Abstract
Cytochrome P450s are oxidative metabolic enzymes that play critical roles in the biotransformation of endogenous compounds and xenobiotics. The expression and activity of P450 enzymes varies considerably throughout human development; the deficit in our understanding of these dynamics limits our ability to predict environmental and pharmaceutical exposure effects. In an effort to develop a more comprehensive understanding of the ontogeny of P450 enzymes, we employed a multi-omic characterization of P450 transcript expression, protein abundance, and functional activity. Modified mechanism-based inhibitors of P450s were used as chemical probes for isolating active P450 proteoforms in human hepatic microsomes with developmental stages ranging from early gestation to late adult. High-resolution liquid chromatography–mass spectrometry was used to identify and quantify probe-labeled P450s, allowing for a functional profile of P450 ontogeny. Total protein abundance profiles and P450 rRNA was also measured, and our results reveal life-stage–dependent variability in P450 expression, abundance, and activity throughout human development and frequent discordant relationships between expression and activity. We have significantly expanded the knowledge of P450 ontogeny, particularly at the level of individual P450 activity. We anticipate that these results will be useful for enabling predictive therapeutic dosing, and for avoiding potentially adverse and harmful reactions during maturation from both therapeutic drugs and environmental xenobiotics.
Introduction
The expression and activity of drug-metabolizing enzymes (DMEs) are major determinants of the overall pharmacokinetic differences observed between adults, children, neonates, and prenatal individuals (Hines and McCarver, 2002; Hines, 2008). Cytochrome P450s (P450s) are heme-containing DMEs with oxidase activity and are found at highest concentrations in the liver. This enzyme superfamily is responsible for the biotransformation of endogenous compounds, pharmacological agents, and environmental xenobiotics (Nelson et al., 1993). However, P450s can also metabolize protoxins to toxins resulting in detrimental health effects. A comprehensive understanding of P450 expression and activity throughout human development is needed to inform predictive therapeutic dosing and enable the avoidance during maturation of potentially adverse and harmful reactions from both therapeutic drugs and environmental toxins (de Wildt et al., 1999; Hines, 2008; Myllynen et al., 2009; Lee et al., 2012).
Cytochrome P450 expression and activity varies dramatically through prenatal development and from neonate maturation to adulthood. Differences between growth stages in DME composition and function can lead to neonate and childhood toxicity owing to inaccurate dosing and dangerous exposure (Lee et al., 2012), as exemplified in the classic historical case of adverse reactions of neonates to chloramphenicol when doses were extrapolated from adult levels (Myllynen et al., 2009). Variability in P450 activity can lead to severe pharmacological effects by altering drug disposition, resulting in drug underexposure, overexposure, or toxic drug interactions (Nebert, 2000; Blake et al., 2005). To account for life-stage-dependent variability in P450 expression throughout human development, P450s have been parsed into three broad classes: 1) those P450s that are expressed at highest levels during the first trimester of gestation; 2) those with fairly constant expression from gestation through adulthood; and 3) those with negligible or low prenatal expression that then increases through postnatal maturation (Hines, 2007; Hines, 2013). Herein, we correlate expression-based measurements to functional activity measurements of individual P450s in the primary xenobiotic metabolizing families 1–3, as well as other families associated with xenobiotic metabolism and steroidogenesis. Throughout human development complete voids of knowledge still exist for P450 ontogeny, in terms of both P450 expression and activity.
The critical paucity of data on the ontogeny of individual P450 isoforms through maturation is the result of the scarcity of tissue samples, which has largely forcing the use of rodents for ontogeny studies. But, significant variability between species in the primary structure, function, and regulation of different P450 isoforms makes it difficult to extrapolate between species (Hines, 2008; Myllynen et al., 2009). Available human data has revealed a number of discrepancies leading to difficulties in characterizing P450 ontogeny; these include limited sample sets, disparate measurement techniques, high interindividual variability observed between different samples sets, and large differences between rodent and human P450 cohorts (Hines, 2008).
Herein, we have sought to expand our knowledge of P450 expression and activity in the human liver from early gestation to late adulthood using classic techniques to measure gene and protein expression, and we have used a chemical probe approach, activity-based protein profiling (ABPP), to interrogate P450 activity. Activity-based protein profiling of P450s uses modified P450 mechanism–based inhibitors as probes to profile the functionally active cohort of enzymes directly in microsomes isolated from human liver tissues (Wright and Cravatt, 2007; Cravatt et al., 2008; Wright et al., 2009; Crowell et al., 2013; Ismail et al., 2013). The approach requires the catalytic activity of the P450s to form a covalent bond between P450 and probe, from which the targeted proteins can be identified by liquid chromatography–mass spectrometry (LC-MS)–based proteomics or fluorescent gel imaging. Together, our results provide a thorough analysis and comparison of P450 expression and activity profiles through human maturation.
Materials and Methods
Reagents and Chemicals.
miRNeasy Mini Kit, QuantiTect Primer Assays, and QuantiTect SYBR Green RT-PCT kit were obtained from Qiagen (Valencia, CA). TURBO DNA-free kit was purchased from Thermo Fisher Scientific (Sunnyvale, CA). Materials for labeling and enrichment include NADPH tetrasodium salt (Enzo Life Science, Farmingdale, NY), streptavidin agarose resin (Thermo Fisher Scientific; 1–3 mg biotinylated bovine serum albumin protein per milliliter resin), sequencing grade trypsin (Promega, Madison, WI ), Pierce BCA Protein Assay Kit (Thermo Scientific), and Bio-Spin chromatography columns (Bio-Rad, Hercules, CA).
Human Tissue Samples.
All prenatal human liver tissue samples (de-identified) were obtained from the Birth Defects Research Laboratory tissue repository at the University of Washington. At time of collection, prenatal samples were split with some of the tissue placed in RNA Later [quantitative reverse transcription–polymerase chain reaction (qRT-PCR) analysis], and the remainder flash frozen (ABPP and proteomic analyses) and stored at –80°C. Additional human liver microsome samples obtained from a commercial source (XenoTech, LLC, Lenexa, KS), and reported by Smith et al. (2011), were included in this study to extend the enzyme analyses through neonatal and adult developmental ages (sample metadata can be found in Supplemental Dataset 1; available metadata for each sample is rather disparate, and therefore we chose not to include it in our statistical analyses or data interpretation). The Institutional Review Board of Pacific Northwest National Laboratory approved all sample transfer procedures. Total number of prenatal liver samples is 30, ranging from gestational day (GD) 87 to 140; five samples are from late first trimester, and 25 samples are from early-to-mid–second trimester of development. Additional samples we evaluated are neonate/infant (age < 1 year; n = 6), juvenile (age > 1 year and < 18 year; n = 10), and adult (age > 18 year; n = 9).
Microsomal Preparation.
