Abstract
The multidrug transporter P-glycoprotein (P-gp, ABCB1) is an ATP-dependent pump that mediates the efflux of structurally diverse drugs and xenobiotics across cell membranes, affecting drug pharmacokinetics and contributing to the development of multidrug resistance. Structural information about the conformational changes in human P-gp during the ATP hydrolysis cycle has not been directly demonstrated, although mechanistic information has been inferred from biochemical and biophysical studies conducted with P-gp and its orthologs, or from structures of other ATP-binding cassette transporters. Using single-particle cryo-electron microscopy, we report the surprising discovery that, in the absence of the transport substrate and nucleotides, human P-gp can exist in both open [nucleotide binding domains (NBDs) apart; inward-facing] and closed (NBDs close; outward-facing) conformations. We also probe conformational states of human P-gp during the catalytic cycle, and demonstrate that, following ATP hydrolysis, P-gp transitions through a complete closed conformation to a complete open conformation in the presence of ADP.
Introduction
P-glycoprotein (P-gp) is a member of the ATP-binding cassette (ABC) protein superfamily, which mediates the detoxification of cells in an ATP-dependent fashion by exporting a wide range of structurally dissimilar compounds across the plasma membrane (Ambudkar et al., 1999). ABC transporters share common structural features: they have a symmetric or pseudosymmetric structure, with two transmembrane domains (TMDs) and two nucleotide-binding domains (NBDs) (Wilkens, 2015). Binding and hydrolysis of ATP is mediated by the NBDs; ATP binding has generally been thought to bring the NBDs into close proximity, although this has not been demonstrated structurally.
A common theme of efflux by ABC transporters is that ATP hydrolysis at the interface of NBDs is required for transition between an inward-facing conformation (which allows drug/substrate binding to a cytoplasmically accessible pocket between the TMDs) and an outward-facing conformation (which would then allow release of the substrate to the extracellular space) (Wilkens, 2015). ATP hydrolysis is known to require the “closure” of the NBDs, where both arms of the cytoplasmic domain come into close contact; the rate-limiting step of this reaction is the release of ADP posthydrolysis (Kerr et al., 2001). Whereas the presence of certain substrates can impact the rate of ATP hydrolysis, human P-gp has a significant basal ATPase activity in the absence of any substrate (Ramachandra et al., 1996; Loo et al., 2012). Thus, although ATPase activity is required for drug transport, the presence or absence of substrate does not alter the fundamental conformational changes involved in the ATP hydrolysis cycle.
Several structures of both eukaryotic and prokaryotic ABC transporters have been reported using X-ray crystallography (Dawson and Locher, 2006; Aller et al., 2009; Jin et al., 2012; Choudhury et al., 2014; Perez et al., 2015). The majority of these structures are found in the open, inward-facing conformation, and no matched outward-facing or closed conformations of any single eukaryotic ABC transporter family member have been reported yet. Because of this, the structural mechanisms underlying either the ATPase activity or the transport activity of P-gp are not adequately defined. An additional layer of complexity arises from the fact that P-gp is highly conformationally flexible, with large movements of the NBD arms in the course of the reaction cycle, making it challenging to determine the range of functionally relevant conformations using X-ray crystallography (Aller et al., 2009; Ward et al., 2013; Moeller et al., 2015; Szewczyk et al., 2015).
In this study, we used cryo-electron microscopy (cryo-EM) to evaluate the conformations of human P-gp (hP-gp) in each distinct state of its ATPase cycle. Since P-gp exhibits basal ATPase activity, we evaluated hP-gp conformations at four different points in the ATPase cycle in the absence of any added substrates. Despite the low resolution of the cryo-EM density maps, likely originating from the flexible nature of P-gp, we observed that hP-gp in its apo state can coexist in two conformations, where the NBDs are either separated (resembling what is likely an inward-facing conformation) or in close proximity (resembling a probable outward-facing conformation). These coexisting conformations are present even in the presence of bound ATP prior to hydrolysis, whereas post-hydrolysis the transporter sequentially transitions through outward-facing and inward-facing states that correspond to conformations before and after release of phosphate from the active site, respectively.
Materials and Methods
Purification of Human P-gp.
Crude membranes from High Five insect cells (Thermo Fisher Scientific, Waltham, MA) expressing wild-type (WT)–hP-gp, or its double mutant EQ–hP-gp (E556Q/E1201Q), with a 6-histidine tag at the C-terminal end were isolated as described previously (Kerr et al., 2001). The membranes (150–250 mg of protein) were solubilized by using 2% n-dodecyl β-d-maltoside in a buffer containing 10 mM Tris-HCl (pH 7.5), 150 mM NaCl, 15% glycerol, 5 mM β-mercaptoethanol, 20 mM imidazole, and UltraCruz EDTA-free protease inhibitor cocktail tablets (Santa Cruz Biotechnology, Dallas, TX). The solubilized extract was centrifuged at 38,000 rpm (Ti-45 rotor; Beckman Coulter, Brea, CA) for 45 minutes at 4°C. The supernatant was incubated with Ni-NTA resin (Qiagen Inc., Valencia, CA) pre-equilibrated in solubilization buffer with 0.09% n-dodecyl β-d-maltoside for 14–16 hours at 4°C. The beads were washed and P-gp was eluted with the same buffer containing 300 mM imidazole. Fab isolated from UIC2 monoclonal antibody was then added at a molar ratio of P-gp:Fab (1:3) and incubated at 4°C for 15 minutes. The hP-gp–Fab complex was centrifuged at 300,000 × g for 45 minutes. The complex formation was evaluated on native PAGE 4–16% Bis-Tris protein gels (Thermo Fisher Scientific, Waltham, MA).
Purification of Fab from UIC2 Monoclonal Antibody.
