Abstract
Perturbations in blood vessels play a critical role in the pathophysiology of brain injury and neurodegeneration. Here, we use a systematic genome-wide transcriptome screening approach to investigate the vasculome after brain trauma in mice. Mice were subjected to controlled cortical impact and brains were extracted for analysis at 24 h post-injury. The core of the traumatic lesion was removed and then cortical microvesels were isolated from nondirectly damaged ipsilateral cortex. Compared to contralateral cortex and normal cortex from sham-operated mice, we identified a wide spectrum of responses in the vasculome after trauma. Up-regulated pathways included those involved in regulation of inflammation and extracellular matrix processes. Decreased pathways included those involved in regulation of metabolism, mitochondrial function, and transport systems. These findings suggest that microvascular perturbations can be widespread and not necessarily localized to core areas of direct injury per se and may further provide a broader gene network context for existing knowledge regarding inflammation, metabolism, and blood–brain barrier alterations after brain trauma. Further efforts are warranted to map the vasculome with higher spatial and temporal resolution from acute to delayed phase post-trauma. Investigating the widespread network responses in the vasculome may reveal potential mechanisms, therapeutic targets, and biomarkers for traumatic brain injury.
Key words: : extracellular matrix, inflammation, microvessels, neurovascular unit, traumatic brain injury
Introduction
Traumatic brain injury (TBI) is a major public health issue.1 Intense research has been directed to identify neuroprotective therapies to improve outcome after trauma. Unfortunately, clinical trials have not yielded any broadly efficacious treatments, and current care for TBI patients remains mostly supportive.2–4
Over the past decade, the concept of the neurovascular unit has emerged to suggest that treatments for stroke and neurodegeneration cannot be found by focusing on neurons alone. Instead, cell-cell signaling between all elements in neuronal, glial, and vascular compartments must be addressed in order to rescue function and improve outcomes in damaged and diseased brain.5–7 Is it possible that for TBI, pure neuroprotection alone is also insufficient? In this study, we hypothesize that widespread vascular responses may occur in the context of TBI pathophysiology.
It was recently proposed that responses in blood microvessels (i.e., perturbations in the vasculome) mediate brain function and dysfunction.7–10 The entire network of microvessels in the brain is no longer viewed as inert pipes. Instead, the cerebral endothelium is now recognized to be an active participant for cell-cell signaling in the neurovascular unit. Under normal conditions, endothelium may provide signals that support neurogenesis,11,12 protect neurons against stress,13,14 and mediate homeostasis in oligodendrocyte precursor pools.15 When activated by injury, the endothelium may produce proinflammatory cytokines that magnify neuroinflammation and secondary injury.16–18 Injured endothelial cells may no longer achieve the appropriate balance between coagulation and fibrinolysis, resulting in hemorrhage, microthrombin formation, and perturbations of cerebral blood flow.19–21 Furthermore, disruption of endothelial tight junctions and basal lamina may lead to breakdown of the blood–brain barrier (BBB) with subsequent brain edema.22,23 Stressed endothelium may also release measurable signals into the bloodstream, thus providing a potential source of biomarkers.24 Ultimately, the entire brain endothelium plays a central role in substrate delivery, BBB function, flow-metabolism coupling, and trophic support for neurons and glia. Hence, mapping responses in the vasculome may offer new insights into mechanisms, therapeutic targets, and potential biomarkers in TBI. In this study, we used systematic genome-wide transcriptome screening approach to map the vasculome after controlled cortical impact (CCI) brain injury in mice.
Methods
Mouse controlled cortical impact
All CCI protocols were approved by the Massachusetts General Hospital Institutional Animal Care and Use Committee and complied with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Male C57Bl/6 mice (10–12 weeks of age) were used for this model, as reported previously.25 Briefly, mice were anesthetized with 4% isoflurane (Anaquest, Memphis, TN) in 70% N2 and 30% O2 and positioned in a stereotaxic frame. A 5-mm craniotomy was performed over the right cerebral hemisphere between the bregma and lambda by the use of a trephine drill, and the bone flap was removed. After craniotomy, mice were subjected to injury by a pneumatic cylinder with a 3-mm flat-tip impounder, 4.8 m/s of velocity, 0.6 mm of depth, and 150 ms of impact duration. Post-CCI, the scalp was sutured closed and mice were returned to their cages to recover from anesthesia. To confirm reproducible injury, mouse brains were cut and standard hematoxylin-eosin (H&E) stains were used to assess the distribution of traumatic injury.
Preparation of brain microvessels
Mice were sacrificed 24 h post-injury. After perfusion with phosphate-buffered saline (PBS), brains were harvested. Two sides of hemispheres (contra- and ipsilateral sides) were separated, removing the core lesion areas (which fall apart quite easily) and underlying white matter, then rolling on the filter paper to get rid of meninges and the remaining loose debris. After this, cortical tissue from each hemisphere was pooled from 4 mice, homogenized in cold PBS on ice with Knote Dounce glass tissue grinder (Part 885300-0002; Kimble Chase Life Science, Vineland, NJ), then centrifuged at 4°C, 500g for 5 min. The tissue pellet was suspended with 18% Dextran solution (molecular weight 60–90 kDa; USB Corporation, Cleveland, OH) in PBS and then centrifuged again at 4°C, 2500g for 20 min. The resulting pellet from this Dextran gradient centrifugation provides the raw sample of brain microvessels, as previously described.26 To increase yield, the supernatant was centrifuged again and the pellets from two centrifugations were pooled, washed once in PBS, and then resuspended in PBS. The final suspension was filtered through a 40-μm cell strainer to get rid of remaining single cells or small cell clumps. The resulting microvessels on the top of the cell strainer were used directly for RNA extraction. To confirm purity, isolated microvessels were subjected to immunostaining for endothelial, smooth muscle cell, and glial markers. Primary antibodies anti-CD31, alpha-smooth muscle actin (α-SMA), and glial fibrillary acidic protein (GFAP) were used. Texas Red–labeled lycopersicon Esculentum (tomato) Lectin (Vector Laboratories, Burlingame, CA) was also used to stain vessel fragments. Note that in this study, we collected brain microvessel fragments using dextran gradient centrifugation, unlike the original vasculome paper where enzyme digestion and CD-31 magnetic bead isolation were required for the structurally tougher tissue from heart and kidney. We acknowledge that the comparative purity of endothelial cells may be different between these two methods. But according to reverse-transcriptase polymerase chain reaction (RT-PCR) verification, sufficient purity of microvessels was still obtained here.
Quantitative real-time polymerase chain reaction
Relative expressions (ipsilateral vs. contralateral [Ipsi/Cont] and ipsilateral vs. normal condition [Ipsi/Norm]) of some selected genes were verified by RT-PCR, a commonly used technique to quantify relative gene expression. First-strand complementary DNA (cDNA) was synthesized with the SuperScript VILO synthesis system (Invitrogen, Grand Island, NY), consisting of SuperScript III reverse transcriptase and Oligo(dT) primers. Specific gene expression level was quantified by real-time PCR on an ABI-7500 (Applied Biosystems, Foster City, CA) thermal cycler using predesigned Taqman primers with FAM fluorescent labeling (Applied Biosystems). Data normalization was performed by quantification of the endogenous 18S ribosomal RNA, and fold change was measured with 2-ΔΔCt method.
Hematoxylin-eosin staining
To check the histomorphological condition of mouse brain hemisphere, standard H&E staining was processed by Massachusetts General Hospital Histopathology Research Core (Boston, MA).
Microarray sample analysis
Total RNA was extracted from the fresh preparation of brain microvasculature with the RNeasy Plus Micro kit (Qiagen, Hilden, Germany). Concentration of RNA was measured by Nanodrop, and integrity of RNA was tested with the RNA integrity number (RIN) score on the Agilent Bioanalyzer 2100 (Agilent Technologies, Inc., Santa Clara, CAQ). All samples had RIN scores larger than 8.0. Microarray hybridization and scanning was done by Expression Analysis Corporation with Affymetrix murine whole genome microarray M430 2.0, with 3-IVT (in vitro transcription) and Encore Biotin Module for amplification, fragmentation, and labeling. Raw expression data from each chip for each sample were summarized and normalized using the Robust Multi-Array algorithm. The quality of each chip was determined by manually checking mean values, variances, and paired scatter plots as well as principal component analysis plots. All chips passed the quality check. All statistical analyses were performed with the statistics software R (version 2.6.2; available from: http://www.r-project.org) and R packages developed by the BioConductor project (available from: http://www.bioconductor.org).
Microarray data analysis
After normalization and quality checking, samples were clustered using an unsupervised clustering method based on Pearson's correlation coefficient-derived distance matrix using all probes on the chip. Distances between clusters were determined using the complete link method. Next, differentially expressed genes were identified, based on both statistical significances, which were determined using the significance analysis of microarrays algorithm (a variant of t-test and specifically designed for microarray data), and fold change threshold. The genes with p < 0.01 and fold change >2 or <0.5 were considered as differentially expressed. The combination of fold change threshold and p values serves to eliminate most false positives, producing a more stringent list. Third, significantly changed gene sets in whole transcriptome were identified using the gene set enrichment analysis (GSEA) method.27 The GSEA method requires two inputs: 1) a master list of genes ranked according to expression differences between two states and 2) a priori–defined gene sets (e.g., pathways) that consist of specific function-related genes. In this study, input 1 is a list of all genes presented on the Affymetrix mouse M430 2.0 platform and ranked according to the fold changes between two groups; input 2 is the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database (http://www.genome.jp/kegg/) or Gene Ontology (GO) database (http://geneontology.org/).28,29 Enrichment scores were calculated with standard parameters.
Results
Purity and cluster analysis of the traumatic brain injury vasculome
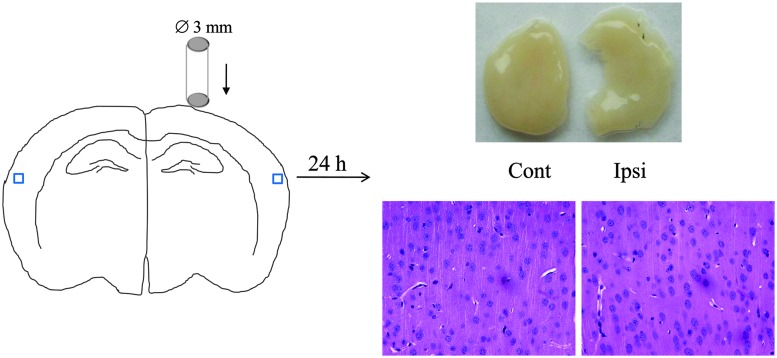
Mice were subjected to CCI (n = 12), and brains were removed at 24 h post-TBI. To prevent contamination of the vasculome by brain debris and nonspecific necrotic tissue, the center core of the TBI lesion was dissected away and tissue samples were isolated from nondirectly damaged ipsi- and contralateral cortex (Fig. 1). H&E staining confirmed that these nondirectly damaged cortical areas had no obvious damage (Fig. 1). For comparison, normal cortex was also obtained from sham-operated mice (n = 6).
FIG. 1.