Liver tissues were processed according to prior methods with some minor changes (Wright and Cravatt, 2007). Tissues were rinsed with ice-cold 1.5% KCl, finely minced with a Tissue Tearor homogenizer (Biospec, Bartlesville, OK) for five 1-second pulses and dounce homogenized with both a loose and tight fitting pestle in 250 mM sucrose in EDTA-free phosphate-buffered saline (PBS; ∼2 ml per tissue). The samples were then centrifuged at 10,000g for 20 minutes at 4°C to remove cellular debris, nuclei, and mitochondria. The supernatant was then centrifuged at 100,000g for 90 minutes at 4°C to separate the microsome from the cytosol. Microsomes were resuspended in 250 mM sucrose in PBS buffer with minimal dounce homogenization, and protein concentrations were determined by a BCA assay. All samples were stored at –80°C until use.
Activity-Based Protein Profiling.
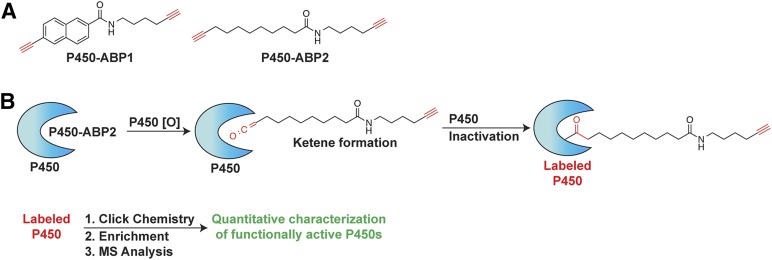
ABPP utilizes chemical probes derived from aryl- or aliphatic-alkyne mechanism–based inhibitors of P450 enzymes, which directly report on the activity of P450 enzymes in subcellular fractions, whole cells, or organisms (Wright and Cravatt, 2007; Cravatt et al., 2008; Hollenberg et al., 2008; Wright et al., 2009; Crowell et al., 2013). Herein, we use a multiplexed labeling approach in which an aryl alkyne probe (P450-ABP1)(Wright and Cravatt, 2007) and an aliphatic alkyne probe (P450-ABP2) (Wright et al., 2009) are added simultaneously to human prenatal tissue microsome samples (Fig. 1). Multiplexed ABPP yields broad coverage of target enzyme families (Wiedner et al., 2012); this is particularly true in the P450 superfamily, in which individual P450 isoforms often display broad substrate specificity (Crowell et al., 2013). Aliphatic and aryl alkynes have been demonstrated to inhibit P450s from all major detoxifying P450 subfamilies (Wright et al., 2009). In the presence of NADPH functionally active microsomal P450s directly oxidize the probes yielding reactive ketene moieties, which subsequently react irreversibly with the functional P450 (Wright and Cravatt, 2007; Wright et al., 2009; Crowell et al., 2013). Each probe also contains a second alkyne group that serves as a reactive handle for copper-catalyzed azide-alkyne cycloadditions (“click chemistry”). Following probe labeling of P450s, click chemistry is used to append fluorescent reporters for gel imaging or biotin for enrichment and LC-MS analyses of probed enzymes (Speers et al., 2003; Wright and Cravatt, 2007).
Fig. 1.
(A) Structure of P450 activity-based probes (P450-ABPs). (B) One alkyne (red) is oxidized by a functionally active P450 enzyme in a mechanism-dependent manner yielding a reactive ketene that irreversibly reacts with and inactivates the now probe-labeled P450. The second alkyne enables click chemistry–mediated addition of a biotin group for subsequent streptavidin resin–based enrichment and quantitative LC-MS analysis of probe-labeled P450s.
Human liver microsomal proteome samples (1 mg/ml protein concentrations, 800 μg total protein) were treated with a mixture of activity-based probes (20 μM each of P450-ABP1 and P450-ABP2). To probed samples was added NADPH (0.63 M), a requisite P450 cofactor for activity, and samples were incubated at 37°C for 60 minutes in the dark. Control samples were prepared by heat-shock pretreatment of the microsome samples (98°C, 10 minutes) prior to addition of activity-based probes (ABPs) and NADPH. Following ABP incubation, the samples were reacted with a biotin-azide (30 μM) tag under click-chemistry conditions as described previously (Wright and Cravatt, 2007; Crowell et al., 2013). Prior to enrichment of probe-labeled proteins, sample protein amounts were normalized to 350 μg protein. Probe-labeled proteins were enriched on streptavidin resin, reduced with Tris(2-carboxyethyl)phosphine (TCEP, 2 mM), and alkylated with iodoacetamide (4 mM). Proteins were digested on-resin with trypsin, and the resulting peptides were collected and analyzed by LC-MS as described previously (Crowell et al., 2013).
Proteomics Data Analysis.
Generated tandem mass spectrometry (MS/MS) spectra were searched using the mass spectrum–generating function plus (MSGF+) algorithm against the publicly available Homo sapiens–translated genome sequence (www.uniprot.org; Sept. 2013 collection; Kim et al., 2008). Identified peptides of at least six amino acids in length and having MSGF scores ≤ 1E–10, which corresponds to an estimated false discovery rate <1% at the peptide level, were used to generate an accurate mass and time (AMT) tag database (Zimmer et al., 2006). Matched peptide features from each LC-MS dataset were then filtered on a false discovery rate of less than or equal to 5% using the Statistical Tools for AMT tag confidence metric (Stanley et al., 2011). When a peptide is identified by the AMT tag approach, its relative abundance can be determined by calculating the area under the curve of the peptide ion peak in the MS measurement. Relative peptide abundance measurements in technical replicates were normalized to the dataset with the least information using linear regression in DAnTE (Polpitiya et al., 2008). Missing peptide abundance values were imputed with half of the minimum peptide abundance value across the entire dataset. Peptide abundance values were then rolled up to proteins using RRollup (Polpitiya et al., 2008); a minimum of three peptides was required for the Grubb test, with a p-value cutoff of 0.05. Only peptides unique in identifying a single protein were used to estimate protein abundances. Additionally, owing to low probe-dependent labeling of P450 activity, enrichment of active P450s resulted in low abundance measurements in the MS; therefore, proteins represented by a single unique peptide were permitted. To identify a protein as specifically labeled by the ABPs, we required the following criteria: 1) peptide measurements for a protein exhibit a significant difference comparing ABP-labeled with NADPH to ABP-labeled heat-shock controls (p < 0.05); and, 2) the protein exhibits ≥4-fold greater abundance in the ABP-labeled with NADPH sample relative to the heat-shock controls.
Proteomics Statistical Analysis.
The primary goal of the statistical analysis was to compare the average expression of the prenatal [trimester one (TM1) and trimester two (TM2)] sample groups to the average expression of the neonate/infant, juvenile, and adult sample groups. To account for missing data, two univariate statistical tests were performed on each protein across all age ranges: 1) a simple two-sample t test, and 2) a G test (Webb-Robertson et al., 2010). The two-sample t test identifies proteins that have a quantitative significant log2 fold-change difference between the two groupings and the G test identifies proteins that have a significant difference in the presence or absence of identifications between the groups. In particular, missing data may be either censored (below the limit-of-detection of the instrument) or missing at random. The G test evaluates the null hypothesis that the data are missing at random, and a significant result indicates that the data are censored from one group or the other. All P values with both the t test and G-test evaluations were corrected with a Bonferroni correction to account for the multiple tests being performed. A Bonferroni was selected because it is very stringent and the number of tests performed is relatively small (<30 in either the global or ABPP datasets) (Webb-Robertson et al., 2015).
