Abstract
Telomerase‐mediated telomere elongation provides cell populations with the ability to proliferate indefinitely. Telomerase is capable of recognizing and extending the shortest telomeres in cells; nevertheless, how this mechanism is executed remains unclear. Here, we show that, in the fission yeast Schizosaccharomyces pombe, shortened telomeres are highly transcribed into the evolutionarily conserved long noncoding RNA TERRA. A fraction of TERRA produced upon telomere shortening is polyadenylated and largely devoid of telomeric repeats, and furthermore, telomerase physically interacts with this polyadenylated TERRA in vivo. We also show that experimentally enhanced transcription of a manipulated telomere promotes its association with telomerase and concomitant elongation. Our data represent the first direct evidence that TERRA stimulates telomerase recruitment and activity at chromosome ends in an organism with human‐like telomeres.
Keywords: fission yeast, telomerase, telomere length regulation, TERRA, transcription
Subject Categories: Chromatin, Epigenetics, Genomics & Functional Genomics; RNA Biology
Introduction
The physical ends of linear eukaryotic chromosomes are transcribed into an intricate array of long noncoding RNA species including the G‐rich telomeric RNA TERRA 1, 2. The C‐rich strand of telomeres is used by RNA polymerase II (RNAPII) to template TERRA transcription starting from subtelomeric promoters, which have been so far characterized only in human and murine cells 3, 4, 5. TERRA is evolutionarily conserved and has been documented in mammals, yeasts, birds, plants, and fish 6, 7, 8, 9. Yet, the complexity of the telomeric transcriptome has been best illustrated in the fission yeast Schizosaccharomyces pombe 10, 11, 12. Besides TERRA, fission yeast cells contain ARIA, which is made exclusively of C‐rich telomeric repeats, and two antiparallel RNA species (ARRET and αARRET), which are transcribed from subtelomeres and are devoid of telomeric sequences 10, 11, 12 (Fig 1A). Further exploration of the S. pombe telomeric transcriptome revealed that deletion of the genes encoding the telomere‐associated proteins Rap1 and Taz1 strongly increases all species of the telomeric transcriptome, whereas loss of the heterochromatin factors Swi6 and Clr4 increases ARRET and αARRET levels, while leaving the levels of TERRA and ARIA largely unchanged 10, 11, 12. At least a fraction of TERRA (approximately 10% in humans and fission yeast and the large majority in budding yeast) is 3′ end polyadenylated 6, 11, 13, 14. The polyadenylation state seems to affect TERRA stability and localization within the nucleus. In human cancer cells, non‐polyadenylated TERRA has a half‐life of < 3 h and associates with telomeric chromatin, whereas polyadenylated TERRA is significantly more stable and is found diffusely localized throughout the nucleoplasm 13.
Figure 1. Polyadenylated TERRA largely devoid of telomeric sequences accumulates upon telomere shortening.
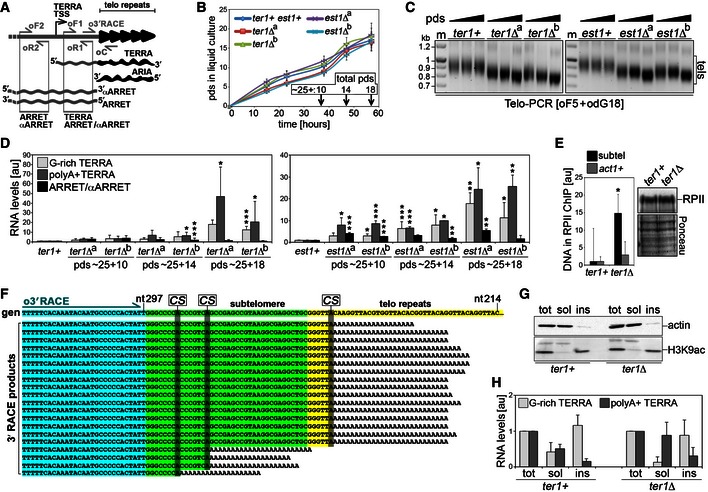
- Schematic representation of the S. pombe telomeric transcriptome. Telomeric repeats are shown in black and subtelomeric sequences in gray. Oligonucleotides used for RT–PCR and for 3′ end RACE are indicated.
- Growth curves of the indicated strains shown as population doublings (pds) on the y‐axis and hours in liquid culture on the x‐axis. Before inoculation in liquid YES medium, single cells underwent approximately 25 pds to produce visible colonies on solid medium. For each strain, averages and SDs (error bars) of three biological replicates are shown. Arrows indicate times of harvesting and corresponding pds for experiments are shown in (C) and (D).
- Telo‐PCR analysis of telomere length in wt (ter1+ and est1+) and two independent clones (a and b) deleted for ter1+ or est1+ grown for ˜25 + 10 (˜35), ˜25 + 14 (˜39), and ˜25 + 18 (˜43) pds in YES medium. PCRs were performed with the subtelomeric oligonucleotide oF5, located 620 bp upstream of the first telomeric repeat, and the oligo(dG)‐containing oligonucleotide odG18. Marker (m) molecular weights are on the left in kilobases. tels: telomere‐containing Telo‐PCR products.
- qRT–PCR analysis of G‐rich TERRA, polyA+ TERRA, and ARRET/αARRET levels in cells as in (B). Values are normalized to ACT1 mRNA and expressed as fold increase over control wt strains (ter1+ and est1+) grown in parallel. Bars and error bars are averages and SD from 4 (ter1Δ clones) or 3 (est1Δ clones) independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 (relative to wt; two‐tailed Student's t‐test).
- ChIP experiments with anti‐RNA polymerase II (RPII) antibodies and extracts from ter1+ and ter1Δ cells grown for ˜43 pds. Quantitative PCRs were performed using oligonucleotides oF1 + oR1 (subtel) or oligonucleotides amplifying a region from the actively transcribed act1+ locus. Values are expressed as fold increase over ter1+. Bars and error bars are averages and SD from three independent experiments. *P < 0.05 (relative to ter1+; two‐tailed Student's t‐test). The Western blot to the right shows no difference in RNPII levels between ter1+ and ter1Δ cells used in ChIP experiments. Ponceau S staining serves as loading control.
- 3′ RACE analysis of polyA+ TERRA in ter1Δ cells grown for ˜43 pds. The sequence at the top is the genomic reference sequence (gen) taken from the pNSU70 plasmid (nucleotide positions are indicated). The sequences below derive from all positive clones isolated. Sequences boxed in blue, green, and yellow correspond to the oligonucleotide used for RACE (o3′RACE), subtelomeric DNA, and telomeric repeats, respectively. Genomic adenines corresponding to putative RNA cleavage sites (CS) are highlighted.
- Western blot analysis of cellular fractions from ter1+ and ter1Δ cells grown for ˜43 pds. Total proteins (tot) and equivalents of soluble (sol) and insoluble (ins) proteins were loaded. Actin serves as a control for soluble proteins and H3K9ac for chromatin‐bound proteins.
- qRT–PCR analysis of total TERRA and polyA+ TERRA in cellular fractions as in (G). Values are expressed after normalization to total cellular extracts. Bars and error bars are averages and SD from 3 independent experiments.
The functions that TERRA exerts at telomeres are still elusive, primarily due to the lack of robust and reproducible laboratory tools that deregulate TERRA levels and/or transcription in cells. Nevertheless, it is becoming evident that TERRA participates in telomere length maintenance in different cellular settings. In cells that elongate telomeres through homologous recombination‐based mechanisms (ALT cells, for alternative lengthening of telomeres), TERRA renders telomeres recombinogenic by forming RNA:DNA hybrids with the C‐rich telomeric strand. Telomeric hybrids are negatively regulated by the RNA endonuclease RNaseH1, and altering the levels of this enzyme in ALT cells is detrimental for telomere maintenance 15, 16, 17, 18, 19. Human and budding yeast TERRA has also been found to physically interact with telomerase 20, 21, the specialized reverse transcriptase that synthesizes de novo telomeric sequences to extend the 3′ ends of chromosomes, suggesting a possible functional interplay between these two factors. However, a unified model for the effect of TERRA on telomerase is still lacking, due in part to seemingly contradictory data from independent groups. In vitro TERRA inhibits human telomerase activity in a way that is counteracted by the RNA‐binding protein hnRNPA1 7, 20, 22. Nevertheless, increased transcription of experimentally modified budding yeast telomeres promotes their shortening in a telomerase‐independent manner 6, 23, 24, 25, challenging the hypothesis that TERRA restricts telomerase activity in vivo. Similar experiments performed in human cells could not reveal any effects exerted by TERRA transcription on telomere length, although subtle changes would be undetectable due to the very long telomeres of the cells employed 26. More recently, live‐cell imaging of tagged TERRA molecules in budding yeast uncovered an intriguing cellular scenario where TERRA transcribed from very short telomeres diffuses in the nucleoplasm where it forms clusters with the telomerase RNA TLC1 in early S phase; later on, TERRA, together with telomerase, moves back to the telomere from which it was transcribed, implying that TERRA might recruit telomerase to telomeres 21. These data would argue that TERRA positively regulates telomerase activity at telomeres, a hypothesis consistent with the observation that cellular levels of TERRA are inversely proportional to telomere length in both budding yeast and human cancer cells 21, 27. Here, we shed light on the functional interaction between TERRA and telomerase using S. pombe, which combines the amenity of yeast manipulation with telomeres that closely resemble the ones of humans.
