Abstract
The CACNA1C gene, encoding a subunit of the L-type voltage-gated calcium channel is one of the best-supported susceptibility genes for bipolar disorder (BD). Genome-wide association studies have identified a cluster of non-coding single-nucleotide polymorphisms (SNPs) in intron 3 to be highly associated with BD and schizophrenia. The mechanism by which these SNPs confer risk of BD appears to be through an altered regulation of CACNA1C expression. The role of CACNA1C DNA methylation in BD has not yet been addressed. The aim of this study was to investigate if CACNA1C DNA methylation is altered in BD. First, the methylation status of five CpG islands (CGIs) across CACNA1C in blood from BD subjects (n=40) and healthy controls (n=38) was determined. Four islands were almost completely methylated or completely unmethylated, while one island (CGI 3) in intron 3 displayed intermediate methylation levels. In the main analysis, the methylation status of CGI 3 was analyzed in a larger sample of BD subjects (n=582) and control individuals (n=319). Out of six CpG sites that were investigated, five sites showed significant hypermethylation in cases (lowest P=1.16 × 10−7 for CpG35). Nearby SNPs were found to influence the methylation level, and we identified rs2238056 in intron 3 as the strongest methylation quantitative trait locus (P=2.6 × 10−7) for CpG35. In addition, we found an increased methylation in females, and no difference between bipolar I and II. In conclusion, we find that CACNA1C methylation is associated with BD and suggest that the regulatory effect of the non-coding risk variants involves a shift in DNA methylation.
Introduction
Bipolar disorder (BD) is a common complex mental disorder characterized by episodes of mania, hypomania and depression. The disorder affects ~1% of the population and the genetic risk component is high with heritability estimates reaching 89%.1, 2 CACNA1C is one of the most consistently associated BD genes. CACNA1C, encodes the pore-forming α1C subunit of the L-type voltage-gated calcium channel also referred to as Cav1.2 α1C subunit. In the central nervous system, Cav1.2 channels are predominantly located in the postsynaptic dendritic processes and somata. On membrane depolarization, the Cav1.2 channels allow postsynaptic influx of calcium ions coupled with activation of neuronal transcription factors involved in dendritic development, neuronal survival, synaptic plasticity, as well as memory formation and learning.3, 4
The first genome-wide association study (GWAS) that suggested CACNA1C to be a susceptibility gene for BD was performed on 1461 BD patients and 2008 control individuals, collected as part of the Systematic Treatment Enhancement Program for Bipolar Disorder (STEP-BD) and the University College London (UCL) sample. This GWAS showed nominally significant association of rs1006737 located in intron 3 of CACNA1C (P=1 × 10−4) with BD.5 Subsequently, a larger GWAS with 4387 BD cases and 6209 controls, including overlapping samples from the first study, further supported the association of rs1006737, now reaching borderline genome-wide significance (7 × 10−8).6 A meta analysis of BD carried out by the Psychiatric Genomic Consortium (PGC) Working Bipolar Group, encompassing 11 974 BD cases and 51 792 controls, identified a nearby single-nucleotide polymorphism (SNP), rs4765913, as the most significant risk SNP in CACNA1C (P=1.52 × 10−8).7 The two SNPs are in moderate linkage disequilibrium (LD) (r2=0.44) and both are positioned within what is today reckoned as a narrow intronic risk locus of CACNA1C. The most recent published GWAS of BD, performed in 9747 BD patients and 14 278 controls, also found association of rs4765913 with BD (P=9.96 × 10−6).8
Shortly after the first GWAS implicated rs1006737 in BD, several studies found association of the same SNP with schizophrenia (SZ).9, 10, 11 A large GWAS of SZ (including 36 989 cases and 113 075 controls) performed by the PGC identified a highly significant association of rs2007044 in intron 3 of CACNA1C with SZ (P=2.63 × 10−17).12 A recent meta analysis further supported association of CACNA1C with SZ, with rs4765905 positioned in intron 3 as the best finding (P=5.14 × 10−17).13 It is clear, that although arrays, imputation methods and the best-associated SNPs may vary between studies, the association with both BP and SZ robustly originates from a relatively narrow set of SNPs positioned in the large intron 3 of CACNA1C.
In healthy subjects, the CACNA1C risk allele (A-allele) at rs1006737 has been associated with increased hippocampal activity during emotional processing and increased prefrontal activity during executive cognition, specific patterns of brain activity that are associated with mental illness.9 In addition, the risk allele have also been associated with higher psychopathology scores for depression and anxiety,14 higher paranoid ideation scores,15 increased activity in right amygdala during fear-face recognition,16 and increased amygdala volume,17 confirming the important role of this risk locus in the etiology of psychiatric traits.