The evaluation of the global data found 17 proteins with a significant increase from the prenatal group to the postnatal group with a Bonferroni correction and an additional three without the correction (Supplemental Table 2). Of these, six were significant only on basis of the G-test (censoring), nine were significant only on the basis of log2 fold-change, and the remaining five were significant by both analyses and with confirmation of an increase in abundance by both tests. The global data had three proteins that decreased with a Bonferroni correction and an additional one without the correction, and of these three were a quantitative change and one was a qualitative change. Supplemental Dataset 2 gives the Bonferroni corrected P values, the percentage of the data within each group observed for significant G-test results and the log2 fold-change for significant quantitative results.
The evaluation of the ABPP data found 11 total proteins with a significant increase from the prenatal group to the postnatal group with a Bonferroni correction and an additional two proteins significant prior to the correction. All of the significantly increasing proteins were identified via a G-test. There were three significantly decreasing proteins with a Bonferroni correction, and an additional two proteins with no correction. Of these five, two were qualitative differences and three were on the basis of fold changes (see Supplemental Table 3 and Supplemental Dataset 3).
Global Proteomic Analysis of Microsomes.
Two replicates of microsomal lysate (100 μg) were denatured, solubilized, and reduced with 8 M urea, 1% CHAPS, and 5 mM dithiothreitol, respectively, and incubated at 60°C for 30 minutes. Samples were then alkylated with 40 mM iodoacetamide at 37°C for 60 minutes. Each sample was diluted 8-fold with NH4HCO3 (100 mM, pH 8.4) to reduce salt concentration. CaCl2 (1 mM) was added to the diluted samples, and proteins were digested using sequencing grade trypsin (Promega) at a ratio of 1 unit of trypsin per 50 units of protein for 3 hours at 37°C. Solid-phase extraction was performed on digested samples using Supelcosil LC-SCX columns (Sigma-Aldrich, St. Louis, MO). Samples were acidified with 20% formic acid to achieve 1% final concentration. Columns were conditioned with 1-ml aliquots of MeOH (2×), 1-ml aliquots of 0.1% trifluoroacetic acid (TFA) in water (2×), and 1-ml aliquots of 1% TFA in water (2×). Samples were then loaded slowly and washed with 1-ml aliquots of 0.1% TFA in MeOH (4×). Samples were eluted with 1 ml of 5% NH4OH in 30% MeOH. Sample volumes were reduced by vacuum centrifugation until dry, and reconstituted in 2% acetonitrile in water and centrifuged at 10,000g for 5 minutes to pellet column residue. Peptide concentrations were determined via the BCA assay. Sample protein concentrations were normalized to 0.25 μg/μl and characterized by LC-MS. Data analysis was consistent with the methods listed above, including statistical confidence metrics.
Quantitative Reverse Transcription-Polymerase Chain Reaction.
All human prenatal tissue samples were removed from RNAlater solution. Tissues were diced into small pieces, and the pieces were transferred into 25-ml tubes containing the QIAzol reagent. The liver pieces were shredded fully and completely with a Tissue Tearor homogenizer and a small-gauge generator. The samples were homogenized at least 3× for 30–45 seconds. Samples were kept on ice to prevent overheating and RNA degradation. Chloroform (0.20 ml) was added to each sample per 1 ml QIAzol lysis reagent, and then the samples were vigorously shaken by hand for 30 seconds and then centrifuged at 10,000g for 20 minutes at 4°C. Further total RNA was extracted using the RNeasy Mini Kit (Qiagen) according to the manufacturer’s directions. Total RNA purity and concentration was determined by NanoDrop (Thermo Scientific). Genomic DNA was removed from the RNA samples by DNA-free DNase treatment (Thermo Fisher Scientific). The accession numbers of orthologous P450 genes were obtained from Choudhary et al. (2005). Primers (see Supplemental Table 1) for amplification of genes were obtained from Qiagen (premade QuantiTect Primer Assays).
A QuantiTect SYBR Green RT-PCR Kit (Qiagen) was used for qRT-PCR following the manufacturer’s recommendations. qRT-PCR was performed on an ABI Prism 7700 sequence detection system (Applied Biosystems/Thermo Fisher Scientific). qRT-PCR was carried out with the QuantiTect SYBR Green RT-PCR master mix (Qiagen) using 50 ng RNA in a 25-μl final reaction mixture (reverse transcription 30 minutes at 50°C; PCR initial activation 15 minutes at 95°C). The average threshold cycle (Ct) for each gene was determined from duplicate reactions and normalized against GAPDH and β-actin (Barber et al., 2005). Data were analyzed by taking the difference between mean threshold of qRT-PCR cycle values for individual P450 genes and the endogenous GAPDH control (Supplemental Datasets 4, 5, and 6).
Results
Transcriptional Ontogeny of P450 Expression in the Developing Prenatal Liver.
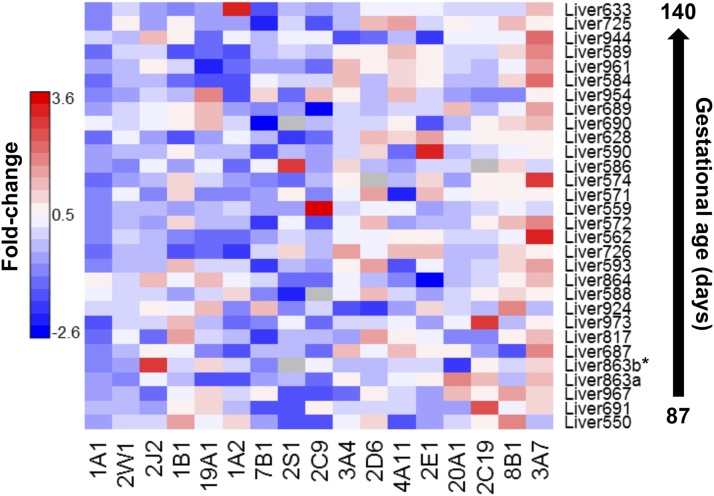
Gene expression of P450s was measured in human prenatal livers. We focused primarily on characterizing the expression of P450 genes representing the primary drug/xenobiotic-metabolizing and steroidogenesis families, which are important, respectively, for predicting chemicals that prenatal individuals may be particularly sensitive to and developmental functions associated with steroid synthesis, respectively. The relative level of P450 gene expression compared with an internal control (GAPDH) are shown for individual prenatal livers in Fig. 2. The overall pattern of P450 expression is fairly similar in the panel of prenatal livers. β-actin was also used as a second internal control and showed nearly identical results in comparison with the GAPDH. β-actin results are shown in Supplemental Dataset 3.
