Abstract
Hydrogels based on triblock copolymers of polyethylene glycol and partially methacrylated poly(N-(2-hydroxypropyl) methacrylamide mono/dilactate) are an attractive class of biomaterials due to their biodegradability, cytocompatibility, and tunable thermo-responsive and mechanical properties. By fine-tuning these properties, the hydrogels can be 3D bioprinted, to generate e.g. constructs for cartilage repair. This study investigated whether hydrogels based on the above mentioned polymer with a 10% degree of methacrylation (M10P10), support cartilage formation by chondrocytes, and whether the incorporation of methacrylated chondroitin sulfate (CSMA) or methacrylated hyaluronic acid (HAMA) can improve the mechanical properties, long-term stability, and printability.
Chondrocyte-laden M10P10 hydrogels were cultured for 42 days to evaluate chondrogenesis. M10P10 hydrogels with or without polysaccharides were evaluated for their mechanical properties (before and after UV photo-cross-linking), degradation kinetics, and printability.
Extensive cartilage matrix production occurred in M10P10 hydrogels, highlighting their potential for cartilage repair strategies. The incorporation of polysaccharides increased the storage modulus of polymer mixtures and decreased the degradation kinetics in cross-linked hydrogels. Addition of HAMA to M10P10 hydrogels improved printability and resulted in 3D constructs with excellent cell viability. Hence, this novel combination of M10P10 with HAMA forms an interesting class of hydrogels for cartilage bioprinting.
Keywords: Thermo-sensitive block copolymers, hyaluronic acid, chondroitin sulfate, hydrogel, cartilage regeneration, 3D bioprinting
Introduction
Articular cartilage is the tissue that covers the extremities of the bones inside the joints. The tissue functions as a damper due to its high osmotic pressure and reduces surface friction due to its smooth surface structure. Articular cartilage contains proteoglycans, collagen type II, water, and cells, the chondrocytes. Since the tissue lacks vasculature and innervation, and contains only few chondrocytes, it has a limited regenerative capacity1,2. The implantation of cell-laden hydrogel scaffolds is regarded as a promising approach to treat cartilage defects. Hydrogels, networks of hydrophilic polymers, have high water content, which supports cell survival and allow homogeneous encapsulation of cells as well as biological and chemical cues. Therefore, cell-laden hydrogel implants can promote new tissue formation while initially providing structural support. For the generation of successful cell-laden constructs, it is essential to have control over the mechanical properties and degradation kinetics of the construct, as it should progressively be replaced by newly-formed tissue after implantation3. The mechanical properties and degradation kinetics of hydrogels can be easily tailored over a broad range and in a highly reproducible manner by a proper design of the building blocks4–6. In addition, thermo-responsive functionalities can be introduced in the building blocks, providing the opportunity to generate injectable and three dimensional (3D) printable hydrogels7.
Copolymers based on a polyethylene glycol (PEG) mid-block flanked by two poly(N-(2-hydroxypropyl) methacrylamide mono/dilactate) (polyHPMA-lac) outer blocks have recently been investigated for pharmaceutical and biomedical applications8–12. Methacrylated polyHPMA-lac-PEG triblock copolymers display lower critical solution temperature (LCST) behavior in aqueous solutions, meaning that these polymers are soluble at low temperatures and form physical gels, by self-assembly due to dehydration of polymer chains, at temperatures above a critical temperature, called the cloud point (CP)13. The thermo-sensitive behavior of methacrylated polyHPMA-lac-PEG triblock copolymers is highly tunable, e.g. to physiologically relevant temperatures, by adapting the content of the lactate groups present in the outer blocks as well as the number of methacrylate groups9,13,14. In addition, the methacrylate groups allow UV light-mediated photo-cross-linking, which prevents rapid disassembly of the polymer networks13. Chemically cross-linked hydrogels with tailored degradation rates and mechanical properties can be obtained by varying the number of methacrylate units per polymer chain, the molecular weight of the PEG mid-block, as well as that of the thermo-sensitive flanking blocks and the polymer concentration in the hydrogel8,9,11,13. The thermo-sensitive behavior of methacrylated polyHPMA-lac-PEG triblock copolymers allows easy handling of the polymer solution at low temperatures, when it behaves as a viscous liquid, to incorporate cells. Previous studies have shown high viability of encapsulated articular chondrocytes in methacrylated polyHPMA-lac-PEG triblock copolymer based hydrogels10. However, long-term culture and actual cartilage matrix formation in these hydrogels has not been investigated so far.
Cell-laden hydrogels can accurately be shaped with 3D biofabrication techniques to mimic the architecture of native tissues e.g. the zonal organization of articular cartilage15, and to generate patient specific construct shapes. 3D bioprinting is a form of biofabrication based on computer-aided layer-by-layer material deposition16–19. As such, bioprinting also allows the incorporation of pores or perfusable channels in the 3D structure, for easy diffusion of nutrients, oxygen and metabolites during (in vitro) construct maturation7. Hydrogels composed of methacrylated polyHPMA-lac-PEG triblock copolymers have already been shown to be printable due to their thermo-sensitive behavior10. However, this required a relatively high polymer concentration and a high degree of methacrylation (DM)10. In general, dense polymer networks due to e.g. high polymer concentrations and high DM, have adverse effects on the matrix production of embedded cells20,21 and are therefore unfavourable for the fabrication of tissue repair constructs. In order to tackle this well-known dilemma in bioprinting7, hybrid materials can be designed, for example by incorporating polysaccharides, which increase the viscosity of the polymer solution and can potentially improve the printability without hampering the matrix production of embedded cells22–26. In this study, the polysaccharides chondroitin sulfate (CS) and hyaluronic acid (HA) were methacrylated to allow UV photo-cross-linking27,28 and blended with low DM (10%) polyHPMA-lac-PEG triblock copolymers, as both are natural polysaccharides abundantly present in native cartilage. In addition, they have demonstrated anabolic effects on extracellular matrix synthesis by chondrocytes and stem cells24,29–34. Therefore, these polysaccharides are attractive candidates to optimize methacrylated polyHPMA-lac-PEG triblock copolymer based hydrogels for cartilage bioprinting. It is hypothesized that the incorporation of methacrylated HA (HAMA) or methacrylated CS (CSMA) in methacrylated polyHPMA-lac-PEG triblock hydrogels will affect the mechanical properties, decrease the degradation rate and improve the 3D printability in comparison to hydrogels made of methacrylated polyHPMA-lac-PEG triblock only. The aim of this study was to characterize methacrylated polyHPMA-lac-PEG triblock copolymer based hydrogels in terms of chondrogenesis, mechanical behavior, degradation kinetics and printability. It was also investigated whether the incorporation of HAMA or CSMA in this synthetic hydrogel can further improve the mechanical properties, affect the degradation rate, and enhance the printability.
Materials and Methods
Materials
All chemicals were obtained from Sigma-Aldrich (Zwijndrecht, the Netherlands) and all solvents from Biosolve (Valkenswaard, the Netherlands) unless indicated otherwise. Chemicals and solvents were used as received. PEG 10 kDa was supplied by Merck (Darmstadt, Germany). HA sodium salt (1560 kDa) was supplied by Lifecore Biomedical (Chaska, MN, USA). CS A sodium salt from bovine trachea (Sigma-Aldrich, Zwijndrecht, the Netherlands) was analyzed with Viscotek Gel Permeation Chromatography (GPC) and showed a bimodal molecular weight distribution (number average molecular weight, Mn 26.9 kDa, 94% mass content and 353.8 kDa, 6% mass content; details are given in Figure S1). L-lactide was purchased from Corbion Purac (Amsterdam, the Netherlands) and Irgacure 2959 was a kind gift from BASF (Ludwigshafen, Germany). N-(2-hydroxypropyl) methacrylamide (HPMA), HPMA mono- and dilactate and PEG10000-4,4’-azobis(cyanopentanoate) macroinitiator were synthesized as previously reported35–37. Phosphate buffered saline (PBS), penicillin/streptomycin (pen/strep; 10,000 units/ml penicillin and 10 mg/ml streptomycin) and picogreen DNA assay were supplied by Invitrogen (Carlsbad, California, USA). Three different types of Dulbecco’s Modified Eagle Medium (DMEM) were used: DMEM 31885 from Gibco (referred to as DMEM), high glucose DMEM D6429 from Sigma-Aldrich (referred to as high glucose DMEM) and DMEM/F-12+GlutaMax-1 31331 from Invitrogen (referred to as DMEM/F-12). Fetal bovine serum (FBS) was obtained from Gibco (Invitrogen corporation) and type II collagenase was purchased from Worthington Biochemical Corp (Lakewood, NJ, USA). ITS+ premix (human recombinant insulin, human transferrin, selenous acid, bovine serum albumin, linoleic acid) was obtained from BD Biosciences (Breda, the Netherlands), recombinant human TGF-β1 from Peprotech (London, UK), pronase (11459643001) from Roche Life Sciences (Indiana, USA), hyaluronidase (H2126) from Sigma-Aldrich and Tissucol Duo S (fibrin and thrombin) from Baxter (Utrecht, the Netherlands). Antibody against collagen type I (1:100; EPR7785, ab138492) was obtained from Abcam (Cambridge, UK). Antibodies against collagen types II and VI (1:100; II-6B3II and 1:5, 5C6, respectively) were obtained from the Developmental Studies Hybridoma Bank (Iowa City, IA, USA). Secondary horse radish-peroxidase conjugated antibodies for collagen type I (EnVision+, K4010), collagen type II (1:100, IgG HRP, P0447), and collagen type VI (EnVision+, K4007) were ordered from DAKO (Heverlee, the Netherlands). Calcein-AM (to stain living cells) and ethidium homodimer-1 (to stain nuclei of dead cells) were obtained from Life Technologies (L3224, Bleiswijk, the Netherlands). Finally, Dye-Trak ’F’ microspheres (Fluorescent Orange) were ordered from Triton Technology Inc. (San Diego, CA, USA).
