An automated pipeline, I-TASSER-MR, has been developed to facilitate structure determination using molecular repacement (MR) for samples for which only distantly related structures are available in the Protein Data Bank. By combining iterative fragmental structure-assembly simulations and progressive trimming of poorly modeled regions, the pipeline improves the success rate of MR.
Keywords: molecular replacement, protein structure prediction, X-ray crystallography, I-TASSER, threading
Abstract
Molecular replacement (MR) often requires templates with high homology to solve the phase problem in X-ray crystallography. I-TASSER-MR has been developed to test whether the success rate for structure determination of distant-homology proteins could be improved by a combination of iterative fragmental structure-assembly simulations with progressive sequence truncation designed to trim regions with high variation. The pipeline was tested on two independent protein sets consisting of 61 proteins from CASP8 and 100 high-resolution proteins from the PDB. After excluding homologous templates, I-TASSER generated full-length models with an average TM-score of 0.773, which is 12% higher than the best threading templates. Using these as search models, I-TASSER-MR found correct MR solutions for 95 of 161 targets as judged by having a TFZ of >8 or with the final structure closer to the native than the initial search models. The success rate was 16% higher than when using the best threading templates. I-TASSER-MR was also applied to 14 protein targets from structure genomics centers. Seven of these were successfully solved by I-TASSER-MR. These results confirm that advanced structure assembly and progressive structural editing can significantly improve the success rate of MR for targets with distant homology to proteins of known structure.
1. Introduction
The electron density of a macromolecular crystal structure can be directly calculated by Fourier transformation of the amplitudes of its diffracted X-rays, provided that the phase of each diffraction maximum is known. However, such phases cannot be directly measured, resulting in the so-called ‘phase problem’. One solution to the phase problem is molecular replacement (MR), which estimates the phase of each diffraction amplitude by placing one or more homologous search models in the unit cell of the crystal (Rossmann, 1990 ▸; Navaza, 1994 ▸). As more structures for diverse protein folds become known, MR has become increasingly useful for providing an initial set of phases that can be used to bootstrap towards a structural model consistent with the primary diffraction data.
One key to successful MR is to identify a search model that is close in structure to a substantial fraction of the scattering mass in the unit cell of the crystal. Following the principle that similar sequences adopt similar structures, traditional MR approaches use the atomic structure of a protein with high sequence identity to the crystallized target. However, the success rate of MR decreases rapidly as sequence identity falls below 30% owing to structural divergence of proteins in this so-called ‘twilight zone’ (Rost, 1999 ▸). Indeed, studies of the relationship between protein homology and backbone conformation have established a strong correlation between percentage identity and root-mean-square deviation (r.m.s.d.) of backbone atoms given by r.m.s.d. (Å) = 0.40exp(1.87H), where H is the fraction of mutated residues of a homologous domain (Chothia & Lesk, 1986 ▸). Thus, at 30% identity homologous domains are expected to have an r.m.s.d. of 1.5 Å in the position of their backbones in comparable regions. Many attempts have been made to push the boundary of homology-based MR. For example, Read and coworkers incorporated the use of maximum-likelihood statistics to enhance the sensitivity of MR searches (Read, 2001 ▸; Storoni et al., 2004 ▸; McCoy et al., 2005 ▸). In addition, automated pipelines such as CaspR (Claude et al., 2004 ▸), MrBUMP (Keegan & Winn, 2008 ▸) and BALBES (Long et al., 2008 ▸) were created to use multiple homologous template search models in order to enhance the success rate of MR. Some methods, including CHAINSAW (Stein, 2008 ▸) and Sculptor (Bunkóczi & Read, 2011 ▸), focused on editing template–target alignments to improve the signal-to-noise ratio of MR. Others explored the possibility of using template models detected by advanced fold-recognition algorithms for MR and found that more sophisticated profile–profile alignment methods can improve distant-homology template detection and increase the success rate in cases of low sequence identity (<35%; Jones, 2001 ▸; Schwarzenbacher et al., 2004 ▸). Later, the relationship between the accuracy of the initial protein model and its suitability as a search model in MR was investigated and it was shown that sequence identity alone is not the only strong determinant for successful MR procedures (Giorgetti et al., 2005 ▸). Low-resolution structural information can also be used to generate higher quality search models, because coordinates based on comparative modeling and nuclear magnetic resonance can be structurally refined at the atomic level and used for successful MR (Qian et al., 2007 ▸). Building on this idea, it has been shown that structural fragments of low-resolution models can be positioned by MR independently and then used to reassemble a full-length model that can be used for successful MR (Shrestha & Zhang, 2015 ▸). These methods open the door to the exploitation of low-resolution structures derived from ab initio modeling for high-resolution structure determination by MR.
With the development of new threading and fold-recognition methods, considerable progress has been made in the detection of distantly homologous templates (Zhang, 2008b ▸). In particular, template reassembly and refinement techniques can generate templates that are considerably closer in structure to that of the target protein, as demonstrated in recent community-wide blind CASP experiments (Kinch et al., 2011 ▸; Tai et al., 2014 ▸). One example, I-TASSER, was designed to construct full-length atomic models by reassembling structural fragments excised from the template structures under an optimized knowledge-based force field (Wu et al., 2007 ▸; Yang, Yan et al., 2015 ▸). The results from CASP10 showed that I-TASSER generated models that were closer to the native structure than the best threading templates for 81% of the target proteins, resulting in an average r.m.s.d. improvement of 1.05 Å in the threaded regions (Zhang, 2014 ▸). Consequently, we hypothesized that the I-TASSER structure-assembly method, combined with state-of-the-art phasing tools, could be used to improve automated MR phasing. To test this, we focused on distantly related protein targets and tested various model-editing procedures to examine the advantages and limitations of the truncation and editing of search models. We report here that we achieved a significant improvement in the success of automated protein structure solution by MR, starting only with the query sequence of the crystallized protein and the experimentally determined X-ray diffraction amplitudes. The resulting pipeline, called I-TASSER-MR, is freely downloadable at http://zhanglab.ccmb.med.umich.edu/I-TASSER-MR.
2. Methods
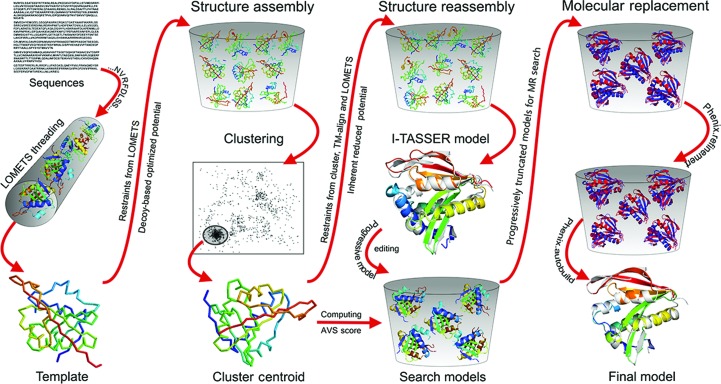
The I-TASSER-MR pipeline consists of three steps of I-TASSER-based protein structure prediction followed by progressive model editing and then by an MR search and automated model refinement (Fig. 1 ▸).
Figure 1.
Flow chart of I-TASSER-MR. The target sequence is first threaded through nonredundant structures from the PDB library to identify structure templates (column 1), with three-dimensional full-length models constructed by iterative fragment-reassembly simulations (column 2). The structure models are progressively edited based on AVS, which demarks poorly predicted regions (column 3), and the resulting models are used in a standard MR search by Phaser followed by automated model building and refinement with phenix.autobuild (column 4).
2.1. I-TASSER
The query sequence is first threaded through a nonredundant structure library derived from the Protein Data Bank (PDB) to search for structural templates and super-secondary-structure motifs using LOMETS (Wu & Zhang, 2007 ▸), a meta-threading algorithm consisting of eight distinct programs: FFAS-3D (Xu et al., 2014 ▸), HHsearch (Söding, 2005 ▸), MUSTER (Wu & Zhang, 2008 ▸), pGenTHREADER (Lobley et al., 2009 ▸), PPAS (Yan et al., 2013 ▸), PROSPECT2 (Xu & Xu, 2000 ▸), SP3 (Zhou & Zhou, 2005 ▸) and SPARKS-X (Yang et al., 2011 ▸). Continuous fragments (i.e. structural motifs without gaps in the query–template alignments) are then excised from the top-ranked templates and used to reassemble full-length models, whereby the structural regions whose sequences were not aligned by LOMETS (mainly loops) are built using a lattice-based ab initio folding procedure (Zhang et al., 2003 ▸). The structural reassembly simulations are performed by replica-exchange Monte Carlo (REMC) simulations, which are guided by a composite knowledge-based potential (Zhang et al., 2002 ▸). The structure conformations generated from REMC are clustered by SPICKER (Zhang & Skolnick, 2004a ▸), with the lowest free-energy conformations selected from the centroids of the largest clusters.
