioLabs), for 6–8 hr at 37°C. Digested DNA was precipitated by sodium acetate–ethanol extraction, cleaned with 70% ethanol, and resuspended in water. mRNA encoding each TALEN arm was in vitro transcribed, using a mMESSAGE mMachine T3 kit (Life Technologies, Grand Island, NY), stopped with the supplied ammonium acetate solution, and cleaned via phenol–chloroform extraction and isopropanol precipitation (see mMESSAGE mMachine T3 kit for protocol). TALEN mRNA pairs were injected into one-cell zebrafish embryos at doses ranging from 20 to 100 pg. Larvae (72 hr postfertilization [hpf]) were collected and screened for somatic nonhomologous end-joining (NHEJ) activity by RFLP assay, described previously.9,28 To quantify NHEJ activity, ImageJ was used to measure band intensity. The undigested band level was divided by the sum of both the undigested and digested bands and multiplied by 100 to calculate the percent somatic NHEJ activity level for each TALEN pair.
Examining germline efficiencies of TALENs
To screen for germline transmission of TALEN-induced mutations, injected fish were out-crossed to obtain F1 embryos. At least eight of the F1 offspring were screened by PCR and subsequent RFLP analyses. First, DNA was isolated from the embryos, using the NaOH extraction method.30 Next, gene-specific PCR was conducted on individual embryo DNA preparations, and the resulting PCR product was digested to assess germline transmission of the TALEN-induced mutation. To obtain sequence information about TALEN-induced mutations, uncut DNA from the digest was excised from the gel and purified with a QIAEX II gel extraction kit (cat. no. 20021; Qiagen, Hilden, Germany). The purified, uncut DNA was then sequenced by Sanger sequencing and analyzed to determine mutations in the gene corresponding to the TALEN spacer region. Any mutation located in this region was recorded, and the surviving embryos from the clutch were raised.
Programming TALENs using an accessible laboratory automation tool, OpenTrons
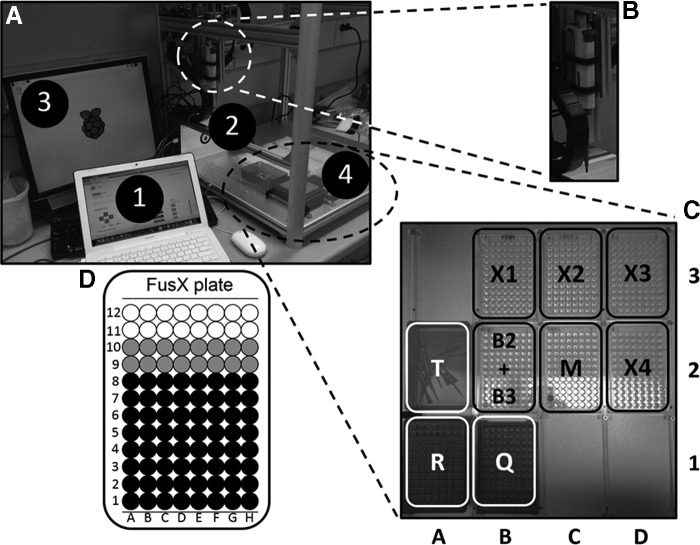
All 64 RVD plasmids of each FusX1–4 trimer library were organized by columns (starting with column A) into four 96-well plates (Fig. 2D). An additional 96-well plate was used to store the 64 Fus_B3 and the 16 Fus_B2 plasmids. These plasmids were mapped onto the appropriate plate in alphabetical order of the nucleotide sequence of their target recognition site (i.e., AAA, AAC, AAG, AAT, ACA, ACC, and so on). A sixth 96-well plate was used to store aliquots of the four pLR plasmids (in the same alphabetical order as described above), the GoldyTALEN receiver plasmid, and the 10× NEBuffer 3.1 (New England BioLabs) from well A1 (NI pLR plasmid) to F1 (10× NEBuffer 3.1) accordingly. An OT-One OpenTrons liquid-handling robot (LHR) (firmware v1.0.0; Fig. 2A), running on a Raspberry Pi 2 single-board computer (Raspbian GNU/Linux 8.0 [Jessie]) and a Smoothieboard 5× numerical controller (SmoothiewareOT version 1.0.5; Fig. 2A [2]), was set up to handle a 10-μl DRAGONLAB micropipette (Fig. 2B) and was connected via Ethernet to a MacBook laptop (2.2 GHz Intel Core Duo processor and 4 GB 667 MHz DDR2 SDRAM memory, operating on Mac OS X version 10.7.5; Fig. 2A [1]), following the instructions provided by the manufacturer (see https://docs.google.com/document/d/199qBoWq3_eKI6HM-k_STKBz4hbdg0s6lK49ye62q1UM/edit). Next, the six flat-bottom 96-well plates described above, a trash bin, a tip rack, and destination rack for the TALEN arm assemblies were placed in designated sockets of the LHR (Fig. 2C).
Figure 2.
