Abstract
Immune activation associated with HIV-1 infection contributes to morbidity and mortality. We studied whether chloroquine, through Toll-like receptor (TLR) antagonist properties, could reduce immune activation thought to be driven by TLR ligands, such as gut-derived bacterial elements and HIV-1 RNAs. AIDS Clinical Trials Group A5258 was a randomized, double-blind, placebo-controlled study in 33 HIV-1-infected participants off antiretroviral therapy (ART) and 37 participants on ART. Study participants in each cohort were randomized 1:1 to receive chloroquine 250 mg orally for the first 12 weeks then cross over to placebo for 12 weeks or placebo first and then chloroquine. Combining the periods of chloroquine use in both arms of the on-ART cohort yielded a modest reduction in the proportions of CD8 T cells co-expressing CD38 and DR (median decrease = 3.0%, p = .003). The effect on immune activation in the off-ART cohort was likely confounded by increased plasma HIV-1 RNA during chloroquine administration (median 0.29 log10 increase, p < .001). Transcriptional analyses in the off-ART cohort showed decreased expression of interferon-stimulated genes in 5 of 10 chloroquine-treated participants and modest decreases in CD38 and CCR5 RNAs in all chloroquine-treated participants. Chloroquine modestly reduced immune activation in ART-treated HIV-infected participants. Clinical Trials Registry Number: NCT00819390.
Introduction
Chronic HIV infection is characterized by increased immune activation,1–3 lymphocyte turnover,1,4 and heightened expression of inflammatory cytokines.5 Immune activation predicts clinical outcome in natural history studies,6–8 and inflammatory indices are linked to morbidity, mortality, and failure to restore circulating CD4 T-cell counts in individuals receiving antiretroviral therapy (ART).9–11 Immune activation and inflammation decrease, but do not completely normalize, when viral suppression is achieved after administration of ART.12,13 The drivers of persistent activation and inflammation in treated HIV infection are not known but plausible candidates include HIV, other copathogens such as cytomegalovirus, and translocation of microbial products from the damaged gut.14,15
Toll-like receptors (TLRs), a family of transmembrane microbial sensors expressed on a variety of cell types,16–18 arm innate defenses and, when ligated, can result in immune activation and inflammation. TLRs recognizing microbial surface components are typically expressed on the cell surface; TLRs recognizing microbial nucleic acids are typically expressed in the endosomal compartment.18
Chloroquine, initially developed as an antimalarial agent, has immunomodulatory properties useful in the treatment of autoimmune diseases such as systemic lupus erythematosus and rheumatoid arthritis.19,20 In both in vitro and in vivo experimental models, chloroquine inhibits proinflammatory cytokine release induced by microbial TLR ligands by downregulating TLR9 and TLR4 mRNA expression, blocking NFκB and activated protein-1 activation, interrupting endosome maturation,21 and inhibiting nucleic acid binding to TLR7, 8, and 9.22 When given to individuals with autoimmune disease, chloroquine decreased the numbers of activated HLA-DR+ cells in the skin of those with systemic lupus erythematosus23 and reduced the elevated levels of the proinflammatory cytokines, interleukin (IL)-6, IL-18, and tumor necrosis factor-α in plasma.23
We investigated whether chloroquine administration could reduce the immune activation in HIV infection posited to be driven by microbial TLR ligands.
Materials and Methods
Study design
AIDS Clinical Trials Group A5258 was a randomized, double-blind, placebo-controlled study comparing 12 weeks of chloroquine to placebo administration for effects on immune activation in two sequential cohorts of HIV-infected individuals: off-ART (cohort 1: arms A and B) and on-ART (cohort 2: arms C and D). Eligible participants were aged 18–55 (cohort 1) (taking no ART for at least 6 months before study entry, with plasma HIV-1 RNA levels ≥1,000 copies/ml and CD4 T-cell counts ≥400 cells/mm3 (off-ART) or (cohort 2) taking ART for at least 24 months before entry, with CD4+ T-cell counts <350 cells/mm3 and plasma HIV-1 RNA levels below the limit of detection using clinical assays (on-ART).
Study participants in each cohort were randomized 1:1 to receive chloroquine 250 mg orally for the first 12 weeks and then to cross over to placebo for 12 weeks (arms A and C) or placebo for 12 weeks then cross over to chloroquine 250 mg for 12 weeks (arms B and D). The design allowed (1) comparison of week 12 change from baseline between arms A and B (off-ART) and between arms C and D (on-ART) and (2) assessment of changes after 12 weeks of chloroquine in both arms by combining first 12 weeks in arm A and crossover period in arm B (off-ART), and similarly in arms C and D.
The protocol was approved by each site's institutional review board. Written informed consent was obtained from all participants. The study was conducted according to human experimentation guidelines of the U.S. Department of Health and Human Services and was monitored by a study monitoring committee of the AIDS Clinical Trial Group (NCT 01266616).
Evaluation of participants and follow-up
After screening and baseline evaluations were completed, study participants had clinical and laboratory safety assessments at weeks 4, 10, 12, 16, 22, 24, and 28. Ophthalmologic evaluations were performed at screening and at weeks 12 and 24; audiometric examinations were done as needed. Plasma levels of HIV-1 RNA and CD4 and CD8 T lymphocyte counts and other immunologic assays were performed at screening, study entry, and weeks 10, 12, 22, 24 (also 14 days before entry, for immunologic assays).
Immunologic evaluations
Flow cytometry
Cryopreserved peripheral blood mononuclear cells were thawed and stained with fluorochrome-labeled monoclonal antibodies (Becton Dickinson [BD]) and examined for expression of CD38 and HLA-DR on CD4 and CD8 T cells in cohort 1 and were examined using a BD FACSCalibur flow cytometer in cohort 1 and a BD LSR II instrument in cohort 2. For both cohorts, viable lymphocyte subpopulations were identified by staining with fluorochrome-labeled antibodies targeting CD45RA, CCR7, CD127, and PD-1. Intracellular expression of Bcl-2 and Ki-67 was also examined after permeabilization. Gates were established using fluorochrome-labeled isotype control antibodies and indices were monitored using a BD LSR II flow cytometer and analyzed with BD FACSDiva software.
Dendritic cells (DCs) were identified as lineage negative, HLA-DR positive, and by either CD123 or CD11c (all antibodies from BD Biosciences). DCs were also characterized for expression of co-stimulatory molecules CD83, CD80, CD86 (BD Biosciences), and the negative regulator PDL-1 (eBioscience). For cohort 2, antibodies recognizing CCR7 (BD Biosciences) and ILT-7 (eBioscience) were added to the staining panel. Fluorescence Minus One controls were used to define positivity of the DC marker antigens. Cells were analyzed on a BD LSR II flow cytometer using FACSDiva software.
Plasma assays
Levels of IL-6, soluble CD14 (sCD14), tumor necrosis factor receptor type 1, and interferon gamma-induced protein 10 (IP10) in EDTA-anticoagulated plasma were measured using Quantikine ELISA kits (R&D Systems). Levels of d-dimers were measured using the Asserachrom D-Di immunoassay (Diagnostica Stago). Plasma levels of lipopolysaccharides (LPS) were quantified using the Limulus Amebocyte Lysate (LAL) assay (QCL-1000; Lonza) per the manufacturer's instructions.
For cohort 1, plasma samples were analyzed for IFNα and IFNβ levels by ELISA, and IFNα bioactivity (ELISA and iLite kits, PBL Interferon Source) per the manufacturer's directions.