Fig. 2.
qRT-PCR of P450 genes involved in drug metabolism and steroidogenesis in prenatal livers. Relative gene expression levels of human P450 genes in prenatal livers were determined by qRT-PCR. For each gene, the average threshold cycle (Ct) was determined from the mean of three technical reactions ±S.D. and normalized against a housekeeping gene (GAPDH). Relative expression values are given as fold-change against the internal reference, GAPDH. Samples below the asterisked sample are all from TM1, all other samples are TM2. The specific values corresponding to the heatmap are provided in Supplemental Dataset 4; unprocessed raw values for the replicates are in Supplemental Dataset 5.
Proteomic Profiling of P450 Protein Expression in the Liver from Gestational through Adult Development.
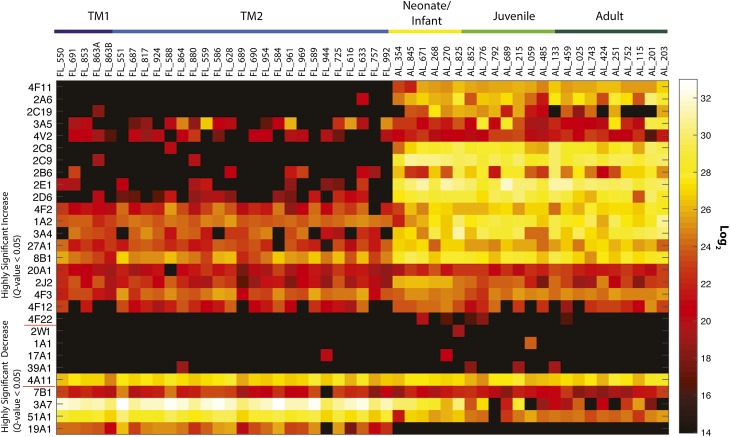
Global quantitative proteome profiling was performed to characterize individual P450 abundance in each prenatal liver sample. Thirty P450s were identified throughout the panel of tissues regardless of age, with 15 from the primary drug-metabolizing subfamilies 1, 2, and 3, and 15 others with minor roles in drug metabolism and/or considerable roles in steroidogenesis. Distinctive patterns of protein expression emerge that are consistent with class-based organization of hepatic drug-metabolizing enzymes (Fig. 3). These proteins were grouped by activity patterns of LC-MS identification and loosely fall into the previously described P450 ontogeny classifications and include class 1 (those with a significant decrease in postnatal samples) 3A7, 7B1, 19A1, 51A1; class 2 (consistent from gestation through adulthood) 1A1, 2W1, 17A1, 39A1; and class 3 (significant increase in postnatal samples) 1A2, 2A6, 2B6, 2C8, 2C9, 2C19, 2D6, 2E1, 2J2, 3A4, 3A5, 4F2, 4F3, 4F11, 4F12, 4F22, 4V2, 8B1, 20A1, 27A1.
Fig. 3.
Global P450 protein expression, quantified by the AMT tag approach. Comparison of the average expression of the prenatal first trimester (TM1) and prenatal second trimester (TM2) groups to the average expression of neonate/infant, juvenile, and adult groups is demonstrated through a Q test value, which represents a significance metric applied to the various statistical analyses. Q test values <0.05 represent high confidence determination that protein abundance values increase or decrease when going from prenatal to postnatal samples. Samples increase in age from left to right. Values correlating to the heatmap are provided in the Supplemental Dataset 7. Quantitative LC-MS values at the peptide level are provided in Supplemental Dataset 8.
Activity-Based Proteomic Profiling of P450 Function in the Liver from Gestational to Adult Development.
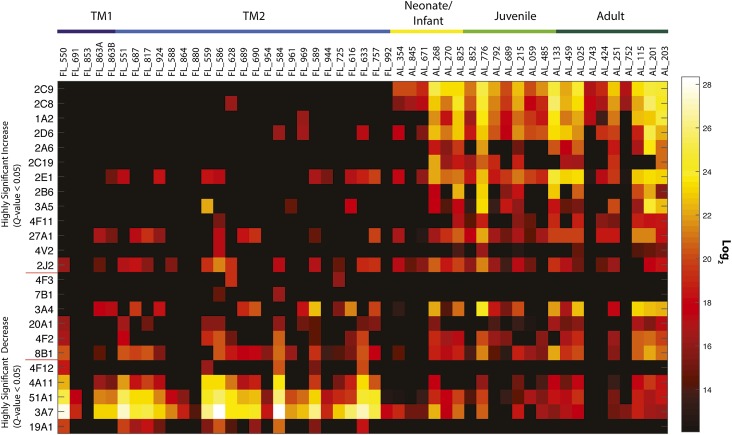
Activity-based protein profiling was applied to the entire complement of samples characterized by global proteome measurements. Using the AMT tag approach for relative quantification, 24 P450 enzymes were identified throughout the array of samples by ABPP (Fig. 4). These proteins were grouped by activity patterns of LC-MS identification and loosely fall into the previously described P450 ontogeny classifications and include class 1 (those with a significant decrease in postnatal samples) 3A7, 4A11, 4F12, 19A1, 51A1; class 2 (consistent from gestation through adulthood) 3A4, 4F2, 4F3, 7B1, 8B1, 20A1; and class 3 (significant increase in postnatal samples) 1A2, 2A6, 2B6, 2C8, 2C9, 2C19, 2D6, 2E1, 2J2, 3A5, 4F11, 4V2, 27A1.
Fig. 4.
Activity-based protein profiling of P450 enzymes, quantified by the AMT tag approach. Comparison of the average probe-enriched P450s from the prenatal first trimester (TM1) and prenatal second trimester (TM2) groups to the average expression of neonate/infant, juvenile, and adult groups is demonstrated through a Q test value, which represents a significance metric applied to the various statistical analyses. Q test values <0.05 represent high confidence determination that protein abundance values increase or decrease when going from prenatal to postnatal samples. Samples increase in age from left to right. Values correlating to the heatmap are provided in Supplemental Dataset 9. Quantitative LC-MS values at the peptide level are provided in Supplemental Dataset 10.
Public Deposition of Data.
The mass spectrometry proteomics data has been deposited with the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifiers PXD002781 and 10.6019/PXD0027.
Correlation of P450 Gene and Protein Expression and Enzyme Activity.