Results
Telomere shortening stimulates TERRA transcription in S. pombe
To test the existence of a cross talk between TERRA and telomere length in fission yeast, we analyzed the levels of TERRA in response to telomere shortening by deleting ter1+, encoding the RNA moiety of telomerase 28, 29, or est1+, encoding a non‐catalytic, accessory subunit of the telomerase holoenzyme 30. Independent colonies from transformation plates were inoculated in liquid medium, and genomic DNA and total RNA were extracted after 10, 14, and 18 population doublings (pds) in liquid culture (Fig 1B). Considering that transformed cells underwent approximately 25 pds on plate, the final number of pds are estimated to be ~25 + 10 (~35), ~25 + 14 (~39), and ~25 + 18 (~43). Telomeric PCR (Telo‐PCR) experiments on genomic DNA confirmed that telomeres became progressively shorter in independent ter1Δ and est1Δ clones (Fig 1C). However, telomeres were still long enough to support normal proliferation because all the strains were growing at similar rates at the times of cell harvesting (Fig 1B). RNA was reverse‐transcribed using an oligonucleotide complementary to the telomeric stretch within TERRA (oligonucleotide oC; Fig 1A) or an oligo(dT) oligonucleotide (odT). While oC primes reverse transcription of all TERRA molecules containing G‐rich telomeric repeats (herein referred to as G‐rich TERRA), odT will specifically reverse transcribe polyadenylated TERRA (referred to as polyA+ TERRA). cDNA was subjected to real‐time quantitative PCR (qPCR) with oligonucleotides spanning a subtelomeric fragment downstream of the TERRA transcription start site (TSS) (oligonucleotides oF1 and oR1; Fig 1A). We detected a progressive signal increase over successive pds in both cDNA samples, leading to a 20‐ to 50‐fold increase in G‐rich and polyA+ TERRA levels over wt controls at the latest passages (Fig 1D). Because ARRET and αARRET species are 3′ polyadenylated and they both span a portion of the subtelomeric region comprised between TERRA TSS and the telomeric tract 11, PCR amplification products obtained using oF1 and oR1 could derive from TERRA, ARRET/αARRET, or both (Fig 1A). We therefore repeated qPCR measurements using odT reverse‐transcribed cDNA and oligonucleotides placed immediately upstream of TERRA TSS (oligonucleotides oF2 and oR2; Fig 1A), which specifically amplify ARRET and αARRET sequences not shared by TERRA, and detected only a very modest increase in PCR products (Fig 1D). Thus, shortening of fission yeast telomeres provokes stabilization of TERRA transcripts, some of which are polyadenylated, without substantially affecting ARRET/αARRET cellular levels. Chromatin immunoprecipitation (ChIP) experiments using cross‐linked extracts prepared from wt and ter1Δ cells grown for ~43 pds indicated that RNAPII bound more avidly to chromosome ends in mutant cells, while RNAPII binding to the act1+ locus remained unchanged (Fig 1E). This indicates that TERRA accumulation in cells with short telomeres derives at least in part from increased transcription.
PolyA+ TERRA is largely devoid of telomeric repeats and is enriched in soluble cellular fractions
To characterize the molecular features of polyA+ TERRA, we sequenced products of 3′ end RACE experiments performed on poly(A)+ cDNA prepared from wt and ter1Δ cells grown for ~43 pds. To minimize the chances to amplify sequences from αARRET RNA, we performed PCRs using a subtelomeric oligonucleotide placed only 54 bp away from the first telomeric repeat (Fig 1A and F). We were unable to identify TERRA species in RACE products from wt cDNA, most likely due to the very low abundance of polyA+ TERRA in wt cells. On the contrary, we identified several products from ter1Δ cDNA containing subtelomeric sequences followed by poly(A) tracts (Fig 1F). The majority of the identified polyA+ TERRA molecules extended for only 6 nucleotides within the telomeric tract on the corresponding genomic sequence, while the remaining molecules terminated more internally (Fig 1F). In all cases, the poly(A) stretch began at genomic adenines, which likely correspond to alternative RNA cleavage sites (Fig 1F). While we do not exclude the existence of polydenylated TERRA molecules containing long telomeric sequences, our analysis strongly implies that, at least in ter1Δ cells, the majority of polyA+ TERRA is largely devoid of telomeric repeats. The fact that we could sequence polyA+ TERRA 3′ ends only in ter1Δ samples is consistent with its accumulation in the mutant (Fig 1D) and further confirms that the sequenced products do not correspond to αARRET, as this RNA did not accumulate in ter1Δ cells (Fig. 1D).
We next analyzed the intracellular distribution of TERRA by fractionating wt and ter1Δ cell extracts into soluble and insoluble components. Chromatin was specifically enriched in insoluble fractions as shown by Western blot detection of histone H3 acetylated at lysine 9 (H3K9ac), while actin was almost exclusively detected in soluble fractions (Fig 1G). qRT–PCR analysis of RNA from such fractions indicated that G‐rich and polyA+ TERRA were enriched in insoluble and soluble fractions, respectively (Fig 1H). We conclude that, similar to what was previously observed in human cancer cells 13, polyadenylated TERRA is primarily released from telomeric chromatin and can likely diffuse throughout the nucleoplasm.
PolyA+ TERRA physically associates with telomerase
Whether TERRA associates with telomerase in fission yeast was not previously explored. We performed RNA immunoprecipitations (RIPs) using cross‐linked extracts from cells expressing a C‐terminally myc‐tagged version of Trt1 (Trt1‐myc), the catalytic subunit of telomerase 31. RNA from input and immunoprecipitated material was reverse‐transcribed with oC or odT oligonucleotides, followed by TERRA qPCRs with oF1 and oR1 oligonucleotides (Fig 1A). While no significant accumulation of PCR products was detected over untagged strain controls in oC samples, we measured a ~10‐fold increase over controls upon reverse transcription with odT (Fig 2A). No enrichment was detected when PCRs were performed using odT cDNA and ARRET/αARRET‐specific oF2 and oR2 oligonucleotides (Figs 1A and 2A). Confirming the specificity of our RIPs, the telomerase RNA TER1 was found to be associated with Trt1‐myc, while the unrelated messenger RNA ACT1 was not (Fig 2A). Together with our 3′ end RACE results (Fig 1F), these data reveal that polyA+ TERRA forms readily detectable complexes with telomerase, while G‐rich TERRA, ARRET, and αARRET do not. DNaseI treatment of immunocomplexes bound to beads prior to RNA extraction did not affect the yields of co‐immunoprecipitated polyA+ TERRA (Fig 2B), strongly suggesting that DNA does not mediate TERRA association with Trt1‐myc.
Figure 2. Polyadenylated TERRA interacts with telomerase.
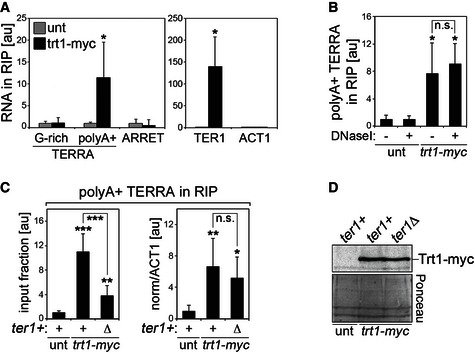
- RIP experiments were performed using anti‐myc antibodies and extracts from Trt1‐myc‐expressing strains followed by qRT–PCR to detect G‐rich TERRA, polyA+ TERRA, ARRET, TER1, and ACT1. Values represent fractions of input RNA detected in immunoprecipitated material expressed as fold increase over untagged (unt) wt strain. Bars and error bars are averages and SD from three independent experiments. *P < 0.05 (relative to unt; two‐tailed Student's t‐test).
- RIP experiments as in (A) where, prior to RNA extraction, washed beads were resuspended in DNaseI buffer (DNbuff) and incubated for 1 hour at 30°C in the presence or absence of DNaseI. Values represent fractions of input polyA+ TERRA detected in immunoprecipitated material expressed as fold increase over an untagged (unt) wt strain. Bars and error bars are averages and SD from three independent experiments. *P < 0.05 (relative to unt; two‐tailed Student's t‐test).
- RIP experiments were performed using anti‐myc antibodies and extracts from Trt1‐myc‐expressing strains (either ter1+ or ter1Δ) cultured in YES medium for ˜43 pds. Values represent polyA+ TERRA detected in immunoprecipitated material expressed as a fraction of the input (graph on the left) or after normalization to ACT1 RNA in the corresponding input (on the right), and expressed as fold increase over an untagged wt strain (unt ter1+). Bars and error bars are averages and SD from three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 (relative to unt ter1+; two‐tailed Student's t‐test).
- Western blot analysis of Trt1‐myc in cells used in (C). Ponceau S staining serves as a loading control.
Deletion of ter1+ decreased the amount of polyA+ TERRA associated with Trt1‐myc when expressed as a fraction of the input (Fig 2C). However, this decrease can be explained as a consequence of the higher amount of polyA+ TERRA found in mutant cells, as normalizing to the stable ACT1 mRNA in the corresponding inputs reversed this effect (Fig 2C). It is worth mentioning that TERRA induction in ter1Δ Trt1‐myc cells was not as strong as in the one observed in ter1Δ cells with unmodified trt1+ locus (Fig 1D). This is likely due to already higher levels of TERRA in ter1+ Trt1‐myc control cells due to the fact that myc tagging of Trt1 is associated with telomeres that are shorter than in wt cells, yet stable and functional 31. Finally, ter1+ deletion did not affect the levels of Trt1‐myc in cells (Fig 2D). We conclude that TER1 is dispensable for the TERRA/Trt1‐myc interaction and that globally increasing the cellular pool of polyA+ TERRA is not sufficient to increase TERRA/Trt1‐myc complexes, suggesting that Trt1 levels may be limiting.
Generation of S. pombe strains carrying “transcriptionally inducible” telomeres (tiTELs)
To directly modulate transcription at fission yeast telomeres, we generated a strain carrying “transcriptionally inducible telomeres” (tiTELs). We inserted a thiamine repressible nmt1 + gene promoter (Pnmt1 32) 91 bp upstream of the endogenous TERRA transcription start site (Fig 3A). Hybridization of NotI‐digested chromosomes separated by pulsed‐field gel electrophoresis with DNA probes spanning the last 150 bp of Pnmt1 (nmt1 probe in Fig 3A) revealed that the manipulated strain contained two Pnmt1 promoters inserted at the subtelomeres of the right arm of chromosome I and of the left arm of chromosome II (Fig EV1A). Clones with single insertions were successively isolated by backcrossing (Fig EV1A). Pnmt1 insertion eliminated the endogenous ApaI restriction site originally present 37 bp upstream of natural telomeres and generated a new HindIII restriction site 492 bp upstream of tiTEL telomeric tracts (Fig 3A). Therefore, hybridization of ApaI‐digested genomic DNA with nmt1 probes detected a band comprised between 4 and 10 kb present only in tiTEL and not in wt samples; this band corresponds to tiTEL‐containing fragments retained in the upper part of the gel (Fig EV1B). Hybridization of the same blots with telomeric probes produced a diffuse hybridization signal comprised between ~400 and 150 bp in both wt and tiTEL strains (compare untreated samples in Fig EV1B); this signal corresponds to Pnmt1‐less natural telomeres (herein referred to as nTELs) released in the lower part of the gel. Hybridizations of HindIII‐digested genomic DNA with nmt1 probes visualized tiTELs as a smear of ~600–900 bp migrating in the lower part of the gel (Fig 3B). Because the newly inserted HindIII site is 492 bp upstream of the tiTEL telomeric tract, the telomeric repeat array within tiTELs is comprised between ~200 and 400 bp in length, similarly to nTELs. Hybridization of the same HindIII‐digested genomic DNA with a telomeric probe visualized not only tiTEL telomeric sequences released in the lower part of the gel, but also telomeric sequences between ~1,000 and 1,500 bp in the tiTEL strain and between ~600 and 900 bp in the wt strain (nTELs REL in Fig 3B). These telomeric sequences, which belong to nTELs carrying a polymorphic HindIII site not present on the other nTELs, allow simultaneous visualization of tiTELs and nTELs on the same membrane as two populations of distinct sizes.