To further refine the SPICKER cluster models, a second round of fragment-assembly simulation is implemented by I-TASSER starting from the cluster-centroid model of the REMC simulation. Spatial restraints derived from the centroids and from PDB structures that have the highest structural similarity to the centroids, as detected by the structure-alignment program TM-align (Zhang & Skolnick, 2005 ▸), are used to guide the second round of simulation. The models obtained in the second-round refinement simulations are further refined using fragment-guided molecular-dynamics all-atom simulations (Zhang et al., 2011 ▸).
2.2. Progressive editing of MR search models
Because I-TASSER models are expected to have local errors that could prevent successful MR, a progressive editing procedure was used to truncate regions that have the highest probability of being incorrect, with the aim of generating search models with minimum r.m.s.d. from the target protein while maintaining high coverage of the target sequence. The idea of trimming unreliably modeled regions is not new. For example, Qian et al. (2007 ▸) used conformational variation to identify low-accuracy regions and performed a structural refinement and rebuilding process focused on the structurally variable regions to improve global model quality. Bibby et al. (2012 ▸) documented a correlation between conformational variation and modeling error, and proposed trimming off residues with large variation after structure clustering by SPICKER (Zhang & Skolnick, 2004a ▸). Sammito et al. (2014 ▸) recently proposed systematically ‘shredding’ residues from a search model and evaluating how they improve the score of a molecular-replacement solution.
Following Bibby et al. (2012 ▸), as well as our observation of a strong correlation between modeling accuracy and structural diversity of I-TASSER simulations (Zhang, 2008a ▸; Zhang et al., 2003 ▸), we defined an average variation score (AVS) for the ith residue as
where M is the number of structure conformations (known as decoys) in a SPICKER cluster generated by the I-TASSER simulation. (x j,i, y j,i, z j,i) and (x C,i, y C,i, z C,i) are the coordinates of the ith residue of the jth decoy and the cluster-centroid model, respectively, after superposition by the TM-score program (Zhang & Skolnick, 2004b ▸). TM-score determines the coordinate transformation between residue pairs, with shorter distances having a stronger weight; the superposition is therefore less sensitive to outliers than superpositions based on r.m.s.d.
Residues are first sorted by their respective AVS scores, and those with the highest AVS scores are progressively truncated to generate a series of search models with progressively more truncations. For each model k, the number of truncated residues is given by
where L is the length of the target sequence. Thus, 96 edited copies are attempted for each I-TASSER model, with the last copy having only 5% of the residues remaining.
2.3. B-factor assignment strategies
Because diffraction amplitudes are sensitive to atomic motion, particularly at high resolution, setting reasonable B factors for different regions of a search model can be important for MR (Bunkóczi & Read, 2011 ▸). B factors, by their nature, also represent uncertainty in the position of individual atoms. It has been suggested that local error prediction be used to estimate and adjust the atomic B factors of search models (Read & Chavali, 2007 ▸). Indeed, successful solutions are increased by 45% (or 101%) by introducing the predicted (or true) local error into the B-factor term (Pawlowski & Bujnicki, 2012 ▸). Most recently, it was shown that B factors based on coordinate-error estimation can improve signal to noise such that weighted search models of low quality give a better signal than analogous unweighted high-quality models (Bunkóczi et al., 2015 ▸). In addition, there are likely to be regions with low B factors in the native structure that are poorly predicted. Assigning high B factors to such poorly modeled regions is also advantageous to MR because it reduces their influence on high-resolution amplitude terms, while preserving their influence on low-resolution terms. In other words, B factors can be used to selectively down-weight inaccurate portions of the protein.
Three different methods were tested for estimating the B factors for the I-TASSER models. The first was setting the B factor of every atom to a constant value of 20 Å2 (this value is actually arbitrary because Phaser normalizes the scattering such that the average scattering intensity in each resolution range is a constant). The second was setting the atomic B factors equal to their accessible surface area (B ASA), as computed by DSSP (Kabsch & Sander, 1983 ▸), with a minimum B factor of 10 Å2. The third was setting the B factor of each residue to the AVS (Read & Chavali, 2007 ▸; Bunkóczi et al., 2015 ▸),
where AVSi is the average variability score of the ith residue in (1).
2.4. MR and automatic model building
Phaser (McCoy et al., 2007 ▸) is used for MR in the MR_AUTO mode, in which the percentage sequence identity of the best template is input to Phaser for the purpose of estimating the r.m.s. error of the search model. Log-likelihood gain (LLG) and translation-function Z-score (TFZ) values are used to evaluate the MR solutions, where LLG indicates how much better the solution is compared with a random solution and TFZ indicates how many standard deviations the LLG value of the solution is above the mean LLG. A recent quantitative study showed that a TFZ of ≥8 indicates success for a nonpolar space group, while a TFZ of ≥6 is sufficient in a polar space group (Oeffner et al., 2013 ▸). The best solution from Phaser is then input to phenix.refine (Afonine et al., 2012 ▸) to perform 20 cycles of refinement. Finally, phenix.autobuild (Terwilliger et al., 2008 ▸) is used for automatic model construction. The procedure of phasing and model construction is fully automated, and the entire process of I-TASSER-MR, including I-TASSER modeling and the automated phasing and model completion, takes 15–20 h for a protein of 200 residues on a 2.8 GHz IBM NeXtScale machine. In this study, MR was only attempted for the first I-TASSER model derived from the largest cluster obtained by SPICKER, although I-TASSER-MR allows users to explore up to the top five models from I-TASSER.
2.5. Assessment of structural models
Structural similarity between the final I-TASSER-MR models and the deposited crystal structures was assessed by r.m.s.d., TM-score, GDT-TS score and the percentile-based spread (PBS). The r.m.s.d. and PBS were calculated by ShakErr (Pozharski, 2010 ▸), whereas the TM-score and GDT-TS score were calculated by the TM-score program (Zhang & Skolnick, 2004b ▸). Calculation of the r.m.s.d. and PBS are based on all atoms, whereas the TM-score and GDT-TS calculations are based on Cα atoms.
The TM-score is defined as
where L is the target sequence length, L built is the number of residues built (or the length aligned for threading templates), di is the distance between the Cα atoms of the ith residue in the model and the experimentally determined structure, and d 0 = 1.24(L − 15)1/3 − 1.8 is a distance scale that ensures that the TM-score is independent of protein length (Zhang & Skolnick, 2004b ▸). The TM-score is a more reliable indicator of the quality of a solution than the r.m.s.d. because the r.m.s.d. only accounts for the error of the structure regions that are compared, whereas the TM-score accounts for both error and coverage. The r.m.s.d. also weights each distance pair equally, and therefore a local variation can generate a large r.m.s.d. even when the global topology is correct. In contrast, the TM-score weights smaller differences more than larger distances, and the value is thus more sensitive to correct global topology. TM-scores range from 0 to 1. A TM-score of <0.17 corresponds to a random structure pair, whereas a TM-score of >0.5 indicates a similar fold (Xu & Zhang, 2010 ▸).
GDT-TS is a metric used in CASP experiments to evaluate the quality of structure predictions (Zemla et al., 1999 ▸), and is defined as the average fraction of residues that have a difference in their superimposed positions within 1, 2, 4 and 8 Å cutoffs after optimal superposition, i.e. GDT-TS = (1/4L)(n d<1 + n d<2 + n d<4 + n d<8), where L is the length of the target sequence and n d<x is the number of residues in the model with a distance of <x Å from the deposited structure. Similar to TM-score, GDT-TS ranges from 0 to 1 and is insensitive to local structural errors, but differs in that it has a power-law dependence on the protein length (Zhang & Skolnick, 2004b ▸). The PBS represents the average variation in atomic positions, which is also insensitive to the presence of outliers (Pozharski, 2010 ▸).
A solution with a TFZ of above 8 has been proposed to be a reliable indicator of successful MR (Oeffner et al., 2013 ▸). Although statistics from Phaser indicate whether a model gives a significant hit, successful structure solutions are also dependent on the automated building and refinement steps of the pipeline. It has been proposed that success is indicated by a substantial fraction of the final structure being closer to the experimentally determined structure than the search model (Giorgetti et al., 2005 ▸). More stringent criteria have been proposed to be that the automated solution should have both a reasonable chemical structure and significant differences from the search model (Terwilliger et al., 2008 ▸). Combining these proposals, we defined success in our automated pipeline as an automatically built final model with a higher TM-score or GDT-score or a lower r.m.s.d. than the search model and/or with a TFZ of >8. Despite its flaw of being sensitive to local errors, r.m.s.d. has been included as part of the assessment here because it is widely used in structural biology and is intuitively easy to understand.