The hybridoma (UIC2/A) producing the UIC2 monoclonal antibody directed against an extracellular domain of a cell surface hP-gp was obtained from Dr. Eugene Mechetner (Mechetner et al., 1997). UIC2 monoclonal antibody was produced from this hybridoma as described previously (Mechetner et al., 1997). Fab fragments were isolated using a Fab Preparation Kit (Thermo Fisher Scientific, Waltham, MA) according to instructions provided by the manufacturer. The purity and the yield of the Fab fragment were evaluated by separating purified Fab on 4–16% Bis-Tris protein gel under reducing and nonreducing conditions, followed by staining with colloidal blue stain.
Grid Preparation for Cryo-Electron Microscopy.
Two and a half microliters of UIC2 Fab-bound hP-gp solution (2–4 mg/ml) in the absence or presence of nucleotides and, where appropriate, with 0.3 mM sodium orthovanadate was deposited on plasma cleaned Quantifoil TEM grids (Quantifoil, Jena, Germany). Blotting of excess liquid and plunge-freezing into liquid ethane were performed with a Leica EM GP (Leica Microsystems, Wetzlar, Germany) set to a blotting time of 3–5 seconds and 95% humidity.
Data Acquisition.
Electron micrographs were recorded as movie frames using a Titan Krios electron microscope equipped with a Falcon II detector (FEI Company, Hillsboro, OR). Specimens were imaged at a nominal magnification of 47,000×, corresponding to a pixel size of 1.41 Å at the specimen plane. Micrographs were acquired automatically with the data collection software EPU (FEI Company) spanning a defocus range of −1.5 to −2.5 µm, at a total dose of ∼70 e-/Å2 distributed over seven intermediate frames.
Image Processing.
Drifts and beam-induced motion were compensated by iterative alignment of the intermediate frames using an expectation-maximization algorithm as previously described (Bartesaghi et al., 2014). The program CTFFIND3 was used to estimate contrast transfer function parameters (Mindell and Grigorieff, 2003). Micrographs were selected by manual inspection, and particles were picked semi-automatically by cross correlation of the images with a Gaussian disk with a radius of 110 Å, followed by a search for local maxima. The particles were extracted in a box size of 224 × 224 pixels and classified into two-dimensional (2D) classes using reference free 2D classification (Tang et al., 2007). Classes with clear structural features from the EQ–hP-gp mutant in the apo conformation data set were used to generate a de novo three-dimensional (3D) initial model using EMAN2 (Tang et al., 2007). The clear density of the Fab bound to the extracellular side of the protein and the distinctive NBD density that resembles the NBDs in crystal structures of ABC transporters validate the initial model. The final model used projection images that only contained the first ∼35 e-/Å2 of the dose, and reconstruction with 3D classification was carried out using the program RELION (version 1.3) (Scheres, 2012). The apo state EQ–hP-gp data were classified with 11, 5, and 3 classes; all other data sets were classified with 5 and 3 classes. In all cases, except for the ADP-bound state, the number of classes did not alter the results. In the case of the ADP data set, an additional class, corresponding to the intermediately open structure, was identified only after classifying with 5 classes, rather than with 3 classes. A summary of the number of particles in each 3D class from each state can be found in Supplemental Table 1. Fourier Shell Correlation curves for the EQ apo open and closed structures (those which were used for docking crystal structures) are presented in Supplemental Fig. 1A. The orientation distributions of the projection images of all maps presented in this study are presented in Supplemental Fig. 1, B-F, indicating well spread orientation distribution.
Fits of X-ray Coordinates into Cryo-EM Maps.
Structures of available X-ray crystallographic models of homologous ABC transporters were placed into maps from our study using UCSF Chimera (Pettersen et al., 2004). Models used in this study include those of mouse P-gp [Protein Data Bank number (PDB) 4M1M and 4KSC], Staphylococcus aureus Sav1866 (PDB 2HYD), and Escherichia coli McjD (PDB 4PL0). Prior to fitting the X-ray structures, maps were thresholded according to the molecular volume of the protein. The threshold values for our experimentally determined maps were selected based on the volume of the Fab region, which was matched to the X-ray coordinates of the UIC2 crystal structure (L. Esser, S. Shukla, et al., unpublished data; PDB 5JUE). As the cytoplasmic domains in the maps of the open conformations are clearly separated from each other, it is possible to compare the volume of the cytoplasmic regions of the filtered 4M1M crystal structure with the respective regions in our cryo-EM maps; the thresholds established by analyzing the volume of the Fab region yielded volumes for the cytoplasmic regions that matched those of 4M1M. We then assessed how well each crystal structure model fit our cryo-EM data by computing the percentage of atoms in the crystal structure that lay outside the cryo-EM map threshold.
ATPase Assay.
Vanadate-sensitive ATP hydrolysis by hP-gp in crude membranes was measured as described previously (Ambudkar, 1998).
Extraction of Nucleotide from Purified Protein.
An equal volume of a phenol:chloroform:isoamyl alcohol (25:24:1, v/v/v) mixture was added to either purified WT–hP-gp (15 μM), purified EQ–hP-gp (15 μM), or 10 μM ATP in elution buffer and vortexed vigorously for 2 minutes. The aqueous layer was then separated from the organic layer by centrifuging at 13,000 rpm for 5 minutes at 4°C. The upper aqueous layer was then resolved using a Vydac C18 column (Fisher Scientific, Waltham, MA) at a flow rate of 0.3 ml/min using gradient elution (solvent A: 100 mM KH2PO4, pH 6.0; solvent B: 90% methanol) with absorbance measured at 254 nm, as described previously (Kim et al., 2015).
Oxidative Cross-Linking of hP-gp Using Copper Sulfate.