Enhanced FusX assembly using an automated liquid-handling laboratory automation tool. (A) OT-One liquid-handling robot (LHR) set up for TALEN (transcription activator-like effector as nuclease) arm assembly via the FusX system. The MacBook laptop (1) operates the graphical user interface of the LHR software running on a Raspberry Pi 2 (RPi) single-board computer. (2) The RPi takes the JSON script of an arbitrary protocol and parses it into a G-code job to be run by the Smoothieboard numerical controller. (3) A monitor connected to the RPi via HDMI to visualize the RPi OS (GNU/Linux 8.0). (4) LHR workspace. (B) DRAGONLAB 10-μl micropipette. (C) Zoom-in on LHR workspace. Black border: six 96-well plates containing the libraries FusX1–4, Fus_B2+B3, and an extra plate (M) with the receiver backbone plasmid and the pLR plasmid stocks. White border: 10-μl tip rack (R), trash bin (T), and destination aliquots for TALEN arm assembly (Q). (D) Organization of the FusX 96-well plates. In black, all 64 possible combinations of RVD array plasmids were arranged on the basis of the output from the Mojo Hand inventory-management software for TALEN arm assembly for libraries FusX1–4 and Fus_B3. In gray, the 16 extra wells for plasmids of the Fus_B2 library in the pFusB2+B3 plate.
The LHR was calibrated as recommended by the manufacturer (see https://github.com/Opentrons/otone_docs/blob/master/Calibrate_Run.md). Briefly, the 3-D coordinates of the position of the A1 well were mapped into the LHR software for all plates. Subsequently, the z coordinates of the functional positions of the pipette plunger were assigned (starting point, liquid blowout point, and tip ejection point). The JSON (JavaScript Object Notation) file encoding the liquid-handling protocol for TALEN arm assembly was generated using the online editor tool provided by the manufacturer (http://editor.mix.bio/). The protocol used in this study consisted of the successive transfer of the appropriate component plasmid from all FusX system libraries and the receiver plasmid into a destination tube per TALEN arm for up to seven different TALEN arm sequences. The JSON file used for this specific experiment will be provided on e-mail request to the corresponding author.
Day 1 of the manual FusX protocol was modified to account for the micropipette used by the LHR. Both the component plasmid stocks and the receiver plasmid GoldyTALEN were diluted 5-fold so that 5 μl was pipetted into the TALEN arm aliquot instead of the standard 1 μl. In parallel, two other aliquots were pipetted manually per TALEN arm as positive controls: one using the LHR-modified protocol and another using the standard protocol. Next, 1 μl of the restriction enzyme BsmBI (cat. no. R0580; New England BioLabs) and 1.2 μl of 10× NEBuffer 3.1 (New England BioLabs) were added to 10 μl of each manual repeat. For each TALEN arm, all three aliquots were treated according to the standard FusX protocol.
Results
Building the FusX system from the popular Golden Gate TALEN kit 2.0
The Golden Gate TALEN (GGT) kit is a commonly used TALE assembly system that requires two rounds of Golden Gate reactions, GG-1 and GG-2, to assemble a full-length TALE domain.17 On day 1, the user assembles two or three partial RVD arrays from monomeric repeat modules into initial arrays termed pFus_A and pFus_B. The generation of pFus_A partial arrays can be particularly problematic for some users, however, because of the relatively complex 11-plasmid digestion–ligation step. On day 3, these two partial arrays are then combined in a second digestion–ligation reaction into the full-length binding domain within a functional TAL receiver vector.
The FusX system uses the same basic assembly principles as the GGT kit but modifies preexisting components to achieve the goals of increasing the success rate, shortening the overall process, and reducing the workload of the end user, while maintaining a Golden Gate compatible platform. This was accomplished by:
1. Reducing the number of component plasmids required to obtain a full-length TALE domain in a single assembly reaction.
2. Establishing a single assembly reaction, thereby reducing the time required to assemble a full-length TALE domain.
3. Minimizing the number of library component (or intermediate) plasmids that make up the FusX system.
The principle of the FusX system is to break down the large partial pFus_A RVD arrays into smaller components with all necessary modifications completed in three steps:
1. Generation of TALE intermediate vectors (pFusX1–4) by modifying pFus_A30B from the GGT kit. The original pFUS_A30A (renamed pFusX1 for nomenclature consistency) is used alongside three newly synthesized derivatives from pFUSA_30B made by subcloning short oligos or PCR products (pFusX2, pFusX3, and pFusX4; Table 1). The four base 5′ and 3′ overhangs produced by BsmBI were changed without frame shifting (Fig. 2 and Table 2) such that these four vectors are now sequentially complementary to each other and also backward compatible with the original GGT kit components: pFus_B# plasmids, pLR and TAL effector backbones.
2. Synthesis of “middle” TALE repeat monomers (pXX-M and pXX-MM). Without any modification, 5′ and 3′ overhangs produced by BsaI in the pFusX1–4 vectors during GG-1 are compatible with TALE repeat monomer in the GGT kit at position 1 (pXX-1) and position 10 (pXX-10) (Fig. 1 and Table 3). To assemble intermediate trimer libraries, two sets of new middle monomers (pXX-M or pXX-MM) were synthesized with compatible 3′ and 5′ overhangs on BsaI digestion to join pXX-1 and pXX-10 in GG-1 (Fig. 1 and Table 3). Silent mutations were introduced in all pXX-MM clones, with these modified-middle repeats only introduced into the FusX3 intermediate library to create a unique priming site in the full-length RVD array.
3. Synthesis of TALE intermediate libraries FusX1–4 and Fus_B2-4. To generate the components of the FusX system, a mixed GG-1 reaction (Fig. 1) was conducted to synthesize seven premade libraries of partial RVD arrays. Each FusX# library component contains 3 TALE repeat monomers (each of 4 carry different RVDs) for a total of 43 = 64 plasmids per library. The Fus_B# libraries contain 2, 3, or 4 RVD array combinations, for a total of 16, 64, and 256 plasmids per library, respectively (Table 3).