Sketch of mouse CCI model and sample collection. Left panel shows the CCI model with controlled dropping impounder. Top right shows representative light images of cortexes for contralateral (Cont) and ipsilateral (Ipsi) samples, indicating the missing core area in Ipsi. Lower panel shows representative images of hematoxylin and eosin staining (lower, 400×), indicating no obvious damage in both Contr and Ipsi cortexes. CCI, controlled cortical impact. Color image is available online at www.liebertpub.com/neu
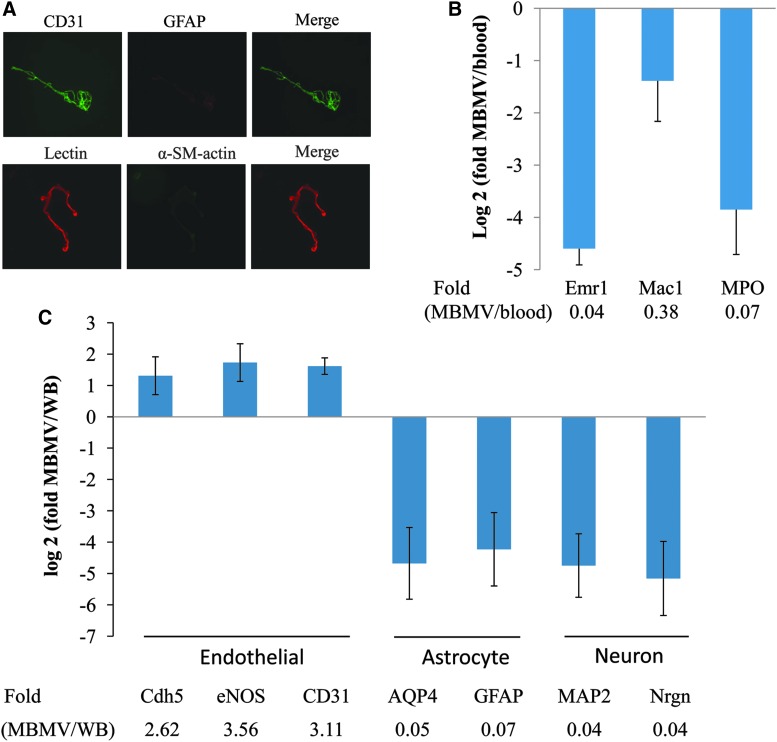
Microvessels were then purified from all samples. Immunostaining demonstrated that isolated microvessel fragments were positive for endothelial CD31 markers and lectin staining and lacked contaminating signals from astrocytic GFAP or smooth muscle/pericyte α-SMA markers (Fig. 2A). RT-PCR confirmed purity of the isolated vasculome, with enriched levels of endothelial genes (Cdh5, eNOS, and CD31) and low levels of astrocyte (AQP4 and GFAP) and neuronal (MAP2 and Nrgn) genes (Fig. 2B), when compared to whole brain cortex RNA samples. There was also no significant contamination from blood; low levels of monocyte (Emr1 and Mac1) and neutrophil (MPO) genes were detected in our vasculome samples, when compared to blood samples (Fig. 2C).
FIG. 2.
Purity of brain microvessel fragments. Partial of fragments were used for fluorescent immunostaining (A), with cell markers for endothelial cells (CD31 and lectin), astrocyte (GFAP), and smooth muscle cells/pericyte (α-SM-actin). (B) Expression levels of brain cell markers are compared between microvessel fragments (MBMV) and whole brain (WB) by reverse-transcriptase polymerase chain reaction, and (C) expression levels of some leukocytes genes are compared between MBMV and mouse WB. Bar represents the mean ± standard deviation from three individual samples. AQP4, aquaporin 4; Emr1, endothelial growth factor–like module-containing mucin-like hormone receptor-like 1; eNOS, endothelial nitric oxide synthase; GFAP, glial fibrillary acidic protein; Mac1, macrophage antigen 1; MAP2, microtubule-associated protein 2; MPO, myeloperoxidase; Nrgn, neurogranin; α-SM-actin, alpha-smooth muscle actin. Color image is available online at www.liebertpub.com/neu
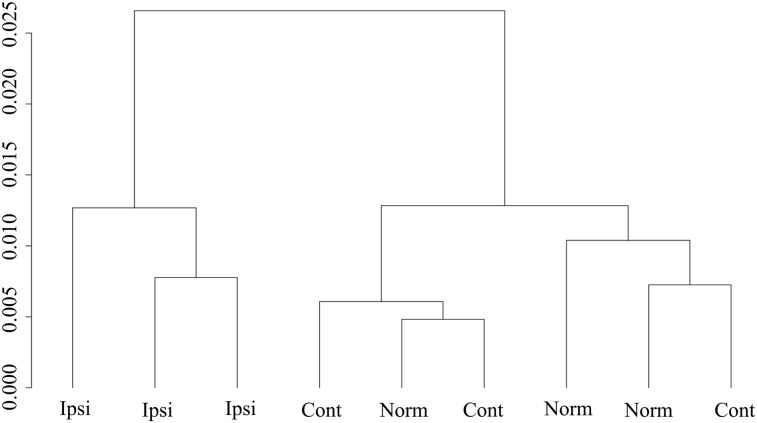
Twelve hemisphere tissue samples per group were obtained for normal shams and contra- and ipsilateral groups, and four samples were pooled into one microarray chip, finally resulting in three chips per study group. Expression profiles were clustered based on the signals of all probes on each chip. Profiles from contralateral and normal cortex were similar to each other, suggesting that this CCI model of TBI did not significantly affect contralateral endothelium by 24 h post-injury (Fig. 3). However, all three samples from ipsilateral cortex were clustered together and clearly separated from normal and contralateral cortex (Fig. 3). This unsupervised clustering result indicated that the vasculome from nondirectly damaged cortical areas was significantly perturbed by TBI.
FIG. 3.
Hierarchical cluster analysis of microarray data. The expression levels of all probes in each chip for individual sample were analyzed, as vasculome from normal (Norm), contralatral (Cont) and non-directly-damaged ipsilatral (Ipsi) samples, using unsupervised clustering algorithm. y-axis indicates sum of absolute distance between samples.
Differentially expressed genes
Applying the standard criteria of p ≤ 0.01 and fold change ≥2 or ≤0.5, there were only 13 genes that were different between samples from contralateral versus normal cortex, again suggesting the lack of large effects in the contralateral mouse brain vasculome post-CCI. However, significant changes were detected in the vasculome from nondirectly damaged ipsilateral cortex; 141 genes were significantly up-regulated and 331 genes were significantly down-regulated compared to contralateral levels, whereas 191 significantly up-regulated genes and 238 significantly down-regulated genes compared to normal samples (see Supplementary Table 1 for full listing) (see online supplementary material at http://www.liebertpub.com). To further ensure that our microarray measurements were reliable, quantitative PCR was used to confirm changes of representative genes from various functional groups, including growth factors, extracellular matrix (ECM) proteinase and components, endothelial response genes, and cytokines. Although there were slight numerical differences in exact levels of quantitation, overall patterns of up- and down-regulation appeared to match between PCR and microarray analyses (Table 1).
Table 1.
Verification of Microarray Data by RT-PCR
| Microarray | qPCR | ||||
|---|---|---|---|---|---|
| Symbol | Ipsi/norm | Ipsi/cont | Ipsi/norm | Ipsi/cont | Description |
| Fmod | 0.08 | 0.07 | 0.06 | 0.05 | Fibromodulin |
| Cxcl12 | 0.25 | 0.24 | 0.51 | 0.39 | Chemokine (C-X-C motif) ligand 12 |
| VCAM-1 | 0.26 | 0.39 | 0.23 | 0.30 | Vascular cell adhesion protein 1 |
| PDGF-d | 0.30 | 0.27 | 0.67 | 0.39 | Platelet-derived growth factor, D polypeptide |
| IGF-1R | 0.52 | 0.47 | 0.69 | 0.71 | Insulin-like growth factor I receptor |
| ZO-1 | 0.71 | 0.63 | 0.62 | 0.44 | Zona occludens protein 1 |
| MMP-9 | 1.10 | 1.07 | 1.51 | 2.91 | Matrix metallopeptidase 9 |
| NOS-3 | 1.42 | 1.00 | 1.31 | 0.75 | Nitric oxide synthase 3 |
| BDNF | 1.81 | 1.92 | 1.05 | 2.12 | Brain-derived neurontrophic factor |
| IL-6 | 3.60 | 3.39 | 20.00 | 10.48 | Interleukin-6 |
| Tnc | 6.41 | 4.89 | 26.01 | 18.41 | Tenascin C |
| Thbs1 | 6.96 | 5.40 | 9.98 | 9.19 | Thrombospondin 1 |
| CD44 | 12.66 | 6.56 | 20.14 | 26.05 | CD44 antigen |
| TIMP-1 | 15.03 | 5.49 | 8.73 | 7.45 | Tissue inhibitor of metalloproteinase 1 |
| Serpine1 | 20.87 | 12.02 | 31.5 | 22.3 | Serine (or cysteine) peptidase inhibitor clade E, member 1 |
| MMP-3 | 25.46 | 15.62 | 65.75 | 1882 | Matrix metallopeptidase 3 |
The values fold change represent the mean of three samples in each group of nondirectly damaged ipsilateral (Ipsi), contralateral (Cont), and normal (Norm) microvessel fragments.
RT-PCR, reverse-transcriptase polymerase chain reaction; qPCR, quantitative polymerase chain reaction.
After general confirmation of these changes in gene expression, the entire TBI vasculome was then assessed to obtain lists of differentially expressed genes in nondirectly damaged ipsilateral cortex. Up-regulated genes (greater than 5.0-fold change vs. sham) and down-regulated genes (less than 0.30-fold change vs. sham) were compiled (Tables 2 and 3, respectively). In general, these differentially expressed genes appeared to belong to prominent functional groups (e.g., matrix proteins, proteinase and their regulators, inflammation response genes and cytokines, and metabolism-related enzymes). Some patterns may also emerge. Genes belonging to matrix and inflammation responses tended to be up-regulated, whereas most of the metabolic genes tended to show reduced expression post-TBI.
Table 2.
List of the Most Increased Genes in Mouse TBI Vasculome Compared to Normal Brain Vasculome
| Fold change | ||||||
|---|---|---|---|---|---|---|
| Probe ID | Gene ID | Symbol | Description | Ipsi/norm | Cont/norm | Function |
| 418666_at | 19288 | Ptx3 | Pentraxin-related gene | 34.40 | 4.88 | Inflammation |
| 1418945_at | 17392 | Mmp3 | Matrix metallopeptidase 3 | 25.46 | 1.63 | ECM |
| 1419149_at | 18787 | Serpine1 | Serine (or cysteine) peptidase inhibitor, clade E, member 1 | 20.87 | 1.73 | ECM |
| 1416431_at | 67951 | Tubb6 | Tubulin, beta 6 | 19.59 | 2.12 | Membrane |
| 1460227_at | 21857 | Timp1 | Tissue inhibitor of metalloproteinase 1 | 15.03 | 2.74 | ECM |
| 1451038_at | 30878 | Apln | Apelin | 14.93 | 1.36 | Metabolism |
| 1418547_at | 21789 | Tfpi2 | Tissue factor pathway inhibitor 2 | 14.93 | 1.96 | ECM |
| 1434376_at | 12505 | Cd44 | CD44 antigen | 12.66 | 1.93 | ECM |
| 1437785_at | 101401 | Adamts9 | A disintegrin-like and metallopeptidase with thrombospondin type 1 motif, 9 | 10.28 | 1.47 | ECM |
| 1434046_at | 433470 | AA467197 | Expressed sequence AA467197 | 8.93 | 1.05 | |
| 1437139_at | 14654 | Glra1 | Glycine receptor, alpha 1 subunit | 8.54 | 1.27 | Transport |
| 1449982_at | 16156 | Il11 | Interleukin-11 | 7.92 | 1.24 | Cytokine |
| 1419100_at | 20716 | Serpina3n | Serine (or cysteine) peptidase inhibitor, clade A, member 3N | 7.20 | 1.94 | ECM |
| 1421811_at | 21825 | Thbs1 | Thrombospondin 1 | 6.96 | 1.29 | ECM |
| 1416342_at | 21923 | Tnc | Tenascin C | 6.41 | 1.31 | ECM |
| 1422053_at | 16323 | Inhba | Inhibin beta-A | 6.35 | 1.02 | Inflammation |
| 1448831_at | 11601 | Angpt2 | Angiopoietin 2 | 6.15 | 1.69 | Peptide |
| 1437279_x_at | 20969 | Sdc1 | syndecan 1 | 5.78 | 1.67 | ECM |
| 1450716_at | 11504 | Adamts1 | A disintegrin-like and metallopeptidase with thrombospondin type 1 motif, 1 | 5.71 | 1.13 | ECM |
| 1416529_at | 13730 | Emp1 | Epithelial membrane protein 1 | 5.66 | 1.20 | Membrane |
| 1447839_x_at | 11535 | Adm | Adrenomedullin | 5.23 | 1.33 | Peptide |
| 1416039_x_at | 16007 | Cyr61 | Cysteine-rich protein 61 | 5.18 | 1.02 | ECM |
| 1421207_at | 16878 | Lif | Leukemia inhibitory factor | 5.03 | 1.30 | Cytokine |
| 1448471_a_at | 13024 | Ctla2a | Cytotoxic T-lymphocyte-associated protein 2 alpha | 5.01 | 1.59 | Inflammation |
TBI, traumatic brain injury; Ipsi, ipsilateral; Norm, normal; Cont, contralateral; ECM extracellular matrix.
