Abstract
Formalin-fixed paraffin-embedded (FFPE) tissue is the predominant preparation for diagnostic histopathological evaluation and increasingly the biospecimen on which molecular diagnostics are performed. However, formalin is carcinogenic and results in cross-linking of proteins and nicking and alterations of nucleic acids. Alternative fixatives, including 70% ethanol, improved biomolecular integrity; however, they have yet to replace neutral-buffered formalin (NBF). Herein, we describe the phosphate-buffered ethanol 70% (BE70) fixative. The histomorphology of BE70-fixed tissue is very similar to that of NBF; however, it is a non-cross-linking fixative and lacks the carcinogenic profile of formaldehyde-based fixatives. RNA isolated from tissue fixed in BE70 was of substantially higher quality and quantity than that was recovered from formalin-fixed tissue. Furthermore, the BE70 fixative showed excellent RNA and DNA integrity compared with that of NBF fixative based on real-time polymerase chain reaction analysis results. Immunohistochemical staining was similar for the antigen tested. In conclusion, BE70 is a non-cross-linking fixative that is superior to NBF and 70% ethanol with reference to biomolecule recovery and quality from paraffin-embedded tissue. Additional studies to compare the histomorphologic and immunohistochemical performance and utility in a clinical setting are required.
Keywords: alcohol, fixation, formalin, histomorphology, paraffin embedded, real-time RT-PCR, RNA integrity, tissue
Introduction
Fixatives are solutions designed to prevent autolysis and degradation of the tissues, render the tissue aseptic, and preserve morphology and cellular details. The 10% neutral-buffered formalin (NBF, 3.7%–4.0% formaldehyde solution in phosphate buffer) has been the gold standard in histopathological studies for decades. NBF is easy to use, inexpensive, and penetrates tissue relatively fast.1 Formalin fixation is excellent for reproducing histomorphology, and biospecimens are relatively stable after paraffin embedding,2 which enables long-term retrospective studies. However, NBF fixation results in random cross-linking of proteins, and fragmented and chemical modifications of nucleic acids,3,4 resulting in unreliable and irreproducible detection of biomolecules in diagnostic assays. Furthermore, formaldehyde is toxic, recently reclassified by the U.S. Environmental Protection Agency from a potential carcinogen to a carcinogen.5,6
The literature is rich with alternative fixatives to address the shortcomings of NBF.7–14 Among alternative fixatives, alcohol-based fixatives are considered the most promising for molecular pathology, as they do not cross-link proteins.7 They also emit no toxic vapors, produce similar histopathological results, and result in less damage to nucleic acids and less antigen retrieval for immunohistochemistry. However, the disadvantages of ethanol fixatives included tissue shrinkage and hardening, lysis of erythrocytes, and flammability.8,15 To overcome these issues, some studies have proposed specific additives to ethanol fixatives3,7,8,16–19 but none of these ethanol-based fixatives have met widespread application. Ideally, it is more preferable to have a nontoxic fixative that allows for morphological analysis, high-quality special histochemical and immunohistochemical staining, and well-preserved macromolecules, and is also economical.8 These economic factors include not only the cost of the reagents but the safety measures required for its use, as well as disposal cost.
In the present study, we evaluated different additives (phosphate-buffered solutions, glycerol, and glacial acetic acid) in a base fixative of 70% ethanol. We evaluated histomorphology, protein, DNA, and RNA quantity and quality compared with those of NBF. By this approach, we can define the contribution of the individual components of the fixative to the performance of the final formulation of the combined reagent. The final formulation is buffered ethanol 70% (BE70), which has many of the properties of formaldehyde-based fixatives, but is “molecularly friendly.” This fixative has good cost, safety, ease of use, and basic congruence with current pathology practice.
Materials and Methods
Fixatives
We used six different fixatives, including 70% ethanol (E), 70% ethanol + 0.5× phosphate-buffered saline (PBS; 1.7 mM KH2PO4, 0.5 mM Na2HPO4, 150 mM NaCl, pH 7.4) (EP), 70% ethanol + 1% glycerol + 0.5× PBS (EGP), 70% ethanol + 0.5% glacial acetic acid + 0.5× PBS (EAP), 70% ethanol + 1% glycerol + 0.5% glacial acetic acid + 0.5× PBS (EGAP; BE70), and 10% NBF. Concentrations of glycerol, glacial acetic acid, and PBS were varied (data not shown). The final fixative formulation was adjusted by volume. Glycerol and glacial acetic acid were purchased from Sigma-Aldrich (St. Louis, MO). Ethanol and NBF were purchased from VWR (Radnor, PA). All fixatives were stored and used at room temperature.
Tissue Samples and Fixation
Mice were acquired from the Small Animals Section, Veterinary Resources Branch, National Institutes of Health (NIH). The animals were housed and euthanized in accordance with NIH guidelines for care and use of laboratory animals.20 The ischemic time period was <5 min. Mouse liver, kidney, pancreas, spleen, small intestine, heart, lung, brain, and skin samples were fixed for 24 hr in 10 ml of different fixatives (exceeding a 1:10 tissue fixative volume). To examine the effects of the BE70 fixative, we compared it with 10% NBF on antigen degradation and molecular quality, respectively. Tissues were then processed using an enclosed automated processor (Tissue-Tek VIP IV; Sakura Finetek, Inc., Torrance, CA) with a 30 min/station protocol.21 In short, the tissues were dehydrated through an ethanol series, cleared with xylene, and infiltrated with molten paraffin. The tissues were then embedded in paraffin and sectioned for histological and molecular evaluation.
Immunohistochemical and Histochemical Evaluation
Hematoxylin (modified Harris Hematoxylin; Thermo Scientific, Waltham, MA) and eosin (Eosin-Y; Thermo Scientific) staining was performed for each tissue sample to examine histomorphologic features. Immunohistochemical staining was performed on 5-µm-thick sections. The tissue sections were deparaffinized with xylene and dehydrated through a graded ethanol series. Endogenous peroxidase activity was quenched with 3% H2O2 in water for 10 min. Non-specific staining was minimized with a protein block (Dako, Carpinteria, CA) for 15 min. The sections were subject to antigen retrieval, as needed, and incubated with primary antibodies, a detailed list of which, along with dilutions, is provided in Table 1. The antigen–antibody reaction was detected with Dako EnVision+ Dual Link System-HRP (Dako) and DAB+ (3,3′-diaminobenzidine; Dako). Negative controls including immunoglobulin G (IgG) and omission of the primary antibody were performed. Positive controls included mouse spleen, kidney, and embryo for CD31, aquaporin 1 (AQP1), and Ki-67 antibodies, respectively. Slides were counterstained with hematoxylin, dehydrated in a graded ethanol series, cleared in xylene, and coverslipped. Photomicrographs were generated on a Zeiss Axioplan 2ie outfitted with an Axicam HRc camera, running under Axiovision software (version 4.5; Zeiss, Thornwood, NY).
Table 1.
Antibodies Used for Immunohistochemistry.
| Antibody | Vendor | Clonality | Catalog No. | Incubation | Dilution | Antigen Retrieval |
|---|---|---|---|---|---|---|
| CD31 | Abcam | Poly rabbit | ab28364 | 30 min at RT | 1/150 | Not applied |
| AQP1 | Santa Cruz Biotechnology | Poly rabbit | Sc-20810 | 30 min at RT | 1/250 | Not applied |
| Ki-67 | Sigma-Aldrich | Mono rabbit (SP6) | 275R | 30 min at RT | Ready to use | 20 min, pressure cooker, pH 6 |
Abbreviations: RT, room temperature; AQP1, aquaporin 1.
Protein Extraction and Western Blotting
Protein was extracted from two 10-µm formalin-fixed paraffin-embedded (FFPE) tissue sections as described previously.22 Briefly, the sections were trimmed of excess wax and homogenized using a Disposable Pellet Mixer in 200 µl protein extraction solution (1× high-pH antigen retrieval buffer [pH 9.9; Dako], 1% NaN3, 1% sodium dodecyl sulfate [SDS], 10% glycerol and protease inhibitor [1 tablet/25 ml; Roche Science, Manheim, Germany]), followed by 15-min incubation at 115C in a pressure cooker (Dako). After incubation, the tissue lysates were centrifuged at 13,000 rpm for 30 min at 4C. The supernatants were collected and stored at −20C. Total protein concentrations were measured with the bicinchoninic acid (BCA) Protein Assay Kit (Pierce Biotechnology, Rockford, IL).
Proteins (10 µg) extracted from the different fixative solutions were resolved by 4% to 12% NuPAGE® Novex Bis-Tris polyacrylamide gel electrophoresis (PAGE), and electroblotted to a nitrocellulose membrane using the iBlot™ Dry Blotting System (Invitrogen, Carlsbad, CA). The membranes were blocked with 5% nonfat dry milk in Tris-buffered saline with Tween (TBST; 50 mM Tris, pH 7.5, 150 mM NaCl, and 0.05% Tween-20) for 1 hr, washed, and incubated overnight at 4C in TBST with rabbit anti-AQP1 polyclonal antibody (Cat. No. sc-20810; dilution 1:100; Santa Cruz Biotechnology, Dallas, TA) and mouse anti-glyceraldehyde 3-phosphate dehydrogenase (GAPDH) monoclonal antibody (clone 6C5; dilution 1:3000; Calbiochem, Gibbstown, NJ). Specific molecules were detected with horseradish peroxidase–labeled anti-mouse secondary antibodies (Chemicon International, Temecula, CA) and enhanced with SuperSignal Chemiluminescence kit (Pierce Biotechnology). Signals were detected on KODAK BIOMAX MR X-ray film (Kodak, Rochester, NY). A quantitative analysis of the western blotting was performed using ImageQuant (Version 5.2; Molecular Dynamics, Sunnyvale, CA).