Microarray assays
RNA extraction, and microarray analysis were conducted at the Yerkes NHP Genomics Core Laboratory (www.yerkes.emory.edu/nhp_genomics_core/). Whole blood was collected into RNA PAXgene tubes (QIAGEN) and purified and assessed as previously described.24 RNA was processed using the Affymetrix IVT Express Kit25 and hybridized to Human Genome U133 Plus 2.0 arrays (Affymetrix). Baseline and week 12 samples from 10 chloroquine-treated and 10 placebo participants were run blinded in cohort 1 and separately in cohort 2.
Microarray data were submitted to the GEO database according to MIAME standards. The accession numbers are Series GSE71065; off-ART dataset GSE71063; and on-ART dataset GSE71064.
Statistical methods
The primary end point was the week 12 change from baseline in percentage of CD8 T cells expressing HLA-DR and CD38. For the immunologic end points, baseline was computed as the mean of the pre-entry and entry values, and the “week 12” value as the mean of the values at weeks 10 and 12. The primary analysis excluded subjects who discontinued study treatment for ≥14 days and (1) started ART (off-ART arms) or (2) stopped ART or had HIV virological rebound (on-ART arms); otherwise, an as-treated approach was taken. Quantitative measures between arms were compared using Wilcoxon rank sum tests. Significant changes from baseline were assessed using sign tests. Spearman's rank correlation (rho) tests were used to measure associations between two measures. Two-sided tests were performed using a 5% level of significance for all but microarray analyses, and no adjustments were made for multiple testing.
Microarray data analysis
Microarray data were preprocessed and normalized using RMA in the bioconductor limma package.26 One chloroquine-treated participant in the on-ART cohort was found to be an outlier and was excluded from downstream analyses. To identify differentially expressed genes, a paired t-test was used on the paired baseline and week 12 samples from each participant in the chloroquine treatment arm of each cohort, with a gene-filtering criterion of an unadjusted p value of p = .01 (Supplementary Tables S1 and S2; Supplementary Data are available online at www.liebertpub.com/aid).
Gene Set Enrichment Analysis (GSEA) was used to detect significantly enriched pathways (www.broadinstitute.org/gsea/index.jsp), in which pathways with a nominal p-value of <.05 were considered significantly enriched. We chose genes to focus further analyses based on the following criteria (1) they were determined to be statistically differentially expressed based on the paired t-test described above (2) the collective genes in a pathway were determined to be significantly enriched by GSEA, or (3) we also ran analyses on genes know to be affected by chloroquine treatment (interferon-stimulated genes [ISGs]) or known markers of HIV-associated immune activation (MKI67, CCR5, and HLA-DR).
Results
Study population
Between March 2009 and July 2010, 33 study participants were enrolled into cohort 1; between December 2010 and November 2012, 37 study participants were enrolled into cohort 2. In the off-ART cohort 1, 16 participants were randomized to arm A and 17, to arm B. In the on-ART cohort 2, 18 and 19 were randomized to arms C and D, respectively (Table 1).
Table 1.
Baseline Demographics
| Off-ART cohort | On-ART cohort | ||||
|---|---|---|---|---|---|
| Characteristic | Total (N = 70) | Arm A (N = 16) | Arm B (N = 17) | Arm C (N = 18) | Arm D (N = 19) |
| Age (in years) | |||||
| Median (Q1, Q3) | 46 (36, 50) | 35 (29, 43) | 39 (35, 44) | 50 (48, 55) | 49 (41, 56) |
| 18–29 | 8 (11%) | 5 (31%) | 2 (12%) | 1 (6%) | 0 (0%) |
| 30–39 | 16 (23%) | 5 (31%) | 7 (41%) | 1 (6%) | 3 (16%) |
| 40–49 | 25 (36%) | 5 (31%) | 8 (47%) | 5 (28%) | 7 (37%) |
| 50–59 | 18 (26%) | 1 (6%) | 0 (0%) | 10 (56%) | 7 (37%) |
| 60–69 | 2 (3%) | 0 (0%) | 0 (0%) | 0 (0%) | 2 (11%) |
| 70–79 | 1 (1%) | 0 (0%) | 0 (0%) | 1 (6%) | 0 (0%) |
| Gender | |||||
| Male | 63 (90%) | 14 (88%) | 15 (88%) | 17 (94%) | 17 (89%) |
| Female | 7 (10%) | 2 (13%) | 2 (12%) | 1 (6%) | 2 (11%) |
| Race/Ethnicity | |||||
| White non-Hispanic | 39 (56%) | 9 (56%) | 11 (65%) | 9 (50%) | 10 (53%) |
| Black non-Hispanic | 28 (40%) | 7 (44%) | 5 (29%) | 8 (44%) | 8 (42%) |
| Hispanic (regardless of race) | 3 (4%) | 0 (0%) | 1 (6%) | 1 (6%) | 1 (5%) |
| Baseline CD4 count (cells/mm3) | |||||
| Median (Q1, Q3) | 386 (260, 541) | 641 (561, 755) | 493 (448, 541) | 259 (224, 281) | 270 (218, 296) |
| Baseline CD8 count (cells/mm3) | |||||
| Median (Q1, Q3) | 740 (540, 1,094) | 920 (632, 1,108) | 1,083 (737, 1,260) | 640 (454, 985) | 656 (414, 842) |
| Entry HIV-1 RNA | |||||
| ≤LLQ | 33 (47%) | 0 (0%) | 0 (0%) | 16 (89%) | 17 (89%) |
| >LLQ | 37 (53%) | 16 (100%) | 17 (100%) | 2 (11%) | 2 (11%) |
| Entry log10 HIV-1 RNA (copies/ml) | |||||
| Median (Q1, Q3) | 4.46 (4.03, 4.75)a | 4.48 (4.02, 4.74) | 4.42 (4.03, 4.83) | N/Ab | N/Ab |
Arms A and B only (N = 33).
HIV-1 RNA values of 30 and 48 copies (LLQ = 20), 129 (LLQ = 40), and 75 (LLQ = 50).
ART, antiretroviral therapy; LLQ, lower limit of quantification.
In the off-ART cohort, 6 of the 33 participants (3 in arm A, 3 in arm B) who started study treatment discontinued study treatment prematurely. In the on-ART cohort, 3 (all from arm C) discontinued prematurely. None of the study treatment discontinuations was related to study treatment-related adverse events. None of the ART-treated participants interrupted ART during the study.
Safety
No adverse events attributable to the study treatment occurred. There were no opportunistic infections or death. One episode of pneumonia in arm C occurred during treatment with chloroquine.
Immunologic evaluations
In the off-ART cohort, 31 participants qualified for the analyses of the first 12 weeks of the study; 28 participants were included in the analyses of changes after 12 weeks of chloroquine treatment in both arms (before or after crossover). In the on-ART cohort, 35 and 34 participants were included in these analyses, respectively.