Table 3.
List of Most Decreased Genes in Mouse Brain TBI Vasculome Compared to Normal Brain Vasculome
| Fold change | ||||||
|---|---|---|---|---|---|---|
| Probe ID | Gene ID | Symbol | Description | Ipsi/norm | Cont/norm | Function group |
| 1456084_x_at | 14264 | Fmod | Fibromodulin | 0.082 | 1.14 | ECM |
| 1450468_at | 17926 | Myoc | Myocilin | 0.092 | 0.85 | Membrane |
| 1429286_at | 68888 | Gkn3 | Gastrokine 3 | 0.16 | 0.96 | ECM |
| 1419663_at | 18295 | Ogn | Osteoglycin | 0.16 | 1.00 | ECM |
| 1418979_at | 105387 | Akr1c14 | Aldo-keto reductase family 1, member C14 | 0.16 | 1.10 | Metabolism |
| 1434734_at | 623474 | Rad54b | RAD54 homolog B (S.cerevisiae) | 0.22 | 0.79 | Cell response |
| 1451006_at | 22436 | Xdh | Xanthine dehydrogenase | 0.24 | 1.00 | Metabolism |
| 1423858_a_at | 15360 | Hmgcs2 | 3-hydroxy-3-methylglutaryl-Coenzyme A synthase 2 | 0.25 | 0.85 | Metabolism |
| 1416966_at | 19879 | Slc22a8 | Solute carrier family 22 (organic anion transporter), member 8 | 0.25 | 1.28 | Transport |
| 1417574_at | 20315 | Cxcl12 | Chemokine (C-X-C motif) ligand 12 | 0.25 | 1.07 | Cytokine |
| 1451322_at | 69574 | Cmbl | Carboxymethylenebutenolidase-like (Pseudomonas) | 0.26 | 1.25 | Metabolism |
| 1415989_at | 22329 | Vcam1 | Vascular cell adhesion molecule 1 | 0.26 | 0.68 | Inflammation |
| 1427345_a_at | 20887 | Sult1a1 | Sulfotransferase family 1A, phenol-preferring, member 1 | 0.26 | 1.44 | Metabolism |
| 1426215_at | 13195 | Ddc | Dopa decarboxylase | 0.26 | 1.52 | Metabolism |
| 1450699_at | 20341 | Selenbp1 | Selenium binding protein 1 | 0.26 | 1.17 | Transport |
| 1438696_at | 13616 | Edn3 | Endothelin 3 | 0.27 | 1.02 | Active peptide |
| 1457867_at | 433323 | Sgpp2 | Sphingosine-1-phosphate phosphotase 2 | 0.27 | 1.02 | Metabolism |
| 1444139_at | 73284 | Ddit4l | DNA-damage-inducible transcript 4-like | 0.28 | 0.82 | Cell response |
| 1422804_at | 20708 | Serpinb6b | Serine (or cysteine) peptidase inhibitor, clade B, member 6b | 0.29 | 0.98 | Cytosolic inhibitor |
| 1451047_at | 16431 | Itm2a | Integral membrane protein 2A | 0.29 | 1.18 | Membrane |
| 1439790_at | 20723 | Serpinb9 | Serine (or cysteine) peptidase inhibitor, clade B, member 9 | 0.29 | 1.00 | Cytosolic inhibitor |
| 1440096_at | 407800 | Ecm2 | Extracellular matrix protein 2 | 0.29 | 0.80 | ECM |
| 1416321_s_at | 116847 | Prelp | Proline arginine-rich end leucine-rich repeat | 0.29 | 1.14 | ECM |
TBI, traumatic brain injury; Ipsi, ipsilateral; Norm, normal; Cont, contralateral; ECM extracellular matrix.
The fact that many vasculome genes were altered in the matrix category may be consistent with the overall concept of the neurovascular unit. Matrix proteins mediate communication between cells, interaction between cells and extracellular environment, and can also modulate the bioavailability of growth factors and other signaling molecules. For BBB regulation, for example, matrix metalloproteinase (MMP) 3 mediates BBB leakage and edema and hemorrhage.30,31 Serpine1 (also known as PAI-1 [plasminogen activator inhibitor type 1]), a tissue plasminogen activator (tPA) inhibitor, is known to regulate the tightness of brain endothelial barriers.32 The increase of angiopoietin (Ang) 2, or the disturbed ratio of Ang1/Ang2, is suspected of contributing to loss of BBB integrity after brain injury.33,34 Tissue inhibitor of metalloproteinase (TIMP-1), as a general inhibitor to MMPs, may also ameliorate BBB leakage post-injury.35 Perhaps these vasculome responses represent, in part, acute pathophysiology as well as the beginnings of endogenous repair post-TBI.
Another prominent aspect of the TBI vasculome may be related to the angiogenesis process36,37; many genes were altered in this category. Thrombospondin 1 (THBS-1) is one of the endogenous protein inhibitors of angiogenesis; Adamts (1 and 9) have potential antiangiogenic effects from their THBS-1 repeat domains.38 Transgenic mice with overexpressed tissue factor pathway inhibitor-2 show reduced neovascularization,39 whereas PAI-1-deficient mice show markedly increased angiogenesis and enhanced wound healing,40 and addition of PAI-1 reduced Sonic hedgehog–induced tube formation.41 Cysteine-rich angiogenic inducer 61 has been identified as a novel, vascular endothelial growth factor (VEGF)-A-independent, clinically relevant proangiogenic factor response to hedgehog signaling, for tumor growth, tissue repair, and normal vascular development.42–44 Syndecan 1 (Sdc1), a major component of the endothelial glycocalyx, is reported to participate in VEGF-induced angiogenesis45 and might also negatively regulate fibroblast growth factor 2 (FGF2)-induced vascular growth,46 suggesting the dual roles of Sdc1 in regulation of angiogenesis. Ang2 is considered as a proangiogenesis factor, facilitates VEGF-induced angiogenesis in the mature mouse brain compared to VEGF alone,47,48 and is involved in brain development and tumor growth.49,50 By human transcriptome and interactome analysis, CD44 was identified as one of six proteins at the center of an angiogenesis-associated network, suggesting its central role of crosstalk between protein families in regulation of angiogenesis.51 Taken together, this broad spectrum of angiogenic genes suggests that the TBI vasculome may be surprisingly trying to remodel, even at this acute stage post-injury.
For metabolic enzymes, the majority of responses trended toward down-regulation, suggesting that the functional status of microvessels may be significantly altered even far away from directly damaged tissue. Apelin (Apln) regulates glucose metabolism and insulin sensitivity, partially through a nitric oxide (NO)-dependent pathway.52,53 It is known to be beneficial to blood vessels by promoting angiogenesis, ensuring vascular integrity,54–57 and regulating blood pressure.58 Further, it is suggested that Apln has protective effects on neurons, by activating several prosurvival signaling and inhibiting excitotoxic signaling in neurons.59,60 Gene xdh encodes for two enzymes, XDH and XO (xanthine oxidase), which could be converted by reversible sulfhydryl oxidation or irreversible proteolytic modification. Both are known for metabolism of purines and as a resource for oxygen radicals and have been implicated in the pathogenesis of endothelial injury and several diseases. There is an association between increased XO activity and negative clinical outcomes,61–64 but the final balance may be complex given that recent reports also suggest that XO could a beneficial factor in those pathological conditions by inducing NO production.65 Sphingosine phosphate phosphatase 2 (Sgpp2) is one of the metabolic enzymes essential for degradation of S1P (sphingosine 1-phosphate), which is a bioactive lipid molecule that acts as both an extracellular signaling mediator and an intracellular second messenger, suggesting that Sgpp2 participates in S1P-related functions, including angiogenesis and BBB regulation.66
For the general category of inflammatory response genes, the TBI vasculome showed a general increase in expression. Pentraxin 3 (PTX3), a hormonal mediator of innate immunity, regulates hyaluronic acid-enriched ECM assembly and inhibits FGF2-mediated angiogenesis.67,68 PTX3 is induced under inflammatory conditions, and deficiency of PTX3 promotes vascular inflammation and atherosclerosis, suggesting the vascular protective effects of PTX3.69 Similarly, PTX3 is reported to have a protective function in seizure-induced neurodegeneration.70 Both interleukin (IL)-11 and leukemia inhibitory factor (LIF) are members of the Il-6 family of cytokines, and both are neuropoietic cytokines with trophic effects on subsets of neurons, with beneficial effects for tissue recovery post-injury.71–74 IL-11 also shows thrombopoietic activity and cytoprotective effects.75–77 It was reported that plasma IL-11 levels in spontaneous intracerebral hemorrhage (ICH) patients are highly associated with mortality and hydrocephalus occurring after ICH.78 LIF is also a key regulator for stem cell proliferation and differentiation, such as for astrocytes, oligodendrocyte progenitor cells, and neural stem cells, in addition to endothelial cells.79–81 Inhba is a subunit to several cytokines, including homodimer of Activin A, and heterodimer of Activin AB or Inhibin A, all belong to the transforming growth factor beta (TGF-β) superfamily, but Activins and Inhibins are functionally antagonists. Activin A is reported to be released early in acute systemic inflammation, increased in many disease conditions,82 and strongly expressed during the repairing process of various tissues and organs, including the cardiovascular system and brain.83 Activin A also exerts antiproliferation and anti-angiogenesis effects, but with neuroprotective effect.84 It mediates the best documented neuroprotective growth factor, FGF2-induced neuroprotection.85 Taken together, many of these up-regulated genes may represent enhancement of the inflammatory status in the TBI vasculome. However, there may be hints of pathway specificity, and not all responses were uniformly up-regulated. Some interesting signals were decreased. For example, chemokine (C-X-C motif) ligand 12 (Cxcl12; also known as SDF-1 [stromal cell-derived factor-1]), a chemoattractant for immune cells in response, regulates leukocyte recruitment and their transendothelial migration.86,87 Vascular cell adhesion molecule (VCAM-1), an adhesion molecule, also functions as a scaffold for leukocyte (and other immune cells) migration and a trigger of endothelial signaling through nicotinamide adenine dinucleotide phosphate oxidase–generated reactive oxygen species.88 Both Cxcl12 and VCAM-1 appeared to be reduced in nondirectly impacted microvessels, perhaps suggesting an endogenous response to help to reduce the leukocyte invasion around this “histologically normal” area. It is well accepted that neuroinflammation with TBI has both detrimental and beneficial effects,89,90 and our vasculome responses may be consistent with this general principle as well.
A key function of cerebral endothelium is the regulation of permeability and hemodynamics. No significant changes were detected for the primary tight junction and adherens junction proteins, consistent with the fact that no explicit BBB damage occurred in the nondirectly damaged cortex. However, the TBI vasculome showed interesting perturbations in some permeability-associated genes. The membrane proteins, integral membrane protein 2A and epithelial membrane protein 1 (EMP1), have been reported to have specific expression in brain microvascular endothelial cells, compared to aortic endothelial cells, suggesting that they might be novel contributors to BBB function. EMP1 was further shown to be colocalized with occludin and this might facilitate its ability to coregulate BBB function under injury conditions.91 For hemodynamic status, a larger list of genes was detected, including adrenomedullin (Adm) and endothelin 3 (Edn3), two autocrine peptides released from endothelial cells. Adm is a potent hypotensive hormone peptide, with high concentration in the cerebral circulation.92 The vasculoprotective effect of Adm may involve vasodilation effects, reduction of oxidative stress, inhibition of endothelial cell apoptosis, promotion of angiogenesis, regulation of permeability, and control of fluid and electrolyte homeostasis.92–96 Adm is also implicated as a promising plasma biomarker, so its presence in the TBI vasculome may warrant further investigation. Administration of exogenous Adm and its binding protein has shown neuroprotective effects against brain injury in experimental disease models.97,98 Edn3, a vasoconstrictor from the endothelin family, is known to induce NO production, promote angiogenesis, regulate synthesis of inflammatory mediators and glutamate transporters, and control fluid and electrolyte homeostasis in the brain.99–104 This broad spectrum of vascular responses again suggests a potentially wide distribution of microvessel reactions to injury in the entire ipsilateral cortex after TBI.