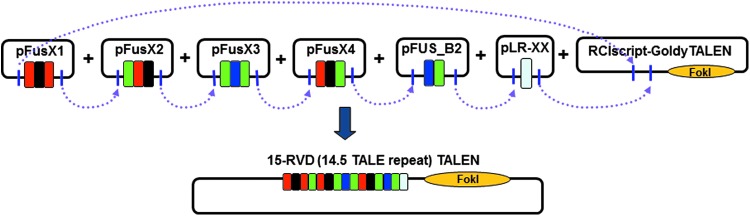
By premaking smaller modular arrays via the GG-1 reaction, the first 12 repeats of the TALE domain are derived from the FusX1–4 libraries in a scheme of: 3 + 3 + 3 + 3. The remaining repeats of the domain are provided by the Fus_B# libraries (or any custom pFus_B# plasmids) and the last, half-repeat comes from the pLR. In addition, FusX3 plasmids incorporate silent mutations to allow internal priming necessary for DNA sequence confirmation of the final assembled TALE domain. Thus, to assemble a full-length DNA-binding domain into the final TAL effector backbone, the user assembles one single reaction containing: pFusX1 (3 repeats) + pFusX2 (3 repeats) + pFusX3 (3 repeats) + pFusX4 (3 repeats) + pFus_B2–4 (2–4 repeats or any custom pFus_B#) + pLR (half-repeat) as shown in Fig. 3. The seven TALE intermediate libraries described previously, combined with the last half-repeat plasmids (pLRs), are capable of generating any combination of 14.5, 15.5, or 16.5 module arrays. By building larger custom pFusX_B# plasmids (containing 5–10 RVD array), the user can increase the final length of the TALE domain to 17.5–22.5 TALE repeats.
Figure 3.
Using the FusX system to assemble custom TALENs in a single reaction. Dotted arrows indicate assembly at the compatible 5′ and 3′ overhangs generated with BsmBI.
Using the FusX system: TALEN assembly testing
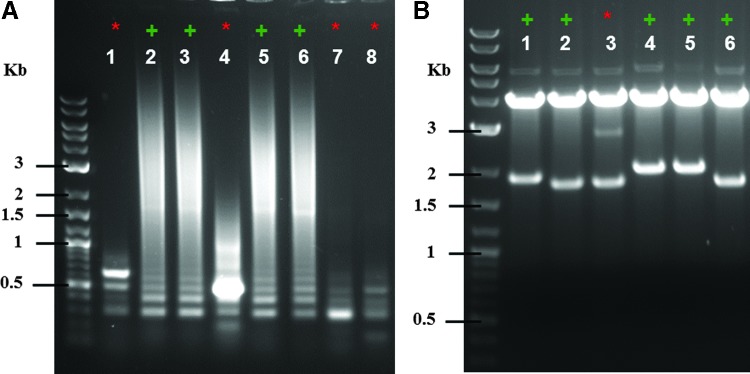
We have assembled more than 160 arrays of 14.5–16.5 TALE repeats via the FusX method. We have not experienced a failure to obtain a clone with the correct sequence. In 90–95% of cases, screening one to four colonies is sufficient to find a positive clone. In about 5–10% of cases, a less favorable assembly is encountered and up to eight colonies are screened to identify a correct clone. In general, if there is a high proportion of white to blue colonies, then a majority (if not all) of the clones tested (white) will be correct (data note shown). Figure 4A shows examples of positive and negative colony PCR results, and Fig. 4B shows the size of RVD arrays on restriction digest with AatII + StuI. On occasion, a colony that appears positive by colony PCR, will, on digest, contain what appears to be a mixed population of plasmids (Fig. 4B, lane 3). We do not use these mixed populations, but simply pick another positive colony from this transformation.
Figure 4.
Quality control of TALEN assembly, using colony PCR and restriction enzyme digestion. (A) Representative colony PCR using primers TAL_F1/TAL_R2. Correct colonies (+) result in characteristic laddering due to the repetitive nature of the incorporated RVDs. (B) Representative restriction enzyme digestion analyses with AatII/StuI are shown [these clones differ from those shown in (A)]. The size of the lower band is dependent on the number of TALE repeats within the TALEN, which can aid in determining correct clones. +, correct TALEN clones; *, incorrect TALEN clones.
TALEN pairs assembled with FusX system have comparable activity to those from GGT kit, and show high somatic and germline activities in vivo
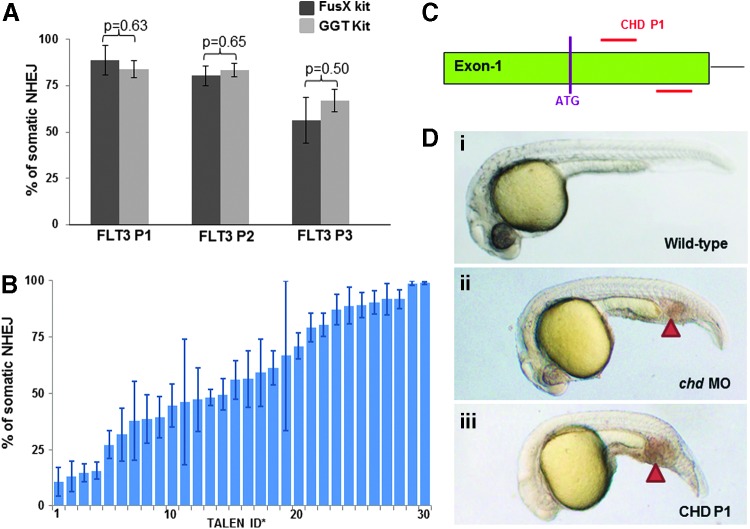
Using the FusX system, we remade three pairs of previously published GoldyTALENs targeting zebrafish flt3: FLT3 P1, P2, and P3.28 All three TALEN pairs (named FLT3 P1X, P2X, and P3X to distinguish them from the original pair) were successfully assembled and sequence verified. To test whether silent mutations introduced in pFusX3 altered TALEN activity, these newly synthesized TALEN pairs were tested in zebrafish embryos, and their somatic activities were comparable to the previously published original TALEN pairs (Fig. 5A). We then tested the somatic efficiency of a larger number of FusX-assembled TALENs in zebrafish. A collection of 30 TALEN pairs at various loci was designed and assembled (including FLT3 P1–3), and the somatic efficiency of each pair was assessed by RFLP analysis (Fig. 5B and Table 4). Importantly, the range of somatic efficiency (approximately 11–99%) is similar to what has been observed using the GoldyTALEN backbone and the original Golden Gate assembly method.28 Twenty-seven TALEN pairs made using the FusX system have been subsequently tested for germline transmission, and all 27 have shown good transmission of locus-specific alterations (Table 4). To demonstrate gene targeting at the phenotypic level, we also synthesized a TALEN pair (CHD P1) targeting zebrafish chordin Exon-1 (Fig. 5C) with the FusX system. Zebrafish chordin is a bone morphogenetic protein (BMP) antagonist. Similar to previously reported homozygous chordin mutant (chordino)31 or morpholino-mediated knockdown morphant,32 TALEN (CHD P1)-injected embryos are strongly ventralized with the characteristic intermediate-cell-mass (ICM) expansion (Fig. 5D).
