Abstract
Mutation of FAM134B (Family with Sequence Similarity 134, Member B) leading to loss of function of its encoded Golgi protein and has been reported induce apoptosis in neurological disorders. FAM134B mutation is still unexplored in cancer. Herein, we studied the DNA copy number variation and novel mutation sites of FAM134B in a large cohort of freshly collected oesophageal squamous cell carcinoma (ESCC) tissue samples. In ESCC tissues, 37% (38/102) showed increased FAM134B DNA copies whereas 35% (36/102) showed loss of FAM134B copies relative to matched non-cancer tissues. Novel mutations were detected in exons 4, 5, 7, 9 as well as introns 2, 4-8 of FAM134B via HRM (High-Resolution Melt) and Sanger sequencing analysis. Overall, thirty-seven FAM134B mutations were noted in which most (31/37) mutations were homozygous. FAM134B mutations were detected in all the cases with metastatic ESCC in the lymph node tested and in 14% (8/57) of the primary ESCC. Genetic alteration of FAM134B is a frequent event in the progression of ESCCs. These findings imply that mutation might be the major driving source of FAM134B genetic modulation in ESCCs.
Oesophageal squamous cell carcinoma (ESCC) is the most common histological subtype of oesophageal carcinomas and it has complex molecular pathology compared to other carcinomas1,2,3. Identification of various genetic and epigenetic changes including mutations of key regulatory genes have a significant role in predicting the biological behaviour of ESCC as well as the prognosis of the patients with ESCC4,5,6,7,8,9,10,11,12,13.
Our previous studies using comparative genomic hybridization analysis revealed that genes in the region of chromosome 5p play a vital role in the pathogenesis of ESCC14,15. FAM134B (Family with sequence similarity 134, member B) also known as JK1is a novel gene placed at chromosome 5p15.1 downstream of δ-catenin16. The oncogenic properties of FAM134B (JK1) were first reported both in ESCC tissues and cell lines, while ESCC tissues also showed altered FAM134B expression16. Also, our recent studies have confirmed the growth related properties of FAM134B by exhibiting multiple tumour suppressor properties of FAM134B (JK1) in colorectal cancer tissues and cell lines17,18,19.
Recently, other researchers have reported that FAM134B encodes a cis-Golgi transmembrane protein, and its mutation can regulate cell apoptosis and long-term survival of nociceptive and autonomic ganglion neurons20,21,22,23,24,25. Homozygous loss of function mutations in FAM134B was reported for the first time in hereditary sensory and autonomic neuropathy type IIB (HSAN IIB) and in vascular dementia20,24. In addition, Khaminets et al. have most recently noted that FAM134B regulated endoplasmic reticulum turnover by selective autophagy25. To the best of our knowledge, at the time of writing, there is no data available on the mutational significance of FAM134B in human cancers. Also, the clinicopathological correlation of FAM134B mutation and clinicopathological parameters has never been reported in human cancer samples. Thus, the current study aims to detect mutations in different exon and intron regions of FAM134B (JK1) in ESCC tissue samples. In addition, the correlations of FAM134B (JK1) mutations with various clinicopathological parameters and natural copy number variations in ESCCs were analysed.
Results
Identification of FAM134B (JK1) DNA in ESCC tissues
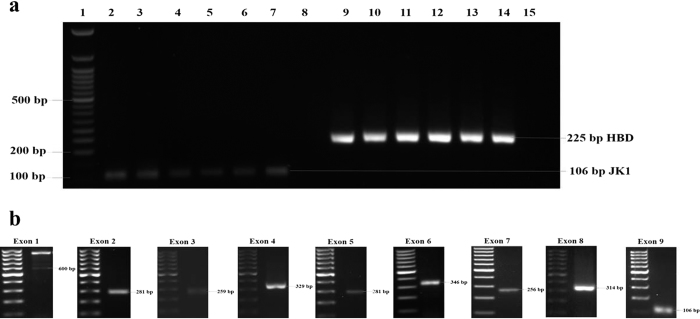
FAM134B (JK1) DNA was identified in all the studied samples. In these samples, 600-base pairs (bp), 281-bp, 259-bp, 329-bp, 282-bp, 346-bp, 256-bp, 314-bp and 106-bp fragments were amplified for exon 1, exon 2, exon 3, exon 4, exon 5, exon 6, exon 7, exon 8 and exon 9 of FAM134B (JK1) respectively. Also, a 225-bp fragment was amplified for the control gene, haemoglobin delta (HBD) (Fig. 1a,b).
Figure 1. FAM134B (JK1) gene amplification in oesophageal squamous cell carcinoma.
(a) Amplified PCR products of FAM134B (JK1) (122bp) and HBD (225bp) in 2% agarose gel. FAM134B (JK1) and HBD were present in all the samples (2–14) except for the water control (15). Fifty-base pair DNA ladder was used for comparison. (b) Representative amplified PCR products of exon 1, exon 2, exon 3, exon 4, exon 5, exon 6, exon 7, exon 8 and exon 9 of FAM134B (JK1) in 1.5% agarose gel. Hundred-base pair DNA ladder was used for comparison.
FAM134B (JK1) copy number variations
In ESCC (n = 102), 37% (n = 38) showed increased FAM134B (JK1) copies whereas 35% (n = 36) showed a loss of FAM134B (JK1) copies relative to matched non-cancer tissue. Meanwhile, the remaining 22% (n = 28) did not show any change in copies. In ESCCs with lymph node metastasis (n = 64), 66% (n = 42) showed high FAM134B (JK1) copies whereas 34% (n = 22) revealed deletion (p = 0.80). The DNA copy number of FAM134B (JK1) in cancer showed no statistical correlations with the grades and pathological stages of ESCC or with the gender and the age of the patient (p > 0.05) (Table 1).