Synthesis of methacrylated poly(N-(2-hydroxypropyl) methacrylamide mono/dilactate)-PEG triblock
The synthesis of a methacrylated thermo-sensitive triblock copolymer, consisting of a hydrophilic PEG-based mid-block flanked by two partially methacrylated pHPMA-lac outer blocks was carried out as previously described by Vermonden et al.13,14. Briefly, a free radical polymerization in acetonitrile was carried out at 70 °C for 40 hours under a N2 atmosphere, using PEG10000-4,4’-azobis(cyanopentanoate) as macroinitiator and HPMA mono- and dilactate (molar ratio mono/dilactate = 75:25) as monomers, with a mass ratio monomers/macroinitiator of 4:1. After precipitation in cold diethyl ether, the polymer was collected and further modified via partial esterification of the hydroxyl groups present on the lactate units with methacrylate groups. This reaction was carried out in dry tetrahydrofuran as solvent and methacrylic anhydride (MA, molar feed of 13.3% of the free hydroxyl groups of the polymer) was used as methacrylating agent in presence of triethylamine and 4-dimethylaminopyridine. The methacrylated polyHPMA-lac-PEG triblock copolymer is further referred to as M10P10 (M10 refers to a DM of 10% and P10 refers to a PEG block with a molecular weight (MW) of 10 kDa) and its precursor as M0P10. A low DM of 10% was chosen to achieve a low network density in the cross-linked hydrogel, which is likely beneficial for cell behavior20.
Methacrylation of polysaccharides
Methacrylation of CS was carried out using a transesterification reaction, as described by Abbadessa et al.38. Briefly, CS A sodium salt was converted into tetrabuthylammonium (TBA) salt (CS-TBA) by using a Dowex® 50WX8 hydrogen form resin, previously saturated with TBA fluoride. Subsequently, 2.7 g (3.08 mmol of disaccharide units) of CS-TBA was dissolved in 100 ml of dry dimethyl sulfoxide (DMSO) under a N2 atmosphere at 50 °C. Next, 4-dimethylaminopyridine (0.495 g) and glycidyl methacrylate (GMA, 195 µl) were added and the reaction mixture was stirred at 50 °C for 48 hours. After the reaction, the mixture was diluted with water and the pH was lowered to 5.5 using a 0.2 M solution of HCl in water. The polymer solution was further dialyzed against a 150 mM NaCl solution in water for 3 days and against water for 4 days. The polymer was finally collected, as Na+ salt, after freeze-drying and it is further referred to as CSMA.
HA was methacrylated using a slightly modified method from the one reported by Hachet et al28. Briefly, 0.5 g (1.25 mmol of disaccharide units) of HA was dissolved in 80 ml of ultrapure water at 4 °C overnight. Subsequently, N,N-dimethylformamide (DMF) was added to obtain a mixture with 1:1 water/DMF volume ratio. Next, 926 µl (6.25 mmol) of MA was added drop-wise at 4 °C to the HA solution while the pH was kept between 8 and 9 by adding 0.5 M NaOH. The pH was monitored for 4 hours and adjusted to 8-9. After overnight stirring at 4 °C, the polymer was precipitated by addition of NaCl (final concentration in the mixture 0.5 M) and cold ethanol (final ethanol/water ratio of 2.3:1), and further purified by means of dialysis (MWCO 10,000-14,000 Da). Purified HAMA was collected after freeze-drying.
The DM of HAMA was investigated using a method based on the detection of methacrylic acid, which is released after basic hydrolysis of the ester bonds present in the methacrylated polysaccharide39. The formed methacrylic acid was detected with a High Performance Liquid Chromatography (HPLC) Waters 2695 separating module equipped with a Waters 2487 dual λ absorbance detector (λ = 210 nm, Waters Corporation, Milford, MA, USA) and with a C18 column (Sunfire). HAMA (15 mg) was dissolved in 10 ml of 0.02 M NaOH at 37 °C for 2 hours. Subsequently, 2 ml of 2 M acetic acid was added. After filtration using a 0.2 µm filter, the samples were injected in the HPLC system and eluted at 1 ml/minute using a mixture of acetonitrile/water (15:85, pH = 2) as mobile phase. Calibration was performed using solutions of methacrylic acid of different concentrations in the same eluent.
Experimental design and hydrogel groups
To investigate if M10P10 hydrogels support chondrogenesis of chondrocytes, UV cross-linked constructs from an equine chondrocyte (passage 1, n = 3 donors) laden M10P10 (18% w/w) polymer mixture were prepared. Constructs were cultured for 42 days and evaluated for evidence of chondrogenesis at days 0 (harvest directly after cell encapsulation), 28 and 42, via quantitative measurements and histology. This gel formulation is further referred to as cell-laden hydrogel M.
To investigate whether the incorporation of HAMA or CSMA in M10P10 can improve the mechanical properties, affect the degradation rate, and enhance the printability, cell-free polymer mixtures based on M10P10 (18% w/w), M10P10 (14% w/w) blended with CSMA (4% w/w), or M10P10 (14% w/w) blended with HAMA (0.9% w/w) were prepared and are further referred to as mixtures M, MCS and MHA, respectively (Table 1). These mixtures were analyzed for their thermo-sensitive properties using rheological measurements. Cell-free UV cross-linked M, MCS and MHA hydrogels were further characterized for their Young’s modulus and their degradation/swelling behavior in PBS (pH 7.4) enriched with 0.02% of NaN3 at 37 °C. Finally, 3D constructs were printed with polymer mixture MHA laden with fluorescent microspheres to assess homogeneous encapsulation, using a 3D bioprinter (regenHU, Villaz-St-Pierre, Switzerland). Additionally, constructs with primary chondrocytes were printed using mixtures M, MCS and MHA to assess viability 1 and 7 days after printing. All measurements were performed in triplicate.
Table 1.
Compositions of the three hydrogel groups
| hydrogel | polymer concentration (w/w%) | ||
|---|---|---|---|
| M10P10 | CSMA | HAMA | |
| M | 18% | - | - |
| MCS | 14% | 4% | - |
| MHA | 14% | - | 0.9% |
Chondrocyte isolation and culture
Primary chondrocytes were isolated from full-thickness cartilage of the stifle joints of fresh equine cadavers (n = 3; 3–10 years old horses), with consent of the owners. Macroscopically healthy cartilage was removed from the joint under aseptic conditions and the cartilage was digested overnight at 37 °C in DMEM supplemented with collagenase II (1.5 µg/ml), hyaluronidase (1 mg/ml), FBS (10%) and pen/strep (1%). After digestion, the cell suspension was filtered through a 40 µm cell strainer. Chondrocytes were washed with PBS and stored in liquid N2 until further use.
In order to prepare cell-laden constructs, the chondrocytes were expanded in monolayer culture for 14 days (seeding density of 5 · 103 cells/cm2) in chondrocyte expansion medium consisting of DMEM, FBS (10%) and pen/strep (1%). The chondrocytes were harvested and mixed with the polymer mixture at passage 1 when they reached 80-90% confluence. Cell-laden constructs were cultured in chondrogenic differentiation medium consisting of high glucose DMEM supplemented with ITS+ premix (1%), dexamethasone (0.1 µM), L-ascorbic acid-2-phosphate (0.2 mM), recombinant human TGF-β1 (10 ng/ml) and pen/strep (1%) to stimulate chondrogensis and redifferentation of the chondrocytes40,41.
Fabrication of cell-laden chemically cross-linked M10P10-based hydrogels
M10P10 was dissolved in PBS at 4 °C and Irgacure was added (concentration: 0.05% w/w). The resulting mixture (M10P10 concentration: 20.5% w/w) was stirred overnight in the dark at 4 °C. The expanded chondrocytes were mixed on ice with the polymer mixture to obtain a concentration of 15-20 · 106 chondrocytes/ml (concentration varied per donor). Correcting for the average weight of the added cells, the final concentrations of Irgacure and M10P10 in the cell-laden polymer mixture were 0.044% w/w and 18% w/w, respectively. The cell-laden suspension was injected into a Teflon mold, which was covered with a glass slide to generate cylindrical samples (sample size: 6 mm in diameter, 2 mm in height). The filled molds were placed at 37 °C for 5 minutes to allow physical gelation of the hydrogel. Subsequently, chemical cross-linking was induced with a UV lamp (CL-1000L Model, UVP, Cambridge, UK, Intensity: 7.2 mW/cm2, irradiation time: 15 minutes). Next, the samples were cultured at 37 °C and 5% CO2 for 42 days in chondrogenic differentiation medium. The medium was refreshed twice a week. Fibrin gels were prepared as a positive control for cell behavior. Chondrocytes were mixed with fibrinogen (Tissucol Duo S, diluted 1:15 in PBS) to get a cell density of 30-40 · 106 cells/ml. Next, 30 µl of thrombin (Tissucol Duo S, diluted 1:50 in PBS, 500 IU) was pipetted into the cylindrical molds and 30 µl of cell-laden fibrinogen suspension was mixed into the thrombin solution to generate a final cell concentration of 15–20 · 106 chondrocytes/ml (same as for cell-laden M hydrogels). Samples were incubated for 15 minutes at room temperature and placed in culture with chondrogenic differentiation medium as described above.
Histology & Immunohistochemistry
At days 0 (harvest directly after cell encapsulation), 28 and 42, three samples of each hydrogel group (M and fibrin) were harvested. Part of each sample was fixed overnight in formalin (37%) and dehydrated through a graded ethanol series. After clearing in xylene, the samples were embedded in paraffin and sectioned at a thickness of 5 µm. Sections were stained with safranin-O to visualize proteoglycans, fast green to visualize collagens, and hematoxylin to stain cell nuclei, as previously described42.
Collagen types I, II and VI were visualized with immunohistochemistry. First, the sections were deparaffinized and hydrated. Next, antigen retrieval was performed with pronase (1 mg/ml in PBS) and hyaluronidase (10 mg/ml in PBS) for 30 minutes at 37 °C, followed by a blocking step of 10 minutes with H2O2 (0.3% in PBS) at room temperature. The primary antibody was incubated overnight at 4 °C. Mouse IgG was used at matched concentrations for negative control staining. After incubation, the matching secondary antibody was added and incubated for 30 minutes for collagen type I and 60 minutes for collagen types II and VI, at room temperature. Finally, all stainings were visualized with 3,3′-diaminobenzidine peroxidase substrate solution for 3-10 minutes and counterstained with Mayer’s hematoxylin. All stained sections were evaluated and photographed using a light microscope (Olympus BX51 microscope, Olympus DP70 camera, Hamburg, Germany).