P450 1–3 family enzymes play major roles in drug metabolism, whereas P450 family 4 enzymes primarily metabolize endogenous compounds such as eicosanoids. Prior reports on the P450 1A subfamily have been contentious, with argument for little to no expression, or for highly variable expression across prenatal development (Omiecinski et al., 1990; Murray et al., 1992; Cazeneuve et al., 1994; Hakkola et al., 1994, 1997; Shimada et al., 1996; Sonnier and Cresteil, 1998; Choudhary et al., 2005; Bieche et al., 2007). By qRT-PCR, we determined that gene expression levels are negligible to very low, but some extreme outliers were identified (e.g., CYP1A2 in liver 633; Fig. 2). Global proteomic analysis showed variable levels of P450 1A2 expression for all prenatal samples at severalfold less than postnatal samples, and detected active 1A2 proteoforms in a single prenatal sample. Active enzyme profiles only resulted in the identification of 1A2 in a single prenatal sample. Our qRT-PCR results also indicate minimal CYP1A1 expression and moderate CYP1B1 expression, yet no measurement of protein abundance was made by proteomics for either P450 1A1 or 1B1. In a prior qRT-PCR analysis, only 3 of 66 samples that were older than 22 weeks were found to express CYP1B1 (Hakkola et al., 1994, 1997; Bieche et al., 2007).
All P450 2A members displayed the general trend of increasing both abundance and activity with increasing age. The P450 2C subfamily is responsible for the metabolism of ∼30% of all clinical drugs in adults. We focused on two 2C enzymes because the application of several therapeutics used for pediatric ailments are metabolized by 2C9 and 2C19, an association exists between 2C9 and sudden infant death syndrome (Treluyer et al., 2000), and there was a lack of prior 2C9 versus 2C19 ontogeny profiling (Treluyer et al., 1997; Koukouritaki et al., 2004). Consistent with previous reports (Hines, 2008), our qRT-PCR analyses reveal elevated CYP2C19 and minimal CYP2C9 expression. We identified P450 2C9 in two prenatal samples and 2C19 in one prenatal sample via global proteomics, yet no active proteoforms were detected by ABPP. This may result from our cohort being primarily early-to-middle second trimester and/or a difference in sensitivity comparing prior antibody-based approaches to LC-MS–based ABPP.
Though minimally expressed in the adult, P450 2D6 accounts for the oxidative metabolism of ∼12% of all clinical drugs (Shimada et al., 1994). Previous studies have not found that prenatal CYP2D6 mRNA levels correlated well with immunochemical protein assays or metabolic activity (Treluyer et al., 1991). In our study, minimal protein expression for 2D6 was measured in ∼50% of prenatal samples and active 2D6 was only measured in three second-trimester prenatal samples. Poor correlation between transcription, translation, and activity in prenatal samples has led to suggestions of a translational control mechanism that may overlay any transcriptional regulation (Hines, 2008).
Aside from the minor role played in xenobiotic metabolism, P450 2J2 converts arachidonic acid to biologically active eicosanoids, which may play integral roles in development (Lee et al., 2010). Expression of CYP2J2 was low but detectable in ∼20% of prenatal samples, but proteomic analysis positively detected P450 2J2 throughout development, but at severalfold higher levels in neonates and infants. ABPP profiles for 2J2 mirrored the global proteomic results but with identification made for only ∼70% of all samples.
Nearly half of all clinical drugs are oxidatively metabolized by the P450 3A subfamily (Hines, 2008). This subfamily dominated our measurements for gene and protein expression and ABPP-determined activity, consistent with many prior reports (Lacroix et al., 1997; de Wildt et al., 1999; Stevens et al., 2003; Choudhary et al., 2005; Bieche et al., 2007). P450 3A7 is a class 1 enzyme with high expression during prenatal development and a rapid waning after birth (Lacroix et al., 1997; Stevens et al., 2003; Hines, 2007; Hines, 2013). In our panel, CYP3A7 is the most highly expressed gene in ∼80% of samples. Protein and activity profiles showed high levels of 3A7 in all prenatal samples and a swift decrease in both measurements after birth. P450 3A4, the major adult isoform, had relatively high gene expression, protein, and activity in the panel but with significant increase in all postnatal measurements (class 3).
The P450 4 family encodes constitutive and inducible isozymes, including the P450 4A subfamily, a hormonally regulated group that is induced by peroxisome proliferating chemicals (Simpson, 1997). In all life stages, we detected consistent gene and protein expression for the fatty acid–oxidizing 4A11, though activity levels significantly decreased postnatally. The P450 4F subfamily is recognized for its dual role in modulating the concentrations of eicosanoids during inflammation as well as in the metabolism of clinically significant drugs (Kalsotra and Strobel, 2006). We did not perform qRT-PCR on this subfamily, but all members identified in our proteomic analysis (4F2, 4F3, 4F11, 4F12, 4F22) were either missing from most prenatal samples (4F11, 4F22) or were measured but had significantly higher levels in postnatal samples. Interestingly, a subset of prenatal samples had moderate levels of 4F12 activity, yet no activity was measured in postnatal samples. P450 4V2 protein expression was detected in many samples with abundance increasing with age, yet activity was only measured in one prenatal sample and ∼30% of postnatal samples.
The primary role of cytochrome P450s from non-CYP1–4 families tends to involve endogenous metabolism and steroid production, processes critical for prenatal development and viability. P450 7B1 catalyzes the 7α-hydroxylation of several steroid substrates, including those involved in hormonal signaling (Tang et al., 2006). Prenatal ontogeny studies have revealed 7B1 to be primarily extrahepatic, with little or no liver expression (Tang et al., 2006). Our qRT-PCR measurements identified detectable levels in only 20% of the samples, consistent with prior analyses. Protein expression levels were measured for all but one prenatal sample, but these levels decreased with age and activity was nonexistent for all but two prenatal samples.
In contrast, P450 8B1, a sterol 12α-hydroxylase that catalyzes cholic acid synthesis and controls soluble cholesterol levels (Gafvels et al., 1999), is highly expressed in nearly all prenatal livers examined in our study, and protein was detected in every sample with a significant increase in postnatal expression. Active P450 8B1 was also detected in most samples, yet there was no significant change in activity between age groups.
P450 19A1 converts androgens to estrogens, and our analysis shows that the gene for this enzyme was expressed in ∼50% of second-trimester prenatal samples, yet proteomic analysis identified 19A1 in all but one prenatal sample and activity in ∼40% of the prenatal samples. No 19A1 identification was made in postnatal samples.
The “orphan” enzyme, P450 20A1, has no known function. Our analysis measured low levels of gene expression in both trimesters. Proteome analysis identified similar levels of 20A1 in all but one sample across all life stages, and activity was measured for this enzyme in ∼40% of samples.
Finally, P450 51A1 stands out because it was identified in the whole sample panel save one juvenile sample and activity was measured in ∼80% of samples. Both measurement profiles mirrored each other in that levels significantly decreased after birth. We note that some samples showed no activity at all: FL_853, FL_880, AL_057, and AL_551. These samples were not removed from analyses because protein expression was identified by global proteomics in the samples. However, it is probable that some aspect of the processing of these samples altered the functional capacity of enzymes.