Table 1. Clinicopathological features of ESCC and JK1 (FAM134B) copy number variation.
| Characteristics | Number of Patients | DNA copy | P value | ||
|---|---|---|---|---|---|
| Amplification Deletion | No change | ||||
| Age | |||||
| <60 years | 34(33.3%) | 15(44.1%) | 7(20.6%) | 12(35.3%) | 0.07 |
| >60 | 68(66.7%) | 23(33.8%) | 29(42.6%) | 16(23.5%) | |
| Gender | |||||
| Male | 84(82.4%) | 32(38.1%) | 28(33.3%) | 24(28.6%) | 0.66 |
| Female | 18(17.6%) | 6(33.3%) | 8(44.4%) | 4(22.2%) | |
| Histological Grade | |||||
| Well | 22(21.6%) | 10(45.5%) | 6(27.3%) | 6(27.3%) | 0.61 |
| Moderate | 59(57.8%) | 23(39.0%) | 21(35.6%) | 15(25.4%) | |
| Poor | 21(20.6%) | 5(23.8%) | 9(42.9%) | 7(33.3%) | |
| Site | |||||
| Upper | 12(11.8%) | 5(41.7%) | 6(50.0%) | 1(8.3%) | 0.12 |
| Middle | 55(53.9%) | 23(41.8%) | 14(25.5%) | 18(32.7%) | |
| Lower | 35(34.3%) | 10(28.6%) | 16(45.7%) | 9(25.7%) | |
| T Stage | |||||
| 1 | 6(5.9%) | 2(33.3%) | 2(33.3%) | 2(33.3%) | 0.85 |
| 2 | 11(10.8%) | 5(45.5%) | 5(45.5%) | 1(9.1%) | |
| 3 | 64(62.7%) | 23(35.9%) | 22(34.4%) | 19(29.7%) | |
| 4 | 21(20.6%) | 8(38.1%) | 7(33.3%) | 6(28.6%) | |
| Lymph node Metastasis | |||||
| Negative | 38(37.3%) | 11(28.9%) | 14(36.8%) | 13(34.2%) | 0.33 |
| Positive | 64(62.7%) | 27(42.2%) | 22(34.4%) | 15(23.4%) | |
| Distant metastasis | |||||
| Negative | 97(95.1%) | 35(36.1%) | 35(36.1%) | 27(27.8%) | 0.55 |
| Positive | 5(4.9%) | 3(60.0%) | 1(20.0%) | 1(20.0%) | |
| TNM Stage | |||||
| Stage I | 6(5.9%) | 4(66.7%) | 0(0.0%) | 2(33.3%) | 0.22 |
| Stage II | 26(25.5%) | 7(26.9%) | 12(46.2%) | 7(26.9%) | |
| Stage III | 65(63.7%) | 24(36.9%) | 23(35.4%) | 18(27.7%) | |
| Stage IV | 5(4.9%) | 3(60.0%) | 1(20.0%) | 1(20.0%) | |
Mutation screening using high-resolution Melt (HRM) curve and Sanger sequencing
FAM134B (JK1) mutations showed no correlation with other clinical and pathological features of ESCC including gender, age of the patient as well as the sites, histological grades and pathological stages of carcinoma (p > 0.05) (Table 2). Also, mutations did not show any correlation with the DNA copy number of FAM134B (JK1) (Tables 3 and 4).
Table 2. Clinicopathological features and JK1 (FAM134B) mutations in oesophageal squamous cell carcinoma.
| Characteristics | Number of patients | Mutation cases | No mutation cases | p value |
|---|---|---|---|---|
| Age | ||||
| <60 year | 20 | 11 | 9 | 0.17 |
| >60 year | 37 | 13 | 24 | |
| Gender | ||||
| Male | 50 | 19 | 31 | 0.12 |
| Female | 7 | 5 | 2 | |
| Site | ||||
| Upper | 5 | 2 | 3 | 0.44 |
| Middle | 40 | 15 | 25 | |
| Lower | 12 | 7 | 5 | |
| Grade | ||||
| Well | 20 | 7 | 13 | 0.58 |
| Moderate | 28 | 12 | 16 | |
| Poor | 9 | 5 | 4 | |
| T Stage | ||||
| 1 | 1 | 0 | 1 | 0.62 |
| 2 | 6 | 3 | 3 | |
| 3 | 38 | 17 | 21 | |
| 4 | 12 | 4 | 8 | |
| Lymph node Metastasis | ||||
| Negative | 20 | 7 | 13 | 0.58 |
| Positive | 37 | 17 | 20 | |
| Distant metastasis | ||||
| Negative | 54 | 22 | 32 | 0.57 |
| Positive | 3 | 2 | 1 | |
| TNM Stage | ||||
| Stage I | 1 | 1 | 0 | 0.55 |
| Stage II | 14 | 5 | 9 | |
| Stage III | 39 | 17 | 22 | |
| Stage IV | 3 | 2 | 1 | |
Table 3. Mutations detected in different exons of FAM134B (JK1) in ESCC.
| Exons | Sample Code | Primary cancer with mutation | Lymph node metastasis with mutation | FAM134B DNA copy number change | DNA change | Type of mutation | Codon change | Protein change | Comments | |
|---|---|---|---|---|---|---|---|---|---|---|
| Primary cancer | Lymph node metastasis | |||||||||
| Exon 9 | 41 | No | Yes | N | N | c. 1128T > A | Substitution | CTT > CTA | p.Leu347Leu | Potentially functional due to splice site modification |
| 30 | No | Yes | D | D | c. 1107G > C c. 1129T > C | Substitution | GAG > GAC TCA > CCA | p.Glu340Asp p.Ser348Pro | pGlu340Asp - Likely functional significance pSer348Pro - Probably lesser effect/polymorphism due to lack of conservation in other species | |
| 14 | Yes | No | A | A | c. 1149A > G | Substitution | GAA > GAG | p.Glu354Glu | Likely functional significance (also affects splice sites) | |
| 19 | No | Yes | D | A | c. 1112T > C c. 1129T > C | Substitution | GTT > GCT TCA > CCA | p.Val342Ala p.Ser348Pro | pVal342Ala - Likely functional significance (also affects splice sites) pSer348Pro - Probably lesser effect/polymorphism due to lack of conservation in other species | |
| 3 | Yes | No | D | D | c. 1112T > C c. 1149A > G c. 1152T > G c. 1153G > C | Substitution | GTT > GCT GAA > GAG AAT > AAG GGC > CGC | p.Val342Ala p.Glu354Glu p.Asn355Lys p.Gly356Arg | pVal342Ala - Likely functional significance (also affects splice sites) pGlu354Glu - Likely functional significance (also affects splice sites) pAsn355Lys - Likely functional significance pGly356Arg - Likely functional significance (also affects splice sites) | |