As the risk SNPs in CACNA1C do not lead to changes in the coding sequence of the protein, the central hypothesis guiding the current research effort is that intron 3 contains important regulatory functions. Results from a recent study suggest that the intron 3 risk SNPs have a regulatory effect on three-dimensional genome architecture associated with chromosomal loopings and thus influence CACNA1C expression through interactions with its transcription start site.18 DNA methylation is a powerful key regulator of gene expression across time, tissue and lifespan,19, 20 and because it is well-established that DNA methylation levels can be influenced by nearby genetic variants, methylation changes can provide a functional mechanism by which non-coding sequence variation can result in phenotypic variability.21
In this study, we investigated if DNA methylation of CACNA1C is altered in BD cases. A systematic study of all five CpG Islands (CGIs) across the gene was first performed to get an overview of its methylation landscape. Subsequently, the DNA methylation of a CGI positioned in intron 3 of CACNA1C was analyzed in a larger case–control sample revealing significant hypermethylation in BD cases, which was partly driven by genotypes in the established intronic risk locus.
Materials and methods
Subjects and sample preparation
BD subjects and control individuals were collected at UCL and collaborating clinical centers. The UCL cases comprised Caucasian individuals who were ascertained and received clinical diagnoses of BD according to UK National Health Service (NHS) psychiatrists at interview using the categories of the International Classification of Disease version 10 (ICD10), as described in a previous BD GWAS.5 The UCL control subjects were recruited from London branches of the National Blood Service, from local NHS family doctor clinics and from university student volunteers also as described before.5 All control subjects were interviewed with the Schizophrenia and Affective Disorders Schedule-Life Time version to exclude all psychiatric disorders.22 The sample is partly overlapping with the UCL sample described in Sklar et al.5 and the PGC mega-analysis of BD.7 All subjects from the UCL sample, where DNA was still available and where the DNA was extracted from whole blood, were used for the study, in total 725 subjects. In addition, 176 new cases and controls were included, collected using the same inclusion criteria, reaching a final sample size of 901 subjects. For a description of the overlap see Supplementary Table 1.
For the UCL BD cases, pedigree and family history data were collected based on self-reports by BD participants. The BD cases were categorized according to whether there was a history of mental illness, suicide or alcohol dependence from only the maternal or paternal side of the family.
The study was approved by the National Research Ethical Committee and all participants provided written informed consent.
Genomic DNA was extracted from frozen whole blood using standard procedures. For methylation analysis, 200 ng genomic DNA from each individuals was bisulfite converted with the EZ DNA Methylation Kit (Zymo Research, Irvine, CA, USA) according to manufacturer's instructions.
EpiTYPER
EpiTYPER assays were designed for a total of 169 CpG sites spread over 5 CGIs using the EpiDesigner software (Sequenom, San Diego, CA, USA; www.EpiDesigner.com). After PCR amplification of bisulfite-converted DNA, the amplicons were treated with Shrimp Alkaline Phosphatase (Sequenom) followed by in vitro transcription. The resulting product was treated with RNase A to cleave the RNA at each U nucleotide using the T-Cleavage MassCLEAVE Kit (Sequenom). The fragments were then conditioned with Clean Resin (Sequenom) with the protocol provided in the kit and dispensed onto a SpectroCHIPII (Sequenom) and then analyzed on a MassARRAY Workstation in EpiTYPER mode. Quality control and initial data analysis were performed with the use of EpiTYPER (version 1.2, Sequenom). Oligonucleotide sequences for the EpiTYPER assays are provided in Supplementary Table 2.
iPLEX
iPLEX assays for six selected CpG sites in CGI 3, located in intron 3 of CACNA1C, were designed using Assay Designer in Typer 4.0 (Sequenom) using bioinformatically bisulfite-converted sequences of the targeted region. Inosine was added to oligonucleotide sequences that contained overlapping CpGs or SNPs. No more than one inosine was allowed in an oligonucleotide to ensure binding specificity. The oligonucleotide sequences for the iPLEX assays are listed in Supplementary Table 3. The amplified products were prepared according to iPLEX manual and dispensed on the same equipment as described above. The data were analyzed in Allelotype mode. Quality control and detection of allele ratios (as a measure of methylation status) were performed using Typer 4.0 (Sequenom). The data were subsequently imported into R (www.r-project.org) for statistical analyses. During all experimental work and QC, the investigator was blinded to affection status. Cases and controls were in addition spread in between each other on each plate during bisulfite conversion and Sequenom analysis to avoid experimental batch effects.
Statistical analyses
A non-parametric Mann–Whitney test was used to compare the levels of DNA methylation between cases and controls, males and females, cases with bipolar I and bipolar II, and cases with a positive versus a negative familial history. For the analysis of familial history, cases with a maternal history of psychiatric disorder (maternal) were compared with cases with no mental disorder in the family (none). A similar analysis was carried out comparing cases with paternal history versus none. Methylation quantitative trait loci (mQTL) analysis was performed using the 725 subjects from the UCL sample with Affymetrix 500 K Array genotypes available through the NIMH Repository (https://www.nimhgenetics.org/available_data/data_biosamples/gwas_data.php).
