Abstract
Objective
To determine clinical and biological variables that predict time to initiation of symptomatic therapy in de novo Parkinson's disease patients.
Methods
Parkinson's Progression Markers Initiative is a longitudinal case–control study of de novo, untreated Parkinson's disease participants at enrolment. Participants contribute a wide range of motor and non‐motor measures, including biofluids and imaging biomarkers. The machine learning method of random survival forests was used to examine the ability of baseline variables to predict time to initiation of symptomatic therapy since study enrollment (baseline).
Results
There were 423 PD participants enrolled in PPMI and 33 initial baseline variables. Cross‐validation results showed that the three‐predictor subset of disease duration (time from diagnosis to enrollment), the modified Schwab and England activities of daily living scale, and the Movement Disorder Society Unified Parkinson's Disease Rating Scale (MDS‐UPDRS) total score modestly predicted time to initiation of symptomatic therapy (C = 0.70, pseudo‐R 2 = 0.13). Prediction using the three variables was similar to using the entire set of 33. None of the biological variables increased accuracy of the prediction. A prognostic index for time to initiation of symptomatic therapy was created using the linear and nonlinear effects of the three top variables based on a post hoc Cox model.
Interpretation
Our findings using a novel machine learning method support previously reported clinical variables that predict time to initiation of symptomatic therapy. However, the inclusion of biological variables did not increase prediction accuracy. Our prognostic index constructed, based on the group‐level survival curve can provide an indication of the risk of initiation of ST for PD patients based on functions of the three top predictors.
Keywords: Biomarkers, Parkinson's disease, symptomatic therapy
Introduction
Parkinson's disease (PD) is a chronic neurodegenerative disease that has a substantial impact on the patient quality of life (QOL). While there is no curative treatment, there is a large armamentarium of symptomatic therapy (ST). Time to initiation of symptomatic therapy (TIST) is considered to be an important milestone in disease progression.1 A number of previously completed large studies that tested putative disease‐modifying therapies used TIST as the primary outcome measure for the efficacy of intervention.2, 3, 4 TIST is a milestone that is subject to a number of subjective modifiers including patient and physician preference. However, TIST reflects the degree of disease‐related disability, and as such, it is likely linked to the degree of underlying neurodegeneration. All previously completed analysis of the TIST were based on the correlation of the baseline demographics and disease‐related clinical characteristics.5, 6, 7 This study aimed to explore clinical and, for the first time, biological predictors of TIST in a large cohort of at‐baseline de novo PD participants using modern machine learning methods.
Methods
Study design
Data used in the preparation of this article were obtained from the Parkinson's Progression Markers Initiative (PPMI) database (www.ppmi-info.org/data). Subjects with newly‐diagnosed untreated PD and matched healthy controls (HCs) were enrolled in the PPMI, a study for which the aims and methods were previously published.8 In brief, PPMI is an ongoing observational, international, multicenter (18 US, 5 European, and one Australian sites) study aimed to identify serological, genetic, spinal fluid, and imaging biomarkers of PD progression in a large cohort of participants with newly‐diagnosed PD compared to matched HC. The study was launched in June 2010 and has successfully completed enrollment of 423 PD participants and 196 HCs. The data used for this paper constitutes the analysis of the baseline and up to 49 months‐follow‐up dataset for the PD cohort as obtained from the PPMI database (www.ppmi-info.org, accessed September 15, 2014).
Participants
At baseline, PD subjects were required to be above age 30 and (1) have two of the following: bradykinesia, rigidity and resting tremor or have an asymmetric resting tremor, or asymmetric bradykinesia; (2) have been recently diagnosed (within 2 years); (3) untreated; and (4) have reduced striatal 123‐I Ioflupane dopamine transporter (DatScan®, GE Healthcare, Arlington Heights, IL) imaging binding consistent with PD. As per protocol, participants were expected not to require initiation of ST for the first 6 months of the study. Afterward the decision of the time and choice of ST were made by the site investigator. Data on the TIST was reported to the coordination center and captured in the database. PPMI participants are assessed with a wide spectrum of clinical measures including Movement Disorders Society Unified Parkinson's Disease Rating Scale (MDS‐UPDRS)9 total and subscale scores.
Standard protocol approvals, registrations, and consents
Each participating PPMI site (1) received approval from an ethical standards committee on human experimentation before study initiation; and (2) obtained written informed consent for research from all individuals participating in the study.
Statistical methods
The outcome was TIST, defined as the years to initiation of ST from study entry (baseline), which was censored for 33% of the sample. Little is known about multivariate prediction of TIST, and predictors might interact in complex ways, might be highly correlated, or might have nonlinear effects. To allow for such possibilities, the machine learning method of random survival forests (RSF) was used for the analysis.10 RSF is a variant of random forests11 for right‐censored data which use randomization in growing recursive regression trees and then averages over the trees, which tends to produce more accurate predictions relative to traditional methods.12 RSF requires minimal data assumptions, automatically accounts for complex relationships, and has previously proven useful in exploratory neurodegenerative research.13 In the analysis, 2000 trees were grown for each group of predictors and averaging was over all the trees to yield estimated survival curves and indexes of the variables’ predictive ability. There was <1% missing data for the predictors (no missing data for TIST), which was dynamically imputed within RSF using an iterative algorithm.10
Two models were planned prior to the analysis and six models were unplanned. The planned models were a reference model with no predictors (Reference‐0) and the model with all 33 predictors (RSF‐33). The 33 predictors were selected based on all the variables collected in the PPMI database and included subject demographics, disease characteristics, cerebrospinal fluid biomarkers, and dopamine transporter imaging striatal binding ratios (DATscan) as summarized in Table 3. The unplanned models were reduced‐variable models (<33 predictors) based on the results of RSF‐33. The last two models were semi‐parametric survival models (Cox models) developed from the 3‐predictor RSF (RSF‐3; justification provided below). The Cox model was included because RSF is a “black‐box” method that does not provide a single regression equation illustrating the nature of the predictor effects. For the full Cox model (Cox‐full), all main effects were included, all nonlinear effects (quadratic polynomials), and interactions among pairs of predictors (product terms). Backward elimination based on the AIC14 was used to select a final reduced model with fewer effects (Cox‐reduced).
Table 3.
Prediction accuracy of Reference model (no predictors), random survival forests (RSF), and Cox regression models
| Planned? | C | Brier | Pseudo R 2 | |
|---|---|---|---|---|
| Reference‐0 | Yes | 0.50 | 0.14 | NA |
| RSF‐33 | Yes | 0.68 | 0.12 | 0.13 |
| RSF‐26 | No | 0.68 | 0.12 | 0.13 |
| RSF‐07 | No | 0.70 | 0.12 | 0.12 |
| RSF‐03 | No | 0.70 | 0.12 | 0.13 |
| RSF‐01 | No | 0.64 | 0.13 | 0.05 |
| Cox‐full | No | 0.71 | 0.11 | 0.18 |
| Cox‐reduced | No | 0.71 | 0.11 | 0.19 |
Label suffix is number of predictors (but not number of effects; see Discussion in the text).