Discussion
Evaluation of gene and protein expression is important to understanding the developing liver but does not provide insight into actual functional metabolic capacity. Enzyme activity must be measured to truly understand metabolic potential. Traditional assays for P450 activity have limitations, such as interpretation of activity data being complicated by the overlapping substrate specificities of P450 enzyme subfamilies. Given the considerable number of liver P450 enzymes, and the limitation of sample resources (particularly with gestational samples), running a suite of traditional assays large enough to evaluate activity of a broad swath of P450s would be intractable. Activity-based protein profiling can overcome these challenges through the use of chemical probes that irreversibly label only functionally active P450 enzymes that can then be isolated. Coupled to LC-MS analyses, relative quantitation of P450 activities can be measured. We have employed a multi-omic analysis to provide a comprehensive comparison of life-stage P450 ontogeny at the gene expression, protein expression, and protein activity levels. Figures 2–4 reveal enzyme activity trends that do not always correlate with protein or gene expression, illuminating the importance of ABPP to profiling the ontogeny of activity in order to get a more accurate measure of the functional capacity of individual P450s in each liver sample.
As discussed throughout, we have characterized P450 enzyme ontogeny on a class basis as delineated by Hines (2013). In evaluating our ABPP data on a class basis and comparing to prior results, we identified P450 3A7 as the dominant active class 1 isoform; however, we also newly classified four P450s (4A11, 4F12, 19A1, and 51A1) into class 1. We also used ABPP to newly classify five class 2 P450s (4F2, 4F3, 7B1, 8B1, and 20A1) and six class 3 P450s (2A6, 2C8, 2J2, 4F11, 4V2, and 27A1). Of those P450s classified by Hines the vast majority correlate with our data, including 1A2, 2C9, 2D6, 2E1, and 3A7. Interestingly, P450s 2B6, 2C19, and 3A5 were previously characterized as class 2 enzymes, but in both our global proteomics and ABPP analyses we observe these as occurring primarily with only postnatal expression. Importantly, new classifications point to the strength of ABPP because activity assays are not available for many individual P450 enzymes, and/or no prior transcriptome or RT-PCR ontogeny studies have been performed which identified these P450s.
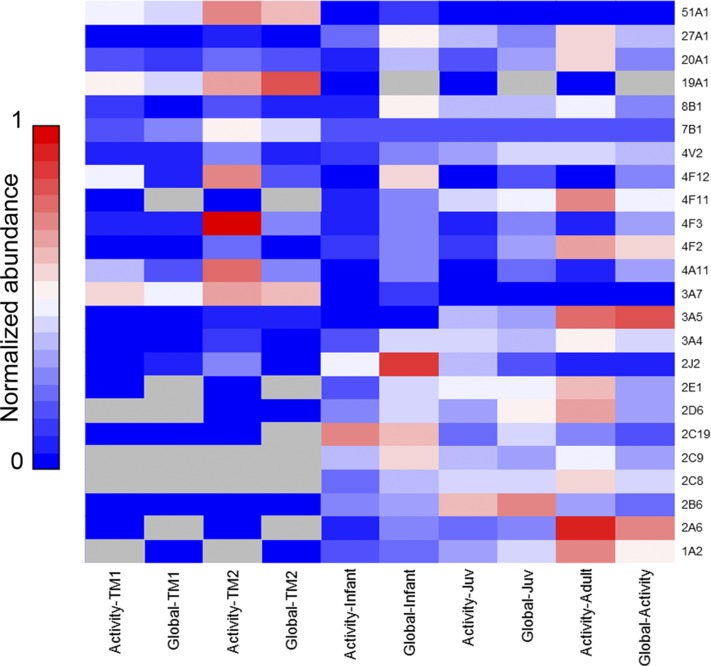
Cytochrome P450 protein expression is typically quantified by antibody-based Western blotting; and protein activity, by substrate-based assays that can be nonselective for single P450 isoforms. An important feature of our global proteome and ABPP measurements is the ability to rapidly yield relative quantification of P450 abundance and activity of numerous individual P450 enzymes (Wright and Cravatt, 2007; Wright et al., 2009). Figure 5 (see also Supplemental Fig. 1) displays the correlation of age-dependent protein expression and activity for P450s measured by both global and ABPP proteomics (note: P450s 1A1, 2W1, 4F22, 17A1, and 39A1 were identified only in global proteome analyses; see Supplemental Datasets 8 and 9). What is quickly apparent is that there is rarely a direct correlation between P450 activity and protein expression; within Fig. 5 some interesting age-dependent features can be readily identified. First, 3A7 aside, the key drug-metabolizing families are far more abundant and active after birth, and both features are somewhat correlated for most family 1–3 enzymes. Our postnatal sample analyses show quite a different trend, in which activity is considerably higher then abundance. This indicates highly active metabolic capacity for xenobiotics and demonstrates that quantifying protein abundance alone inadequately illustrates P450 metabolic capacity. Additionally, with regard to therapeutic drug dosing, it is clear that developmental stages are greatly altered in the activity of P450 enzymes involved in drug metabolism. During infancy and juvenile stages, P450 activities are generally lower than expression. This suggests that drug dosing must be carefully considered for pediatric therapies.
Fig. 5.
Comparison of P450 protein abundance to P450 activity (as determined by ABPP). Activity and protein expression values are averaged by life stage, and normalized on a 0–1 scale for each P450. Thereby, points of highest expression or activity for each P450 are observed as increasingly red. See also Supplemental Figure 1.
In summation, profound changes occur in P450 gene and protein expression and activity throughout human development. Life stage–dependent differences in P450 activity probably play a pivotal role in not only the disposition and efficacy of therapeutic drugs but also the metabolism of other xenobiotics and environmental agents. These results combined with prior findings can lead to more judicious predictions of drug metabolism that will result in safer dosing. We do note that ABPP is not absent of potential issues that should be carefully evaluated, such as, the method as performed relies on the native expression and activity of the requisite NADPH-dependent P450 reductase within each sample, and relies also on the individual P450’s catalytic activity toward the activity-based probes. In light of these potential issues, it is most appropriate to compare the probe-determined activity of each P450 across samples, as opposed to direct comparison of individual P450 activity, since the probes do not specifically measure a catalytic rate. Differences when correlating global proteomic abundances to ABPP measurements may be attributable to differences in expression of electron-transferring accessory enzymes such as NADPH-dependent P450 reductase (POR) or cytochrome b5. Differing POR/P450 ratios can alter activities, as can protein-protein interactions that result in P450 inhibition or cooperativity (Denisov et al., 2009; Reed and Backes, 2012). In the near future we anticipate that ABPP will be applied to broader cohorts of samples to expand our findings, and enhance our understanding of P450-mediated oxidative metabolism throughout human life stages.
Supplementary Material
Abbreviations
- ABP
activity-based probe
- ABPP
activity-based protein profiling
- AMT
accurate mass and time
- DME
drug-metabolizing enzyme
- LC-MS
liquid chromatography–mass spectrometry
- P450
cytochrome P450
- qRT-PCR
quantitative reverse transcription–polymerase chain reaction
- TFA
trifluoroacetic acid
Authorship Contributions
Participated in research design: Sadler, Smith, Corley, Wright.
Conducted experiments: Sadler, Nandhikonda.
Performed data analysis: Sadler, Nandhikonda, Webb-Robertson, Ansong, Anderson, Wright.