Figure 3. Characterization of transcriptionally inducible telomeres (“tiTELs”).
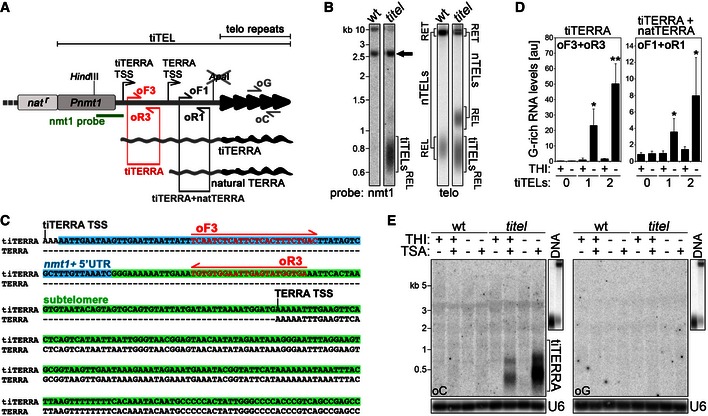
- Schematic representation of tiTELs. An nmt1 promoter (Pnmt1) was inserted upstream of the natural TERRA TSS. Telomeric repeats are in black and the subtelomeric tract in gray. A cassette conferring resistance to nourseothricin (nat r) was used to select for positively transformed cells. The inserted cassette generates a novel HindIII restriction site, while the endogenous ApaI restriction site close to the telomeric tract is disrupted upon integration. Probes and oligonucleotides used for Northern blot, Southern blot, and RT–PCR analyses are indicated. The sketch is not to scale.
- Genomic DNA from wt cells (CAF13) and cells carrying two tiTELs (CAF110) maintained in YES medium was digested with HindIII and Southern blot‐hybridized consecutively with the indicated probes. RET refers to telomeric sequences (tiTELs or natural telomeres, nTELs) retained in the upper part of the gel; REL refers to telomeric sequences released in the lower part of the gel. The black arrow points to the natural nmt1+ sequence on chromosome III. Marker molecular weights are on the left in kilobases.
- 5′ RACE analysis of the tiTERRA transcription start site using total RNA from cells carrying two tiTELs (CAF110) maintained in YES medium. Nucleotides boxed in blue overlap with the 5′ UTR of the nmt1+ transcript, while the ones in green correspond to subtelomeric sequences. tiTERRA and TERRA transcription start sites (TSS) are indicated.
- qRT–PCR analysis of tiTERRA in cells carrying no tiTEL (CAF13), one tiTEL (CAF545), or two tiTELs (CAF110) cultured in EMM in the presence or absence of thiamine (THI; repressed and induced condition, respectively) for 24 h. RNA was reverse‐transcribed with oC, and cDNA was amplified with oF3 and oR3 oligonucleotides to detect only tiTERRA (graph on the left) or with oF1 and oR1 oligonucleotides to detect simultaneously tiTERRA and natural TERRA (on the right). Values are normalized to ACT1 mRNA and expressed as fold increase over CAF545 in uninduced conditions (THI+). Bars and error bars are averages and SD from 3 independent experiments. *P < 0.05, **P < 0.01 (relative to CAF545 THI+; two‐tailed Student's t‐test).
- Northern blot analysis of total RNA from wt cells and cells carrying two tiTELs (CAF110) treated with combinations of THI and trichostatin A (TSA) for 24 h. Two identical membranes were hybridized in parallel with oligonucleotides corresponding to C‐rich (oC) or G‐rich (oG) telomeric repeats. To confirm similar hybridization efficiencies of the two oligonucleotides, Northern blot membranes were simultaneously hybridized and exposed along with blots of digested genomic DNA (upper right insets). After signal detection, RNA membranes were stripped and rehybridized with U6 probes to assure equal loading. Marker molecular weights are on the left in kilobases.
Figure EV1. Genomic organization and functional characterization of tiTELs.
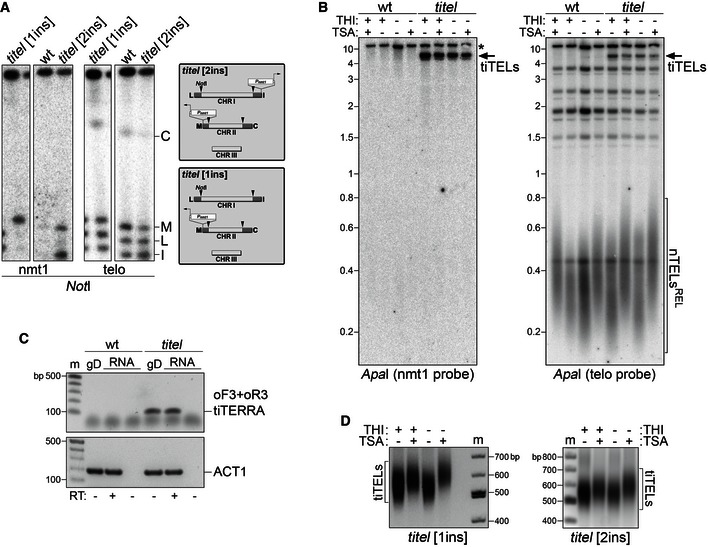
- Pulsed‐field gel electrophoresis (PFGE) analysis of Pnmt1 insertion (ins) in tiTEL strains. Genomic DNA from wt cells (CAF13) and cells carrying one tiTEL (CF545) or two tiTELs (CAF110) maintained in YES medium was digested with NotI, separated by PFGE, and hybridized sequentially using nmt1 and telomeric probes. The positions and nomenclature of the four NotI terminal restriction fragments entering the gel are indicated on the right. Genomic organization of tiTEL strains is sketched on the right.
- Genomic DNA was extracted from wt (CAF13) cells and cells carrying two tiTELs (CAF110) cultured for 24 h in EMM with THI and TSA as indicated. DNA was digested with ApaI and hybridized sequentially using nmt1 and telomeric probes. The asterisk indicates a band corresponding the endogenous nmt1+. TiTELs and natural telomeres (nTELs) are indicated. REL refers to telomeric sequences released in the lower part of the gel. Marker molecular weights are on the left in kilobases.
- Control RT–PCR experiments showing the specificity of oF3 + oR3 oligonucleotides. Nucleic acids were prepared from wt cells (CAF13) and cells carrying one tiTEL (CAF545) cultured for 24 h in EMM without THI. gD indicates genomic DNA, RT indicates whether or not samples were treated with reverse transcriptase. ACT1 PCR was performed to assure presence of genomic DNA and cDNA templates. Marker (m) molecular weights are on the left in base pairs.
- Telo‐PCR analysis of tiTELs using genomic DNA from cells carrying one tiTEL (CAF545, on the left) or two tiTELs (CAF110, on the right) cultured for 24 h in EMM with THI and TSA as indicated. Marker (m) molecular weights are shown in base pairs.
The transcriptional activity of tiTELs was first monitored by 5′ end RACE using total RNA from cells grown in complete YES medium. Sequencing of independent RACE products identified the previously described TERRA TSS 11 as well as a novel transcript containing an additional 164 bases at its 5′ end. Of these 164 bases, the first 72 are derived from the nmt1+ mRNA 5′ UTR and the remaining 92 bases are identical to the subtelomeric sequence immediately upstream of natural TERRA TSS (Fig 3C). We named this novel transcript tiTERRA, for “transcriptionally inducible TERRA”. TERRA and tiTERRA TSSs corresponded to adenines followed by the sequence AAAATT (Fig 3C). We then extracted RNA from wt and tiTEL cells grown in EMM in the presence or absence of thiamine (THI; repressed and induced condition, respectively) for approximately 8 pds and performed qRT–PCRs using oC oligonucleotides for reverse transcription. To specifically detect G‐rich tiTERRA and not endogenous TERRA, we used PCR oligonucleotides encompassing the junction between the 5′ UTR of nmt1+ and the downstream subtelomeric sequence (oligonucleotides oF3 and oR3; Fig 3A and C). Control experiments confirmed the specificity of oF3 + oR3 PCR (Fig EV1C). Compared to a strain carrying one tiTEL grown in THI+ medium, THI withdrawal induced a ~20‐fold increase in G‐rich tiTERRA in the same strain and a ~50‐fold increase in a clone carrying two tiTELs (Fig 3D). As expected, no amplification signal was detected in wt cells (Fig 3D). We also amplified the same cDNA with oF1 and oR1 oligonucleotides to simultaneously measure G‐rich tiTERRA and TERRA from natural chromosome ends (Fig 3A). In uninduced conditions, G‐rich TERRA levels were similar in wt and tiTEL samples; THI withdrawal increased G‐rich TERRA by ~four‐ and eightfold in cells with 1 and 2 tiTELs, respectively, over wt (Fig 3D). Altogether, these results show that (i) tiTEL transcription can be induced simply by withdrawing THI; (ii) each tiTEL likely produces G‐rich tiTERRA with similar efficiencies in strains with one or two tiTELs; (iii) cellular levels of G‐rich tiTERRA are comparable to endogenous TERRA in uninduced conditions; and (iv) considering that fission yeast has 6 chromosome ends, full tiTEL induction increases telomere transcription within fairly physiological ranges.