Cellular immune activation markers in off-ART cohort
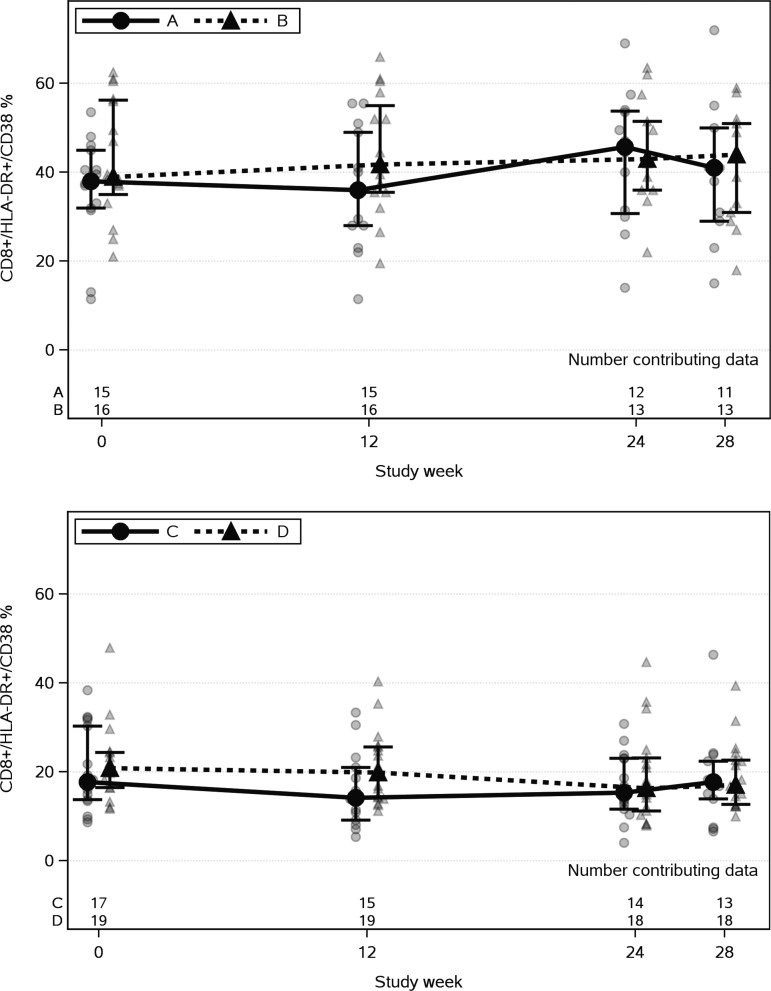
The median week-12 decrease from baseline CD8 T-cell activation (% HLA-DR and CD38+) was larger in arm A (chloroquine) than in arm B (placebo), but not significantly (−2.0% vs. −0.5%, p = .428, Table 2 and Fig. 1). There was not a significant week 12 change from baseline in arm A. Combining the changes after 12 weeks of chloroquine in both arms also showed no significant change. Similarly, no significant change from baseline in the percentage of CD4 T cells expressing HLA-DR and CD38 was seen after 12 weeks of chloroquine.
Table 2.
Immunologic and Virological End Points
| Off-ART cohort | On-ART cohort | |||||
|---|---|---|---|---|---|---|
| Week 12 change from baseline | Arm A | Arm B | p* | Arm C | Arm D | p* |
| CD8+/HLA-DR+/CD38+ % | −2.0 (−10.0, 9.0) | −0.5 (−2.3, 2.8) | .428 | −3.1 (−4.3, 0.4) | −1.2 (−3.3, 1.4) | .247 |
| CD8+/CD38+ % | 0.0 (−9.5, 3.5) | 0.3 (−2.0, 2.0) | .961 | −0.8 (−3.7, 3.3) | −2.5 (−4.5, 4.2) | .800 |
| CD8+/HLA-DR+ % | −3.5 (−9.0, 8.0) | 1.8 (−1.5, 3.5) | .313 | −3.1 (−6.9, −1.0) | −1.4 (−4.6, 0.2) | .093 |
| CD4+/HLA-DR+/CD38+ % | 0.0 (−3.0, 1.5) | 0.5 (−1.8, 3.0) | .428 | −0.9 (−2.1, 0.0) | −0.1 (−1.8, 0.6) | .436 |
| CD4+/CD38+ % | 1.0 (−6.0, 4.0) | 0.5 (−2.8, 2.5) | .946 | −0.6 (−2.4, 2.3) | −1.1 (−3.1, 1.4) | .876 |
| CD4+/HLA-DR+ % | 0.0 (−3.0, 1.0) | 0.0 (−1.8, 2.8) | .500 | −1.6 (−3.5, −0.5) | −0.5 (−2.4, 1.4) | .107 |
| CD8+ count (cells/mm3) | −36 (−83, 43) | 3 (−120, 86) | .740 | −18 (−67, 6) | 19 (−39, 66) | .208 |
| CD8+ % | 2.0 (−1.0, 2.5) | −0.3 (−1.3, 1.3) | .080 | 0.5 (−1.0, 1.8) | −0.3 (−1.0, 1.0) | .567 |
| CD4+ count (cells/mm3) | −27 (−150, 4) | −11 (−38, 71) | .206 | −6 (−21, 10) | 7 (−17, 23) | .247 |
| CD4+ % | 0.0 (−2.0, 0.5) | 1.0 (0.0, 1.5) | .035 | 1.0 (−1.0, 1.0) | −0.2 (−2.0, 0.5) | .418 |
| Log10 HIV-1 RNA (c/ml) | 0.29 (0.15, 0.35) | −0.20 (−0.27, −0.02) | <.001 | N/A | N/A | N/A |
| Central memory | ||||||
| CD8+ CM% | 0.10 (−0.10, 0.60) | 0.10 (−0.13, 0.48) | .868 | 0.38 (−0.05, 0.95) | 0.10 (−0.35, 0.40) | .282 |
| CD8+ CM Bcl2+% | −0.58 (−1.15, 3.50) | 0.62 (−1.10, 2.15) | .735 | 0.04 (−1.98, 1.20) | 0.01 (−1.52, 1.33) | .756 |
| CD8+ CM ki-67+ % | 0.15 (−0.25, 2.55) | 0.05 (−1.02, 1.17) | .394 | −0.07 (−0.37, 0.30) | 0.12 (−0.49, 0.45) | .935 |
| CD8+ CM Bcl2+/ki-67+ % | −0.05 (−0.20, 1.55) | 0.30 (−0.02, 0.73) | .814 | −0.09 (−0.30, 0.29) | 0.06 (−0.49, 0.38) | .731 |
| CD8+ CM CD127+ % | −3.20 (−8.20, 0.85) | 0.63 (−5.43, 5.38) | .277 | 0.96 (0.22, 3.63) | 0.41 (−1.86, 1.80) | .321 |
| CD8+ CM PD-1+ % | 1.75 (−5.65, 7.35) | −3.05 (−5.25, 1.98) | .440 | −1.28 (−4.31, 0.94) | 0.01 (−2.97, 2.23) | .843 |
| CD8+ CM CD127+/PD-1+ % | −0.60 (−6.50, 3.10) | 0.53 (−3.80, 4.18) | .464 | −0.37 (−2.34, 1.61) | 0.26 (−3.53, 2.22) | .957 |
| CD4+ CM% | 0.40 (−2.05, 2.15) | 1.20 (−0.75, 3.27) | .572 | −0.05 (−4.50, 1.90) | −0.65 (−3.00, 1.25) | .767 |
| CD4+ CM Bcl2+ % | −0.25 (−0.60, 1.80) | −0.20 (−0.55, 1.35) | .629 | −0.48 (−3.03, 1.23) | 0.36 (−3.00, 1.03) | .589 |
| CD4+ CM ki-67+ % | 0.25 (0.00, 0.40) | −0.40 (−0.65, 0.60) | .382 | −0.29 (−0.61, 0.56) | 0.03 (−0.50, 0.66) | .832 |
| CD4+ CM Bcl2+/ki-67+ % | 0.02 (−0.30, 0.35) | −0.05 (−0.40, 0.43) | .782 | −0.15 (−0.57, 0.30) | −0.28 (−0.50, 0.29) | .935 |
| CD4+ CM CD127+ % | −1.55 (−4.15, −0.05) | 0.95 (−2.85, 2.20) | .160 | 0.74 (−1.74, 4.06) | 0.06 (−3.36, 1.30) | .627 |
| CD4+ CM PD-1+ % | −0.15 (−2.50, 6.35) | −1.15 (−4.60, 2.38) | .281 | −0.48 (−3.72, 2.24) | −0.39 (−5.05, 0.93) | .304 |
| CD4+ CM CD127+/PD-1+ % | −0.65 (−3.45, 3.30) | −0.65 (−3.45, 1.85) | .830 | 0.34 (−1.30, 3.34) | −0.95 (−4.23, 1.12) | .358 |
| Dendritic cells | ||||||
| % mDC of total DCs | 5.25 (−1.23, 7.70) | −1.68 (−4.78, 2.18) | .060 | 1.14 (−4.02, 7.40) | 0.47 (−5.37, 3.72) | .523 |
| % mDC of total viable singlets | 0.14 (−0.003, 0.647) | −0.004 (−0.076, 0.076) | .033 | −0.01 (−0.09, 0.10) | −0.02 (−0.12, 0.04) | .589 |
| % mDC CD80+ | 0.10 (−0.73, 0.28) | 0.30 (0.01, 0.70) | .045 | 0.27 (−0.37, 0.55) | −0.07 (−0.34, 0.30) | .317 |
| % mDC CD83+ | 5.78 (−6.21, 8.44) | 3.26 (−5.61, 10.36) | .740 | −11.30 (−21.47, 16.78) | −3.02 (−6.55, 6.33) | .461 |
| % mDC CD86+ | 0.63 (−1.78, 4.08) | −0.82 (−2.26, 0.58) | .220 | −0.86 (−2.78, 0.90) | 0.14 (−3.25, 1.90) | .731 |