3. Results
3.1. Benchmark data sets
Two sets of proteins were assembled to benchmark the I-TASSER-MR approach. The first (the CASP8 set) consists of 61 proteins from the CASP8 experiment with structures determined by X-ray crystallography and fewer than 300 residues and ≤4 copies in the asymmetric unit. According to assignment by the CASP8 assessors (Tress et al., 2009 ▸), 22 belong to the TBM-HA category (template-based modeling – high accuracy; the targets usually have easily detectable homology templates), 37 targets belong to the TBM category (template-based modeling targets that have templates existing in the PDB but the detection of which is usually more difficult and often with substantial alignment errors) and two belong to the FM category (no similar structures in the PDB). The second set (the High-Res set) consists of 100 nonredundant proteins selected from the PDB by PISCES (Wang & Dunbrack, 2003 ▸) using a pairwise sequence-identity cutoff of 25%, a sequence length of <300 residues and a resolution of ≤1.5 Å to explore the effect of high resolution on the performance of I-TASSER-MR. In contrast, the CASP8 set has no resolution cutoff. No proteins in the High-Res set are homologous to any in the CASP8 set.
To ensure distant-homology modeling, any I-TASSER templates that had a sequence identity of >30% to a target protein or one detectable by PSI-BLAST with an E-value of <0.5 (referred to in the following as ‘closely related templates’) were excluded from the I-TASSER template library. To apply the latter filter, we used PSI-BLAST to build a sequence profile by searching the NCBI NR database (ftp://ftp.ncbi.nih.gov/blast/db) in three iterations with an E-value cutoff of 0.001. The resulting profile was then used to match all structures in the PDB to identify closely related templates, which were excluded if they had an E-value of <0.5. A similar template cutoff was applied to the second step of I-TASSER iteration when TM-align was used to search for fragments within the PDB library. For the CASP8 set, an additional filter was imposed to exclude all templates solved after CASP8. After application of all of the filters, the average sequence identity between the I-TASSER templates and the target proteins was 15%.
Multiple B-factor assignment schemes were tested for each set, and individual targets were deemed to be successfully determined by I-TASSER-MR if at least one of these schemes generated a correct solution.
3.2. MR solutions for the CASP8 set
3.2.1. Overall results
The I-TASSER-MR pipeline (Fig. 1 ▸) was first tested on the CASP8 set. Using the criterion specified above, 33 out of the 61 targets were successfully solved. A summary of the MR results for each of the 61 targets, including the range and mean values for both successful and failed cases, is presented in the upper part of Table 1 ▸. Details of these 33 successful targets and the search models are listed in Supplementary Tables S1 and S2, respectively, and those for the unsuccessful targets in Supplementary Table S3. The average coverage of the final refined structures is 81 ± 16%, with an average PBS of 0.26 ± 0.14 Å. The average TM-score and PBS of the initial I-TASSER models for all 61 test targets were 0.73 ± 0.17 and 2.16 ± 0.82 Å, respectively. The average TM-score and PBS were 0.84 ± 0.05 and 1.74 ± 0.32 Å, respectively, for the 33 successful targets, as opposed to 0.62 ± 0.19 and 2.65 ± 0.95 Å for the 28 unsuccessful targets. This indicates that the quality of the initial I-TASSER model is a major contributor to success or failure.
Table 1. Summary of the results of I-TASSER-MR on the two protein sets.
| PDB structure | I-TASSER | Phaser | I-TASSER-MR structure | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| D min † (Å) | L ‡ | PBS§ (Å) | TM¶ | GT†† | LLG‡‡ | TFZ§§ | R free ¶¶ | TM††† | PBS (Å) | |
| Benchmark I: 61 CASP8 targets | ||||||||||
| Success | ||||||||||
| Range | 0.98–2.90 | 95–286 | 1.15–2.73 | 0.70–0.94 | 0.65–0.90 | 27–366 | 4.5–40.8 | 0.25–0.41 | 0.50–0.97 | 0.09–0.75 |
| Mean | 2.03 ± 0.39 | 179 ± 58 | 1.74 ± 0.32 | 0.84 ± 0.05 | 0.77 ± 0.07 | 103 ± 95 | 9.5 ± 6.9 | 0.31 ± 0.05 | 0.82 ± 0.14 | 0.26 ± 0.14 |
| Failure | ||||||||||
| Range | 1.15–2.81 | 103–292 | 1.31–5.32 | 0.19–0.84 | 0.10–0.79 | 3–124 | 3.5–7.0 | 0.48–0.58 | 0.04–0.61 | 1.93–14.96 |
| Mean | 1.92 ± 0.39 | 203 ± 59 | 2.65 ± 0.95 | 0.62 ± 0.19 | 0.52 ± 0.19 | 35 ± 27 | 5.4 ± 0.8 | 0.54 ± 0.02 | 0.25 ± 0.16 | 2.88 ± 0.87 |
| Benchmark II: 100 High-Res targets | ||||||||||
| Success | ||||||||||
| Range | 0.98–1.50 | 104–296 | 1.03–2.85 | 0.68–0.95 | 0.59–0.94 | 12–352 | 0–17.3 | 0.21–0.44 | 0.31–0.99 | 0.05–0.67 |
| Mean | 1.29 ± 0.15 | 183 ± 53 | 1.70 ± 0.34 | 0.85 ± 0.07 | 0.78 ± 0.08 | 69 ± 62 | 7.3 ± 2.8 | 0.28 ± 0.04 | 0.84 ± 0.17 | 0.17 ± 0.12 |
| Failure | ||||||||||
| Range | 0.83–1.50 | 101–295 | 1.18–3.10 | 0.46–0.83 | 0.36–0.83 | 9–52 | 0–6.9 | 0.45–0.58 | 0.11–0.61 | 1.35–5.01 |
| Mean | 1.27 ± 0.20 | 171 ± 57 | 2.27 ± 0.42 | 0.71 ± 0.10 | 0.63 ± 0.10 | 23 ± 11 | 5.1 ± 1.1 | 0.54 ± 0.03 | 0.37 ± 0.15 | 2.75 ± 0.89 |
Resolution of the deposited X-ray structure.
Length of the target sequence.
Percentile-based spread of the first I-TASSER search model relative to the experimental model.
TM-score of the first I-TASSER model.
GDT-TS score of the first I-TASSER model.
LLG of the Phaser solution relative to random.
TFZ of the Phaser solution.
R free of the final I-TASSER-MR model.
TM-score of the final I-TASSER-MR model versus the experimental model.
PBS of the final I-TASSER-MR model versus the experimental model.
3.2.2. Impact of truncation editing
The residues in the I-TASSER models with the highest uncertainty in the structural assembly simulation were progressively truncated based on their AVS score (1) to create 96 variants of each search model (see §2). For simplicity of analysis, if more than one search model succeeded in MR, we only report the results for that with the least truncation. In the CASP8 set of targets, a successful MR solution was achieved without any truncation editing for ten of the structures. The average TM-score and PBS score of the initial I-TASSER models before truncation for these proteins were 0.88 ± 0.04 and 1.51 ± 0.21 Å, respectively. For the remaining 23 cases in which some truncation editing had to be conducted, the average TM-scores and PBS scores were 0.81 ± 0.04 and 1.84 ± 0.31 Å, respectively. Thus, the initial models that needed to be truncated were, as hypothesized, of lower quality, and truncation of the unreliably modeled regions led to a significant increase in the number of correct solutions.