Endogenous TERRA in wt cells is hardly detectable by Northern blot hybridization due to its low abundance 11. We previously showed that human TERRA levels are suppressed by histone deacetylases (HDACs) 14. Moreover, fission yeast strains deleted for rap1+ or its regulator cay1+ accumulate histone H3K9ac at the TERRA TSS and fail to properly suppress TERRA transcription 33. To test how histone acetylation impacts tiTERRA production, we treated tiTEL cells with combinations of THI and the HDAC inhibitor trichostatin A (TSA) and performed Northern blot hybridization of total RNA with strand‐specific oC and oG telomeric oligonucleotide probes (Fig 3A). No clear signal was detected in the absence of TSA treatment (Fig 3E), indicating that tiTERRA levels remain low even in induced conditions. Addition of TSA in the presence of THI produced the appearance of G‐rich RNA species shorter than 500 bases in length, while TSA treatment in the absence of THI induced a further (> fivefold) accumulation of slightly longer G‐rich RNA molecules (Fig 3E). No specific hybridization signal was obtained in wt cells in any of the tested conditions, indicating that the G‐rich RNA detected in tiTEL strains treated with TSA derived from tiTERRA. TiTERRA induction was not accompanied by detectable stabilization of ARIA molecules in hybridizations with oG oligonucleotides (Fig 3E).
tiTERRA transcription leads to telomerase‐mediated tiTEL elongation
To determine whether tiTERRA transcription impacts telomere elongation, we analyzed tiTEL lengths in cells cultured with different combinations of THI and TSA for 24 h. Southern blot hybridizations of HindIII‐digested DNA did not disclose any obvious differences in tiTEL and nTEL lengths among samples grown with or without THI in the absence of TSA (Fig 4A, compare lanes 5 and 7). Similarly, no apparent effect was exerted by THI on the length of ApaI‐released nTELs in both wt and tiTEL strains (Fig EV1B). By contrast, the length of HindIII‐ and ApaI‐released tiTELs and nTELs increased by approximately 100 bp in THI+ TSA+ samples (Fig 4A, compare lanes 5 and 6; Fig EV1B). In THI− TSA+ samples, tiTELs, but not nTELs, underwent a further ~100‐bp elongation as compared to THI+ TSA+ cells (Fig 4A, compare lanes 6 and 8; Fig EV1B). Analogous elongation dynamics in response to THI and TSA were observed by Telo‐PCR in strains with 1 or 2 integrated tiTELs ruling out the possibility of clone‐specific responses (Fig EV1D). These data show that tiTEL transcription promotes its elongation in cis. Importantly, transcription‐dependent elongation of tiTELs in THI− TSA+ samples was telomerase dependent as it was prevented by deletion of trt1+ (Fig 4B, compare lanes 6 and 8). On the contrary, tiTELs and nTELs were still slightly longer in trt1Δ cells treated with TSA, regardless of the presence of THI (Fig 4B, compare lanes 5, 6, and 8). This could indicate that TSA alone promotes telomere elongation independently of telomerase or that replicative telomere shortening in TSA‐treated trt1Δ cells occurred more slowly than in untreated counterparts. Indeed, while THI did not affect cell growth with all cells undergoing ~8 pds in liquid culture in 24 h in the absence of TSA regardless of ter1+ status, TSA treatment decreased this to ~5 pds. Deletion of the genes coding for the homologous recombination factor Rad51 or the DNA exonuclease Exo1 did not suppress TSA‐induced telomere elongation nor transcription‐induced tiTEL elongation in cells grown in THI− TSA+ medium (Fig EV2). Hence, Rad51 and Exo1 do not play major roles in these molecular pathways of telomere lengthening.
Figure 4. TiTERRA transcription induces telomerase‐dependent tiTEL lengthening.
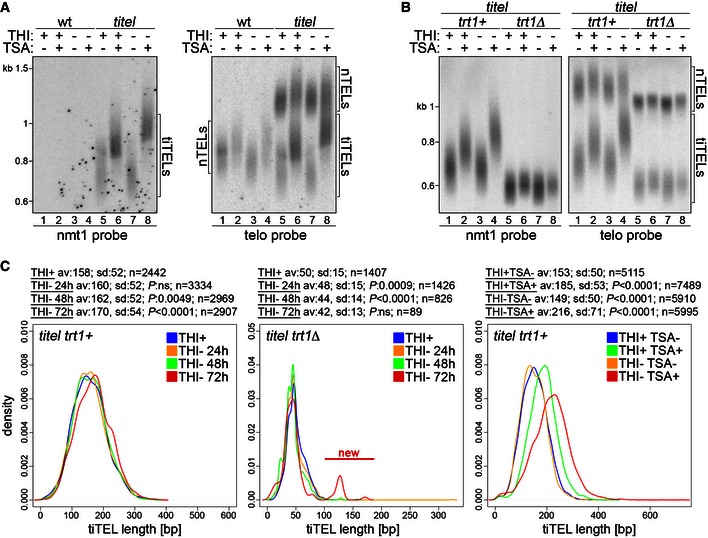
- Telomere restriction fragment analysis of genomic DNA from wt cells and cells carrying two tiTELs (CAF110) grown for 24 h in EMM containing THI and TSA as indicated. DNA was digested with HindIII and hybridized first with a nmt1 probe and successively with a telomeric probe. Marker molecular weights are on the left in kilobases. Numbers at the bottom indicate gel lanes.
- Genomic DNA from cells carrying two tiTELs and deleted for trt1+ (CAF113) was analyzed as in (A). Marker molecular weights are on the left in kilobases. Numbers at the bottom indicate gel lanes.
- ter1+ cells carrying two tiTELs (CAF110, left panel) or trt1Δ cells carrying one tiTEL (MKSP2104, central panel) were cultured in EMM in the presence of THI for 24 h (THI+) or in the absence of THI for 24, 48, or 72 h. In the right panel, cells carrying two tiTELs (CAF110) were treated with TSA for 24 h in the presence or absence of THI. TiTEL telomeric sequences were amplified from genomic DNA and sequenced using a PacBio platform. The statistical analysis for each set of conditions is displayed above each panel. P‐values for each comparison were derived from the Student's t‐test between each condition and the THI+ sample using Welch's correction when appropriate. The new population of longer tiTELs appearing in trt1Δ cells upon prolonged transcription induction is indicated in red (center panel).
Figure EV2. Rad51 and Exo1 are not required for tiTEL elongation.
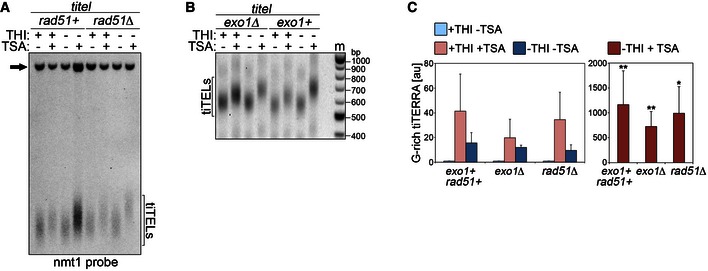
- Genomic DNA was extracted from cells carrying one tiTEL either rad51+ (CAF545) or rad51Δ (CAF550) cultured for 24 h in EMM with THI and TSA as indicated. DNA was digested with HindIII and Southern blot‐hybridized with nmt1 probes. The black arrow points to the nmt1+ sequence on chromosome III.
- Telo‐PCR analysis of tiTELs in cells carrying one tiTEL either exo1+ (CAF545) or exo1Δ (CAF655) cultured for 24 h in EMM with THI and TSA as indicated. Marker (m) molecular weights are on the right in base pairs.
- qRT–PCR analysis of total tiTERRA levels in cells as in (A) and (B). Values are expressed as fold increase over uninduced exo1+ rad51+ strain (CAF545). Bars and error bars are averages and SD from 3 independent experiments. *P < 0.05, **P < 0.01 (relative to uninduced parental; two‐tailed Student's t‐test).
The lack of visible effects on tiTEL length in induced samples in the absence of TSA could derive from the inability of our Southern blot‐based analysis to score mild changes in lengths. We therefore PCR‐amplified and sequenced tens of tiTELs and nTELs from strains grown in the absence of THI for 1 week by adapting the single telomere extension (STEX) assay originally developed in budding yeast 34, 35 for use in fission yeast (Fig EV3A–C). TiTELs from cells grown in the absence of THI were on average 24 bp longer than the ones from cells grown in the presence of THI (n ≥ 65 telomeres, Fig EV3D). On the contrary, the lengths of nTELs from induced and uninduced cells were not significantly different (n ≥ 35 telomeres, Fig EV3D). This shows that tiTERRA transcription promotes tiTEL elongation also in the absence of TSA without affecting the length of other telomeres in trans. To precisely follow the dynamics of tiTEL elongation upon transcription, we grew cells for 24, 48, and 72 h in the absence of THI and sequenced tiTEL STEX products using a PacBio platform. In trt1+ strains, tiTELs progressively lengthened from an average of 158 bp in uninduced conditions to 170 bp after 72 h of induction (n > 2,400 telomeres, Fig 4C). Thus, tiTEL elongation upon tiTERRA induction is a dynamic process established progressively over successive pds. In trt1Δ strains, transcription induction provoked a precipitous loss of telomeric sequences, which limited the number of analyzable tiTELs to only a few hundred in samples grown in the absence of THI for 48 and 72 h. Nevertheless, we did not observe any elongation of bulk tiTELs (Fig 4C), further confirming that telomerase elongates tiTELs in induced conditions. These data also show that telomerase counteracts rapid loss of telomeric sequences when transcription is increased, consistent with previous observations in budding yeast 25. At 72 h post‐induction, we also observed the appearance of a novel small population of tiTELs at approximately 130 bp, while the bulk of tiTELs was around 42 bp (Fig 4C). We suspect that these longer telomeres result from late recombination events triggered by transcription of very short telomeres, as previously shown in budding yeast 18. Finally, next‐generation sequencing of trt1+ tiTELs from cells treated with combinations of THI and TSA for 24 h confirmed the results obtained by Southern blotting (Fig 4A and C): TSA elongated tiTELs already when administrated in uninduced conditions and even more in the absence of THI (Fig 4C).
Figure EV3. Sequencing of tiTELs.
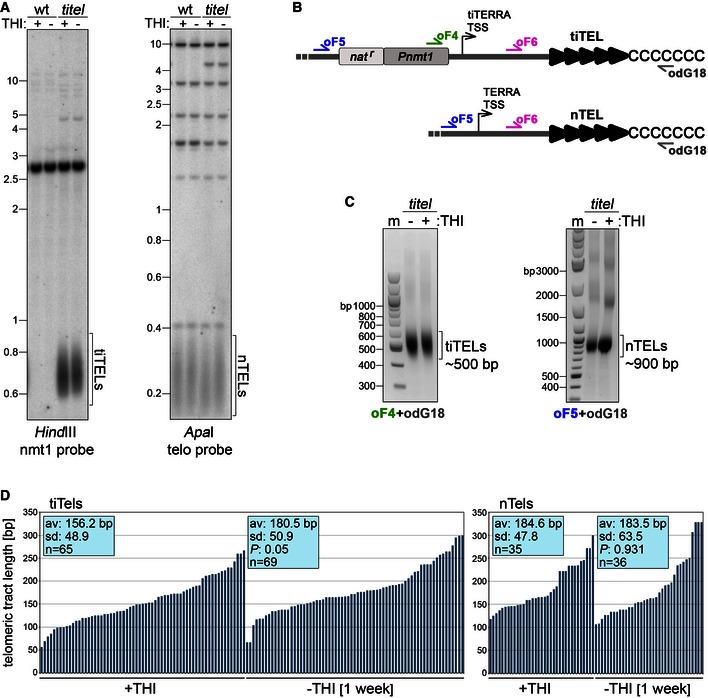
- Genomic DNA was isolated from wt cells (CAF13) and cells carrying two tiTELs (CAF110) cultured for 7 days in EMM with or without THI. DNA was digested and hybridized as indicated. Marker molecular weights are on the left in kilobases.