All models were developed on the full dataset, and the concordance index, C, was used to index in‐sample prediction accuracy, which indicates the extent to which the predicted survival for a pair of patients correctly orders them in terms of their actual TIST. Cross‐validation was used to help account for optimism due to variable selection and other biases.15 Because C is not a strictly proper scoring rule, the integral of the time‐dependent Brier score (BS) was used to assess predictive accuracy in the cross–validation over the survival time (smaller % indicating less prediction error)16 and a pseudo‐R 2 was computed to index the relative BS for each model compared to Reference‐0 (no predictors). A recent survey of oncology and cardio‐vascular research found mean C = 0.78 with the lower and higher quartiles of 0.69 and 0.88, respectively. Therefore, we consider C = 0.78 to represent average effect size and the quartiles to represent “small” and “large” effects.17
Results
Demographics
Baseline demographic and disease characteristics of the cohort divided by the ST− (censored event) versus ST+ (observed event) subgroups are presented in Table 1. At the time of data analysis, the mean duration of study participation was 2.11 years (SD = 0.8; min 0.0/max 4.1), and 284/423 subjects started ST. The mean (SD) of TIST was 0.78 (0.5) years (N = 92 before 6 months). Table 2 shows demographic and PD characteristics of the cohort separated by the time to initiation of ST (N = 118 between 6 and 12 months, N = 65 between 1 and 2 years, and N = 9 after 2 years since enrollment). Based on t‐tests and chi‐squared tests comparing the ST+ and ST− groups, the following variables were significantly associated with the initiation of ST: higher level of education (P = 0.0149), higher (worse) baseline MDS‐UPDRS scores (P < 0.0001), lower (worse) Schwab and England activities of daily living (SE‐ADL) scores18(P < 0.0001), higher (worse) degree of postural instability and gait disorder subscore (PIGD) and tremor subscore of the MDS‐UPDRS (P = 0.0141 and P = 0.0026, respectively) calculated based on the algorithm published by Stebbins et al.,19 worse cognition as measured by Montreal Cognitive Assessment Scale (MOCA)20 (P = 0.0031) and longer study participation (<0.0001) .
Table 1.
Baseline demographics and PD characteristics
| Variable | All PD | PD Subjects | PD Subjects | P‐value (ST− vs. ST+) |
|---|---|---|---|---|
| Subjects | ST‐ (censored event) | ST+ (observed event) | ||
| (N = 423) | (N = 139) | (N = 284) | ||
| Age | 0.9664 | |||
| Mean (SD) | 61.66 (9.7) | 61.69 (10.3) | 61.65 (9.4) | |
| (Min, Max) | (33.5, 84.9) | (33.5, 82.3) | (33.7, 84.9) | |
| Missing | 0 | 0 | 0 | |
| Gender | 0.3810 | |||
| Male | 277 (65.48%) | 87 (62.59%) | 190 (66.90%) | |
| Female | 146 (34.52%) | 52 (37.41%) | 94 (33.10%) | |
| Missing | 0 | 0 | 0 | |
| Education | 0.0149 | |||
| <13 Years | 76 (17.97%) | 34 (24.46%) | 42 (14.79%) | |
| 13–23 Years | 344 (81.32%) | 104 (74.82%) | 240 (84.51%) | |
| >23 Years | 3 (0.71%) | 1 (0.72%) | 2 (0.70%) | |
| Missing | 0 | 0 | 0 | |
| Ethnicity | 0.4922 | |||
| Hispanic/latino | 9 (2.13%) | 2 (1.44%) | 7 (2.46%) | |
| Not Hispanic/latino | 414 (97.87%) | 137 (98.56%) | 277 (97.54%) | |
| Missing | 0 | 0 | 0 | |
| Race | 0.5530 | |||
| White | 391 (92.43%) | 130 (93.53%) | 261 (91.90%) | |
| Black/African‐American | 6 (1.42%) | 2 (1.44%) | 4 (1.41%) | |
| Asian | 8 (1.89%) | 1 (0.72%) | 7 (2.46%) | |
| Other | 18 (4.26%) | 6 (4.32%) | 12 (4.23%) | |
| Missing | 0 | 0 | 0 | |
| Family history of PD | 0.4161 | |||
| Family members w/PD | 102 (24.17%) | 30 (21.74%) | 72 (25.35%) | |
| No family members w/PD | 320 (75.83%) | 108 (78.26%) | 212 (74.65%) | |
| Missing | 1 | 1 | 0 | |
| MDS‐UPDRS mean (SD) score and subscores | ||||
| MDS‐UPDRS total score | 32.36 (13.1) | 27.90 (12.0) | 34.54 (13.1) | <.0001 |
| MDS‐UPDRS part I | 5.57 (4.1) | 4.94 (4.1) | 5.88 (4.0) | 0.0252 |
| MDS‐UPDRS part II | 5.90 (4.2) | 4.64 (3.4) | 6.52 (4.4) | <.0001 |
| MDS‐UPDRS part III (Motor Exam) | 20.89 (8.9) | 18.32 (8.4) | 22.15 (8.8) | <.0001 |
| Missing | 1 | 0 | 1 | |
| Hoehn and Yahr | 0.2371 | |||
| Stage 1 | 186 (43.97%) | 68 (48.92%) | 118 (41.55%) | |
| Stage 2 | 235 (55.56%) | 71 (51.08%) | 164 (57.75%) | |
| Stage 3–5 | 2 (0.47%) | 0 (0.00%) | 2 (0.70%) | |
| Missing | 0 | 0 | 0 | |
| Modified Schwab and England ADL (SE‐ADL) | <.0001 | |||
| Mean (SD) | 93.15 (5.9) | 95.35 (5.5) | 92.08 (5.8) | |
| (Min, Max) | (70.0, 100.0) | (75.0, 100.0) | (70.0, 100.0) | |