| % mDC PDL-1+ | −0.24 (−1.42, 1.30) | 1.79 (−2.27, 7.63) | .299 | 0.63 (−0.80, 2.64) | −0.42 (−1.30, 3.38) | .832 |
| % pDC of total DCs | −4.55 (−6.03, −0.95) | 2.35 (−3.04, 4.60) | .064 | −1.14 (−7.40, 4.02) | −0.47 (−3.72, 5.37) | .523 |
| % pDC of total viable singlets | 0.02 (−0.02, 0.21) | −0.01 (−0.05, 0.09) | .453 | −0.01 (−0.06, 0.06) | 0.01 (−0.05, 0.07) | .446 |
| % pDC CD80+ | −0.004 (−0.05, 0.00) | 0.001 (0.000, 0.053) | .056 | 0.003 (−0.07, 0.08) | −0.001 (−0.05, 0.14) | .838 |
| % pDC CD83+ | 0.05 (−11.20, 13.61) | 1.21 (−10.28, 6.45) | .984 | −4.95 (−14.59, 0.53) | −0.23 (−11.36, 2.43) | .523 |
| % pDC CD86+ | −1.09 (−2.49, 0.50) | 0.08 (−1.14, 0.83) | .446 | 0.91 (−3.40, 4.04) | 0.75 (−2.35, 3.84) | .781 |
| % pDC PDL-1+ | −0.36, −0.86, 0.73) | −0.17 (−1.28, 2.22) | .892 | 0.08 (−2.92, 1.57) | −0.13 (−1.17, 2.86) | .756 |
| Cytokines | ||||||
| IL-6 (pg/ml) | 0.18 (−0.18, 0.57) | 0.11 (−0.07, 0.60) | .953 | 0.08 (−0.43, 0.30) | 0.01 (−0.40, 0.48) | .779 |
| LPS (pg/ml) | 3.14 (0.32, 12.33) | 0.00 (−0.86, 7.23) | .173 | 0.00 (−3.50, 1.00) | 0.25 (0.00, 1.50) | .095 |
| Type I IFN (U/ml) | 0.00 (0.00, 0.00) | 0.00 (−0.06, 0.00) | .752 | Assay not done | Assay not done | N/A |
| IFN-Beta (pg/ml) | 0.02 (−0.12, 0.28) | −0.03 (−0.08, 0.00) | .056 | Assay not done | Assay not done | N/A |
| IFN-Gamma IP-10 (pg/ml) | −9.72 (−74.37, 55.89) | −9.98 (−28.32, 32.62) | .861 | −20.60 (−42.43, −6.10) | −8.68 (−21.31, 12.76) | .168 |
| sTNFr-I (pg/ml) | −58.85 (−79.25, 28.21) | −56.37 (−173.40, 146.95) | .599 | 3.42 (−256.99, 57.07) | 55.57 (−58.84, 138.36) | .125 |
| sCD14 (million pg/ml) | 0.02 (−0.13, 0.21) | −0.06 (−0.23, 0.25) | .520 | 0.03 (−0.11, 0.13) | 0.03 (−0.09, 0.10) | .866 |
| d-Dimer (ng/ml) | 28.29 (−65.86, 61.66) | 6.45 (−45.61, 72.65) | .892 | 17.01 (−0.57, 50.92) | 6.92 (−3.63, 17.46) | .283 |
Wilcoxon rank sum test p values to compare chloroquine and placebo arms within each cohort.
CM, central memory; DC, dendritic cells; IL, interleukin; LPS, lipopolysaccharides; mDC, myeloid DC; pDC, plasmacytoid DC.
FIG. 1.
CD8+/HLA-DR+/CD38+ % for off-ART (arms A and B) and on-ART (arms C and D) cohorts at study weeks. Connected bold filled symbols represent median, and the bars represent interquartile ranges. Arms A and C received chloroquine for the first 12 weeks, then placebo. Arms B and D received placebo for the first 12 weeks, then chloroquine. The number of participants varied over the study weeks, due to missed visits or inadequate samples. ART, antiretroviral therapy.
Plasma HIV RNA changes and relationship to cellular immune activation markers in the Off-ART cohort
There was a significant increase from baseline in plasma HIV-1 RNA in arm A after 12 weeks of chloroquine (median 0.29 log10 increase, p < .001) that was greater than the change in the placebo arm B (median 0.20 log10 decrease, p = .077); the difference between the arms was statistically significant (p < .001, Table 2). The combined changes in both arms after 12 weeks of chloroquine also showed a significant increase from baselines (median 0.30 log10HIV-1 RNA, p < .001). HIV RNA levels fell 12 weeks after stopping chloroquine (arm A) back to levels not significantly different from baseline (p = .388).
Combining data from both arms yielded a significant positive association between changes in log10HIV-1 RNA and the proportion of CD8 T cells expressing HLA-DR and CD38 (rho = 0.485, p = .009). Thus, we suspected that the increase in HIV levels as a result of chloroquine use may have confounded any effect of chloroquine on immune activation in the off-ART cohort.
Cellular immune activation markers in the on-ART cohort
There was a trend of a week 12 decrease from baseline in the proportions of CD8 T cells expressing HLA-DR and CD38 in arm C during chloroquine treatment (median −3.1%, p = .077). The week-12 changes from baseline were not significantly different between arms C and D (p = .247, Table 2 and Fig. 1). Combining the changes in the proportion of CD8 T cells expressing HLA-DR and CD38 after 12 weeks of chloroquine treatment in both arms showed a significant decrease in CD8 T-cell activation (median decrease = 3.0%, p = .003). A separate analysis within arm D gave similar results (median decrease = 2.9% during chloroquine treatment, p = .031).