Pathway analysis
Although statistically probing the TBI vasculome for individually altered genes is a logical first step, another complementary way to assess changes over entire transcriptome is via pathway enrichment analysis by GSEA. Using GSEA analysis based on KEGG and GO terms, many statistically enriched pathways were found in nondirectly damaged cortex (Table 4). These clusters of gene responses included increased pathways of ECM, matrix peptidases and inhibitors, and cell communication, cytokine-receptor interactions, and inflammatory responses. Reduced pathways included those comprising mitochondrial function, oxidative phosphorylation, fatty acid metabolism, glutathione metabolism, gap junctions, and transport processes. Specific signaling pathways were also identified, including TGF-β and Wnt, both of which have important roles in the neurovascular unit, such as BBB permeability, angiogenesis, and neurogenesis.
Table 4.
Enriched Pathways in Mouse Brain TBI Vasculome Compared to Normal Brain Vasculome
| Ipsi/norm | Ipsi/norm | ||||
|---|---|---|---|---|---|
| p value | ES | Pathway (from KEGG) | p value | ES | Pathway (GO term) |
| 1.67E-12 | −0.25 | Oxidative phosphorylation | 0.00E+00 | −0.10 | Integral to membrane |
| 1.96E-08 | −0.16 | Wnt-signaling pathway | 1.55E-15 | −0.13 | Mitochondrion |
| 9.32E-08 | −0.32 | Valine, leucine, and isoleucine degradation | 2.76E-07 | −0.08 | Transport |
| 5.75E-07 | −0.15 | Axon guidance | 4.03E-07 | −0.07 | Zinc ion binding |
| 3.95E-06 | −0.29 | Fatty acid metabolism | 1.64E-06 | −0.28 | Electron transport chain |
| 4.62E-06 | −0.33 | Propanoate metabolism | 3.76E-06 | −0.12 | Modification-dependent protein catabolic |
| 1.04E-05 | −0.13 | Calcium-signaling pathway | 6.44E-04 | −0.04 | Protein binding |
| 1.42E-05 | −0.17 | Gap junction | 1.20E-03 | −0.48 | Glutathione metabolic process |
| 1.17E-04 | −0.09 | MAPK-signaling pathway | 4.59E-03 | −0.07 | Metabolic process |
| 9.34E-04 | −0.10 | Ubiquitin-mediated proteolysis | |||
| 2.28E-03 | −0.13 | TGF-β-signaling pathway | 2.25E-10 | 0.10 | Extracellular region |
| 2.64E-03 | −0.25 | Glutathione metabolism | 7.82E-08 | 0.19 | Proteinaceous extracellular matrix |
| 2.86E-03 | −0.44 | Thiamine metabolism | 3.40E-06 | 0.21 | Cytokine activity |
| 3.68E-03 | −0.29 | beta-Alanine metabolism | 7.51E-06 | 0.23 | Inflammatory response |
| 1.30E-04 | 0.19 | Serine-type endopeptidase activity | |||
| 1.41E-05 | 0.17 | Cell communication | 1.60E-04 | 0.25 | Chemotaxis |
| 3.33E-03 | 0.09 | Cytokine-cytokine receptor interaction | 5.97E-04 | 0.21 | Peptidase inhibitor activity |
Analysis here is based on vasculomes from nondirectly damaged ipsilateral 24 h post-TBI and normal brains, based on KEGG pathway database or GO database. Negative ES (enrichment score) for decreased genes expression of such pathways; positive ES for increased genes expression of such pathways in TBI vasculome compared to normal brain.
TBI, traumatic brain injury; Ipsi, ipsilateral; Norm, normal; KEGG, Kyoto Encyclopedia of Genes and Genomes; GO, Gene Ontology; MAPK, mitogen-activated protein kinase; TGF, transforming growth factor.
Increased terms belonging to the cytokine signaling categories are consistent with known effects of TBI on neuroinflammation.90,105,106 Reductions in mitochondria activity and oxidative phosphorylation are consistent with known detrimental effects of TBI on energy metabolism.107,108 The present findings suggest that inflammatory and metabolic responses in endothelial cells may contribute to vascular dysfunction after TBI. Further, the reduced transport, tight junction, and gap junction categories all suggest a compromised BBB homeostasis in nondirectly damaged areas. Overall, GSEA analysis revealed alterations in pathways that correlate well with the single-gene up-/down-regulation analyses described earlier. Taken together, these mean that the deleterious vascular effects of TBI might be surprisingly diffuse, beyond core regions of directly impacted cortex.
Overlap with genome-wide association study genes
It has long been suggested that past TBI increases the subsequent incidence of chronic neurodegenerative disorders, including Alzheimer's disease (AD), Parkinson's disease (PD), and amyotrophic lateral sclerosis.109,110 Previously, we reported that the mouse brain vasculome contained many genes that are found in genome-wide association study (GWAS) datasets for human central nervous system (CNS) disorders, such as stroke, AD, and PD.111 Is it possible that the TBI vasculome may also involve GWAS genes that would contribute to the vascular pathophysiology of brain injury? To assess this hypothesis, we compared our first draft of the TBI vacsulome with the three GWAS datasets. At 24 h post-trauma, differentially expressed genes in nondirectly damaged microvessels were significantly associated with stroke GWAS genes (p value of 8.25E-05 and odds ratio of 2.50). Similar linkages were detected for GWAS datasets in PD and AD as well. The list of significantly differentially expressed genes in the TBI vasculome overlapped with these disease GWAS databases are shown in Table 5.
Table 5.
List of Disease GWAS Genes Significantly Changed in TBI Vasculome
| Gene ID | Symbol | Related disease | Ipsi/norm | Description |
|---|---|---|---|---|
| 76459 | Car12 | Stroke | + | Carbonic anyhydrase 12 |
| 12505 | Cd44 | Stroke | + | CD44 antigen |
| 17064 | Cd93 | Stroke, PD | + | CD93 antigen |
| 14654 | Glra1 | Stroke | + | Glycine receptor, alpha 1 subunit |
| 195646 | Hs3st2 | Stroke | + | Heparan sulfate (glucosamine) 3-O-sulfotransferase 2 |
| 18793 | Plaur | Stroke | + | Plasminogen activator, urokinase receptor |
| 65112 | Pmepa1 | Stroke, PD | + | Prostate transmembrane protein, androgen induced 1 |
| 277360 | Prex1 | Stroke | + | Phosphatidylinositol-3,4,5-trisphosphate–dependent Rac exchange factor 1 |
| 12702 | Socs3 | Stroke | + | Suppressor of cytokine signaling 3 |
| 72475 | Ssbp3 | Stroke | + | Single-stranded DNA-binding protein 3 |
| 217262 | Abca9 | Stroke | − | ATP-binding cassette, subfamily A (ABC1), member 9 |
| 330941 | AI593442 | Stroke | − | Expressed sequence AI593442 |
| 11982 | Atp10a | Stroke | − | ATPase, class V, type 10A |
| 320405 | Cadps2 | Stroke | − | Ca2+-dependent activator protein for secretion 2 |
| 13176 | Dcc | Stroke | − | Deleted in colorectal carcinoma |
| 269109 | Dpp10 | Stroke | − | Dipeptidylpeptidase 10 |
| 74559 | Elovl7 | Stroke | − | ELOVL family member 7, elongation of long chain fatty acids (yeast) |
| 217082 | Hlf | Stroke | − | Hepatic leukemia factor |
| 209378 | Itih5 | Stroke | − | Inter-alpha (globulin) inhibitor H5 |
| 243382 | Ppm1k | Stroke | − | Protein phosphatase 1K (PP2C domain containing) |
| 227545 | Proser2 | Stroke | − | Proline and serine rich 2 |
| 11514 | Adcy8 | AD | + | Adenylate cyclase 8 |
| 67801 | Pllp | AD | + | Plasma membrane proteolipid |
| 12826 | Col4a1 | PD | + | Collagen, type IV, alpha 1 |
| 15900 | Irf8 | PD | + | Interferon-regulatory factor 8 |
| 11994 | Pcdh15 | PD | + | Protocadherin 15 |
| 219257 | Pcdh20 | PD | − | Protocadherin 20 |
| 19279 | Ptprr | PD | − | Protein tyrosine phosphatase, receptor type, R |
The significantly changed gene list in mouse TBI vasculome is searched for disease GWAS gene, including stroke, AD, and PD. Plus sign (+) indicates significantly increased gene; minus sign (–) indicates significantly decreased gene in mouse TBI vasculome compared to normal brain.
GWAS, genome-wide association studies; TBI, traumatic brain injury; Ipsi, ipsilateral; Norm, normal; PD, Parkinson's disease; AD, Alzheimer's disease; ATP, adenosine triphosphate.
Although the links between the TBI vasculome and GWAS data sets are intriguing, specific mechanisms remain to be defined. Nevertheless, a few genes may possess signaling roles that potentially underlie TBI pathophysiology. For example, stroke GWAS gene Cd44 is one with a significant increase after trauma. As an adhesion molecule as well as a signaling regulator, Cd44 may be involved in endothelial cell recognition, lymphocyte trafficking, actin cytoskeleton organization, angiogenesis, and vascular integrity.51,112–114 Cd44 is also known to increase in the brain after ischemia,115 and CD44 knockout mice showed reduced ischemic infarct area with improved neurological functions compared to that of wild-type mice.116 Whether and how CD44 underlies vascular pathology after TBI should be rigorously explored.
Stroke GWAS gene suppressor of cytokine signaling 3 (SOCS3) is an inducible negative regulator of signaling by the Janus kinase/signal transduction and activator of transcription 3 pathway, and it is activated in many CNS injury conditions,117–119 as part of an endogenous response to regulate endothelial cell death,120,121 pathological angiogenesis, and inflammation response.122–125 Together with CD93 and Apelin, SOCS3 is down-regulated in the brain after water deprivation.126 At the same time, SOCS3 can also act as an intrinsic inhibitor for neural axon regeneration. Hence, crosstalk between the vasculome and neural plasticity remains a possibility.
Prostate transmembrane protein, androgen-induced 1 (PMEPA1) and CD93 are related with both stroke and PD. There are a few reports demonstrating that PMEPA1 can bind with E3 ubiquitin-protein ligase NEDD4 (neural precursor cell expressed developmentally down-regulated protein 4) to negatively regulate cell growth.127 PMEPA1 is also a direct target gene of TGF-β signaling and participates in a negative feedback loop to control the duration and intensity of TGF-β/SMDH (mothers of decapentaplegic homologue) signaling.128 Given that TGF-β signaling has been identified as a major pathway in the TBI vasculome, this linkage with GWAS genes further suggests that it may play a role in the pathophysiology of widespread vascular dysfunction in noninjured tissue.
CD93 is a membrane-associated glycoprotein, expressed in many types of cells, regulates phagocytosis of apoptotic cells during organ development, tissue repair, and maintenance of tissue homeostasis, and also regulates leukocyte migration and endothelial cell adhesion.129 Recently, it is reported that as a novel regulator of inflammation, CD93 was highly induced in mouse brain after focal cerebral ischemia, especially in endothelial cells and selected macrophages and microglia.130 CD93 knockout mice showed increased leukocyte infiltration into the brain, increased edema, and larger infarct volumes after ischemia and reperfusion, suggesting the potential neuroprotection from CD93.130 Although human CD93, an epidermal growth factor (EGF)-like domain-containing transmembrane protein, is predominantly expressed in the vascular endothelium; soluble CD93 (with an EGF-like domain) has been detected in plasma.131 Membrane-bound CD93 is proteolytically cleaved to soluble form in response to inflammatory signals, and soluble CD93 released from endothelium is a novel angiogenic factor.131,132 The level of soluble CD93 in plasma has been proposed as a novel biomarker for coronary artery disease and myocardial infarction.133 Whether soluble CD93 released from the TBI vasculome may also act as a biomarker remains to be investigated.
Discussion
The vasculome may provide a potentially useful conceptual framework for investigating how the vascular phenomenon may contribute to the pathophysiology of CNS injury and disease. Here, we mapped the initial draft of the TBI vasculome in a mouse model of CCI. Overall, we found that many genes and pathways were perturbed in ipsilateral cortex, even in remote areas far from the core lesion site.
