Abstract
An increasing number of academic and private biobanks store faecal samples for research and stool transplantation purposes. As stool samples are unique from other biological tissues and materials, stool biobanks require appropriate regulation and additional research on gut microbiota to ensure safe application in the clinic.

Subject Categories: Microbiology, Virology & Host Pathogen Interaction; S&S: Health & Disease; S&S: Politics, Policy & Law
A “microbiome” is the entire collection of microorganisms, their genomes and their metabolic interactions in a specifically defined environment. For example, the microorganisms in the human digestive tract are often collectively referred to as the “gut microbiome”. It is a vast microbial “ecosystem” in terms of biological diversity—the number of bacterial cells in the gut outnumber the cells in the human body by 10‐fold and their genes outnumber human genes by 100‐fold—and its composition and dynamics are influenced by interactions with the host, including the host genome and metabolism, diet, lifestyle, health complications, and antibiotic and environmental exposure 1, 2. The gut microbiome in turn influences many human metabolic and other functions such as energy production, body temperature, reproduction and tissue growth; it is a “hidden organ” because the many functions it serves are equivalent or in some cases more important than other organs 1. Microbes in the gut can also affect human behaviour and health by controlling the development and function of immune, gastrointestinal, and neurological systems through a gut–brain link 3, 4, 5.
Interruption of the gut microbial system is called “dysbiosis”, and it may increase the risk of various diseases including inflammatory bowel diseases, colon cancer, irritable bowel syndrome, liver disease, obesity and metabolic syndromes 2. The proper function of the gut microbiota therefore depends on a “healthy” balance of its members and their interaction with the host. The presence or absence of key species—mostly Bacteroidetes and Firmicutes—will not only elicit specific metabolic responses, but also play an important role in ensuring homeostasis of the entire gut 2. Moreover, the gut microbiome is highly dynamic: Wu et al 6 observed that some genera such as Bacteroides, Prevotela and Ruminococcus have the ability to form clusters that can quickly change and adapt in response to dietary changes.
Various clinical studies have demonstrated that transplanting microbial communities from healthy individuals to patients with a dysbiotic gut microbiota (commonly known as faecal microbiota transplantation; FMT) could become a viable therapeutic option for treating some diseases, including infection with antibiotic‐resistant Clostridium difficile or for restoring cancer patients' gut microbiota after chemotherapy or radiation 7.
The collection and storage of gut microbes along with their genetic and metabolic profiles is therefore becoming increasingly important for therapeutic approaches to a wide range of diseases. This can be termed “biobanking of gut microbes”, which is similar to the banking of biological samples and tissues such as blood, sperm, organs and even gene banks. Such “poo banks” or “stool banks” have already shown some promise in treating patients with gut disorders, especially those infected with Clostridium difficile (http://www.openbiome.org/about). This approach has not only eliminated practical barriers in FMT but also contributes to translational research into the human gut microbiome. Going further, Khoruts et al 7 argue that the dynamic nature of the human microbiome determines its metabolic applications; manipulating the microbiome in a deliberate and controlled fashion may eliminate disease and/or restore health.
In addition to the OpenBiome project (Massachusetts, USA), various research institutes and clinics have started banking patients' own stool for future medical use. Some of these stool banks include AdvancingBio (Mather, CA, USA), Stool Bank East (Leiden, The Netherlands), Taymount Clinic (Hitchin, UK) and Melbourne FMT (Melbourne, Australia). The knowledge gained from these banks, their donors and patients will help to optimise and streamline the therapeutic use of stool samples. The availability of next‐generation sequencing, high‐throughput techniques and sophisticated modelling technologies will also make it increasingly feasible to identify and analyse all the microorganisms associated with the human gut together with host genetics and metabolism and to understand how these contribute to changes in the gut environment. This knowledge may then enable medical practitioners to customise therapeutic applications by modulating patients' microbiome and specific host–microbe interactions.
Contemporary methods for manipulating the microbiome include the use of probiotics, prebiotics and a blend of both with symbiotic microorganisms in addition to specific antibiotics. However, given the complexity of the gut environment, there is scepticism in the clinical community about the validity of such practices. In immune‐compromised individuals, probiotics can even inflict poorer health outcomes through bacteremia, fungemia and sepsis 8. Hence, it is promising to use “faecal biobanks” to select clusters of specific genera for each individual patient and his or her condition rather than using general approaches based on administering probiotics or antibiotics.