Biochemical assays
The remaining part of each harvested cell-laden hydrogel was weighed, freeze dried, and weighed again to determine the sample dry weight and water content. Next, the dried hydrogels were digested overnight at 56 °C in 200 µL papain digestion buffer (0.2 M NaH2PO4 + 0.01 M EDTA · 2 H2O in milliQ, pH = 6.0) supplemented with 250 µL/ml papain solution (16-40 units/mg protein) and 0.01 M cysteine. To determine the glycosaminoglycan (GAG) content, as a measure for proteoglycan, a dimethylmethylene blue (DMMB)43 assay was used with known concentrations of chondroitin sulfate C as a reference. The amount of GAG was normalized to the dry weight and DNA content of the samples, as measured by the Quant-iT PicoGreen dsDNA kit and read on a spectrofluorometer (Biorad, Hercules, California, USA), all according to the manufacturer’s protocols.
Fabrication of chemically cross-linked hydrogels modified with polysaccharides
Defined amounts of M10P10 and CSMA or HAMA (Table 1) were dissolved in PBS at 4 °C and Irgacure was added as the last component (final concentration: 0.044% w/w). The polymer mixture containing CSMA was stirred overnight while the mixture containing HAMA was stirred for 48 hours at 4 °C to allow complete dissolution. Subsequently, the polymer mixtures were injected into Teflon molds (sample size: 6 mm in diameter, 2 mm in height), incubated for 5 minutes at 37 °C and UV irradiated as described for the cell-laden cross-linked M hydrogels (section ‘Fabrication of cell-laden chemically cross-linked M10P10-based hydrogels’). Two different hydrogel compositions, MCS and MHA were prepared, in which M10P10 was partially replaced by either CSMA or HAMA, respectively. Finally, hydrogels containing only M10P10 in the maximum total polymer concentration used for hybrid gels were prepared as a control group (18% w/w, hydrogels M). The total polymer concentration in MHA hydrogels was slightly lower compared to the other two hydrogels, since it was not possible to dissolve more than 0.9% w/w of this polysaccharide due to its high MW.
Mechanical analysis
Thermo-responsive properties of the polymer mixtures (M, MCS and MHA) before chemical cross-linking were studied using an AR G-2 rheometer (TA-Instruments, Etten-Leur, The Netherlands), equipped with a cone-plate measuring geometry (cone diameter: 20 mm, angle: 1°). All polymer mixtures were tested under oscillation temperature sweeps from 4 to 50 °C employing a frequency of 1 Hz and a strain of 1%, which was found to be within the linear viscoelastic range of all formulations (Figure S2). Values of storage and loss moduli (G’ and G”, respectively) were recorded for each sweep and the resulting rheograms were reported showing the lines interconnecting all data points for each run.
To investigate the stiffness of hydrogel constructs after UV cross-linking, all polymer mixtures (M, MCS and MHA) were molded as described in section ‘Fabrication of chemically cross-linked hydrogels modified with polysaccharides’ and allowed to swell for 3 hours in PBS at room temperature. Next, hydrogels were examined under unconfined compression test using a Dynamic Mechanical Analyzer, DMA (2980 DMA, TA Instruments, Etten-Leur, The Netherlands). The hydrogels were subjected to a preload force of 0.001 N and subsequently compressed with a force ramp rate of 0.25 N/minute and an upper force limit of 1 N13. The Young’s Modulus was calculated as the slope of the initial linear segment of the stress/strain curve22.
In vitro swelling-degradation study
For all polymer mixtures (M, MCS and MHA) cross-linked samples (6 mm of diameter, 2 mm of height, 56.5 µl of volume) prepared as described in section ‘Fabrication of chemically cross-linked hydrogels modified with polysaccharides’ were placed in glass vials (diameter: 1.75 cm) with 1 ml of PBS (pH 7.4), supplemented with 0.02% of NaN3. The vials were incubated at 37 °C and the solutions were refreshed twice per week. At multiple time points, the hydrogels were weighed and the swelling ratio (SR) was calculated as follows:
| Equation 1 |
in which mday x represents the hydrogel mass after x days of incubation and mday 0 the hydrogel mass before the hydrogel was placed in PBS.
Printing of hydrogels
A 3DDiscovery bioprinter (regenHU, Villaz-St-Pierre, Switzerland) equipped with a Bluepoint 4 UV lamp (point light source, wavelength range: 300-600 nm, UV-A intensity at 5 cm = 103 mW/cm2, Hönle UV Technology AG, Gräfelfing, Germany) was used for the 3D printing of hydrogels. Filaments were generated with a micro valve (CF300H) print head, for optimal control over volume deposition rates, using optimized printer settings (Table S1). To generate porous constructs, alternating layers of vertical and horizontal filaments were deposited in the x,y-plane. Cross-linking was performed in a layer-by-layer fashion, exposing each deposited layer for 3 seconds to UV light from a distance of 5 cm. After printing, the constructs were irradiated for an additional 9 seconds.
Printing of hydrogels loaded with fluorescent microspheres and cells
To evaluate the feasibility of homogeneous cell encapsulation, polymer mixture MHA was supplemented with fluorescently labeled microspheres (Fluorescent Orange Dye-Trak ‘F’ microspheres, Triton Technology, diameter 15 µm similar as a single cell, concentration in the polymer mixture 0.8 million/ml) and constructs were 3D printed using optimized print settings (Table S1). To visualize the distribution of the microspheres in the constructs, an Olympus BX51 microscope was used.
To evaluate cell viability after printing, primary chondrocytes (harvested and expanded as described in section ‘Chondrocyte isolation and culture’) were encapsulated in mixtures M, MCS and MHA. The cell-laden mixtures were heated to 37 °C and three constructs were subsequently printed using the aforementioned print method reported in section ‘Printing of hydrogels’. As a positive control, cast hydrogels were prepared for each mixture using the same method as for the equine chondrocyte laden hydrogels (section ‘Fabrication of cell-laden chemically cross-linked M10P10-based hydrogels’). Each printed construct was cut into four pieces, which were cultured in separate wells with chondrocyte expansion medium. Viability was checked on two pieces at day 1 and for the other pieces after 7 days of culture. To check cell viability, the hydrogels were stained for 20 minutes with calcein-AM (4 µM in PBS) and ethidium homodimer-1 (2 µM in PBS) at 37 °C. After washing three times in PBS, the red and green fluorescent signals were visualized using an Olympus BX51 microscope and three images of each hydrogel quarter were analyzed.
Statistics
Statistical analyses were performed using SPSS software (version 20, IBM Corporation, USA). Differences in Young’s modulus between the hydrogel groups (M, MHA, MCS) and differences in chondrocyte viability after printing at each time point, were determined with a One-Way ANOVA test. For GAG values normalized to the DNA content, both hydrogels (M and fib) at all time-points (6 groups in total) were compared with each other using a Randomized Block Design ANOVA to correct for donor variability. The GAG, DNA, and water contents normalized to the dry weight at the different time points were compared to each other within each hydrogel formulation by a Randomized Block Design ANOVA. A significance level of 0.05 and a Tukey’s Post-hoc analysis were used for all tests.
Results and Discussion
Synthesis and characterization of thermo-sensitive polymers and methacrylated polysaccharides
M0P10 and M10P10 (Figure 1) were obtained in a high yield (80% and 96%, respectively). Their chemical structures, confirmed by 1H-NMR, were in accordance to previously reported data13,14. The Mn and DM of M10P10 determined by 1H-NMR were 42.4 kDa and 10.7%, respectively, whereas the Mn according to GPC was 34.6 kDa with a PDI value of 2.0. The cloud points of M0P10 and M10P10 were 35 °C and 20 °C, respectively. Table 2 summarizes the polymer characteristics for M0P10 and M10P10.
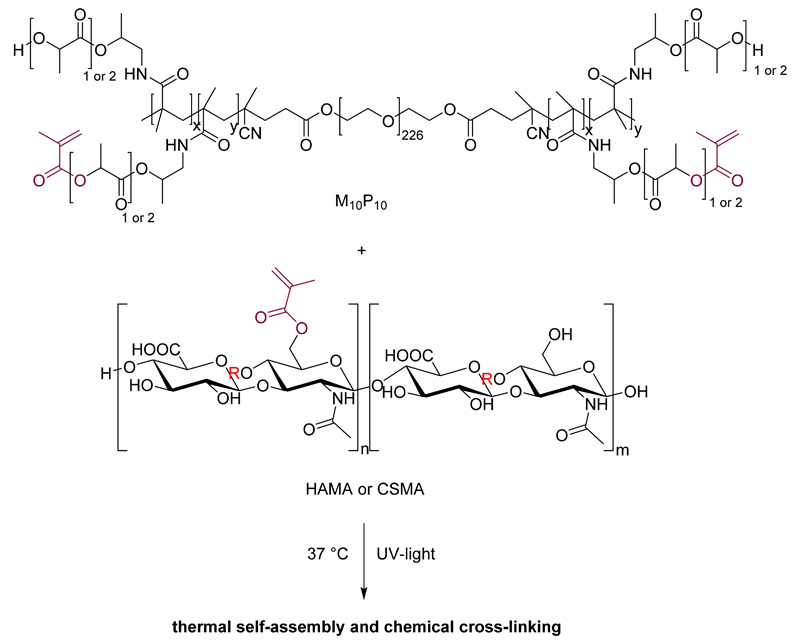
Figure 1.
Chemical structure of M10P10 (top) and methacrylated HA (bottom, R = H in equatorial position) or CS (bottom, R = SO3H in axial position). M10P10 confers thermo-sensitive properties to the gel, whereas the presence of methacrylate groups in both polymers allows UV-mediated chemical cross-linking.
Table 2.