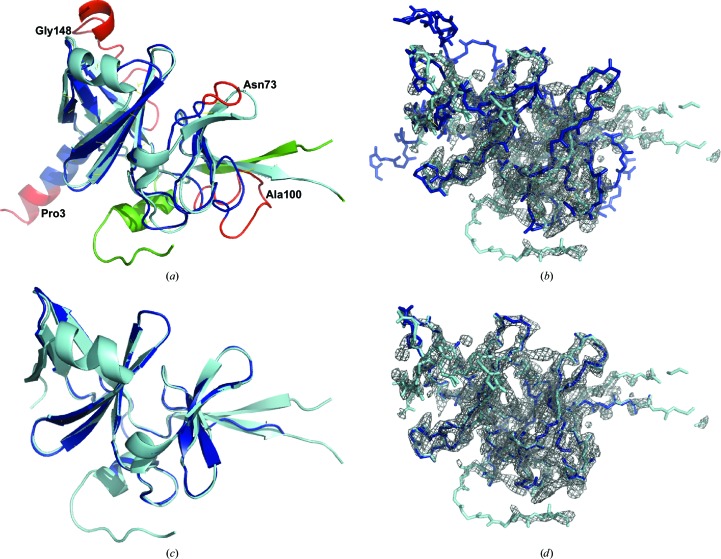
Fig. 2 ▸ presents an example of how progressive truncation impacted success for a Rieske-type ferredoxin protein (PDB entry 3d89; Levin et al., 2008 ▸), which has a three-layer β-sandwich fold (157 residues, resolution 2.07 Å). The TM-score of the initial I-TASSER model was 0.7. Although the overall topology of the I-TASSER model is similar to the deposited structure, there are several regions in the termini and loops that exhibit very high variability in the I-TASSER simulations, resulting in a relatively high PBS of 1.78 Å (Fig. 2 ▸ a). When the full-length I-TASSER model was used as the search model, the LLG and TFZ from Phaser were 20 and 4.1, respectively. The final structure generated from this MR solution had an R free from phenix.autobuild (Adams et al., 2010 ▸) of 0.6, which is close to random (Kleywegt & Jones, 1997 ▸), and a PBS of 2.56 Å, which is even higher than that of the starting model. As shown in Fig. 2 ▸(b), there are a number of regions that do not correlate well with the experimental 2m|F o| − D|F c| σA-weighted map calculated by the Electron Density Server (EDS; Kleywegt et al., 2004 ▸).
Figure 2.
Illustration of progressive truncation in I-TASSER-MR. (a) Superposition of the initial I-TASSER model (blue) and the experimental structure (cyan; PDB entry 3d89). The structure regions with a high AVS score selected for truncation by I-TASSER-MR are marked in red. The r.m.s.d., PBS and GDT-TS score of the two structures were 2.76 Å (821 atoms), 1.78 Å (821 atoms) and 0.66, respectively. (b) Overlay of the I-TASSER-MR solution (blue) generated without model truncation on the experimental 2m|F o| − D|F c| σA-weighted map calculated by the EDS contoured at 2σ. (c) Superposition of the final I-TASSER-MR structure solved using model truncation (blue) and the experimental structure. The r.m.s.d., PBS and GDT-TS scores of the structure were 0.95 Å (809 atoms), 0.24 Å (809 atoms) and 0.74, respectively. (d) Overlay of the successful truncated I-TASSER-MR structure (blue) and the experimental electron density.
The first successful I-TASSER-MR solution in the progressive truncation scheme had 46 high-AVS residues trimmed at positions 1–7, 71–75, 101–114 and 138–157. These amino acids are located at the N- or C-termini or in loops and have a high deviation from the experimental structure (Fig. 2 ▸ a), demonstrating a close correlation between the AVS and errors in local structure. After trimming, the PBS of the I-TASSER search model was essentially the same (1.68 Å), but the LLG and TFZ of the Phaser solution increased to 41 and 5.3, respectively. This improved solution led to a final structure with an R free of 0.36 and a PBS of 0.24 Å (Fig. 2 ▸ c). Accordingly, much better correlation between the I-TASSER-MR solution and the electron-density map is observed (Fig. 2 ▸ d). In this example, the global structure of the initial I-TASSER model was close to the target (TM-score = 0.7). Therefore, although the truncation of a few high-AVS residues did not dramatically change the PBS value of the search models, it did significantly improve the MR statistics and the final structure that could be built.
3.3. MR solutions for the High-Res set
To explore the effect of high resolution on the performance of I-TASSER-MR, we tested the pipeline on a second set of 100 nonredundant proteins determined at a higher resolution on average (1.28 Å) than the CASP8 set (1.98 Å). A summary of the MR results for the 100 PDB targets is given at the bottom of Table 1 ▸. Details of the successful runs are listed in Supplementary Tables S4 and S5 and those of unsuccessful runs in Supplementary Table S6. Overall, the success rate of I-TASSER-MR on the High-Res set was 62%. Of these, 38 required AVS-based truncation to achieve a successful solution. The average number of truncated residues was 20.5 ± 19, which reduced the PBS of the initial I-TASSER search models from 1.77 ± 0.36 to 1.68 ± 0.33 Å on average. The average PBS for the final 62 successfully rebuilt solutions was 0.17 ± 0.12 Å, compared with 1.68 ± 0.30 Å achieved by superimposing I-TASSER models based on prediction (Table 1 ▸). On average, 83% of the residues in these solutions were successfully constructed by I-TASSER-MR. Thus, the results once again demonstrate the usefulness of progressive model editing and the automated I-TASSER-MR pipeline in general.
The overall success rate of I-TASSER-MR in the High-Res set (62%) is slightly higher than that for the CASP8 targets (54%). One reason may be that the I-TASSER search models are of higher quality in the High-Res set (TM-score = 0.80) than in the CASP8 set (TM-score = 0.73). Secondly, the higher resolution diffraction data of the High-Res targets, with a resolution that is on average 0.7 Å better than the CASP8 set, may also positively impact the success rate of the High-Res set by providing higher quality data for Phaser and autobuilding.
Combining the two sets together, there are 69 cases in which I-TASSER-MR failed to generate a correct atomic structure. In 66 of these cases the I-TASSER models were incorrectly placed by MR (i.e. failed at the Phaser step). In the three remaining cases the models were correctly placed by Phaser (TFZ > 8) but phenix.autobuild did not create a model closer to the initial model. Thus, there is room for improvement in both the search and model-building stages of the pipeline.
3.4. Impact of B factor on MR
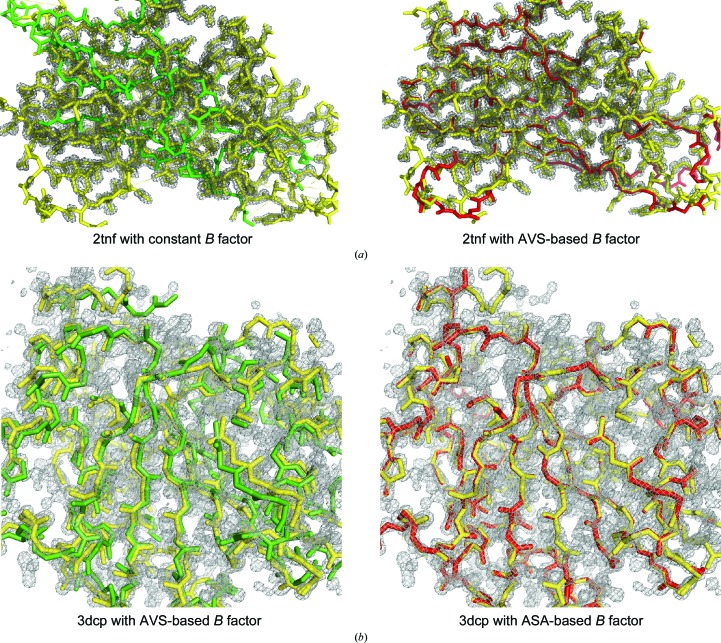
To test the effect of temperature factors on the success of MR by I-TASSER-MR, we tried three different B-factor schemes for the CASP8 and High-Res sets: setting the B factors to a constant value, setting the B factors according to the accessible surface area (ASA) or setting the B factors according to AVS (see §2). If the normalized accessible surface area is used as a B factor, four additional targets (PDB entries 1tu9, 2bbr, 2hc1 and 3dcp) could be solved relative to constant B factors. Two more targets (PDB entries 1i12 and 2tnf) could be solved using AVS-based B factors relative to the other two schemes. Fig. 3 ▸ compares the final MR models overlaid on the electron-density maps when using different B factors for PDB entries 2tnf and 3dcp, where a much closer match to the electron density using the AVS-based or ASA-based B factors was observed. In a recent study, we proposed a machine-learning method, ResQ (Yang, Wang et al., 2015 ▸), to estimate B factors based on the threading templates and sequence profiles. The results showed that additional targets could be solved using the new B-factor predictor. Thus, different B-factor assignment schemes can be complementary, and success rates can be improved if one uses multiple B-factor predictors.
Figure 3.
Impact of B-factor assignment on MR solutions. (a) Superposition of PDB entry 2tnf (yellow) with I-TASSER-MR-generated solutions with constant (green, left panel) or AVS-based (red, right panel) B-factor schemes, which are overlaid on a 2m|F o| − D|F c| σA-weighted map calculated by the EDS contoured at 2σ. (b) Superposition of the native structure of PDB entry 3dcp (yellow) with I-TASSER-MR-generated solutions with a constant (green, left panel) or ASA-based (red, right panel) B-factor setting, which are overlaid on the corresponding electron-density map.