In our study, the number of down-regulated genes was larger than up-regulated genes. This stands in contrast to previous TBI studies, where increased gene expression was more commonly reported. Von Gertten and colleagues used a cDNA microarray of 6200 gene probes to study expression changes induced by CCI in rat ipsilateral cortex, specifically within the impact area and surrounding cortex.134 This study identified 150 genes that were significantly increased and 61 genes that were down-regulated compared to contralateral cortex at 24 h post-injury. In another relevant study, Natale and colleagues assessed gene expression in a rat model of fluid percussion and in mice with cortical impact injury, with measurements obtained within a 4-mm-diameter disk of parieto-occipital cortex containing core damaged tissue and surrounding contusion areas at 24 h post-injury.135 In this study, 64 up-regulated genes and only 10 down-regulated genes were found in rat ipsilateral cortex after fluid percussion, and 207 up-regulated genes and 76 down-regulated genes in mouse ipsilateral cortex after cortical impact. Though the precise numbers may be without biological significance, the predominance of up- or down-regulation in gene expression may suggest different states of responses in the perturbed brain. There may be two reasons why our data differ. First, almost all previous studies examined total gene expression in macroscopically dissected brain. For a heterogenous multi-cell organ such as the brain, this aggregate response may be misleading. Because our TBI vasculome should be cell specific for cerebral endothelium in microvessels, the predominance of down-regulated genes may tell us something about the state of microvessels and vascular homeostasis in perturbed brain. A second potential issue may be related to the location of analysis. Previous studies tended to emphasize lesion cores and perilesional areas, whereas our study mapped the vasculome in the entire nondirectly damaged ipsilateral cortex. But at least one previous study attempted to examine TBI responses in nondamaged areas. Truettner and colleagues measured the expression of heat shock protein 70, brain-derived neurotrophic factor (BDNF), and nerve growth factor in rat brain after moderate fluid percussion injury. All three genes were rapidly up-regulated post-injury, even in cortex far from areas with histopathological damage. This study concluded that these gene expression patterns may be related with neuronal excitation and plasticity.136 Our TBI vasculome suggests that hemispheric changes may be more common than previously thought, and the broad spectrum of down-regulated genes may suggest a widespread suppression of normal function in all microvessels. These findings may be consistent with the emerging idea of diffuse responses in the brain post-TBI. Axonal perturbations in white matter are now thought to underline overall alternations in neuronal connectivity post-TBI.137,138 The same may be true in terms of diffuse perturbations in vascular homeostasis. Altogether, the entire neurovascular unit may be affected throughout the hemisphere, and investigations into these network responses should be warranted.139
A major overall feature of the TBI vasculome may relate to the convergence of individual gene analysis and pathway analysis. Overall, genes and pathways related to matrix, cytokines, and inflammation were up-regulated, whereas those related to metabolism, transport, and barrier/hemodynamic function tended to be down-regulated. The decrease of metabolic processes in microvessels from nondirectly damaged cortex involved a wide array of effects, including reduced mitochondrion-related functions, oxidative phosphorylation for energy supply, and glutathione metabolic process. There also appeared to be a general reduction in tight junction and gap junction regulation, as well as transport systems at the neurovascular interface. At the same time, these “remote” microvessels showed an amplified response to trauma, including augmented inflammation response, cytokine activity, and receptor activities. Furthermore, many alterations were detected in ECM categories, all of which may influence cell-cell communication within the entire neurovascular unit. Taken together, these pathway findings suggest that microvessels from nondirectly damaged cortex are not quiescent post-injury, and widespread perturbations in vascular homeostasis may underlie an important mechanism throughout the TBI hemisphere.
The concept of the neurovascular unit emphasizes that cellular responses cannot occur in isolation and crosstalk between all neural, glial, and vascular compartments may be unavoidable. The same may apply to the TBI vasculome. Although, by definition, the vasculome is centered on vascular phenomena, many of our detected signals may connect with other cell types. For example, changes were observed in genes from the semaphorin family. Sema4A is an angiogenesis inhibitor, but it can also affect axon guidance. Fmod and Prelp (from a class II small leucine-rich repeat proteoglycans family) and TnC comprise important matricellular proteins in the neurovascular matrix, but all three are reported to regulate the environmental niche of stem cells, suggesting that they could promote angiogenesis as well as neurogenesis during tissue remodeling post-TBI.140–143 Et-3 (Edn3) and its receptor, EDNRB, are found in endothelial systems, but may also regulate proliferation, differentiation, and migration of neural crest cells during development.144 How the vasculome affects endogenous responses in other cell types post-TBI may be an interesting future direction.
Although our individual gene and pathway analyses revealed many cascades and potential targets, it is not easy to functionally interpret how the entire gene network is balanced between deleterious versus beneficial responses. For example, increases in THBS-1 and MMP-3, the decrease in the anti-inflammation gene Cxcl12, and the overall decrease of glutathione metabolism and oxidative phosphorylation may all point toward vascular dysfunction. In contrast, there was also up-regulation in protection signals, such as the anti-inflammatory IL-6 family (IL-6, IL-11, and LIF), and an elevation of TIMP-1 and growth factors, such as BDNF, VEGF, osteoglycin, and platelet-derived growth factor D. The same is true for some matrix-related signals, including tPA/PAI-1/MMP3. How the various ECM components and proteases coalesce to regulate neurovascular proteolysis versus homeostasis, angiogenesis versus edema, and other dynamic phenomena will have to be rigorously examined in future studies.
Taken together, our initial findings suggest that the TBI vasculome is broadly perturbed even in histological normal nondirectly damaged areas. However, there are a few caveats to keep in mind for this proof-of-concept study. First, for the sake of signal to noise, we mapped the vasculome in the entire cortex. It remains possible that even away from the core or perilesional rim, there may still exist gradients in vascular response. Further efforts to map the vasculome with more spatial precision, perhaps with laser-capture methods, may be interesting, especially for the purposes of distinguishing critical responses in the pericontusional rim from more distal profiles in the entire ipsilateral vasculome. Second, we only focused on acute 24-h post-TBI data in this proof-of-concept study. How the vasculome evolves over time may provide new insights into the progressive pathophysiology as tissue deteriorate and recovers. More detailed investigations are required to map how the TBI vasculome evolves from days to weeks post-trauma. Third, our technical approach concentrated on microvessels because these comprised the vast majority of vessels in the brain. However, changes in larger vessels may also be important, especially in terms of collateral supplies and vasculogenesis during recovery. How the vasculome differs at different levels of the vascular tree remains unknown. Fourth, beyond endothelial cells, multiple cell types also contribute to the neurovascular unit in terms of dysfunction and endogenous repair. For example, microglia cells are major participants in TBI response; mapping the microgliome and other cellular responses should also be considered. Finally, the TBI vasculome is expressed at many levels. We started by mapping the transcriptome, but whether gene patterns truly match protein patterns remains unknown. Our transcriptome findings may be quite relevant because recent studies now estimate that transcription can account for up to 80% of protein variance.145–147 Nevertheless, future efforts to study the proteomic and metabolomic profiles of the TBI vasculome may also be warranted.
TBI is a highly complex and challenging problem. Increasingly, it is recognized that vascular responses may play an essential role in its pathophysiology.148 In this proof-of-concept study, we mapped the 24-h post-TBI vasculome in a mouse model of CCI and showed that widespread microvessel perturbations are present even in nondirectly damaged areas. Further investigations into the vasculome may reveal new opportunities for pursuing targets and biomarkers for TBI.
Supplementary Material
Acknowledgments
This work was supported, in part, by grants from the National Institutes of Health grant K08NS057339 and Department of Defense (DoD) office of the Congressionally Directed Medical Research Programs. The content is solely the responsibility of the authors and does not necessarily represent the official views of the DoD.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Centers for Disease Control and Prevention. (2013). CDC grand rounds: reducing severe traumatic brain injury in the United States. MMWR Morb. Mortal. Wkly. Rep. 62, 549–552 [PMC free article] [PubMed] [Google Scholar]
- 2.Narayan R.K., Michel M.E., Ansell B., Baethmann A., Biegon A., Bracken M.B., Bullock M.R., Choi S.C., Clifton G.L., Contant C.F., Coplin W.M., Dietrich W.D., Ghajar J., Grady S.M., Grossman R.G., Hall E.D., Heetderks W., Hovda D.A., Jallo J., Katz R.L., Knoller N., Kochanek P.M., Maas A.I., Majde J., Marion D.W., Marmarou A., Marshall L.F., McIntosh T.K., Miller E., Mohberg N., Muizelaar J.P., Pitts L.H., Quinn P., Riesenfeld G., Robertson C.S., Strauss K.I., Teasdale G., Temkin N., Tuma R., Wade C., Walker M.D., Weinrich M., Whyte J., Wilberger J., Young A.B., and Yurkewicz L. (2002). Clinical trials in head injury. J. Neurotrauma 19, 503–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kabadi S.V., and Faden A.I. (2014). Neuroprotective strategies for traumatic brain injury: improving clinical translation. Int. J. Mol. Sci. 15, 1216–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Algattas H., and Huang J.H. (2014). Traumatic brain injury pathophysiology and treatments: early, intermediate, and late phases post-injury. Int. J. Mol. Sci. 15, 309–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arai K., Lok J., Guo S., Hayakawa K., Xing C., and Lo E.H. (2011). Cellular mechanisms of neurovascular damage and repair after stroke. J. Child Neurol. 26, 1193–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lo E.H., Dalkara T., and Moskowitz M.A. (2003). Mechanisms, challenges and opportunities in stroke. Nat. Rev. Neurosci. 4, 399–415 [DOI] [PubMed] [Google Scholar]
- 7.Lecrux C., and Hamel E. (2011). The neurovascular unit in brain function and disease. Acta Physiol. (Oxf.) 203, 47–59 [DOI] [PubMed] [Google Scholar]