The mQTL analysis was performed using linear regression in PLINK23 including genotyped SNPs located in a region that encompassed the CACNA1C gene plus 5 kb up- and downstream (focused analysis) or all genotyped SNPs on chromosome 12 (full analysis). For the most significant CpG site in CGI 3, a beta regression analysis was performed to assess the contribution of case–control status, genotype and gender on DNA methylation level. A genetic association analysis in the UCL sample was performed for 109 genotyped SNPs in CACNA1C and a 5 kb up- and downstream region using a linear regression in PLINK and an additive model.
Results
Association of CACNA1C methylation with BD
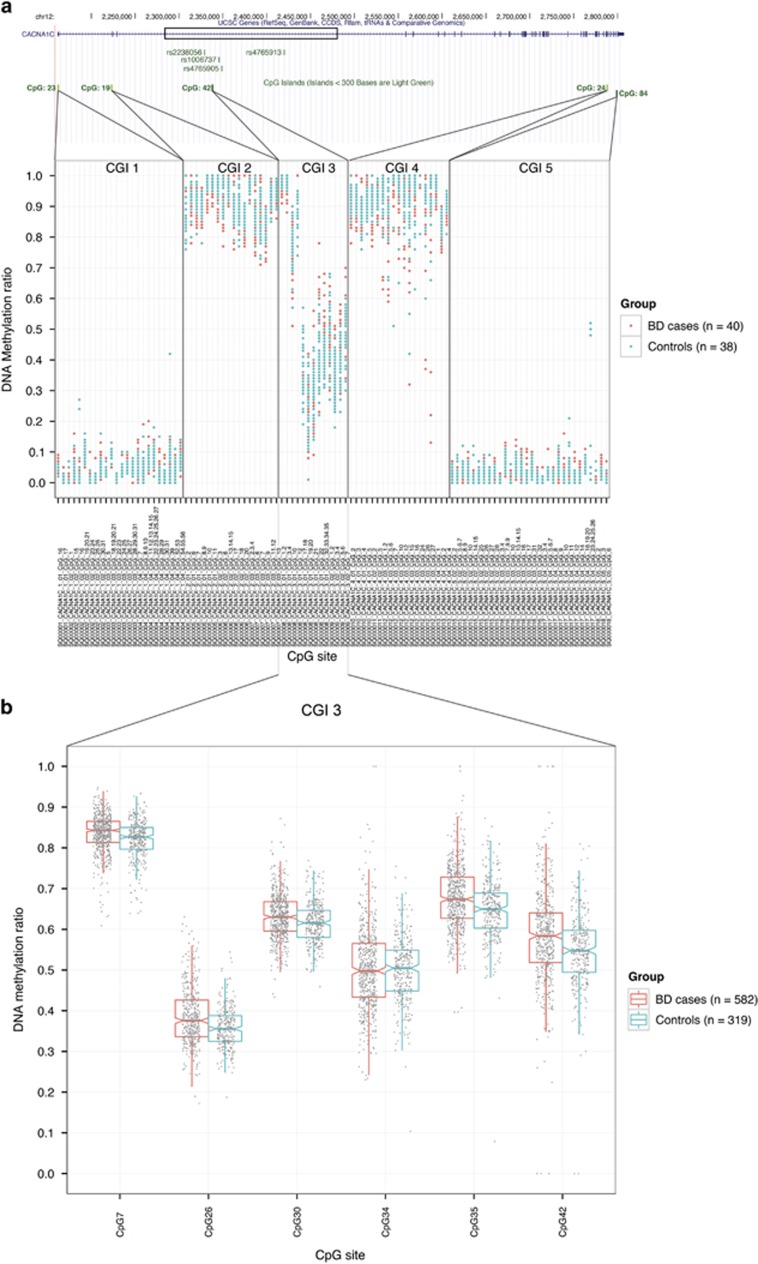
The initial methylation profile of the five CACNA1C CGIs in DNA derived from the blood of BD subjects (n=40) and controls (n=38) using EpiTYPER identified a large difference in DNA methylation between the islands with relatively little overall difference between individuals. In general, CGI 1 and CGI 5 were found to have low methylation levels, CGI 2 and CGI 4 were highly methylated, while CGI 3 displayed intermediate methylation (Figure 1a).
Figure 1.
DNA methylation of CACNA1C in BD cases and controls. (a) Shows the gene structure of CACNA1C and methylation landscape across its all five CpG islands in BD cases and controls; (b) shows detailed analysis of DNA methylation levels of CpG island 3 in CACNA1C in BD cases in comparison to controls. BD, bipolar disorder. CGI, CpG island.