RNA Extraction and Complementary DNA (cDNA) Synthesis
RNA was extracted from two 10-µm-thick tissue sections as described previously.20 Briefly, the sections were trimmed of excess wax and deparaffinized by three incubations in PROTOCOL (Fisher Scientific; Kalamazoo, MI) for 15 min at 95C with shaking, followed by three centrifugations at room temperature for 2 min at 10,000 × g. The specimens were rinsed briefly once in 100% ethanol and resuspended and ground in a solution of 4 M guanidine isothiocyanate, 20 mM sodium acetate, and 25 mM β-mercaptoethanol (pH 5.5), followed by 72-hr incubation at 65C with mild shaking. After incubation, RNA was isolated by phenol/chloroform extraction. The extracted RNA was treated with 2 µl TURBO DNase buffer, 4 units TURBO DNase (Invitrogen), and 40 units of RNase inhibitor (Promega, Madison, WI) in a 100-µl reaction volume to remove possible contaminating genomic DNA. The mixture was incubated at 37C for 30 min and purified by phenol/chloroform extraction. Total RNA was obtained from frozen tissue using TRIzol reagent (Invitrogen) and further purified with the RNeasy Mini Kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions.21
Approximately 5 µg total RNA from each sample was transcribed into cDNA. The extracted RNA, random hexamers (Promega), and the SuperScript-II RT kit (Invitrogen) were used to synthesize cDNA.20,21 All samples were reverse transcribed under the same conditions. The synthesized cDNA was stored at −20C and used in multiplex reverse transcription–polymerase chain reaction (RT-PCR) reactions as a template.
RNA Quantity and Quality
The quantity of RNA was determined using the NanoDrop ND-1000 UV spectrophotometer (NanoDrop Technologies, Wilmington, DE). RNA quality was assessed using the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA) with the RNA 6000 LabChip kit (Agilent Technologies). We measured RNA integrity number (RIN) using the Agilent 2100 expert software (Agilent Technologies) and assessed RNA integrity using paraffin-embedded RNA metric (PERM) number which is a novel metric for RNA extracted from fixed tissues23 based on a formula that approximates a weighed area-under-the-curve approach.
Multiplex RT-PCR
Multiplex RT-PCR was performed using the Multiplex Polymerase Chain Reaction (MPCR) kit (Maxim Biotech, San Francisco, CA) for mouse tumor necrosis factor (TNF) signaling genes set-3. This kit was designed to detect nine genes with amplicons of 189 to 658 bp. We performed multiplex RT-PCR according to the manufacturer’s instructions in a 50-µl reaction mixture. An initial pre-PCR step of 96C for 5 min was performed in the Bio-Rad iCycler PCR Thermal Cycler (Bio-Rad Laboratories, Hercules, CA), followed by 37 PCR cycles under the following conditions: two cycles of 94C for 1 min, and 62C for 4 min, and 35 cycles of 94C for 1 min and 62C for 2 min. The final cycles was followed by an additional 70C for 10 min to complete partial polymerization. A MPCR positive control (Maxim Biotech) was used in each run. The positive control included frozen mouse kidney RNA. A negative control containing no nucleic acid was also included in each run.
A 1-µl aliquot of the multiplex RT-PCR product was loaded into the DNA 1000 kit (Agilent Technologies) and capillary electrophoresed in the Agilent 2100 Bioanalyzer. We used the Agilent 2100 expert software to compare the electropherograms.
Real-Time Quantitative RT-PCR
After removing genomic DNA with a DNA eliminator column (Qiagen), 4 µg total RNA was reverse transcribed into first-strand cDNA using a QuantiTect Reverse Transcription kit (Qiagen). cDNA was generated from each of three replicates derived from different fixative solutions and frozen mouse kidney RNA. We performed quantitative real-time PCR using TaqMan® Gene Expression reagent (Applied Biosystems, Foster City, CA). Briefly, quantitative real-time PCR was performed with 1 and 2 µg cDNA for housekeeping genes and toll-like receptor-4 (TLR-4), respectively. We examined the cycle threshold (Ct) values for TLR-4, 18S rRNA, and hypoxanthine-guanine phosphoribosyltransferase (HPRT) genes to assess RNA integrity in triplicate. The Mann–Whitney U test was used to evaluate RNA integrity for each fixative condition.
Assessment of DNA Quantity and Quality
DNA was extracted from a 1-mm tissue core24 using the QIAamp DNA FFPE Tissue kit (Qiagen), according to the manufacturer’s instructions. Prior to the final elution step, 40 µl of elution buffer was applied to the column and incubated at room temperature for 2 min, followed by centrifugation. DNA yield was determined using a NanoDrop ND-1000 UV spectrophotometer. DNA quality assessed using a BioScore Screening and Amplification kit (Enzo Life Sciences, Farmingdale, NY).24 In addition, we also performed real-time PCR using the TaqMan® Gene Expression reagent (Applied Biosystems). Briefly, quantitative real-time PCR was performed with 1 µg DNA assayed in a 20 µl reaction volume. The reactions were incubated for 2 min at 50C, 10 min at 95C for initial denaturing, followed by 50 cycles of 95C for 15 sec and 60C for 1 min in the 7500 real-time PCR system (Applied Biosystems). We determined the Ct-value for the HPRT gene to assess DNA integrity in triplicates. The Mann–Whitney U test was used to evaluate DNA integrity for each fixative condition.
Results
Histomorphology
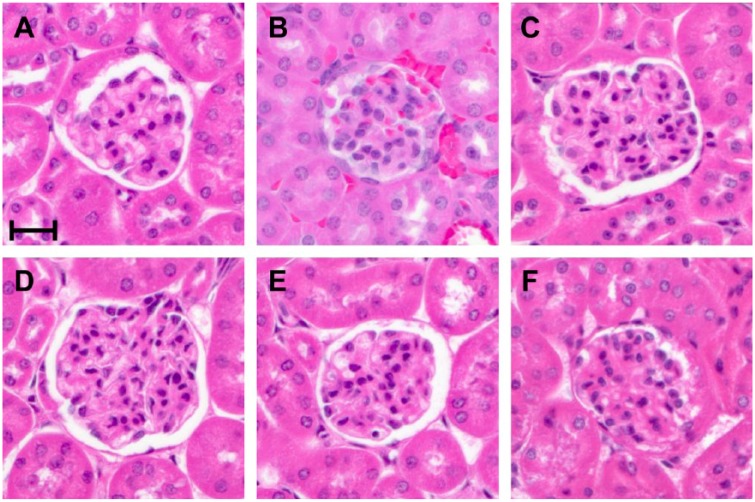
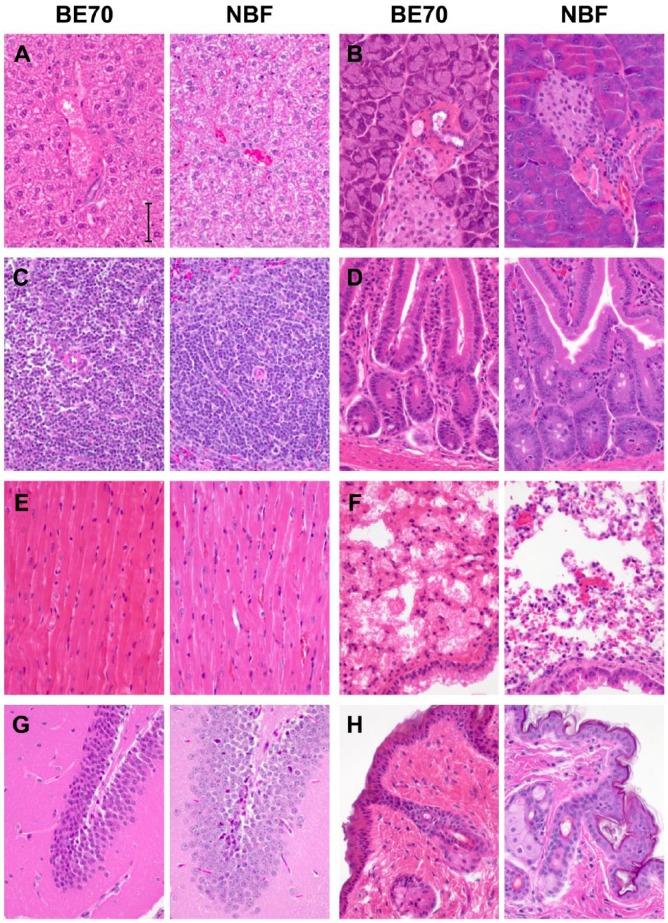
Fixatives are applied to mediate the preservation of tissue; however in their application, they are generally combined with some form of impregnation to allow for microtomy and staining of microscopic sections. Most commonly fixatives are combined with paraffin impregnation (embedding), and frequently stained with hematoxylin and eosin (H&E) as contrasting agents. Figure 1 demonstrates the histomorphologic features of a mouse glomerulus comparing NBF with BE70, as well as the sequential evolution of BE70 from a base fixative of 70% ethanol, by the addition of PBS, glacial acetic acid, and glycerol. Mouse tissues were chosen as a continuation of our prior studies in pre-analytic variables.2,21 Ethanol (70%) results in greater shrinkage with poorer cytologic features, compared with NBF. The introduction of PBS as a buffering solution offsets these changes, including less shrinkage and improved cytologic detail. Figure 2 demonstrates the histomorphology observed with H&E staining, comparing BE70 and NBF in a panel of mouse tissues, including liver, spleen muscle, brain, pancreas, colon, lung, and skin. Overall BE70 results in histomorphologic features similar to 70% ethanol, and not dissimilar to NBF. Eosin staining is more intense when stained under the same conditions. Chromatin is more condensed with BE70 fixation, and nucleoli are more prominent. Red blood cells lose their hemoglobin and are “ghost like” after BE70 fixation, and eosinophils are not recognizable on H&E.