- 8.Iadecola C. (2010). The overlap between neurodegenerative and vascular factors in the pathogenesis of dementia. Acta Neuropathol. 120, 287–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zlokovic B.V. (2011). Neurovascular pathways to neurodegeneration in Alzheimer's disease and other disorders. Nat. Rev. Neurosci. 12, 723–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zacchigna S., Lambrechts D., and Carmeliet P. (2008). Neurovascular signalling defects in neurodegeneration. Nat. Rev. Neurosci. 9, 169–181 [DOI] [PubMed] [Google Scholar]
- 11.Zacchigna S., Ruiz de Almodovar C., and Carmeliet P. (2008). Similarities between angiogenesis and neural development: what small animal models can tell us. Curr. Top. Dev. Biol. 80, 1–55 [DOI] [PubMed] [Google Scholar]
- 12.Shen Q., Goderie S.K., Jin L., Karanth N., Sun Y., Abramova N., Vincent P., Pumiglia K., and Temple S. (2004). Endothelial cells stimulate self-renewal and expand neurogenesis of neural stem cells. Science 304, 1338–1340 [DOI] [PubMed] [Google Scholar]
- 13.Dugas J.C., Mandemakers W., Rogers M., Ibrahim A., Daneman R., and Barres B.A. (2008). A novel purification method for CNS projection neurons leads to the identification of brain vascular cells as a source of trophic support for corticospinal motor neurons. J. Neurosci. 28, 8294–8305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo S., Kim W.J., Lok J., Lee S.R., Besancon E., Luo B.H., Stins M.F., Wang X., Dedhar S., and Lo E.H. (2008). Neuroprotection via matrix-trophic coupling between cerebral endothelial cells and neurons. Proc. Natl. Acad. Sci. U. S. A. 105, 7582–7587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arai K., and Lo E.H. (2009). An oligovascular niche: cerebral endothelial cells promote the survival and proliferation of oligodendrocyte precursor cells. J. Neurosci. 29, 4351–4355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pober J.S., and Sessa W.C. (2007). Evolving functions of endothelial cells in inflammation. Nat. Rev. Immunol. 7, 803–815 [DOI] [PubMed] [Google Scholar]
- 17.Rossi B., Angiari S., Zenaro E., Budui S.L., and Constantin G. (2011). Vascular inflammation in central nervous system diseases: adhesion receptors controlling leukocyte-endothelial interactions. J. Leukoc. Biol. 89, 539–556 [DOI] [PubMed] [Google Scholar]
- 18.Pober J.S., Min W., and Bradley J.R. (2009). Mechanisms of endothelial dysfunction, injury, and death. Annu. Rev. Pathol. 4, 71–95 [DOI] [PubMed] [Google Scholar]
- 19.van Hinsbergh V.W. (2012). Endothelium—role in regulation of coagulation and inflammation. Semin. Immunopathol. 34, 93–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wallez Y., and Huber P. (2008). Endothelial adherens and tight junctions in vascular homeostasis, inflammation and angiogenesis. Biochim. Biophys. Acta 1778, 794–809 [DOI] [PubMed] [Google Scholar]
- 21.Schouten M., Wiersinga W.J., Levi M., and van der Poll T. (2008). Inflammation, endothelium, and coagulation in sepsis. J. Leukoc. Biol. 83, 536–545 [DOI] [PubMed] [Google Scholar]
- 22.Rosenberg G.A. (2012). Neurological diseases in relation to the blood-brain barrier. J. Cereb. Blood Flow Metab. 32, 1139–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ayata C., and Ropper A.H. (2002). Ischaemic brain oedema. J. Clin. Neurosci. 9, 113–124 [DOI] [PubMed] [Google Scholar]
- 24.Ning M., Sarracino D.A., Kho A.T., Guo S., Lee S.R., Krastins B., Buonanno F.S., Vizcaino J.A., Orchard S., McMullin D., Wang X., and Lo E.H. (2011). Proteomic temporal profile of human brain endothelium after oxidative stress. Stroke 42, 37–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mori T., Wang X., Aoki T., and Lo E.H. (2002). Downregulation of matrix metalloproteinase-9 and attenuation of edema via inhibition of ERK mitogen activated protein kinase in traumatic brain injury. J. Neurotrauma 19, 1411–1419 [DOI] [PubMed] [Google Scholar]
- 26.Song L., and Pachter J.S. (2003). Culture of murine brain microvascular endothelial cells that maintain expression and cytoskeletal association of tight junction-associated proteins. In Vitro Cell. Dev. Biol. Anim. 39, 313–320 [DOI] [PubMed] [Google Scholar]
- 27.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., and Mesirov J.P. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102, 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanehisa M., and Goto S. (2000). KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harris M.A., Clark J., Ireland A., Lomax J., Ashburner M., Foulger R., Eilbeck K., Lewis S., Marshall B., Mungall C., Richter J., Rubin G.M., Blake J.A., Bult C., Dolan M., Drabkin H., Eppig J.T., Hill D.P., Ni L., Ringwald M., Balakrishnan R., Cherry J.M., Christie K.R., Costanzo M.C., Dwight S.S., Engel S., Fisk D.G., Hirschman J.E., Hong E.L., Nash R.S., Sethuraman A., Theesfeld C.L., Botstein D., Dolinski K., Feierbach B., Berardini T., Mundodi S., Rhee S.Y., Apweiler R., Barrell D., Camon E., Dimmer E., Lee V., Chisholm R., Gaudet P., Kibbe W., Kishore R., Schwarz E.M., Sternberg P., Gwinn M., Hannick L., Wortman J., Berriman M., Wood V., de la Cruz N., Tonellato P., Jaiswal P., Seigfried T., and White R. (2004). The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 32, D258–D261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suzuki Y., Nagai N., Yamakawa K., Kawakami J., Lijnen H.R., and Umemura K. (2009). Tissue-type plasminogen activator (t-PA) induces stromelysin-1 (MMP-3) in endothelial cells through activation of lipoprotein receptor-related protein. Blood 114, 3352–3358 [DOI] [PubMed] [Google Scholar]
- 31.Rosenberg G.A., and Yang Y. (2007). Vasogenic edema due to tight junction disruption by matrix metalloproteinases in cerebral ischemia. Neurosurg. Focus 22, E4. [DOI] [PubMed] [Google Scholar]
- 32.Dohgu S., Takata F., Matsumoto J., Oda M., Harada E., Watanabe T., Nishioku T., Shuto H., Yamauchi A., and Kataoka Y. (2011). Autocrine and paracrine up-regulation of blood-brain barrier function by plasminogen activator inhibitor-1. Microvasc. Res. 81, 103–107 [DOI] [PubMed] [Google Scholar]
- 33.Avraham H.K., Jiang S., Fu Y., Nakshatri H., Ovadia H., and Avraham S. (2014). Angiopoietin-2 mediates blood-brain barrier impairment and colonization of triple-negative breast cancer cells in brain. J. Pathol. 232, 369–381 [DOI] [PubMed] [Google Scholar]
- 34.Chittiboina P., Ganta V., Monceaux C.P., Scott L.K., Nanda A., and Alexander J.S. (2013). Angiopoietins as promising biomarkers and potential therapeutic targets in brain injury. Pathophysiology 20, 15–21 [DOI] [PubMed] [Google Scholar]
- 35.Fujimoto M., Takagi Y., Aoki T., Hayase M., Marumo T., Gomi M., Nishimura M., Kataoka H., Hashimoto N., and Nozaki K. (2008). Tissue inhibitor of metalloproteinases protect blood-brain barrier disruption in focal cerebral ischemia. J. Cereb. Blood Flow Metab. 28, 1674–1685 [DOI] [PubMed] [Google Scholar]
- 36.Haass C., Kaether C., Thinakaran G., and Sisodia S. (2012). Trafficking and proteolytic processing of APP. Cold Spring Harb. Perspect. Med. 2, a006270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang X., Kazerounian S., Duquette M., Perruzzi C., Nagy J.A., Dvorak H.F., Parangi S., and Lawler J. (2009). Thrombospondin-1 modulates vascular endothelial growth factor activity at the receptor level. FASEB J. 23, 3368–3376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iruela-Arispe M.L., Carpizo D., and Luque A. (2003). ADAMTS1: a matrix metalloprotease with angioinhibitory properties. Ann. N. Y. Acad. Sci. 995, 183–190 [DOI] [PubMed] [Google Scholar]
- 39.Ivanciu L., Gerard R.D., Tang H., Lupu F., and Lupu C. (2007). Adenovirus-mediated expression of tissue factor pathway inhibitor-2 inhibits endothelial cell migration and angiogenesis. Arterioscler. Thromb. Vasc. Biol. 27, 310–316 [DOI] [PubMed] [Google Scholar]
- 40.Ebrahimian T.G., Squiban C., Roque T., Lugo-Martinez H., Hneino M., Buard V., Gourmelon P., Benderitter M., Milliat F., and Tamarat R. (2012). Plasminogen activator inhibitor-1 controls bone marrow-derived cells therapeutic effect through MMP9 signaling: role in physiological and pathological wound healing. Stem Cells 30, 1436–1446 [DOI] [PubMed] [Google Scholar]
- 41.Teng H., Chopp M., Hozeska-Solgot A., Shen L., Lu M., Tang C., and Zhang Z.G. (2012). Tissue plasminogen activator and plasminogen activator inhibitor 1 contribute to sonic hedgehog-induced in vitro cerebral angiogenesis. PLoS One 7, e33444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harris L.G., Pannell L.K., Singh S., Samant R.S., and Shevde L.A. (2012). Increased vascularity and spontaneous metastasis of breast cancer by hedgehog signaling mediated upregulation of cyr61. Oncogene 31, 3370–3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hilfiker-Kleiner D., Kaminski K., Kaminska A., Fuchs M., Klein G., Podewski E., Grote K., Kiian I., Wollert K.C., Hilfiker A., and Drexler H. (2004). Regulation of proangiogenic factor CCN1 in cardiac muscle: impact of ischemia, pressure overload, and neurohumoral activation. Circulation 109, 2227–2233 [DOI] [PubMed] [Google Scholar]
- 44.Mo F.E., Muntean A.G., Chen C.C., Stolz D.B., Watkins S.C., and Lau L.F. (2002). CYR61 (CCN1) is essential for placental development and vascular integrity. Mol Cell Biol 22, 8709–8720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rapraeger A.C., Ell B.J., Roy M., Li X., Morrison O.R., Thomas G.M., and Beauvais D.M. (2013). Vascular endothelial-cadherin stimulates syndecan-1-coupled insulin-like growth factor-1 receptor and cross-talk between alphaVbeta3 integrin and vascular endothelial growth factor receptor 2 at the onset of endothelial cell dissemination during angiogenesis. FEBS J. 280, 2194–2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yuan K., Hong T.M., Chen J.J., Tsai W.H., and Lin M.T. (2004). Syndecan-1 up-regulated by ephrinB2/EphB4 plays dual roles in inflammatory angiogenesis. Blood 104, 1025–1033 [DOI] [PubMed] [Google Scholar]
- 47.Zhu Y., Lee C., Shen F., Du R., Young W.L., and Yang G.Y. (2005). Angiopoietin-2 facilitates vascular endothelial growth factor-induced angiogenesis in the mature mouse brain. Stroke 36, 1533–1537 [DOI] [PubMed] [Google Scholar]
- 48.Mochizuki Y., Nakamura T., Kanetake H., and Kanda S. (2002). Angiopoietin 2 stimulates migration and tube-like structure formation of murine brain capillary endothelial cells through c-Fes and c-Fyn. J. Cell Sci. 115, 175–183 [DOI] [PubMed] [Google Scholar]
- 49.Vates G.E., Hashimoto T., Young W.L., and Lawton M.T. (2005). Angiogenesis in the brain during development: the effects of vascular endothelial growth factor and angiopoietin-2 in an animal model. J. Neurosurg. 103, 136–145 [DOI] [PubMed] [Google Scholar]
- 50.Zagzag D., Hooper A., Friedlander D.R., Chan W., Holash J., Wiegand S.J., Yancopoulos G. D., and Grumet M. (1999). In situ expression of angiopoietins in astrocytomas identifies angiopoietin-2 as an early marker of tumor angiogenesis. Exp. Neurol. 159, 391–400 [DOI] [PubMed] [Google Scholar]
- 51.Rivera C.G., Bader J.S., and Popel A.S. (2011). Angiogenesis-associated crosstalk between collagens, CXC chemokines, and thrombospondin domain-containing proteins. Ann. Biomed. Eng. 39, 2213–2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Duparc T., Colom A., Cani P.D., Massaly N., Rastrelli S., Drougard A., Le Gonidec S., Mouledous L., Frances B., Leclercq I., Llorens-Cortes C., Pospisilik J.A., Delzenne N.M., Valet P., Castan-Laurell I., and Knauf C. (2011). Central apelin controls glucose homeostasis via a nitric oxide-dependent pathway in mice. Antioxid. Redox Signal. 15, 1477–1496 [DOI] [PubMed] [Google Scholar]