3.5. Location of pruned residues
To explore what kinds of residues needed to be deleted in order to succeed in MR, we analyzed 60 of the 161 successful targets for which some residue truncation was needed to find a solution. Three secondary-structure types (helix, strand and coil) were specified by DSSP from the PDB structure. The residues missing from the original PDB entries are considered to be unstructured residues, which are often intrinsically disordered in folded proteins (Dunker et al., 2000 ▸). The most frequently truncated residues were located in coil regions (45%), followed by unstructured (27%), helical (20%) and strand (7.3%) regions, where the percentage values are normalized by the length of the targets. We further computed the percentage of truncated residues, which is normalized by the number of residues in each secondary-structure type. The highest ratio was found in unstructured regions (78%), followed by coil (14%), helical (6%) and strand (3%) regions. These results reflect the fact that coil and unstructured regions tend to be more difficult to predict and thus have a higher AVS fluctuation in the I-TASSER simulations.
We also divided each protein sequence into ten equal parts to examine the frequency of truncation as a function of position (Supplementary Fig. S1). The largest fractions of truncated residues are N-terminal (28%) and C-terminal (19%). The average number of truncated residues in the middle parts is far below the terminal regions (∼7 ± 1%), which is not surprising given that core residues are in general more tightly packed and therefore have a lower AVS in I-TASSER simulations. The modeling accuracy is also often higher in the core part, which is essential to MR.
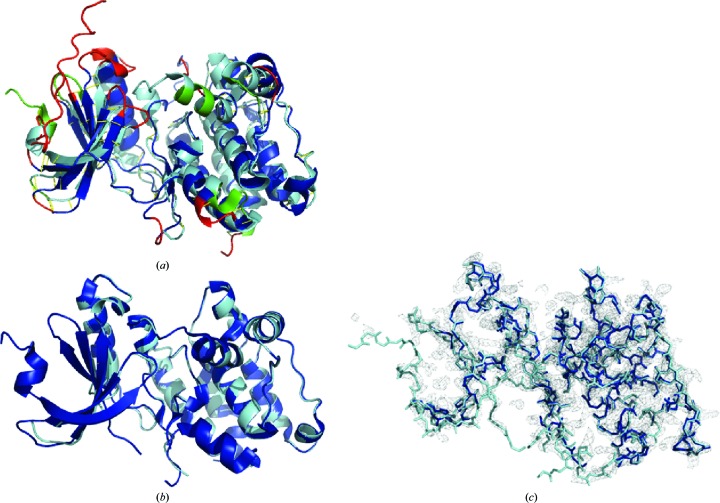
Fig. 4 ▸ shows the distribution of truncated residues in a typical example: the kinase domain of the calcium-dependent protein kinase cgd3_920 (PDB entry 3dfa, resolution 2.45 Å; Structural Genomics Consortium, unpublished work). The first truncated model that led to a successful solution had 60 residues truncated (Fig. 4 ▸ a). Of the high-AVS residues, six are in helical regions, two in strands, 34 in coils and 18 in unstructured regions. Despite the relatively high fraction of residue truncation (23% of the total residues were trimmed off), 97.5% of the structure was successfully built with a final PBS of 0.24 Å (Fig. 4 ▸ b). The final I-TASSER-MR model has an R free of 0.36 and thus high correlation with the corresponding electron-density map (Fig. 4 ▸ c).
Figure 4.
Typical distribution of residue truncations. (a) Superposition of the initial I-TASSER model (blue) and the native structure (cyan) of PDB entry 3dfa. Truncated residues are highlighted in red for the initial I-TASSER model; the analogous residues in the deposited structure are in green. The r.m.s.d., PBS and GDT-TS scores of the two structures are 2.98 Å (1761 atoms), 2.36 Å (1761 atoms) and 0.68, respectively. (b) Superposition of the I-TASSER-MR model and the experimental structure. The r.m.s.d., PBS and GDT-TS score of the two structures are 0.74 Å (1430 atoms), 0.24 Å (1430 atoms) and 0.63, respectively. (c) Overlay of the I-TASSER-MR structure (blue) on the 2m|F o| − D|F c| σA-weighted map contoured at 2σ.
3.6. Analysis of how many residues need to be truncated
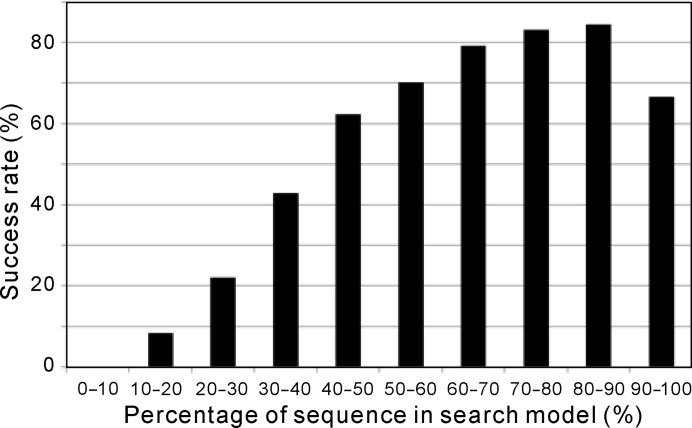
The I-TASSER models were progressively edited, with the number of remaining residues in the search models ranging from 100 to 5% of the full length of the target protein. The highest success rate was achieved for search models that retained 60–90% of their target sequence (Fig. 5 ▸). Previous studies suggested that the highest success rate is achieved when 21–40% of the sequence is used for MR (Bibby et al., 2012 ▸). Although the progressive editing procedure used by I-TASSER-MR is similar to the cluster-and-truncate approach used in this previous work, the test sets and the preparation of the models are very different. The proteins used by Bibby and coworkers contained 40–120 residues, whereas the proteins used here ranged from 95 to 298 residues. In addition, Bibby and coworkers used ab initio models, whereas I-TASSER-MR is based on I-TASSER, which is a template-based modeling method. Current ab initio structure predictions can most reliably fold proteins with a short length (Zhang, 2008b ▸). Therefore, the average PBS and r.m.s.d. values of the ab initio models will become greatly reduced as more residues are truncated, which will improve the MR results. I-TASSER models are more sensitive to the availability and quality of the templates than to the length of the target sequence. In fact, the average PBS and r.m.s.d. of the I-TASSER models are almost unchanged after the initial truncation of ∼10% of the residues. Thus, retaining more residues will lead to the retention of more and higher quality structural information, which explains the shift of the success-rate peak towards larger coverage of the search model compared with the observations of Bibby and coworkers. Nevertheless, exploring a wide range of truncations helped to increase the overall yield of the I-TASSER-MR pipeline. When the truncation threshold was extended to higher than 40%, two more targets were solved: PDB entries 2vsw (at 43% truncation) and 3e03 (at 58% truncation).
Figure 5.
Success rate of I-TASSER-MR at different levels of truncation. The x axis indicates the percentage of residues in the search model after model truncation, normalized by the target length.
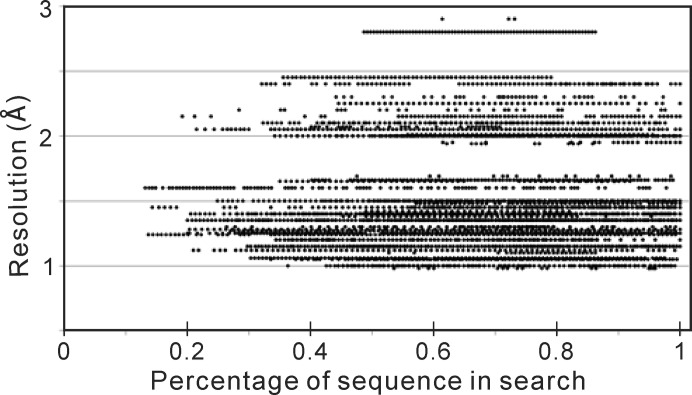
Plotting the resolution of the target PDB entry against the truncation fraction in successful I-TASSER-MR solutions indicates that high-resolution targets can tolerate deeper truncation (Fig. 6 ▸). There are eight targets that can tolerate up to 80% truncation. Of these, six have a resolution of less than 1.5 Å. The two remaining targets (PDB entries 3czx and 3db5) have resolutions of 1.6 and 2.15 Å, respectively. For the ten targets that have a diffraction resolution of >2.15 Å, only one target (PDB entry 3d8p, 2.2 Å resolution) can achieve a successful MR solution with a deepest tolerated truncation of 71.6%.
Figure 6.
Scatter plots of the resolution of the diffraction data for the 95 successful protein targets. The x axis shows the percentage of the sequence remaining in the search models for the successful MR solution. Each point represents one successful solution from a target protein at a specific truncation, whereby one protein can have multiple successful MR solutions at different truncations.