No significant change from baseline occurred in the percentage of CD4 T cells expressing HLA-DR and CD38 after 12 weeks of chloroquine (arm C). Although there was a significant decrease from baseline at week 12 in this percentage within arm C (p = .021), changes were not significantly different between the two arms (p = .107, Table 2). There were no significant changes in either the percentage of CD4 T cells expressing HLA-DR and CD38 (p = .121) or the percentage of CD4 T cells expressing CD38 (p = 1.000) after 12 weeks of chloroquine in both arms combined, but there was a significant change in the proportion of CD4+ T cells expressing HLA-DR (median decrease of 1.5%, p = .003).
CD4 T-cell counts
There were no significant changes from baseline in CD4 T-cell counts after 12 weeks of chloroquine in either arm A (off-ART) or arm C (on-ART) and no significant differences in the changes between the chloroquine arm and the placebo arm in either cohort (Table 2). Combining the changes in CD4 T-cell count after 12 weeks of chloroquine in both arms in each cohort indicated a significant median decrease in each during chloroquine administration (off-ART: −39 cells/mm3, p = .036; on-ART: −15 cells/mm3, p = .024).
T-cell memory subsets
Among the markers measured on CD4 and CD8 central memory T cells, arm A (off-ART) and the combined off-ART arms showed a significant median decrease after 12 weeks of chloroquine in the proportions of CD4+ cells expressing the IL-7 receptor alpha chain, CD127 (−1.55%, p = .035, Table 2; −1.18%, p = .036, respectively).
The chloroquine arm in both cohorts showed no significant changes in any of the myeloid DC (mDC) and plasmacytoid DC (pDC) markers measured (Table 2). Combined data from both arms after 12 weeks of chloroquine showed a significant median change from baseline only in the proportion of pDCs expressing CD83 in the on-ART cohort (median decrease = 2.52%, p = .009).
Cytokines and soluble activation markers
Of the cytokines and soluble activation markers measured, arm A (chloroquine) in the off-ART cohort showed a significant week 12 median change only in LPS (+3.14 pg/ml, p = .013) levels. There was a trend toward a decrease in IP10 after 12 weeks of chloroquine treatment in the on-ART arm C (−20.6 pg/ml, p = .057). The differences between the two arms within either cohort in week 12 changes from baseline in all markers were not significant (p > .05, Table 2). When both arms within each cohort were combined, after 12 weeks of chloroquine there was a significant median change only in IP10 in the on-ART cohort (−12.77 pg/ml, p = .011).
Microarray analyses
Off-ART cohort (arms A and B)
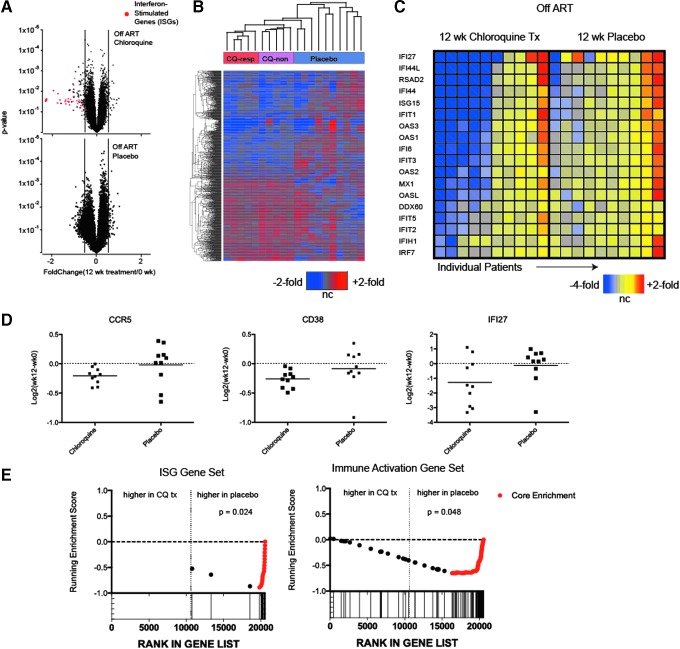
The effect of chloroquine on the blood transcriptome was modest, and a low number of differentially expressed genes was observed. Nevertheless, a handful of genes were downregulated after chloroquine treatment beyond the technical and clinical variability predicted by the placebo analysis (Fig. 2A). Manual inspection, volcano plots, and hierarchical clustering of the top 1,000 genes with the highest variance after chloroquine treatment showed that the majority of the downregulated genes were ISGs. Five of 10 (50%) of the chloroquine-treated participants had marked downregulation of ISGs compared to none in the control group (Fig. 2B and Supplementary Table S1).
FIG. 2.
Transcriptome response in whole blood to chloroquine treatment in therapy-naïve (off-ART) HIV-infected participants. RNA from whole blood was obtained before treatment and after 12 weeks of chloroquine or placebo administration and was hybridized to human Affymetrix arrays. Differential expression was assessed using paired t-tests of the chloroquine and placebo data sets independently. (A) Volcano plots showing distribution of fold-changes (x-axis) compared to paired t-test p-values (y-axis) of all genes on the array. Red dots on the upper panel indicate probe sets specific for ISGs. (B) Two-way hierarchical clustering of 1,000 genes exhibiting most variance in the chloroquine-treated participants. The color scale is shown at the bottom of the panel; ratios are relative to pretreatment samples. (C) Two-way clustering and heat map of ISGs. Each column represents individual participants. The color scale is shown at the bottom of the panel; ratios are relative to pretreatment samples. (D) Plot of fold-changes for individual participants, for select immune activation genes. Data points depict the log ratio of fold-changes for individual participants; bars indicate means. (E) Gene set enrichment plots depicting enrichment testing of ISGs and immune activation gene sets defined in studies of simian immunodeficiency virus infection (details in the Materials and Methods section). The microarray data from week 12 postchloroquine treatment samples were directly compared to data from week 12 placebo treatment; the x-axis depicts the ranking of nonredundant, annotated genes contained on the array according their signal-to-noise ratio; genes were ranked from those upregulated in the chloroquine-treated samples on the left to those upregulated in placebo samples on the right. Individual genes from each gene set are depicted by both dots and vertical lines at the bottom. Genes in the leading edge that contribute most significantly to the enrichment are depicted in red. The significance of the enrichment statistic is shown in the upper right quadrant. ISGs, interferon-stimulated genes.
Of genes reflecting immune activation, CD38 (−1.198-fold change; p = .0003) and CCR5 (−1.15 average fold change, p < .001) were moderately but significantly reduced in 10/10 participants (Fig. 2A, D). In contrast, expression of these genes in the placebo-treated participants was randomly distributed.
We also compared the week 12 chloroquine and placebo data sets for signatures of ISGs and immune activation using gene-set enrichment analysis with gene sets we had established previously using primary data27; we found a strong and statistically significant enrichment of both pathways in the placebo group, with lowered expression of these pathways in the chloroquine group (Fig. 2E). This complementary analysis strategy provided additional evidence that chloroquine had a suppressive effect on ISG and immune activation gene expression in the off-ART cohort.