- 53.Yue P., Jin H., Aillaud M., Deng A.C., Azuma J., Asagami T., Kundu R.K., Reaven G.M., Quertermous T., and Tsao P.S. (2010). Apelin is necessary for the maintenance of insulin sensitivity. Am. J. Physiol. Endocrinol. Metab. 298, E59–E67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sawane M., Kidoya H., Muramatsu F., Takakura N., and Kajiya K. (2011). Apelin attenuates UVB-induced edema and inflammation by promoting vessel function. Am. J. Pathol. 179, 2691–2697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kidoya H., Naito H., and Takakura N. (2010). Apelin induces enlarged and nonleaky blood vessels for functional recovery from ischemia. Blood 115, 3166–3174 [DOI] [PubMed] [Google Scholar]
- 56.Eyries M., Siegfried G., Ciumas M., Montagne K., Agrapart M., Lebrin F., and Soubrier F. (2008). Hypoxia-induced apelin expression regulates endothelial cell proliferation and regenerative angiogenesis. Circ. Res. 103, 432–440 [DOI] [PubMed] [Google Scholar]
- 57.Cox C.M., D'Agostino S.L., Miller M.K., Heimark R.L., and Krieg P.A. (2006). Apelin, the ligand for the endothelial G-protein-coupled receptor, APJ, is a potent angiogenic factor required for normal vascular development of the frog embryo. Dev. Biol. 296, 177–189 [DOI] [PubMed] [Google Scholar]
- 58.Sonmez A., Celebi G., Erdem G., Tapan S., Genc H., Tasci I., Ercin C.N., Dogru T., Kilic S., Uckaya G., Yilmaz M.I., Erbil M.K., and Kutlu M. (2010). Plasma apelin and ADMA Levels in patients with essential hypertension. Clin. Exp. Hypertens. 32, 179–183 [DOI] [PubMed] [Google Scholar]
- 59.Cook D.R., Gleichman A.J., Cross S.A., Doshi S., Ho W., Jordan-Sciutto K.L., Lynch D.R., and Kolson D.L. (2011). NMDA receptor modulation by the neuropeptide apelin: implications for excitotoxic injury. J. Neurochem. 118, 1113–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zeng X.J., Yu S.P., Zhang L., and Wei L. (2010). Neuroprotective effect of the endogenous neural peptide apelin in cultured mouse cortical neurons. Exp. Cell Res. 316, 1773–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang G., Qian P., Jackson F.R., Qian G., and Wu G. (2008). Sequential activation of JAKs, STATs and xanthine dehydrogenase/oxidase by hypoxia in lung microvascular endothelial cells. Int. J. Biochem. Cell Biol. 40, 461–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schroder K., Vecchione C., Jung O., Schreiber J. G., Shiri-Sverdlov R., van Gorp P.J., Busse R., and Brandes R.P. (2006). Xanthine oxidase inhibitor tungsten prevents the development of atherosclerosis in ApoE knockout mice fed a Western-type diet. Free Radic. Biol. Med. 41, 1353–1360 [DOI] [PubMed] [Google Scholar]
- 63.Meneshian A., and Bulkley G.B. (2002). The physiology of endothelial xanthine oxidase: from urate catabolism to reperfusion injury to inflammatory signal transduction. Microcirculation 9, 161–175 [DOI] [PubMed] [Google Scholar]
- 64.Yokoyama Y., Beckman J.S., Beckman T.K., Wheat J.K., Cash T.G., Freeman B.A., and Parks D.A. (1990). Circulating xanthine oxidase: potential mediator of ischemic injury. Am. J. Physiol. 258, G564–G570 [DOI] [PubMed] [Google Scholar]
- 65.Cantu-Medellin N., and Kelley E.E. (2013). Xanthine oxidoreductase-catalyzed reactive species generation: A process in critical need of reevaluation. Redox Biol. 1, 353–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ogawa C., Kihara A., Gokoh M., and Igarashi Y. (2003). Identification and characterization of a novel human sphingosine-1-phosphate phosphohydrolase, hSPP2. J. Biol. Chem. 278, 1268–1272 [DOI] [PubMed] [Google Scholar]
- 67.Leali D., Inforzato A., Ronca R., Bianchi R., Belleri M., Coltrini D., Di Salle E., Sironi M., Norata G.D., Bottazzi B., Garlanda C., Day A.J., and Presta M. (2012). Long pentraxin 3/tumor necrosis factor-stimulated gene-6 interaction: a biological rheostat for fibroblast growth factor 2-mediated angiogenesis. Arterioscler. Thromb. Vasc. Biol. 32, 696–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Doni A., Mantovani G., Porta C., Tuckermann J., Reichardt H.M., Kleiman A., Sironi M., Rubino L., Pasqualini F., Nebuloni M., Signorini S., Peri G., Sica A., Beck-Peccoz P., Bottazzi B., and Mantovani A. (2008). Cell-specific regulation of PTX3 by glucocorticoid hormones in hematopoietic and nonhematopoietic cells. J. Biol. Chem. 283, 29983–29992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Norata G.D., Marchesi P., Pulakazhi Venu V.K., Pasqualini F., Anselmo A., Moalli F., Pizzitola I., Garlanda C., Mantovani A., and Catapano A.L. (2009). Deficiency of the long pentraxin PTX3 promotes vascular inflammation and atherosclerosis. Circulation 120, 699–708 [DOI] [PubMed] [Google Scholar]
- 70.Ravizza T., Moneta D., Bottazzi B., Peri G., Garlanda C., Hirsch E., Richards G.J., Mantovani A., and Vezzani A. (2001). Dynamic induction of the long pentraxin PTX3 in the CNS after limbic seizures: evidence for a protective role in seizure-induced neurodegeneration. Neuroscience 105, 43–53 [DOI] [PubMed] [Google Scholar]
- 71.Fujio Y., Maeda M., Mohri T., Obana M., Iwakura T., Hayama A., Yamashita T., Nakayama H., and Azuma J. (2011). Glycoprotein 130 cytokine signal as a therapeutic target against cardiovascular diseases. J. Pharmacol. Sci. 117, 213–222 [DOI] [PubMed] [Google Scholar]
- 72.Cakir M., Altunbas H., and Karayalcin U. (2003). Hyperglycemia: an independent marker of in-hospital mortality in patients with undiagnosed diabetes. J. Clin. Endocrinol. Metab. 88, 1402; author reply, 1402 [DOI] [PubMed] [Google Scholar]
- 73.Mahboubi K., Biedermann B.C., Carroll J.M., and Pober J.S. (2000). IL-11 activates human endothelial cells to resist immune-mediated injury. J. Immunol. 164, 3837–3846 [DOI] [PubMed] [Google Scholar]
- 74.Thier M., Hall M., Heath J.K., Pennica D., and Weis J. (1999). Trophic effects of cardiotrophin-1 and interleukin-11 on rat dorsal root ganglion neurons in vitro. Brain Res. Mol. Brain Res. 64, 80–84 [DOI] [PubMed] [Google Scholar]
- 75.Kirkiles-Smith N.C., Mahboubi K., Plescia J., McNiff J.M., Karras J., Schechner J.S., Altieri D.C., and Pober J.S. (2004). IL-11 protects human microvascular endothelium from alloinjury in vivo by induction of survivin expression. J. Immunol. 172, 1391–1396 [DOI] [PubMed] [Google Scholar]
- 76.Denis C.V., Kwack K., Saffaripour S., Maganti S., Andre P., Schaub R.G., and Wagner D.D. (2001). Interleukin 11 significantly increases plasma von Willebrand factor and factor VIII in wild type and von Willebrand disease mouse models. Blood 97, 465–472 [DOI] [PubMed] [Google Scholar]
- 77.Opal S.M., and DePalo V.A. (2000). Anti-inflammatory cytokines. Chest 117, 1162–1172 [DOI] [PubMed] [Google Scholar]
- 78.Fang H.Y., Ko W.J., and Lin C.Y. (2005). Plasma interleukin 11 levels correlate with outcome of spontaneous intracerebral hemorrhage. Surg. Neurol. 64, 511–517; discussion, 517–518 [DOI] [PubMed] [Google Scholar]
- 79.Deverman B.E., and Patterson P.H. (2012). Exogenous leukemia inhibitory factor stimulates oligodendrocyte progenitor cell proliferation and enhances hippocampal remyelination. J. Neurosci. 32, 2100–2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bauer S., and Patterson P.H. (2006). Leukemia inhibitory factor promotes neural stem cell self-renewal in the adult brain. J. Neurosci. 26, 12089–12099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mi H., Haeberle H., and Barres B.A. (2001). Induction of astrocyte differentiation by endothelial cells. J. Neurosci. 21, 1538–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jones K.L., de Kretser D.M., Patella S., and Phillips D.J. (2004). Activin A and follistatin in systemic inflammation. Mol. Cell. Endocrinol. 225, 119–125 [DOI] [PubMed] [Google Scholar]
- 83.Hubner G., Alzheimer C., and Werner S. (1999). Activin: a novel player in tissue repair processes. Histol. Histopathol. 14, 295–304 [DOI] [PubMed] [Google Scholar]
- 84.Kaneda H., Arao T., Matsumoto K., De Velasco M.A., Tamura D., Aomatsu K., Kudo K., Sakai K., Nagai T., Fujita Y., Tanaka K., Yanagihara K., Yamada Y., Okamoto I., Nakagawa K., and Nishio K. (2011). Activin A inhibits vascular endothelial cell growth and suppresses tumour angiogenesis in gastric cancer. Br. J. Cancer 105, 1210–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tretter Y.P., Hertel M., Munz B., ten Bruggencate G., Werner S., and Alzheimer C. (2000). Induction of activin A is essential for the neuroprotective action of basic fibroblast growth factor in vivo. Nat. Med. 6, 812–815 [DOI] [PubMed] [Google Scholar]
- 86.Man S., Tucky B., Cotleur A., Drazba J., Takeshita Y., and Ransohoff R.M. (2012). CXCL12-induced monocyte-endothelial interactions promote lymphocyte transmigration across an in vitro blood-brain barrier. Sci. Transl. Med. 4, 119ra114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu K.K., and Dorovini-Zis K. (2009). Regulation of CXCL12 and CXCR4 expression by human brain endothelial cells and their role in CD4+ and CD8+ T cell adhesion and transendothelial migration. J. Neuroimmunol. 215, 49–64 [DOI] [PubMed] [Google Scholar]
- 88.Cook-Mills J.M., Marchese M.E., and Abdala-Valencia H. (2011). Vascular cell adhesion molecule-1 expression and signaling during disease: regulation by reactive oxygen species and antioxidants. Antioxid. Redox Signal. 15, 1607–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Woodcock T., and Morganti-Kossmann M.C. (2013). The role of markers of inflammation in traumatic brain injury. Front. Neurol. 4, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kumar A., and Loane D. J. (2012). Neuroinflammation after traumatic brain injury: opportunities for therapeutic intervention. Brain Behav. Immun. 26, 1191–1201 [DOI] [PubMed] [Google Scholar]
- 91.Bangsow T., Baumann E., Bangsow C., Jaeger M.H., Pelzer B., Gruhn P., Wolf S., von Melchner H., and Stanimirovic D.B. (2008). The epithelial membrane protein 1 is a novel tight junction protein of the blood-brain barrier. J. Cereb. Blood Flow Metab. 28, 1249–1260 [DOI] [PubMed] [Google Scholar]
- 92.Kis B., Abraham C.S., Deli M.A., Kobayashi H., Niwa M., Yamashita H., Busija D.W., and Ueta Y. (2003). Adrenomedullin, an autocrine mediator of blood-brain barrier function. Hypertens. Res. 26,Suppl., S61–S70 [DOI] [PubMed] [Google Scholar]
- 93.Kato J., Tsuruda T., Kita T., Kitamura K., and Eto T. (2005). Adrenomedullin: a protective factor for blood vessels. Arterioscler. Thromb. Vasc. Biol. 25, 2480–2487 [DOI] [PubMed] [Google Scholar]
- 94.Ribatti D., Nico B., Spinazzi R., Vacca A., and Nussdorfer G.G. (2005). The role of adrenomedullin in angiogenesis. Peptides 26, 1670–1675 [DOI] [PubMed] [Google Scholar]
- 95.Temmesfeld-Wollbruck B., Hocke A.C., Suttorp N., and Hippenstiel S. (2007). Adrenomedullin and endothelial barrier function. Thromb. Haemost. 98, 944–951 [DOI] [PubMed] [Google Scholar]
- 96.Samson W.K. (1999). Adrenomedullin and the control of fluid and electrolyte homeostasis. Annu. Rev. Physiol. 61, 363–389 [DOI] [PubMed] [Google Scholar]
- 97.Risso F.M., Sannia A., Gavilanes D.A., Vles H.J., Colivicchi M., Ricotti A., Li Volti, G., and Gazzolo D. (2012). Biomarkers of brain damage in preterm infants. J. Matern. Fetal Neonatal Med. 25, Suppl. 4, 101–104 [DOI] [PubMed] [Google Scholar]
- 98.Cheyuo C., Yang W.L., and Wang P. (2012). The critical role of adrenomedullin and its binding protein, AMBP-1, in neuroprotection. Biol. Chem. 393, 429–439 [DOI] [PubMed] [Google Scholar]
- 99.Minami M., Yokokawa K., Kohno M., Yasunari K., and Yoshikawa J. (1998). Suppression of endothelin-3-induced nitric oxide synthesis by triglyceride in human endothelial cells. J. Cardiovasc. Pharmacol. 31,Suppl. 1, S467–S469 [DOI] [PubMed] [Google Scholar]