3.7. Improvement of MR using I-TASSER models compared with LOMETS templates
Many MR search models are built from homologous templates that are identified from the PDB library based on sequence comparison or fold recognition (Schwarzenbacher et al., 2004 ▸; Stein, 2008 ▸; Bunkóczi & Read, 2011 ▸). However, it has been demonstrated in various benchmarking and blind protein structure-prediction experiments that structure-reassembly methods such as I-TASSER have the ability to create templates closer to the native structure (Wu et al., 2007 ▸; Zhang, 2014 ▸). To test whether the structure-assembly simulation followed by AVS truncation performs better than more traditional MR approaches, LOMETS was used in conjunction with CHAINSAW (Stein, 2008 ▸) and Sculptor (Bunkóczi & Read, 2011 ▸), two widely used programs for preparing homologous models for MR that prune nonconserved residues from the target–template alignments, to generate search models for the CASP8 and High-Res data sets. We used the default settings of CHAINSAW and 12 different predefined protocols of Sculptor (consisting of different combinations of main-chain deletion, side-chain pruning and B-factor modification) to edit the same set of LOMETS templates that were used by I-TASSER for structure assembly. Supplementary Tables S7 and S8 list the MR results using LOMETS templates for the CASP8 and High-Res data sets, respectively, where the best from the top 20 templates with the highest TM-score was adopted; these 20 templates were also used as input templates for the I-TASSER simulations.
Results show that 70 (21 of 61 CASP8 and 49 of 100 High-Res data-set) targets had a successful MR solution when using LOMETS templates. There were therefore 29 targets that were solved by I-TASSER-MR but not by LOMETS models. The average TM-score and PBS of the I-TASSER models for these 29 targets, before truncation editing, were 0.81 ± 0.05 and 1.79 ± 0.31 Å, respectively, in comparison to 0.70 ± 0.08 and 1.84 ± 0.31 Å for the corresponding LOMETS models. Here, the difference in TM-score (0.81 versus 0.70) is more significant than that in PBS (1.79 versus 1.84 Å), mainly because TM-score accounts for both accuracy and coverage of the models, whereas PBS only accounts for modeling accuracy. Because the I-TASSER model is full-length and LOMETS models only contain part of the conserved residues, one could argue that the high TM-score of the I-TASSER model was owing to the addition of unaligned structures. Even when considering the same threading aligned regions (i.e. ignoring the contribution from the I-TASSER ab initio folding for the unaligned regions), the average TM-score and PBS of the I-TASSER models are 0.76 ± 0.13 and 1.49 ± 0.53 Å, respectively, which are still better than the LOMETs models.
Among these 29 targets, there were 15 targets for which the first successful truncated I-TASSER model had less coverage than the corresponding LOMETS search models. Except for PDB entry 1tu9, all of the targets have a higher TM-score than the best LOMETS template when considering the same threading aligned regions. These data suggest that structure refinement by the I-TASSER fragment-assembly simulations represents a major improvement for generating search models.
Supplementary Fig. S2 shows a comparison of the MR results based on LOMETS and I-TASSER models from the target with PDB code 1tu9, where the I-TASSER model has a lower TM-score than the LOMETS template but I-TASSER-MR generated a solution with a much lower R free. The first successful I-TASSER search model has an r.m.s.d. of 1.76 Å and a TM-score of 0.69, while the r.m.s.d. and TM-score of the best of 13 LOMETS search models produced by CHAINSAW and Sculptor are 2.0 Å and 0.62, respectively. Although 24% of the residues were deleted in I-TASSER-MR, 96% of the residues were finally built with an r.m.s.d. of 0.07 Å (and a PBS of 0.08 Å). In this example, the success of I-TASSER-MR over LOMETS is mainly owing to the AVS-based progressive editing procedure, which correctly identified the structural regions with a higher modeling accuracy in the search model.
Among the 161 test targets, there were four (PDB entries 1nnx, 2o1q, 3b79 and 3d0j) for which LOMETS-based MR succeeded but I-TASSER-MR failed. In these cases, the TM-score or PBS of the I-TASSER search models was worse than that of the LOMETS templates. The first I-TASSER model is often the closest to the correct structure, but sometimes the lower ranked models from different SPICKER clusters represent better search models. The fifth I-TASSER model for PDB entry 1nnx and the second I-TASSER model for PDB entry 2olq have higher TM-scores and were able to find correct MR solutions. However, no better models were found for PDB entries 3b79 and 3d0j even when we used the top five I-TASSER models because I-TASSER deteriorated the LOMETS templates in these two cases.
3.8. Application to PSI targets
Structural genomics (SG) is a community-wide effort initiated at the end of the last century with the goal of solving as many nonhomologous protein structures as possible in order to increase diversity in the PDB and to facilitate genome-wide comparative structure predictions (Burley et al., 1999 ▸). Owing to a lack of homologous protein templates, molecular replacement is often not applicable to SG targets. Experimental phasing methods, such as isomorphous replacement and anomalous diffraction, must often be employed.
In Supplementary Table S9, we collect the statistics for I-TASSER-MR runs for a set of 14 SG targets that were solved by Protein Structure Initiative (PSI) centers and deposited in the PDB in March 2014. In columns 7 and 8, we list the PBS and TM-score of the full-length models created by I-TASSER, where all templates solved after the deposition date of each target were excluded from the LOMETS template library used by I-TASSER. To confirm that closely related homologous proteins were not used, a routine filter (i.e. a sequence identity of <30% and a PSI-BLAST E-value of >0.5) was also used to eliminate additional homologous templates. We list the experimental methods that the original authors used to phase the structures in column 6 of Supplementary Table S9. It is unknown whether MR was attempted for most of these entries.
I-TASSER generated full-length models of reasonably good quality with an average TM-score of 0.74 ± 0.21 and PBS of 2.24 ± 0.75 Å for the 14 PSI targets. There were eight targets with a TM-score above 0.8. Among these, six (PDB entries 4p47, 4ps6, 4pux, 4pw0, 4pxy and 4pz0 at the top of Supplementary Table S9) were solved by I-TASSER-MR. PDB entry 4pqx had what seemed to be a good MR solution (LLG = 109, TFZ = 9.2) but failed to be rebuilt. Overall, the average PBS and TM-scores of the I-TASSER-MR structures were 0.09 ± 0.04 Å and 0.89 ± 0.09, respectively, for the six successfully built cases.
3.9. When will I-TASSER-MR work?
The quality of the initial model is the most important factor in determining whether MR will be successful. Interestingly, among the 95 successful MR solutions, more than half (58) have initial I-TASSER models, before truncation, with a GDT-TS score of below 0.8. The lowest GDT-TS score (for PDB entry 3frh) was 0.59. Previously, a GDT-TS score of >0.8 was thought to be required for successful MR (Giorgetti et al., 2005 ▸). The major reason for this difference is probably owing to differences in implementation, as Giorgetti and coworkers performed searches only on full-chain models, whereas a progressive phasing search using various optimal edits of the probe models was conducted in I-TASSER-MR. Considering the data from Supplementary Tables S1–S6, we can conclude that a GDT-TS of above 0.83 should guarantee success, whereas models with a GDT-TS of <0.59 never succeed in I-TASSER-MR. If judging the initial models by TM-score, a TM-score of above 0.84 should guarantee success, whereas models with a TM-score of <0.68 never succeed. If judging the models by PBS, we found that a PBS of below 1.18 Å is sufficient for success, whereas models with a PBS of >2.85 never succeed. These cutoffs exhibit no clear dependency on the resolution of the diffraction data.
The GDT-TS-based and TM-score-based criteria require knowledge of the experimental structure, and thus cannot be used when the target structure is unknown. In I-TASSER, the quality of the structure models can however be estimated by the confidence score (C-score), which is calculated by a combination of the significance of threading alignments and the convergence of the structural assembly simulations (Zhang, 2008a ▸),
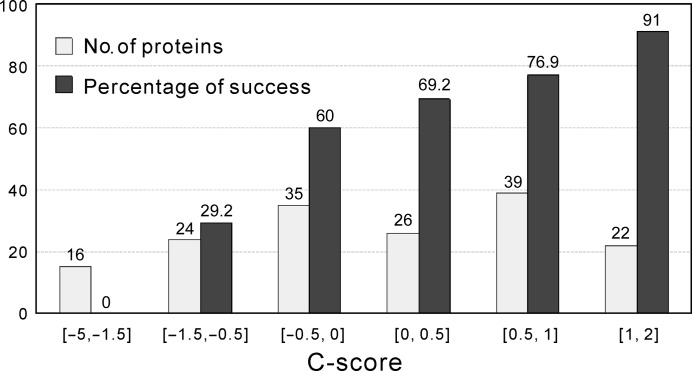
where M is the multiplicity of the decoy structures in the SPICKER cluster, M tot is the total number of decoys generated by I-TASSER, 〈r.m.s.d.〉 is the average r.m.s.d. of the decoy structures to the centroid of the cluster, N is the number of threading programs used in LOMETS and Z(i)/Z 0(i) is the normalized Z-score of the first templates by the ith threading program. The C-score generally ranges from −5 to 2, with a higher value indicating better quality. Large-scale benchmarking data built on 500 nonredundant proteins (Zhang, 2008a ▸) have shown a strong correlation between the C-score and TM-score values of I-TASSER models (Pearson correlation coefficient = 0.91). In Fig. 7 ▸, we show the success rate of MR versus the C-score of the I-TASSER predictions, which shows an almost linear correlation in the region with C-score > −1.5. The success rate reaches 91% in the region with C-score > 1. However, when the C-score is below −1.5, which roughly corresponds to the minimum C-score required for a model prediction of correct fold (TM-score > 0.5; Zhang, 2008a ▸), MR has not been observed to succeed.