Characteristics of thermo-sensitive polymers and polysaccharides
| Polymer | DM (%) | Mn (kDa) | PDI | CP (°C) |
|---|---|---|---|---|
| M0P10 | 0a | 43.9a | 1.9b | 35c |
| 36.2b | ||||
| M10P10 | 10.7a | 42.4a | 2.0b | 20c |
| 34.6b | ||||
| CS | 0a | 26.9 (94%)d | 1.4d | n.a. |
| 353.8 (6%)d | 1.3d | |||
| CSMA | 15.2a | n.d. | n.d. | n.a. |
| HA | 0a | 1560e | n.d. | n.a. |
| HAMA | 23.4f | n.d. | n.d. | n.a. |
Determined by 1H-NMR
Determined by GPC
Determined by UV-VIS spectrophotometry
Determined by Viscotek
Average MW determined by Multi-Angle Light Scattering Size Exclusion Chromatography (MALS-SEC) as reported from the supplier
Determined by HPLC
n.d.: not determined
n.a.: not applicable
The methods employed for the methacrylation of CS and HA resulted in high yields of CSMA and HAMA (>84% for both polysaccharides). The methacrylated polysaccharides (chemical structures shown in Figure 1) were analyzed by 1H-NMR. The presence of the signals at 6.2 and 5.8 ppm, representative of the two vinyl protons present in the methacrylate groups, and the signal at 2.0 ppm, typical of the protons belonging to its methyl group, confirmed the partial functionalization of the hydroxyl groups with methacrylate groups.
The methacrylation of CS was performed in DMSO using GMA as methacrylating agent, and a molar feed of GMA and CS-TBA repeating units of 0.48:1 resulted in a DM of 15.2% (Table 2), calculated according to 1H-NMR. Moreover, the absence in the 1H-NMR spectrum of the signals at 5.5 and 5.2 ppm representative of a possible glyceryl spacer between the methacrylate group and the disaccharide unit, excluded the presence of products originating from ring opening reaction44. Thus, the reaction mechanism follows a transesterification mechanism, which is in line with our previous findings38.
For the synthesis of HAMA, we selected the method reported by Hachet et al.28. This reaction was performed in a mixture of water and DMF using a large excess of MA (molar ratio of 5:1 between MA and repeating units of HA). This high feed ratio is generally used for methacrylation reactions in aqueous environment because it is necessary to compensate for the amount of MA lost as methacrylic acid due to hydrolysis45,46. A lower polymer concentration, 3.1 versus 12.0 mg/ml was used compared to previously reported reactions, which were performed using a lower MW HA28,47. The use of relatively low concentration was necessary to facilitate pH monitoring and general handling of the reaction mixture, considering the high viscosity of high MW HA solutions. This low HA concentration likely explains our lower methacrylate incorporation (5%) compared with previous reports (≥ 14%)28,47. Because of the poor resolution of the 1H-NMR spectra for high MW HAMA, an HPLC-based method was employed to accurately determine the DM, which was found to be 23.4% (Table 2).
Matrix production of embedded chondrocytes
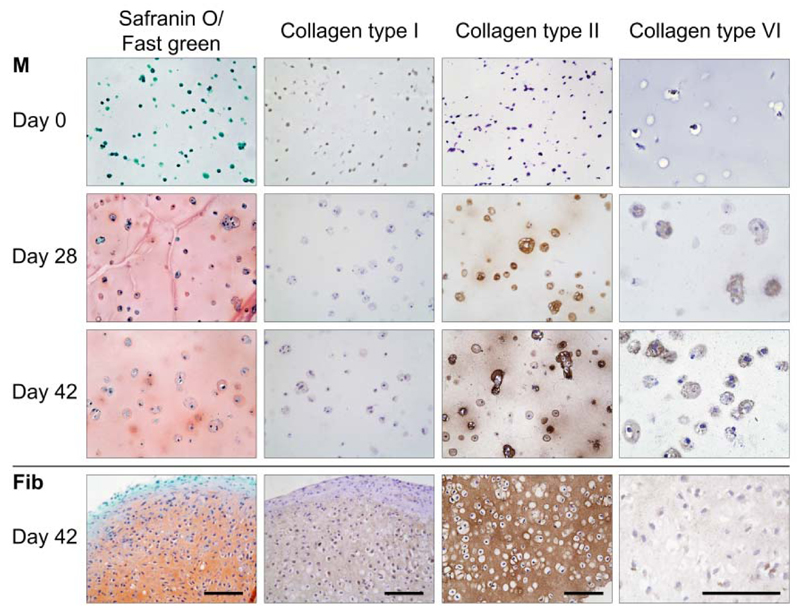
Hydrogels composed of methacrylated polyHPMA-lac-PEG triblock copolymers have been shown to support the short-term survival of chondrocytes, however, the effect on the matrix production was not yet reported10. In this study, equine chondrocytes were encapsulated into an 18% M10P10-based hydrogel (hydrogel M) and cultured up to 42 days in chondrogenic differentiation medium. The matrix production in this hydrogel was compared to that of chondrocytes embedded in fibrin gel (positive control), which is the golden standard for clinical delivery of cells for cartilage repair procedures and is known to support chondrogenesis due to its bioactive peptide sequences48,49. Hydrogel M supported cartilage-like tissue formation of the encapsulated chondrocyte and Safranin-O staining revealed a homogeneous deposition of proteoglycans after 28 and 42 days of culture (Figure 2). In addition, immunolocalization of collagen type II revealed that its deposition was limited to distinct areas around the cells at day 28. However, after 42 days a more homogeneous distribution was observed. Both stainings were more intense in the fibrin gels at day 28 and 42 compared to hydrogel M samples at these time points (Figure 2). An explanation for this effect is the compaction of the fibrin gels during the first days of culture50–52. Because of this, the relative cell density and amount of matrix per gel volume increased as can be observed in the high DNA/dwt and GAG/dwt values for fibrin samples (Figure 3e, f). The sample dry weight was ten times higher for hydrogels M compared to fibrin gels and this difference remained over time (data not shown). Water volume normalized to the dry weight of M hydrogels increased at day 28 and 42 compared to day 0 (Figure 3d, 250% and 330% respectively). Although hydrogel compaction after implantation in a defect may localize the cells at the bottom of the defect, it will on the other hand result in an incomplete defect-fill. Moreover, contracting materials may be difficult to combine in hybrid scaffolds, e.g. hydrogel constructs reinforced with polymeric fibers, aimed to increase construct stiffness7,22,53. In these hydrid constructs, shrinking is a major drawback since it may cause stress at the interface and lead to loss of construct integrity.
Figure 2.
Histology and immunohistochemistry of chondrocytes differentiated in M10P10-based hydrogels (M) with fibrin (fib) as a positive control. From left to right: safranin-O staining, collagen types I, II and VI staining. Scale bars represents 100 µm and is the same for all images of the same staining (column).
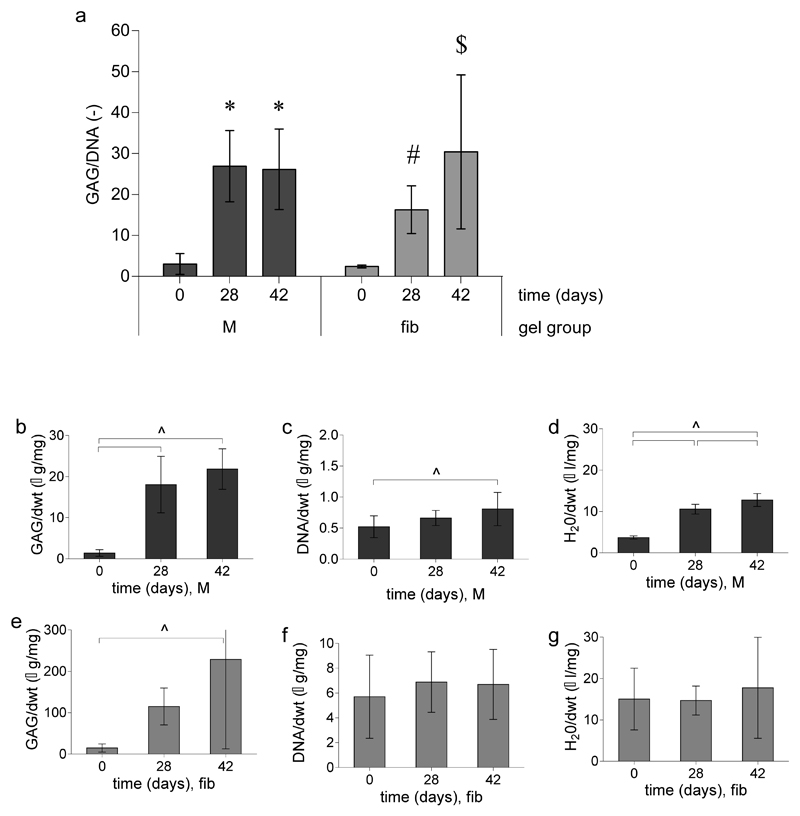
Figure 3.
Quantitative GAG, DNA, and water measurements for equine chondrocytes encapsulated in M10P10-based hydrogels (M) and fibrin (fib) gels. a) GAG content normalized to DNA for both hydrogels over time. * denotes significant differences compared to day 0; # indicates that the group is significantly higher than the day 0 controls but lower compared to fibrin day 42. $ indicates that the group is significantly higher than the day 0 controls and day 28 fibrin samples but equal to the M hydrogels at days 28 and 42. b, c, d) GAG, DNA and water content normalized to the dry weight (dwt) for M hydrogels over time, respectively. e, f, g) GAG, DNA, and water content normalized to the dry weight (dwt) for fibrin gels over time. ^ significant difference between groups, respectively.
A collagen type VI staining was performed to visualize chondron formation. Chondrons are chondrocytes with their pericellular matrix, consisting of proteoglycans, collagen types II and VI54, and are known to be more active in matrix deposition than chondrocytes55. In hydrogels M, collagen type VI positive areas were found around the cells after 28 and 42 days of culture, indicating that chondrocytes formed chondron-like structures during culture. In fibrin samples a slight overall positive collagen type VI staining was found. Further, only limited positive staining for collagen type I was observed in all hydrogel samples, suggesting limited dedifferentiation of the embedded chondrocytes.