On-ART cohort (arms C and D)
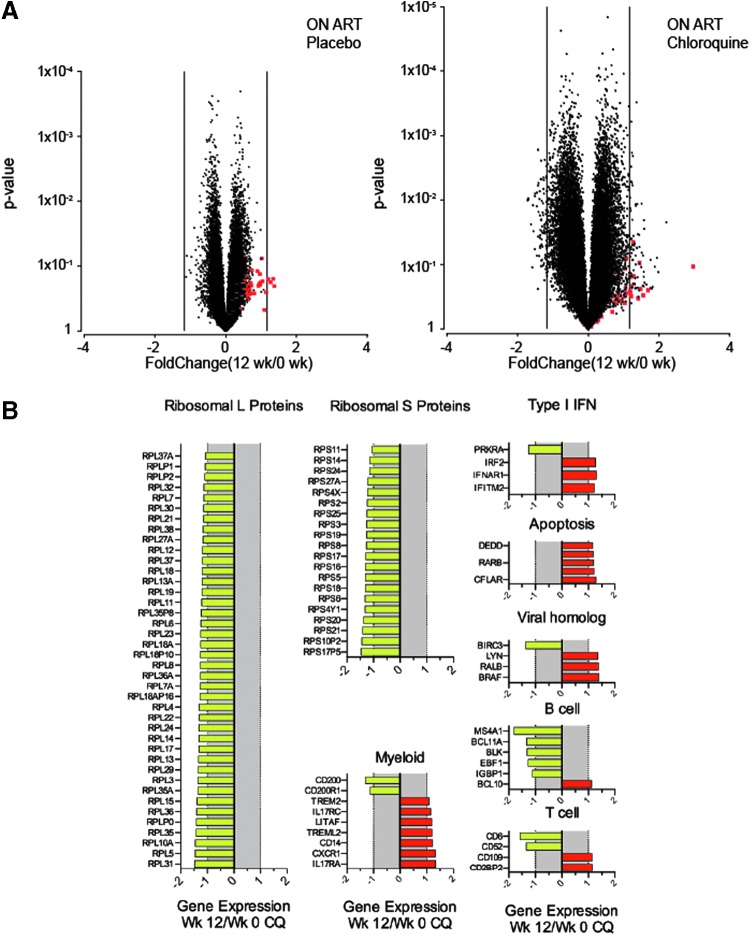
A large number of genes associated with the 60S (ribosomal L proteins, RPL genes) and 40S (ribosomal S proteins, RPS genes) subunits, 72 probe sets representing 40 unique RPL genes and 32 probe sets (20 unique genes) of RPS genes, were downregulated after chloroquine treatment. None of these probe sets was downmodulated after placebo administration. In contrast to our observations in the off-ART cohort, we did not observe any effect of chloroquine on expression of classical immune activation marker genes (MKI67, CD38, or HLA-DR), although there was strong downregulation of CD6, which has been considered a putative co-stimulatory molecule on T cells (Fig. 3A, B and Supplementary Table S2).28
FIG. 3.
Gene expression profiling of clinical response to chloroquine treatment in HIV-infected participants receiving ART (on-ART). Microarray analysis was conducted as described in Figure 2 on RNA from whole blood samples collected at pretreatment and after 12 weeks of chloroquine or placebo. Significance of statistical differential expression induced by the 12-week treatment phase was tested by performing paired t-tests separately for the chloroquine and placebo data sets. (A) Volcano plots depicting fold-changes (x-axis) versus paired t-test p-values (y-axis) of all genes on the array. Red dots depict probe sets specific for ISGs. (B) Breakout expression plots of gene families with differential expression after 12 weeks of chloroquine treatment.
Discussion
In this randomized, double-blind, placebo-controlled, crossover study, chloroquine modestly reduced immune activation in ART-treated HIV-infected participants. The primary analyses compared the changes across treatment arms in the proportion of CD8 T cells co-expressing CD38 and HLA-DR in the first 12 weeks before crossover, but the crossover design provided greater statistical power in secondary analyses. When the periods of chloroquine use before and after crossover in both arms of the on-ART study were combined, there was a significant reduction in the proportions of CD8 T cells co-expressing CD38 and HLA-DR and in CD4 T cells expressing HLA-DR. The effect was modest, however, representing only a reduction of 3% in the percentage of activated CD8 T cells. Furthermore, no consistent effect on soluble markers of inflammation was observed, and these markers have a stronger association with clinical outcomes in patients on ART than do cellular markers of immune activation.11,29
Any effect on immune activation in the off-ART study was confounded by enhancement of viral replication by chloroquine, which itself may have driven greater immune activation as suggested by a significant correlation between changes in plasma viremia and CD8 T-cell activation. Assessment in the on-ART cohort removed HIV replication as a potentially confounding driver of activation, although it is possible that chloroquine could have induced low-level viral replication, only detectable by single copy assay, in this cohort.
Transcriptional analyses revealed that approximately half of those receiving chloroquine in the off-ART cohort had decreased expression of ISGs, despite an increase in plasma viremia. Typically, ISG expression increases with HIV-1 replication. We have observed in the nonhuman primate model after depletion of CD8 T cells that even modest (0.5 log) increases in viremia increase ISG expression.30 It is interesting that, in this study, we observed an “uncoupling” of ISG expression and plasma viremia. We also observed a modest but consistent decrease in CD38 and CCR5 RNAs. In the on-ART cohort, no effect of chloroquine on genes associated with immune activation was discerned, but suppressive ART appeared to unmask a suppressive effect of chloroquine on genes encoding proteins associated with the 40 S and 60 S ribosomal subunits. The significance of this finding is unclear, but it could indicate that chloroquine influences ribosomal activity and the control of gene expression at the translation step.
Although earlier studies found a reduction in or no effect on viremia during chloroquine or hydrochloroquine administration,31–33 a modest increase was seen in one controlled trial of hydroxychloroquine in persons off ART.34 Similar increases in plasma HIV levels had been seen in earlier studies of immunosuppressive agents: cyclosporine A,35 prednisone,36 and thalidomide37 in untreated HIV infection.
The mechanism whereby chloroquine enhances HIV replication is unclear. Conceivably, blockade of TLR7/TLR8 signaling may limit induction of host antiviral defenses such as those mediated by type 1 interferons. Alternatively, the effect on plasma viremia could have been the result of a direct enhancement of infectivity by chloroquine, a finding in some in vitro studies,38,39 while others have shown chloroquine to inhibit HIV replication.40,41
Other researchers have assessed the effects of chloroquine or hydroxychloroquine, a related compound, on immune activation in HIV-infected participants. One small study among participants not receiving ART reported a reduction in the frequency of CD38+HLA-DR+CD8 T cells and Ki-67 expression in both CD8 and CD4 T cells after 2 months of chloroquine treatment (six participants received 250 mg/day, three received 500 mg/day) that was not seen in the three placebo recipients.42 Two earlier randomized controlled trials using hydroxychloroquine 800 mg/day (bioequivalent to 500 mg/day of chloroquine) found reduced plasma levels of IL-6, but not IL-1α, IL-1β, or TNF-α, after 8 weeks of treatment.31,32 In comparison, a randomized, double-blind, placebo-controlled study of 48 weeks of hydroxychloroquine 400 mg/day in 83 participants not receiving ART found no effect on the frequency of CD38+HLA-DR+CD8 T cells or CD38+HLA-DR+CD4 T cells, or on Ki67 expression in CD8 or CD4 T cells, or on plasma levels of inflammatory cytokines or d-dimers.34
In an open-label study, administering hydroxychloroquine 400 mg/day for 6 months to 20 ART-treated participants led to a nonsignificant decrease in the frequency of CD38+HLA-DR+CD8 T cells. CD4 T-cell cycling (Ki67) and monocyte activation (CD69) fell. T regulatory cells increased; an increased pDC population produced less interferon-α; and plasma levels of LPS, IL-6, and TNF-α fell.43 Another uncontrolled study of chloroquine 250 mg/day for 24 weeks in 19 ART-treated participants found no reduction in T-cell activation, the frequency of pDCs; or plasma levels of d-dimer, C-reactive protein, or inflammatory cytokines.44
CD4 T-cell counts decreased slightly with chloroquine use in our study. The decline was lower in participants receiving ART (−15 vs. −39 cells/mm3, combining chloroquine periods across cohort arms), suggesting that at least some of the decrease might have been the result of enhanced viral replication whereas decreases in the on-ART arm might reflect other effects of the drug. One recent study of chloroquine treatment in ART-naïve participants reported an additional decline of CD4 T cells of 62 cells/mm3 relative to the placebo group,32 whereas a second study of therapy-naïve chloroquine treatment reported no effect on CD4 T-cell levels.42 Whether this effect of chloroquine on CD4 T cell counts would persist or increase over time and place persons at risk for morbidity is unknown.