Crude membranes from High-Five insect cells [50 μg of protein in 50 μl of buffer (10 mM Tris-HCl, 150 mM NaCl, pH 7.5)] were incubated with 5 mM ATP (Sigma-Aldrich, St. Louis, MO) and 10 mM MgCl2 in the presence or absence of 0.3 mM vanadate for 10 minutes at 37°C. CuSO4 (0.5 mM) was then added for various time points ranging from 15 seconds to 15 minutes at 37°C to complete the cross-linking. The reactions were stopped by adding 10 mM N-ethyl maleimide, 20 mM EDTA and SDS-PAGE sample buffer without reducing agent. The samples (0.25–0.5 μg of protein) were subjected to SDS-PAGE (7% Tris-acetate gel; Thermo Fisher Scientific, Waltham, MA) at a constant voltage of 150 V for 2 hours, and immunoblot analysis was performed with P-gp–specific mouse monoclonal antibody C219 (1:2000 dilution) and enhanced chemiluminescence (GE Healthcare Bio-Sciences, Pittsburgh, PA).
Results
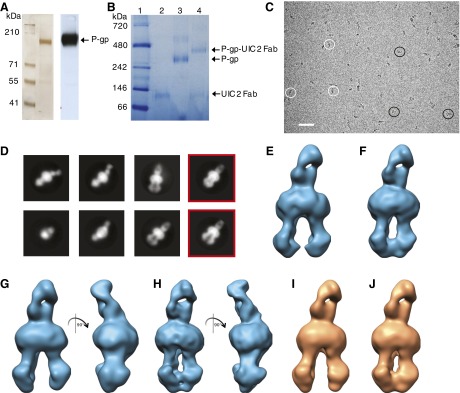
We carried out cryo-EM studies of hP-gp in each of the four pivotal states in the ATPase cycle: the “apo,” unbound state (P-gp); the prehydrolysis ATP-bound state (P-gp•ATP); the post-hydrolytic ADP-trapped state obtained in the presence of sodium orthovanadate (Vi) [P-gp•ADP•Vi, equivalent to the phosphate (Pi) bound form, P-gp•ADP•Pi]; and the ADP-bound post-Pi release state (P-gp•ADP). Because P-gp is small and pseudosymmetric, it can be challenging to properly assign orientations of particles during cryo-EM analysis. To facilitate more accurate assignment of particle orientations in projection images, we determined structures of the complex of hP-gp and the Fab fragment of the hP-gp–specific monoclonal antibody UIC2, which binds to the extracellular regions of hP-gp (Mechetner et al., 1997), forming a stable complex (hP-gp•UIC2) with purified P-gp (Fig. 1, A and B). UIC2 has been shown not to affect the ATPase activity of human P-gp (Ritchie et al., 2011), making this a useful tool to determine P-gp structure. Projection electron micrographs of hP-gp dispersed in vitreous ice display a range of orientations, including side and top views (Fig. 1C).
Fig. 1.
Cryo-EM analysis of nucleotide-free and ATP-bound prehydrolytic states of hP-gp. (A) Purified P-gp (2 μg of protein) was separated on a 7% Tris-acetate gel and detected by silver stain (left panel) or Western blot analysis (right panel) using the C219 antibody. The protein gel and the Western blot are representative of four independent experiments. (B) Two micrograms of purified Fab, hP-gp, and hP-gp–Fab complex were separated on a native 4–16% Bis-Tris gel and stained with colloidal blue stain. Lanes display molecular weight standards (lane 1), purified UIC2 Fab fragment (lane 2), purified hP-gp (lane 3), and hP-gp–Fab complex (lane 4). The native gel is representative of three independent experiments. (C) A representative micrograph of EQ mutant hP-gp in the apo (nucleotide-free) state taken at an approximate defocus value of −3.4 µm. Selected front- and side-view projections of the protein are indicated with black and white circles, respectively. Scale bar = 500 Å. (D) Representative 2D image classes from apo EQ–hP-gp bound to UIC2. The NBD regions in the classes display variations in arrangement relative to the bound Fab, with the NBDs closer in some and further apart in others (two far-right red squares). (E and F) 3D cryo-EM reconstructions of hP-gp bound to UIC2 antibody in open (E) and closed (F) conformations. (G–J) The EQ–hP-gp mutant displays open (G and I) and closed (H and J) conformations in both apo (G and H) and ATP-bound (I and J) states. All maps presented here have resolutions of ∼15 Å based on the 0.5 Fourier Shell Correlation criterion from comparing two half-maps, and display features consistent with cryo-EM density maps at this resolution (Supplemental Fig. 2A).
2D class averages show clear evidence of distinct P-gp conformations with “front view” classes (Fig. 1D) that differ primarily with respect to the proximity of the two NBDs: in one, they are separated with a classic inverted V-shape formation corresponding to the shape of the “inward-facing” conformation of the transporter, whereas in the other, they are in close contact, with a well connected “bridge” of density between the two NBD arms. These differences were also confirmed with 3D classification, demonstrating the presence of two prominent classes with roughly similar proportions, consistent with open and closed hP-gp conformations (Fig. 1, E and F, respectively). To our knowledge, the presence of a closed conformation in the apo (nucleotide- and substrate-free) state where the NBDs are in close contact has not yet been reported for any member of the ABC transporter family.
To verify that the observed closed-state conformation was not an artifact due to trapped nucleotides that were carried with P-gp through the purification procedures, purified hP-gp samples (15 μM) were subjected to nucleotide extraction with phenol:chloroform:isoamylalcohol as described previously (Kim et al., 2015); in this analysis, trapped carryover ATP in the purified protein would result in micromolar concentrations of ATP. Even though our assay could detect micromolar concentrations (10 μM) of ATP, the concentration of any ATP trapped in the purified protein sample was too low to be detected (Supplemental Fig. 2A). These chemical analyses thus confirm that the closed conformation can be sampled even in the absence of bound nucleotide.