Quantitative measurements were performed for GAG, DNA and water content. However, a large variation in cell performance of the three different equine donors (age 3–10 years old) was observed (Figure 3), which is in line with previous reported studies56. GAG content normalized to DNA content (GAG/DNA) was similar in M hydrogels at days 28 and 42 (27±9 µg/µg and 26±10 µg/µg, respectively, Figure 3a). At day 28, GAG/DNA was statistically higher compared to the fibrin control gels (16±6 µg/µg, Figure 3a) at this time point. After 42 days of culture both hydrogel formulations performed equally. The GAG content normalized to the dry weight of both the M and fibrin hydrogels increased with time (Figure 3b and 3e). However, DNA levels normalized to the dry weight only showed a significant increase for the M hydrogels over time (0.52±0.18 µg/mg at day 0 and 0.81±0.30 µg/mg at day 42, Figure 3c), indicating cell proliferation. Finally, higher GAG/dry weight and DNA/dry weight values were found for fibrin gels compared to hydrogels with formulation M, which can be explained by the compaction and relatively fast degradation of the fibrin gels. In addition, M hydrogels seemed to swell during cultures as the H2O/dry weight increased during culture.
Thus, chondrocytes in hydrogels with formulation M produced similar levels of cartilage-like matrix compared to chondrocytes in fibrin gels. In addition, no compaction occurred for M hydrogels. Encouraged by these results, hydrogels with formulation M were further evaluated and CSMA and HAMA were incorporated to optimize the mechanical properties, degradation kinetics, and printability.
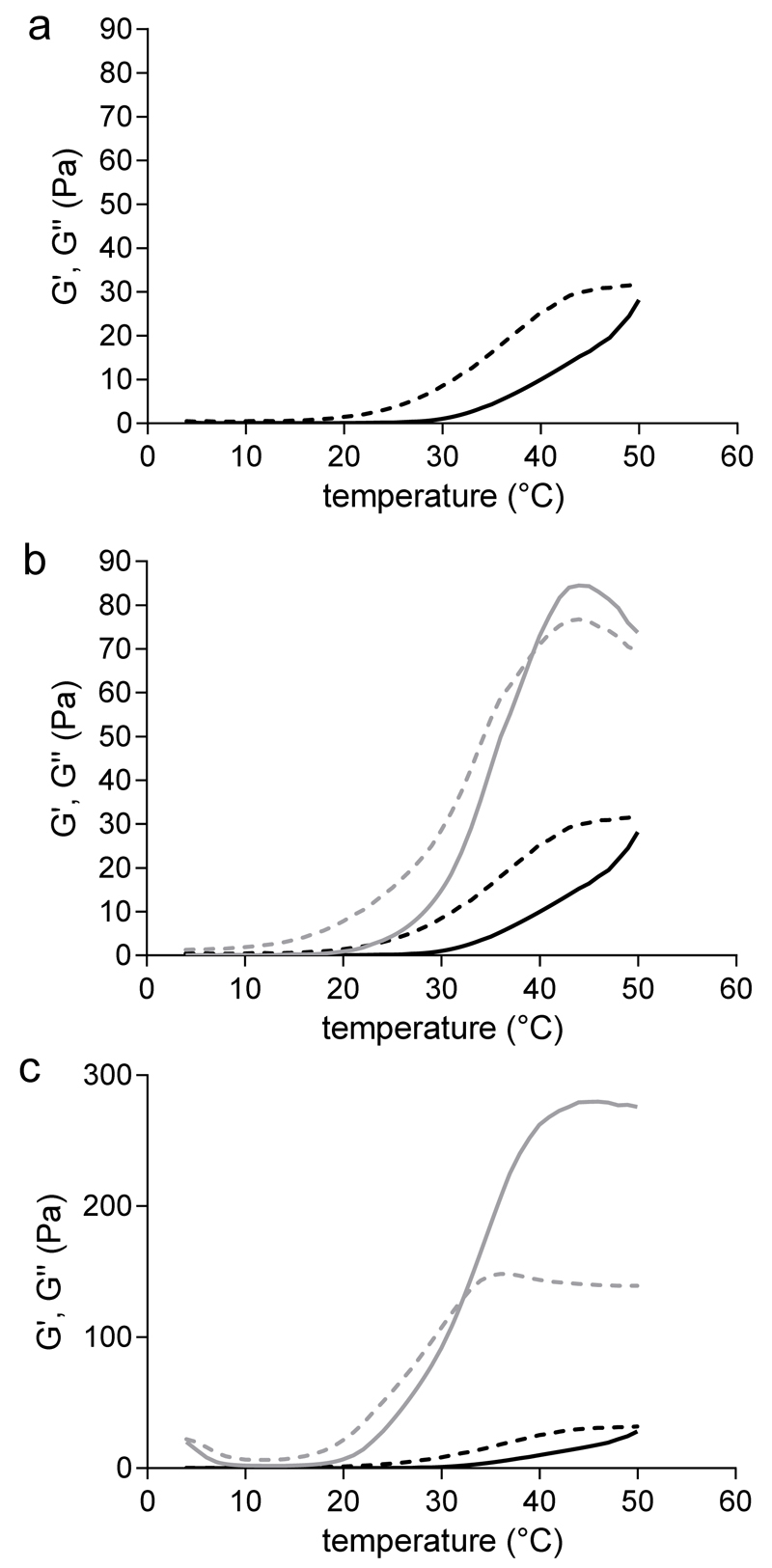
Thermo-gelation of polymer mixtures before chemical cross-linking
Figure 4 shows storage and loss moduli, G’ and G”, as a function of temperature for all polymer mixtures. Mixtures based only on M10P10, exhibited an increase of G’ when increasing the temperature, up to 29±2 Pa at 50 °C, while G” displayed higher values over the whole temperature range (Figure 4a). M10P10 is a thermo-sensitive polymer capable to self-assemble and to form hydrophobic domains above defined temperatures, leading to a physical gel within a certain range of concentrations13. The absence of a gelation temperature (Tgel), here defined as the temperature at which G’ crosses G”, as well as the low value of G’ reached upon rising the temperature for polymer mixture M, is due to the relatively low concentration and high CP (20 °C) of the thermo-sensitive polymer used in this study.
Figure 4.
Rheograms of polymer mixtures. G’ (solid line) and G” (dotted line) moduli as a function of temperature, recorded during a temperature sweep experiment from 4 to 50 °C. a) hydrogels based on 18% (w/w) M10P10 (M hydrogels). b) hydrogels based on 14% (w/w) M10P10 and 4% (w/w) CSMA (MCS hydrogels, grey lines) compared with M hydrogels (black lines). c) hydrogels based on 14% (w/w) M10P10 and 0.9% (w/w) HAMA (MHA hydrogels, grey lines) compared with M hydrogels (black lines).
Figures 4b and 4c show that a continuous increase in G’ as a function of temperature was observed for aqueous systems of MCS and MHA. The values of the storage modulus at 37 and 50 °C were 56±6 and 84±24 Pa, respectively, for MCS hydrogels, and 216±14 and 263±12 Pa, respectively, for MHA hydrogels. For both MCS and MHA mixtures a Tgel was found (39 °C for MCS hydrogels and 32 °C for MHA hydrogels). In line with previous findings, it can be observed that the partial replacement of M10P10 with CSMA or HAMA resulted in the formation of physical gels with much higher G’ values above 20 °C than polymer mixtures only composed of M10P1038. The beneficial role of the added polysaccharide on the mechanical properties of the hydrogel is more remarkable for MHA hydrogels, where an even lower total polymer concentration (Table 1) led to the formation of the stiffest hydrogel (G’ = 216±14 at 37 °C). The rheological behavior of the polysaccharide-enriched formulations clearly shows that the elastic properties of hydrogels based on M10P10 can be improved by the addition of polysaccharides, without increasing the total polymer concentration.
Mechanical properties and in vitro swelling-degradation behavior of chemically cross-linked hydrogels
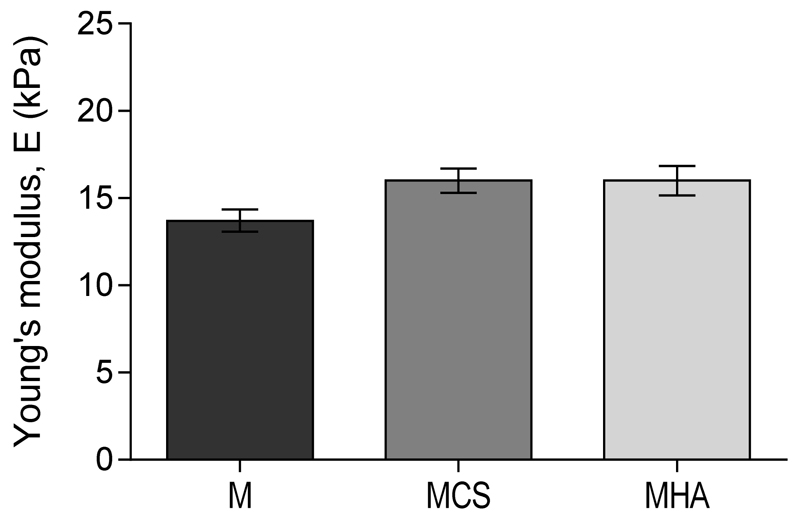
The injection of polymer mixtures in a Teflon mold at 4 °C, followed by a temperature increase to 37 °C and UV irradiation for 15 min, resulted in the formation of cylindrically shaped constructs. Figure 5 shows the Young’s moduli for the different hydrogel constructs after 3 hours of swelling in PBS. The Young’s modulus values were 13.7±1.1, 16.0±1.4 and 16.0±1.9 kPa, for M, MCS and MHA hydrogels, respectively. No significant differences between the three hydrogel formulations were found. Hence, no differences in cell response due to different mechanical stimuli can be expected in the three hydrogels. The influence of polysaccharide molecular weight on the final stiffness can be illustrated by comparing MCS and MHA hydrogels. Hydrogels with comparable Young’s moduli were obtained, despite the much lower concentration of the higher MW polysaccharide (0.9% vs. 4%) and the lower number of methacrylate groups in MHA hydrogels, calculated considering the slight difference in DM of the two polysaccharides (Figure 5). In line, the positive influence of HA with higher MW has been reported previously for hybrid hydrogel systems based on acrylated HA and thiol-modified 4-arm PEG or thiol-derivatives of HA and PEG-vinylsulfones, cross-linked via Michael addition-type reaction57,58. As can be expected for hydrogel materials, the stiffness of these hydrogel constructs is significantly lower than that of native cartilage (400–800 kPa59–61).
Figure 5.