- 100.Filipovich T., and Fleisher-Berkovich S. (2008). Regulation of glial inflammatory mediators synthesis: possible role of endothelins. Peptides 29, 2250–2256 [DOI] [PubMed] [Google Scholar]
- 101.Rozyczka J., Figiel M., and Engele J. (2004). Endothelins negatively regulate glial glutamate transporter expression. Brain Pathol. 14, 406–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bek E.L., and McMillen M.A. (2000). Endothelins are angiogenic. J. Cardiovasc. Pharmacol. 36, S135–S139 [DOI] [PubMed] [Google Scholar]
- 103.Hiyama T.Y., Yoshida M., Matsumoto M., Suzuki R., Matsuda T., Watanabe E., and Noda M. (2013). Endothelin-3 expression in the subfornical organ enhances the sensitivity of Na(x), the brain sodium-level sensor, to suppress salt intake. Cell Metab. 17, 507–519 [DOI] [PubMed] [Google Scholar]
- 104.Samson W.K., Skala K., Huang F.L., Gluntz S., Alexander B., and Gomez-Sanchez C.E. (1991). Central nervous system action of endothelin-3 to inhibit water drinking in the rat. Brain Res. 539, 347–351 [DOI] [PubMed] [Google Scholar]
- 105.Lozano D., Gonzales-Portillo G.S., Acosta S., de la Pena I., Tajiri N., Kaneko Y., and Borlongan C.V. (2015). Neuroinflammatory responses to traumatic brain injury: etiology, clinical consequences, and therapeutic opportunities. Neuropsychiatr. Dis. Treat. 11, 97–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bigler E.D. (2013). Neuroinflammation and the dynamic lesion in traumatic brain injury. Brain 136, 9–11 [DOI] [PubMed] [Google Scholar]
- 107.Dai W., Cheng H.L., Huang R.Q., Zhuang Z., and Shi J.X. (2009). Quantitative detection of the expression of mitochondrial cytochrome c oxidase subunits mRNA in the cerebral cortex after experimental traumatic brain injury. Brain Res. 1251, 287–295 [DOI] [PubMed] [Google Scholar]
- 108.Davidson S.M., and Duchen M.R. (2007). Endothelial mitochondria: contributing to vascular function and disease. Circ. Res. 100, 1128–1141 [DOI] [PubMed] [Google Scholar]
- 109.Faden A.I., and Loane D.J. (2015). Chronic neurodegeneration after traumatic brain injury: Alzheimer disease, chronic traumatic encephalopathy, or persistent neuroinflammation? Neurotherapeutics 12, 143–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Breunig J.J., Guillot-Sestier M.V., and Town T. (2013). Brain injury, neuroinflammation and Alzheimer's disease. Front. Aging Neurosci. 5, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Guo S., Zhou Y., Xing C., Lok J., Som A.T., Ning M., Ji X., and Lo E.H. (2012). The vasculome of the mouse brain. PLoS One 7, e52665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Flynn K.M., Michaud M., Canosa S., and Madri J.A. (2013). CD44 regulates vascular endothelial barrier integrity via a PECAM-1 dependent mechanism. Angiogenesis 16, 689–705 [DOI] [PubMed] [Google Scholar]
- 113.Lennon F.E., and Singleton P.A. (2011). Hyaluronan regulation of vascular integrity. Am. J. Cardiovasc. Dis. 1, 200–213 [PMC free article] [PubMed] [Google Scholar]
- 114.Ponta H., Sherman L., and Herrlich P.A. (2003). CD44: from adhesion molecules to signalling regulators. Nat. Rev. Mol. Cell Biol. 4, 33–45 [DOI] [PubMed] [Google Scholar]
- 115.Wang H., Zhan Y., Xu L., Feuerstein G.Z., and Wang X. (2001). Use of suppression subtractive hybridization for differential gene expression in stroke: discovery of CD44 gene expression and localization in permanent focal stroke in rats. Stroke 32, 1020–1027 [DOI] [PubMed] [Google Scholar]
- 116.Wang X., Xu L., Wang H., Zhan Y., Pure E., and Feuerstein G.Z. (2002). CD44 deficiency in mice protects brain from cerebral ischemia injury. J. Neurochem. 83, 1172–1179 [DOI] [PubMed] [Google Scholar]
- 117.Carow B., and Rottenberg M.E. (2014). SOCS3, a major regulator of infection and inflammation. Front. Immunol. 5, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.White G.E., Cotterill A., Addley M.R., Soilleux E.J., and Greaves D.R. (2011). Suppressor of cytokine signalling protein SOCS3 expression is increased at sites of acute and chronic inflammation. J. Mol. Histol. 42, 137–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Baker B.J., Akhtar L.N., and Benveniste E.N. (2009). SOCS1 and SOCS3 in the control of CNS immunity. Trends Immunol. 30, 392–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yin Y., Liu W., Ji G., and Dai Y. (2013). The essential role of p38 MAPK in mediating the interplay of oxLDL and IL-10 in regulating endothelial cell apoptosis. Eur. J. Cell Biol. 92, 150–159 [DOI] [PubMed] [Google Scholar]
- 121.Townsend S.F., Dallman M.F., and Miller W.L. (1990). Rat insulin-like growth factor-I and -II mRNAs are unchanged during compensatory adrenal growth but decrease during ACTH-induced adrenal growth. J. Biol. Chem. 265, 22117–22122 [PubMed] [Google Scholar]
- 122.Stahl A., Joyal J.S., Chen J., Sapieha P., Juan A.M., Hatton C.J., Pei D.T., Hurst C.G., Seaward M.R., Krah N.M., Dennison R.J., Greene E.R., Boscolo E., Panigrahy D., and Smith L.E. (2012). SOCS3 is an endogenous inhibitor of pathologic angiogenesis. Blood 120, 2925–2929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Stratton C.W., Weeks L.S., and Tausk F. (1987). Beta-lactamase induction and aminoglycoside susceptibility in Pseudomonas aeruginosa. J. Antimicrob. Chemother. 19, 21–25 [DOI] [PubMed] [Google Scholar]
- 124.Sands W.A., Woolson H.D., Milne G.R., Rutherford C., and Palmer T.M. (2006). Exchange protein activated by cyclic AMP (Epac)-mediated induction of suppressor of cytokine signaling 3 (SOCS-3) in vascular endothelial cells. Mol. Cell. Biol. 26, 6333–6346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wong P.K., Egan P.J., Croker B.A., O'Donnell K., Sims N.A., Drake S., Kiu H., McManus E.J., Alexander W.S., Roberts A.W., and Wicks I.P. (2006). SOCS-3 negatively regulates innate and adaptive immune mechanisms in acute IL-1-dependent inflammatory arthritis. J. Clin. Invest. 116, 1571–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tang C., Zelenak C., Volkl J., Eichenmuller M., Regel I., Frohlich H., Kempe D., Jimenez L., Le Bellego L., Vergne S., and Lang F. (2011). Hydration-sensitive gene expression in brain. Cell. Physiol. Biochem. 27, 757–768 [DOI] [PubMed] [Google Scholar]
- 127.Xu L.L., Shi Y., Petrovics G., Sun C., Makarem M., Zhang W., Sesterhenn I.A., McLeod D.G., Sun L., Moul J.W., and Srivastava S. (2003). PMEPA1, an androgen-regulated NEDD4-binding protein, exhibits cell growth inhibitory function and decreased expression during prostate cancer progression. Cancer Res. 63, 4299–4304 [PubMed] [Google Scholar]
- 128.Watanabe Y., Itoh S., Goto T., Ohnishi E., Inamitsu M., Itoh F., Satoh K., Wiercinska E., Yang W., Shi L., Tanaka A., Nakano N., Mommaas A.M., Shibuya H., Ten Dijke, P., and Kato M. (2010). TMEPAI, a transmembrane TGF-beta-inducible protein, sequesters Smad proteins from active participation in TGF-beta signaling. Mol. Cell. 37, 123–134 [DOI] [PubMed] [Google Scholar]
- 129.Greenlee M.C., Sullivan S.A., and Bohlson S.S. (2008). CD93 and related family members: their role in innate immunity. Curr. Drug Targets 9, 130–138 [DOI] [PubMed] [Google Scholar]
- 130.Harhausen D., Prinz V., Ziegler G., Gertz K., Endres M., Lehrach H., Gasque P., Botto M., Stahel P.F., Dirnagl U., Nietfeld W., and Trendelenburg G. (2010). CD93/AA4.1: a novel regulator of inflammation in murine focal cerebral ischemia. J. Immunol. 184, 6407–6417 [DOI] [PubMed] [Google Scholar]
- 131.Kao Y.C., Jiang S.J., Pan W.A., Wang K.C., Chen P.K., Wei H.J., Chen W.S., Chang B.I., Shi G.Y., and Wu H.L. (2012). The epidermal growth factor-like domain of CD93 is a potent angiogenic factor. PLoS One 7, e51647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Greenlee M.C., Sullivan S.A., and Bohlson S.S. (2009). Detection and characterization of soluble CD93 released during inflammation. Inflamm. Res. 58, 909–919 [DOI] [PubMed] [Google Scholar]
- 133.Malarstig A., Silveira A., Wagsater D., Ohrvik J., Backlund A., Samnegard A., Khademi M., Hellenius M.L., Leander K., Olsson T., Uhlen M., de Faire U., Eriksson P., and Hamsten A. (2011). Plasma CD93 concentration is a potential novel biomarker for coronary artery disease. J. Intern. Med. 270, 229–236 [DOI] [PubMed] [Google Scholar]
- 134.von Gertten C., Flores Morales A., Holmin S., Mathiesen T., and Nordqvist A.C. (2005). Genomic responses in rat cerebral cortex after traumatic brain injury. BMC Neurosci. 6, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Natale J.E., Ahmed F., Cernak I., Stoica B., and Faden A.I. (2003). Gene expression profile changes are commonly modulated across models and species after traumatic brain injury. J. Neurotrauma 20, 907–927 [DOI] [PubMed] [Google Scholar]
- 136.Truettner J., Schmidt-Kastner R., Busto R., Alonso O.F., Loor J.Y., Dietrich W.D., and Ginsberg M.D. (1999). Expression of brain-derived neurotrophic factor, nerve growth factor, and heat shock protein HSP70 following fluid percussion brain injury in rats. J. Neurotrauma 16, 471–486 [DOI] [PubMed] [Google Scholar]
- 137.Sharp D.J., Scott G., and Leech R. (2014). Network dysfunction after traumatic brain injury. Nat. Rev. Neurol. 10, 156–166 [DOI] [PubMed] [Google Scholar]
- 138.Smith D.H., Meaney D. F., and Shull W. H. (2003). Diffuse axonal injury in head trauma. J. Head Trauma Rehabil. 18, 307–316 [DOI] [PubMed] [Google Scholar]
- 139.Werner C., and Engelhard K. (2007). Pathophysiology of traumatic brain injury. Br. J. Anaesth. 99, 4–9 [DOI] [PubMed] [Google Scholar]
- 140.Tsuru M., Soejima T., Shiba N., Kimura K., Sato K., Toyama Y., and Nagata K. (2013). Proline/arginine-rich end leucine-rich repeat protein converts stem cells to ligament tissue and Zn(II) influences its nuclear expression. Stem Cells Dev. 22, 2057–2070 [DOI] [PubMed] [Google Scholar]
- 141.Bi Y., Ehirchiou D., Kilts T.M., Inkson C.A., Embree M.C., Sonoyama W., Li L., Leet A.I., Seo B.M., Zhang L., Shi S., and Young M.F. (2007). Identification of tendon stem/progenitor cells and the role of the extracellular matrix in their niche. Nat. Med. 13, 1219–1227 [DOI] [PubMed] [Google Scholar]
- 142.Garcion E., Halilagic A., Faissner A., and ffrench-Constant C. (2004). Generation of an environmental niche for neural stem cell development by the extracellular matrix molecule tenascin C. Development 131, 3423–3432 [DOI] [PubMed] [Google Scholar]
- 143.Zagzag D., Friedlander D.R., Dosik J., Chikramane S., Chan W., Greco M.A., Allen J.C., Dorovini-Zis K., and Grumet M. (1996). Tenascin-C expression by angiogenic vessels in human astrocytomas and by human brain endothelial cells in vitro. Cancer Res. 56, 182–189 [PubMed] [Google Scholar]
- 144.Heuckeroth R.O. (2003). Finding your way to the end: a tale of GDNF and endothelin-3. Neuron 40, 871–873 [DOI] [PubMed] [Google Scholar]
- 145.Li J.J., Bickel P.J., and Biggin M.D. (2014). System wide analyses have underestimated protein abundances and the importance of transcription in mammals. PeerJ 2, e270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Li J.J., and Biggin M.D. (2015). Gene expression. Statistics requantitates the central dogma. Science 347, 1066–1067 [DOI] [PubMed] [Google Scholar]
- 147.Schwanhausser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., and Selbach M. (2011). Global quantification of mammalian gene expression control. Nature 473, 337–342 [DOI] [PubMed] [Google Scholar]
- 148.Lo E.H., Lok J., Ning M., and Whalen M.J. (eds). (2014). Vascular Mechanisms in CNS Trauma. Springer series in translational stroke research 5. Springer Science+Business Media: New York [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.