Because WT hP-gp immediately hydrolyses ATP, to explore the transient, ATP-bound state of hP-gp, we used a mutant form of hP-gp, E556Q/E1201Q. In the E556Q/E1201Q (EQ) mutant, the catalytic glutamate residues within the Walker B domain in both NBDs are substituted with glutamine, resulting in occlusion of ATP in the prehydrolytic state (Sauna et al., 2006, 2007). As observed with WT–hP-gp, both open and closed conformations also exist in the apo state of the EQ–hP-gp mutant (Fig. 1, G and H). The overall shapes of both open and closed conformations of the apo (nucleotide-free) EQ mutant are essentially superimposable with those observed for the WT protein, indicating that the EQ mutation does not significantly alter the structural landscape of hP-gp. Essentially the same mixture of closed and open conformations was observed upon analysis of the EQ mutant in the presence of ATP, providing a structural snapshot of the next step in the reaction cycle, which precedes ATP hydrolysis (the EQ mutant lacks the ability to hydrolyze ATP) (Fig. 1, I and J). The presence of trapped ATP in the EQ–hP-gp under the conditions used for cryo-EM analysis was confirmed by high-performance liquid chromatography of the extracted nucleotides from this sample (Supplemental Fig. 2B).
Hydrolysis of ATP by P-gp leads to a transient post-hydrolytic state, in which ADP and phosphate remain in the active site. To capture the post-hydrolytic state, WT–hP-gp was incubated with ATP and Vi at 37°C for 15 minutes. Vi has been used to arrest hP-gp and other ABC transporters in the nucleotide-trapped transition state (P-gp•ADP•Vi) (Urbatsch et al., 1995; Senior, 2011). This state is known to have low affinity for transport substrates, and has been proposed to be outward-facing (Sauna and Ambudkar, 2000). Our structural analysis of the post-hydrolytic state shows that only the closed conformation with bridged NBDs is present. In this state, Vi binding enables the ADP generated by hydrolysis to stay bound to the protein. Our findings that hP-gp is in a closed conformation in the presence of ATP and Vi under conditions in which ATP is hydrolyzed is at variance with a recent report (Moeller et al., 2015) on ABC transporter structures, in which the closed state for mouse P-gp was not observed under these conditions. It is possible that the use of cryo-EM rather than negative stain and the higher resolution of our maps may explain our observation of the closed conformation in this step of the P-gp reaction cycle.
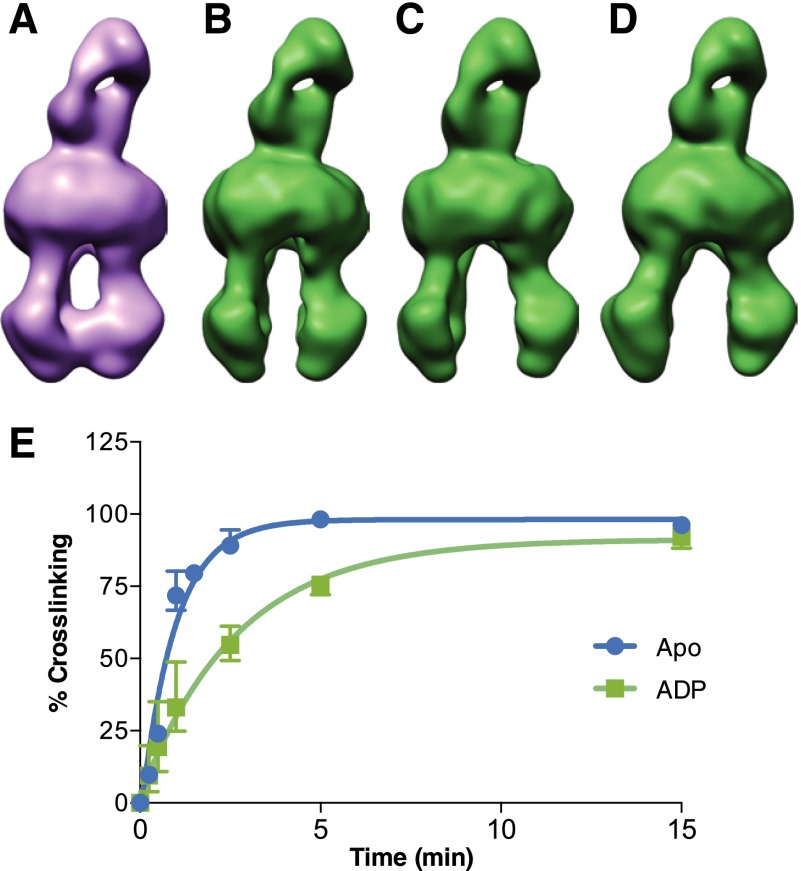
To determine the conformation of the post-hydrolytic ADP-bound state of hP-gp, the final step in the cycle, we incubated WT–hP-gp with 5 mM ADP and 5 mM MgCl2. Under these conditions, 3D classification revealed several open conformations (Fig. 2, B–D), suggesting that the presence of ADP prevents closure of the NBDs. Whereas we present only three different density maps for this state, our analysis cannot rule out the existence of a continuum of conformations that are sampled by the transporter in the presence of ADP. To independently test the conformational landscape derived by cryo-EM analysis, we compared the extent of cysteine cross-linking in the presence of ADP with that observed for apo hP-gp. Previous studies have established that, under physiologically relevant conditions, the NBDs of P-gp can be cross-linked by a disulfide bond between C431 and C1074 in apo, but not in the ADP•Vi-bound state (Supplemental Fig. 2C). Additionally, the apo EQ–hP-gp mutant likewise is amenable to cross-linking, whereas the ATP-bound form is somewhat less so (Supplemental Fig. 2D). The precise conformational mechanism by which C431 and C1074 come into close enough proximity (>5 Å) to cross-link is not yet clear, but some level of conformational flexibility seems to be required for this cross-linking to occur. The conformational flexibility of these residues has also been supported by cross-linking in apo conformation with the bifunctional cross-linker M17M, which has a 24-Å linker length (Sim et al., 2013). Evaluation of the relative extent of cross-linking for the apo and ADP-bound conditions for hP-gp over time showed that the time required for 50% CuSO4-induced cross-linking (T1/2) of C431 and C1074 in the NBDs changed from 0.65 (+0.20/−0.12) minutes for the apo form to 1.86 (+0.88/−0.45) minutes in the presence of ADP (Fig. 2E). This significant slowing of NBD cross-linking supports our conclusion that ADP-bound hP-gp exists primarily in an open conformation.