Dynamic mechanical analysis on chemically cross-linked hydrogels. Young’s moduli for hydrogels based on M10P10 (M), hydrogels based on M10P10 and CSMA (MCS) and hydrogels based on M10P10 and HAMA (MHA), measured under unconfined compression (n = 3).
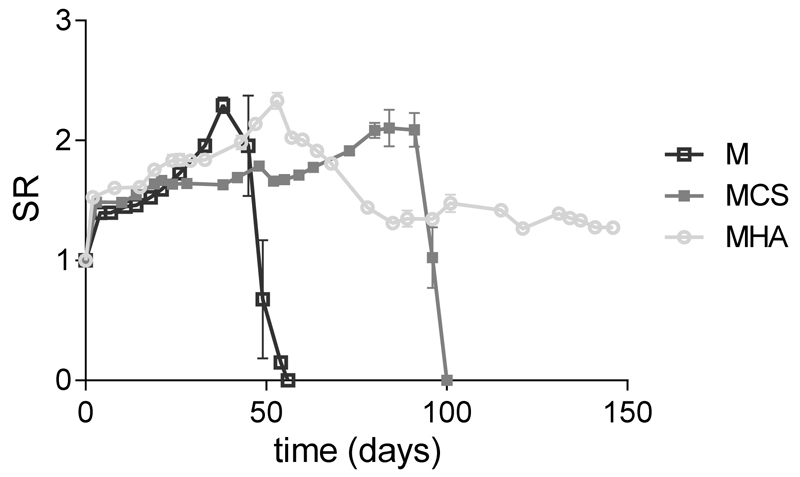
Figure 6 shows that M hydrogels initially swelled for 38 days during which the SR reached a maximum of 2.3±0.1. Complete degradation occurred in 56 days of incubation at 37 °C. This degradation profile is in line with previously reported studies8,13. The degradability of hydrogels based on chemically cross-linked polyHPMA-lac-PEG triblock copolymers at pH 7.4 and 37 °C is due to the hydrolysis of several ester bonds9. The first soluble degradation products are lactic acid units obtained by the hydrolysis of OH-terminated lactate side chains. Consequently, the remaining gel matrix exhibits an increased hydrophilic character with a higher water-uptake capacity, leading to the typical swelling phase. Mass loss is seen when the elimination of the water-soluble degradation products from the matrix exceeds the water uptake. This swelling-degradation behavior might also explain the absence of GAG increase in the chondrocyte laden M hydrogels between 28 and 42 days of culture. The swelling process and the presence of a partially degraded and thus less dense hydrogel matrix between day 28 and 42 may have contributed to the leaching of newly formed GAGs out of the gel62.
Figure 6.
Swelling and degradation profiles for hydrogels based on M10P10 (M), hydrogels based on M10P10 and CSMA (MCS), and hydrogels based on M10P10 and HAMA (MHA) in PBS buffer at 37 °C. Error bars represent the standard deviation of experiments performed in triplicate. SR represents the swelling ratio and was calculated according to equation 1.
In contrast to M hydrogels, the hydrogels containing polysaccharides degraded much slower (Figure 6). More specifically, MCS hydrogels swelled for 91 days with a maximum SR of 2.1±0.2 and underwent complete disintegration in 100 days, whereas the degradation profile of MHA hydrogels showed a maximum in the SR of 2.3±0.1 at day 53, followed by partial mass loss during the subsequent 32 days and reached a plateau in SR of 1.4 for the subsequent 61 days of monitoring. Thus, the presence of the two polysaccharides, increased the stability of the hydrogels under the tested conditions. In fact, the loss of polysaccharides from these hydrogels can only occur after the polysaccharide molecules diffuse out of the hydrogel matrix and are dissolved in the surrounding buffer. This phenomenon can take place only after complete hydrolysis of the ester bonds of the polymerized methacrylate groups, which connect a polysaccharide chain to another polysaccharide or M10P10 chain. However, it has been reported that polymerized methacrylate groups directly attached to polysaccharide chains are very stable at pH 7.4 and 37 °C63,64. Therefore, it was not surprising that no complete degradation of MHA hydrogels was observed under the applied conditions. Taking this in mind, the full mass loss observed for MCS hydrogels after 100 days can be ascribed to disintegration of the macroscopic hydrogel in smaller fragments, which is confirmed by the observation that the PBS buffer was slightly turbid during the last days of the study.
In general, the highest stability of the hydrogels is observed when M10P10 is partially replaced by HA (MHA hydrogels) at the tested concentrations. Nevertheless, it should be taken into consideration that the degradation profile of the polysaccharide-enriched hydrogels would likely be different if tested in vivo, because of the role played by enzymatic degradation via e.g. hyaluronidase65.
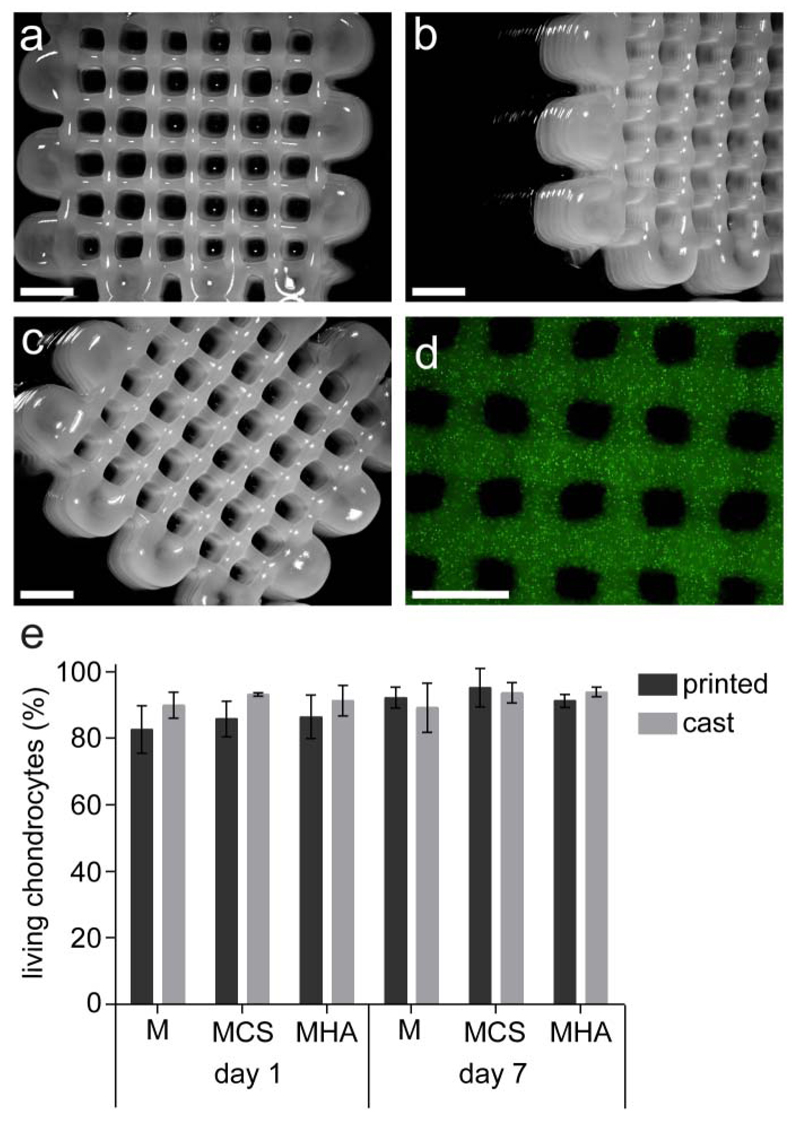
Three dimensional printing of hydrogels
Shape stable, 3D printed hydrogel constructs with highly regular internal porosity were obtained, when printing MHA hydrogels, above the Tgel (Figure 7a-c). Polymer mixtures M and MSC could not be printed with high shape fidelity at cell friendly temperature, as polymer mixture M did not form a stable physical gel below 40 °C and the MCS polymer mixture had a too low viscosity at 37 °C, forming only a weak physical gel at cell friendly temperatures.
Figure 7.
3D printed porous constructs based on MHA. a) top view. b) top-side view. c) top-corner view. d) top view showing a homogeneous distribution of encapsulated green fluorescent beads. e) percentage of living chondrocytes in printed and cast (control) constructs for each hydrogel formulation after 1 and 7 days of culture. No statistical differences were observed between hydrogel formulations. Scale bar represents 2 mm.
In line with previous observations, polymer mixtures exhibiting physical hydrogel formation and a relatively high G’ (216±14 Pa) at 37 °C allowed adequate stability of the extruded filaments on the deposition plate (pre-heated at 40 °C), and thus 3D printing with high shape fidelity (MHA hydrogels)38. On the contrary, the rheological properties of MCS polymer mixture were found insufficient for successful 3D printing.
Fluorescent microbeads with similar sizes as cells (diameter = 15 µm), were homogeneously dispersed in the MHA polymer mixture before printing. This homogeneous distribution was maintained during the printing process (Figure 7d). To investigate the influence of printing on cell viability, primary chondrocytes were dispersed in the three polymer mixtures (M, MCS and MHA) and 3D constructs were printed. The cell viability was found to be between 85% and 95%, at both 1 and 7 days after printing, similar to those of the cast hydrogel controls (Figure 7e) indicating good biocompatibility for all three hydrogel formulations and no adverse effects due to the printing procedure.
In a previous study, a hydrogel based on cross-linkable pHPMA-lac-PEG triblock copolymers was used to print porous 3D structures. However, this required a relatively high polymer concentration (25% w/w) and DM (30%)10. The addition of HAMA has led to a hydrogel platform that could be printed at a considerable lower concentration (14% M10P10 + 0.9% HAMA) and DM of the thermo-sensitive polymer (10%), which is likely beneficial for the cartilage-like matrix deposition of incorporated cells20,21. In addition, the presence of HAMA itself is likely to improve the cartilage-like tissue production and remodeling by embedded chondrocytes23,24,29–34,66. In fact, the differentiation potential of chondrocytes in hydrogels with formulation MHA (and MCS) was confirmed by collagen type II detection after 42 days of culture (Figure S3). Nevertheless, the exact concentration of HAMA still needs further attention for this aspect, as studies have reported a dose-dependent effect in which high HA(MA) concentrations exhibit a less stimulating effect or even a reduction in cartilage-like tissue formation of chondrocytes compared to a lower HA(MA) concentration24,67–71. Taken together, the partial replacement of pHPMA-lac-PEG triblock copolymer with a low amount of HAMA, in combination with a layer-by-layer UV irradiation strategy during the printing process, is a promising approach for cell-friendly additive manufacturing of these hydrogels.