The main methylation analysis of CGI 3 included six CpG sites that were evaluated in BD subjects (n=582) and control individuals (n=319). From this analysis, a relatively small but highly significant degree of hypermethylation in BD subjects was identified for five of the six CpG sites (Figure 1b and Table 1). The most significant difference in methylation between BD subjects and controls was found for CpG35 (P=1.16 × 10−7, Δβ=0.03) (Figure 1b and Table 1). The DNA methylation levels were highly correlated between the six sites (Supplementary Table 4).
Table 1. Overview of association between DNA methylation and BD and mQTL for CGI 3 of CACNA1C.
|
Methylation difference between BD cases and controls P-value |
Focused mQTL analysis |
Full mQTL analysis |
rs1006737 as mQTL | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CpG site | All individuals | Females only | Males only | mQTL (most significant) | P-value | P-value (Bonferroni-corrected 24 markers) | mQTL (most significant) | P-value | P-value (Bonferroni-corrected 2634 markers) | P-value |
| 7 | 6.16E−07 | 1.39E−04 | 9.06E−03 | rs2239030 | 4.36E−05 | 1.05E−03 | rs2239030 | 4.36E−05 | 1.15E−01 | 9.52E−03 |
| 26 | 7.15E−07 | 1.60E−04 | 1.23E−02 | rs2239030 | 4.74E−04 | 1.14E−02 | rs4026015 | 1.49E−05 | 3.92E−02 | 2.85E−02 |
| 30 | 2.00E−05 | 7.20E−03 | 7.17E−03 | rs2238056 | 2.90E−07 | 6.96E−06 | rs2238056 | 2.90E−07 | 7.64E−04 | 1.36E−04 |
| 34 | 7.70E−01 | 3.54E−01 | 7.68E−01 | rs2238056 | 1.38E−04 | 3.32E−03 | rs2238056 | 1.38E−04 | 3.65E−01 | 6.73E−02 |
| 35 | 1.16E−07 | 1.01E−03 | 6.41E−04 | rs2238056 | 2.63E−07 | 6.32E−06 | rs2238056 | 2.63E−07 | 6.94E−04 | 3.02E−04 |
| 42 | 8.66E−07 | 1.54E−04 | 3.09E−02 | rs10848634 | 2.14E−07 | 5.13E−06 | rs10848634 | 2.14E−07 | 5.63E−04 | 8.47E−05 |
Abbreviations: BD, bipolar disorder; CGI, CpG island; mQTL, methylation quantitative trait loci.
Entries marked in bold indicate findings with P-value <0.05.
Identification of mQTL
Because it is well-known that genetic variation can influence methylation levels24, 25 we speculated whether methylation at CGI 3 was influenced by nearby SNPs. A correlation analysis including methylation levels at the six CpG sites and SNPs on chromosome 12 was performed for all 725 individuals for whom genotype information was available (Supplementary Table 1). This analysis was performed by including all genotyped SNPs within CACNA1C, as well as 5 kb upstream and downstream of the gene (109 SNPs, 24 of whom were independent). This analysis will henceforth be referred to as the focused analysis. In addition, an analysis including all SNPs on chromosome 12 (15 882 SNPs, 2634 independent) was performed, referred to as the full analysis.
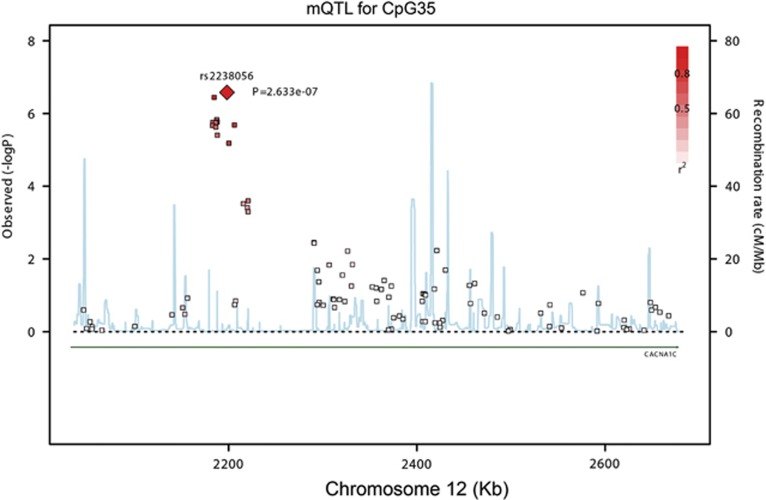
In the focused analysis, we found multiple SNPs that were acting as significant mQTL for sites in CGI 3 (Table 1). All of these SNPs were located within the relatively narrow risk locus in intron 3. SNP rs2238056 was identified as the strongest mQTL (P=2.6 × 10−7) for CpG35, the most significantly associated CpG site in BD subjects (Figure 2). In the full analysis, all the SNPs that were identified as mQTL in the focused analysis were re-identified as the best mQTL SNPs, with exception of CpG26, where rs2239030 was replaced with rs4026015 as the most associated mQTL SNP (Table 1). Overall the analyses demonstrate that CGI 3 methylation is influenced primarily by SNPs within the intron 3 risk locus of CACNA1C. The mQTL for CpG35 remained significant after Bonferroni correction in both the focused and the full mQTL analyses (Table 1). Interestingly, the mQTL SNPs identified in this study were in moderate LD with the bipolar GWAS top hit rs4765913 (index SNP) with r2=0.32 for rs2239030, r2=0.3 for rs2238056 and r2=0.4 for rs10848634 according to results from the PGC Bipolar Disorder Working Group.7 This finding supports the hypothesis that the well-established risk SNPs in CACNA1C for BD may have a role in DNA methylation status.