Fig. 2.
Cryo-EM analysis of hP-gp in post-hydrolytic and ADP-bound states. (A) WT–hP-gp bound to UIC2 Fab and trapped with ADP•Vi in the post-hydrolytic state reveals only the closed conformation, even with 3D classification. (B–D) WT–hP-gp bound to UIC2 Fab and incubated with 5 mM ADP is found in multiple open conformations, but not in the closed conformation. (E) WT–hP-gp–expressing crude membranes were incubated in the absence (apo, circle, blue) or presence of 5 mM ADP (square, green) at 37°C for 10 minutes. CuSO4-induced cross-linking was performed for the indicated times (x-axis). The extent of cross-linking expressed as a percentage (y-axis) was calculated by densitometric analysis of cross-linked P-gp band on Western blots. The error bars denote standard error (n = 3), and the time required for 50% cross-linking (T1/2) was calculated by nonlinear regression analysis to an exponential decay using GraphPad Prism 6.0 (GraphPad Software, La Jolla, CA).
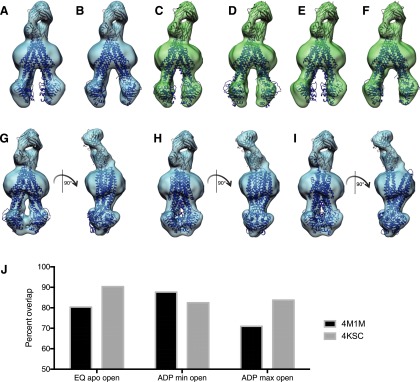
Numerous crystal structures for ABC transporter family members have previously been reported; however, the majority of these are found in an open, inward-facing conformation. Because the resolution of our cryo-EM maps is not sufficient to allow us to create a model for hP-gp directly, we have generated molecular envelopes by fitting existing models of ABC transporters into the density maps. The models that most resemble the open hP-gp cryo-EM conformations are those of mouse P-gp in the open, inward-facing conformation (PDB 4M1M and 4KSC) (Ward et al., 2013; Li et al., 2014). By rigidly fitting each of these into the open conformation of apo EQ–hP-gp (Fig. 3, A and B) and into the open structure of WT-ADP hP-gp with the greatest distance (Fig. 3, E and F) or least distance (Fig. 3, C and D) between NBDs, we can see that the previously determined open conformations for mouse P-gp closely match the spectrum of open conformations that we observed. Quantitation of the relative fit of each of these models into our maps suggests that the 4KSC mouse P-gp structure fits better to the apo open state or to the open conformation of hP-gp•ADP with greater distance between the NBDs, whereas the 4M1M mouse P-gp model is a better match for the structure of hP-gp•ADP with the least distance between NBDs (Fig. 3J).
Fig. 3.
Cryo-EM reconstructions of open and closed conformations of hP-gp fit with X-ray crystallographic models. X-ray crystallographic models of mouse P-gp in the open conformation (PDB entries 4M1M and 4KSC) were fit into hP-gp cryo-EM density maps along with coordinates of the UIC2 Fab (obtained from X-ray crystallographic analysis; Esser L., et al., unpublished data; PBD 5JUE) using the program Chimera (Pettersen et al., 2004). (A and B) Map of the nucleotide-free EQ–hP-gp mutant in the open-conformation fit with 4M1M (A) or 4KSC (B). (C and D) Map of WT–hP-gp bound to ADP with most-separated NBDs fit with 4M1M (C) or 4KSC (D). (E and F) Map of WT–hP-gp bound to ADP with least-separated NBDs fit with 4M1M (E) or 4KSC (F). (G–I) Map of WT–hP-gp bound to ADP•Vi fit with 4M1M (G), the model of S. aureus ABC transporter Sav1866 (PBD 2HYD) (H), or the model of E. coli ABC transporter McjD (PBD 4PL0) (I). (J) Quantitation of best fit of open conformations with 4M1M and 4KSC, showing the total percentage of atoms in the X-ray models that fall within the density of our cryo-EM–derived map. Maps were identically thresholded to the volume of the UIC2 Fab crystal structure (PDB 5JUE) filtered to 15-Å resolution.
We fit the EQ apo closed map with the only available structures reported to be of an occluded or outward-facing conformation, those of McjD (PDB 4PLO) from E. coli and Sav1866 (PBD 2HYD) from S. aureus, respectively, as well as the structure of mouse P-gp in the open conformation (PBD 4M1M; Fig. 3G). As shown in Fig. 3, H and I, neither 4PLO nor 2HYD adequately fit our closed structure, as the angles between the NBDs and the remainder of the protein are not consistent with our data. In contrast to the open (apo) conformation, none of the available crystal structures, including those reported to capture a closed state, show a good match to the density map of the closed hP-gp form that we observed, suggesting that human P-gp exists in a unique conformation in its closed (outside open) state, which is different from reported closed conformations of other ABC transporters.