| 35 | Yes | No | A | – | c. 1092T > G c. 1093G > C c. 1097GA > AC c. 1107G > T c. 1129T > C | Substitution | CTT > CTG GAC > CAC CGA > CAC GAG > GAC TCA > CCA | p.Leu335Leu p.Asp336His p.Arg337His p.Glu340Asp p.Ser348Pro | pLeu335Leu - Likely functional significance (also affects splice sites) pAsp336His- Likely functional significance (also affects splice sites) pArg337His - Likely functional significance (also affects splice sites) pGlu340Asp - Likely functional significance pSer348Pro - Probably lesser effect/polymorphism due to lack of conservation in other species | |
| 43 | Yes | Yes | A | N | c. 1112T > C c. 1129T > C c. 1092T > G c. 1093G > C | Substitution | GTT > GCT TCA > CCA CTT > CTG GAC > CAC | p.Val342Ala p.Ser348Pro p.Leu335Leu p.Asp336His | pVal342Ala - Likely functional significance (also affects splice sites) pSer348Pro - Probably lesser effect/polymorphism due to lack of conservation in other species pLeu335Leu - Likely functional significance (also affects splice sites) pAsp336His- Likely functional significance (also affects splice sites) | |
| 47 | Yes | No | D | – | c. 1107G > C c. 1129T > C | Substitution | GAG > GAC TCA > CCA | p.Glu340Asp p.Ser348Pro | pGlu340Asp - Likely functional significance pSer348Pro - Probably lesser effect/polymorphism due to lack of conservation in other species | |
| 50 | Yes | Yes | D | A | c. 1107G > T c. 1129T > C c. 1164A > G | Substitution | GAG > GAC TCA > CCA ACA > ACG | p.Glu340Asp p.Ser348Pro p.Thr360Thr | pGlu340Asp - Likely functional significance pSer348Pro - Probably lesser effect/polymorphism due to lack of conservation in other species pThr360Thr -Likely functional significance (also affects splice sites) | |
| 51 | Yes | No | N | – | c. 1112T > C c. 1129T > C | Substitution | GTT > GCT TCA > CCA | p.Val342Ala p.Ser348Pro | pVal342Ala - Likely functional significance (also affects splice sites) pSer348Pro - Probably lesser effect/polymorphism due to lack of conservation in other species | |
| 34 | Yes | No | A | – | c. 1137delT | Deletion | TTT > TTC | p.Phe350Phe | pPhe350Phe- Likely functional significance (frameshift mutation, truncates protein, may change splicing) | |
| Exon 7 | 39 | No | Yes | A | N | c. 816C > T or c. 903C > T | Substitution | GAT > GAC | p.Asp272Asp | pAsp272Asp-Likely functional significance (also affects splice sites) |
| 19 | No | Yes | D | A | c. 816C > T or c. 903C > T | Substitution | GAT > GAC | p.Asp272Asp | pAsp272Asp -Likely functional significance (also affects splice sites) | |
| Exon 5 | 14 | Yes | No | A | A | c. 711G > Ac. 744C > T | Substitution Substitution | ACG > ACA CTC > CTT | p.Thr208Thr p.Leu219Leu | Has no functional significance Has no functional significance |
| 6 | No | Yes | A | A | c. 711G > A | Substitution | ACG > ACA | p.Thr208Thr | Has no functional significance | |
| 12 | No | Yes | N | N | c. 711G > A | Substitution | ACG > ACA | p.Thr208Thr | Has no functional significance | |
| 41 | No | Yes | N | N | c. 711G > A | Substitution | ACG > ACA | p.Thr208Thr | Has no functional significance | |
| 20 | No | Yes | N | N | c. 711G > A | Substitution | ACG > ACA | p.Thr208Thr | Has no functional significance | |
| 11 | No | Yes | A | N | c. 711G > A | Substitution | ACG > ACA | p.Thr208Thr | Has no functional significance | |
| 3 | Yes | No | D | D | c. 711G > A | Substitution | ACG > ACA | p.Thr208Thr | Has no functional significance | |
| 15 | No | Yes | D | N | c. 711G > A | Substitution | ACG > ACA | p.Thr208Thr | Has no functional significance | |
| Exon 4 | 6 | No | Yes | A | A | c. 546–547CT > GG c. 660G > A | Substitution Substitution | AGC > AGG and TGG > GGG GAA > AAA | p.Ser153Arg p.Trp154Gly p.Glu163Lys | pSer153Arg and pTrp154Gly-Likely functional significance. (also affects splice sites) pGlu163Lys-Likely functional significance (also affects splice sites). |
| 38 | No | Yes | N | D | c. 660G > A | Substitution | GAA > AAA | p.Glu163Lys | pGlu163Lys-Likely functional significance (also affects splice sites). | |
| 30 | No | Yes | D | D | c. 660G > A | Substitution | GAA > AAA | p.Glu163Lys | pGlu163Lys-Likely functional significance (also affects splice sites). | |
| 43 | No | Yes | A | N | c. 660G > A | Substitution | GAA > AAA | p.Glu163Lys | pGlu163Lys-Likely functional significance (also affects splice sites). | |
| 59 | No | Yes | N | D | c. 660G > A | Substitution | GAA > AAA | p.Glu163Lys | pGlu163Lys-Likely functional significance (also affects splice sites). | |
| 19 | No | Yes | D | A | c. 660G > A | Substitution | GAA > AAA | p.Glu163Lys | pGlu163Lys-Likely functional significance (also affects splice sites). | |
Table 4. Mutations detected in different introns of FAM134B (JK1) in ESCC.