- Sketch showing the positions of oligonucleotides used for Telo‐PCR amplification and sequencing of tiTELs and natural telomeres from the same cell. Telo‐PCRs were performed using oF4 or oF5 in combination with the G‐rich oligonucleotide odG18. Sequencing was performed with oF6.
- Examples of Telo‐PCR products using genomic DNA from tiTEL cells as in (A) and amplified using the indicated oligonucleotides. Marker (m) molecular weights are on the left in base pairs.
- Telomere PCR products corresponding to tiTELs (on the left) or nTELs (on the right) from tiTEL cells as in (A) were cloned and sequenced. Blue bars represent individual telomeres plotted against their length in base pairs (y‐axis). Average telomere length of the sequenced population (av) and standard deviations (SD) are indicated along with P‐values (−THI vs. +THI) calculated with the two‐tailed Student's t‐test.
TiTERRA transcription stimulates telomerase association with tiTERRA and tiTELs
To better understand how telomerase specifically elongates hyper‐transcribing tiTELs, we generated a strain carrying one tiTEL and expressing Trt1‐myc. In this strain, compared to uninduced THI+ samples, THI withdrawal led to a ~sixfold increase in G‐rich tiTERRA and a ~50‐fold increase in polyA+ tiTERRA; treatments with TSA in THI+ medium led to a ~20‐fold and ~eightfold increase in G‐rich and polyA+ tiTERRA, respectively; combining TSA treatment with THI withdrawal led to a ~150‐fold and ~250‐fold increase in G‐rich and polyA+ tiTERRA, respectively (Fig 5A). Nevertheless, when comparing THI− TSA− with THI− TSA+ samples, G‐rich tiTERRA increased ~25 fold, while polyA+ tiTERRA only ~fivefold (Fig 5A). This suggests that combining forced RNAPII transcription (THI withdrawal) in the presence of TSA might lead to production of polyA+ tiTERRA containing telomeric stretches long enough to permit oC reverse transcription. Telo‐PCR analysis confirmed that tiTELs elongated in THI+ TSA+ and even further in THI− TSA+ conditions also in this strain background (Fig 5B).
Figure 5. TiTERRA transcription induction stimulates telomerase association with tiTERRA and with tiTELs.
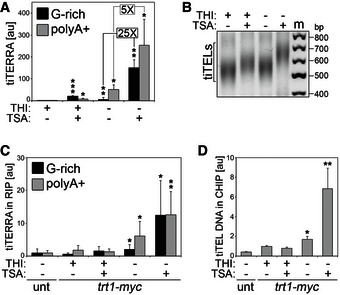
- qRT–PCR analysis of G‐rich and polyA+ tiTERRA in cells carrying one tiTEL and expressing Trt1‐myc (CAF610) cultured in EMM with the indicated combinations of THI and TSA for 24 h. Values are normalized to ACT1 mRNA and expressed as fold increase over THI+ TSA− samples. Bars and error bars are averages and SD from 4 independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 (relative to THI+ TSA−; two‐tailed Student's t‐test).
- Telo‐PCR analysis of tiTELs using genomic DNA extracted from cells as in (A). Marker (m) molecular weights are on the right in base pairs.
- RIP experiments performed using anti‐myc antibodies and extracts from cells as in (A) followed by qRT–PCR analysis of G‐rich and polyA+ tiTERRA. Values correspond to fraction of input RNA expressed as fold increase over an untagged (unt) control tiTEL strain (CAF545). Bars and error bars are averages and SD from three independent experiments. *P < 0.05, **P < 0.01 (relative to unt; two‐tailed Student's t‐test).
- ChIP experiments performed using anti‐myc antibodies and extracts from cells as in (A) followed by qPCR using tiTEL‐specific oligonucleotides. Values correspond to fraction of input DNA expressed as fold increase over THI+ TSA−. Unt: untagged strain (CAF545). Bars and error bars are averages and SD from at least three independent experiments. *P < 0.05, **P < 0.01 (relative to THI+ TSA−; two‐tailed Student's t‐test).
We next performed RIP experiments to examine the interaction between tiTERRA and telomerase. We found that the physical interaction between tiTERRA and Trt1‐myc was enhanced in THI− TSA− conditions; addition of TSA in the absence of THI (but not TSA in THI+ media) promoted a further increase in the tiTERRA‐Trt1‐myc interaction (Fig 5C). We conclude that increasing tiTEL transcription leads to a preferential binding of telomerase to tiTERRA, likely disfavoring binding to natural TERRA from other telomeres. PolyA+ tiTERRA was clearly more enriched than G‐rich tiTERRA in Trt1‐myc pull‐down fractions in THI− TSA− conditions, while in THI− TSA+ the enrichment of the two RNA species was similar (Fig 5C). This is consistent with accumulation of polyA+ tiTERRA containing telomeric repeats in THI− TSA+‐treated cells. As expected, TER1 RNA was found to interact with Trt1‐myc while ACT1 was not (Fig EV4A). ChIP experiments revealed that, coincident with the dynamics of tiTEL lengthening (Figs 4A and C, and 5B), Trt1‐myc bound more avidly to tiTELs in the absence of THI, with a ~twofold increase in binding in THI− TSA− and a ~sevenfold increase in THI− TSA+ conditions as compared to THI+ TSA− samples (Fig 5D). TSA treatment alone did not cause a statistically significant change in Trt1‐myc binding to tiTELs (Fig 5D). Trt1‐myc protein levels remained fairly constant in the different samples (Fig EV4B), ruling out the possibility that the increased binding of Trt1‐myc to tiTERRA and tiTELs is an indirect consequence of Trt1‐myc upregulation in cells. Finally, we generated a tiTEL strain expressing a GFP‐tagged version of the telomeric protein Taz1 and performed ChIPs using anti‐GFP antibodies. Taz1 binding to tiTELs was slightly diminished, although not in a statistically significant manner, when cells were grown in the presence of TSA while it was completely unaffected by THI levels (Fig EV4C and D). It is therefore unlikely that tiTERRA‐mediated tiTEL elongation is a consequence of diminished binding of Taz1 to tiTELs.
Figure EV4. Control experiments for tiTERRA RIPs and binding of Taz1‐GFP to tiTELs.
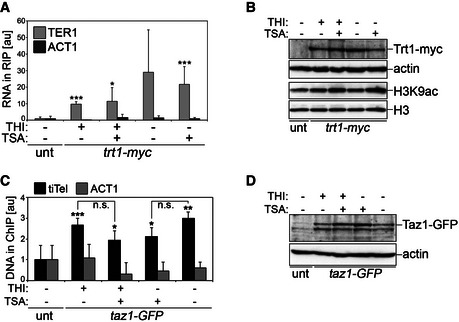
- RIP experiments performed using anti‐myc antibodies and extracts from cells carrying one tiTEL and expressing Trt1‐myc (CAF610) cultured for 24 h in EMM with THI and TSA as indicated. TER1 (positive control) and ACT1 (negative control) in input and immunoprecipitated material were quantified by qRT–PCR. Values correspond to fraction of input RNA expressed as fold increase over an untagged (unt) control tiTEL strain (CAF545). Bars and error bars are averages and SD from three independent experiments. *P < 0.05, ***P < 0.001 (relative to unt; two‐tailed Student's t‐test).
- Western blot analysis of Trt1‐myc and histone H3 acetylated at lysine 9 (H3K9ac) levels in cells as in (A). Actin and total H3 serve as loading controls. The accumulation of H3K9ac in TSA+ cells confirms the effectiveness of TSA. The two upper panels and the two lower panels are from two independent membranes.
- ChIP experiments were performed using anti‐GFP antibodies and extracts from cells carrying one tiTEL and expressing Taz1‐GFP (MKSP1781) cultured for 24 h in EMM with THI and TSA as indicated followed by tiTEL qPCR. ACT1 qPCRs were performed to control for specificity. Values correspond to fraction of input DNA expressed as fold increase over an untagged control tiTEL strain (CAF545). Bars and error bars are averages and SD from three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 (relative to unt; two‐tailed Student's t‐test).
- Western blot analysis of Taz1‐GFP protein levels in cells as in (C). Actin serves as a loading control.
Discussion
Although a connection between TERRA and telomerase has been suggested previously 6, 7, 20, 21, 22, here we offer the first direct evidence that telomere transcription stimulates telomerase‐mediated telomere elongation in cis. Combining our data with previously published work, we propose a unifying model illustrating how telomerase can discriminate between telomeres of different lengths, thereby preferentially elongating the shortest ones 35. In this model (Fig 6), a long telomere is maintained in a “closed state” and produces only limited amounts of TERRA due to low H3K9ac at TERRA promoters and thus low RNAPII‐mediated transcription. TERRA generated by a telomere in a close state is largely non‐polyadenylated and remains associated with telomeric heterochromatin. As long as a closed state is maintained, telomerase accessibility is restricted and the telomere progressively shortens as the cell divides. Once shortened to a critical length, the telomere would shift to an “open state”, characterized by higher H3K9ac levels, increased RNAPII‐mediated transcription and production of polyadenylated TERRA molecules that can freely diffuse in the nucleoplasm. Based on work in budding yeast 21, we propose that nucleoplasmic polyA+ TERRA associates with telomerase and chaperones it back to its telomere of origin (Fig 6). TERRA transcription per se could also increase the accessibility of the telomere to telomerase, for example, by diminishing the density of telomere‐bound telomerase inhibitors, by altering the landscape of telomeric chromatin modifications, or by promoting telomere dynamics within the nucleus 36. Once engaged, telomerase would elongate the telomere and thus re‐establish a silent, closed state (Fig 6).
Figure 6. Speculative model for TERRA's role in mediating telomerase‐dependent elongation of short telomeres.
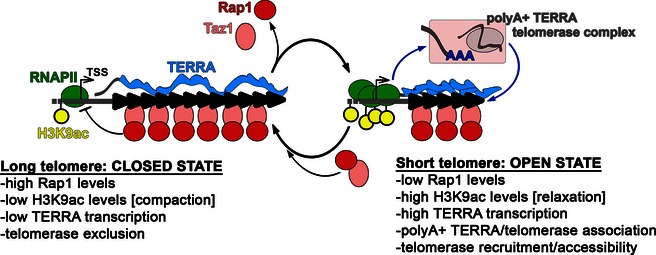
See main text for details.