Figure 2.
Overview for mQTL analysis between all SNPs genotyped in CACNA1C in the UCL sample and DNA methylation levels at the most significantly hypermethylated CpG site (CpG35). mQTL, methylation quantitative trait loci; SNP, single-nucleotide polymorphism; UCL, University College London.
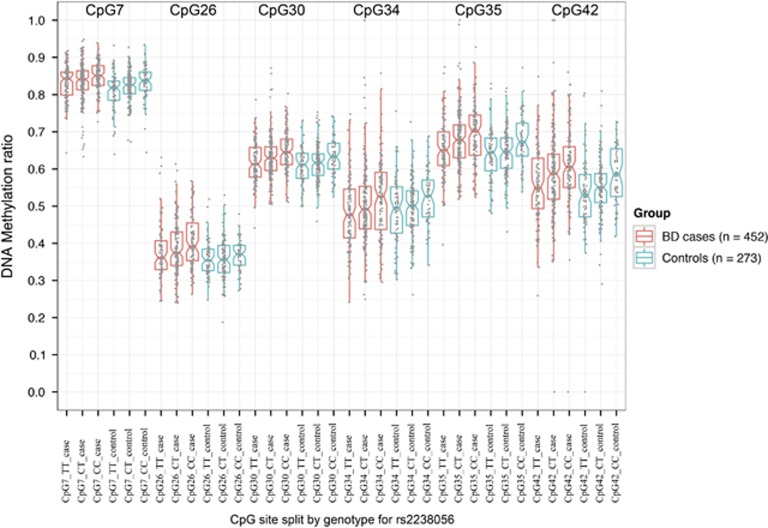
For the best CGI 3 mQTL in our study (rs2238056), the minor C-allele was associated with increased DNA methylation levels for all of the examined CpG sites in CGI 3 (Figure 3). A similar analysis for rs1006737 showed that the risk minor A-allele was associated with increased DNA methylation for all CGI 3 CpG sites in a dose-dependent manner, which is consistent with these two minor alleles being in LD. This effect was found in both cases and controls (Table 1 and Supplementary Figure 1).
Figure 3.
DNA methylation levels at six CpG sites in CpG island 3 of CACNA1C split by genotype of rs2238056, identified as most significant mQTL for the most significantly hypermethylated CpG site (CpG35). BD, bipolar disorder; mQTL, methylation quantitative trait loci.
To test if the identified top mQTL could be risk SNPs themselves, we carried out a genetic association analysis in the UCL sample, including rs1006737 for comparison. Two of these top mQTL and rs1006737 were borderline significantly associated with BD in the UCL sample, with better P-values in the PGC-BP data set, most likely because of larger sample size (Supplementary Table 5).26 Interestingly, the mQTL rs2238056 is highly associated with SZ in the PGC SCZ 52 study (P=1.45 × 10−13).12 The data suggest that the mQTL may be risk SNPs themselves, although the high degree of LD in the region makes the identification of the true causal risk SNP(s) complex.
Effect of gender, bipolar I/II and familial history
We finally investigated if any of the other known variables examined in the BD cases (Supplementary Table 1) could explain the variation in methylation among cases and controls. We found a hypermethylation in females compared with males. A gender-specific analysis confirmed a significant hypermethylation in cases both among females and males (Table 1). A beta regression analysis of CpG35 showed significant correlation of DNA methylation level with affection status (P=1.61 × 10−5), gender (P=2.27 × 10−5) and genotype of rs2238056 (P=6.10 × 10−5). All these covariates influenced methylation level to approximately the same extent. Odds of methylation increased by 15% for being a case, 15% for being a female and by 9% for each copy of the minor allele of rs2238056. No difference was found between cases with or without a parental history of psychiatric disease (P>0.05). Also, no difference was found between bipolar I or bipolar II patients (P>0.05); however, due to a small number of individuals with BD II diagnosis these results may not be generalizable. We conclude that the CGI 3 hypermethylation in cases is driven partly, but not entirely, by genotypes and by gender.