Figure 7.
Correlation of MR with the C-score of the I-TASSER models. Histogram of the C-score of the I-TASSER models (light bars) and the average success rate of I-TASSER-MR in each C-score range (dark bars).
Evaluation of the success of I-TASSER-MR versus protein size (Supplementary Fig. S3) reveals no clear relationship between these parameters (at least for proteins of up to 300 residues), a different result from the experiments of Bibby et al. (2012 ▸), who observed that smaller proteins with a length of <100 residues are more likely to succeed in MR than longer proteins. Again, this difference is probably owing to the fact that Bibby and coworkers used models from ab initio modeling, which has the greatest success for small proteins (Zhang, 2008b ▸). Because I-TASSER is based on the reassembly of template structures, the quality of the search models depend far more on the availability of templates than on protein size. Finally, we investigated the relationship of the number of molecules in the asymmetric unit to the success rate. The success rates of targets with one subunit in the asymmetric unit and of targets with more than one subunit were 59 and 60%, respectively. Therefore, the number of chains does not seem to affect the success rate of I-TASSER-MR.
4. Conclusion
We have developed an integrated pipeline for automated molecular-replacement structure determination starting from the amino-acid sequence of the target protein, which is built on I-TASSER protein-structure predictions. A progressive model-editing procedure was introduced to truncate unreliably modeled residues based on an intermediate structural variation score of the I-TASSER assembly simulations. These optimized structure models were transferred to widely used tools (Phaser and phenix.autobuild) for the MR search, automated model binding and refinement. Whereas many of the strategies used in this study, including truncation-based model editing and modeling-error-based B-factor estimation, have been explored by previous investigators, I-TASSER-MR here provides a single efficient interface that connects cutting-edge structure assembly, MR phasing and autobuilding tools for protein structure determination.
The I-TASSER-MR pipeline was tested on two independent protein sets that consisted of 61 targets from CASP8 and of 100 nonredundant high-resolution proteins from the PDB. Considering the two data sets together, a success rate of 59% was achieved based on the first I-TASSER model, which is derived from the largest SPICKER cluster after closely related templates are excluded from the structure-assembly process. In 60 of the 95 successful runs, some level of model-truncation editing was required. The model editing reduced the sequence length by 12% (or 22 amino acids) on average, which improved the average r.m.s.d. of the search models by 1.29 Å. The majority of the truncated residues are located in the loop/termini (45%) and unstructured regions (27%) of the PDB entries, where I-TASSER models are known to have lower accuracy. The overall PBS and r.m.s.d. of the 92 final I-TASSER-MR models built after MR were 0.2 and 0.94 Å, respectively, which are 1.5 and 1.47 Å lower, respectively, than those for the initial I-TASSER search model. I-TASSER generates the structure prediction by a combination of multiple threading alignments, in which the first models (from the largest SPICKER cluster) are often closest to the native structure. Nevertheless, if all top five models from I-TASSER were used instead of just the first, six additional solutions for the 161 targets could be achieved. Furthermore, we could explore the effect of target diffraction resolution on MR success because the two test sets have different average resolutions. The data suggest a potentially positive impact of target resolution on the results, as demonstrated by the slightly higher success rate for the High-Res set over the CASP8 set. It was also observed that models for high-resolution targets tend to tolerate deeper structural truncations (Fig. 6 ▸).
To show its practical utility towards solving proteins with distant homology or potentially unknown fold, the I-TASSER-MR pipeline was also applied to 14 SG targets solved by the PSI centers in March 2014 based on other experimental methods. Using I-TASSER-MR six of these PSI targets were solved, with the final models having an average PBS of 0.09 Å and TM-score of 0.89. Although correct template identification is essential to most MR experiments, the follow-up structure-reassembly and refinement simulations had a strong impact on improving the success rate. In our tests, 70 of the 161 targets could be solved using the best threading templates with traditional pruning approaches, but an additional 29 targets could successfully be solved by the combination of I-TASSER fragment-structure assembly and progressive model-editing procedures. There are only four cases in which threading-based MR procedures in LOMETS succeeded but I-TASSER-MR failed based on the first I-TASSER model; two of them could be remedied by considering lower-rank I-TASSER models.
Finally, we propose exploiting the C-score of the I-TASSER simulations, which is strongly correlated with the accuracy of the I-TASSER models, to estimate the likelihood of success of the I-TASSER-MR pipeline for a given target. It was shown that the success rate of MR increases almost linearly with the C-score, with nearly 91% of targets being solvable if the C-score is above 1.0. However, when the C-score drops below −1.5 there is little hope of achieving a successful solution.
Despite this success, we note that for 40% of the proteins in our test sets (or 57% of the PSI targets) the pipeline failed to achieve a successful MR solution. The major reason for failure is owing to the relative low similarity of the search model to the targets. Given that the fold of the I-TASSER models is approximately correct for almost all of the proteins tested (i.e. a TM-score of >0.4), including the unsuccessful MR proteins, it may be possible to use the diffraction data to improve the accuracy of the I-TASSER structure predictions. Here, structure decoys generated by the folding simulations are used as a target model for MR. Because good models have a better chance of generating a better match with the diffraction data (for example, as assessed by R free, LLG or TFZ), correlation with the diffraction data can be used as an energy term combined with the I-TASSER force field to guide the structural assembly simulations, provided that the higher quality models can be successfully placed in the unit cell. A similar idea has already been implemented by other laboratories via the integration of electron-density information with Rosetta modeling (DiMaio et al., 2011 ▸; Terwilliger et al., 2012 ▸). A hybrid approach combining diffraction data with cutting-edge structure-modeling techniques should therefore extend the scope of MR even further in solving nonhomologous or distant-homology targets that are traditionally considered to be unsolvable using traditional MR strategies.
Supplementary Material
Supporting Information.. DOI: 10.1107/S2059798316003016/rr5107sup1.pdf
Acknowledgments
The authors are grateful to Drs Renxiang Yan and Jianyi Yang for helpful discussion. Thanks are also due to Dr Randy J. Read and the three anonymous reviewers for their helpful suggestions. This work was supported in part by the National Institutes of General Medical Sciences (GM083107 and GM084222) and of Heart, Lung and Blood (HL086865, HL122416 and HL071818) and the National Natural Science Foundation of China (30700162). YW and YZ conceived the project. YW, JV and ZX conducted the calculations and data analysis and developed the web service. YW, JJGT and YZ wrote the manuscript.