| Missing | 0 | 0 | 0 | |
| Duration of disease since diagnosis (Mon) | 0.4597 | |||
| Mean (SD) | 6.65 (6.5) | 6.32 (5.6) | 6.82 (6.9) | |
| (Min, Max) | (0.4, 35.8) | (0.9, 32.3) | (0.4, 35.8) | |
| Missing | 0 | 0 | 0 | |
| Age of PD diagnosis | 0.9332 | |||
| Mean (SD) | 61.11 (9.7) | 61.17 (10.2) | 61.08 (9.4) | |
| (Min, Max) | (31.8, 84.8) | (33.0, 81.8) | (31.8, 84.8) | |
| Missing | 0 | 0 | 0 | |
| TD/Non‐TD classification | 0.9118 | |||
| TD | 299 (70.85%) | 98 (70.50%) | 201 (71.02%) | |
| PIGD or Indeterminate | 123 (29.15%) | 41 (29.50%) | 82 (28.98%) | |
| Missing | 1 | 0 | 1 | |
| MDS‐UPDRS PIGD sum score | 0.0141 | |||
| Mean (SD) | 0.23 (0.2) | 0.19 (0.2) | 0.24 (0.2) | |
| (Min, Max) | (0.0, 1.4) | (0.0, 1.2) | (0.0, 1.4) | |
| Missing | 1 | 0 | 1 | |
| MDS‐UPDRS tremor sum score | 0.0026 | |||
| Mean (SD) | 0.49 (0.3) | 0.43 (0.3) | 0.52 (0.3) | |
| (Min, Max) | (0.0, 1.8) | (0.0, 1.3) | (0.0, 1.8) | |
| Missing | 1 | 0 | 1 | |
| Side most affected | 0.8743 | |||
| Left | 180 (42.55%) | 57 (41.01%) | 123 (43.31%) | |
| Right | 233 (55.08%) | 79 (56.83%) | 154 (54.23%) | |
| Symmetric | 10 (2.36%) | 3 (2.16%) | 7 (2.46%) | |
| Missing | 0 | 0 | 0 | |
| Time enrolled in study (Years) | <.0001 | |||
| Mean (SD) | 2.11 (0.8) | 1.79 (0.7) | 2.27 (0.8) | |
| (Min, Max) | (0.0, 4.1) | (0.0, 3.5) | (0.0, 4.1) | |
| Missing | 0 | 0 | 0 | |
| Non‐motor disease characteristics | ||||
| MOCA | 0.0031 | |||
| Mean (SD) | 27.13 (2.3) | 27.60 (2.2) | 26.90 (2.3) | |
| (Min, Max) | (17.0, 30.0) | (21.0, 30.0) | (17.0, 30.0) | |
| Missing | 0 | 0 | 0 | |
| GDS | 0.2904 | |||
| Mean (SD) | 2.32 (2.4) | 2.50 (2.8) | 2.24 (2.3) | |
| (Min, Max) | (0.0, 14.0) | (0.0, 14.0) | (0.0, 12.0) | |
| Missing | 0 | 0 | 0 | |
| SCOPA‐AUT | 0.4498 | |||
| Mean (SD) | 9.50 (6.2) | 9.17 (6.1) | 9.65 (6.2) | |
| (Min, Max) | (0.0, 39.0) | (0.0, 28.0) | (0.0, 39.0) | |
| Missing | 0 | 0 | 0 | |
| STAI ‐ state subscore | 0.7182 | |||
| Mean (SD) | 32.96 (10.2) | 32.71 (10.0) | 33.09 (10.4) | |
| (Min, Max) | (20.0, 76.0) | (20.0, 60.0) | (20.0, 76.0) | |
| Missing | 1 | 0 | 1 | |
| STAI ‐ trait subscore | 0.3958 | |||
| Mean (SD) | 32.37 (9.5) | 32.93 (10.1) | 32.10 (9.1) | |
| (Min, Max) | (20.0, 63.0) | (20.0, 63.0) | (20.0, 62.0) | |
| Missing | 1 | 0 | 1 | |
| QUIP | 0.7675 | |||
| Mean (SD) | 0.28 (0.6) | 0.29 (0.6) | 0.28 (0.6) | |
| (Min, Max) | (0.0, 4.0) | (0.0, 4.0) | (0.0, 4.0) | |
| Missing | 1 | 0 | 1 | |
| RBD Q | 0.1453 | |||
| Mean (SD) | 4.12 (2.7) | 3.85 (2.7) | 4.25 (2.7) | |
| (Min, Max) | (0.0, 12.0) | (0.0, 12.0) | (0.0, 12.0) | |
| Missing | 0 | 0 | 0 | |
| Epworth sleepiness scale | 0.7141 | |||
| Mean (SD) | 5.80 (3.5) | 5.89 (3.3) | 5.76 (3.5) | |
| (Min, Max) | (0.0, 20.0) | (0.0, 17.0) | (0.0, 20.0) | |
| Missing | 0 | 0 | 0 | |
Modified Schwab and England activities of daily living scale (SE‐ADL); MDS‐UPDRS Tremor Sum Score is calculated as a mean of 11 tremor items (2.10 and 3.15–3.18) MDS‐UPDRS Postural instability Gait score (PIGD) Sum Score is calculated as a mean of five items (2.12, 2.13 and 3.10–3.12). TD versus PIGD subtype is calculated as a ratio of the tremor versus PIGD mean scores.
MOCA, montreal cognitive assessment scale; GDS‐15, 15‐item geriatric depression scale; SCOPA‐AUT, the scale for outcomes for PD–autonomic function; STAI, state and trait anxiety scale, QUIP, the questionnaire for impulsive‐compulsive disorders in parkinson's disease; RBDQ‐REM, sleep behavior disorder questionnaire.
Table 2.
Demographics and PD characteristics by time to initiation of ST
| Variable | All PD | PD Subjects | PD Subjects | PD Subjects | PD Subjects | PD Subjects |
|---|---|---|---|---|---|---|
| Subjects | ST− | ST+ ≤6 Month | ST+ 6 Mo‐1 Year | ST+ 1–2 Years | ST+ >2 Years | |
| (N = 423) | (N = 139) | (N = 92) | (N = 118) | (N = 65) | (N = 9) | |
| Age | ||||||
| Mean (SD) | 61.66 (9.7) | 61.69 (10.3) | 60.94 (9.9) | 62.06 (9.3) | 61.92 (9.6) | 61.52 (5.0) |
| (Min, Max) | (33.5, 84.9) | (33.5, 82.3) | (38.5, 80.2) | (33.7, 84.9) | (36.6, 83.0) | (51.7, 66.6) |
| Missing | 0 | 0 | 0 | 0 | 0 | 0 |
| Gender | ||||||
| Male | 277 (65.48%) | 87 (62.59%) | 60 (65.22%) | 84 (71.19%) | 39 (60.00%) | 7 (77.78%) |
| Female | 146 (34.52%) | 52 (37.41%) | 32 (34.78%) | 34 (28.81%) | 26 (40.00%) | 2 (22.22%) |
| Missing | 0 | 0 | 0 | 0 | 0 | 0 |
| Education | ||||||
| <13 Years | 76 (17.97%) | 34 (24.46%) | 18 (19.57%) | 15 (12.71%) | 9 (13.85%) | 0 (0.00%) |
| 13–23 Years | 344 (81.32%) | 104 (74.82%) | 74 (80.43%) | 102 (86.44%) | 55 (84.62%) | 9 (100.00%) |