Discussion
To our knowledge, this is the first study to investigate DNA methylation of CACNA1C in context of BD and with BD-associated genetic risk variants. We profiled the DNA methylation landscape of five CGIs in CACNA1C in blood-derived DNA and found CGI 3 to be significantly hypermethylated in BD subjects compared with controls. We also found that methylation was influenced by gender as well as by nearby genotypes.
Because of LD in the human genome, it is in general difficult to pinpoint the causal variation in a locus identified in a GWAS. For complex traits specifically it may be impossible to demonstrate that an implicated SNP is the causal variant. However, as pointed out in an editorial comment in Nature Genetics,27 there should at least be evidence that the variants identified in a GWAS show a consistent correlation with gene expression or gene regulation that is also highly correlated with the human trait originally assayed. In our study, we found an overall hypermethylation in bipolar subjects, as well as increased methylation associated with the well-established risk A-allele for susceptibility SNP rs1006737. Our study opens up the possibility that the non-coding CACNA1C risk SNPs may confer risk of disease through altered methylation in intron 3.
Altered DNA methylation within a gene, as observed in this study, has been linked to abnormal gene expression, alternative mRNA splicing and use of alternative promoters.28, 29, 30, 31 The exact relationship between DNA methylation and gene expression is, however, greatly dependent on the genomic location and tissue.29 It is therefore not straightforward to predict the effect and origin of the increased methylation found in blood of bipolar subjects in this study.
Previous studies investigating the correlation between CACNA1C risk genotypes and gene expression have found opposite effects of the risk allele depending on the investigated tissue. In human cerebellum, the risk A-allele of rs1006737 was correlated with decreased CACNA1C mRNA expression,18, 32 while the same allele was correlated with increased expression in human post-mortem dorsolateral prefrontal cortex9 and in induced human neurons.33 In the GTEx portal (V6 Release)34 a single significant eQTL for CACNA1C expression (rs7297582 in CACNA1C intron 3) was identified in cerebellum, with the minor allele correlating with reduced expression. As LD exist between rs7297582 and rs1006737 (r2=0.71), these data are in line with the previously published studies in cerebellum.18, 32 No shared eQTLs exist between blood and brain in GTEx with the current sample size (338 whole blood).
Whether the CGI 3 hypermethylation observed in our study was driven directly or indirectly by the nearby risk alleles is unknown. A recent study investigating the mechanistic relationship between the SNPs, DNA methylation and gene expression found that genetic variation can act either directly (SNP affects methylation, which change gene expression) or indirectly (SNP affects gene expression, which affects methylation level) on DNA methylation, and that the effect may vary with tissue.35 Studies addressing methylation and CACNA1C expression across different brain regions are necessary to fully uncover the molecular effect of increased CACNA1C methylation.
Increased CACNA1C CGI 3 methylation was detected in females. It is unclear if this gender-specific methylation effect is related to disease because this difference was also observed in the control subjects. Gender differences have previously been reported in CACNA1C genetic association studies, for example, Dao et al.,36 found female-only association between mood disorders and multiple intron 3 SNPs, that included rs1006737. Gender-specific association was also observed by Strohmaier et al.,37 who reported gender-specific impacts of rs1006737 in CACNA1C on a range of personality traits, resilience factors and depressive symptoms in 3793 healthy subjects. These studies differ in the rs1006737 alleles that were associated with mood disorders and their endophenotypes and so the impact of gender on genetic association at this locus remains unclear.37 It is possible that increased baseline DNA methylation of CGI 3 in females may provide a mechanism through which the risk allele of CACNA1C could determine sex-specific endophenotypes of BD.
A potential limitation of this study is the use of blood samples for methylation analysis. One argument for using blood samples for DNA methylation in psychiatric disorders is the common accessibility to such samples, providing adequate statistical power to detect small effects and mQTL. Moreover, recent studies have indicated that the DNA methylation status of many CpG sites in the brain are mirrored in the blood,38 and that common mQTL also exist between these two tissues, supporting the use of blood as a surrogate tissue to delineate disease-relevant processes within the brain.39
It is possible that medication influences DNA methylation status of CACNA1C. Two recent studies investigated the effect of commonly used mood stabilizers (including lithium, valproate and carbamazepine) on human methylome in neuroblastoma cells and whole blood of BD patients and reported significant drug-related changes in DNA methylation at several genes.40, 41 However, both of these studies used the Infinium HumanMethylation27 BeadChip array (Illumina, San Diego, CA, USA), where CGI 3 of CACNA1C is not covered, making a more comprehensive analysis of the effects of mood stabilizers on CACNA1C methylation of high interest in the future.