Discussion
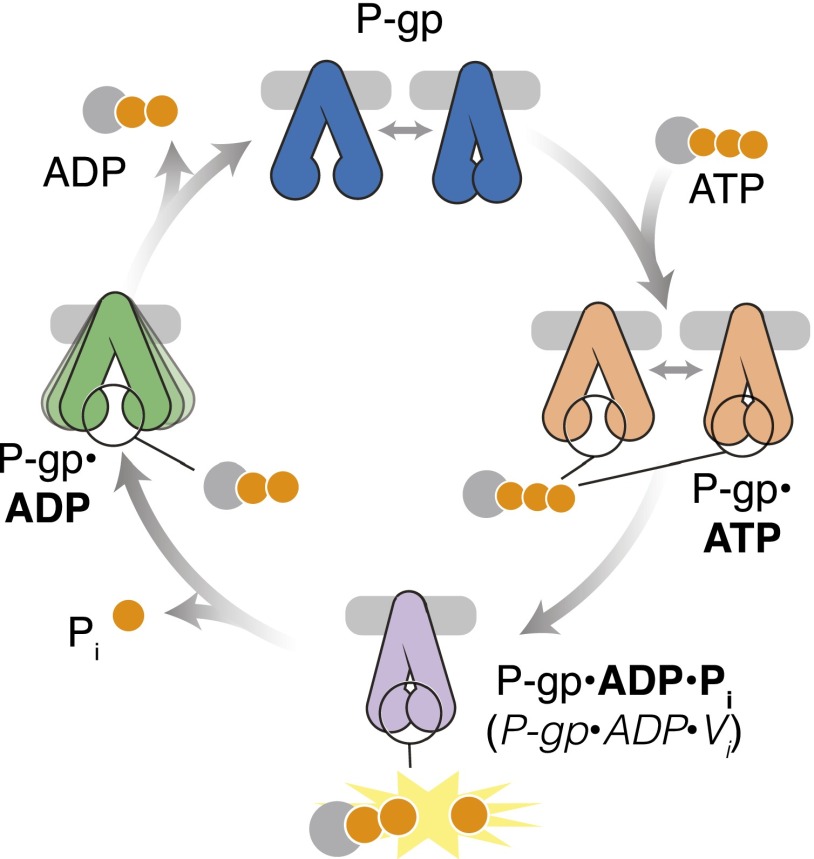
The ATPase cycle of mammalian P-gp has been biochemically well characterized (Sauna and Ambudkar, 2007; Al-Shawi, 2011; Sharom, 2014). The reaction cycle is the same in the absence or presence of transport substrate, except that substrates alter the maximal velocity without affecting the Km for ATP (Ambudkar et al., 1992; Sarkadi et al., 1992). The substrates or modulators affect the rate of ADP release, as this is the rate-limiting step in the ATP hydrolysis cycle (Kerr et al., 2001). In this study, we used cryo-EM to analyze the conformational landscape of the hP-gp ATPase cycle (Fig. 4), demonstrating a dynamic equilibrium between an open conformation and a novel closed conformation in the nucleotide-free and prehydrolytic ATP-bound states. Following ATP hydrolysis, and the subsequent release of phosphate (due to its significantly lower affinity) (Urbatsch et al., 1995), the hP-gp•ADP complex is predominantly in an open, inward-facing conformation. Upon dissociation of ADP from the catalytic site, hP-gp is reset for the next cycle. At the current resolution of these maps, we cannot unequivocally demonstrate whether the open and closed conformations correspond to inward-facing or outward-facing in the TMD. Nevertheless, we have previously shown that the ADP•Vi-bound state is an outward-facing conformation, as this posthydrolysis state has relatively low affinity for drug substrate (Sauna and Ambudkar, 2000), suggesting that the closed conformation observed here is outward-facing, and that the open conformations are inward-facing. Although substrate binding to the drug-binding pocket in the transmembrane region is not required for the basal ATPase activity, the discovery of the uniformly closed conformation in the post-hydrolytic state and the predominantly open conformation in the ADP-bound state also supports the finding that drug-substrate transport is coupled to ATPase activity (Ambudkar et al., 1997).
Fig. 4.
Schematic of hP-gp at major steps during the ATPase reaction cycle. Our data indicate that, in the nucleotide free state (top, dark blue), hP-gp exists in a dynamic equilibrium between open and closed conformations. Binding of ATP (right, orange) also allows both open and closed conformations. After hydrolysis of ATP to ADP•Pi (as trapped by ADP•Vi; bottom, purple), hP-gp is found exclusively in the closed conformation. Release of Pi allows the ADP-bound state of hP-gp (left, green) to open. The ADP-bound state is highly flexible, but remains in the open state. Release of ADP allows hP-gp to sample both open and closed conformations (top).