| >23 Years | 3 (0.71%) | 1 (0.72%) | 0 (0.00%) | 1 (0.85%) | 1 (1.54%) | 0 (0.00%) |
| Missing | 0 | 0 | 0 | 0 | 0 | 0 |
| Ethnicity | ||||||
| Hispanic/latino | 9 (2.13%) | 2 (1.44%) | 1 (1.09%) | 3 (2.54%) | 1 (1.54%) | 2 (22.22%) |
| Not Hispanic/latino | 414 (97.87%) | 137 (98.56%) | 91 (98.91%) | 115 (97.46%) | 64 (98.46%) | 7 (77.78%) |
| Missing | 0 | 0 | 0 | 0 | 0 | 0 |
| Race | ||||||
| White | 391 (92.43%) | 130 (93.53%) | 85 (92.39%) | 109 (92.37%) | 58 (89.23%) | 9 (100.00%) |
| Black/African‐American | 6 (1.42%) | 2 (1.44%) | 1 (1.09%) | 3 (2.54%) | 0 (0.00%) | 0 (0.00%) |
| Asian | 8 (1.89%) | 1 (0.72%) | 4 (4.35%) | 1 (0.85%) | 2 (3.08%) | 0 (0.00%) |
| Other | 18 (4.26%) | 6 (4.32%) | 2 (2.17%) | 5 (4.24%) | 5 (7.69%) | 0 (0.00%) |
| Missing | 0 | 0 | 0 | 0 | 0 | 0 |
| Family history of PD | ||||||
| Family members w/PD | 102 (24.17%) | 30 (21.74%) | 24 (26.09%) | 26 (22.03%) | 20 (30.77%) | 2 (22.22%) |
| No Family members w/PD | 320 (75.83%) | 108 (78.26%) | 68 (73.91%) | 92 (77.97%) | 45 (69.23%) | 7 (77.78%) |
| Missing | 1 | 1 | 0 | 0 | 0 | 0 |
| MDS‐UPDRS mean (SD) score and subscores | ||||||
| MDS‐UPDRS total score | 32.36 (13.1) | 27.90 (12.0) | 36.58 (14.2) | 34.97 (13.0) | 31.51 (11.5) | 30.11 (10.9) |
| MDS‐UPDRS part I | 5.57 (4.1) | 4.94 (4.1) | 6.27 (4.0) | 5.86 (4.0) | 5.62 (4.1) | 4.11 (2.9) |
| MDS‐UPDRS part II | 5.90 (4.2) | 4.64 (3.4) | 7.74 (4.5) | 6.58 (4.7) | 5.02 (3.4) | 4.22 (2.9) |
| MDS‐UPDRS part III (motor exam) | 20.89 (8.9) | 18.32 (8.4) | 22.57 (10.0) | 22.57 (8.2) | 20.88 (8.4) | 21.78 (7.4) |
| Missing | 1 | 0 | 0 | 1 | 0 | 0 |
| Hoehn and Yahr | ||||||
| Stage 1 | 186 (43.97%) | 68 (48.92%) | 40 (43.48%) | 42 (35.59%) | 30 (46.15%) | 6 (66.67%) |
| Stage 2 | 235 (55.56%) | 71 (51.08%) | 51 (55.43%) | 75 (63.56%) | 35 (53.85%) | 3 (33.33%) |
| Stage 3–5 | 2 (0.47%) | 0 (0.00%) | 1 (1.09%) | 1 (0.85%) | 0 (0.00%) | 0 (0.00%) |
| Missing | 0 | 0 | 0 | 0 | 0 | 0 |
| Modified Schwab and England ADL | ||||||
| Mean (SD) | 93.15 (5.9) | 95.35 (5.5) | 90.22 (6.1) | 92.54 (5.3) | 93.85 (5.6) | 92.22 (3.6) |
| (Min, Max) | (70.0, 100.0) | (75.0, 100.0) | (70.0, 100.0) | (80.0, 100.0) | (80.0, 100.0) | (90.0, 100.0) |
| Missing | 0 | 0 | 0 | 0 | 0 | 0 |
| Duration of disease since diagnosis (Mon) | ||||||
| Mean (SD) | 6.65 (6.5) | 6.32 (5.6) | 5.56 (4.7) | 6.59 (7.0) | 8.56 (8.7) | 10.14 (8.2) |
| (Min, Max) | (0.4, 35.8) | (0.9, 32.3) | (0.9, 25.7) | (0.7, 31.9) | (0.4, 35.8) | (1.0, 23.0) |
| Missing | 0 | 0 | 0 | 0 | 0 | 0 |
| Age of PD Diagnosis | ||||||
| Mean (SD) | 61.11 (9.7) | 61.17 (10.2) | 60.48 (9.9) | 61.51 (9.3) | 61.21 (9.6) | 60.68 (5.6) |
| (Min, Max) | (31.8, 84.8) | (33.0, 81.8) | (38.4, 80.0) | (31.8, 84.8) | (35.8, 81.7) | (49.8, 65.7) |
| Missing | 0 | 0 | 0 | 0 | 0 | 0 |
| TD/Non‐TD classification | ||||||
| TD | 299 (70.85%) | 98 (70.50%) | 58 (63.04%) | 85 (72.65%) | 52 (80.00%) | 6 (66.67%) |
| PIGD or Indeterminate | 123 (29.15%) | 41 (29.50%) | 34 (36.96%) | 32 (27.35%) | 13 (20.00%) | 3 (33.33%) |
| Missing | 1 | 0 | 0 | 1 | 0 | 0 |
| PIGD sum score | ||||||
| Mean (SD) | 0.23 (0.2) | 0.19 (0.2) | 0.29 (0.3) | 0.23 (0.2) | 0.20 (0.2) | 0.24 (0.2) |
| (Min, Max) | (0.0, 1.4) | (0.0, 1.2) | (0.0, 1.4) | (0.0, 1.0) | (0.0, 0.6) | (0.0, 0.6) |
| Missing | 1 | 0 | 0 | 1 | 0 | 0 |
| Tremor Sum Score | ||||||
| Mean (SD) | 0.49 (0.3) | 0.43 (0.3) | 0.49 (0.4) | 0.55 (0.3) | 0.54 (0.3) | 0.52 (0.4) |
| (Min, Max) | (0.0, 1.8) | (0.0, 1.3) | (0.0, 1.8) | (0.0, 1.6) | (0.0, 1.3) | (0.0, 1.1) |
| Missing | 1 | 0 | 0 | 1 | 0 | 0 |
| Side most affected | ||||||
| Left | 180 (42.55%) | 57 (41.01%) | 42 (45.65%) | 52 (44.07%) | 26 (40.00%) | 3 (33.33%) |
| Right | 233 (55.08%) | 79 (56.83%) | 47 (51.09%) | 64 (54.24%) | 37 (56.92%) | 6 (66.67%) |
| Symmetric | 10 (2.36%) | 3 (2.16%) | 3 (3.26%) | 2 (1.69%) | 2 (3.08%) | 0 (0.00%) |
| Missing | 0 | 0 | 0 | 0 | 0 | 0 |
| Time enrolled in study (Years) | ||||||
| Mean (SD) | 2.11 (0.8) | 1.79 (0.7) | 2.03 (0.8) | 2.29 (0.8) | 2.45 (0.7) | 3.08 (0.5) |
| (Min, Max) | (0.0, 4.1) | (0.0, 3.5) | (0.0, 4.1) | (1.0, 4.1) | (1.2, 3.8) | (2.3, 3.7) |
| Missing | 0 | 0 | 0 | 0 | 0 | 0 |
| Time to initiate ST (Years) | ||||||
| Mean (SD) | NA | NA | 0.36 (0.1) | 0.66 (0.1) | 1.37 (0.3) | 2.53 (0.5) |
| (Min, Max) | NA | NA | (0.0, 0.5) | (0.5, 1.0) | (1.0, 2.0) | (2.0, 3.3) |
| Missing | NA | NA | 0 | 0 | 0 | 0 |
Report generated on data submitted as of: 15 September 2014.
Please refer to Table 1 for abbreviations. See Discussion in the text.