Figure 5.
In vivo mutagenic activity of TALEN pairs assembled with the FusX system. (A) Comparison of somatic efficiency between TALEN pairs (with identical RVD arrays) assembled via the FusX system (FLT3 P1X, P2X, and P3X) or via the original GGT method (FLT3 P1, P2, and P3). Average results of 3 separate experiments analyzing groups of 10 embryos are shown. Error bars represent the SEM, and efficiencies of TALEN pairs were statistically analyzed with unpaired t tests. (B) Somatic efficiencies of 30 TALEN pairs assembled with the FusX system. Error bars represent the SEM. Detailed descriptions of these TALEN pairs are found in Table 4. (C) Design of CHD P1 (TALEN 13) targeting zebrafish chordin Exon-1. (D) Embryos injected with CHD P1 (panel iii) showed significant ICM expansion (arrowhead) at 24 hr postfertilization (hpf) compared with wild type (panel i), phenocopying the morpholino-mediated chordin knockdown (panel ii).
Table 4.
Somatic activities and germline efficiencies of 30 TALEN pairs synthesized with FusX system
| RVD array of TALE domain (5 ′ to 3 ′) | |||||
|---|---|---|---|---|---|
| TALEN ID | Target gene | Left arm | Right arm | RFLP-based somatic efficiency | Germline efficiencya |
| 1 | bcs1l | NN NN NI NN NG NN NN HD NN NN NI NN NI NN NN | NN HD HD NG NI NG NN NI NI HD NG HD HD NG N | 10.6 ± 6.3% | 3/7 |
| 2 | aars2 | NI NG NI NI NI NG NG NI NG NI NI NG NN HD NI NN | NI NI NI NI NN NI HD NN HD HD NI NN NN HD HD NI | 13.1 ± 6.8% | 2/3 |
| 3 | coq7 | NG NN NG NG NN HD HD NN HD NG NN NG NN NN NI | NI NI NN NI NI HD NN NG NI HD HD NG NI NI NG | 14.5 ± 4.0% | 1/6 |
| 4 | micu3a | NI HD NG HD HD NI HD HD NI NN NG NN NG NN NN NI | HD NN NI NI NN NN NG NG HD HD NG NN NI NI HD | 15.6 ± 3.9% | 1/7 |
| 5 | tk2 | HD NI HD HD NI HD NI NN NI HD HD NG NN HD HD | HD HD NG HD NG HD NG NN HD NI HD HD NG HD NG | 27.2 ± 6.2% | 2/2 |
| 6 | ndufab1b | HD HD NI HD HD HD HD NG HD NI HD NI NG NG NI NN | NI NI NN NN NI HD NI NG NI HD NI NN NI NI HD NG | 31.6 ± 11.6% | N/A |
| 7 | micu1 | HD NG NI NN NI NN NG HD HD NI NN HD NI NN HD | HD NG NG HD NG NN NI NN HD HD HD NN HD HD NG HD | 37.8 ± 17.6% | 2/2 |
| 8 | atp5e | NI HD NI NG HD NI NN NN NG NI HD NG HD NG NN HD | HD NI NN NG NN HD NI NN HD HD HD NN HD NI HD NN | 38.7 ± 10.8% | 1/4 |
| 9 | coq2 | HD HD NG HD NN NN NI NN NN NI HD NI NN HD NG HD | NN NG HD HD NG NI HD NG HD NI NN HD HD NG NG NI | 39.3 ± 9.3% | 4/7 |
| 10 | fxn | NI NI NI HD NI NI NI NI NN NI NN NI HD HD NG | NN HD HD NG NG NG NG HD NG NG HD NG HD HD NI | 44.5 ± 9.5% | 2/6 |
| 11 | smdt1 | NG HD NN NG NN NG NN HD HD NN NN NG HD NN NN NG | NN HD NI NN HD NG NN HD NG NN NI NI NN HD HD HD | 46.2 ± 27.9% | 1/2 |
| 12 | cox412b | HD NI NI NI NN NI NN NI NI NN NN NI NN NI NI | HD NG HD HD NG HD HD NG NG NN HD NG NN NI NN NG | 47.2 ± 14.1% | 2/7 |
| 13 | chd (CHD P1) | HD NN HD NI NG HD NG NN NG NN HD NI HD NN NN | NN NN NI NG NN NN NN HD NI NN HD NN HD NN NN | 48.2 ± 3.6% | N/A |
| 14 | micu2 | NG NG NN HD HD NG HD HD NI NG NN NI NG NI NG | HD NG HD NG NN NN NN NG NN NG HD NI NG NN NG | 49.5 ± 7.0% | N/A |
| 15 | micu3b | NG NG HD NG NG NG HD NN NG HD NG NG HD HD NI NN | NG NN NI NI NI NN HD NG NI NG NG NG NG NI NI | 55.9 ± 8.6% | 3/8 |
| 16 | flt3 (FLT P3X) | NG NI NN NG NN NG NN HD NI HD NG NG HD NG NN | NI HD HD NI HD HD NG NN NI NN NI NN NG NG NN | 56.3 ± 12.5% | 2/4 |
| 17 | ndufaf6 | NN NN NI NN NN NI HD NI NN NI NN HD NN NG NI HD | HD NG HD HD NI NG NI NN NN HD NG NG HD HD NI | 59.2 ± 14.9% | 1/2 |