In summary, short-term administration of chloroquine 250 mg/day to persons with chronic HIV infection, treated or not with ART, had only modest effects on cellular indices of immune activation, activities unlikely to elicit substantial clinical benefit. It is conceivable that higher doses of chloroquine would have greater activity in this regard, but might also be more toxic.45 In those not receiving ART, HIV RNA levels in plasma rose during chloroquine administration, likely reflecting the adverse effects of immune suppression on viral control. Our results suggest that chloroquine and related agents are not likely to confer meaningful clinical benefit in HIV infection. It remains to be seen in treated HIV infection, where immune activation, coagulation, and inflammation have been linked to morbid outcomes, whether other interventions that can attenuate these indices are associated with lower risks of morbid events.
Supplementary Material
Acknowledgments
We are indebted to Pamela Fried for editorial assistance, the clinicians who referred patients to the study, and patients who participated. This work was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (Award Numbers UM1 AI068634, AI068636, AI76174, AI36219, and UM1 AI106701). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. AIDS Clinical Trial Group Site Grant Numbers: Case CRS (Site 2501) Grant UM1 AI069501; MetroHealth CRS (Site 2503) Grant UM1 AI 6950742; Penn Therapeutics CRS (Site 6201) Grant UM1 AI069534-09; Georgetown University CRS (Site 1008) Grant UM1 AI069494, CFAR P30-AI045008-17; Washington University in St. Louis CRS (Site 2101) Grant U01 AI69439; Johns Hopkins University CRS (Site 0201) Grant 2UM1 AI069465, UL1TR001079, Institute for Clinical and Translational Research; University of Pittsburgh CRS (Site 1001) Grant UM1AI069494; Vanderbilt University CRS (Site 3652) Grant AI069439, TR000445; University of Colorado CRS (Site 6101) Grant AI069432, UL1 TR001082; Chapel Hill CRS (Site 3201) Grants UM1 AI069423, CTSA: UL1TR001111, CFAR: P30 AI050410; Alabama CRS (Site 31788) Grant 5UM1AI069452-10; UC San Diego CRS (Site 701) Grant AI069432; Weill Cornell-Chelsea CRS (Site 7804) Grant UM1AI069419, UL1TR000457; Massachusetts General Hospital CRS (Site 101) Grant 2UM1AI069412-08; University of Cincinnati CRS (Site 2401) Grant UM1AI068636. We would also like to acknowledge the following additional investigators at participating sites: Case CRS: Kristen Allen, RN and Jane Baum RN; MetroHealth: Kim Whitely, RN and Traci Davis, RN; CRS–Penn Therapeutics: Wayne Wagner, RN; Georgetown University CRS: Joseph G Timpone, Jr, MD; Washington University in St Louis CRS: Michael Klebert, PhD and Michael Royal, RPh; Johns Hopkins University CRS: Ilene Wiggins, RN and Andrea Weiss, BPharm; University of Pittsburgh CRS: Deborah McMahon, MD and Renee Weinman, BS CCRP; Vanderbilt University CRS: Marcia Free, RN and Michael Morgan, RN, FNP; University of Colorado CRS: M Graham Ray, RN and Thomas B Campbell, MD; Chapel Hill CRS: Jonathan Oakes, BA and Susan Blevins, MS ANP; Alabama CRS: Elizabeth Lindsey, RN and Kamellia Safavy, MSc; UC San Diego CRS: Kathleen Nuffer, NP and Dee Dee Pacheco; Weill Cornell-Chelsea CRS: Valery Hughes, NP and Todd Stroberg, RN; Massachusetts General Hospital CRS: Teri Flynn, ANP-BC and Amy Sbrolla, RN BSN; University of Cincinnati CRS: Eva Whitehead, RN, BSN and Carl J. Fichtenbaum, MD.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Brenchley JM, Price DA, Schacker TW, et al. : Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med 2006;12:1365–1371 [DOI] [PubMed] [Google Scholar]
- 2.Malaspina A, Moir S, Kottilil S, et al. : Deleterious effect of HIV-1 plasma viremia on B cell costimulatory function. J Immunol 2003;170:5965–5972 [DOI] [PubMed] [Google Scholar]
- 3.Hazenberg MD, Stuart JW, Otto SA, et al. : T-cell division in human immunodeficiency virus (HIV)-1 infection is mainly due to immune activation: a longitudinal analysis in participants before and during highly active antiretroviral therapy (HAART). Blood 2000;95:249–255 [PubMed] [Google Scholar]
- 4.Connolly NC, Riddler SA, Rinaldo CR: Proinflammatory cytokines in HIV disease-a review and rationale for new therapeutic approaches. AIDS Rev 2005;7:168–180 [PubMed] [Google Scholar]
- 5.Valdez H, Lederman MM: Cytokines and cytokine therapies in HIV infection. AIDS Clin Rev 1997–1998:187–228 [PubMed] [Google Scholar]
- 6.Deeks SG, Kitchen CM, Liu L, et al. : Immune activation set point during early HIV infection predicts subsequent CD4+ T-cell changes independent of viral load. Blood 2004;104:942–947 [DOI] [PubMed] [Google Scholar]
- 7.Giorgi JV, Lyles RH, Matud JL, et al. : Predictive value of immunologic and virologic markers after long or short duration of HIV-1 infection. J Acquir Immune Defic Syndr 2002;29:346–355 [DOI] [PubMed] [Google Scholar]
- 8.Fahey JL, Taylor JM, Manna B, et al. : Prognostic significance of plasma markers of immune activation, HIV viral load and CD4 T-cell measurements. AIDS 1998;12:1581–1590 [DOI] [PubMed] [Google Scholar]
- 9.Hunt PW, Martin JN, Sinclair E, et al. : T cell activation is associated with lower CD4+ T cell gains in human immunodeficiency virus-infected participants with sustained viral suppression during antiretroviral therapy. J Infect Dis 2003;187:1534–1543 [DOI] [PubMed] [Google Scholar]
- 10.Lederman MM, Calabrese L, Funderburg NT, et al. : Immunologic failure despite suppressive antiretroviral therapy is related to activation and turnover of memory CD4 cells. J Infect Dis 2011;204:1217–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tenorio AR, Zheng Y, Bosch RJ, et al. : Soluble markers of inflammation and coagulation but not T-cell activation predict non-AIDS-defining morbid events during suppressive antiretroviral treatment. J Infect Dis 2014;210:1248–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Emu B, Sinclair E, Favre D, et al. : Phenotypic, functional, and kinetic parameters associated with apparent T-cell control of human immunodeficiency virus replication in individuals with and without antiretroviral treatment. J Virol 2005;79:14169–14178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benito JM, Lopez M, Lozano S, et al. : Differential upregulation of CD38 on different T-cell subsets may influence the ability to reconstitute CD4+ T cells under successful highly active antiretroviral therapy. J Acquir Immune Defic Syndr 2005;38:373–381 [DOI] [PubMed] [Google Scholar]
- 14.Brenchley JM, Price DA, Douek DC: HIV disease: fallout from a mucosal catastrophe? Nat Immunol 2006;7:235–239 [DOI] [PubMed] [Google Scholar]
- 15.Sandler NG, Douek DC: Microbial translocation in HIV infection: causes, consequences and treatment opportunities. Nat Rev Microbiol 2012;10:655–666 [DOI] [PubMed] [Google Scholar]