Figure 1.
Histomorphological assessment of different fixatives on mouse kidney. Mouse kidney tissue section stained with hematoxylin and eosin (H&E) after buffered ethanol 70% (BE70) (A), neutral-buffered formalin (NBF) (B), 70% ethanol (E) (C), 70% ethanol + 0.5× PBS (EP) (D), 70% ethanol + 1% glycerol + 0.5× PBS (EGP) (E), or 70% ethanol + 0.5% glacial acetic acid + 0.5× PBS (EAP) (F) fixation. Scale bar, 20 µm. Abbreviation: PBS, phosphate-buffered saline.
Figure 2.
Comparison between buffered ethanol 70% (BE70) and neutral-buffered formalin (NBF) fixation. Tissue sections were stained with hematoxylin and eosin (H&E) after BE70 or NBF fixation. H&E images of liver (A), pancreas (B), spleen (C), duodenum (D), heart (E), lung (F), brain (G), and skin (H). Scale bar, 50 µm.
Immunohistochemical Staining
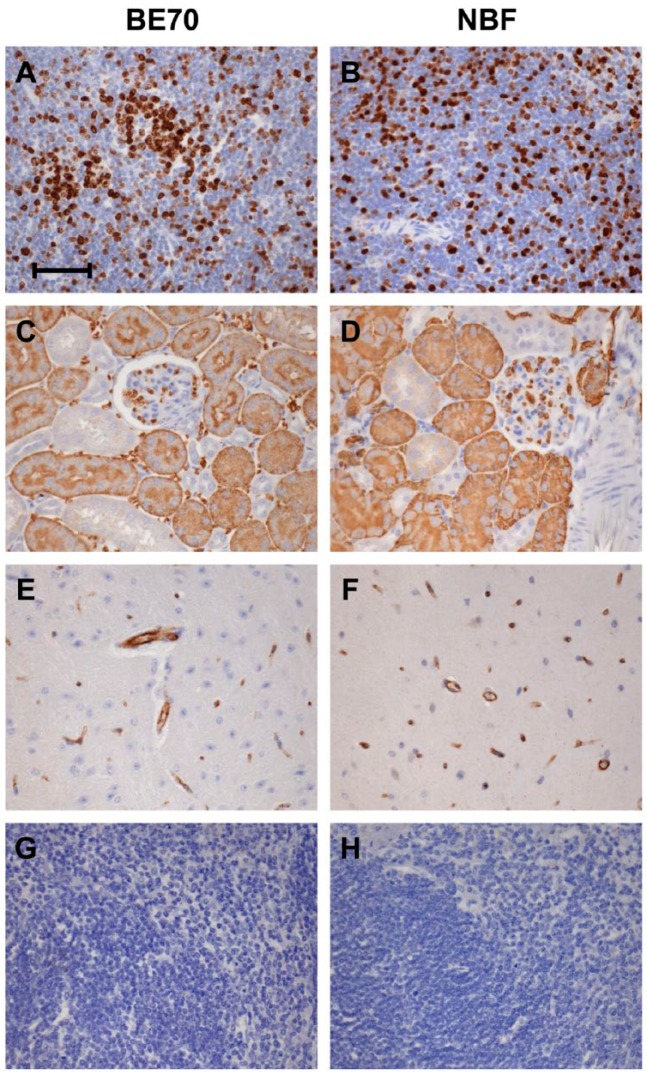
Immunohistochemistry is a critical element of modern histopathology. As such, we performed immunohistochemistry for Ki-67, AQP1, and CD31 on normal mouse tissue to demonstrate the retention of quality of staining in the nuclear, cytoplasmic, and membranous compartments, respectively (Fig. 3). The staining pattern observed for these antigens was entirely retained, with the same staining conditions. Ki-67 stained in proliferating lymphocytes in the spleen. AQP1 stained in both the proximal tubule and the glomeruli in the kidney,25 and CD31 stained in vessels in the brain.
Figure 3.
Immunohistochemical staining between buffered ethanol 70% (BE70) and neutral-buffered formalin (NBF) fixation. Immunohistochemistry staining with anti-Ki-67 in the mouse spleen (A and B), anti-aquaporin 1 (AQP1) in the mouse kidney (C and D), anti-CD31 in the mouse brain (E and F), rabbit immunoglobulin G (IgG) isotype control (G), and no primary antibody control (H). Scale bar, 50 µm.
Protein Quantity and Quality
Proteins were extracted from mouse kidney tissues fixed with different ethanol fixative formulations or NBF fixatives, and subjected to western blotting with anti-AQP1 and anti-GAPDH antibodies. NBF-fixed mouse kidney tissue was used as a negative control (Fig. 4). Although the protein extraction yield of the BE70-fixed tissue was the highest (5.25 ± 0.490 µg/mm3) among the tested fixatives, no significant difference was detected between the BE70 and NBF fixatives (4.42 ± 0. 918 µg/mm3).
Figure 4.
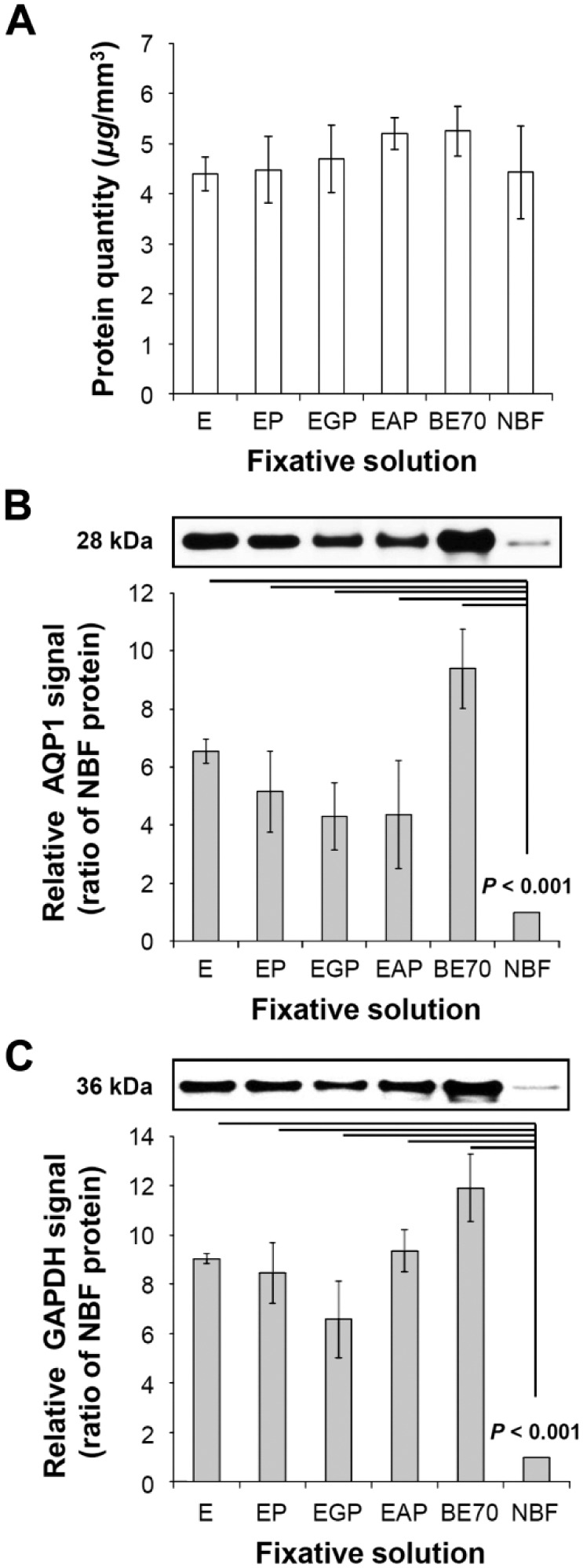
Protein quantity and quality of buffered ethanol 70% (BE70) fixative. (A) Amount of protein extracted from each condition was measured using the bicinchoninic acid (BCA) Protein Assay Kit. The protein extraction yield was expressed as the mean of three replicated samples (mean ± SD). (B) Protein integrity of different fixative solutions was assessed by western blotting. Proteins extracted from different fixative solutions were separated by 4% to 12% reducing sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), electroblotted to nitrocellulose membrane, and probed with anti-aquaporin 1 (AQP1; 1:1000). (C) Western blotting by anti-glyceraldehyde 3-phosphate dehydrogenase (GAPDH) antibody. Relative GAPDH signal of each entity was normalized to neutral-buffered formalin (NBF). Abbreviations: E, 70% ethanol.