| 18 | ndufa9 | NG NN NG NN NI NG HD HD HD NG NG NI HD HD NN | HD NG NN NI NN NN NG NI HD NI NG NN NI NN NI | 61.1 ± 7.6% | 3/8 |
| 19 | pdss2 | NG NG NN NN NN NI NI HD NI NI NN NI NG NN NN | NG NG NN HD HD NI NI HD NI NN NI NI NI NI NG | 66.7 ± 33.3% | 2/12 |
| 20 | cox4i2 | NG NG NN HD HD NI NI HD NI NN NI NI NI NI NG | HD HD NG NG HD NG NN HD NG NG HD NI NN NN HD NG | 70.9 ± 5.8% | 2/2 |
| 21 | sdha | HD NG NN NG HD NG NN NI NN NN HD HD NN NN HD | NN NN NN NI NI NI NI NN NG NG NG NN NN NG NN NI | 79.2 ± 6.3% | 1/2 |
| 22 | flt3 (FLT P2X) | NI HD HD HD NI NN HD NI NI NN NI NN HD NG NG | NG NI NN NG NN NG NN HD NI HD NG NG HD NG NN | 80.3 ± 5.2% | 3/3 |
| 23 | mcu | NG HD HD NG NN HD NI NN NN NI NN NN HD NN HD NG | NN NI HD NI NN NG HD NG NN NN NG NN NI HD HD | 87.0 ± 6.7% | 3/3 |
| 24 | flt3 (FLT P1X) | NN NG NG NI NG NN NG NI NG NG HD NI HD NN HD | NI HD HD HD NI NN HD NI NI NN NI NN HD NG NG | 88.8 ± 8.1% | 2/2 |
| 25 | tmem70 | NN NG NI NI NN NI NG NN NN NG NI HD NI NI NG NG | NI NN NI NG HD NG HD HD NI NG HD NG NG HD NG NN | 89.1 ± 5.5% | 1/2 |
| 26 | cox10 | NN NN NG NI HD NN NI HD NI NN NN NG NG HD NI NN | NI HD HD NI NN NN HD NG HD NI NN NN HD NG NG | 90.4 ± 5.1% | 1/3 |
| 27 | ndufs4 | NI HD HD NI NN NN HD NG HD NI NN NN HD NG NG | HD NG NG NI NI NG NI NG NN HD NG HD HD NG HD NG | 91.8 ± 6.9% | 2/2 |
| 28 | mftmt | HD HD NN NI NI NN NI NI HD NG NN HD NI HD NG | NI NG HD NG NN HD HD NI NI NI NN NG NI NN HD | 91.8 ± 4.2% | 1/2 |
| 29 | cox6b1 | NI NG NI NN NN NN NI NN HD NN NG HD NI NG NN NN | NI NN NG NG HD NG HD HD NI NN HD NG NG HD NG | 98.8 ± 0.9% | 2/4 |
| 30 | surf1 | NG NI NG NG HD NI NN NI HD NN NG HD NI NN NN | NI NI NI NI NG HD NG NN NI NI NG NG NI HD NI HD | 99.0 ± 0.6% | 1/2 |
In vivo somatic activities (percentage of NEHJ-mediated mutations at the somatic level, based on RFLP assay) and germline transmission efficiencies of 30 TALEN pairs synthesized with the FusX system. FLT P1X, P2X, and P3X are independent TALEN pairs targeting different regions of the flt3 gene, and somatic activities are determined by RFLP assay (n = 3) and presented with the SEM.
Germline efficiency is shown as the number of F0 that gave mutant offspring over the total number of F0 screened embryos.
N/A, not applicable as germline testing was not completed for these three TALEN pairs; RFLP, restriction fragment length polymorphism; RVD, repeat-variable diresidue; TALE, transcription activator-like effector; TALEN, TALE as nuclease.
TALEN programming using an accessible laboratory automation tool
The FusX system is designed to support robotic liquid handling to minimize human-derived technical errors and for enhanced scalability (Fig. 2). All plasmids in FusX1–4, Fus_B2, and Fus_B3 were mapped onto 96-well plates as shown in Fig. 2D. The mapping configuration can be synced with an inventory management and/or TALEN design software of preference (e.g., Mojo Hand29). In turn, the position of each 96-well plate was mapped in a grid onto the workspace of an OT-One liquid-handling robot (LHR; Fig. 2C). Thus, any desired RVD array sequence can be systematically consolidated into a single aliquot, using an LHR device, if the coordinates of every well in every plate are known. Liquid-handling robotics was used to aliquot the RVD array sequence for 16 TALEN arms. Positive controls were made by manually assembling the RVDs of 10 of the TALEN arms in parallel to the robot assemblies. Sequencing data confirmed the successful assembly of all TALEN arms, whether pipetted manually or robotically (data not shown).