Our study raises a number of key questions. For example, how is TERRA transcription controlled by telomere length? Rap1 likely plays a major role in this process through regulation of heterochromatin establishment/maintenance at telomeres. Indeed, Rap1 suppresses TERRA transcription at least in part by directly or indirectly restricting H3K9ac at TERRA promoters 11, 33, 37. In normal cells, telomere shortening leads to the release of Rap1 (through release of the Rap1 recruiting factor Taz1), which would alleviate repression of TERRA transcription and allow switching from a closed to an open state. Conversely, telomerase‐mediated telomere elongation would guarantee enough reloading of Taz1 and thus Rap1 molecules to suppress TERRA production (Fig 6). Consistent with this model, rap1Δ cells possess extremely long telomeres, which are maintained by telomerase 38, 39. However, our data indicate that solely increasing TERRA transcription is not sufficient to stimulate telomerase recruitment and activity at telomeres as long as TERRA is not polyadenylated. Indeed, telomerase association with tiTELs and tiTERRA, as well as telomerase‐mediated tiTEL elongation, was induced only when polyA+ tiTERRA production was increased (i.e., in THI− medium). Identifying the nuclear machineries that execute TERRA cleavage and polyadenylation will help to define not only how TERRA and telomerase functionally interact but also how TERRA 3′ end modifications and telomere length are coordinated. In this context, Rap1 could serve an important role, as polyA+ TERRA accumulates in rap1Δ cells 11. Because polyadenylated RNAs can be degraded by the nuclear exosome 40, one could speculate that telomere shortening stabilizes polyA+ TERRA by inhibiting exosome activity. Regardless, the concomitant accumulation of TERRA promoter‐bound RNAPII and cellular polyA+ TERRA in cells with short telomeres argues that an increase in transcription strongly contributes to polyA+ TERRA production.
How short polyadenylated TERRA devoid of telomeric repeats interacts with telomerase is another open question. TERRA was found to be associated with telomerase in both human and budding yeast cell extracts 20, 21. Moreover, in vitro studies proved that human TERRA‐like oligonucleotides comprising UUAGGG repeats bind telomerase by base pairing with the template region of hTR (the RNA component of human telomerase) and by interacting with the hTERT polypeptide (the catalytic subunit) 20, 21. The lack of telomeric repeats in polyA+ TERRA and the fact that telomerase/polyA+ TERRA complexes are not affected by ter1+ deletion suggest that TERRA subtelomeric sequences could also establish interactions with the catalytic subunit of telomerase. Furthermore, the potent inhibition exerted by TERRA on telomerase activity in vitro 7, 20, 22 should not be preserved by polyA+ TERRA that is devoid of telomeric repeats. This suggests the intriguing possibility that different cellular pools of TERRA could have opposing effects on telomerase activity. Finally, induced transcription of tiTELs established in other organisms was not observed to lead to telomere elongation 6, 24, 25, 26. Rather, tiTEL transcription in budding yeast provoked telomerase‐independent, Exo1‐dependent tiTEL shortening 24, 25. Here, we have found that fission yeast Exo1 is not involved in regulating tiTEL length dynamics. This might explain why tiTELs behave differently in budding and fission yeasts and suggests species‐specific roles of TERRA in modulating activities at chromosome ends. Whether TERRA supports Exo1 action at telomeres in organisms other than budding yeast remains to be addressed. Although these and many other riddles await solutions, this study and other reports have established that TERRA is an essential and versatile player in telomere length regulation in different eukaryotes both in telomerase‐positive and in telomerase‐negative (ALT) immortal cells 15, 16, 17, 18, 19. Therapeutic suppression of TERRA and/or telomere transcription may offer the unique opportunity to fight proliferation of cancer cells irrespective of how they achieve telomere maintenance.
Materials and Methods
Schizosaccharomyces pombe strains
Schizosaccharomyces pombe strains used in this study are listed in Table 1 and indicated as CAF or MKSP strains. CAF13 and CAF146 were kind gifts from J. P. Cooper. CAF305 was a kind gift from P. Baumann. CAF656 and CAF657 were generated by replacing the SPNCRNA.214 gene in CAF13 with a natMX6 cassette amplified from the pFA6‐natMX6 plasmid (Euroscarf). CAF658 and CAF659 were generated by replacing SPBC2D10.13 in CAF146 with a natMX6 cassette. CAF110 was generated by subtelomeric insertion of a P3 (full strength) nmt1+ gene promoter amplified from the pFA6a‐natMX6‐P3nmt1 plasmid (Euroscarf). CAF545 was generated by crossing CAF110 to CAF146. CAF610 was generated by crossing CAF545 to CAF305. CAF113 was generated by replacing SPBC29A3.14c in CAF110 with a kanMX6 cassette amplified from the pFA6‐kanMX6 plasmid (Euroscarf). CAF550 was generated by crossing CAF110 to a strain where SPAC644.14c was replaced by a natMX6 cassette. CAF660 was generated by replacing SPNCRNA.214 in CAF305 with a natMX6 cassette. CAF655 was generated by crossing CAF110 to a strain where SPBC29A10.05 was replaced by a ura4 cassette. MKSP1781 was generated by integrating a GFP‐KanR cassette at the SPAC16A10.07c locus in CAF110. All strains were validated for correct genomic locus arrangement by PCR, Southern blotting and/or Western blotting, and appropriate phenotypic analysis.
Table 1.
Schizosaccharomyces pombe strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| CAF13 | h− (strain 972) | J.P. Cooper lab |
| CAF110 | h− natMX6‐P3nmt1‐TEL‐I‐R natMX6‐P3nmt1‐TEL‐II‐L | this study |
| CAF113 | h− natMX6‐P3nmt1‐TEL‐I‐R P3nmt1‐TEL‐II‐L trt1Δ::kanMX6 | this study |
| CAF146 | h+ (strain 975) | J. P. Cooper lab |
| CAF305 | h− ade6‐M210 leu1‐32 ura4‐D18 his3‐D1 trt1‐myc9 | 31 |
| CAF545 | h+ natMX6‐P3nmt‐TEL‐II‐L | this study |
| CAF550 | h− rad51Δ::natMX6 natMX6‐P3nmt1‐TEL‐I‐R | this study |
| CAF655 | h? natMX6‐P3nmt1‐TEL‐I‐R exo1Δ::ura4 | this study |
| CAF610 | h+ trt1‐myc9 natMX6‐P3nmt‐TEL‐II‐L | this study |
| CAF656 | h− ter1Δ::natMX6 clone A | this study |
| CAF657 | h− ter1Δ::natMX6 clone B | this study |
| CAF658 | h+ est1Δ::natMX6 clone A | this study |
| CAF659 | h+ est1Δ::natMX6 clone B | this study |
| CAF660 | h− ade6‐M210 leu1‐32 ura4‐D18 his3‐D1 trt1‐myc9 ter1Δ::natMX6 | this study |
| MKSP2104 | h− natMX6‐P3nmt1‐TEL‐I‐R trt1Δ::kanMX6 | this study |
| MKSP1781 | h− natMX6‐P3nmt1‐TEL‐I‐R natMX6‐P3nmt1‐TEL‐II‐L Taz1‐GFP::kanMX6 | this study |
Yeast culture conditions
Yeasts were grown in yeast extract medium (YES) supplemented with amino acids or Edinburgh minimal medium (EMM) at 30°C. When required, 5 μM thiamine (Sigma‐Aldrich) or 30 μg/ml trichostatin A (Selleckchem) was added to the medium. For growth curves, single colonies from plates were inoculated in liquid medium, and 24 h later, optical density at 600 nm was measured using an Ultrospec 2100 pro spectrophotometer (GE Healthcare). Growth curves were started by diluting cultures to OD600 = 0.05, which corresponded to population doubling (pd) 0. Cells were then grown for up to 60 h with intermediate dilutions to prevent accumulation in stationary phase. OD600 was measured at the indicated time points, and pds were calculated as log2(OD600/0.05) after adjusting OD600 for dilutions. Approximately 25 pds were estimated to be necessary for a single cell to produce a visible colony on solid medium. Graphs were assembled using Microsoft Excel.
RNA isolation and analysis
Total RNA was isolated from exponentially growing cells (OD600: 0.5–1.0) using the hot phenol method 41. For Northern blot analysis, RNA was treated twice with RNase‐free DNaseI (Qiagen), electrophoresed in 1.2% formaldehyde agarose gels, transferred to positively charged nylon membranes (GE Osmonics), and hybridized to radioactively labeled probes at 45°C for 18 h. Oligonucleotide oC and oG probes (Table 2) were 5′ end‐labeled with T4 polynucleotide kinase (New England Biolabs) and [γ‐32P]ATP. Membranes were washed twice in 2× SSC, 0.2% SDS for 20 min and once in 1× SSC, 0.2% SDS at 45°C. Radioactive signals were detected using a Typhoon FLA 9000 Biomolecular Imager (GE Healthcare) and analyzed using ImageJ. Control hybridizations using samples treated with RNase A confirmed that radioactive signals originated exclusively from RNA. For RT–PCR experiments, total RNA was treated three times with RNase‐free DNase I (Qiagen) and reverse‐transcribed using the SuperScript III First‐Strand Synthesis System (Invitrogen) with 10 μM odT or oC plus 1 μM ACT1 or ter1_reverse oligonucleotides (Table 2) as indicated. Quantitative PCR amplification of cDNA was carried out using the KAPA SYBR FAST qPCR Master Mix (Kapa Biosystems) and oligonucleotides indicated in Table 2 with the following cycling parameters: 1 cycle at 98°C for 5 min, followed by 45 cycles at 98°C for 10 s, 60°C for 15 s, and 72°C for 20 s, and a final extension step at 72°C for 5 min. No RT and no template reactions were always included to control for DNA contaminations. 5′ and 3′ RACE experiments were performed using the FirstChoice RLM‐RACE kit (Ambion) as previously described 11. Total RNA was used as starting material, and PCRs were performed using RACE adaptor oligonucleotides supplied with the kit in combination with o5′RACE or o3′RACE oligonucleotides spanning S. pombe subtelomeric sequences (Table 2). PCR products were cloned into the pDrive vector using the Qiagen PCR Cloning Kit. Inserts were PCR‐amplified from independent colonies using the universal primers M13 forward and reverse, which flank the cloning site in pDrive. PCR products were Sanger‐sequenced at Microsynth (Balgach, Switzerland).
Table 2.