Gut microbiota biobanks and the accompanying molecular information about its bacterial members may therefore grow into a valuable health resource, but it also presents various unique problems that are distinct from biobanks that store tissue samples, organs or biological information. These challenges include selecting healthy donors, managing and interpreting the considerable amount of biological data that is needed to analyse and select appropriate samples for therapy, externalisation and collaboration with other banks and clinics, and appropriate regulation. The first three tasks are relatively straightforward and would require information and communication technology along with bioinformatics, but the regulatory issues related to donors and patients require more careful consideration. It would require, for instance, a reliable and widely accepted definition of “healthy” microbiota so as to select appropriate donors and prevent inadvertent cross‐infection (e.g. hepatitis or HIV). It raises the question of compensating donors: Should volunteers be rewarded financially or otherwise for giving stool samples? Finally, there are privacy issues for both donors and patients during collection, storage, information management and clinical use (Fig 1). These concerns are potentially more challenging than for tissue or organ banks: Franzosa et al 9 analysed microbial communities from the human body and observed that more than 80% of human individuals could be uniquely identified by their gut microbiome.
Figure 1. Biobanking gut microbiome samples: an overview of processes and regulation.
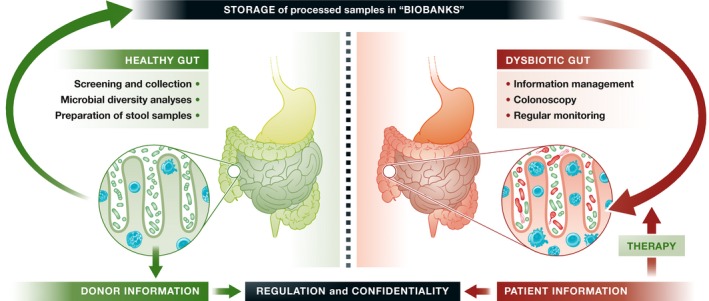
Nonetheless, biobanking the gut microbiome has great potential to safeguard an individual's overall health by restoring a balanced and healthy microbiota. It is therefore a unique and much broader therapeutic approach compared to other biobanks. Broader acceptance by patients and the public at large will, however, depend on proper regulation by health agencies and proper handling of samples and information by both public and private operators. The US Food and Drug Administration (FDA) has recently issued a draft guidance document for using FMT to treat infections with C. difficile that do not respond to standard therapies 10. It defines a stool bank as an establishment that collects, prepares and stores FMT products for distribution to other establishments, healthcare providers or other entities for therapy or clinical research, whereas an establishment that collects or prepares FMT products solely treating their own patients, such as a hospital laboratory, would not be considered a stool bank. This should be the starting point for adequate regulation—not just in the USA but elsewhere—that is needed to both guarantee safety and improve the efficiency of FMT products for therapeutic use.
References
- 1. O'Hara AM, Shanahan F (2006) EMBO Rep 7: 688–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sekirov I, Russell SL, Antunes LCM et al (2010) Physiol Rev 90: 859–904 [DOI] [PubMed] [Google Scholar]
- 3. Paun A, Danska JS (2015) Curr Opin Immunol 37: 34–39 [DOI] [PubMed] [Google Scholar]
- 4. Geurts L, Neyrinck AM, Delzenne NM et al (2014) Beneficial Microbes 5: 3–17 [DOI] [PubMed] [Google Scholar]
- 5. Burokas A, Moloney RD, Dinan TG et al (2015) Adv Appl Microbiol 91: 1–62 [DOI] [PubMed] [Google Scholar]
- 6. Wu GD, Chen J, Hoffmann C et al (2011) Science 334: 105–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Khoruts A, Dicksved J, Jansson JK et al (2010) J Clin Gastroenterol 44: 354–360 [DOI] [PubMed] [Google Scholar]
- 8. Theodorakopoulou M, Perros E, Giamarellos‐Bourboulis EJ et al (2013) Int J Antimicrob Agents 42: S41–S44 [DOI] [PubMed] [Google Scholar]
- 9. Franzosa EA, Huang K, Meadow JF et al (2015) Proc Natl Acad Sci USA 112: E2930–E2938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Federal Register 10632 (2016) FDA Notices 81(40) March 1 2016, Maryland, USA