The presence of possible pseudosymmetry in certain projection views could be a source for misalignment, thereby limiting map resolution. In addition, it is possible that the presence of the UIC2 Fab and our use of detergent micelles may influence the relative proportion of molecules in each state in our structural analysis. Further structural studies of hP-gp in a native membrane environment may shed more light on whether antibody binding affects the equilibrium between open and closed conformations. Nevertheless, the conformational heterogeneity we report here is consistent with earlier findings, and potentially explains the apparently conflicting results reported previously with various other members of the ABC transporter family. For example, the ATP analog adenylyl-imidodiphosphate (AMP-PNP)–bound form of the S. aureus transporter Sav1866 was reported to be present in an outward-facing conformation (Dawson and Locher, 2006), whereas that of the mitochondrial transporter ABCB10 bound to AMP-PNP was reported to be present in an inward-facing conformation (Shintre et al., 2013). For the bacterial heterodimeric ABC transporter TM287/288, NBDs were observed in close contact (but still in an inward-facing conformation) in the presence or absence of AMP-PNP (Hohl et al., 2014). Similarly, a recent study suggested that a bacterial ABC lipid-linked oligosaccharide flippase (PglK) can adopt an inward-facing conformation in the absence of nucleotide, and an outward-facing conformation in the presence of ATP in vitro, generating a “futile cycle.” Nevertheless, the functional state required for flippase activity was found to be only the outward-facing state (see Fig. 5 in Perez et al., 2015). We propose that the equilibrium between the inward- and outward-facing conformations may vary for different transporters, thus rationalizing these seemingly divergent observations across various members of this protein family.
Supplementary Material
Acknowledgments
The authors thank Drs. Jason Pierson and Lingbo Yu for assistance with data collection, Michael Maurizi and Luke Diorio for help with separation of nucleotides, Dr. Soojay Banerjee for many helpful discussions, George Leiman for assistance in preparing the manuscript, and Veronica Falconieri for assistance with preparing the schematic illustration. This study used the computational resources of the National Institutes of Health High-Performance Computing Biowulf cluster (http://hpc.nih.gov). The density maps presented here have been deposited with the Electron Microscopy Data Bank (accession numbers EMD-3421, 3422, 3423, 3424, 3425, 3426, 3427, 3428, 3429, and 3430), and the refined atomic model for UIC2 has been deposited in the Protein Data Bank (accession number PDB-5JUE).
Abbreviations
- ABC
ATP-binding cassette
- AMP-PNP
adenylyl-imidodiphosphate
- cryo-EM
cryo-electron microscopy
- hP-gp
human P-glycoprotein
- NBD
nucleotide-binding domain
- PDB
Protein Data Bank
- P-gp
P-glycoprotein
- 2D
two-dimensional
- 3D
three-dimensional
- TMD
transmembrane domain
- Vi
sodium orthovanadate
- WT
wild type
Authorship Contributions
Participated in research design: Frank, Shukla, Ambudkar, Subramaniam.
Conducted experiments: Frank, Shukla, Rao, Merk, Mobin.
Contributed new reagents or analytic tools: Esser, Gottesman, Xia.
Performed data analysis: Frank, Shukla, Borgnia, Bartesaghi, Ambudkar, Subramaniam.
Wrote or contributed to the writing of the manuscript: Frank, Shukla, Earl, Ambudkar, Subramaniam.
Footnotes
This work was supported by funds from the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research, the NIH-FEI Living Laboratory for Structural Biology, and the Center for Cancer Research Center for Molecular Microscopy.
 This article has supplemental material available at molpharm.aspetjournals.org.
This article has supplemental material available at molpharm.aspetjournals.org.
References
- Aller SG, Yu J, Ward A, Weng Y, Chittaboina S, Zhuo R, Harrell PM, Trinh YT, Zhang Q, Urbatsch IL, et al. (2009) Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding. Science 323:1718–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Shawi MK. (2011) Catalytic and transport cycles of ABC exporters. Essays Biochem 50:63–83. [DOI] [PubMed] [Google Scholar]
- Ambudkar SV. (1998) Drug-stimulatable ATPase activity in crude membranes of human MDR1-transfected mammalian cells. Methods Enzymol 292:504–514. [DOI] [PubMed] [Google Scholar]
- Ambudkar SV, Cardarelli CO, Pashinsky I, Stein WD. (1997) Relation between the turnover number for vinblastine transport and for vinblastine-stimulated ATP hydrolysis by human P-glycoprotein. J Biol Chem 272:21160–21166. [DOI] [PubMed] [Google Scholar]
- Ambudkar SV, Dey S, Hrycyna CA, Ramachandra M, Pastan I, Gottesman MM. (1999) Biochemical, cellular, and pharmacological aspects of the multidrug transporter. Annu Rev Pharmacol Toxicol 39:361–398. [DOI] [PubMed] [Google Scholar]
- Ambudkar SV, Lelong IH, Zhang J, Cardarelli CO, Gottesman MM, Pastan I. (1992) Partial purification and reconstitution of the human multidrug-resistance pump: characterization of the drug-stimulatable ATP hydrolysis. Proc Natl Acad Sci USA 89:8472–8476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartesaghi A, Matthies D, Banerjee S, Merk A, Subramaniam S. (2014) Structure of β-galactosidase at 3.2-Å resolution obtained by cryo-electron microscopy. Proc Natl Acad Sci USA 111:11709–11714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury HG, Tong Z, Mathavan I, Li Y, Iwata S, Zirah S, Rebuffat S, van Veen HW, Beis K. (2014) Structure of an antibacterial peptide ATP-binding cassette transporter in a novel outward occluded state. Proc Natl Acad Sci USA 111:9145–9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson RJ, Locher KP. (2006) Structure of a bacterial multidrug ABC transporter. Nature 443:180–185. [DOI] [PubMed] [Google Scholar]