Discussion
Genome science is a rapidly evolving field, with many exciting and innovative tools now in common use today (see Peng and colleagues,33 Kim and Kim,34 and Chandrasegaran and Carroll35 for reviews). One area for new iteration of technologies is that of functional genomics, especially in the era of personalized medicine. For example, off-target and other less desirable features of “knockdown” strategies have been shown to limit the applications of RNA interference36,37 and morpholino-based methods.38,39 These RNA-targeted technologies are still popular today, due in part to the time-sensitive requirements for assessment of gene function for applications such as personalized medicine that can obviate the use of traditional and time-consuming germline genetic assessment approaches.
To our knowledge, the highest described germline activity for a collection of CRISPR/Cas9 reagents in zebrafish was reported by the Burgess laboratory,40 and the results described here for TALENs used similar protocols for germline activity assessment. In Varshney and colleagues,40 15% of the gRNAs failed to exhibit any activity, with 53% of the tested gRNAs showing germline transmission (n = 7 F1 embryos tested). All of the TALENs used here showed good somatic activity, and 45% of the TALEN-injected fish transmitted offspring with mutations through the germline (n = 8 F1 embryos tested).
In addition to working well for germline functional studies, we report that the FusX platform can be a suitable alternative to RNA-based genomic tools for rapid gene function assessment. For example, TALENs are highly active in zebrafish, with a subset of TALENs regularly generating animals with nearly all somatic chromosomes generating an insertion/deletion at the target site.9,28,33,41 We note that for such high-activity reagents, biallelic targeting of a lethal locus could result in reduced fitness of the injected animals and lower germline propagation rates. In such circumstances, injecting lower doses of the TALENs so that heterozygous animals will be generated would be appropriate. As demonstrated by the chordin locus, active TALENs can rapidly replicate germline knockout data in somatic samples. The rate of obtaining this somatic genetic assessment is comparable to that which can be collected using morpholinos in zebrafish. High-activity TALENs could also make possible multigene-targeting approaches, in which many genes are mutated simultaneously. This general approach may also be viable using other custom nuclease platforms.
The biggest challenge for widespread use of TALENs has been limited accessibility, in particular, the cost, efficiency, and compatibility of the assembly processes. The relatively complex assembly method remains a major concern compared with other genome-editing tools such as the newly developed CRISPR/Cas system.42–46 The original Golden Gate kit 2.0 is a highly flexible and adaptable method.17 Nonetheless, the requirement for two rounds of Golden Gate reactions, including the assembly of 10 RVD-containing intermediate plasmids in the first round, is somewhat laborious for high-throughput TALEN production. Alternative assembly methods consisting of 1- to 3-day, single-step protocols have been developed.19,20,47 However, high setup cost (requiring special equipment), inflexibility (TALE domains of only certain lengths can be synthesized), and incompatibility with the GGT kit limit the popularity of these methods (reviewed in Campbell and colleagues1). Therefore, an improved method with short assembly time, low cost, and high compatibility without compromising TALEN activity is needed.
The FusX system is basically a modified version of the GGT kit. Instead of 10-RVD repeat TALE intermediates, preassembled trimers were used and a TALE domain between 14.5 and 16.5 repeats can be assembled in a single Golden Gate reaction. Previously reported as well as newly designed TALEN pairs were assembled using the FusX system and tested in vivo in zebrafish embryos. Correct sequences of all 30 pairs of TALENs synthesized verified the assembly system, and a comparable range of somatic efficiencies, in particular between FLT3 P1, P2, and P3 and FLT3 P1X, P2X, and P3X, respectively, suggested that silent mutations introduced in pFusX3 do not affect TALEN activity.
The one-step Golden Gate reaction with the FusX system is almost identical to the second-round Golden Gate reaction in the GGT kit. Both kits use common, inexpensive molecular reagents and beyond the FusX plasmids, no alternative reagents are required when switching from the GGT kit to the FusX system. Importantly, the FusX system is backward compatible with all TAL effector scaffolds previously constructed for the GGT kit. Moreover, because the FusX system is functional with all pFus_B# vectors, it retains the flexibility to assemble TALE domains with up to 22.5 repeats. However, pFus_B5 to pFus_B10 are required to assemble TAL domains longer than 16.5 repeats and would need to be made separately using the original GGT kit. Finally, sequencing primers can be designed to target the silent mutations introduced in FusX3 at TALE repeat 8, ensuring complete sequencing of the final TALE domain.
In addition to TALEN-mediated genome editing, TAL effectors are also used as programmable transcription activator/repressors,48,49 DNA and histone methylation modifiers,50,51 and fluorescent DNA visualizers.52 Similar to TALENs, all these applications involve the fusion of the TALE DNA-binding domain with particular functional domains (e.g., VP64, Hairy/SID/KRAB Repression domain, lysine-specific demethylase, TET1 hydroxylase catalytic domain, GFP, etc.). The FusX system can also be used to synthesize a wide range of these nonnuclease genome science tools.