The quality of protein extracted from the ethanol fixative formulations and NBF-fixed tissues was evaluated by western blotting. The quantitative image analyses of western blot intensities were evaluated by the Mann–Whitney U test. As shown in Fig. 4B and C, the AQP1 and GAPDH signals were more intense in the BE70-fixed tissue samples (all ps < 0.001) than that in the NBF-fixed tissue samples. We found that AQP1 and GAPDH were better preserved in the BE70-fixed tissue (9.8- and 11.9-fold increase, respectively) than in NBF-fixed tissue, whereas the EGP fixative (4.3- and 6.5-fold increase for AQP1 and GAPDH, respectively) showed the lowest signals among the ethanol fixatives evaluated. However, no significant difference was detected between the BE70 and EGP fixatives. These results suggest that the BE70 fixative provided significant advantages in protein quality over NBF, but did not significantly impact the amount of protein recovered.
RNA Quantity and Quality
After RNA extraction from mouse kidneys fixed with the different fixatives and impregnation with paraffin, RNA quantity was assessed by UV spectrophotometry. BE70 (mean 7.35 ± 0.69 µg/mm3) showed a beneficial effect on RNA recovery, compared with that of the NBF fixative (mean 3.32 ± 0.15 µg/mm3; p = 0.005; Fig. 5A). The A260/A280 ratio of BE70 (mean 2.01) was also similar to that of NBF (mean 1.89).
Figure 5.
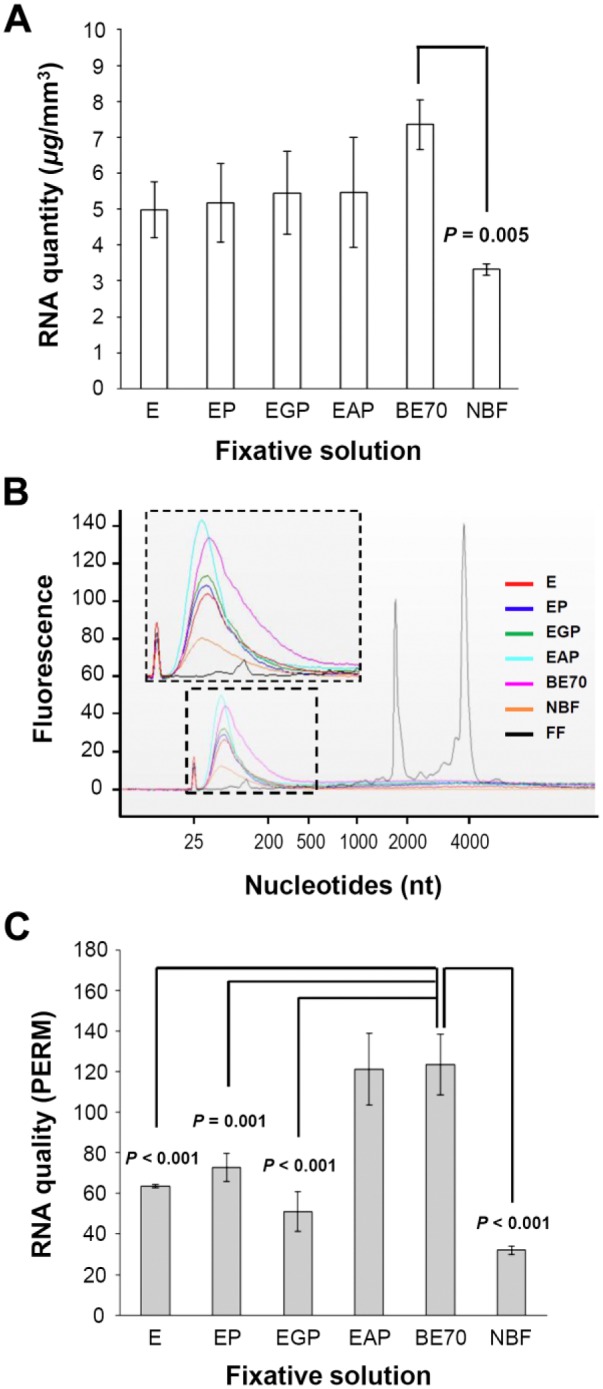
RNA quantity and quality from tissue fixed in different fixative solutions. (A) RNA quantity extracted from each specimen was measured by the Nanodrop spectrophotometer. RNA extraction yield is expressed as the mean of three replicates (mean ± standard deviation [SD]). (B) Representative data are presented as an electropherogram. To compare the quality of RNA extracted under each condition, we used an electropherogram to overlay all seven different conditions. (C) RNA integrity is presented as paraffin-embedded RNA metric (PERM). Abbreviations: E, 70% ethanol; BE70, buffered ethanol 70%; NBF, neutral-buffered formalin; FF, fresh frozen mouse kidney (positive control).
In addition, we analyzed the quality of RNA extracted using a Bioanalyzer. Although more RNA was recovered from the BE70 fixative, the electropherogram demonstrated that the RNA was shorter in fragment length than obtained from the frozen tissue (Fig. 5B). The RNA pattern observed from the FFPE tissues indicated shorter fragments of 100 to 200 nucleotides. BE70 leads to increased RNA fragment length, supporting the finding that the BE70 fixative preserves higher quality of RNA than that of NBF.
The RIN has been widely adopted as a measure of the quality of RNA isolated from fresh and frozen tissues. The RIN is an imperfect measure of quality, lacks a strong correlation with gene-specific measurements,26 and cannot be accurately applied to RNA isolated from FFPE tissue.21 In this context, we applied the PERM value that is a novel metric for FFPE RNA.23 This metric is based on a formula that approximates a weighed area-under-the-curve approach. As shown in Fig. 5C, we were able to dissect the impact of individual components of ethanol-based fixatives on RNA quality. BE70 fixative (mean PERM 123.40) and EAP fixative (mean PERM 121.23) showed the highest PERM. Interestingly, the effect of adding PBS and glycerol was minimal. As expected, NBF showed the lowest number (mean PERM 32.02) among those of the tested fixatives (Fig. 5C).
RNA Integrity
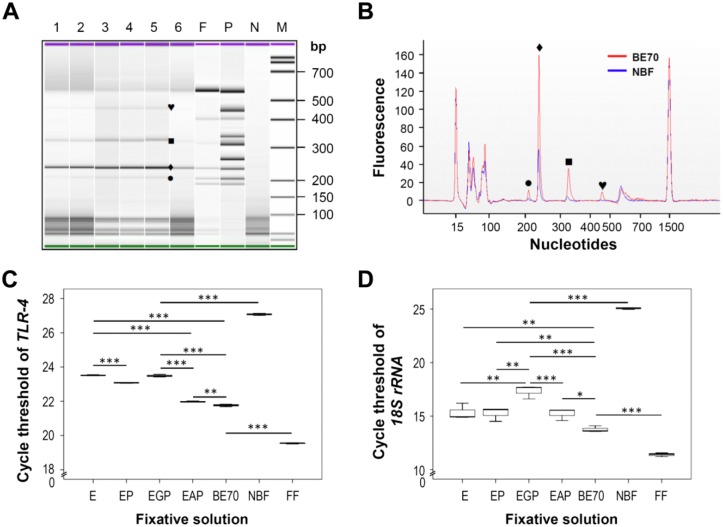
To evaluate the size limitation of RNA amplicons extracted from tissue fixed with the panel of fixatives, and impregnated with paraffin, we performed multiplex RT-PCR using the MPCR kit for mouse TNF signaling genes set-3. As shown in Fig. 6A, four bands (205, 235, 316, and 449 bp) corresponding with the exact sizes of the targets in the BE70 fixative were detected, whereas two bands (235 and 316 bp) were detected in NBF-fixed tissues with relatively weak signals (Fig. 6A and 6B).
Figure 6.
RNA integrity profile of RNA samples derived from experimental mouse kidney tissues. Gene expression profile of mouse kidney tissue was assessed by multiplex reverse transcription–polymerase reaction (RT-PCR). Tumor necrosis factor signaling genes profiling was measured using the Multiplex Polymerase Chain Reaction (MPCR) kit (Maxim Biotech). One microliter of the PCR reaction was run on the Agilent 2100 Bioanalyzer using the DNA 1000 chip. Representative data are presented as a gel-like image (A) and an electropherogram (B). The fixative conditions of each sample are indicated on the gel-like image above each lane: 1, E (70% ethanol); 2, EP (70% ethanol + 0.5× PBS); 3, EGP (70% ethanol + 1% glycerol + 0.5× PBS); 4, EAP (70% ethanol + 0.5% glacial acetic acid + 0.5× PBS); 5, BE70 (buffered ethanol 70%); 6, NBF (neutral buffered formalin); F, fresh frozen mouse kidney; P, positive control; N, negative control (water). To compare the quality of PCR amplicon, we used an electropherogram to overlay the BE70 and NBF fixative conditions. Each symbol represents the difference between BE70 and NBF. (C) The mean cycle threshold (Ct) value of the toll-like receptor-4 (TLR-4) was determined in kidney tissue under different fixative conditions. Gene expression levels are shown as a box plots. The values are the average quantitative real-time RT-PCR cycle threshold numbers (Ct-values). (D) The mean Ct-value of the housekeeping gene (18S rRNA) was determined in kidney tissue under different fixative conditions. Bars indicate standard deviation. Fresh frozen (FF) mouse kidney is used as a positive control. *p < 0.05, **p < 0.01, ***p < 0.001.