| Introns | Sample Code | Primary cancer | Lymph node metastasis |
FAM134B
DNA copy number change |
DNA change | Type of mutation | |
|---|---|---|---|---|---|---|---|
| Primary cancer | Lymph node metastasis | ||||||
| Intron 2 | 39 | No | Yes | A | N | c. 514 + 144delA c. 514 + 37 − 38delTT | Deletion Deletion |
| 41 | No | Yes | N | N | c. 408 − 27delA c. 514 + 37 − 38delTT | Deletion Deletion | |
| 20 | No | Yes | N | N | c. 408 − 27delA c. 514 + 34T > A c. 514 + 37 − 38delTT | Deletion Substitution Deletion | |
| 73 | No | Yes | A | A | c. 408 − 27delA | Deletion | |
| Intron 4 | 16 | No | Yes | N | D | c. 408 − 27delA c. 514 + 37 − 38delTT | Deletion Deletion |
| 39 | No | Yes | A | N | c. 546 − 64delA c. 672 + 46 − 47delTG | Deletion Deletion | |
| 6 | No | Yes | A | A | c. 546 − 64delA c. 546 − 56delA c. 672 + 46 − 47delTG | Deletion Deletion Deletion | |
| 38 | No | Yes | N | D | c. 546 − 56delA c. 672 + 26delT | Deletion Deletion | |
| 30 | No | Yes | D | D | c. 546 − 64delA c. 546 − 56delA | Deletion Deletion | |
| Intron 5 | 43 | No | Yes | A | N | c. 546 − 56delA c. 672 + 26delT | Deletion Deletion |
| 59 | No | Yes | N | D | c. 546 − 56delA c. 672 + 26delT c. 672 + 46 − 47delTG | Deletion Deletion Deletion | |
| 19 | No | Yes | D | A | c. 546 − 56delA | Deletion | |
| 41 | No | Yes | N | N | c. 673 − 54delC | Deletion | |
| 11 | No | Yes | A | N | c. 673 − 54C > A c. 757 + 56G > A | Substitution Substitution | |
| 3 | Yes | No | D | D | c. 673 − 65delA | Deletion | |
| 15 | No | Yes | D | N | c. 757 + 56G > A c. 673 − 52 − 53GA > AG | Substitution Substitution | |
| Intron 6 | 38 | No | Yes | N | D | c. 758 − 61 − 62TG > CTc. 758 − 71G > T | Substitution Substitution |
| 30 | No | Yes | D | D | c. 758 − 71G > T | Substitution | |
| Intron 7 | 19 | No | Yes | D | A | c. 873 + 23T > C or c. 960 + 23T > C | Substitution |
| Intron 8 | 14 | Yes | No | A | A | c. 1087 + 97C > A | Substitution |
| 6 | No | Yes | A | A | c. 1087 + 97C > A | Substitution | |
| 30 | No | Yes | D | D | c. 961 − 33delA c. 1087 + 97C > A | Deletion Substitution | |
| 41 | No | Yes | N | N | c. 961 − 33delA c. 1087 + 97C > A | Deletion Substitution | |
| 31 | No | Yes | A | A | c. 961 − 33delA c. 1087 + 97C > A | Deletion Substitution | |
Overall, 37 FAM134B (JK1) mutations were found in ESCC (Tables 3 and 4). Of these, 36 FAM134B (JK1) mutations were noted in metastatic ESCC in lymph node and 16 mutations were detected in primary cancer. Of these 37 mutations, 6 were heterozygous mutations and 31 were homozygous mutations. On the other hand, only 14% (8/57) of the primary carcinoma harboured mutations of FAM134B (JK1). Mutations were noted in both the primary cancer and the lymph node metastasis with ESCC in 2 cases. In these 2 cases (cases 43 and 50), the mutations noted were identical. Also, of the 37 FAM134B (JK1) mutations, 11 mutations result in alteration of the amino acid sequences in FAM134B (JK1) protein while 26 mutations were synonymous.
Among the 9 exons of FAM134B (JK1) examined, mutations were detected in exons 4, 5, 7 and 9 as well as introns 2, 4, 5, 6, 7 and 8 for FAM134B (JK1) in ESCC (Figs 2 and 3). In exon 9 of FAM134B (JK1), thirteen substitution and one frameshift (deletion) mutations were documented in ESCCs. The most common substitution variants in exon 9 are c. 1129T > C, c. 1107G > C, c. 1112T > C which alters the codons and subsequently amino acids - TCA > CCA (p. Ser348Pro), GAG > GAC (p. Glu340Asp) and GTT > GCT (p. Val342Ala) respectively. One frameshift (deletion) FAM134B (JK1) mutation, c. 1137delT, which alters the codon TTT > TTC resulting in no change in amino acid, but altered amino acids following the frameshift (p. Phe379fs) was noted in ESCC. For exon 4 of FAM134B (JK1), two mutations, c. 546-547CT > GG and c. 660G > A, were detected in ESCC tissues (Table 3). The mutation in c. 546-547CT > GG change the codons in amino acids - AGC > AGG (p. Ser153Arg) and TGG > GGG (p. Trp154Gly). Meanwhile the mutation in c. 660G > A change the codons in amino acids- GAA > AAA (p. Glu163Lys). Furthermore, four polymorphisms were in identified intron 4 FAM134B (JK1) in ESCC.
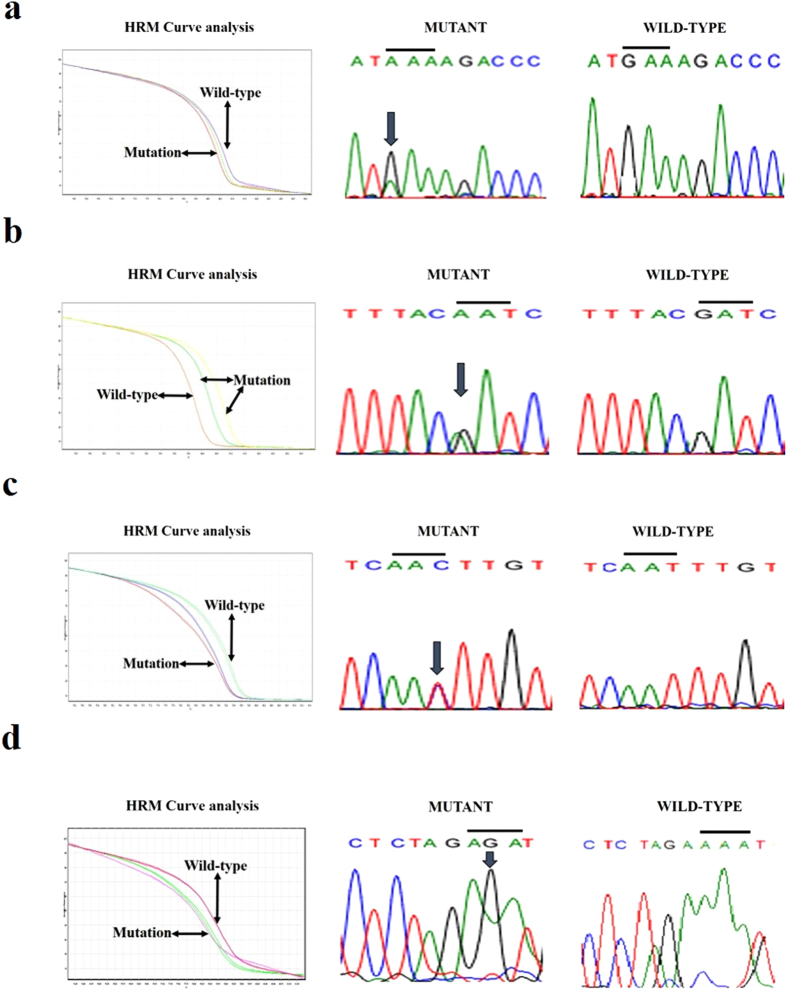
Figure 2. Validation of FAM134B (JK1) different exons sequence variant HRM curve analysis results by Sanger sequencing.