Random survival forests
Table 3 shows the in‐sample (C) and cross‐validation (Brier score, pseudo‐R 2) indexes for all the models examined in RSF. The concordance for RSF‐33 (C = 0.68) was small according to the benchmarks discussed above, as was the reduction in the Brier score relative to the reference model (pseudo‐R 2 = 0.13). Table 4 ranks each variable with two measures of predictive ability: minimal depth (MD) and variable importance (Vimp). MD indexes how deep into the regression trees a predictor tends to occur.21 A deeper occurrence means a predictor is less important in the regression trees, so smaller values of MD indicate greater predictive ability. Vimp indexes how much prediction changes when the split in the regression trees for a predictor is random.10 Larger values indicate greater predictive ability. The variables are rank‐ordered by MD. The most important predictor was duration (time from diagnosis to enrollment) [1], and the least important predictor was ethnicity [33]. Several subgroups of predictors were identified based on the separation in space among clusters in a plot of MD by Vimp (see Fig. 1). Based on the initial results, we further examined subsets of the best 26 predictors (duration [1] to Epworth sleepiness scale (ESS) [26]), the best seven predictors (duration [1] to MOCA total score [7]), and the best three predictors (duration [1], SE‐ADL [2], MDS‐UPDRS Total Score [3]). Table 3 suggests a preference for RSF‐3 because it was the most parsimonious model that tied for the largest concordance (C = 0.70) and the largest reduction in the Brier score (pseudo‐R 2 = 0.13). Subsequently, the initial full Cox model was based on the set of three predictors and had all interactions and nonlinear (quadratic) effects included. Backward elimination selected the final reduced model with the predictors of duration, duration2, SE‐ADL, SE‐ADL2 , and MDS‐UPDRS total score. The reduced Cox model appeared to well‐characterize the important effects, as the model had the largest concordance (C = 0.71 ) and the largest reduction in Brier score (pseudo‐R 2 = 0.13) of all the models; however, it is considered a below average effect according to our benchmarks. Table 5 shows the parameter estimates for the reduced Cox model. In order to summarize the reduced model Cox effects, a prognostic index (PI)22 was computed for each patient using the composite of the scores of the predictors in the first column of Table 5 weighted by their estimated coefficients. Four risk groups of nearly equal size were formed based on the computed PI distribution. The distribution was arbitrarily scaled to mean = 20 and SD = 5. Figure 2 shows survival probabilities based on the Cox model of Table 5. The first three graphs (A–C) show the curves for the first and third quartiles of the predictor of interest holding the other predictors at their mean values (nonlinear terms were included). The graph D at the bottom right shows the survival curves of the four PI groups and the ranges of the PI that define the groups. As the figure shows, higher relative risk of ST (lower survival probability) was associated with shorter disease duration, lower SE‐ADL, and higher MDS‐UPDRS scores. The PI graph (bottom right) indicates that greater risk was associated with a higher PI score (similar to the MDS‐UPDRS).
Table 4.
Results of the 33 variable random survival forests analysis. Predictors are ranked by minimal depth (MD)
| Rank | Predictor | MD | Vimp | Rank | Predictor | MD | Vimp |
|---|---|---|---|---|---|---|---|
| 1 | Disease duration | 2.2175 | 0.0508 | 17 | Age | 5.7695 | 0.0006 |
| 2 | SE‐ADL | 3.4480 | 0.0129 | 18 | Serum urate | 5.8085 | 0.0009 |
| 3 | MDS‐UPDRS.total | 3.7695 | 0.0078 | 19 | Age.at.Diagnosis | 5.8190 | −0.0001 |
| 4 | Contralateral putamen* | 5.1750 | 0.0027 | 20 | CSF P.tau/.A‐beta | 5.8505 | 0.0010 |
| 5 | CSF P.tau | 5.2765 | 0.0018 | 21 | CSF t‐tau/A‐beta | 5.8530 | 0.0003 |
| 6 | Ipsilatearal .putamen* | 5.3680 | 0.0016 | 22 | SCOPA.total | 5.9180 | 0.0014 |
| 7 | MOCA.total | 5.4015 | 0.0036 | 23 | CSF t.tau | 5.9440 | 0.0008 |
| 8 | CSF P‐tau/t‐tau | 5.6150 | 0.0021 | 24 | RBD Q | 5.9500 | 0.0013 |
| 9 | Education | 5.6220 | 0.0021 | 25 | GDS.total | 6.0290 | 0.0013 |
| 10 | STAI.state | 5.6290 | 0.0021 | 26 | Epworth.SS | 6.0515 | 0.0009 |
| 11 | STAI.trait | 5.6530 | 0.0018 | 27 | Side Most Affected | 7.2495 | −0.0002 |
| 12 | CSF A‐beta 1–42 | 5.6825 | 0.0004 | 28 | Gender | 7.8210 | 0.0003 |
| 13 | Contralateral caudate* | 5.6990 | 0.0013 | 29 | Hoehn and Yahr. | 7.8570 | −0.0005 |
| 14 | Ipsilatearal.caudate* | 5.7080 | 0.0013 | 30 | TD/ non‐TD | 7.9525 | 0.0005 |
| 15 | PIGD sum score | 5.7325 | 0.0031 | 31 | QUIP | 8.3090 | 0.0014 |
| 16 | CSF Alpha.Synuclein | 5.7645 | 0.0002 | 32 | Family history | 8.4520 | −0.0007 |
| 33 | Ethnicity | 9.6355 | −0.0000 |
Note: *Ipsilateral and contralateral refer to DAT tracer uptake and are defined in relation to the predominant side of clinical symptoms. For PD subjects with symmetrical presentation, ipsilateral and contralateral sides are defined as the mean of the left and right values.
Please, refer to Table 1 for the explanation of the abbreviations.
Figure 1.
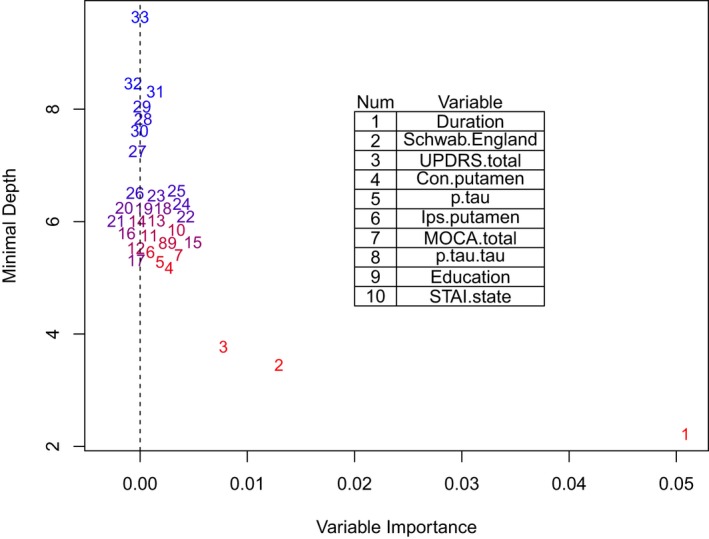
Minimal depth by variable importance for the 33 variable random survival forests analysis.
Table 5.
Cox regression results
| Coef | Exp(Coef) | SE(Coef) | Z‐value | |
|---|---|---|---|---|
| Disease duration | −0.1476 | 0.8628 | 0.0286 | −5.1564 |
| SE‐ADL | 0.3158 | 1.3714 | 0.2336 | 1.3521 |
| UPDRS | 0.0212 | 1.0214 | 0.0052 | 4.0804 |
| (Duration)2 | 0.0024 | 1.0024 | 0.0010 | 2.4637 |
| (SE‐ADL)2 | −0.0020 | 0.9980 | 0.0013 | −1.5781 |
See Discussion in the text.
Figure 2.