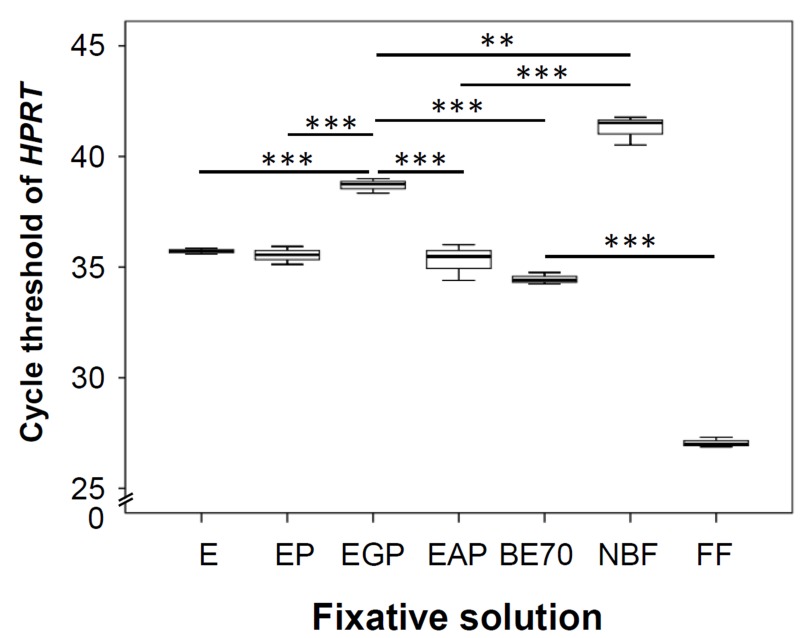
The effect of fixatives on RNA integrity was evaluated by real-time quantitative RT-PCR using TLR-4, 18S rRNA, and HPRT primers. As shown in Fig. 6C, the TLR-4 showed lower Ct-value in the BE70-fixed tissue samples than that in the NBF-fixed tissue samples. Endogenous control 18S rRNA generally showed lower Ct-values (Fig. 6D), whereas the HPRT gene showed higher Ct-values (Supplementary Fig. 1). The Ct-values of the qRT-PCR amplifications were 19.54, 11.40, and 27.06 for TLR-4, 18S rRNA, and HPRT, respectively, in preparations from fresh frozen tissues. Among the tested fixatives, BE70 resulted in Ct-values of 21.76, 13.76, and 34.46 for TLR-4, 18S rRNA, and HPRT, whereas EGP demonstrated 23.49, 17.33, and 38.69 for TLR-4, 18S rRNA, and HPRT, respectively. In contrast, Ct-values were higher in samples generated from NBF, with mean of 27.07, 25.06, and 41.27 for TLR-4, 18S rRNA, and HPRT, respectively. Thus, the Ct-value of BE70 was the lowest among all fixed samples (Fig. 6C and Supplementary Fig. 1).
DNA Quantity and Quality
DNA was successfully extracted from all fixed tissues. The DNA extraction yield of BE70-fixed tissue was similar to that of NBF-fixed tissue (mean 1.09-fold; Fig. 7A). The A260/A280 ratio of BE70-fixed tissue (mean 1.86) was also similar to that of NBF-fixed tissue (mean 1.80). In addition, we tested DNA obtained from fixed tissues for suitability in an array analysis using the BioScore Screening and Amplification kit.24 We amplified approximately 9.73 ± 0.23 µg of high-quality DNA for nucleic acid array analysis using a 100-ng DNA template extracted from all combination of ethanol-fixed tissues. However, the DNA prepared from NBF-fixed tissues was of intermediate quality (mean 2.81 ± 0.13 µg) for microarray application (Fig. 7B).
Figure 7.
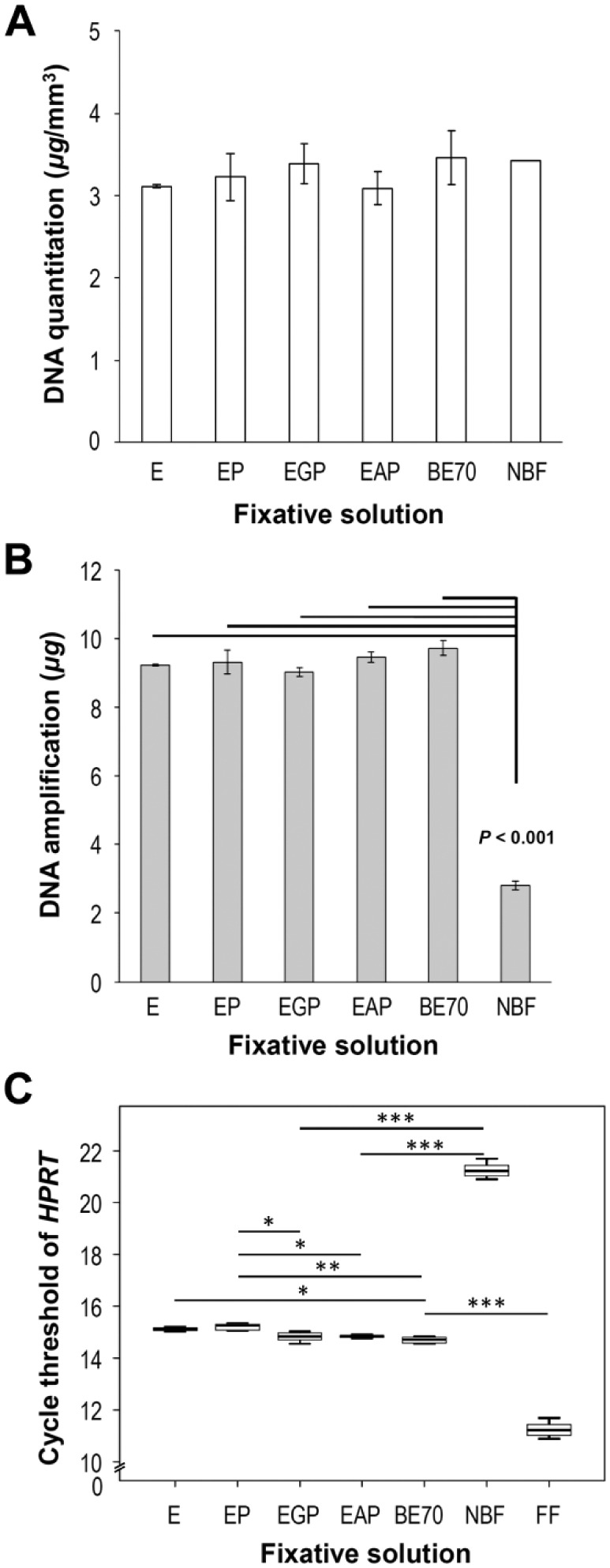
DNA quantity and quality from tissue fixed in different fixative solutions. (A) Quantity of RNA extracted from each specimen was measured with the Nanodrop spectrophotometer. DNA extraction yield is expressed as the mean of three replicates (mean ± SD). (B) DNA quality was assessed using the BioScore™ Screening and Amplification kit. We amplified approximately 10 µg DNA using a 100-ng DNA template from buffered ethanol 70% (BE70). (C) Mean cycle threshold (Ct) values of the housekeeping gene hypoxanthine-guanine phosphoribosyltransferase (HPRT) were determined in kidney under different fixative conditions. Gene expression levels are shown as box plots. The values are the mean polymerase chain reaction Ct-values. Bars indicate standard deviations. Abbreviations: E, 70% ethanol; NBF, neutral-buffered formalin; FF, fresh frozen mouse kidney (positive control). *p < 0.05, **p < 0.01, ***p < 0.001.
To examine the impact of the different fixatives on DNA integrity, we performed real-time PCR using HPRT primers. As shown Fig. 7C, the Ct-value of BE70-fixed tissue (mean Ct-value 14.70) was lower than that of NBF-fixed tissue (mean Ct-value 21.25). In addition, the Ct-value of BE70-fixed tissue was similar to 70% ethanol-fixed tissue (mean Ct-value 15.11). As expected, the Ct-value of fresh frozen (mean Ct-value 11.25) tissue was lower than that of ethanol-fixed tissue (Fig. 7C).
Discussion
The use of formaldehyde solutions, followed by paraffin impregnation, has been used in research and clinical applications for over a century. The primary issues have been quality, economic factors, and reproducibility. Formalin fixation is widely assumed the gold standard of histomorphology, and diagnostic pathology is based on the substantial fund of knowledge from its use worldwide.27 Tissue repositories of pathology specimens of the material fixed in formalin are extensive. Both of these factors are assumed to be barriers to the introduction of alternative fixatives; however, economic factors and safety concerns, as well as the long history of alternative fixatives in both research and clinical practice support the proposition that formalin may not remain the primary fixative of the future. The long-standing models of fixation and impregnation were observational and lacked integration with molecular data. The familiarity of investigators with FFPE tissue results in a blindness to the artifacts introduced by fixation and impregnation.28 All methods of preservation result in artifact; however, it is the reproducibility of these artifacts that are the hallmark of successful fixatives. With the declaration that formaldehyde is a carcinogen, and the desire to move diagnostics toward a molecular basis, the desire for a fixative to replace formalin has never been greater. The process of fixation and impregnation has advanced over the last century in a “fit for purpose” manner, lacking scientifically validated models of the process of contribution of individual reagents or process.2,21 There is now the understanding of the contribution of buffered to formaldehyde-based fixatives, as well as a better understanding of the impregnation process demonstrated that fixation is a necessary precursor to impregnation, and that retained water results in biomolecular degradation.2 This information allows a directed approach to the development of fixatives, rather than a trial-and-error approach.