(a) The presence of variant (GAA > AAA) in exon 4 of FAM134B (JK1) is demonstrated via normalized melting curves and sequencing (mutant versus wildtype). (b) HRM analysis shows the polymorphism (GAT > AAT) in exon 5 of FAM134B (JK1) as evident by normalized melting curves and sequencing (mutant versus wide type). (c) The presence of variant (AAT > AAC) in exon 7 of FAM134B (JK1) is confirmed via normalized melting curves and sequencing (mutant versus wide type). (d) The evidence of mutant (AAA > AGA) in exon 9 of FAM134B (JK1) is showed using normalized melting curves and sequencing (mutant versus wide type).
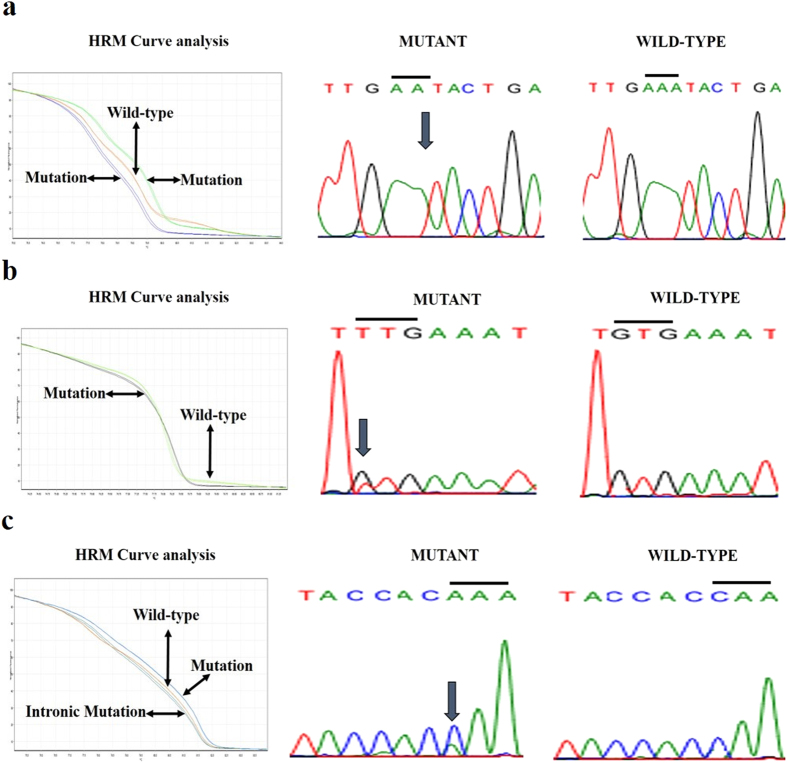
Figure 3. Confirmation of FAM134B (JK1) different introns sequence variant HRM curve analysis results via Sanger sequencing.
(a) HRM curve analysis shows characteristic melting pattern of FAM134B (JK1) intron 2 amplicons using normalized melting curves and Sanger sequencing corroborates the mutation as being c. 14 + 144delA. (b) The evidence of mutant (GTG > TTG) in intron 6 of FAM134B (JK1) is shown using normalized melting curves and sequencing (mutant versus wild type). (c) HRM analysis shows the polymorphism (CAA > AAA) in intron 8 of FAM134B (JK1) as evident by normalized melting curves and sequencing (mutant versus wide type).
Mutations in other exons and introns of FAM134B (JK1) were less common in ESCC. There was no mutation noted in exon 8 of FAM134B (JK1) in ESCC. However, two types of variants, c961-33delA and c1087 + 97C > A, were detected in intron 8. Also, one silent mutation (c. 816C > T) and a mutation (c. 873 + 23T > C) were noted for exon 7 and intron 7 of FAM134B (JK1) in ESCC respectively. In addition, two synonymous variants (c. 319G > A and c. 352C > T) were noted in exon 5 of FAM134B in ESCC. Lastly, five polymorphisms were detected in intron 2 of FAM134B (with the most frequent variant being c. 408-27delA).
Discussion
Chromosomal copy number changes can indicate activation of oncogenes and inactivation of tumour suppressor genes in human cancers26. In our previous studies, FAM134B (JK1) copy number alterations were found as a frequent event in colorectal adenoma and adenocarcinoma19. The current study is the first to investigate FAM134B (JK1) copy number changes in a large cohort of ESCC. Approximately one third of cancers studied revealed amplification and one third revealed deletion of FAM134B in ESCC. This altered DNA copy number changes of FAM134B indicate its varied modulation potential in different ESCC patients.
This current study is the first systematic study to investigate mutation sites in FAM134B (JK1) gene in ESCC. Until now, five homozygous mutations of FAM134B (JK1) were detected in neurological diseases. They are three nonsense mutations, one frame-shift mutation, and one is a splice-site mutation20,21,22,23. These mutations were located in exon 1 and intron 7 of the gene. In addition, there were 2 heterozygous mutations reported in exon 1. In ESCC, we have noted 37 distinct mutations of FAM134B (JK1). Thirty-one are homozygous mutations and 6 were heterozygous mutations. These mutations were found in exons 4, 5, 7, 9 and introns 2, 4, 5, 6, 7 and 8 of FAM134B (JK1). None of these mutation sites are identical to those noted reported in the neurological diseases. Thus, the mutations related to the pathogenesis of ESCC appear to be are unique. The roles of FAM134B (JK1) mutations in the pathogenesis of the cancer should therefore be different from their roles in neurological diseases. The mechanisms and presence of any downstream effects of the FAM134B (JK1) mutations need to be further investigated in ESCC to confirm their significance.
Of the 37 novel mutation sites in FAM134B (JK1) detected in ESCC, 11 mutations changed the amino acid of the resulting protein while 26 mutation sites do not alter the amino acid composition of the protein. The most common mutations are present in exon 9. Amongst the 11 mutations that changed the amino acid structure of JK1 are three relatively common variants in exon 9 (c. 1129T > C, c. 1107G > C, c. 1112T > C). Of these, c. 1129T > C, is predicted by in silico methods as being of minimal effect due to lack of conservation of this amino acid in other species while the other two have potentially higher functional significance due to their predicted ability to also cause splice site modification in the protein. In addition, in exon 9 of FAM134B (JK1), one frame-shift (deletion) mutation, c. 1137delT, is likely to have increased functional significance as it radically alters and can truncate the protein as well as potentially changing exon splicing of the RNA. The other important mutation that lead to an amino acid change was noted in exon 4 resulting in an exon change GAA > AAA (protein change-p.Glu163Lys). The novel mutations detected in this study that alters the amino acid structure of FAM134B (JK1) have potential to affect the function of FAM134B (JK1).