Overall, we have identified CACNA1C hypermethylation in whole blood of BD patients. Our study indicates that the regulatory effect of risk alleles in intron 3 is accompanied by an upwards shift in DNA methylation of the intron 3 CGI. In conclusion, CACNA1C DNA methylation may play a role in BD.
Acknowledgments
This study was funded by grants from the Toyota-Foundation and the Lundbeck Foundation, Denmark. The UCL clinical and control samples were collected with support from the Bipolar Organization, the Neuroscience Research Charitable Trust, the Central London NHS Blood Transfusion Service, the Camden and Islington NHS Foundation Trust and 10 other NHS Mental Health Trusts, the Stanley Foundation, the National Institute for Health Research Mental Health Research Network and the NIHR supported Primary Care Research Network. Genetic analysis of the UCL cohort has been supported by UK Medical Research Council project grants G9623693N, G0500791, G0701007 and G1000708. The Stanley Foundation and the Stanley Psychiatric Research Center at the Broad Institute, Boston, Massachusetts funded the genome-wide association genotyping. The funding bodies had no involvement in any aspect of the study.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Translational Psychiatry website (http://www.nature.com/tp)
Supplementary Material
References
- Craddock N, Sklar P. Genetics of bipolar disorder: successful start to a long journey. Trends Genet 2009; 25: 99–105. [DOI] [PubMed] [Google Scholar]
- Craddock N, Jones I. Genetics of bipolar disorder. J Med Genet 1999; 36: 585–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch B, Szostek A, Lin R, Shevtsova O. Subcellular distribution of L-type calcium channel subtypes in rat hippocampal neurons. Neuroscience 2009; 164: 641–657. [DOI] [PubMed] [Google Scholar]
- Bhat S, Dao DT, Terrillion CE, Arad M, Smith RJ, Soldatov NM et al. CACNA1C (Cav1.2) in the pathophysiology of psychiatric disease. Prog Neurobiol 2012; 99: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklar P, Smoller JW, Fan J, Ferreira MAR, Perlis RH, Chambert K et al. Whole-genome association study of bipolar disorder. Mol Psychiatry 2008; 13: 558–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira MAR, O'Donovan MC, Meng YA, Jones IR, Ruderfer DM, Jones L et al. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat Genet 2008; 40: 1056–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psychiatric GWAS Consortium Bipolar Disorder Working Group.. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet 2011; 43: 977–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühleisen TW, Leber M, Schulze TG, Strohmaier J, Degenhardt F, Treutlein J et al. Genome-wide association study reveals two new risk loci for bipolar disorder. Nat Commun 2014; 5: 3339. [DOI] [PubMed] [Google Scholar]
- Bigos KL, Mattay VS, Callicott JH, Straub RE, Vakkalanka R, Kolachana B et al. Genetic variation in CACNA1C affects brain circuitries related to mental illness. Arch Gen Psychiatry 2010; 67: 939–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyegaard M, Demontis D, Foldager L, Hedemand A, Flint TJ, Sørensen KM et al. CACNA1C (rs1006737) is associated with schizophrenia. Mol Psychiatry 2010; 15: 119–121. [DOI] [PubMed] [Google Scholar]
- Green EK, Grozeva D, Jones I, Jones L, Kirov G, Caesar S et al. The bipolar disorder risk allele at CACNA1C also confers risk of recurrent major depression and of schizophrenia. Mol Psychiatry 2010; 15: 1016–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke S, Neale BM, Corvin A, Walters JTR, Farh K-H, Holmans PA et al. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014; 511: 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Glatt SJ, Uchiyama M, Faraone SV, Tsuang MT. Meta-analysis of data from the Psychiatric Genomics Consortium and additional samples supports association of CACNA1C with risk for schizophrenia. Schizophr Res 2015; 168: 429–433. [DOI] [PubMed] [Google Scholar]
- Erk S, Meyer-Lindenberg A, Schnell K, Opitz von Boberfeld C, Esslinger C, Kirsch P et al. Brain function in carriers of a genome-wide supported bipolar disorder variant. Arch Gen Psychiatry 2010; 67: 803–811. [DOI] [PubMed] [Google Scholar]
- Roussos P, Giakoumaki SG, Georgakopoulos A, Robakis NK, Bitsios P. The CACNA1C and ANK3 risk alleles impact on affective personality traits and startle reactivity but not on cognition or gating in healthy males. Bipolar Disord 2011; 13: 250–259. [DOI] [PubMed] [Google Scholar]
- Jogia J, Ruberto G, Lelli-Chiesa G, Vassos E, Maierú M, Tatarelli R et al. The impact of the CACNA1C gene polymorphism on frontolimbic function in bipolar disorder. Mol Psychiatry 2011; 16: 1070–1071. [DOI] [PubMed] [Google Scholar]