In the present work, the initial report of a buffered ethanol-based fixative is a departure from prior efforts in alternative fixatives. We have chosen characterization of the fixative with reference to biomolecule integrity as the primary endpoints of this study. Full characterization and potential validation of BE70 as an alternative fixative for clinical application require substantial additional effort; however, BE70 may have utility in a research setting at this time. The development of a buffered ethanol fixative came about in an effort to evaluate the contribution of methanol in formaldehyde fixatives. Initial studies were aimed at comparing formaldehyde fixatives with and without methanol and buffers. As an added control, we tested a buffered ethanol solution, which demonstrated superior results to the prior control of 70% ethanol. The studies of the impact of methanol in formaldehyde fixatives demonstrated that the presence of phosphate buffer was substantially greater than the impact of methanol (data not shown).
In an effort to reproduce a histomorphology as close to that obtained with NBF, and overcome the negative chemical effects of formaldehyde, we started with a buffered ethanol fixative. The choice of 70% ethanol is rather clear, as it is commonly the next step in tissue processing and is a widely used fixative in the research, veterinary, and clinical setting. Alcoholic formaldehyde solutions, which appear to lack standardization in formulation, are commonly used as fixatives as well. The introduction of PBS into the aqueous elements of the ethanol solution is unique and helps overcome some of the negative effects of pure ethanol fixatives, most notably shrinkage and hardening of tissue.8,15 Rather than the phosphate buffers utilized in NBF, BE70 is formulated with PBS. We evaluated different formulations of PBS, but were unable to quantify differences in quality. The concentration of PBS was carefully evaluated to prevent salting out. The introduction of the additional components of glycerol and acid offers discrete improvements in performance, as evident in histomorphology as well as molecular metrics. The impact of glycerol may be to replace “non-freezable water” that is tightly bound to the biomolecules,29 while the addition of acid may aid in cell membrane penetration.
We evaluated BE70 with a 24-hr fixation time to be congruent with our previous studies.21 This is also congruent with the clinical recommendations for immunohistochemistry.30 If the two-phase model of formalin fixation28 is correct, then BE70 would penetrate and fix faster than formalin. Although we did not evaluate the rate of penetration of BE70, we anticipate it is similar to that of 70% ethanol, which, according to Gillespie et al., is slower than that of formalin.14 We have not evaluated shorter fixation times; however, anecdotal evidence demonstrates no harm from prolonged fixation times. This is not surprising, given that 70% ethanol solutions are routinely used for archiving of specimens.12
Overall, the histomorphology of BE70 is a blend of the histomorphology obtained with 70% ethanol and NBF. The tissue is generally more eosinophilic, when stained under the same conditions, and red cell lysis.31 In contrast, the addition of PBS offsets some tissue shrinkage associated with 70% ethanol, likely from osmotic effects, when compared with NBF. Nuclei have a crisp, well-defined histomorphology, commonly associated with coagulative fixatives.14 Definition of optimal histomorphology is subjective, and alternative fixatives have commonly been used to address specific histomorphologic features.32
Immunohistochemistry on BE70-fixed tissue is easily optimized and generates staining patterns of nearly identical features compared with NBF (Fig. 3). To evaluate the capacity to perform immunohistochemistry, we evaluated Ki-67, AQP1, and CD31, staining the nuclear, cytoplasmic, and membranous compartments, respectively. This limited evaluation demonstrates similar patterns and intensity of staining using identical staining conditions with reference to all factors including antigen retrieval and antibody dilution. We did not perform a systematic review of immunohistochemical conditions nor perform a rigorous validation; however, we were able to demonstrate identical antigen retrieval generated identical staining patterns between BE70 and NBF for the three antigens tested. Non-cross-linking fixatives routinely require less antigen retrieval,13 and it has been suggested nuclear proteins are preferentially lost during alcohol fixation.33–36 Historically, some antigens may not be detectable in alcohol-based fixatives with antibodies selected for detection by immunohistochemistry on NBF-fixed tissue. In these instances, simply dipping the deparaffinized slide, prior to antigen retrieval into NBF, for 10 sec, is documented to induce sufficient formalin-mediated chemical changes so as to make the antigen detectable.37 Alternatively, the coagulative nature of alcohol fixatives may impede denaturation of proteins with routine methods of antigen retrieval; however, the addition of SDS appears to overcome this issue.22 The adoption of BE70 in a clinical setting in which immunohistochemistry is commonly applied would require the re-validation of the immunohistochemical assays.30
Beyond immunohistochemistry, fixed, paraffin-embedded tissues are routinely used for proteomic studies, including western blot, immunoblotting, and mass spectrophotometer investigations.38 Although there is little difference in the quantity of protein extracted, based on different fixatives, the detection of specific proteins is significantly enhanced (Fig. 4). Our results agree with other published reports,39–41 in which protein yield and immunoreactivity of formalin-fixed tissue are compromised due to cross-linking and may render them of limited quality for proteomic studies.
It is well described that formaldehyde fixation limits the quality of nucleic acids available for extraction from the FFPE tissue. Thus, a number of alternative fixatives and approaches have been applied to the diagnostic pathology. The majority of alternative fixatives use alcohol- or acetone-based solutions with the addition of stabilizing agents. Prior studies on alternative fixatives have reported varying degree of success. Notably, these alternative fixatives significantly improved the quality of RNA or DNA compared with the NBF fixative. These fixatives resulted in longer lengths of RNA than the NBF fixative (average amplicon size, approximately 200 bp), for example, Hepes-glutamic acid buffer-mediated organic solvent protection effect (HOPE; 300 bp),42 Methacarn (850 and 463 bp),43 PAXgene (712 bp),41 RCL2® (377 and 463 bp),44,45 universal molecular fixative (UMFIX; 816, 700, and 450 bp),13,14,46 or other fixatives11 by PCR amplification. In addition, alternative fixatives demonstrate improved quality of DNA as reflected by relatively longer length amplicons by PCR amplification, ranging from 500 bp to 2.4 kbp.11,13,41–46 In agreement with prior studies, we also demonstrated that the RNA and DNA extracted from BE70-fixed tissue are applicable to PCR amplification with the advantage of relatively longer RNA or DNA fragments in comparison with NBF. As shown in Fig. 5B, DNA extracted from BE70-fixed tissue has superior quality (3.46-fold increase in the PCR amplification yield) as that recovered from NBF-fixed tissue. We have applied quantitative real-time PCR to the housekeeping gene and have demonstrated that the Ct-value of BE70 is approximately 1.31-fold higher compared with DNA from the matched, fresh frozen specimen (Fig. 7C). These data suggest that the DNA-based assay with BE70-fixed tissue is improved compared with NBF, but will require an additional DNA starting material, compared with frozen tissue.
In RNA analysis, we demonstrated that the RNA extracted from BE70-fixed and paraffin-embedded tissue is applicable to multiplex RT-PCR. In addition, we found a size limitation of amplicons of about 449 bp through multiplex RT-PCR. The BE70 fixative resulted in superior RNA quantity and quality compared with the NBF fixative (Fig. 6). We previously demonstrated that the RNA quality is affected by the fixation process and warm ischemia times.21 In addition, we also reconfirmed that the process of formalin fixation further damaged the quality of RNA and prolonged fixation resulted in a significant deterioration of the RNA strand length.21,27,47 In this context, not only does the non-cross-linking property of the alcohol-based BE70 fixative improves recovery of RNA from paraffin-embedded tissue but also the phosphate-buffered acid solution results in longer RNA fragments than 70% ethanol solutions of water alone. Although we found that the acidic conditions of a fixative solution resulted in higher RNA integrity compared with an alkaline solution (unpublished observation), the exact mechanism of this observation remains to be defined.
The literature is rich in alternative fixatives; however, their widespread adoption has not been seen for a number of reasons: economic factors, safety, and histomorphology being the primary obstacles. Of the alternative fixatives, the simplest is 70% ethanol and currently the most common. Over the last 125 years, formaldehyde solutions have remained the dominant fixative, with an ever-changing series of alternative fixatives used for specific purposes. Historically, these alternative fixatives were selected for specific histomorphologic applications in specific tissues31,32 or to support now uncommon histology applications. Over the last decade, a number of “molecular friendly” fixatives have been developed10,11,17,41,42,44 to address the current research and clinical applications applied to tissue; however, none have seen widespread adoption for a diversity of reasons.
The replacement of water with 0.5× PBS offers a surprising benefit at all levels of evaluation: fixed tissue, histomorphology, and biomolecular assay levels. The introduction of glycerol and acetic acid has less, but still measurable, impacts on fixative performance, at the biomolecular level. The PBS, glycerol, and acetic acid components of the fixative can be formulated as a concentrate that can be added to 100% or 95% ethanol to generate a working fixative. The fixative is stable for months at room temperature, as well as at 4C. In our studies, we have not investigated the performance of BE70 for the long-term storage of “wet” specimens; however, 70% ethanol has long been used for this purpose, even with specimens originally fixed in formaldehyde solutions. Alternative methods of impregnation are encountered, most commonly in the research setting. Although not tested, we anticipate their performance would be unaltered. Preliminary data (not shown) suggest that BE70-fixed paraffin-embedded specimens have functionally the same degradation profile as FFPE specimens, as degradation is mediated by entrapped water.2
The goals of a fixative are to render the tissue aseptic, prevent putrefaction, and enable examination. The first and second goals are generally met by a large number of reagents, including 70% ethanol (www.cdc.gov/hicpac/Disinfection_Sterilization/6_0disinfection.html).14 The challenge is the goal of the examination, which can be phrased as “fit for purpose.”48 The current state of histopathologic investigation is to focus on DNA, RNA, and proteins. The purpose for which a fixative is applied and the end goals of the examination are the ultimate determinants to the success of a fixative. Clearly, formaldehyde-based fixatives have been the dominant fixative over a century, with incremental improvement, especially the widespread introduction of buffers. The future of how tissue is handled in both the research and clinical settings is unclear. Proposals to split specimens in a clinical setting, either into different fixatives, or frozen versus NBF, have failed to gain traction due to issues of size, heterogeneity, and cost.27 The advancement of an alternative to NBF will depend on two factors: (1) limitations of NBF due to the cost of use and limitations of downstream applications, and (2) validation of a fixative that addresses the current needs of the research and diagnostic community.