There are two known major limitations of the FusX system, with the first being the requirement to establish a collection of component plasmids before initiating any synthesis reactions. With all seven libraries to assemble TALE domains from 14.5 to 16.5 repeats, there are 592 plasmids in total excluding the 4 pLR and TAL effector backbones. However, the core components for assembling highly effective TALE domains with 14.5 repeats28 (FusX1–4 and Fus_B2) consist of only 272 plasmids. Second, the presented FusX system includes a single set of standard RVD ciphers; alternative RVDs, such as NH and N*, are not included. For a comprehensive switch to an alternative RVD for the entire collection, such as replacing all NN with NH, libraries could be recreated using receiver plasmids available in the FusX system. However, if an alternative RVD(s) is required only in certain repeat(s), one could assemble the corresponding pFusX# or pFus_B# clone(s) separately. This latter alternative assembling process is still advantageous over the older platform as it can be accomplished within the same time frame, and the efficiency of assembling trimer intermediates is much higher than that of a 10-repeat intermediate.
The present study combines tools for TALEN design with inventory management and a mechanistic system for TALEN arm assembly. The first significant advantage of this approach is the immediate reduction of the risk of technical errors by human handling (e.g., pipetting variability), enhancing the reproducibility of the method. Starting with the price of the OT-One LHR, we envision that this approach will increase the accessibility of TALEN design and assembly for genome editors. In addition, open source software (OT-One and Mojo Hand) enable users to rapidly customize their protocols for their specific laboratory needs. Another important benefit of open source equipment is its inherent facilitation of community building around TALEN design. Custom protocols can be easily shared between users, implemented in any laboratory, and tailored to a specific project.
Acknowledgments
The authors thank Will Canine and the OpenTrons team for help in troubleshooting hardware and software glitches with the new liquid-handling robot system; Chris Ward, Colby Starker, Dan Carlson, and Dan Voytas for critical suggestions; and all the staff in the Mayo Clinic Zebrafish Facility and HKU Zebrafish Core Facility (FCF, LKS Faculty of Medicine) for excellent zebrafish husbandry. This work is supported by the State of Minnesota (University of Minnesota Mayo Clinic Partnership grant [H001274506] to SCE), National Institutes of Health grants [GM63904, P30DK084567] to SCE, [DA032194] grant to KJC, the Mayo Foundation, NIH Training grant [UL1 TR000135] to PRB and JMC, Marriott Mitochondrial Network, FHB HMRF [02132326] to ACM, and HKU Seed Funding Programme for Basic Research [201401159004, 201411159098] to ACM.
Author Disclosure
No competing financial interests exist.
References
- 1.Campbell JM, Hartjes KA, Nelson TJ, et al. New and TALENted Genome Engineering Toolbox. Circ Res 2013;113:571–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scott JN, Kupinski AP, and Boyes J. Targeted genome regulation and modification using transcription activator-like effectors FEBS J 2014;281:4583–4597 [DOI] [PubMed] [Google Scholar]
- 3.Zhang F, Cong L, Lodato S, et al. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat Biotechnol 2011;29:149–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller JC, Tan S, Qiao G, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol 2011;29:143–148 [DOI] [PubMed] [Google Scholar]
- 5.Hockemeyer D, Wang H, Kiani S, et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol 2011;29:731–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tesson L, Usal C, Ménoret S, et al. Knockout rats generated by embryo microinjection of TALENs. Nat Biotechnol 2011;29:695–696 [DOI] [PubMed] [Google Scholar]
- 7.Carlson DF, Tan W, Lillico SG, et al. Efficient TALEN-mediated gene knockout in livestock. Proc Natl Acad Sci U S A 2012;109:17382-17287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Y, Zhang F, Li X, et al. Transcription activator-like effector nucleases enable efficient plant genome engineering. Plant Physiol 2013;161:20–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bedell VM, Wang Y, Campbell JM, et al. In vivo genome editing using a high-efficiency TALEN system. Nature 2012;491:114–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma S, Zhang S, Wang F, et al. Highly efficient and specific genome editing in silkworm using custom TALENs. PLoS One 2012;7:e45035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sung YH, Baek IJ, Kim DH, et al. Knockout mice created by TALEN-mediated gene targeting. Nat Biotechnol 2013;31:23–24 [DOI] [PubMed] [Google Scholar]
- 12.Beumer KJ, Trautman JK, Mukherjee K, et al. Donor DNA utilization during gene targeting with zinc-finger nucleases. G3 (Bethesda) 2013;10:1534/g3.112.005439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gu K, Yang B, Tian D, et al. R gene expression induced by a type-III effector triggers disease resistance in rice. Nature 2005;435:1122–1125 [DOI] [PubMed] [Google Scholar]
- 14.White FF, and Yang B. Host and pathogen factors controlling the rice–Xanthomonas oryzae interaction. Plant Physiol 2009;150:1677–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boch J, Scholze H, Schornack S, et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science 2009;326:1509–1512 [DOI] [PubMed] [Google Scholar]
- 16.Moscou MJ, and Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science 2009;326:1501. [DOI] [PubMed] [Google Scholar]
- 17.Cermak T, Doyle EL, Christian M, et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res 2011;39:e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reyon D, Tsai SQ, Khayter C, et al. FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol 2012;30:460–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Briggs AW, Rios X, Chari R, et al. Iterative capped assembly: rapid and scalable synthesis of repeat-module DNA such as TAL effectors from individual monomers. Nucleic Acids Res 2012;40:e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Z, Li J, Huang H, et al. An integrated chip for the high-throughput synthesis of transcription activator-like effectors. Angew Chem Int Ed Engl 2012; 51:8505–8508 [DOI] [PubMed] [Google Scholar]