- Lancaster TM, Foley S, Tansey KE, Linden DEJ, Caseras X. CACNA1C risk variant is associated with increased amygdala volume. Eur Arch Psychiatry Clin Neurosci 2015; 266: 269–275. [DOI] [PubMed] [Google Scholar]
- Roussos P, Mitchell AC, Voloudakis G, Fullard JF, Pothula VM, Tsang J et al. A role for noncoding variation in schizophrenia. Cell Rep 2014; 9: 1417–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev 2002; 16: 6–21. [DOI] [PubMed] [Google Scholar]
- Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature 2007; 447: 433–440. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Rohde C, Reinhardt R, Voelcker-Rehage C, Jeltsch A. Non-imprinted allele-specific DNA methylation on human autosomes. Genome Biol 2009; 10: R138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endicott J, Spitzer RL. A diagnostic interview: the schedule for affective disorders and schizophrenia. Arch Gen Psychiatry 1978; 35: 837–844. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MJ, Fejes AP, Kobor MS. DNA methylation, genotype and gene expression: who is driving and who is along for the ride? Genome Biol 2013; 14: 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs JR, van der Brug MP, Hernandez DG, Traynor BJ, Nalls MA, Lai S-L et al. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet 2010; 6: e1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirooznia M, Seifuddin F, Judy J, Goes FS, Potash JB, Zandi PP. Metamoodics: meta-analysis and bioinformatics resource for mood disorders. Mol Psychiatry 2014; 19: 748–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- On beyond GWAS. Nat Genet 2010; 42: 551. [DOI] [PubMed] [Google Scholar]
- Lev Maor G, Yearim A, Ast G. The alternative role of DNA methylation in splicing regulation. Trends Genet 2015; 31: 274–280. [DOI] [PubMed] [Google Scholar]
- Hellman A, Chess A. Gene body-specific methylation on the active X chromosome. Science 2007; 315: 1141–1143. [DOI] [PubMed] [Google Scholar]
- Laurent L, Wong E, Li G, Huynh T, Tsirigos A, Ong CT et al. Dynamic changes in the human methylome during differentiation. Genome Res 2010; 20: 320–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D'Souza C, Fouse SD et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature 2010; 466: 253–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershon ES, Grennan K, Busnello J, Badner JA, Ovsiew F, Memon S et al. A rare mutation of CACNA1C in a patient with bipolar disorder, and decreased gene expression associated with a bipolar-associated common SNP of CACNA1C in brain. Mol Psychiatry 2014; 19: 890–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimizu T, Pan JQ, Mungenast AE, Madison JM, Su S, Ketterman J et al. Functional implications of a psychiatric risk variant within CACNA1C in induced human neurons. Mol Psychiatry 2015; 20: 162–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonsdale J, Thomas J, Salvatore M, Phillips R, Lo E, Shad S et al. The genotype-tissue expression (GTEx) project. Nat Genet 2013; 45: 580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Arcelus M, Lappalainen T, Montgomery SB, Buil A, Ongen H, Yurovsky A et al. Passive and active DNA methylation and the interplay with genetic variation in gene regulation. Elife 2013; 2: e00523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao DT, Mahon PB, Cai X, Kovacsics CE, Blackwell RA, Arad M et al. Mood disorder susceptibility gene CACNA1C modifies mood-related behaviors in mice and interacts with sex to influence behavior in mice and diagnosis in humans. Biol Psychiatry 2010; 68: 801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohmaier J, Amelang M, Hothorn LA, Witt SH, Nieratschker V, Gerhard D et al. The psychiatric vulnerability gene CACNA1C and its sex-specific relationship with personality traits, resilience factors and depressive symptoms in the general population. Mol Psychiatry 2013; 18: 607–613. [DOI] [PubMed] [Google Scholar]
- Aberg KA, Xie LY, McClay JL, Nerella S, Vunck S, Snider S et al. Testing two models describing how methylome-wide studies in blood are informative for psychiatric conditions. Epigenomics 2013; 5: 367–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AK, Kilaru V, Kocak M, Almli LM, Mercer KB, Ressler KJ et al. Methylation quantitative trait loci (meQTLs) are consistently detected across ancestry, developmental stage, and tissue type. BMC Genomics 2014; 15: 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai T, Bundo M, Sugawara H, Sunaga F, Ueda J, Tanaka G et al. Effect of mood stabilizers on DNA methylation in human neuroblastoma cells. Int J Neuropsychopharmacol 2013; 16: 2285–2294. [DOI] [PubMed] [Google Scholar]
- Houtepen LC, van Bergen AH, Vinkers CH, Boks MP. DNA methylation signatures of mood stabilizers and antipsychotics in bipolar disorder. Epigenomics 2016; 8: 197–208. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.