In conclusion, we have developed a new non-aldehyde-based fixative that is compatible with existing protocols, offers improved biomolecular preservation, and is histomorphologically similar to the current fixatives. This fixative is a unique improvement on 70% ethanol and can be formulated by the addition of a concentrate to stock ethanol for simplicity of use. With additional evaluation, the BE70 fixative may be a potential replacement for NBF in both research and clinical settings, with the benefit of better biomolecule preservation, without the trade-off of impaired histomorphology. Substantial additional study and validation is required to elucidate the potential of BE70 in the research and clinical histopathology environment.
Supplementary Material
Supplementary Material
Acknowledgments
SMH thanks Richard Burry, PhD, for his useful discussions concerning the contribution of methanol to formaldehyde-based fixatives, which initiated this study.
Footnotes
Author Contributions: CP, J-YC, and SMH conceived of the study and devised the experimental design. KY, CHC, AS, KTM, and WAS performed experiments. J-YC, CHC, and SMH performed data analysis for experiments. CP and J-YC drafted the final version of the manuscript. SMH revised the manuscript and figures, and added critical content to the discussion. All authors have read and approved the final manuscript.
Competing Interests: The authors declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: The National Institutes of Health has filed a provisional patent application (62/255030) for the fixative described herein. Ms. Perry, Dr. Chung, and Dr. Hewitt are listed as inventors on this patent application; however, the application is assigned to the U.S. Department of Health and Human Services, as the work was performed under contract (C.P.) or as official duty (J.-Y.C. and S.M.H.).
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This research was supported by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, National Institutes of Health.
Literature Cited
- 1. Nykanen M, Kuopio T. Protein and gene expression of estrogen receptor alpha and nuclear morphology of two breast cancer cell lines after different fixation methods. Exp Mol Pathol. 2010;88(2):265–71. doi: 10.1016/j.yexmp.2009.12.003. PubMed PMID: 20025868. [DOI] [PubMed] [Google Scholar]
- 2. Xie R, Chung JY, Ylaya K, Williams RL, Guerrero N, Nakatsuka N, Badie C, Hewitt SM. Factors influencing the degradation of archival formalin-fixed paraffin-embedded tissue sections. J Histochem Cytochem. 2011;59(4):356–65. doi: 10.1369/0022155411398488. PubMed PMID: 21411807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dotti I, Bonin S, Basili G, Nardon E, Balani A, Siracusano S, Zanconati F, Palmisano S, De Manzini N, Stanta G. Effects of formalin, methacarn, and fineFIX fixatives on RNA preservation. Diagn Mol Pathol. 2010;19(2):112–22. doi: 10.1097/PDM.0b013e3181b520f8. PubMed PMID: 20502189. [DOI] [PubMed] [Google Scholar]
- 4. Masuda N, Ohnishi T, Kawamoto S, Monden M, Okubo K. Analysis of chemical modification of RNA from formalin-fixed samples and optimization of molecular biology applications for such samples. Nucleic Acids Res. 1999;27(22):4436–43. PubMed PMID: 10536153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cogliano VJ, Grosse Y, Baan RA, Straif K, Secretan MB, El Ghissassi F. Meeting report: summary of IARC monographs on formaldehyde, 2-butoxyethanol, and 1-tert-butoxy-2-propanol. Environ Health Perspect. 2005;113(9):1205–8. PubMed PMID: 16140628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Berg AO, Bailar JC, Gandolfi AJ, Kriebel D, Morris JB, Pinkerton KE, Rusyn I, Shioda T, Smith TJ, Wetzler M, Zeise L, Zweidler-McKay P, Murray-Smith H, Stoever K, Grossblatt N, Karalic-Loncarevic M, Rose R, Payne R. Review of the formaldehyde assessment in the National Toxicology Program 12th report on carcinogens. Washington, DC: The National Academy of Sciences; 2014. p. 1–246. [Google Scholar]
- 7. Stanta G, Mucelli SP, Petrera F, Bonin S, Bussolati G. A novel fixative improves opportunities of nucleic acids and proteomic analysis in human archive’s tissues. Diagn Mol Pathol. 2006;15(2):115–23. PubMed PMID: 16778593. [DOI] [PubMed] [Google Scholar]
- 8. Moelans CB, ter Hoeve N, van Ginkel JW, ten Kate FJ, van Diest PJ. Formaldehyde substitute fixatives. Analysis of macroscopy, morphologic analysis, and immunohistochemical analysis. Am J Clin Pathol. 2011;136(4):548–56. doi: 10.1309/AJCPHH1B0COCBGOM. PubMed PMID: 21917676. [DOI] [PubMed] [Google Scholar]
- 9. Pereira MA, Dias AR, Faraj SF, Cirqueira Cdos S, Tomitao MT, Nahas SC, Ribeiro U, Jr, de Mello ES. Carnoy’s solution is an adequate tissue fixative for routine surgical pathology, preserving cell morphology and molecular integrity. Histopathology. 2015;66(3):388–97. doi: 10.1111/his.12532. PubMed PMID: 25307771. [DOI] [PubMed] [Google Scholar]
- 10. Olert J, Wiedorn KH, Goldmann T, Kuhl H, Mehraein Y, Scherthan H, Niketeghad F, Vollmer E, Müller AM, Müller-Navia J. HOPE fixation: a novel fixing method and paraffin-embedding technique for human soft tissues. Pathol Res Pract. 2001;197(12):823–6. doi: 10.1078/0344-0338-00166. PubMed PMID: 11795830. [DOI] [PubMed] [Google Scholar]
- 11. Lykidis D, Van Noorden S, Armstrong A, Spencer-Dene B, Li J, Zhuang Z, Stamp GWH. Novel zinc-based fixative for high quality DNA, RNA and protein analysis. Nucleic Acids Res. 2007;35(12):e85. doi: 10.1093/nar/gkm433. PubMed PMID: 17576663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hostein I, Stock N, Soubeyran I, Marty M, De Mascarel I, Bui M, Geneste G, Petersen MC, Coindre JM, Macgrogan G. Nucleic acid quality preservation by an alcohol-based fixative: comparison with frozen tumors in a routine pathology setting. Diagn Mol Pathol. 2011;20(1):52–62. doi: 10.1097/PDM.0b013e3181e71ba5. PubMed PMID: 21326040. [DOI] [PubMed] [Google Scholar]
- 13. Vincek V, Nassiri M, Nadji M, Morales AR. A tissue fixative that protects macromolecules (DNA, RNA, and protein) and histomorphology in clinical samples. Lab Invest. 2003;83(10):1427–35. PubMed PMID: 14563944. [DOI] [PubMed] [Google Scholar]
- 14. Gillespie JW, Best CJ, Bichsel VE, Cole KA, Greenhut SF, Hewitt SM, Ahram M, Gathright YB, Merino MJ, Strausberg RL, Epstein JI, Hamilton SR, Gannot G, Baibakova GV, Calvert VS, Flaig MJ, Chuaqui RF, Herring JC, Pfeifer J, Petricoin EF, Linehan WM, Duray PH, Bova GS, Emmert-Buck MR. Evaluation of non-formalin tissue fixation for molecular profiling studies. Am J Pathol. 2002;160(2):449–57. doi: 10.1016/S0002-9440(10)64864-X. PubMed PMID: 11839565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bostwick DG, al Annouf N, Choi C. Establishment of the formalin-free surgical pathology laboratory. Utility of an alcohol-based fixative. Arch Pathol Lab Med. 1994;118(3):298–302. PubMed PMID: 8135636. [PubMed] [Google Scholar]
- 16. Lassalle S, Hofman V, Marius I, Gavric-Tanga V, Brest P, Havet K, Butori C, Selva E, Santini J, Mograbi B, Hofman P. Assessment of morphology, antigenicity, and nucleic acid integrity for diagnostic thyroid pathology using formalin substitute fixatives. Thyroid. 2009;19(11):1239–48. doi: 10.1089/thy.2009.0095. PubMed PMID: 19888862. [DOI] [PubMed] [Google Scholar]
- 17. Moelans CB, Oostenrijk D, Moons MJ, van Diest PJ. Formaldehyde substitute fixatives: effects on nucleic acid preservation. J Clin Pathol. 2011;64(11):960–7. doi: 10.1136/jclinpath-2011-200152. PubMed PMID: 21715573. [DOI] [PubMed] [Google Scholar]
- 18. Masir N, Ghoddoosi M, Mansor S, Abdul-Rahman F, Florence CS, Mohamed-Ismail NA, Tamby MR, Md-Latar NH. RCL2, a potential formalin substitute for tissue fixation in routine pathological specimens. Histopathology. 2012;60(5):804–15. doi: 10.1111/j.1365-2559.2011.04127.x. PubMed PMID: 22320393. [DOI] [PubMed] [Google Scholar]
- 19. Boissiere-Michot F, Denouel A, Boulle N, Guillaume C, Orsetti B, Lopez-Crapez E, Chateau MC, Bibeau F. The non-crosslinking fixative RCL2®-CS100 is compatible with both pathology diagnosis and molecular analyses. Pathol Oncol Res. 2013;19(1):41–53. doi: 10.1007/s12253-012-9556-2. PubMed PMID: 22893391. [DOI] [PubMed] [Google Scholar]