Wrote or contributed to the writing of the manuscript: Sadler, Nandhikonda, Ansong, Corley, Wright.
Footnotes
This research was supported by the National Institutes of Health National Institute of Environmental Health Sciences [P42 ES016465]. Mass spectrometry analyses were performed in the Environmental Molecular Sciences Laboratory, a US Department of Energy Biological and Environmental Research national scientific user facility at Pacific Northwest National Laboratory. Additionally, this work used instrumentation and capabilities developed under support from the National Institute for General Medical Sciences [P41 GM103493-11]. This work was supported by the Intramural Research Program of the National Institutes of Health National Institute of Environmental Health Sciences.
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
References
- Barber RD, Harmer DW, Coleman RA, Clark BJ. (2005) GAPDH as a housekeeping gene: analysis of GAPDH mRNA expression in a panel of 72 human tissues. Physiology Genomics 21:389–395. [DOI] [PubMed] [Google Scholar]
- Bièche I, Narjoz C, Asselah T, Vacher S, Marcellin P, Lidereau R, Beaune P, de Waziers I. (2007) Reverse transcriptase-PCR quantification of mRNA levels from cytochrome (CYP)1, CYP2 and CYP3 families in 22 different human tissues. Pharmacogenet Genomics 17:731–742. [DOI] [PubMed] [Google Scholar]
- Blake MJ, Castro L, Leeder JS, Kearns GL. (2005) Ontogeny of drug metabolizing enzymes in the neonate. Semin Fetal Neonatal Med 10:123–138. [DOI] [PubMed] [Google Scholar]
- Cazeneuve C, Pons G, Rey E, Treluyer JM, Cresteil T, Thiroux G, D’Athis P, Olive G. (1994) Biotransformation of caffeine in human liver microsomes from foetuses, neonates, infants and adults. Br J Clin Pharmacol 37:405–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary D, Jansson I, Stoilov I, Sarfarazi M, Schenkman JB. (2005) Expression patterns of mouse and human CYP orthologs (families 1-4) during development and in different adult tissues. Arch Biochem Biophys 436:50–61. [DOI] [PubMed] [Google Scholar]
- Cravatt BF, Wright AT, Kozarich JW. (2008) Activity-based protein profiling: from enzyme chemistry to proteomic chemistry. Annu Rev Biochem 77:383–414. [DOI] [PubMed] [Google Scholar]
- Crowell SR, Sharma AK, Amin S, Soelberg JJ, Sadler NC, Wright AT, Baird WM, Williams DE, Corley RA. (2013) Impact of pregnancy on the pharmacokinetics of dibenzo[def,p]chrysene in mice. Toxicol Sci 135:48–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denisov IG, Frank DJ, Sligar SG. (2009) Cooperative properties of cytochromes P450. Pharmacol Ther 124:151–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wildt SN, Kearns GL, Leeder JS, van den Anker JN. (1999) Cytochrome P450 3A: ontogeny and drug disposition. Clin Pharmacokinet 37:485–505. [DOI] [PubMed] [Google Scholar]
- Gåfvels M, Olin M, Chowdhary BP, Raudsepp T, Andersson U, Persson B, Jansson M, Björkhem I, Eggertsen G. (1999) Structure and chromosomal assignment of the sterol 12alpha-hydroxylase gene (CYP8B1) in human and mouse: eukaryotic cytochrome P-450 gene devoid of introns. Genomics 56:184–196. [DOI] [PubMed] [Google Scholar]
- Hakkola J, Pasanen M, Purkunen R, Saarikoski S, Pelkonen O, Mäenpää J, Rane A, Raunio H. (1994) Expression of xenobiotic-metabolizing cytochrome P450 forms in human adult and fetal liver. Biochem Pharmacol 48:59–64. [DOI] [PubMed] [Google Scholar]
- Hakkola J, Pasanen M, Pelkonen O, Hukkanen J, Evisalmi S, Anttila S, Rane A, Mäntylä M, Purkunen R, Saarikoski S, et al. (1997) Expression of CYP1B1 in human adult and fetal tissues and differential inducibility of CYP1B1 and CYP1A1 by Ah receptor ligands in human placenta and cultured cells. Carcinogenesis 18:391–397. [DOI] [PubMed] [Google Scholar]
- Hines RN. (2007) Ontogeny of human hepatic cytochromes P450. J Biochem Mol Toxicol 21:169–175. [DOI] [PubMed] [Google Scholar]
- Hines RN. (2008) The ontogeny of drug metabolism enzymes and implications for adverse drug events. Pharmacol Ther 118:250–267. [DOI] [PubMed] [Google Scholar]
- Hines RN. (2013) Developmental expression of drug metabolizing enzymes: impact on disposition in neonates and young children. Int J Pharm 452:3–7. [DOI] [PubMed] [Google Scholar]
- Hines RN, McCarver DG. (2002) The ontogeny of human drug-metabolizing enzymes: phase I oxidative enzymes. J Pharmacol Exp Ther 300:355–360. [DOI] [PubMed] [Google Scholar]
- Hollenberg PF, Kent UM, Bumpus NN. (2008) Mechanism-based inactivation of human cytochromes p450s: experimental characterization, reactive intermediates, and clinical implications. Chem Res Toxicol 21:189–205. [DOI] [PubMed] [Google Scholar]
- Ismail HM, O’Neill PM, Hong DW, Finn RD, Henderson CJ, Wright AT, Cravatt BF, Hemingway J, Paine MJ. (2013) Pyrethroid activity-based probes for profiling cytochrome P450 activities associated with insecticide interactions. Proc Natl Acad Sci USA 110:19766–19771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalsotra A, Strobel HW. (2006) Cytochrome P450 4F subfamily: at the crossroads of eicosanoid and drug metabolism. Pharmacol Ther 112:589–611. [DOI] [PubMed] [Google Scholar]
- Kim S, Gupta N, Pevzner PA. (2008) Spectral probabilities and generating functions of tandem mass spectra: a strike against decoy databases. J Proteome Res 7:3354–3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koukouritaki SB, Manro JR, Marsh SA, Stevens JC, Rettie AE, McCarver DG, Hines RN. (2004) Developmental expression of human hepatic CYP2C9 and CYP2C19. J Pharmacol Exp Ther 308:965–974. [DOI] [PubMed] [Google Scholar]
- Lacroix D, Sonnier M, Moncion A, Cheron G, Cresteil T. (1997) Expression of CYP3A in the human liver--evidence that the shift between CYP3A7 and CYP3A4 occurs immediately after birth. Eur J Biochem 247:625–634. [DOI] [PubMed] [Google Scholar]
- Lee CA, Neul D, Clouser-Roche A, Dalvie D, Wester MR, Jiang Y, Jones JP, 3rd, Freiwald S, Zientek M, Totah RA. (2010) Identification of novel substrates for human cytochrome P450 2J2. Drug Metab Dispos 38:347–356. [DOI] [PubMed] [Google Scholar]