Conclusions
In this study, UV cross-linked hydrogels based on thermo-sensitive methacrylated pHPMA-lac-PEG triblock copolymer, laden with equine chondrocytes showed potential for significant cartilage-like tissue formation in vitro. Additionally, mechanical analysis and swelling/degradation studies proved that the partial replacement of methacrylated pHPMA-lac-PEG triblock copolymer with CSMA or HAMA can lead to the design of hydrogels with an improved thermo-sensitive profile, a similar stiffness after UV cross-linking, and a slower degradation rate compared to hydrogels consisting of only pHPMA-lac-PEG triblock copolymers. Moreover, hydrogels containing HAMA (MHA hydrogels) were used to 3D bioprint porous structures without adversely affecting cell viability. Taken together, MHA hydrogels are attractive systems for the design of 3D cell-laden constructs for cartilage regeneration.
Supplementary Material
Supporting Information. GPC characterization of chondroitin sulfate (CS), identification of the linear viscoelastic range (LVR) of polymer mixtures, differentiation potential of chondrocytes in MCS and MHA hydrogels, and input parameters for creating 3D printed constructs. This material is available free of charge via the Internet at http://pubs.acs.org
Acknowledgment
The authors would like to thank Mattie H. P. van Rijen and Caroline C. Tippett for their assistance with the histology and biochemical assays, as well as Paola Marica for her contribution to the synthesis of methacrylated chondroitin sulfate. The primary antibody against collagen type II (II-II6B3) and collagen type VI (5C6), developed by T. F. Linsenmayer and E. S. Engvall respectively, were obtained from the DSHB developed under the auspices of the NICHD and maintained by The University of lowa, Department of Biology, lowa City, IA 52242.
Funding Sources
The research leading to these results has received funding from the Dutch Arthritis Foundation (LLP-12), the European Community’s Seventh Framework Programme (FP7/2007-2013) under grant agreement n°309962 (HydroZONES) and the European Research Council under grant agreement n°647426 (3D-JOINT).
Abbreviations
- 1H-NMR
1H-nuclear magnetic resonance
- 3D
three dimensional
- CP
cloud point
- CS
chondroitin sulfate
- CSMA
methacrylated chondroitin sulfate
- CS-TBA
chondroitin sulfate tetrabuthylammonium salt
- DM
degree of methacrylation
- DMA
dynamic mechanical analyzer
- DMEM
dulbecco’s modified eagle medium
- DMF
N,N-dimethylformamide
- DMMB
dimethyl methylene blue
- DMSO
dimethyl sulfoxide
- dwt
dry weight
- FBS
fetal bovine serum
- GAG
glycosaminoglycan
- GMA
glycidyl methacrylate
- GPC
gel permeation chromatography
- HA
hyaluronic acid
- HAMA
methacrylated hyaluronic acid
- HPLC
high performance liquid chromatography
- HPMA
N-(2-hydroxypropyl) methacrylamide
- LCST
lower critical solution temperature
- M
polymer mixture/hydrogel composed of 18% (w/w) M10P10
- M0P10
not methacrylated polyHPMA-lac-PEG triblock copolymer
- M10P10
partially methacrylated polyHPMA-lac-PEG triblock copolymer
- MA
methacrylic anhydride
- MCS
polymer mixture/hydrogel composed of 14% (w/w) M10P10 and 4% (w/w) CSMA
- MHA
polymer mixture/hydrogel composed of 14% (w/w) M10P10 and 0.9% (w/w) HAMA
- Mn
number average molecular weight
- MW
molecular weight
- PBS
phosphate buffered saline
- pen/strep
penicillin/streptomycin
- PDI
polydispersity index
- PEG
polyethylene glycol
- polyHPMA-lac
poly (N-(2-hydroxypropyl) methacrylamide mono/dilactate
- SR
swelling ratio
- TBA
tetrabuthylammonium
- Tgel
gelation temperature
Footnotes
Notes
Any additional relevant notes should be placed here.
References
- (1).Almarza AJ, Athanasiou KA. Ann Biomed Eng. 2004;32(1):2–17. doi: 10.1023/b:abme.0000007786.37957.65. [DOI] [PubMed] [Google Scholar]
- (2).Prakash D, Learmonth D. Knee. 2002;9(1):7–10. doi: 10.1016/s0968-0160(01)00133-8. [DOI] [PubMed] [Google Scholar]
- (3).Hutmacher DW. J Biomater Sci Polym Ed. 2001;12(1):107–124. doi: 10.1163/156856201744489. [DOI] [PubMed] [Google Scholar]
- (4).Vermonden T, Censi R, Hennink WE. Chem Rev. 2012;112(5):2853–2888. doi: 10.1021/cr200157d. [DOI] [PubMed] [Google Scholar]
- (5).Appel EA, del Barrio J, Loh XJ, Scherman OA. Chem Soc Rev. 2012;41(18):6195–6214. doi: 10.1039/c2cs35264h. [DOI] [PubMed] [Google Scholar]
- (6).Annabi N, Tamayol A, Uquillas JA, Akbari M, Bertassoni LE, Cha C, Camci-Unal G, Dokmeci MR, Peppas NA, Khademhosseini A. Adv Mater. 2014;26(1):85–124. doi: 10.1002/adma.201303233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Malda J, Visser J, Melchels FP, Jüngst T, Hennink WE, Dhert WJA, Groll J, Hutmacher DW. Adv Mater. 2013;25(36):5011–5028. doi: 10.1002/adma.201302042. [DOI] [PubMed] [Google Scholar]
- (8).Censi R, Vermonden T, van Steenbergen MJ, Deschout H, Braeckmans K, De Smedt SC, van Nostrum CF, di Martino P, Hennink WE. J Control Release. 2009;140(3):230–236. doi: 10.1016/j.jconrel.2009.06.003. [DOI] [PubMed] [Google Scholar]
- (9).Censi R, Vermonden T, Deschout H, Braeckmans K, Di Martino P, De Smedt SC, Van Nostrum CF, Hennink WE. Biomacromolecules. 2010;11(8):2143–2151. doi: 10.1021/bm100514p. [DOI] [PubMed] [Google Scholar]
- (10).Censi R, Schuurman W, Malda J, di Dato G, Burgisser PE, Dhert WJA, van Nostrum CF, di Martino P, Vermonden T, Hennink WE. Adv Funct Mater. 2011;21(10):1833–1842. [Google Scholar]
- (11).Vermonden T, Jena SS, Barriet D, Censi R, Van Der Gucht J, Hennink WE, Siegel RA. Macromolecules. 2010;43(2):782–789. doi: 10.1021/ma902186e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Censi R, van Putten S, Vermonden T, di Martino P, van Nostrum CF, Harmsen MC, Bank RA, Hennink WE. J Biomed Mater Res A. 2011;97(3):219–229. doi: 10.1002/jbm.a.33048. [DOI] [PubMed] [Google Scholar]
- (13).Vermonden T, Fedorovich NE, van Geemen D, Alblas J, van Nostrum CF, Dhert WJA, Hennink WE. Biomacromolecules. 2008;9(3):919–926. doi: 10.1021/bm7013075. [DOI] [PubMed] [Google Scholar]
- (14).Vermonden T, Besseling NAM, van Steenbergen MJ, Hennink WE. Langmuir. 2006;22(24):10180–10184. doi: 10.1021/la062224m. [DOI] [PubMed] [Google Scholar]
- (15).Klein TJ, Rizzi SC, Reichert JC, Georgi N, Malda J, Schuurman W, Crawford RW, Hutmacher DW. Macromol Biosci. 2009;9(11):1049–1058. doi: 10.1002/mabi.200900176. [DOI] [PubMed] [Google Scholar]
- (16).Visser J, Peters B, Burger TJ, Boomstra J, Dhert WJA, Melchels FPW, Malda J. Biofabrication. 2013;5(3):035007. doi: 10.1088/1758-5082/5/3/035007. [DOI] [PubMed] [Google Scholar]
- (17).Melchels FPW, Domingos MAN, Klein TJ, Malda J, Bartolo PJ, Hutmacher DW. Prog Polym Sci. 2012;37(8):1079–1104. [Google Scholar]
- (18).Levato R, Visser J, Planell JA, Engel E, Malda J, Mateos-Timoneda MA. Biofabrication. 2014;6(3):035020. doi: 10.1088/1758-5082/6/3/035020. [DOI] [PubMed] [Google Scholar]
- (19).Groll J, Boland T, Blunk T, Burdick JA, Cho D-W, Paul D, Derby B, Forgacs G, Li Q, Mironov VA, Moroni L. Biofabrication. 2016;8(1):013001. doi: 10.1088/1758-5090/8/1/013001. [DOI] [PubMed] [Google Scholar]
- (20).Seliktar D. Science. 2012;336(6085):1124–1128. doi: 10.1126/science.1214804. [DOI] [PubMed] [Google Scholar]
- (21).Bryant SJ, Anseth KS. J Biomed Mater Res. 2001;59(1):63–71. doi: 10.1002/jbm.1217. [DOI] [PubMed] [Google Scholar]
- (22).Schuurman W, Levett PA, Pot MW, van Weeren PR, Dhert WJA, Hutmacher DW, Melchels FPW, Klein TJ, Malda J. Macromol Biosci. 2013;13(5):551–561. doi: 10.1002/mabi.201200471. [DOI] [PubMed] [Google Scholar]