- 20. Chung JY, Braunschweig T, Hewitt SM. Optimization of recovery of RNA from formalin-fixed, paraffin-embedded tissue. Diagn Mol Pathol. 2006;15(4):229–36. Epub 2006. November 24. doi: 10.1097/01.pdm.0000213468.91139.2d. PubMed PMID: 17122651. [DOI] [PubMed] [Google Scholar]
- 21. Chung JY, Braunschweig T, Williams R, Guerrero N, Hoffmann KM, Kwon M, Song YK, Libutti SK, Hewitt SM. Factors in tissue handling and processing that impact RNA obtained from formalin-fixed, paraffin-embedded tissue. J Histochem Cytochem. 2008;56(11):1033–42. Epub 2008. August 20. doi: 10.1369/jhc.2008.951863. PubMed PMID: 18711211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chung JY, Lee SJ, Kris Y, Braunschweig T, Traicoff JL, Hewitt SM. A well-based reverse-phase protein array applicable to extracts from formalin-fixed paraffin-embedded tissue. Proteomics Clin Appl. 2008;2(10–11):1539–47. Epub 2008. October 1. doi: 10.1002/prca.200800005. PubMed PMID: 21136801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chung JY, Cho H, Hewitt SM. A quality metric for RNA isolated from formalin fixed, paraffin embedded tissue. BioTechniques. 2016; 60 (5): 239-244. doi: 10.2144/000114415. PubMed PMID: 27177816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chung JY, Yi JM, Xie R, Brown V, Lee O, Ahuja N, Braunschweig T, Hewitt SM. A pressure cooking-based DNA extraction from archival formalin-fixed, paraffin-embedded tissue. Anal Biochem. 2012;425(2):128–34. Epub 2012. March 28. doi: 10.1016/j.ab.2012.03.012. PubMed PMID: 22449494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Maunsbach AB, Marples D, Chin E, Ning G, Bondy C, Agre P, Nielsen S. Aquaporin-1 water channel expression in human kidney. J Am Soc Nephrol. 1997;8(1):1–14. PubMed PMID: 9013443. [DOI] [PubMed] [Google Scholar]
- 26. Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, Lightfoot S, Menzel W, Granzow M, Ragg T. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol. 2006;7:3. doi: 10.1186/1471-2199-7-3. PubMed PMID: 16448564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hewitt SM, Lewis FA, Cao Y, Conrad RC, Cronin M, Danenberg KD, Goralski TJ, Langmore JP, Raja RG, Williams PM, Palma JF, Warrington JA. Tissue handling and specimen preparation in surgical pathology: issues concerning the recovery of nucleic acids from formalin-fixed, paraffin-embedded tissue. Arch Pathol Lab Med. 2008;132(12):1929–35. doi: 10.1043/1543-2165-132.12.1929. PubMed PMID: 19061293. [DOI] [PubMed] [Google Scholar]
- 28. Fox CH, Johnson FB, Whiting J, Roller PP. Formaldehyde fixation. J Histochem Cytochem. 1985;33(8):845–53. PubMed PMID: 3894502. [DOI] [PubMed] [Google Scholar]
- 29. Boi G, Scalia CR, Gendusa R, Ronchi S, Cattoretti G. Disaccharides protect antigens from drying-induced damage in routinely processed tissue sections. J Histochem Cytochem. 2016;64(1):18–31. doi: 10.1369/0022155415616162. PubMed PMID: 26487185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hewitt SM, Robinowitz M, Bogen SA, Gown AM, Kalra KL, Otis CN, Spaulding B, Taylor CR. Quality assurance for design control and implementation of immunohistochemistry assays; approved guideline-CLSI document I/LA28-A2. 2nd ed. Wayne, PA: Clinical and Laboratory Standards Institute; 2011. p. 139. [Google Scholar]
- 31. Lillie JD. Histopathological technique. Philadelphia: Blakiston Co.; 1948. [Google Scholar]
- 32. Bancroft JD, Stevens A. Theory and practice of histological techniques. Edinburgh: Churchill Livingstone; distributed in the U.S. of America by Longman; 1977. 436 p., 7 leaves of plates. [Google Scholar]
- 33. Clever U, Ellgaard EG. Puffing and histone acetylation in polytene chromosomes. Science. 1970;169(3943):373–4. PubMed PMID: 5450368. [DOI] [PubMed] [Google Scholar]
- 34. Prento P, Lyon H. Commercial formalin substitutes for histopathology. Biotech Histochem. 1997;72(5):273–82. PubMed PMID: 9408588. [DOI] [PubMed] [Google Scholar]
- 35. Shi SR, Liu C, Pootrakul L, Tang L, Young A, Chen R, Cote RJ, Taylor CR. Evaluation of the value of frozen tissue section used as “gold standard” for immunohistochemistry. Am J Clin Pathol. 2008;129(3):358–66. doi: 10.1309/7CXUYXT23E5AL8KQ. PubMed PMID: 18285257. [DOI] [PubMed] [Google Scholar]
- 36. Eltoum I, Fredenburgh J, Myers RB, Grizzle WE. Introduction to the theory and practice of fixation of tissues. J Histotechnol. 2001;24(3):173–90. PubMed PMID: WOS:000170884500005. [Google Scholar]
- 37. Panzacchi S, Boiani S, Mandrioli D, Piccioli M, Belpoggi F. Applying immunohistochemistry to alcohol-fixed paraffin-embedded tissues: an innovative technique to reduce use of formaldehyde. Eur J Oncol. 2013;18(2):75–83. PubMed PMID: WOS:000330522900003. [Google Scholar]
- 38. Matsuda KM, Chung JY, Hewitt SM. Histo-proteomic profiling of formalin-fixed, paraffin-embedded tissue. Expert Rev Proteomics. 2010;7(2):227–37. doi: 10.1586/epr.09.106. PubMed PMID: 20377389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nietner T, Jarutat T, Mertens A. Systematic comparison of tissue fixation with alternative fixatives to conventional tissue fixation with buffered formalin in a xenograft-based model. Virchows Arch. 2012;461(3):259–69. doi: 10.1007/s00428-012-1248-5. PubMed PMID: 22814649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Paavilainen L, Edvinsson A, Asplund A, Hober S, Kampf C, Ponten F, Wester K. The impact of tissue fixatives on morphology and antibody-based protein profiling in tissues and cells. J Histochem Cytochem. 2010;58(3):237–46. doi: 10.1369/jhc.2009.954321. PubMed PMID: 19901271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Belloni B, Lambertini C, Nuciforo P, Phillips J, Bruening E, Wong S, Dummer R. Will PAXgene substitute formalin? A morphological and molecular comparative study using a new fixative system. J Clin Pathol. 2013;66(2):124–35. doi: 10.1136/jclinpath-2012-200983. PubMed PMID: 23125305. [DOI] [PubMed] [Google Scholar]
- 42. Braun M, Menon R, Nikolov P, Kirsten R, Petersen K, Schilling D, Schott C, Gündisch S, Fend F, Becker KF, Perner S. The HOPE fixation technique—a promising alternative to common prostate cancer biobanking approaches. BMC Cancer. 2011;11:511. doi: 10.1186/1471-2407-11-511. PubMed PMID: 22151117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shibutani M, Uneyama C, Miyazaki K, Toyoda K, Hirose M. Methacarn fixation: a novel tool for analysis of gene expressions in paraffin-embedded tissue specimens. Lab Invest. 2000;80(2):199–208. PubMed PMID: 10701689. [DOI] [PubMed] [Google Scholar]
- 44. Delfour C, Roger P, Bret C, Berthe ML, Rochaix P, Kalfa N, Raynaud P, Bibeau F, Maudelonde T, Boulle N. RCL2, a new fixative, preserves morphology and nucleic acid integrity in paraffin-embedded breast carcinoma and microdissected breast tumor cells. J Mol Diagn. 2006;8(2):157–69. doi: 10.2353/jmoldx.2006.050105. PubMed PMID: 16645201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Preusser M, Plumer S, Dirnberger E, Hainfellner JA, Mannhalter C. Fixation of brain tumor biopsy specimens with RCL2 results in well-preserved histomorphology, immunohistochemistry and nucleic acids. Brain Pathol. 2010;20(6):1010–20. doi: 10.1111/j.1750-3639.2010.00400.x. PubMed PMID: 20477829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Turashvili G, Yang W, McKinney S, Kalloger S, Gale N, Ng Y, Chow K, Bell L, Lorette J, Carrier M, Luk M, Aparicio S, Huntsman D, Yip S. Nucleic acid quantity and quality from paraffin blocks: defining optimal fixation, processing and DNA/RNA extraction techniques. Exp Mol Pathol. 2012;92(1):33–43. doi: 10.1016/j.yexmp.2011.09.013. PubMed PMID: 21963600. [DOI] [PubMed] [Google Scholar]
- 47. Wu L, Patten N, Yamashiro CT, Chui B. Extraction and amplification of DNA from formalin-fixed, paraffin-embedded tissues. Appl Immunohistochem Mol Morphol. 2002;10(3):269–74. PubMed PMID: 12373156. [DOI] [PubMed] [Google Scholar]
- 48. Hewitt SM, Badve SS, True LD. Impact of preanalytic factors on the design and application of integral biomarkers for directing patient therapy. Clin Cancer Res. 2012;18(6):1524–30. doi: 10.1158/1078-0432.CCR-11-2204. PubMed PMID: 22422404. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.