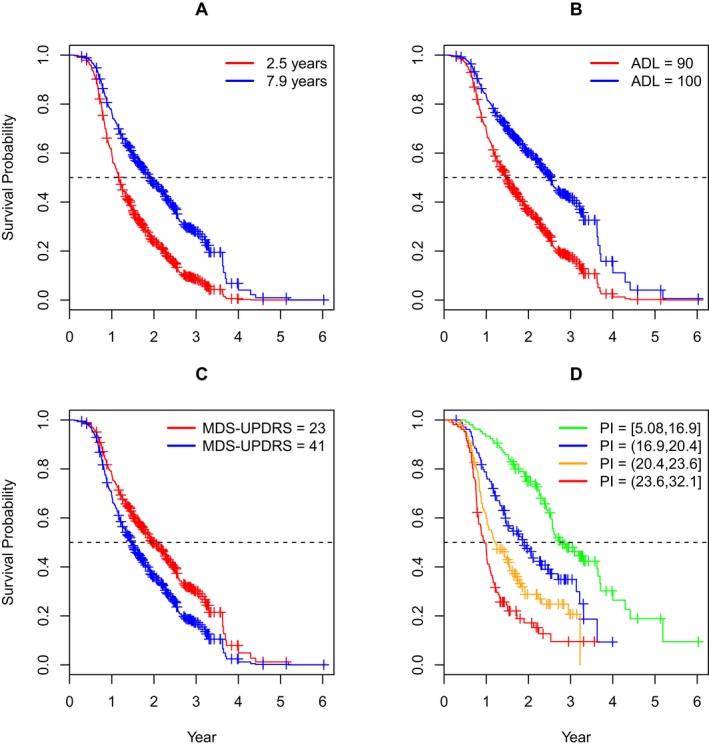
Survival curves for individual predictors (top row and bottom left), and the prognostic index (PI) groups (bottom right). Figure 2 shows survival probabilities based on the Cox model of Table 5. The first three graphs (left to right) show the curves for the first and third quartiles of the predictor of interest holding the other predictors at their mean values (nonlinear terms were included). The last graph at the bottom right shows the survival curves of the four PI groups and the ranges of the PI that define the groups (see text for Discussion).
Additional descriptive statistics of imaging and biologic variables in the ST+ versus ST− groups are provided in Tables 6, 7. Table 6 provides data on the DAT tracer uptake in the ST+ versus ST− group by the region of interest. While there was a significant difference in the ipsilateral and contralateral putamen DAT tracer uptake, these variables ranked # 4 and # 6 in the prediction model (see Table 4) and did not increase prediction accuracy over the three top variables discussed above. Table 7 provides data of the cerebrospinal fluid Biologics in the ST+ versus ST− group. Similar to DAT data, while P‐tau was significantly lower in the ST+ group (P = 0.0063), it ranked # 5 in the overall prediction model (Table 4). Lastly, Table 8 summarizes the classes of the dopaminergic therapy started in the ST+ group.
Table 6.
Results of the DAT tracer uptake in the ST+ versus ST−group
| Variable | All PD | PD Subjects | PD Subjects | P‐value(ST− vs. ST+) |
|---|---|---|---|---|
| Subjects | ST− | ST+ | ||
| (N = 423) | (N = 139) | (N = 284) | ||
| Contralateral caudate | 0.2511 | |||
| Mean (SD) | 1.84 (0.6) | 1.89 (0.6) | 1.82 (0.5) | |
| (Min, Max) | (0.4, 3.7) | (0.5, 3.7) | (0.4, 3.4) | |
| Missing | 7 | 3 | 4 | |
| Ipsilateralcaudate | 0.9076 | |||
| Mean (SD) | 2.16 (0.6) | 2.17 (0.6) | 2.16 (0.6) | |
| (Min, Max) | (0.4, 4.0) | (0.6, 3.8) | (0.4, 4.0) | |
| Missing | 7 | 3 | 4 | |
| Contralateral putamen | 0.0004 | |||
| Mean (SD) | 0.69 (0.3) | 0.76 (0.3) | 0.66 (0.2) | |
| (Min, Max) | (0.1, 2.2) | (0.1, 2.2) | (0.1, 1.4) | |
| Missing | 7 | 3 | 4 | |
| Ipsilateral putamen | 0.0028 | |||
| Mean (SD) | 0.96 (0.4) | 1.04 (0.4) | 0.93 (0.4) | |
| (Min, Max) | (0.3, 2.6) | (0.4, 2.3) | (0.3, 2.6) | |
| Missing | 7 | 3 | 4 |
Report generated on data submitted as of: 15 September 2014.
Note: Ipsilateral and contralateral refer to DAT tracer uptake and are defined in relation to the predominant side of clinical symptoms. For PD subjects with symmetrical presentation, ipsilateral and contralateral sides are defined as the mean of the left and right values.
Table 7.
Results of the CSF Biologics in the ST+ versus ST− group
| Variable | All PD | PD Subjects | PD Subjects | P‐valuea (ST− vs. ST+) |
|---|---|---|---|---|
| Subjects | ST− | ST+ | ||
| (N = 423) | (N = 139) | (N = 284) | ||
| A‐beta 1–42 | 0.1269 | |||
| Mean (SD) | 370.56 (100.4) | 380.01 (102.3) | 366.06 (99.3) | |
| (Min, Max) | (129.2, 796.5) | (129.2, 670.0) | (139.9, 796.5) | |
| Missing | 11 | 6 | 5 | |
| t‐tau | 0.3417 | |||
| Mean (SD) | 44.69 (18.3) | 45.53 (18.0) | 44.29 (18.4) | |
| (Min, Max) | (14.4, 121.0) | (14.4, 107.8) | (15.6, 121.0) | |
| Missing | 15 | 7 | 8 | |
| P‐tau | 0.0063 | |||
| Mean (SD) | 15.64 (10.0) | 16.83 (9.4) | 15.07 (10.3) | |
| (Min, Max) | (4.7, 94.1) | (6.0, 51.3) | (4.7, 94.1) | |
| Missing | 13 | 7 | 6 | |
| t‐tau/A‐beta 1‐42 | 0.9868 | |||
| Mean (SD) | 0.126 (0.064) | 0.126 (0.063) | 0.126 (0.065) | |
| (Min, Max) | (0.045, 0.525) | (0.054, 0.525) | (0.045, 0.487) | |
| Missing | 15 | 7 | 8 | |
| P‐tau/A‐beta 1‐42 | 0.0422 | |||
| Mean (SD) | 0.044 (0.034) | 0.046 (0.025) | 0.043 (0.038) | |
| (Min, Max) | (0.013, 0.509) | (0.018, 0.171) | (0.013, 0.509) | |
| Missing | 13 | 7 | 6 | |
| P‐tau/t‐tau | 0.0838 | |||
| Mean (SD) | 0.371 (0.225) | 0.402 (0.232) | 0.357 (0.220) | |
| (Min, Max) | (0.083, 2.139) | (0.143, 1.285) | (0.083, 2.139) | |
| Missing | 17 | 8 | 9 | |
| Alpha‐Synuclein | 0.7095 | |||
| Mean (SD) | 1844.68 (786.1) | 1846.62 (759.1) | 1843.76 (800.0) | |
| (Min, Max) | (332.9, 6694.6) | (363.1, 4709.8) | (332.9, 6694.6) | |
| Missing | 11 | 6 | 5 | |
| Urate | 0.4407 | |||
| Mean (SD) | 317.68 (79.0) | 312.69 (80.0) | 320.10 (78.5) | |
| (Min, Max) | (167.0, 541.0) | (167.0, 523.0) | (167.0, 541.0) | |
| Missing | 6 | 3 | 3 |
Report generated on data submitted as of: 15 September 2014.