In the current study, FAM134B (JK1) mutations were noted predominately in metastatic ESCCs present in lymph nodes, implying that they have a role in the pathogenesis of lymph node metastases in ESCC. Mutation in the majority of metastatic lymph node tissues compared to primary tumours (Table 3) indicates that FAM134B (JK1) mutation may offer a survival advantage in different tumour microenvironments, or assist in colonisation of those particular microenvironments. Differing FAM134B (JK1) sequences imply that, the molecular interactions and characteristics would differ between primary and metastatic tumours. Furthermore, the classical model for metastatic processes suggests that the majority of the cancer cells in the primary tumour have low metastatic potential and only a few cells acquire enough somatic mutations to become metastatic27. Thus, the absence of FAM134 (JK1) mutation in the bulk of primary ESCC tissues in this study might be due to the harvesting of cancer cells with fewer somatic mutations. Hence, it can be hypothesised that FAM134B (JK1) might act as a potential target for predicting lymph node metastasis in ESCC patients. Further research with a larger series of ESCC patients with matched metastatic tissues and potentially functional studies on particular mutations are needed to confirm the metastatic potential of FAM134B (JK1).
In conclusion, we identified for the first time the mutation of FAM134B (JK1) in ESCC tissue samples. The altered expression patterns and copy number variations of FAM134B in ESCCs might be modulated by these mutation changes in FAM134B. In this study, FAM134B mutations were frequently observed in metastatic lymph node tissues indicating its use as a potential predictor for metastasis in ESCCs. Multiple different genetic alterations incorporating mutations in coding sequences of FAM134B were observed and each might entail either alterations to expression or functional changes of this gene that could play a fundamental role in the progression of ESCCs.
Methods
Recruitment of tissue samples and clinicopathological data
Matched tumour samples and non-cancer tissue (near the surgical resection margin) from the same patient who underwent resection of ESCC were prospectively collected, snapped frozen in liquid nitrogen and stored in minus 80 °C by the author (AKL). Informed consent was obtained from all subjects. In addition, in each case, macroscopically enlarged lymph nodes suspicious for lymph node metastasis were also sampled, snap frozen and stored. After the collection, additional tissues blocks were taken, fixed in formalin and embedded in paraffin for pathological examination. Sections were then cut from these blocks and haematoxylin and eosin stained. They were studied by the author (AKL). The ESCCs were graded according to the World Health Organization (WHO) criteria27. The carcinomas were staged as per the TNM (tumour, lymph node and metastases) classification adopted in the American Joint Committee on Cancer28. Overall, tissues from 102 patients with ESCC were recruited. Eighteen patients had lymph nodes with metastatic ESCC sampled. For each patient, clinical and pathological parameters including gender and age of patients as well as the sites, grades, pathological stages of the ESCC were recorded.
Genomic DNA extraction
Ethical approval was obtained for the use of these samples (GU Ref No: MED/19/08/HREC) by the Griffith University human research ethics committee. The methods were carried out in accordance with the approved guidelines.
The selected samples were sectioned using a cryostat (Leica CM 1850 UV, Wetzlar, Germany) and stained by haematoxylin and eosin. Light microscopic examination was performed by author, AKL, to confirm the presence of non-cancer and cancerous tissues for genomic DNA extraction. Five 10 μm slices was sectioned from the frozen tissue samples for DNA extraction. DNA was extracted and purified from frozen tissue samples using all prep DNA/RNA mini kit (Qiagen, Hilder, Germany), following manufacturer’s instructions. DNA quantification was accomplished via Nanodrop Spectrophotometer (BioLab, Ipswich, MA, USA) and purity was measured using 260/280 ratio. Concentration of DNA was noted in ng/μl and then stored at −20 °C until use.
Primer design
Primers specific for determining FAM134B mutation and copy number changes were designed based on GenBank accession number for variant 1 NM_001034850 and for variant 2 NM_019000 as well as for a control gene, HBD (GenBank accession number NM_000519) using Primer3 version 0.4.0 (http://frodo.wi.mit.edu/primer3). All primers were analysed for specificity using Primer Blast (http://www.ncbi.nlm.nih.gov/tools/primer-blast) and Primer Premier program version 5 (Premier Biosoft, Palo Alto, CA, USA) to check for primer parameters like GC content, annealing temperature of primer and ΔG (Gibbs free energy change - energy required to break the secondary structure) and to forecast any possible mismatching, primer dimmer or hairpin formation.
Primers for FAM134B (JK1) gene were designed within the intronic regions on either side of the exon of interest to ensure coverage of the entire exon 1 to exon 9 to amplify and direct sequencing both isoforms of the gene for screening of mutations in ESCC. Primers were also designed for FAM134B (JK1) and control gene - haemoglobin delta (HBD) to identify DNA copy number variations in ESCC. The primer pairs were obtained from Sigma-Aldrich (St Louis, MO, USA). The list of chosen primer sets are summarized in Table 5.
Table 5. List of Exon primers designed for qRT-PCR and sequencing of FAM134B.