Oligonucleotides used in this study
| Name | Sequence (5′ to 3′) | Application |
|---|---|---|
| ACT1 | CATCTGTTGGAAAGTAGAAAGAGAAGCAAG | RT |
| act1_forward | CCGGTATTCATGAGGCTACT | qPCR |
| act1_reverse | GGAGGAGCAACAATCTTGAC | qPCR |
| dG18 | CGGGATCCGGGGGGGGGGGGGGGGGG | Telo‐PCR |
| o5′RACE | GGGCCCAATAGTGGGGGCATTGTATTTGTG | TERRA 5′ RACE |
| o3′RACE | TTTTTCACAAATACAATGCCCCCACTATT | TERRA 3′ RACE |
| oG | GGGTTACAAGGTTACGTGGTTACACGGTTACA | NB |
| oC | TGTAACCGTGTAACCACGTAACCTTGTAACCC | RT, NB |
| odT | TTTTTTTTTTTTTTTTTTTT | RT |
| oF1 | GAAGTTCACTCAGTCATAATTAATTGGGTAACGGAG | qPCR |
| oF2 | GGTTGAATTGAGCGTGGTAGG | qPCR |
| oF3 | TCAATCTCATTCTCACTTTCTGAC | qPCR |
| oF4 | CATTCTCACTTTCTGACTTATAGTCGC | Telo‐PCR |
| oF5 | TGAGTGTGCTGGAGTACGTT | Telo‐PCR |
| oF6 | GTGTGGAATTGAGTATGGTGAA | Sequencing |
| oR1 | GGGCCCAATAGTGGGGGCATTGTATTTGTG | qPCR |
| oR2 | ACTTACTGCACCCTAACGCA | qPCR |
| oR3 | TCACCATACTCAATTCCACACA | qPCR |
| Pnmt1_HindIII_fw | GCGACTATAAGTCAGAAAGTGAGAATG | PCR |
| Pnmt1_HindIII_rev | GCAATGTGCAGCGAAACTAAAA | PCR |
| ter1_forward | CAACGCCCATGCTTAGAAGGTTGA | RT |
| ter1_reverse | TTTCAAATCAATCACCAAGCCCTCA | qPCR |
| U6 | ATGTCGCAGTGTCATCCTTG | NB |
RT, reverse transcription; NB, Northern blot.
Genomic DNA isolation and analysis
Genomic DNA was extracted from exponentially growing yeast cells according to standard protocols. DNA was digested with HindIII or ApaI (New England Biolabs) for at least 16 h at 37°C and electrophoresed in 0.8–1.2% agarose gels. After electrophoresis, DNA was denatured in gel, transferred to nylon membranes (GE Osmonics), and hybridized at 60°C overnight using random primer labeled telomeric or nmt1 probes. The telomeric probe was a 0.5‐kb DNA fragment containing S. pombe telomeric repeats excised from the pIRT2 plasmid (kind gift from J.P. Cooper). The nmt1 probe was a PCR product obtained by amplification of wt genomic DNA using Pnmt1_HindIII_fwd and Pnmt1_HindIII_rev oligonucleotides (Table 2). Pulsed‐field gel electrophoresis analysis was performed as previously described 38 using a CHEF‐DR III apparatus (Bio‐Rad).
Protein preparation and Western blot analysis
Western blot analysis was performed according to standard protocols using total protein extracts prepared by mechanical lysis in the presence of 20% cold TCA. Primary antibodies were as follows: a mouse monoclonal anti‐myc (Cell Signaling Technology, #2276); a rabbit polyclonal anti‐H3K9ac and a mouse monoclonal anti‐RNAPII CTD (Millipore, #07‐352 and #17‐672); a rabbit polyclonal anti‐H3 and a mouse monoclonal anti‐actin (Abcam, #ab1791 and #ab8224); and a mouse monoclonal anti‐GFP (Roche, #11814460001). Secondary antibodies were HRP‐conjugated goat anti‐mouse and anti‐rabbit IgGs (Bethyl Laboratories). Images were acquired using a FluorChem HD2 apparatus (Alpha Innotech).
Cell fractionation
Cells were harvested from 50 ml cultures (OD600: ~1) and washed sequentially with 25 ml of STOP buffer (150 mM NaCl, 50 mM NaF, 10 mM EDTA, 1 mM NaN3, pH 8), with 25 ml of sterile water and with 5 ml of 1.2 M sorbitol. Cells were pelleted at 2,000 g for 4 min and spheroplasted in 2.5 ml of solution A (50 mM (Na)3citrate, 40 mM EDTA, 1.2 M sorbitol) supplemented with 0.2% beta‐mercaptoethanol and 10 mg of lysing enzyme (Sigma‐Aldrich) at room temperature for 30 min. 2.5 ml of solution B (1.2 M sorbitol, 10 mM Tris–HCl, pH 7.5) was then added, and cells were spun at 2,000 g for 4 min. Supernatant was discarded and the pellet was washed twice in 2.5 ml of 1.2 M sorbitol and resuspended in 860 μl of 1.2 M sorbitol plus 100 μl of 10× lysis buffer [500 mM KOAc, 20 mM Mg(OAc)2, 200 mM Pipes‐KOH, pH 6.8, containing RnaseOUT (Invitrogen) and protease inhibitors (Roche)]. After adding 40 μl of 25% Triton X‐100 (final concentration 1%), cells were resuspended gently and a 100‐μl aliquot was set aside for total cell extracts (tot). The remaining suspension was incubated on ice for 15 min and spun at 15,000 g for 20 min at 4°C. 100 μl of supernatant was recovered and used as soluble fraction (sol). The cell pellet was washed once with 1× lysis buffer, spun again, and resuspended in 800 μl of solution C (10 mM HEPES pH 7.9, 1 mM EDTA, 0.5 M NaCl, containing RnaseOUT and protease inhibitors), and a 100‐μl aliquot was used as insoluble fraction (ins). Proteins from the different fractions were prepared by mixing extracts 1:1 with 2× Laemmli buffer and boiling at 95°C for 10 min. RNA was prepared using the TRIzol reagent (Invitrogen).
Chromatin immunoprecipitation (ChIP)
Yeast cells were harvested from 150 ml cultures (OD600: ~1), cross‐linked in 1% formaldehyde for 30 min at 30°C, and successively quenched in 125 mM glycine for 5 min. Cross‐linked material was resuspended in 400 μl of lysis buffer [50 mM HEPES‐KOH pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% Triton X‐100, 0.1% sodium deoxycholate, protease inhibitor cocktail (Roche)] and subjected to mechanical lysis with glass beads using a FastPrep FP220 apparatus (8 times 4.5 m/s for 30 s). Lysates were centrifuged for 30 min at 16,000 g, and pellets were resuspended in 500 μl of lysis buffer followed by sonication in a Bioruptor UCD‐200 apparatus 3 times for 15 min in high mode with 30‐s on and 30‐s off intervals. Sonicated material was centrifuged for 5 min at 10,000 g, and supernatant containing fragmented chromatin was recovered and quantified using the Lowry protein assay. For each IP, 2 mg of proteins was mixed with 2.5 μl of anti‐myc antibody (Cell Signaling Technology, #2276), 2.5 μl of anti‐RNAPII antibody (Millipore, #17‐672), or 1 μg of anti‐GFP antibody (Roche, #11814460001) and incubated on a rotating wheel at 4°C overnight in the presence of 25 μl of protein A‐ and G‐Sepharose beads (GE Healthcare) blocked with bovine serum albumin and sonicated E. coli genomic DNA. Beads were washed 3 times in lysis buffer, once in lysis buffer containing 500 mM NaCl, once in wash buffer (10 mM Tris–HCl, 250 mM LiCl, 0.5% Nonidet P‐40, 0.5% sodium deoxycholate), and once in lysis buffer. Immunocomplexes were eluted in elution buffer (1% SDS, 100 mM NaHCO3, 40 μg/ml RNAse A) and incubated at 37°C for 1 h. After reversing cross‐links at 65°C for 16 h, DNA was purified with the Wizard SV Gel and PCR Clean‐Up System (Promega).
RNA immunoprecipitation (RIP)
Yeast cells were harvested from 150 ml cultures (OD600: ~1), cross‐linked, and lysed as for ChIP in the presence of RnaseOUT (Invitrogen). Cell extracts were sonicated in a Bioruptor UCD‐200 apparatus for 10 min in high mode with 30‐s on and 30‐s off intervals. Immunoprecipitations were performed using 2 mg of soluble extracts and 2.5 μl of anti‐myc antibody (Cell Signaling Technology, #2276) on a rotating wheel at 4°C for 4 h, followed by incubation at 4°C for 2 h with 25 μl of protein A and G beads (Santa Cruz Biotechnology) blocked with bovine serum albumin and tRNAs (Sigma‐Aldrich). Beads were washed twice with lysis buffer containing 140 mM NaCl, twice with lysis buffer containing 500 mM NaCl, and once with wash buffer (10 mM Tris–HCl, 250 mM LiCl, 0.5% Nonidet P‐40, 0.5% sodium deoxycholate). Immunocomplexes were eluted in 100 mM NaCl, 1% SDS and incubated at 65° C for 2 h to reverse cross‐links. RNA was purified using the TRIzol reagent (Invitrogen). For DNase sensitivity experiments, washed beads were incubated in 300 μl of 1× DNaseI buffer with or without 30 U of RNase‐free DNaseI (Qiagen) and incubated at 30°C for 1 h, spun and treated as above.
Telomere PCR (Telo‐PCR)
Genomic DNA (200 ng) was denatured at 96°C for 5 min and poly(C) tailed by incubation with 1 U of terminal deoxynucleotidyl transferase (TdT, New England Biolabs), 1 μM dCTP, and 1× All‐Phosphor buffer (Amersham) in a final volume of 9 μl at 37°C for 30 min. TdT was inactivated at 65°C for 10 min, and DNA was again denatured at 94°C for 5 min. PCRs were assembled in a final volume of 30 μl and consisted of dG18 oligonucleotide in combination with oF4 for tiTELs and oF5 for nTELs (10 μM each; Table 2), 4 μl of M‐buffer (750 mM Tris–HCl pH 8.8, 200 mM (NH4)2SO4, 0.1% Tween‐20), 0.8 μl of 10 mM dNTPs (Thermo Scientific), 2.5 U of Taq polymerase (Thermo Scientific), and 0.25 U of Pwo DNA polymerase (Roche). PCRs were carried out using the following cycling parameters: 1 cycle at 94°C for 3 min, followed by 38 cycles at 94°C for 20 s, 65°C for 20 s, 72°C for 30 s, and a final extension step at 72°C for 5 min. PCR products were separated in agarose gels, excided, cloned into the pDrive vector using the Qiagen PCR Cloning Kit, and transformed into Stbl2 cells (Thermo Fisher Scientific). Several inserts were PCR‐amplified from independent colonies using M13 primers and Sanger‐sequenced at Microsynth (Balgach, Switzerland) using the oligonucleotide oF6 (Table 2).