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Afonine, P. V., Grosse-Kunstleve, R. W., Echols, N., Headd, J. J., Moriarty, N. W., Mustyakimov, M., Terwilliger, T. C., Urzhumtsev, A., Zwart, P. H. & Adams, P. D. (2012). Acta Cryst. D68, 352–367. [DOI] [PMC free article] [PubMed]
- Bibby, J., Keegan, R. M., Mayans, O., Winn, M. D. & Rigden, D. J. (2012). Acta Cryst. D68, 1622–1631. [DOI] [PubMed]
- Bunkóczi, G. & Read, R. J. (2011). Acta Cryst. D67, 303–312. [DOI] [PMC free article] [PubMed]
- Bunkóczi, G., Wallner, B. & Read, R. J. (2015). Structure, 23, 397–406. [DOI] [PMC free article] [PubMed]
- Burley, S. K., Almo, S. C., Bonanno, J. B., Capel, M., Chance, M. R., Gaasterland, T., Lin, D., Šali, A., Studier, F. W. & Swaminathan, S. (1999). Nature Genet. 23, 151–157. [DOI] [PubMed]
- Chothia, C. & Lesk, A. M. (1986). EMBO J. 5, 823–826. [DOI] [PMC free article] [PubMed]
- Claude, J.-B., Suhre, K., Notredame, C., Claverie, J.-M. & Abergel, C. (2004). Nucleic Acids Res. 32, W606–W609. [DOI] [PMC free article] [PubMed]
- DiMaio, F., Terwilliger, T. C., Read, R. J., Wlodawer, A., Oberdorfer, G., Wagner, U., Valkov, E., Alon, A., Fass, D., Axelrod, H. L., Das, D., Vorobiev, S. M., Iwaï, H., Pokkuluri, P. R. & Baker, D. (2011). Nature (London), 473, 540–543. [DOI] [PMC free article] [PubMed]
- Dunker, A. K., Obradovic, Z., Romero, P., Garner, E. C. & Brown, C. J. (2000). Genome Inform. Ser. Workshop Genome Inform. 11, 161–171. [PubMed]
- Giorgetti, A., Raimondo, D., Miele, A. E. & Tramontano, A. (2005). Bioinformatics, 21, ii72–ii76. [DOI] [PubMed]
- Jones, D. T. (2001). Acta Cryst. D57, 1428–1434. [DOI] [PubMed]
- Kabsch, W. & Sander, C. (1983). Biopolymers, 22, 2577–2637. [DOI] [PubMed]
- Keegan, R. M. & Winn, M. D. (2008). Acta Cryst. D64, 119–124. [DOI] [PMC free article] [PubMed]
- Kinch, L., Yong Shi, S., Cong, Q., Cheng, H., Liao, Y. & Grishin, N. V. (2011). Proteins, 79, Suppl. 10, 59–73. [DOI] [PMC free article] [PubMed]
- Kleywegt, G. J., Harris, M. R., Zou, J., Taylor, T. C., Wählby, A. & Jones, T. A. (2004). Acta Cryst. D60, 2240–2249. [DOI] [PubMed]
- Kleywegt, G. J. & Jones, T. A. (1997). Methods Enzymol. 277, 208–230. [DOI] [PubMed]
- Levin, E. J., Elsen, N. L., Seder, K. D., McCoy, J. G., Fox, B. G. & Phillips, G. N. Jr (2008). Acta Cryst. D64, 933–940. [DOI] [PMC free article] [PubMed]
- Lobley, A., Sadowski, M. I. & Jones, D. T. (2009). Bioinformatics, 25, 1761–1767. [DOI] [PubMed]
- Long, F., Vagin, A. A., Young, P. & Murshudov, G. N. (2008). Acta Cryst. D64, 125–132. [DOI] [PMC free article] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Storoni, L. C. & Read, R. J. (2005). Acta Cryst. D61, 458–464. [DOI] [PubMed]
- Navaza, J. (1994). Acta Cryst. A50, 157–163.
- Oeffner, R. D., Bunkóczi, G., McCoy, A. J. & Read, R. J. (2013). Acta Cryst. D69, 2209–2215. [DOI] [PMC free article] [PubMed]
- Pawlowski, M. & Bujnicki, J. M. (2012). BMC Bioinformatics, 13, 289. [DOI] [PMC free article] [PubMed]
- Pozharski, E. (2010). Acta Cryst. D66, 970–978. [DOI] [PubMed]
- Qian, B., Raman, S., Das, R., Bradley, P., McCoy, A. J., Read, R. J. & Baker, D. (2007). Nature (London), 450, 259–264. [DOI] [PMC free article] [PubMed]
- Read, R. J. (2001). Acta Cryst. D57, 1373–1382. [DOI] [PubMed]
- Read, R. J. & Chavali, G. (2007). Proteins, 69, Suppl. 8, 27–37. [DOI] [PubMed]
- Rossmann, M. G. (1990). Acta Cryst. A46, 73–82. [DOI] [PubMed]
- Rost, B. (1999). Protein Eng. 12, 85–94. [DOI] [PubMed]
- Sammito, M., Meindl, K., de Ilarduya, I. M., Millán, C., Artola-Recolons, C., Hermoso, J. A. & Usón, I. (2014). FEBS J. 281, 4029–4045. [DOI] [PubMed]
- Schwarzenbacher, R., Godzik, A., Grzechnik, S. K. & Jaroszewski, L. (2004). Acta Cryst. D60, 1229–1236. [DOI] [PubMed]
- Shrestha, R. & Zhang, K. Y. J. (2015). Acta Cryst. D71, 304–312. [DOI] [PubMed]
- Söding, J. (2005). Bioinformatics, 21, 951–960. [DOI] [PubMed]
- Stein, N. (2008). J. Appl. Cryst. 41, 641–643.
- Storoni, L. C., McCoy, A. J. & Read, R. J. (2004). Acta Cryst. D60, 432–438. [DOI] [PubMed]
- Tai, C.-H., Bai, H., Taylor, T. J. & Lee, B. (2014). Proteins, 82, Suppl. 2, 57–83. [DOI] [PMC free article] [PubMed]
- Terwilliger, T. C., DiMaio, F., Read, R. J., Baker, D., Bunkóczi, G., Adams, P. D., Grosse-Kunstleve, R. W., Afonine, P. V. & Echols, N. (2012). J. Struct. Funct. Genomics, 13, 81–90. [DOI] [PMC free article] [PubMed]
- Terwilliger, T. C., Grosse-Kunstleve, R. W., Afonine, P. V., Moriarty, N. W., Zwart, P. H., Hung, L.-W., Read, R. J. & Adams, P. D. (2008). Acta Cryst. D64, 61–69. [DOI] [PMC free article] [PubMed]
- Tress, M. L., Ezkurdia, I. & Richardson, J. S. (2009). Proteins, 77, Suppl. 9, 10–17. [DOI] [PMC free article] [PubMed]
- Wang, G. & Dunbrack, R. L. Jr (2003). Bioinformatics, 19, 1589–1591. [DOI] [PubMed]
- Wu, S., Skolnick, J. & Zhang, Y. (2007). BMC Biol. 5, 17. [DOI] [PMC free article] [PubMed]
- Wu, S. & Zhang, Y. (2007). Nucleic Acids. Res. 35, 3375–3382. [DOI] [PMC free article] [PubMed]
- Wu, S. & Zhang, Y. (2008). Proteins, 72, 547–556. [DOI] [PMC free article] [PubMed]
- Xu, D., Jaroszewski, L., Li, Z. & Godzik, A. (2014). Bioinformatics, 30, 660–667. [DOI] [PMC free article] [PubMed]
- Xu, J. & Zhang, Y. (2010). Bioinformatics, 26, 889–895. [DOI] [PMC free article] [PubMed]
- Xu, Y. & Xu, D. (2000). Proteins, 40, 343–354. [PubMed]
- Yan, R., Xu, D., Yang, J., Walker, S. & Zhang, Y. (2013). Sci. Rep. 3, 2619. [DOI] [PMC free article] [PubMed]
- Yang, J., Wang, Y. & Zhang, Y. (2016). J. Mol. Biol. 428, 693–701. [DOI] [PMC free article] [PubMed]
- Yang, J., Yan, R., Roy, A., Xu, D., Poisson, J. & Zhang, Y. (2015). Nature Methods, 12, 7–8. [DOI] [PMC free article] [PubMed]
- Yang, Y., Faraggi, E., Zhao, H. & Zhou, Y. (2011). Bioinformatics, 27, 2076–2082. [DOI] [PMC free article] [PubMed]
- Zemla, A., Venclovas, Č., Moult, J. & Fidelis, K. (1999). Proteins, 37, Suppl. S3, 22–29. [DOI] [PubMed]
- Zhang, J., Liang, Y. & Zhang, Y. (2011). Structure, 19, 1784–1795. [DOI] [PMC free article] [PubMed]
- Zhang, Y. (2008a). BMC Bioinformatics, 9, 40. [DOI] [PMC free article] [PubMed]
- Zhang, Y. (2008b). Curr. Opin. Struct. Biol. 18, 342–348. [DOI] [PMC free article] [PubMed]
- Zhang, Y. (2014). Proteins, 82, Suppl. 2, 175–187. [DOI] [PMC free article] [PubMed]
- Zhang, Y., Kihara, D. & Skolnick, J. (2002). Proteins, 48, 192–201. [DOI] [PubMed]
- Zhang, Y., Kolinski, A. & Skolnick, J. (2003). Biophys. J. 85, 1145–1164. [DOI] [PMC free article] [PubMed]
- Zhang, Y. & Skolnick, J. (2004a). J. Comput. Chem. 25, 865–871. [DOI] [PubMed]
- Zhang, Y. & Skolnick, J. (2004b). Proteins, 57, 702–710. [DOI] [PubMed]
- Zhang, Y. & Skolnick, J. (2005). Nucleic Acids Res. 33, 2302–2309. [DOI] [PMC free article] [PubMed]
- Zhou, H. & Zhou, Y. (2005). Proteins, 58, 321–328. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information.. DOI: 10.1107/S2059798316003016/rr5107sup1.pdf