| Target Genes | Primer sequence (Forward and Reverse) | Amplicon size |
|---|---|---|
| FAM134B Exon 1 | 5′-CGGCACCCACACCCAGGCGCGCCC-3′ | 600 bp |
| 5′-GCACTGGGTCCCCGGGGCCCCG-3′ | ||
| FAM134B Exon 2 | 5′-GTTTCTGTGGCAGGAAGTAAACCC-3′ | 281 bp |
| 5′-GGACTAATTGGCTAATATGCCTAC-3′ | ||
| FAM134B Exon 3 | 5′-GTGTTAGAGATCTGAGCATTCCAC-3′ | 259 bp |
| 5′-CTTGGATTTAGATTCCTGTCAC-3′ | ||
| FAM134B Exon 4 | 5′-CCAGAGGGTGTGGCCAACAGTAG-3′ | 329 bp |
| 5′-TGGAGAAATCTGACAAGCTG-3′ | ||
| FAM134B Exon 5 | 5′-GTCACTTATACCGGTCACTATAG-3′ | 282 bp |
| 5′-CTCATCCCCCCTCTTCAAAC-3′ | ||
| FAM134B Exon 6 | 5′-GTGAAATACTGAAATGTACGTAGC-3′ | 346 bp |
| 5′-CTTTGAGGGAGATTAGCTTC-3′ | ||
| FAM134B Exon 7 | 5′-GAATATGAGAAATGTGGGGTAAG-3′ | 256 bp |
| 5′-GGAGTTTATTAGGAAGATCATTCAGC-3′ | ||
| FAM134B Exon 8 | 5′-CTGTCATTTTGGGGGTTCATATGG-3′ | 314 bp |
| 5′-TGGTGGTAACATGTTATTTACCC-3′ | 106 bp | |
| FAM134B Exon 9 | 5′-TGACCGACCCAGTGAGGA-3′ | |
| 5′-GGGCAAACCAAGGCTTAA-3′ | ||
| Haemoglobin delta | 5′-TGGATGAAGTTGGTGGTGAG-3′ | 225 bp |
| (HBD) | 5′-CAGCATCAGGAGTGGACAGA-3′ |
Real-time quantitative polymerase chain reaction (qPCR)
DNA copy number changes of FAM134B (JK1) were determined using a rotor Gene Q real-time PCR (RT-PCR) Detection system (Qiagen). RT-PCR was achieved in a total volume of 10 μl comprising 5 μl of 2XSensiMix SYBR No-ROX master mix (Bioline, London, UK), 1 μl of each 10 picomole/μl primer, 1 μl of cDNA/ genomic DNA at 20–50 ng/μl and 2 μl of Nuclease-free water. PCR cycling programs encompassed initial denaturation and activated the hot start DNA polymerase in one cycle of 7 minutes at 95 °C followed by 40 cycles of 10 seconds at 95 °C (denaturation), 30 seconds at 60 °C (annealing) and 20 seconds at 72 °C (extension). Melting curve analysis was carried out using 80 cycles of 30 seconds increasing from 55 °C. The melting curves of all final real-time PCR products were analysed for determination of genuine products and contamination by nonspecific products and primer dimers. All samples were also run on 2% agarose gel electrophoresis to ensure that the correct product was amplified in the reaction. To increase the reliability of the results, assays were accomplished in multiple replicates and a non-template control was included in all the experiment. The results of the quantitative real-time polymerase change reaction were analysed using methods published previously29.
High-Resolution Melt (HRM) curve analysis
Fifty-seven matched cancer and non-cancer tissues (including 18 cases with lymph node metastases) were used for HRM analysis. Sections of the lymph nodes were confirmed to have lymph nodes metastases by histological examination as above. HRM curve analysis was accomplished by amplifying target sequences on the Rotor-Gene Q detection system (Qiagen) using the software Rotor-Gene ScreenClust HRM Software. The exon 1, exon 2, exon 3, exon 4, exon 5, exon 6, exon 7, exon 8, exon 9 of FAM134B (JK1) were PCR amplified in a total reaction volume of 10 μl comprising 5 μl of 2Xsensimix HRM master mix, 1 μl of 30 ng/μl genomic DNA, diethylpyrocarbonate (DEPC, RNase free) treated water 2 μl and 1 μl of 5 μmol/L JK-1 primer. The thermal cycling protocol started with one cycle of 98 °C for 4 minutes. Full activation of the SensiFAST DNA polymerase occurs within 30 seconds at 95 °C. This was followed by 40 cycles of 98 °C for 5 seconds. Then, the reaction mix was at annealed at 60 °C for 15 seconds in exon 2, exon 5, exon 6, exon 7, exon 8, exon 9; 64.5 °C for 15 seconds for exon 3 and exon 4 and at 72 °C for 15 seconds for exon 1. Each PCR run included a negative (no template) control. The melt curve data were generated by increasing the temperature from 65 °C to 85 °C for all assays, with a temperature increase rate of 0.05 °C/seconds and recording fluorescence at each step. All the reactions were done in duplicate to increase the reliability of the results. Each mutant allele had its own distinctive melting curve while compared to the wild-type allele. HRM results were interpreted as mutant when both replicates showed a variant compared to wild type. In cases of uncategorized samples, one replicate showed a variant and the other matched the wild type curve.
Purification of PCR products and Sanger sequencing analysis
All the possible mutations detected by HRM analysis were further confirmed by Sanger sequencing. After HRM curve analysis, successful and specific PCR products showing one melting peak were purified according to the manufacturer’s protocols from the NucleoSpin Gel and PCR Clean-up kit (Macherey- Nagel, Bethlehem, PA, USA). The purified PCR products were subjected to sequence by corresponding forward and reverse primer using the Big Dye Terminator (BDT) chemistry Version 3.1 (Applied Biosystems, Foster City, CA, USA) under standardised cycling PCR conditions and analysed by the Australian Genome Research Facility (AGRF) using a 3730xl Capillary sequencer (Applied Biosystems). The composition of DNA sequencing reactions was followed according the samples preparation guide from AGRF. Sequence analysis was performed with Chromas 2.31 software.
Statistical analysis
Correlations of FAM134B (JK1) copy number change and mutations with clinicopathological parameters were performed. Comparisons between groups were implemented using the chi-square test, likelihood ratio and Fisher’s exact test. All the data was entered into a computer data base and the statistical analysis was executed using the Statistical Package for Social Sciences for Windows (version 22.0, IBM SPSS Inc., New York, NY, USA). Significance level was taken at p < 0.05.
Additional Information
How to cite this article: Haque, M. H. et al. Identification of Novel FAM134B (JK1) Mutations in Oesophageal Squamous Cell Carcinoma. Sci. Rep. 6, 29173; doi: 10.1038/srep29173 (2016).
Acknowledgments
The authors would like to acknowledge the funding from higher degree research student (GUIPRS and GUPRS scholarship) from the Griffith University. The project was also supported by the project funding of Menzies Heath Institute Queensland and joint funding by Griffith University and the Gold Coast University Hospital Foundation.
Footnotes
Author Contributions V.G. and A.K.L. developed the idea of novel mutations and supervised the projects. M.H.H. and V.G. designed the experiments; M.H.H. and V.G. conducted the experiments; M.H.H., V.G. and R.A.S. analysed the raw data; K.C. and A.K.L. collected the samples and managed the pathological data. M.H.H., V.G., M.J.A.S. and A.K.L. discussed the results and co-wrote the manuscript.