- Hohl M, Hürlimann LM, Böhm S, Schöppe J, Grütter MG, Bordignon E, Seeger MA. (2014) Structural basis for allosteric cross-talk between the asymmetric nucleotide binding sites of a heterodimeric ABC exporter. Proc Natl Acad Sci USA 111:11025–11030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin MS, Oldham ML, Zhang Q, Chen J. (2012) Crystal structure of the multidrug transporter P-glycoprotein from Caenorhabditis elegans. Nature 490:566–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr KM, Sauna ZE, Ambudkar SV. (2001) Correlation between steady-state ATP hydrolysis and vanadate-induced ADP trapping in Human P-glycoprotein. Evidence for ADP release as the rate-limiting step in the catalytic cycle and its modulation by substrates. J Biol Chem 276:8657–8664. [DOI] [PubMed] [Google Scholar]
- Kim J, Wu S, Tomasiak TM, Mergel C, Winter MB, Stiller SB, Robles-Colmanares Y, Stroud RM, Tampé R, Craik CS, et al. (2015) Subnanometre-resolution electron cryomicroscopy structure of a heterodimeric ABC exporter. Nature 517:396–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Jaimes KF, Aller SG. (2014) Refined structures of mouse P-glycoprotein. Protein Sci 23:34–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo TW, Bartlett MC, Detty MR, Clarke DM. (2012) The ATPase activity of the P-glycoprotein drug pump is highly activated when the N-terminal and central regions of the nucleotide-binding domains are linked closely together. J Biol Chem 287:26806–26816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechetner EB, Schott B, Morse BS, Stein WD, Druley T, Davis KA, Tsuruo T, Roninson IB. (1997) P-glycoprotein function involves conformational transitions detectable by differential immunoreactivity. Proc Natl Acad Sci USA 94:12908–12913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindell JA, Grigorieff N. (2003) Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol 142:334–347. [DOI] [PubMed] [Google Scholar]
- Moeller A, Lee SC, Tao H, Speir JA, Chang G, Urbatsch IL, Potter CS, Carragher B, Zhang Q. (2015) Distinct conformational spectrum of homologous multidrug ABC transporters. Structure 23:450–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez C, Gerber S, Boilevin J, Bucher M, Darbre T, Aebi M, Reymond J-L, Locher KP. (2015) Structure and mechanism of an active lipid-linked oligosaccharide flippase. Nature 524:433–438. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. (2004) UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- Ramachandra M, Ambudkar SV, Gottesman MM, Pastan I, Hrycyna CA. (1996) Functional characterization of a glycine 185-to-valine substitution in human P-glycoprotein by using a vaccinia-based transient expression system. Mol Biol Cell 7:1485–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie TK, Kwon H, Atkins WM. (2011) Conformational analysis of human ATP-binding cassette transporter ABCB1 in lipid nanodiscs and inhibition by the antibodies MRK16 and UIC2. J Biol Chem 286:39489–39496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkadi B, Price EM, Boucher RC, Germann UA, Scarborough GA. (1992) Expression of the human multidrug resistance cDNA in insect cells generates a high activity drug-stimulated membrane ATPase. J Biol Chem 267:4854–4858. [PubMed] [Google Scholar]
- Sauna ZE, Ambudkar SV. (2000) Evidence for a requirement for ATP hydrolysis at two distinct steps during a single turnover of the catalytic cycle of human P-glycoprotein. Proc Natl Acad Sci USA 97:2515–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauna ZE, Ambudkar SV. (2007) About a switch: how P-glycoprotein (ABCB1) harnesses the energy of ATP binding and hydrolysis to do mechanical work. Mol Cancer Ther 6:13–23. [DOI] [PubMed] [Google Scholar]
- Sauna ZE, Kim IW, Nandigama K, Kopp S, Chiba P, Ambudkar SV. (2007) Catalytic cycle of ATP hydrolysis by P-glycoprotein: evidence for formation of the E.S reaction intermediate with ATP-gamma-S, a nonhydrolyzable analogue of ATP. Biochemistry 46:13787–13799. [DOI] [PubMed] [Google Scholar]
- Sauna ZE, Nandigama K, Ambudkar SV. (2006) Exploiting reaction intermediates of the ATPase reaction to elucidate the mechanism of transport by P-glycoprotein (ABCB1). J Biol Chem 281:26501–26511. [DOI] [PubMed] [Google Scholar]
- Scheres SH. (2012) RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180:519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senior AE. (2011) Reaction chemistry ABC-style. Proc Natl Acad Sci USA 108:15015–15016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharom FJ. (2014) Complex Interplay between the P-Glycoprotein Multidrug Efflux Pump and the Membrane: Its Role in Modulating Protein Function. Front Oncol 4:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintre CA, Pike AC, Li Q, Kim JI, Barr AJ, Goubin S, Shrestha L, Yang J, Berridge G, Ross J, et al. (2013) Structures of ABCB10, a human ATP-binding cassette transporter in apo- and nucleotide-bound states. Proc Natl Acad Sci USA 110:9710–9715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim HM, Bhatnagar J, Chufan EE, Kapoor K, Ambudkar SV. (2013) Conserved Walker A cysteines 431 and 1074 in human P-glycoprotein are accessible to thiol-specific agents in the apo and ADP-vanadate trapped conformations. Biochemistry 52:7327–7338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szewczyk P, Tao H, McGrath AP, Villaluz M, Rees SD, Lee SC, Doshi R, Urbatsch IL, Zhang Q, Chang G. (2015) Snapshots of ligand entry, malleable binding and induced helical movement in P-glycoprotein. Acta Crystallogr D Biol Crystallogr 71:732–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. (2007) EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol 157:38–46. [DOI] [PubMed] [Google Scholar]
- Urbatsch IL, Sankaran B, Weber J, Senior AE. (1995) P-glycoprotein is stably inhibited by vanadate-induced trapping of nucleotide at a single catalytic site. J Biol Chem 270:19383–19390. [DOI] [PubMed] [Google Scholar]
- Ward AB, Szewczyk P, Grimard V, Lee CW, Martinez L, Doshi R, Caya A, Villaluz M, Pardon E, Cregger C, et al. (2013) Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain. Proc Natl Acad Sci USA 110:13386–13391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkens S. (2015) Structure and mechanism of ABC transporters. F1000Prime Rep 7:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.