- 21.Sanjana NE, Cong L, Zhou Y, et al. A transcription activator-like effector toolbox for genome engineering. Nat Protoc 2012;7:171–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmid-Burg JL, Schmidt T, Kaiser V, et al. A ligation-independent cloning technique for high-throughput assembly of transcription activator-like effector genes. Nat Biotechnol 2012;31:76–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sakuma T, Hosoi S, Woltjen K, et al. Efficient TALEN construction and evaluation methods for human cell and animal applications. Genes Cells 2013;18:315–326 [DOI] [PubMed] [Google Scholar]
- 24.Uhde-Stone C, Gor N, Chin T, et al. A do-it-yourself protocol for simple transcription activator-like effector assembly. Biol Proced Online 2013;15:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ding Q, Lee YK, Schaefer EA, et al. A TALEN genome-editing system for generating human stem cell-based disease models. Cell Stem Cell 2013;12:238–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang J, Chao R, Abil Z, et al. FairyTALE: a high-throughput TAL effector synthesis platform. ACS Synth Biol 2014;3:67–73 [DOI] [PubMed] [Google Scholar]
- 27.Nakajima K, and Yaoita Y. Comparison of TALEN scaffolds in Xenopus tropicalis. Biol Open 2013;2:1364–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma AC, Lee HB, Clark KJ, et al. High efficiency in vivo genome engineering with a simplified 15-RVD GoldyTALEN design. PLoS One 2013;8:e65259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neff KL, Argue DP, Ma AC, et al. Mojo Hand, a TALEN design tool for genome editing applications. BMC Bioinformatics 2013;14:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meeker ND, Hutchinson SA, Ho L, et al. Method for isolation of PCR-ready genomic DNA from zebrafish tissues. BioTechniques 2007;43:610–614 [DOI] [PubMed] [Google Scholar]
- 31.Schulte-Merker S, Lee KJ, McMahon AP, et al. The zebrafish organizer requires chordino. Nature 1997;387:862–863 [DOI] [PubMed] [Google Scholar]
- 32.Leung AY, Mendenhall EM, Kwan TT, et al. Characterization of expanded intermediate cell mass in zebrafish chordin morphant embryos. Dev Biol 2005;277: 235–254 [DOI] [PubMed] [Google Scholar]
- 33.Peng Y, Clark KJ, Campbell JM, et al. Making designer mutants in model organisms. Development 2014;141:4042–4054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim H, and Kim JS. A guide to genome engineering with programmable nucleases. Nat Rev Genet 2014;15:321–334 [DOI] [PubMed] [Google Scholar]
- 35.Chandrasegaran S, and Carroll DJ. Origins of programmable nucleases for genome engineering. J Mol Biol 2015;http://dx.doi.org/10.1016/j.jmb.2015.10.014 [DOI] [PMC free article] [PubMed]
- 36.Ma Y, Creanga A, Lum L, et al. Prevalence of off-target effects in Drosophila RNA interference screens. Nature 2006;443:359–363 [DOI] [PubMed] [Google Scholar]
- 37.Scacheri PC, Rozenblatt-Rosen O, Caplen NJ, et al. Short interfering RNAs can induce unexpected and divergent changes in the levels of untargeted proteins in mammalian cells. Proc Natl Acad Sci U S A 2004;101:1892–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robu ME, Larson JD, Nasevicius A, et al. p53 activation by knockdown technologies. PLoS Genet 2007;3:e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kok FO, Shin M, Ni CW, et al. Reverse genetic screening reveals poor correlation between morpholino-induced and mutant phenotypes in zebrafish. Dev Cell 2015;32:97–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Varshney GK, Pei W, LaFave MC, et al. High-throughput gene targeting and phenotyping in zebrafish using CRISPR/Cas9. Genome Res 2015;25:1030–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dahlem TJ, Hoshijima K, Jurynec MJ, et al. Simple methods for generating and detecting locus-specific mutations induced with TALENs in the zebrafish genome. PLoS Genet 2012;8:e1002861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jinek M, Chylinski K, Fonfara I, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012;337:816–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mali P, Yang L, Esvelt KM, et al. RNA-guided human genome engineering via Cas9. Science 2013;339:823–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cho SW, Kim S, Kim JM, et al. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol 2013;31:230–232 [DOI] [PubMed] [Google Scholar]
- 45.Hwang WY, Fu Y, Reyon D, et al. Heritable and precise zebrafish genome editing using a CRISPR–Cas system. PLoS One 2013;8:e68708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang N, Sun C, Gao L, et al. Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res 2013;23:465–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reyon D, Khayter C, Regan MR, et al. Engineering designer transcription activator-like effector nucleases (TALENs) by REAL or REAL-Fast assembly. Curr Protoc Mol Biol 2012;Chapter 12:Unit 12.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Crocker J, and Stern DL. TALE-mediated modulation of transcriptional enhancers in vivo. Nat Methods 2013;10:762–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cong L, Zhou R, Kuo YC, et al. Comprehensive interrogation of natural TALE DNA-binding modules and transcriptional repressor domains. Nat Commun 2012;3:968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maeder ML, Angstman JF, Richardson ME, et al. Targeted DNA demethylation and activation of endogenous genes using programmable TALE–TET1 fusion proteins. Nat Biotechnol 2013;31:1137–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mendenhall EM, Williamson KE, Reyon D, et al. Locus-specific editing of histone modifications at endogenous enhancers. Nat Biotechnol 2013;31:1133–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thanisch K, Schneider K, Morbitzer R, et al. Targeting and tracing of specific DNA sequences with dTALEs in living cells. Nucleic Acids Res 2014;42:e38. [DOI] [PMC free article] [PubMed] [Google Scholar]