References
- Lam K. Y. & Ma L. Pathology of oesophageal cancers: local experience and current in-sights. Chin. Med. J. 110, 459–464 (1997). [PubMed] [Google Scholar]
- Lam A. K. Molecular biology of esophageal squamous cell carcinoma. Crit. Rev. Oncol. Hematol. 33, 71–90 (2000). [DOI] [PubMed] [Google Scholar]
- Lam A. K. Cellular and molecular biology of esophageal cancer. In: Esophageal cancer: prevention, diagnosis and therapy. (eds Saba N. F. & El-Rayes B.), Ch. 2, 25–40 (Springer, 2015). [Google Scholar]
- Yu V. Z. et al. Nuclear localization of DNAJB6 is associated with survival of patients with esophageal cancer and reduces AKT Signaling and Proliferation of Cancer Cells. Gastroenterology 149, 1825–1836 (2015). [DOI] [PubMed] [Google Scholar]
- Xu W. W. et al. Targeting VEGFR1- and VEGFR2-expressing non-tumor cells is essential for esophageal cancer therapy. Oncotarget 6, 1790–1805 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan D. et al. Oncogene GAEC1 regulates CAPN10 expression which predicts survival in esophageal squamous cell carcinoma. World J. Gastroenterol. 19, 2772–2780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung Y. et al. Altered E-cadherin expression and p120 catenin localization in esophageal squamous cell carcinoma. Ann. Surg. Oncol. 14, 3260–3267 (2007). [DOI] [PubMed] [Google Scholar]
- Wong M. L. et al. Aberrant promoter hypermethylation and silencing of the critical 3p21 tumour suppressor gene, RASSF1A, in Chinese oesophageal squamous cell carcinoma. Int. J. Oncol. 28, 767–773 (2006). [PubMed] [Google Scholar]
- Fatima. S. et al. Transforming capacity of two novel genes JS-1 and JS-2 located in chromosome 5p and their overexpression in human esophageal squamous cell carcinoma. Int. J. Mol. Med. 17, 159–170 (2006). [PubMed] [Google Scholar]
- Si H. X. et al. E-cadherin expression is commonly downregulated by CpG island hypermethylation in esophageal carcinoma cells. Cancer Lett. 173, 71–78 (2001). [DOI] [PubMed] [Google Scholar]
- Hu Y. C., Lam K. Y., Law. S., Wong J. & Srivastava G. Identification of differentially expressed genes in esophageal squamous cell carcinoma (ESCC) by cDNA expression array: overexpression of Fra-1, Neogenin, Id-1, and CDC25B genes in ESCC. Clin. Cancer Res. 7, 2213–2221 (2001). [PubMed] [Google Scholar]
- Chow V., Law S., Lam K. Y., Luk J. M. & Wong J. Telomerase activity in small cell esophageal carcinoma. Dis. Esophagus 14, 139–142 (2001). [DOI] [PubMed] [Google Scholar]
- Lam K. Y., Law S., Tin L., Tung P. H. & Wong J. The clinicopathological significance of p21 and p53 expression in esophageal squamous cell carcinoma: an analysis of 153 patients. Am. J. Gastroenterol. 94, 2060–2068 (1999). [DOI] [PubMed] [Google Scholar]
- Tang J. C., Lam K. Y., Law S., Wong J. & Srivastava G. Detection of genetic alterations in esophageal squamous cell carcinomas and adjacent normal epithelia by comparative DNA fingerprinting using inter-simple sequence repeat PCR. Clin. Cancer Res. 7, 1539–1545 (2001). [PubMed] [Google Scholar]
- Kwong D. et al. Chromosomal aberrations in esophageal squamous cell carcinoma among Chinese: gain of 12p predicts poor prognosis after surgery. Hum. Pathol. 35, 309–316 (2004). [DOI] [PubMed] [Google Scholar]
- Tang W. K. et al. Oncogenic properties of a novel gene JK-1 located in chromosome 5p and its overexpression in human esophageal squamous cell carcinoma. Int. J. Mol. Med. 19, 915–923 (2007). [PubMed] [Google Scholar]
- Kasem K. et al. The roles of JK-1 (FAM134B) expressions in colorectal cancer. Exp. Cell. Res. 326, 166–173 (2014). [DOI] [PubMed] [Google Scholar]
- Kasem K. et al. JK1 (FAM134B) represses cell migration in colon cancer: a functional study of a novel gene. Exp. Mol. Pathol. 97, 99–104 (2014). [DOI] [PubMed] [Google Scholar]
- Kasem K. et al. JK1 (FAM134B) gene and colorectal cancer: a pilot study on the gene copy number alterations and correlations with clinicopathological parameters. Exp. Mol. Pathol. 97, 31–36 (2014). [DOI] [PubMed] [Google Scholar]
- Kurth I. et al. Mutations in FAM134B, encoding a newly identified Golgi protein, cause severe sensory and autonomic neuropathy. Nat. Genet. 41, 1179–1181 (2009). [DOI] [PubMed] [Google Scholar]
- Davidson G. et al. Frequency of mutations in the genes associated with hereditary sensory and autonomic neuropathy in a UK cohort. J. Neurol. 259, 1673–1685 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy S. M., Davidson G. L., Brandner S., Houlden H. & Reilly M. M. Mutation in FAM134B causing severe hereditary sensory neuropathy. J. Neurol. Neurosurg. Psychiatry. 83, 119–120 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilgaz Aydinlar E. et al. Mutation in FAM134B causing hereditary sensory neuropathy with spasticity in a Turkish family. Muscle Nerve. 49, 774–775 (2014). [DOI] [PubMed] [Google Scholar]
- Kong M., Kim Y. & Lee C. A strong synergistic epistasis between FAM134B and TNFRSF19 on the susceptibility to vascular dementia. Psychiatr Genet. 2, 37–41 (2011). [DOI] [PubMed] [Google Scholar]
- Khaminets A. et al. Regulation of endoplasmic reticulum turnover by selective autophagy. Nature, 522, 354–358 (2015). [DOI] [PubMed] [Google Scholar]
- Hanahan D. & Weinberg R. A. The hallmarks of cancer. Cell 100, 57–70, (2000). [DOI] [PubMed] [Google Scholar]
- Bernards R. & Weinberg R. A. A progression puzzle. Nature, 418, 823–810 (2002). [DOI] [PubMed] [Google Scholar]
- Hamilton S. R. et al. Carcinoma of the colon and rectum. (eds Bosman F. T. et al.) WHO classification of tumours of the digestive system. IARC Press, Lyon, France, 134–146 (2010). [Google Scholar]
- Edge S. B. et al. American Joint Committee on Cancer, American Cancer Society, 7th edn. AJCC cancer staging manual, III. Springer-Verlag, New York, 143–159 (2010). [Google Scholar]
- Gopalan V. et al. GAEC1 and colorectal cancer: a study of the relationships between a novel oncogene and clinicopathologic features. Hum. Pathol. 41, 1009–1015 (2010). [DOI] [PubMed] [Google Scholar]