Of note, P‐tau ranked #5 in the overall prediction model (Table 4).
P‐values from Mann–Whitney U tests.
Table 8.
Classes of dopaminergic therapy in treated PD subjects
| Variable | Treated PD subjects (N = 284) |
|---|---|
| Class of dopaminergic therapya | |
| Dopamine replacements | 85 (31.60%) |
| Dopamine agonists | 65 (24.16%) |
| COMT inhibitors | 0 (0.00%) |
| Mao‐B inhibitors | 106 (39.41%) |
| Anti‐cholinergics | 2 (0.74%) |
| Other (amantadine, apomorphine SQ) | 21 (7.81%) |
| Missing | 15 |
Note: Subjects may have more than one class of DT listed.
Report generated on data submitted as of: 15 September 2014.
Discussion
Our analysis of the predictors of initiation of ST in a cohort of 423 early untreated at enrollment PD participants using a novel machine learning method of random survival forests showed that the 3‐predictor subset of disease duration (time from diagnosis to enrollment), the SE‐ADL, and the MDS‐UPDRS total score modestly predicted time to initiation of ST (C = 0.70, pseudo‐R 2 = 0.13) . Greater risk of initiation of ST was associated with shorter disease duration, lower SE‐ADL score, and higher MDS‐UPDRS score (see Fig. 2). Prediction using the three variables was equal to using the entire set of 33. None of the tested biomarkers or imaging variables improved the prediction accuracy. The two top predictors (SE‐ADL score and disease severity as measured by MDS‐UPDRS) are consistent with the previously published reports from analysis of the two largest studies that used TIST as the primary outcome measure. 23, 24 The finding that shorter disease duration was predictive of greater risk of initiation of ST is at first glance counterintuitive. However, at the time of study enrollment, those individuals with early initiation of ST were not eligible for enrollment into PPMI and did not have a time to the event. Compared to the previous analysis,23 TD/PIGD score was not a strong predictor adjusting for the effect of all the other variables (RSF is a multivariate prediction method). The lack of effect was possibly due to the fact that the PPMI cohort represents subjects with very early disease and as such only 30% are classified into the PIGD subtype and even within that group there is a substantial shift into TD or indeterminate within the first year.
The major novel aspect of this analysis was the inclusion of the biological and imaging variables as predictors. While P‐tau and putaminal DAT uptake were rank‐ordered 4–6 in predictive strength (see Table 4), their addition to the top three predictors did not improve the overall cross‐validation predictive accuracy (see Table 3). While it is disappointing that the biologically based predictors did not improve upon the three top variables, it is not unexpected. The decision to initiate ST is based on a constellation of objective factors like disease severity, as well as subjective factors like patient perceived disability and preference. It remains to be determined with the longer follow‐up of the cohort if earlier initiation of ST correlates with the earlier onset of such major disease progression milestones as onset of postural instability and cognitive impairment. There is an ongoing analysis of the PPMI data on the correlation of the DAT uptake and biologic variables with different measures of PD progression. Another novel aspect of this analysis is the fact that it used machine‐based learning paradigm rather that a priory selection of the predictors. It is reassuring to see that the RSF analysis selected the top variables that are consistent with the clinician's paradigm of the decision process of the TIST. It should be emphasized that, while the three top predictors performed as well as the set of 33, the accuracy of the prediction was modest, with the effect size (C = 0.70) being considered small according to benchmarks.17 The small effect size suggests there are other variables not captured in our dataset that play a role in the prediction of symptomatic therapy initiation.
There are a number of clinical and research implications of our results. In the clinical domain, our prediction model can be used to council patients regarding the timeframe to initiation of ST. For an individual PD patient, based on the scores of the top three predictors (linear and nonlinear effects), a group‐level survival curve can be constructed to provide an indication of the risk of initiation of ST (see graph D of Fig. 2). In the clinical research domain, one of the major limitations of the design of the previously conducted clinical trials testing putative disease‐modifying interventions is the fact that on average, 50% of the participants require initiation of ST in the first year of study, which means that the data are imputed from that point on. Selection of the participants based on our PI model can reduce that number. However, it should be recognized that predictions for a single person can be very inaccurate, as survival curves are inherently group‐level indexes. As such, predictions apply to a person's cohort, with the cohort being defined by the same 3‐predictor scores at baseline that form the basis of the PI‐weighted composite. In addition, studies using time to initiation of ST as the primary outcome should ensure that the groups are balanced on these three top variables.
There are a number of study limitations that have to be highlighted. The 33 predictors were selected based on the scope of the data available at the time of analysis and did not include a number of potentially relevant variables like genetic status, environmental, occupational, and employment history, all of which can be of relevance. Furthermore, the usefulness of the selected model and the PI needs to be confirmed with an external validation study on an independent dataset. Follow‐up data analysis at the point when all subjects start ST may address these limitations. Our results might not be generalizable to the PD population at large, as based on the nature of the study, participants had very early PD, were younger and were expected not to initiate ST for the first 6 months of the study. In addition, our cohort is predominantly White Non Hispanic and as such our data cannot be generalizable to other racial and ethnic groups.
In conclusion, our findings using a novel machine learning method support previously reported clinical variables that predict time to initiation of ST. However, the inclusion of biological variables did not increase prediction accuracy. These data can guide clinicians in counseling the patients and in the selection of the participants for the clinical trials. Further longitudinal analysis will establish correlation of the time to initiation of ST with the time to onset of the major disease disability milestones including postural instability and cognitive impairment.
ClinicalTrials.gov
ClinicalTrials.gov Identifier: NCT01141023.