- 16.Equils O, Schito ML, Karahashi H, et al. : Toll-like receptor 2 (TLR2) and TLR9 signaling results in HIV-long terminal repeat trans-activation and HIV replication in HIV-1 transgenic mouse spleen cells: implications of simultaneous activation of TLRs on HIV replication. J Immunol 2003;170:5159–5164 [DOI] [PubMed] [Google Scholar]
- 17.Iwasaki A, Medzhitov R: Toll-like receptor control of the adaptive immune responses. Nat Immunol 2004;5:987–995 [DOI] [PubMed] [Google Scholar]
- 18.Trinchieri G, Sher A: Cooperation of Toll-like receptor signals in innate immune defence. Nat Rev Immunol 2007;7:179–190 [DOI] [PubMed] [Google Scholar]
- 19.Lenert PS: Targeting Toll-like receptor signaling in plasmacytoid dendritic cells and autoreactive B cells as a therapy for lupus. Arthritis Res Ther 2006;8:203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Savarino A, Boelaert JR, Cassone A, Majori G, Cauda R: Effects of chloroquine on viral infections: an old drug against today's diseases? Lancet Infect Dis 2003;3:722–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hong Z, Jiang Z, Liangxi W, et al. : Chloroquine protects mice from challenge with CpG ODN and LPS by decreasing proinflammatory cytokine release. Int Immunopharmacol 2004;4:223–234 [DOI] [PubMed] [Google Scholar]
- 22.Rutz M, Metzger J, Gellert T, et al. : Toll-like receptor 9 binds single-stranded CpG-DNA in a sequence- and pH-dependent manner. Eur J Immunol 2004;34:2541–2550 [DOI] [PubMed] [Google Scholar]
- 23.Wozniacka A, Lesiak A, Narbutt J, Kobos J, Pavel S, Sysa-Jedrzejowska A: Chloroquine treatment reduces the number of cutaneous HLA-DR+ and CD1a+ cells in participants with systemic lupus erythematosus. Lupus 2007;16:89–94 [DOI] [PubMed] [Google Scholar]
- 24.Bosinger SE, Li Q, Gordon SN, et al. : Global genomic analysis reveals rapid control of a robust innate response in SIV-infected sooty mangabeys. J Clin Invest 2009;119:3556–3572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taaffe JE, Bosinger SE, Del Prete GQ, et al. : CCR5 blockade is well tolerated and induces changes in the tissue distribution of CCR5+ and CD25+ T cells in healthy, SIV-uninfected rhesus macaques. J Med Primatol 2012;41:24–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klatt NR, Bosinger SE, Peck M, et al. : Limited HIV infection of central memory and stem cell memory CD4+ T cells is associated with lack of progression in viremic individuals. PLoS Pathog 2014;10:e1004345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rotger M, Dalmau J, Rauch A, et al. : Comparative transcriptomics of extreme phenotypes of human HIV-1 infection and SIV infection in sooty mangabey and rhesus macaque. J Clin Invest 2011;121:2391–2400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oliveira MI, Goncalves CM, Pinto M, et al. : CD6 attenuates early and late signaling events, setting thresholds for T-cell activation. Eur J Immunol 2012;42:195–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hunt PW, Sinclair E, Rodriguez B, et al. : Gut epithelial barrier dysfunction and innate immune activation predict mortality in treated HIV infection. J Infect Dis 2014;210:1228–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bosinger SE, Jochems SP, Folkner KA, Hayes TL, Klatt NR, Silvestri G: Transcriptional profiling of experimental CD8(+) lymphocyte depletion in rhesus macaques infected with simian immunodeficiency virus SIVmac239. J Virol 2013;87:433–443.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sperber K, Louie M, Kraus T, et al. : Hydroxychloroquine treatment of participants with human immunodeficiency virus type 1. Clin Ther 1995;17:622–636 [DOI] [PubMed] [Google Scholar]
- 32.Sperber K, Chiang G, Chen H, et al. : Comparison of hydroxychloroquine with zidovudine in asymptomatic participants infected with human immunodeficiency virus type 1. Clin Ther 1997;19:913–923 [DOI] [PubMed] [Google Scholar]
- 33.Engchanil C, Kosalaraksa P, Lumbiganon P, et al. : Therapeutic potential of chloroquine added to zidovudine plus didanosine for HIV-1 infected children. J Med Assoc Thai 2006;89:1229–1236 [PubMed] [Google Scholar]
- 34.Paton NI, Goodall RL, Dunn DT, et al. : Effects of hydroxychloroquine on immune activation and disease progression among HIV-infected participants not receiving antiretroviral therapy: a randomized controlled trial. JAMA 2012;308:353–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Calabrese LH, Lederman MM, Spritzler J, et al. : Placebo-controlled trial of cyclosporin-A in HIV-1 disease: implications for solid organ transplantation. J Acquir Immune Defic Syndr 2002;29:356–362 [DOI] [PubMed] [Google Scholar]
- 36.McComsey GA, Whalen CC, Mawhorter SD, et al. : Placebo-controlled trial of prednisone in advanced HIV-1 infection. AIDS 2001;15:321–327 [DOI] [PubMed] [Google Scholar]
- 37.Jacobson JM, Greenspan JS, Spritzler J, et al. : Thalidomide for the treatment of oral aphthous ulcers in participants with human immunodeficiency virus infection. National Institute of Allergy and Infectious Diseases AIDS Clinical Trials Group. N Engl J Med 1997;336:1487–1493 [DOI] [PubMed] [Google Scholar]
- 38.Fredericksen BL, Wei BL, Yao J, Luo T, Garcia JV: Inhibition of endosomal/lysosomal degradation increases the infectivity of human immunodeficiency virus. J Virol 2002;76:11440–11446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vijaykumar TS, Nath A, Chauhan A: Chloroquine mediated molecular tuning of astrocytes for enhanced permissiveness to HIV infection. Virology 2008;381:1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sperber K, Kalb TH, Stecher VJ, Banerjee R, Mayer L: Inhibition of human immunodeficiency virus type 1 replication by hydroxychloroquine in T cells and monocytes. AIDS Res Hum Retroviruses 1993;9:91–98 [DOI] [PubMed] [Google Scholar]
- 41.Savarino A, Lucia MB, Rastrelli E, et al. : Anti-HIV effects of chloroquine: inhibition of viral particle glycosylation and synergism with protease inhibitors. J Acquir Immune Defic Syndr 2004;35:223–232 [DOI] [PubMed] [Google Scholar]
- 42.Murray SM, Down CM, Boulware DR, et al. : Reduction of immune activation with chloroquine therapy during chronic HIV infection. J Virol 2010;84:12082–12086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Piconi S, Parisotto S, Rizzardini G, et al. : Hydroxychloroquine drastically reduces immune activation in HIV-infected, antiretroviral therapy-treated immunologic nonresponders. Blood 2011;118:3263–3272 [DOI] [PubMed] [Google Scholar]
- 44.Routy JP, Angel JB, Patel M, et al. : Assessment of chloroquine as a modulator of immune activation to improve CD4 recovery in immune nonresponding HIV-infected participants receiving antiretroviral therapy. HIV Med 2015;16:48–56 [DOI] [PubMed] [Google Scholar]
- 45.Savarino A, Shytaj IL: Chloroquine and beyond: exploring anti-rheumatic drugs to reduce immune hyperactivation in HIV/AIDS. Retrovirology 2015;12:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.