- (23).Levett PA, Hutmacher DW, Malda J, Klein TJ. PLoS One. 2014;9(12):e113216. doi: 10.1371/journal.pone.0113216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Levett PA, Melchels FPW, Schrobback K, Hutmacher DW, Malda J, Klein TJ. Acta Biomater. 2014;10(1):214–223. doi: 10.1016/j.actbio.2013.10.005. [DOI] [PubMed] [Google Scholar]
- (25).Choi B, Kim S, Lin B, Wu BM, Lee M. ACS Appl Mater Interfaces. 2014;6(22):20110–20121. doi: 10.1021/am505723k. [DOI] [PubMed] [Google Scholar]
- (26).Shim J-H, Jang K-M, Hahn SK, Park JY, Jung H, Oh K, Park KM, Yeom J, Park SH, Kim SW, Wang JH, et al. Biofabrication. 2016;8(1):014102. doi: 10.1088/1758-5090/8/1/014102. [DOI] [PubMed] [Google Scholar]
- (27).Oudshoorn MHM, Rissmann R, Bouwstra JA, Hennink WE. Polymer. 2007;48(7):1915–1920. [Google Scholar]
- (28).Hachet E, Van Den Berghe H, Bayma E, Block MR, Auzély-Velty R. Biomacromolecules. 2012;13(6):1818–1827. doi: 10.1021/bm300324m. [DOI] [PubMed] [Google Scholar]
- (29).Lesley J, Hascall VC, Tammi M, Hyman R. J Biol Chem. 2000;275(35):26967–26975. doi: 10.1074/jbc.M002527200. [DOI] [PubMed] [Google Scholar]
- (30).Chung C, Erickson IE, Mauck RL, Burdick JA. Tissue Eng Part A. 2008;14(7):1121–1131. doi: 10.1089/ten.tea.2007.0291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Park S-H, Park SR, Chung SIl, Pai KS, Min B-H. Artif Organs. 2005;29(10):838–845. doi: 10.1111/j.1525-1594.2005.00137.x. [DOI] [PubMed] [Google Scholar]
- (32).Dinescu S, Gălăţeanu B, Albu M, Lungu A, Radu E, Hermenean A, Costache M. Biomed Res Int. 2013;2013:1–11. doi: 10.1155/2013/598056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Park H, Choi B, Hu J, Lee M. Acta Biomater. 2013;9(1):4779–4786. doi: 10.1016/j.actbio.2012.08.033. [DOI] [PubMed] [Google Scholar]
- (34).Roberts JJ, Nicodemus GD, Ciunta S, Bryant SJ. J biomed Mater Res A. 2011;97(3):281–291. doi: 10.1002/jbm.a.33057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Oupický D, Konák C, Ulbrich K. J Biomater Sci Polym Ed. 1999;10(5):573–590. doi: 10.1163/156856299x00496. [DOI] [PubMed] [Google Scholar]
- (36).Neradovic D, van Steenbergen MJ, Vansteelant L, Meijer YJ, van Nostrum CF, Hennink WE. Macromolecules. 2003;36(20):7491–7498. [Google Scholar]
- (37).Neradovic D, van Nostrum CF, Hennink WE. Macromolecules. 2001;34(22):7589–7591. [Google Scholar]
- (38).Abbadessa A, Blokzijl MM, Mouser VHM, Marica P, Malda J, Hennink WE, Vermonden T. Carbohydr Polym. 2016 doi: 10.1016/j.carbpol.2016.04.080. [DOI] [PubMed] [Google Scholar]
- (39).Stenekes RJ, Hennink W. Polymer. 2000;41(15):5563–5569. [Google Scholar]
- (40).Benya PD, Shaffer JD. Cell. 1982;30(1):215–224. doi: 10.1016/0092-8674(82)90027-7. [DOI] [PubMed] [Google Scholar]
- (41).Guo J, Jourdian GW, Maccallum DK. Connect Tissue Res. 1989;19(2–4):277–297. doi: 10.3109/03008208909043901. [DOI] [PubMed] [Google Scholar]
- (42).Rosenberg L. J Bone Joint Surg Am. 1971;53(1):69–82. [PubMed] [Google Scholar]
- (43).Farndale RW, Sayers CA, Barrett AJ. Connect Tissue Res. 1982;9(4):247–248. doi: 10.3109/03008208209160269. [DOI] [PubMed] [Google Scholar]
- (44).Li Q, Wang D, Elisseeff JH. Macromolecules. 2003;36(7):2556–2562. [Google Scholar]
- (45).Burdick JA, Chung C, Jia X, Randolph MA, Langer R. Biomacromolecules. 2005;6(1):386–391. doi: 10.1021/bm049508a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Smeds KA, Grinstaff MW. J Biomed Mater Res. 2001;54(1):115–121. doi: 10.1002/1097-4636(200101)54:1<115::aid-jbm14>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- (47).Messager L, Portecop N, Hachet E, Lapeyre V, Pignot-Paintrand I, Catargi B, Auzély-Velty R, Ravaine V. J Mater Chem B. 2013;1(27):3369. doi: 10.1039/c3tb20300j. [DOI] [PubMed] [Google Scholar]
- (48).Mastbergen SC, Saris DB, Lafeber FP. Nat Rev Rheumatol. 2013;9(5):277–290. doi: 10.1038/nrrheum.2013.29. [DOI] [PubMed] [Google Scholar]
- (49).Brittberg M. Am J Sports Med. 2010;38(6):1259–1271. doi: 10.1177/0363546509346395. [DOI] [PubMed] [Google Scholar]
- (50).Cummings CL, Gawlitta D, Nerem RM, Stegemann JP. Biomaterials. 2004;25(17):3699–3706. doi: 10.1016/j.biomaterials.2003.10.073. [DOI] [PubMed] [Google Scholar]
- (51).Ahmed TAE, Dare EV, Hincke M. Tissue Eng Part B Rev. 2008;14(2):199–215. doi: 10.1089/ten.teb.2007.0435. [DOI] [PubMed] [Google Scholar]
- (52).Eyrich D, Brandl F, Appel B, Wiese H, Maier G, Wenzel M, Staudenmaier R, Goepferich A, Blunk T. Biomaterials. 2007;28(1):55–65. doi: 10.1016/j.biomaterials.2006.08.027. [DOI] [PubMed] [Google Scholar]
- (53).Visser J, Melchels FPW, Jeon JE, van Bussel EM, Kimpton LS, Byrne HM, Dhert WJA, Dalton PD, Hutmacher DW, Malda J. Nat Commun. 2015;6:6933. doi: 10.1038/ncomms7933. [DOI] [PubMed] [Google Scholar]
- (54).Poole CA, Ayad S, Schofield JR. J Cell Sci. 1988;90:635–643. doi: 10.1242/jcs.90.4.635. [DOI] [PubMed] [Google Scholar]
- (55).Zhang Z. Tissue Eng Part B Rev. 2015;21(3):267–277. doi: 10.1089/ten.teb.2014.0286. [DOI] [PubMed] [Google Scholar]
- (56).Visser J, Levett PA, te Moller NCR, Besems J, Boere KWM, van Rijen MHP, de Grauw JC, Dhert WJA, van Weeren PR, Malda J. Tissue Eng Part A. 2015;21(7–8):1195–1206. doi: 10.1089/ten.tea.2014.0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Kim J, Park Y, Tae G, Kyu BL, Chang MH, Soon JH, In SK, Noh I, Sun K. J Biomed Mater Res - Part A. 2009;88(4):967–975. doi: 10.1002/jbm.a.31947. [DOI] [PubMed] [Google Scholar]
- (58).Jeong CG, Francisco AT, Niu Z, Mancino RL, Craig SL, Setton LA. Acta Biomater. 2014;10(8):3421–3430. doi: 10.1016/j.actbio.2014.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Chen AC, Bae WC, Schinagl RM, Sah RL. J Biomech. 2001;34(1):1–12. doi: 10.1016/s0021-9290(00)00170-6. [DOI] [PubMed] [Google Scholar]
- (60).Athanasiou KA, Agarwal A, Dzida FJ. J Orthop Res. 1994;12(3):340–349. doi: 10.1002/jor.1100120306. [DOI] [PubMed] [Google Scholar]
- (61).Jurvelin JS, Buschmann MD, Hunziker EB. J Biomech. 1997;30(3):235–241. doi: 10.1016/s0021-9290(96)00133-9. [DOI] [PubMed] [Google Scholar]
- (62).Bolis S, Handley CJ, Cornper WD. Biochim Biophys Acta - Gen Subj. 1989;993(2–3):157–167. doi: 10.1016/0304-4165(89)90158-x. [DOI] [PubMed] [Google Scholar]
- (63).Van Dijk-Wolthuis WNE, Hoogeboom JAM, van Steenbergen MJ, Tsang SKY, Hennink WE. Macromolecules. 1997;30(16):4639–4645. [Google Scholar]
- (64).Van de Wetering P, Zuidam NJ, van Steenbergen MJ, van der Houwen OAGJ, Underberg WJM, Hennink WE. Macromolecules. 1998;31(23):8063–8068. [Google Scholar]
- (65).Kurisawa M, Chung JE, Yang YY, Gao SJ, Uyama H. Chem Commun. 2005;(34):4312. doi: 10.1039/b506989k. [DOI] [PubMed] [Google Scholar]
- (66).Hwang NS, Varghese S, Lee HJ, Theprungsirikul P, Sharma B, Elisseeff J, Canver A, Sharma B, Elisseeff J. FEBS Lett. 2007;581(22):4172–4178. doi: 10.1016/j.febslet.2007.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Akmal M, Singh A, Anand A, Kesani A, Aslam N, Goodship A, Bentley G. J Bone Jt Surg - Br Vol. 2005;87-B(8):1143–1149. doi: 10.1302/0301-620X.87B8.15083. [DOI] [PubMed] [Google Scholar]
- (68).Allemann F, Mizuno S, Eid K, Yates KE, Zaleske D, Glowacki J. J Biomed Mater Res. 2001 doi: 10.1002/1097-4636(200104)55:1<13::aid-jbm20>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- (69).Callahan LAS, Ganios AM, McBurney DL, Dilisio MF, Weiner SD, Horton WE, Becker ML. Biomacromolecules. 2012;13(5):1625–1631. doi: 10.1021/bm3003336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Kawasaki K, Ochi M, Uchio Y, Adachi N, Matsusaki M. J Cell Physiol. 1999;179(2):142–148. doi: 10.1002/(SICI)1097-4652(199905)179:2<142::AID-JCP4>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- (71).Villanueva I, Gladem SK, Kessler J, Bryant SJ. Matrix Biol. 2010;29(1):51–62. doi: 10.1016/j.matbio.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.