Conflict of Interest
Tanya Simuni has received research funding from Michael J. Fox Foundation for Parkinson's Research, National Institutes of Health, National Parkinson Foundation, and consulting honoraria from National Parkinson Foundation, Teva Pharmaceuticals, Pfizer, Harbor, UCB, IMPAX, Eli Lilly and Company, Allergan, Merz Inc, and US Worldmeds. Jeffrey D. Long has a consulting agreement with Neurophage Inc. and is a paid consultant for Roche Pharma (F. Hoffmann‐La Roche Ltd) and Azevan Pharmaceuticals. Chelsea Caspell‐Garcia reports no disclosures. Christopher Coffey‐ Serves on the scientific advisory board for data safety and monitoring for NINDS and NIA, received a speaker honorarium for presenting a short course at Rho, Inc., is a consultant for ZZ Biotech, LLC, received research support from the Michael J. Fox Foundation, and is supported by NIH/NINDS, U01 NS077352, PI, 10/01/11‐09/30/18 (2) NIH/NINDS, U01 NS077108, PI, 10/01/11‐09/30/16(3) NIH/NHLBI, U01 HL091843, PI, 08/01/09‐02/28/15(4) NIH/NHLBI, U01 NS038529, PI, 12/01/09‐12/31/13 NIH/NINDS,(5) U01 NS079163, 08/05/2012‐07/31/2015 (6) NIH/NINDS, U01 NS082329, 07/15/2013‐06/30/2018 (7) NIH/NINDS, U01 NS084495, 09/15/2013‐07/31/2018. Shirley Lasch is employed by Molecular NeuroImaging, LLC. Caroline M. Tanner, MD PhD, University of California, San Francisco, San Francisco Veterans Affairs Medical Center. She serves on the Scientific Advisory Boards of the Michael J. Fox Foundation and the National Spasmodic Dysphonia Association as a voluntary consultant, and has provided paid consulting services to Pfizer Pharmaceuticals. She receives grant support from the Michael J. Fox Foundation, the Parkinson's Disease Foundation, the Department of Defense and the National Institutes of Health. Danna Jennings is employed by Molecular NeuroImaging, LLC and is on the speaker bureau for UCB. Karl Kieburtz, MD, MPH – Consultant: National Institutes of Health (NIH, NINDS), Acorda, Astellas Pharma, AstraZeneca, Auspex, Biotie, Britannia, Cangene, CHDI,Civitas,Clearpoint Strategy Group, Clintrex, Cynapsus, INC Research, IntecIsis, Lilly, Lundbeck, Medavante, Medivation, Melior Discovery, Neuroderm, Neurmedix, Omeros, Otsuka, Pfizer, Pharma2B, Prothena/Neotope/Elan Pharmaceutical, Raptor Pharmaceuticals, Roche/Genentech, Sage Bionetworks, Serina, Stealth Peptides, Synagile, Teikoku Pharma, Titan, Turing Pharmaceuticals, Upsher‐Smith, US WorldMeds, Vaccinex, Voyager, Weston Brain Institute Grants/Research Support: National Institutes of Health (NEI, NINDS, NIA, NICHD), Michael J Fox Foundation, Teva. Kenneth Marek – Consultant for Pfizer, GE Healthcare, Merck, Lilly, BMS, Piramal, Prothena, Neurophage, nLife, Roche, and receives funding for the following grants: W81XWH‐06‐1‐0678 Establishing an ‘at risk’ cohort for Parkinson Disease Neuroprevention using olfactory testing and DAT imaging, DOD, Investigator 10/1/06 – 09/30/15; Parkinson Progression Marker Initiative (PPMI), Michael J. Fox Foundation, Principal Investigator 6/15/09 – 6/14/18; DAT imaging in LRRK2 family members, the Michael J. Fox Foundation, Principal Investigator 1/15/10 – 1/14/15. Ownership in Molecular NeuroImaging, LLC.
Supporting information
Data S1. Statistical analysis model.
References
- 1. Miyasaki JM, Martin W, Suchowersky O, et al. Practice parameter: initiation of treatment for Parkinson's disease: an evidence‐based review: report of the Quality Standards Subcommittee of the American Academy of Neurology. Neurology 2002;58:11–17. [DOI] [PubMed] [Google Scholar]
- 2. DATATOP: a multicenter controlled clinical trial in early Parkinson's disease . Parkinson Study Group. Arch Neurol. 1989; 46:1052–1060. [DOI] [PubMed] [Google Scholar]
- 3. Elm JJ, Goetz CG, Ravina B, et al. A responsive outcome for Parkinson's disease neuroprotection futility studies. Ann Neurol 2005;57:197–203. [DOI] [PubMed] [Google Scholar]
- 4. LeWitt P, Oakes D, Cui L. The need for levodopa as an end point of Parkinson's disease progression in a clinical trial of selegiline and alpha‐tocopherol. Parkinson Study Group. Mov Disord 1997;12:183–189. [DOI] [PubMed] [Google Scholar]
- 5. Parashos SA, Swearingen CJ, Biglan KM, et al. Determinants of the timing of symptomatic treatment in early Parkinson disease: the National Institutes of Health Exploratory Trials in Parkinson Disease (NET‐PD) Experience. Arch Neurol 2009;66:1099–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ravina B, Camicioli R, Como PG, et al. The impact of depressive symptoms in early Parkinson disease. Neurology 2007;69:342–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Guimaraes P, Kieburtz K, Goetz CG, et al. Non‐linearity of Parkinson's disease progression: implications for sample size calculations in clinical trials. Clin Trials 2005;2:509–518. [DOI] [PubMed] [Google Scholar]
- 8. Kuhta T, Zadikoff C, Simuni T, et al. Brain donation–what do patients with movement disorders know and how do they feel about it? Parkinsonism Relat Disord 2011;17:204–207. [DOI] [PubMed] [Google Scholar]
- 9. Goetz CG. Movement disorder society‐unified Parkinson's disease rating scale (MDS‐UPDRS): a new scale for the evaluation of Parkinson's disease. Rev Neurol (Paris) 2010;166:1–4. [DOI] [PubMed] [Google Scholar]
- 10. Ishwaran H, Kogalur UB, Blackstone EH, Lauer MS. Random Survival Forests. Ann Appl Stat 2008; 2: 841–860. [Google Scholar]
- 11. Breiman L. Random Forests. Mach Learn 2001;45:5–32. [Google Scholar]
- 12. Breiman L. Statistical modeling: the two cultures. Stat Sci 2001;16:199–231. [Google Scholar]
- 13. Long JD, Paulsen JS. Multivariate prediction of motor diagnosis in huntington disease: 12 years of predict‐HD. Mov Disord. 2015;30(12):1664–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Royston P, Sauerbrei W. Multivariate Model‐Building. New York: Wiley, 2008. [Google Scholar]
- 15. Gneiting T, Raftery AE. Strictly proper scoring rules, prediction, and estimation. J Am Statitical Assoc 2007;102:359–378. [Google Scholar]
- 16. Graf E, Schmoor C, Sauerbrei W, Schumacher M. Assessment and comparison of prognostic classification schemes for survival data. Stat Med 1999;18(17–18):2529–2545. [DOI] [PubMed] [Google Scholar]
- 17. Collins GS, de Groot JA, Dutton S, et al. External validation of multivariable prediction models: a systematic review of methodological conduct and reporting. BMC Med Res Methodol 2014;14:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schwab RSEA. Projection technique for evaluating surgery in Parkinson's disease In: DI Gillingham FJ, ed. Third Symposium on Parkinson's Disease. Edinburgh, Scotland: E & S Livingstone, 1969:152–157. [Google Scholar]
- 19. Stebbins GT, Goetz CG, Burn DJ, et al. How to identify tremor dominant and postural instability/gait difficulty groups with the movement disorder society unified Parkinson's disease rating scale: comparison with the unified Parkinson's disease rating scale. Mov Disord 2013;28:668–670. [DOI] [PubMed] [Google Scholar]
- 20. Nasreddine ZS, Phillips NA, Bedirian V, et al. The Montreal cognitive assessment, MoCA: a brief screening tool for mild cognitive impairment. J Am Geriatr Soc 2005;53:695–699. [DOI] [PubMed] [Google Scholar]
- 21. Ishwaran H, Kogalur UB, Gorodeski EZ , et al. High‐dimensional variable selection for survival data. J Am Statitical Assoc 2010;105:205–217. [Google Scholar]
- 22. Mallett S, Royston P, Waters R, et al. Reporting performance of prognostic models in cancer: a review. BMC Med 2010;8:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. McDermott MP, Jankovic J, Carter J, et al. Factors predictive of the need for levodopa therapy in early, untreated Parkinson's disease. The Parkinson Study Group. Arch Neurol 1995;52:565–570. [DOI] [PubMed] [Google Scholar]
- 24. Marras C, McDermott MP, Marek K, et al. Predictors of time to requiring dopaminergic treatment in 2 Parkinson's disease cohorts. Mov Disord 2011;26:608–613. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Statistical analysis model.