PacBio sequencing and bioinformatic analysis
Six tubes (40 μl each) of Telo‐PCR samples prepared as described above were pooled together, brought to a final volume of 450 μl with sterile water, and loaded onto a Amicon Ultra Centrifugal Filter 0.5 ml 100 K (Millipore). The sample was concentrated by centrifugation at 21,000 g for 10 min at 4° C and then immediately inverted to a new collection tube and spun at 100 g for 2 min. The eluate was diluted with 100 μl of sterile water and then purified using the Qiagen PCR purification kit. The concentration of purified Telo‐PCR products was measured using a NanoDrop 2000c (Thermo Scientific). For each sample, at least 1 μl of purified products was used to generate libraries followed by PacBio sequencing according to the manufacturer (Pacific Biosystems) using the protocol for ~250‐bp libraries carried out at the Yale Center for Genome Analysis. The circular consensus sequence was determined using the manufacturer's analysis pipeline to give raw telomere sequences. Sequences that did not contain the amplifying primers were excluded, with up to two mismatches allowed during the primer search. Sequences were further filtered based on the presence of at least one GGTTAC repeat. The subtelomeric primer and poly(C) tract were trimmed off, and telomere length was determined from nt 560 of the cloned pNSU70 sequence. The analysis pipeline was written in Perl, and plots were generated using R. A more detailed method detailing this general approach will be published elsewhere. Sequences were deposited at the Sequence Read Archive (SRA; accession number: PRJNA307606).
Author contributions
MM, AB, and CMA designed the experiments; MM, HW, AB, and LL performed the experiments and analyzed the data; YH generated strains and prepared Telo‐PCR samples for PacBio sequencing; NL designed and carried out bioinformatics analysis of Telo‐PCR PacBio sequences; MCK oversaw the Telo‐PCR‐Seq sample preparation and analysis; CMA, MCK, and MM wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Expanded View Figures PDF
Review Process File
Acknowledgements
We thank Peter Baumann and Julie P. Cooper for strains and reagents, Rajika Arora for critical reading of the manuscript, and members of the Azzalin laboratory for helpful discussions. This work was supported by the European Research Council (BFTERRA), the Swiss National Science Foundation (PP00P3‐123356/144917), ETH Zurich (ETH‐01 12‐2 and ETH‐07 14‐1), and the National Institutes of Health (NIH, Office of the Director; DP2OD008429).
EMBO Reports (2016) 17: 999–1012
See also: S Coulon & V Géli (July 2016)
References
- 1. Azzalin CM, Lingner J (2015) Telomere functions grounding on TERRA firma. Trends Cell Biol 25: 29–36 [DOI] [PubMed] [Google Scholar]
- 2. Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J (2007) Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318: 798–801 [DOI] [PubMed] [Google Scholar]
- 3. Nergadze SG, Farnung BO, Wischnewski H, Khoriauli L, Vitelli V, Chawla R, Giulotto E, Azzalin CM (2009) CpG‐island promoters drive transcription of human telomeres. RNA 15: 2186–2194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Farnung BO, Giulotto E, Azzalin CM (2010) Promoting transcription of chromosome ends. Transcription 1: 140–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lopez de Silanes I, Grana O, De Bonis ML, Dominguez O, Pisano DG, Blasco MA (2014) Identification of TERRA locus unveils a telomere protection role through association to nearly all chromosomes. Nat Commun 5: 4723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Luke B, Panza A, Redon S, Iglesias N, Li Z, Lingner J (2008) The Rat1p 5′ to 3′ exonuclease degrades telomeric repeat‐containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol Cell 32: 465–477 [DOI] [PubMed] [Google Scholar]
- 7. Schoeftner S, Blasco MA (2008) Developmentally regulated transcription of mammalian telomeres by DNA‐dependent RNA polymerase II. Nat Cell Biol 10: 228–236 [DOI] [PubMed] [Google Scholar]
- 8. Vrbsky J, Akimcheva S, Watson JM, Turner TL, Daxinger L, Vyskot B, Aufsatz W, Riha K (2010) siRNA‐mediated methylation of Arabidopsis telomeres. PLoS Genet 6: e1000986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Solovei I, Gaginskaya ER, Macgregor HC (1994) The arrangement and transcription of telomere DNA sequences at the ends of lampbrush chromosomes of birds. Chromosome Res 2: 460–470 [DOI] [PubMed] [Google Scholar]
- 10. Bah A, Azzalin CM (2012) The telomeric transcriptome: from fission yeast to mammals. Int J Biochem Cell Biol 44: 1055–1059 [DOI] [PubMed] [Google Scholar]
- 11. Bah A, Wischnewski H, Shchepachev V, Azzalin CM (2012) The telomeric transcriptome of Schizosaccharomyces pombe. Nucleic Acids Res 40: 2995–3005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Greenwood J, Cooper JP (2012) Non‐coding telomeric and subtelomeric transcripts are differentially regulated by telomeric and heterochromatin assembly factors in fission yeast. Nucleic Acids Res 40: 2956–2963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Porro A, Feuerhahn S, Reichenbach P, Lingner J (2010) Molecular dissection of telomeric repeat‐containing RNA biogenesis unveils the presence of distinct and multiple regulatory pathways. Mol Cell Biol 30: 4808–4817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Azzalin CM, Lingner J (2008) Telomeres: the silence is broken. Cell Cycle 7: 1161–1165 [DOI] [PubMed] [Google Scholar]
- 15. Arora R, Azzalin CM (2015) Telomere elongation chooses TERRA ALTernatives. RNA Biol 12: 938–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Arora R, Lee Y, Wischnewski H, Brun CM, Schwarz T, Azzalin CM (2014) RNaseH1 regulates TERRA‐telomeric DNA hybrids and telomere maintenance in ALT tumour cells. Nat Commun 5: 5220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pfeiffer V, Crittin J, Grolimund L, Lingner J (2013) The THO complex component Thp2 counteracts telomeric R‐loops and telomere shortening. EMBO J 32: 2861–2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Balk B, Maicher A, Dees M, Klermund J, Luke‐Glaser S, Bender K, Luke B (2013) Telomeric RNA‐DNA hybrids affect telomere‐length dynamics and senescence. Nat Struct Mol Biol 20: 1199–1205 [DOI] [PubMed] [Google Scholar]
- 19. Yu TY, Kao YW, Lin JJ (2014) Telomeric transcripts stimulate telomere recombination to suppress senescence in cells lacking telomerase. Proc Natl Acad Sci USA 111: 3377–3382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Redon S, Reichenbach P, Lingner J (2010) The non‐coding RNA TERRA is a natural ligand and direct inhibitor of human telomerase. Nucleic Acids Res 38: 5797–5806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cusanelli E, Romero CA, Chartrand P (2013) Telomeric noncoding RNA TERRA is induced by telomere shortening to nucleate telomerase molecules at short telomeres. Mol Cell 51: 780–791 [DOI] [PubMed] [Google Scholar]
- 22. Redon S, Zemp I, Lingner J (2013) A three‐state model for the regulation of telomerase by TERRA and hnRNPA1. Nucleic Acids Res 41: 9117–9128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sandell LL, Gottschling DE, Zakian VA (1994) Transcription of a yeast telomere alleviates telomere position effect without affecting chromosome stability. Proc Natl Acad Sci USA 91: 12061–12065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pfeiffer V, Lingner J (2012) TERRA promotes telomere shortening through exonuclease 1‐mediated resection of chromosome ends. PLoS Genet 8: e1002747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Maicher A, Kastner L, Dees M, Luke B (2012) Deregulated telomere transcription causes replication‐dependent telomere shortening and promotes cellular senescence. Nucleic Acids Res 40: 6649–6659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Farnung BO, Brun CM, Arora R, Lorenzi LE, Azzalin CM (2012) Telomerase efficiently elongates highly transcribing telomeres in human cancer cells. PLoS ONE 7: e35714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Arnoult N, Van Beneden A, Decottignies A (2012) Telomere length regulates TERRA levels through increased trimethylation of telomeric H3K9 and HP1alpha. Nat Struct Mol Biol 19: 948–956 [DOI] [PubMed] [Google Scholar]
- 28. Leonardi J, Box JA, Bunch JT, Baumann P (2008) TER1, the RNA subunit of fission yeast telomerase. Nat Struct Mol Biol 15: 26–33 [DOI] [PubMed] [Google Scholar]
- 29. Webb CJ, Zakian VA (2008) Identification and characterization of the Schizosaccharomyces pombe TER1 telomerase RNA. Nat Struct Mol Biol 15: 34–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Beernink HT, Miller K, Deshpande A, Bucher P, Cooper JP (2003) Telomere maintenance in fission yeast requires an Est1 ortholog. Curr Biol 13: 575–580 [DOI] [PubMed] [Google Scholar]
- 31. Tang W, Kannan R, Blanchette M, Baumann P (2012) Telomerase RNA biogenesis involves sequential binding by Sm and Lsm complexes. Nature 484: 260–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Maundrell K (1990) nmt1 of fission yeast. A highly transcribed gene completely repressed by thiamine. J Biol Chem 265: 10857–10864 [PubMed] [Google Scholar]
- 33. Lorenzi LE, Bah A, Wischnewski H, Shchepachev V, Soneson C, Santagostino M, Azzalin CM (2015) Fission yeast Cactin restricts telomere transcription and elongation by controlling Rap1 levels. EMBO J 34: 115–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Forstemann K, Hoss M, Lingner J (2000) Telomerase‐dependent repeat divergence at the 3′ ends of yeast telomeres. Nucleic Acids Res 28: 2690–2694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Teixeira MT, Arneric M, Sperisen P, Lingner J (2004) Telomere length homeostasis is achieved via a switch between telomerase‐ extendible and ‐nonextendible states. Cell 117: 323–335 [DOI] [PubMed] [Google Scholar]
- 36. Arora R, Brun CM, Azzalin CM (2012) Transcription regulates telomere dynamics in human cancer cells. RNA 18: 684–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Iglesias N, Redon S, Pfeiffer V, Dees M, Lingner J, Luke B (2011) Subtelomeric repetitive elements determine TERRA regulation by Rap1/Rif and Rap1/Sir complexes in yeast. EMBO Rep 12: 587–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Miller KM, Ferreira MG, Cooper JP (2005) Taz1, Rap1 and Rif1 act both interdependently and independently to maintain telomeres. EMBO J 24: 3128–3135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Miller KM, Rog O, Cooper JP (2006) Semi‐conservative DNA replication through telomeres requires Taz1. Nature 440: 824–828 [DOI] [PubMed] [Google Scholar]
- 40. Schmidt K, Butler JS (2013) Nuclear RNA surveillance: role of TRAMP in controlling exosome specificity. Wiley Interdiscip Rev RNA 4: 217–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schmitt ME, Brown TA, Trumpower BL (1990) A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res 18: 3091–3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expanded View Figures PDF
Review Process File