- Lee JS, Ward WO, Knapp G, Ren H, Vallanat B, Abbott B, Ho K, Karp SJ, Corton JC. (2012) Transcriptional ontogeny of the developing liver. BMC Genomics 13:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray GI, Foster CO, Barnes TS, Weaver RJ, Snyder CP, Ewen SWB, Melvin WT, Burke MD. (1992) Cytochrome P450IA expression in adult and fetal human liver. Carcinogenesis 13:165–169. [DOI] [PubMed] [Google Scholar]
- Myllynen P, Immonen E, Kummu M, Vähäkangas K. (2009) Developmental expression of drug metabolizing enzymes and transporter proteins in human placenta and fetal tissues. Expert Opin Drug Metab Toxicol 5:1483–1499. [DOI] [PubMed] [Google Scholar]
- Nebert DW. (2000) Drug-metabolizing enzymes, polymorphisms and interindividual response to environmental toxicants. Clin Chem Lab Med 38:857–861. [DOI] [PubMed] [Google Scholar]
- Nelson DR, Kamataki T, Waxman DJ, Guengerich FP, Estabrook RW, Feyereisen R, Gonzalez FJ, Coon MJ, Gunsalus IC, Gotoh O, et al. (1993) The P450 superfamily: update on new sequences, gene mapping, accession numbers, early trivial names of enzymes, and nomenclature. DNA Cell Biol 12:1–51. [DOI] [PubMed] [Google Scholar]
- Omiecinski CJ, Redlich CA, Costa P. (1990) Induction and developmental expression of cytochrome P450IA1 messenger RNA in rat and human tissues: detection by the polymerase chain reaction. Cancer Res 50:4315–4321. [PubMed] [Google Scholar]
- Polpitiya AD, Qian WJ, Jaitly N, Petyuk VA, Adkins JN, Camp DG, 2nd, Anderson GA, Smith RD. (2008) DAnTE: a statistical tool for quantitative analysis of -omics data. Bioinformatics 24:1556–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JR, Backes WL. (2012) Formation of P450 · P450 complexes and their effect on P450 function. Pharmacol Ther 133:299–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T, Yamazaki H, Mimura M, Inui Y, Guengerich FP. (1994) Interindividual variations in human liver cytochrome P-450 enzymes involved in the oxidation of drugs, carcinogens and toxic chemicals: studies with liver microsomes of 30 Japanese and 30 Caucasians. J Pharmacol Exp Ther 270:414–423. [PubMed] [Google Scholar]
- Shimada T, Yamazaki H, Mimura M, Wakamiya N, Ueng YF, Guengerich FP, Inui Y. (1996) Characterization of microsomal cytochrome P450 enzymes involved in the oxidation of xenobiotic chemicals in human fetal liver and adult lungs. Drug Metab Dispos 24:515–522. [PubMed] [Google Scholar]
- Simpson AE. (1997) The cytochrome P450 4 (CYP4) family. Gen Pharmacol 28:351–359. [DOI] [PubMed] [Google Scholar]
- Smith JN, Timchalk C, Bartels MJ, Poet TS. (2011) In vitro age-dependent enzymatic metabolism of chlorpyrifos and chlorpyrifos-oxon in human hepatic microsomes and chlorpyrifos-oxon in plasma. Drug Metab Dispos 39:1353–1362. [DOI] [PubMed] [Google Scholar]
- Sonnier M, Cresteil T. (1998) Delayed ontogenesis of CYP1A2 in the human liver. Eur J Biochem 251:893–898. [DOI] [PubMed] [Google Scholar]
- Speers AE, Adam GC, Cravatt BF. (2003) Activity-based protein profiling in vivo using a copper(i)-catalyzed azide-alkyne [3 + 2] cycloaddition. J Am Chem Soc 125:4686–4687. [DOI] [PubMed] [Google Scholar]
- Stanley JR, Adkins JN, Slysz GW, Monroe ME, Purvine SO, Karpievitch YV, Anderson GA, Smith RD, Dabney AR. (2011) A statistical method for assessing peptide identification confidence in accurate mass and time tag proteomics. Anal Chem 83:6135–6140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens JC, Hines RN, Gu C, Koukouritaki SB, Manro JR, Tandler PJ, Zaya MJ. (2003) Developmental expression of the major human hepatic CYP3A enzymes. J Pharmacol Exp Ther 307:573–582. [DOI] [PubMed] [Google Scholar]
- Tang W, Eggertsen G, Chiang JY, Norlin M. (2006) Estrogen-mediated regulation of CYP7B1: a possible role for controlling DHEA levels in human tissues. J Steroid Biochem Mol Biol 100:42–51. [DOI] [PubMed] [Google Scholar]
- Treluyer JM, Jacqz-Aigrain E, Alvarez F, Cresteil T. (1991) Expression of CYP2D6 in developing human liver. Eur J Biochem 202:583–588. [DOI] [PubMed] [Google Scholar]
- Treluyer JM, Gueret G, Cheron G, Sonnier M, Cresteil T. (1997) Developmental expression of CYP2C and CYP2C-dependent activities in the human liver: in-vivo/in-vitro correlation and inducibility. Pharmacogenetics 7:441–452. [DOI] [PubMed] [Google Scholar]
- Tréluyer JM, Benech H, Colin I, Pruvost A, Chéron G, Cresteil T. (2000) Ontogenesis of CYP2C-dependent arachidonic acid metabolism in the human liver: relationship with sudden infant death syndrome. Pediatr Res 47:677–683. [DOI] [PubMed] [Google Scholar]
- Webb-Robertson BJ, McCue LA, Waters KM, Matzke MM, Jacobs JM, Metz TO, Varnum SM, Pounds JG. (2010) Combined statistical analyses of peptide intensities and peptide occurrences improves identification of significant peptides from MS-based proteomics data. J Proteome Res 9:5748–5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb-Robertson BJ, Wiberg HK, Matzke MM, Brown JN, Wang J, McDermott JE, Smith RD, Rodland KD, Metz TO, Pounds JG, et al. (2015) Review, evaluation, and discussion of the challenges of missing value imputation for mass spectrometry-based label-free global proteomics. J Proteome Res 14:1993–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedner SD, Burnum KE, Pederson LM, Anderson LN, Fortuin S, Chauvigné-Hines LM, Shukla AK, Ansong C, Panisko EA, Smith RD, et al. (2012) Multiplexed activity-based protein profiling of the human pathogen Aspergillus fumigatus reveals large functional changes upon exposure to human serum. J Biol Chem 287:33447–33459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AT, Cravatt BF. (2007) Chemical proteomic probes for profiling cytochrome p450 activities and drug interactions in vivo. Chem Biol 14:1043–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AT, Song JD, Cravatt BF. (2009) A suite of activity-based probes for human cytochrome P450 enzymes. J Am Chem Soc 131:10692–10700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer JS, Monroe ME, Qian WJ, Smith RD. (2006) Advances in proteomics data analysis and display using an accurate mass and time tag approach. Mass Spectrom Rev 25:450–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.